Abstract
The envelope-associated glycoprotein B (gB) is highly conserved within the Herpesviridae and plays a critical role in viral entry. We analyzed the evolutionary conservation of sequence and structural motifs within the Kaposi’s sarcoma-associated herpesvirus (KSHV) gB and homologs of Old World primate rhadinoviruses belonging to the distinct RV1 and RV2 rhadinovirus lineages. In addition to gB homologs of rhadinoviruses infecting the pig-tailed and rhesus macaques, we cloned and sequenced gB homologs of RV1 and RV2 rhadinoviruses infecting chimpanzees. A structural model of the KSHV gB was determined, and functional motifs and sequence variants were mapped to the model structure. Conserved domains and motifs were identified, including an “RGD” motif that plays a critical role in KSHV binding and entry through the cellular integrin αVβ3. The RGD motif was only detected in RV1 rhadinoviruses suggesting an important difference in cell tropism between the two rhadinovirus lineages.
Keywords: glycoprotein, rhadinovirus, Kaposi’s sarcoma herpesvirus, retroperitoneal fibromatosis herpesvirus, structure, integrin, phylogeny
INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV) is the most recently discovered member of the Herpesviridae that infect humans. KSHV was identified as a member of the Rhadinovirus genus of the gammaherpesvirus subfamily based on its sequence similarity with herpesvirus saimiri (HVS), the prototype of this genus found in the New World squirrel monkey (Chang et al., 1994). KSHV was originally detected in Kaposi’s sarcoma (KS) lesions and is now believed to be the causative agent of KS and two lymphoproliferative diseases, primary effusion lymphoma (PEL) and a subset of multicentric Castleman’s disease (Antman and Chang, 2000). We previously identified macaque homologs of KSHV in retroperitoneal fibromatosis (RF) tumor lesions, a vascular fibroproliferative disease that shares morphological and histological properties with KS (Rose et al., 1997). Related strains of the macaque herpesvirus were identified in RF lesions of different macaque species at the Washington National Primate Research Center (WaNPRC), including M. mulatta (rhesus), M. nemestrina (pig-tailed), and M. fascicularis (cynomolgus), and were named retroperitoneal fibromatosis herpesviruses, RFHVMm, RFHVMn and RFHVMf, respectively. Subsequently, other distinct herpesviruses related to KSHV were identified in the same macaque species and were designated rhesus rhadinovirus (RRV-2695) (Desrosiers et al., 1997) and pig-tailed macaque rhadinovirus (PMRV) (Mansfield et al., 1999) at the New England National Primate Research Center (NENPRC) and M. fascicularis gammaherpesvirus (MGVMf), also known as MfaRV2, at the WaNPRC (Bruce et al., 2009; Strand et al., 2000). An additional RRV variant, RRV17577, was identified in a rhesus macaque at the Oregon National Primate Research Center (ONPRC), and an additional pig-tailed rhadinovirus, MneRV2, was identified at the WaNPRC (Schultz et al., 2000). A phylogenetic comparison of the DNA polymerase sequences revealed that the macaque viruses segregated into two distinct lineages of KSHV-like rhadinoviruses. RFHVMm, RFHVMn and RFHVMf clustered together with KSHV in the Rhadinovirus-1 (RV-1) lineage of Old World primate rhadinoviruses, while RRV17577/RRV2695, MneRV2/PMRV and MGVMf/MfaRV2 clustered in a distinct Rhadinovirus-2 (RV-2) lineage (Schultz et al., 2000). Additional studies identified RV1 and RV2 rhadinovirus species in other Old World primate species, including chimpanzee, gorilla, African green monkey, drill and mandrill (Greensill and Schulz, 2000; Greensill et al., 2000b; Lacoste et al., 2000a, 2001; Lacoste et al., 2000b). An RV2 rhadinovirus has not been identified in humans.
Previous studies on the life cycle and pathogenesis of the Herpesviridae, have focused on Epstein-Barr virus (EBV), a human gammaherpesvirus within the Lymphocryptovirus genus, and on the prototypes of the two other Herpesviridae subfamilies, herpes simplex virus 1 (HSV-1), an alphaherpesvirus, and cytomegalovirus (CMV), a betaherpesvirus. These studies have shown that members of the Herpesviridae are enveloped DNA viruses that share a large complement of conserved genes encoding important viral proteins involved in virion formation and virus replication. Among these is glycoprotein B (gB), a protein of the enveloped virion, which is essential for virus replication (Pereira, 1994). Glycoprotein B is the most highly conserved herpesvirus glycoprotein suggesting that gB homologs from different viral species would have similar structures and serve similar functions within the life cycle of the Herpesviridae.
Studies of the gB prototypes from the different Herpesviridae subfamilies have revealed a number of common features. Glycoprotein B consists of a large disulfide-linked external ectodomain that is glycosylated, a major hydrophobic transmembrane domain, and a smaller internal domain (Pereira, 1994). Ten cysteine residues are conserved among gB homologs and form five conserved disulphide linkages giving a common gB structure (Norais et al., 1996). Among the alpha- and betaherpesviruses, gB is a major antigenic protein in the virion envelope and is also found in the endoplasmic reticulum and in the plasma and nuclear membranes of infected cells (Britt, 1984; Claesson-Welsh and Spear, 1987; Gretch et al., 1988; Pereira et al., 1982; Spear, 1976). Fully-processed virion and cell membrane-associated gB are characterized by the presence of complex carbohydrates, whereas immature precursor forms contain only high mannose-type oligosaccharides (Britt and Vugler, 1989; Gretch et al., 1988; Pereira et al., 1984; Person et al., 1982; Wenske et al., 1982). Glycoprotein B was initially detected as a heat-dissociable homodimer (Britt and Vugler, 1992; Cai et al., 1988; Haffey and Spear, 1980; Sarmiento and Spear, 1979). In most viral species, the gB monomeric subunit is proteolytically cleaved into N- and C-terminal fragments linked by disulphide bonds (Britt, 1984; Britt and Vugler, 1989; Johannsen et al., 2004; Pereira et al., 1982; Rasmussen et al., 1985; Spaete et al., 1988).
During the process of infection, herpesviruses enter cells as their envelope fuses with the membrane of the target cell. Glycoprotein B plays a critical role in the fusion machinery where it interacts with a heterodimeric complex of glycoproteins gH and gL to promote fusion (Spear and Longnecker, 2003). The crystal structures of HSV1, CMV and EBV gB have been determined revealing a trimeric structure that resembles the postfusion structure of the class III fusion proteins in vesicular stomatitis virus glycoprotein G and baculovirus gp64 (Backovic et al., 2009; Burke and Heldwein, 2015; Heldwein et al., 2006). The fusion domains of both HSV1 and EBV gB each contain two putative fusion loops with hydrophobic residues that are believed to insert into the target membrane through a conformational change inducing fusion of the envelope and cellular membrane (Backovic et al., 2007; Hannah et al., 2007). A membrane proximal region (MPR) with significant hydrophobic character has been identified immediately upstream of the hydrophobic transmembrane domain that anchors gB in the virion envelope. The MPR is believed to interact with both the fusion loops and the virion envelope modulating the association of gB with target membranes (Shelly et al., 2012).
Relatively little is known regarding the structure or function of the KSHV gB. Initial studies showed that KSHV gB was expressed in CHO cells as a full-length uncleaved form with high mannose-type oligosaccharides, characteristic of immature precursor forms of gB (Pertel et al., 1998). The gB was present in the endoplasmic reticulum and nuclear membrane of transfected CHO cells, but not in the plasma membrane. An uncleaved form of baculovirus-expressed gB from Sf9 insect cells was found to interact with DC-SIGN through high mannose carbohydrate structures (Hensler et al., 2014). Subsequently, contradictory results were obtained in which KSHV gB was detected in the virion and in the plasma membrane of latently-infected BCBL-1 cells which were induced to initiate KSHV replication using phorbol esters (Akula et al., 2001; Baghian et al., 2000). In these studies, KSHV virion-associated gB was shown to be proteolytically cleaved into N- and C-terminal fragments of ~75 kDa and 54–59 kDa that are linked by disulfide bonds to form a ~140 kDa monomer. A hydrophobic signal sequence was detected at the N-terminus of the gB sequence and a signal peptide cleavage site was predicted between Ala23 and Ala24 (Pertel et al., 1998). We identified an arginine-glycine-aspartic acid (RGD) motif immediately downstream of the putative mature N-terminus of KSHV gB, which was predicted to function in binding to cell surface integrin receptors (Rose, 1999, 2000). RGD motifs are present in extracellular matrix proteins that interact with specific members of the integrin receptor family (Hynes, 2002). Subsequently, we demonstrated that the KSHV gB RGD motif binds specifically to the integrin αVβ3, which functions as an RGD-dependent entry receptor for KSHV (Garrigues et al., 2008). Like the gB homologs of HSV1 and hCMV, a heparin binding domain was identified in KSHV gB, suggesting that gB may play a role in the initial binding to target cells through interactions with cell-surface heparan sulfate (Akula et al., 2001). Whereas the heparin binding domains of HSV1 and hCMV gB mapped to the N-terminal region of gB (Laquerre et al., 1998; Silvestri and Sundqvist, 2001), the heparin binding domain of KSHV gB was not homologous and mapped further downstream in the gB sequence. Disintegrin-like domains have been identified in the gB homologs of both hCMV and KSHV, mapping to a motif “RX8D/ELX2FX5C”, which is conserved in the gB homologs of the beta- and gammaherpesviruses (Feire et al., 2004; Walker et al., 2014). These domains may play important conserved roles in herpesvirus binding and entry.
In the present study, we analyzed the evolutionary conservation of structural and functional motifs within KSHV gB and the closely related gB homologs of Old World primate rhadinoviruses. In addition to the gB homologs of the known macaque RV1 and RV2 rhadinoviruses infecting the pig-tailed and rhesus macaques, we have determined the sequence of two distinct gB homologs of rhadinoviruses infecting chimpanzees. Phylogenetic analysis revealed that the chimpanzee rhadinoviruses represented novel members of the RV1 and RV2 rhadinovirus lineages. A structural model of KSHV gB was determined by computer modeling of the EBV gB structure, and known motifs and sequence variants were mapped to the model structure. Highly conserved domains and motifs were identified and characterized, including an N-terminal “RGD” motif. The RGD motif was only detected in the gB homologs of the RV1 rhadinoviruses, indicating a potentially important difference in cell tropism and virus entry between the two rhadinovirus lineages.
RESULTS
Identification of the gB homologs of the chimpanzee RV1 and RV2 rhadinoviruses. We and others have previously shown that there are two distinct RV1 and RV2 rhadinovirus lineages infecting Old World primates (Greensill et al., 2000b; Schultz et al., 2000). Whereas members of both RV1 and RV2 lineages have been identified in numerous Old World primate species, including macaques, gorillas and chimpanzees (Desrosiers et al., 1997; Greensill et al., 2000a; Lacoste et al., 2000a, 2001; Rose et al., 1997), only the RV1 rhadinovirus, KSHV, has been identified in humans. We previously cloned and sequenced the gB genes of RV1 rhadinovirus species from pig-tailed macaques, including RFHVMn-M78114 from the Washington National Primate Research Center (WaNPRC) (Bruce et al., 2013) and RFHVMn-442N from the NIH primate facility, and RFHVMm-YN91 from a rhesus macaque at the Yerkes National Primate Research Center (Rose et al., 2003). We have also cloned and sequenced the gB gene of the pigtailed macaque RV2 rhadinovirus (MneRV2-J97167) from the WaNPRC (Bruce et al., 2015). The sequence of the gB gene of an additional pig-tailed macaque RV2 rhadinovirus (PMRV-98126) was sequenced from a pig-tailed macaque at the New England National Primate Research Center (NENPRC) (Auerbach et al., 2000). Previously, others have cloned and sequenced the gB genes of two variants of the rhesus macaque RV2 rhadinovirus from the Oregon National Primate Research Center (ONPRC) (RRV-17577) (Searles et al., 1999) and NENPRC (RRV-H26-95) (Alexander et al., 2000). The gB genes of several very closely related variants of KSHV, the human RV1 rhadinovirus, have been sequenced from different PEL cell lines, including the KSHV strain GK18 of Caucasian origin representing the NCBI reference sequence (NP_009333), and VG1, a more distantly related African B-subtype virus. Recently, the gB sequences from KSHV variants from sixteen Zambian KS biopsy specimens were obtained from the complete genome sequences (Olp et al., 2015). Whereas human herpesviruses have been identified in the RV1 (KSHV) and LCV (EBV) lineages, a human RV2 lineage herpesvirus has not been identified to date.
To compare the sequence conservation and evolution of the gB genes of Old World primate RV1 and RV2 rhadinoviruses, we sequenced the gB genes of the RV1 and RV2 rhadinoviruses from chimpanzee saliva using consensus degenerate hybrid oligonucleotide primer (CODEHOP)-based PCR primers (Rose et al., 1998) essentially as described previously (Rose, 2005). Two related but distinct rhadinovirus gB sequences were obtained. One sequence, termed PtrRV1, contained sequences overlapping with a partial gB sequence identified previously (ABU62806). The composite PtrRV1 gB sequence was closely related to the gB sequence of KSHV (81.6/87.7% identity) and RFHVMm (66.8/71.2% identity) for both nucleotides and amino acids, respectively (see Table 1 for amino acid comparisons). The other sequence, termed PtrRV2, contained sequences overlapping another partial gB sequence identified previously (EU085378). The composite PtrRV2 gB sequence was closely related to the gB sequence of RRV (64.3/70.8% identity) and more distantly related to PtrRV1 (61.0/58.9% identity), RFHVMn-M78114 (66.8/58.6% identity) and KSHV (54.1/59.2% identity), for both nucleotides and amino acids, respectively. The partial PtRV1 and PtRV2 glycoprotein B sequences have been deposited in the Genbank database with accession numbers KU984977 and KU984976, respectively.
Table 1.
gB amino acid sequence comparisons
| KSHV | PtrRV1 | RFHVMn | RFHVMm | PtrRV2 | MneRV2 | RRV | |
|---|---|---|---|---|---|---|---|
| KSHV | |||||||
| PtrRV1 | 81.6%1 | ||||||
| RFHVMn | 66.9% | 69.8% | |||||
| RFHVMm | 68.8% | 71.2% | 95.0% | ||||
| PtrRV2 | 63.1% | 62.9% | 61.9% | 63.0% | |||
| MneRV2 | 64.2% | 64.3% | 62.4% | 62.5% | 74.8% | ||
| RRV | 64.2% | 63.8% | 62.5% | 62.6% | 74.9% | 91.4% | |
| EBV | 37.4% | 35.1% | 35.4% | 35.0% | 32.4% | 25.2% | 30.5% |
Sequence identity from the alignment in Figure 3 is shown.
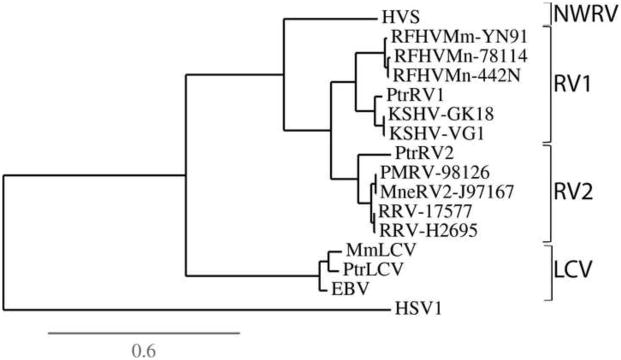
The gB homologs of the Old World primate RV1 and RV2 rhadinoviruses are phylogenetically distinct. The phylogeny of the Old World rhadinovirus and lymphocryptovirus gB homologs was determined by maximum-likelihood analysis of the entire set of gB protein sequences, using the gB sequence of the alphaherpesvirus, herpes simplex virus, as the outgroup. Figure 1 clearly shows the phylogenetic clustering of the KSHV, chimpanzee PtrRV1, macaque RFHVMn and RFHVMm gB sequences within the RV1 lineage. The chimpanzee PtrRV2 gB clustered with the MneRV2/PMRV and RRV gB sequences within the RV2 lineage. The two strains of KSHV (GK18 and VG-1) clustered closely together, as did the two strains of RFHVMn (M78114 and 442N), the two strains of MneRV2 (J97167 and PMRV98126) and the two strains of RRV (17577 and H26-95). The close phylogenetic relationship between the chimpanzee PtrRV1 and KSHV gB sequences, and the more distant relationship with the macaque RFHVMn/Mm gB sequences is consistent with a long term synchronous evolution with their host species. This contrasts with the LCV gB sequences in which the chimpanzee LCV sequence clustered more closely with the macaque MmLCV than with the human EBV (Fig. 1).
Figure 1.
Phylogenetic analysis of the gB homologs of the human, chimpanzee and macaque gammaherpesviruses. The complete gB sequences of the human, chimpanzee and macaque RV1 and RV2 Old World rhadinovirus lineages were aligned with the gB sequences of the corresponding lymphocryptoviruses (LCV) and the phylogenetic relationship was determined by protein maximum likelihood analysis. The clustering within the RV1, RV2 and LCV subgroups is indicated. The gB sequences analyzed include RV-1 lineage: Human (KSHV-GK18, #YP_001129354; KSHV-VG-1,#ADB08179), chimpanzee (PtrRV1, this study), pig-tail macaque (RFHVMn-78114, #AGY30687; RFHVMn-442N, #AF204166); rhesus macaque (RFHVMm-YN91, #AAF78826); RV-2 lineage: chimpanzee (PtrRV2, this study), pig-tail macaque (MneRV2-J97167, #AJE29647; PMRV-98126(Auerbach et al., 2000)), rhesus macaque (RRV-17577, #NP_570749; RRV-H2695, #AF210726); New World rhadinovirus (NWRV): squirrel monkey (HVS, # Q04464); Old World lymphocryptovirus: human (EBV, # NP_039909), chimpanzee (PtrLCV, CAE46448), rhesus macaque (MmLCV, #YP_068009). The gB sequences of human herpes simplex virus 1 (HSV1, #NP_044629) and the squirrel monkey New World rhadinovirus (NWRV) (HVS, #CAC84303) were used as outgroups. The scale for substitutions per site is shown.
An unconventional “TATTTAAA” TATA-like promoter element is conserved in the rhadinovirus gB homologs. To determine the sequence conservation of promoter elements upstream of the translation start site of gB, the nucleotide sequences flanking the start site of the gB homologs of the Old and New World rhadinoviruses were aligned and compared to the sequence of EBV, an Old World lymphocryptovirus. A conserved “TATTTAAA” TATA-like element, 46bp upstream of the AUG initiator, was present in the gB homologs of KSHV, RFHVMn, MneRV2, RRV and herpesvirus saimiri (HVS), a New World primate rhadinovirus (Fig. 2, shaded black). This element contains the unconventional “TATT” sequence that has been identified in various herpesvirus late genes and found to be required for late progression through the viral life cycle (Serio et al., 1998; Wong-Ho et al., 2014) Conservation of a “GGXTGG” sequence upstream and a “GAC” sequence downstream of the “TATTTAAA” was also observed (Fig. 2, shaded red). The EBV gB sequence contained a shorter “TATTTAA” motif. Interestingly, both HVS and EBV contained a “TATAT” TATA box motif immediately downstream of the “TATTTAAA” motif, while EBV contained an additional TATA motif “TATAA” upstream (Fig. 2). Thus, transcription of EBV and HVS gB homologs may be more complex than that seen in the Old World rhadinoviruses.
Figure 2.
Conservation an unconventional TATA-like promoter element in the rhadinovirus gB homologs. The nucleotide sequences upstream of the translation initiation site of KSHV, RFHVMn, MneRV2, RRV, HVS and EBV gB homologs were aligned. TATA-like promoter elements and the ATG translation initiation codons are highlighted in black. Conserved sequences upstream and downstream of the TATA-like elements are highlighted in red. Sequences are derived from the genomes of KSHV (NC_009333), RFHVMn-M78814 (KF703446), MneRV2-J97167 (KP265674), RRV-H2695 (AF210726), HVS (X64346), EBV (NC_007605).
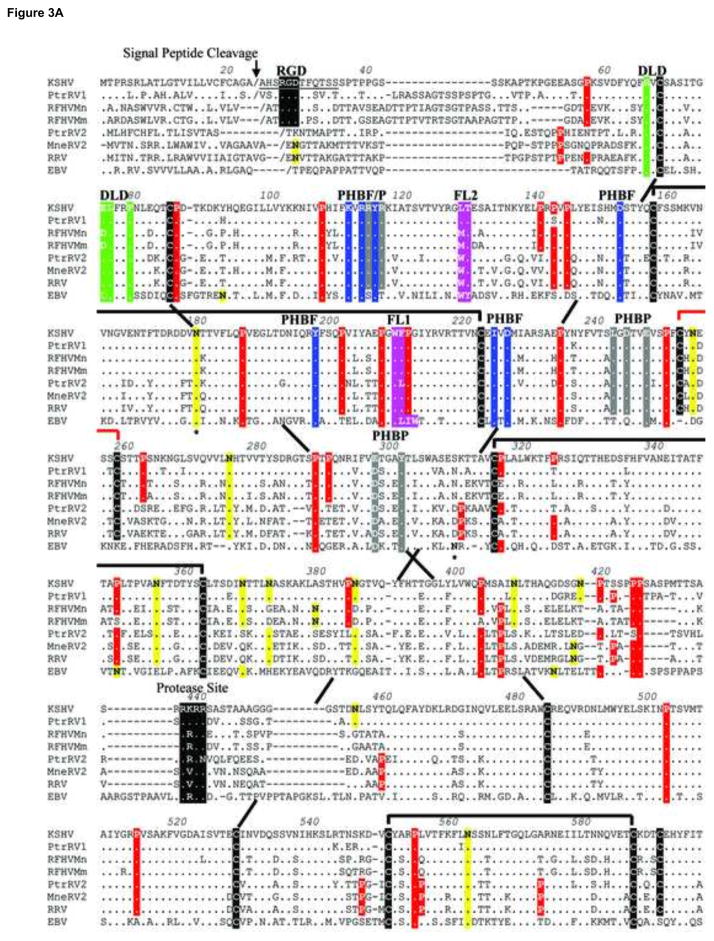
The gB homologs of the Old World primate RV1 and RV2 rhadinoviruses are conserved. The amino acid sequences of the gB homologs of KSHV, PtrRV1, RFHVMn and RFHVMm were aligned with the sequences of PtrRV2, RRV and MneRV2. The EBV gB sequence was aligned for comparison (Fig. 3). Significant similarity was seen throughout the entire gB sequences with thirteen cysteine (Fig. 3; black highlight) and twenty proline (Fig. 3: red highlight) residues conserved across the seven rhadinovirus species. Ten of the cysteines are conserved within the gB homologs of EBV and other herpesviruses, where they form structurally conserved disulphide linkages (Norais et al., 1996). Based on this, we predict a similar linkage pattern for the 10 analogous cysteines in the gB homologs of KSHV and the macaque rhadinoviruses (Fig. 3, black linkages). Of the remaining 3 cysteines conserved among the rhadinovirus sequences, two are predicted to form a small loop in the middle of the extracellular domain between Cys252 and Cys258 (Fig. 3, red linkage). The remaining cysteine (Cys592) is shown as unpaired. Also conserved within the ectodomains of the seven rhadinovirus sequences are 8 potential N-linked glycosylation sites (N-X-S/T, where X is any amino acid except proline)(Fig. 3; yellow highlight). Three of these glycosylation sites, N179, N562, and N628 (KSHV numbering) are also conserved in the EBV gB sequence. The asparagine sites in EBV gB corresponding to N179 and N628 are known to be glycosylated (Fig. 3).
Figure 3.
Amino acid sequence alignment of gB homologs from human, chimpanzee and macaque rhadinoviruses compared to EBV gB. The amino acid sequences of the gB homologs of the RV1 lineage of Old World rhadinoviruses, including KSHV (human), PtrRV1 (chimpanzee), RFHVMn (pig-tailed macaque), and RFHVMm (rhesus macaque), were aligned with the gB homologs of the RV2 rhadinovirus lineage, including PtrRV2 (chimpanzee), MneRV2 (pig-tailed macaque), and RRV-26-95 (rhesus macaque) and the gB homolog of the human lymphocryptovirus EBV. Amino acids conserved with KSHV gB are shown as a (.), except for the conserved cysteines, which are highlighted black. Conserved prolines are highlighted red and conserved NXS/T putative N-linked glycosylation sites are highlighted yellow. Asterices mark the known N-linked glycosylation sites in EBV gB. The disulphide bonds conserved with EBV gB are shown as black brackets or long-range dashed lines, and the putative rhadinovirus-specific disulphide bond between Cys252 and Cys258 is shown as a red bracket. The putative signal peptide cleavage site is indicated. The N-terminal RGD motif in the RV1 rhadinovirus lineage and the conserved internal furin protease cleavage sites are shown, highlighted in black. Hydrophobic residues conserved with the FL1 and FL2 fusion loops of EBV gB are highlighted purple. Conserved residues within a disintegrin-like domain (DLD) are highlighted in green. Conserved residues within the pleckstrin homology binding face (PHBF) or pocket (PHBP) are highlighted in blue and grey, respectively. The MPRa and MPRb domains of the amphipathic membrane-proximal region and the transmembrane helix (TMH) domain are indicated. A localization signal for inner nuclear membrane targeting (LSINM), a C-terminal “acidic cluster” and “YXXϕ” domains implicated in endocytosis are indicated and highlighted in black. The sources of the gB sequences are shown in the legend to Fig. 1.
Two major regions of strong sequence similarity between the gB homologs of the RV1 and RV2 lineages were identified in the ectodomain. The most highly conserved region extends from Gln460 to Arg543 downstream of the protease cleavage site (aa437-440) (numbered relative to the KSHV sequence) (Figs. 3 and 4). This region contains the two cysteine residues predicted to link the N- and C-terminal domains through disulfide bonds. Within this stretch of 84 amino acids, 88% are identical between RFHVMm and RRV, 89% are identical between RFHVMn and MneRHV2, while 94% are identical between RFHV and KSHV. The second most highly conserved domain extends from Arg141 to Cys258. Within this stretch of 191 amino acids, 79% are identical between RFHVMm and RRV, 81% are identical between RFHVMn and MneRHV2, and 88% are identical between RFHV and KSHV, with ten proline residues conserved in all five sequences. In general, the cysteine-defined loop structures were more highly conserved than the linking regions and each loop structure contained a conserved N-linked glycosylation site (Fig. 4).
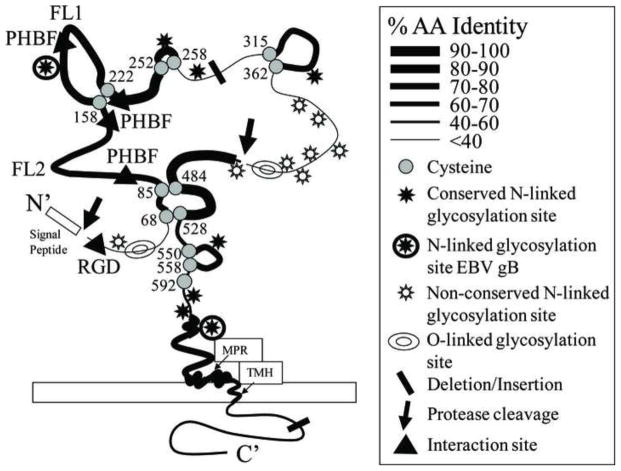
Figure 4.
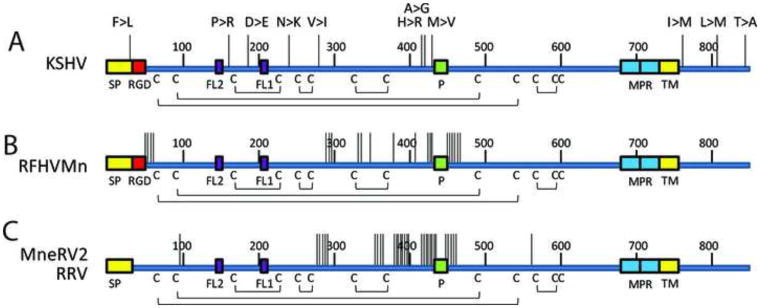
Cartoon of the KSHV gB structure. A two-dimensional cartoon of the monomeric KSHV gB structure is shown with conserved cysteine residues and putative disulfide linkages and loop structures in which the approximate percentage of amino acid identity between the gB homologs of the RV1 and RV2 rhadinovirus subgroups (from Figure 3) is depicted by the thickness of the line representing the polypeptide sequence. Both conserved and non-conserved putative N-linked glycosylation sites (NXS/T) are also shown and the position of known glycosylation sites in EBV gB are indicated; sites are considered conserved if they are in identical positions in all of the KSHV and macaque rhadinovirus gB homologs. Predicted sites of O-linked glycosylation are shown. In addition, the internal signal peptidase and protease cleavage sites, the novel pleckstrin homology binding face (PHBF) and the RGD interaction motif are indicated. The positions of the fusion loops (FL1, FL2) predicted to function in gB induced membrane fusion, the hydrophobic transmembrane helix (TMH) and amphipathic membrane proximal helical regions (MPR) are shown.
The regions that were least conserved across all seven rhadinovirus sequences were the N-terminal region up to aa66, the central region between aa362 and aa460 flanking the protease cleavage site, and the majority of the C-terminal domain from aa770 to the terminus (KSHV numbering). The nonconserved N-terminal and central regions were predicted to be sites of O-linked glycosylation (Fig. 4). Immediately upstream of the predicted O-linked glycosylation sites in the central region is a conserved endoproteolytic cleavage site. All eight gB homologs matched the consensus sequence for the furin protease cleavage site, RX(R/K)R (Figs. 3 and 4), where the cleavage is predicted to occur after the paired basic residues (Hosaka et al., 1991). This site is similarly positioned to sites within a number of other gB homologs that undergo proteolytic cleavage to yield N- and C-terminal gB fragments that remain in close association due to disulfide bonds (Pereira, 1994).
Hydropathy analysis identified a non-conserved N-terminal hydrophobic signal peptide in each of the seven rhadinovirus gB sequences, which could direct the protein to a membrane location. Cleavage of the signal peptide of the KSHV gB was predicted to occur between amino acids Ala23 and Ala24 (Fig. 3). Similar cleavage sites are predicted in the other rhadinovirus sequences. Hydropathy analysis also revealed the presence of a potential hydrophobic transmembrane helix (TMH) in all sequences (Fig. 3, KSHV: aa733-752). This domain has a similar hydrophobic character to domains previously identified in other herpesvirus gB homologs, which anchor gB within cellular membranes and virion envelopes. Two membrane proximal regions MPRa (aa688-705) and MPRb (aa707-730), immediately upstream of the transmembrane domain, were highly conserved between KSHV and the macaque rhadinovirus gB homologs (Fig. 3). MPRa and MPRb correspond to amphipathic domains that were previously predicted to be membrane-spanning in the gB homologs of HSV-1 (Pellett et al., 1985b) and EBV (Pellett et al., 1985a). In those studies, the authors proposed that gB traverses the membrane three times using the MPRa, MPRb and TMH domains. Subsequent studies identified a single membrane spanning domain in the HSV-1 gB (Rasile et al., 1993), while the domains corresponding to MPRa and MPRb within CMV gB were predicted to associate with the lipid membrane but not to span it (Spaete et al., 1988).
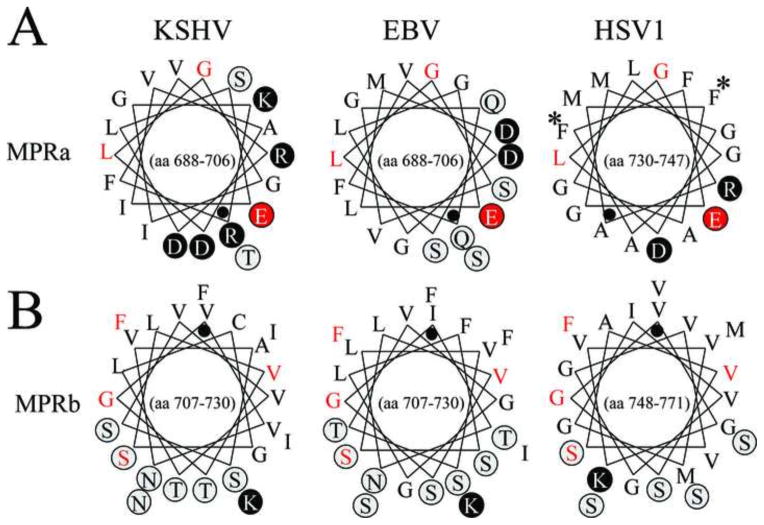
The structure of the MPR is unknown but is presumed to be in close proximity to the fusion loops and functions during interactions with target cell lipid membranes. More recently, the MPR domains were suggested to regulate exposure of hydrophobic fusion loops that mediate fusion of the viral and host lipid membranes during virus entry. The aromatic residues, Phe732 and Phe738 within the MPRa domain of HSV1 gB are predicted to interact with corresponding aromatic residues in the fusion loops, shielding them prior to membrane fusion (Shelly et al., 2012). We compared the MPR domains of KSHV with those of HSV-1 and EBV. Figure 5 shows helical wheel diagrams of the corresponding amphipathic domains, using the KSHV gB sequence as a prototype for the Old World rhadinoviruses. Comparison of the MPRa domains of KSHV, EBV and HSV1 revealed a strong conservation of the hydrophobic face of the predicted amphipathic helices containing hydrophobic residues Leu, Ile, Met, Val, and Phe (Fig. 5A). The residues Glu695, Leu700, and Gly701 in KSHV gB were conserved in EBV and HSV1 gB, anchoring the amphipathic wheel structure. However, both KSHV and EBV gB lacked the aromatic homologs of Phe732 and Phe738 found in HSV1 gB (Fig. 5, labeled with asterix), although conserved leucines were present in positions homologous to HSV1 Phe738. The MPRb domain, which is positioned immediately upstream of TMH, has the most hydrophobic character with approximately two-thirds of the helical face composed of hydrophobic amino acids. The amino acid sequence and amphipathic nature of the MPRb domain of KSHV gB were very similar to the corresponding domains in the other viral sequences. Residues Ser720, Val722, Gly724 and Phe728 in MPRb of KSHV gB are completely conserved with corresponding residues in EBV and HSV1 gB homologs, and anchor the amphipathic wheel structure (Fig. 5, red highlight).
Figure 5.
Conserved structure of the gB membrane proximal regions (MPR). The helical wheel structures of the predicted amphipathic helices of the membrane proximal regions MPRa and MPRb (see Fig. 3), are shown for the gB homologs of the human herpesviruses KSHV, EBV and HSV-1. Polar and charged amino acids are circled and highlighted in grey and black, respectively. A black dot indicates the N-terminal residue of the helical domain which continues counterclockwise for the sequence range indicated within the wheel. Amino acid numbering is derived from the gB precursors where the initiating methionine is the first amino acid. Amino acids conserved across the three sequences are colored red.
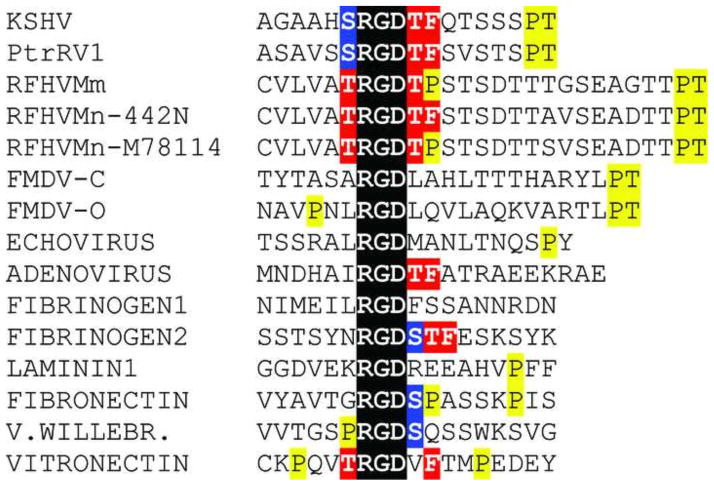
Identification of an RGD motif conserved within the gB homologs of the RV1 lineage of KSHV-like rhadinoviruses. A search for conserved sequence motifs revealed the presence of an “Arg-Gly-Asp” (RGD) motif within the N-terminal region of the KSHV, PtrRV1, RFHVMm and RFHVMn gB sequences (Fig. 3, KSHV: aa27-29). This motif was not present in the gB sequences of any of the members of the RV2 lineage of rhadinoviruses, EBV or other known herpesviruses. The RGD motif was located immediately downstream of the predicted signal peptidase cleavage site in a region that is otherwise very poorly conserved. RGD motifs have been detected in a number of extracellular matrix proteins including vitronectin, fibrinogen, laminin, fibronectin and von Willebrand factor and mediate binding to members of the superfamily of integrin receptors which are involved in cell adhesion, growth, differentiation, and motility (Ruoslahti and Pierschbacher, 1987). Integrin αVβ3, the vitronectin receptor, is the prototype RGD-binding integrin, and its RGD binding domain has been structurally characterized (Xiong et al., 2002). We have shown that the KSHV gB RGD motif binds specifically with purified integrin αVβ3. Furthermore, RGD mimetics, including cyclic and dicyclic RGD peptides, as well as antibodies to αVβ3 block KSHV infection (Garrigues et al., 2008).
The RGD motifs and flanking regions of the gB homologs of KSHV, PtrRV1, RFHVMm and two strains of RFHVMn: 442N and M78114 were compared to those of several viral and extracellular matrix proteins which bind αVβ3 and other RGD-binding integrins. The RGD flanking sequences of KSHV, RFHVMm and RFHVMn were quite similar to the sequences flanking the RGD motifs in the human and macaque extracellular matrix proteins, especially vitronectin (Fig. 6). These similarities included serine and threonine residues immediately upstream and downstream of the RGD, a phenylalanine residue positioned closely downstream and a proline residue positioned further downstream. Importantly, the sequence “TRGDXF in vitronectin is identical to the RGD motif of RFHVMn-442N, strongly suggesting that the RFHVMn gB RGD motif would bind αVβ3. Several viruses, including adenovirus 2, foot-and-mouth disease virus (FMDV) and human parechovirus 1 (HPEV1) encode RGD-containing viral proteins that interact with αVβ3 during virus binding and internalization (Neff et al., 1998; Triantafilou et al., 2000; Wickham et al., 1993). Strong similarities were detected between the RGD and flanking sequences of the adenovirus 2 penton protein and the sequences of KSHV, PtrRV1, and RFHVMn-442N where the sequence “RGDTF” was conserved. Comparison of the RGD motifs in the RV1 rhadinoviruses revealed an identical motif “SRGDTF” in the KSHV and chimpanzee PtrRV1 sequences, which was closely related to the “TRGDTF” motif in RFHVMn strain 442N. RFHVMm and RFHVMn-M78114 contained the conserved sequence “TRGDTP”, which was similar to the “RGDSP” motif present in fibronectin, whose binding to αVβ3 has been characterized by electron microscopy (Adair et al., 2005).
Figure 6.
Comparison of integrin-binding RGD domains. The RGD motif and flanking sequences of the gB homologs of the human, chimpanzee and macaque RV1 rhadinoviruses are compared to a number of other viral and cellular integrin binding proteins. The RGD motif is highlighted in black and conserved serine, threonine, proline and phenylalanine residues are indicated. The gB sequences of KSHV, PtrRV1, RFHVMn-M78114, and RFHVMn-442N are from the accession records indicated in Figure 1. The remainder of the protein sequences were obtained from the following accession records: gB sequence of FMDV-C = Foot and mouth disease virus, serotype C1 - viral protein 1 (VP1) (JC1329); FMDV-O = Foot and mouth disease virus, serotype O - VP1 (P03305); Parechovirus 1 - VP1 (Q66578); Adenovirus 2, penton base (NP_040521); fibrinogen1 and 2 - alpha chain (AAA52426); laminin 1 – alpha-1 precursor (NP_032506); fibronectin (AAD00019); von Willebrand’s factor (NP_000543); vitronectin (P04004). The sequences of the macaque homologs of fibronectin, von Willebrand’s factor and vitronectin were identical to the human sequence.
Identification of additional sequence motifs conserved between the gB homologs of the RV1 and RV2 rhadinoviruses. Further analysis of amino acid alignments in Figure 3 revealed the conservation of several sequence motifs that have been previously identified in other herpesvirus gB homologs. A clustering of acidic residues (glutamic and aspartic acids) was detected at the C-terminus of the seven rhadinovirus sequences, centered at aa840 in KSHV gB (Fig. 3). This region is equivalently positioned and homologous to an acidic cluster in human CMV gB that has been shown to be a signal for endocytosis from the plasma membrane (Tugizov et al., 1999). Two C-terminal Yxxϕ motifs, where ϕ indicates a hydrophobic residue, were in an equivalent position to similar motifs within the varicella zoster (VZV) herpesvirus gB that mediate endocytosis and Golgi localization (Heineman and Hall, 2001). The one Yxxϕ motif conserved with EBV gB is positionally equivalent to a Yxxϕ motif in HSV-1 gB that does not function in endocytosis (Fan et al., 2002). Furthermore, a region upstream of the C-terminal Yxxϕ motif is similar to an equivalently positioned inner nuclear membrane localization signal (LSINM) shown to be functional in human CMV gB (Meyer and Radsak, 2000). The CMV sequence, “DRLRHR”, is sufficient for nuclear envelope translocation with the two arginine residues in positions 4 and 6 essential for function. A close sequence similarity to this motif was detected within the seven rhadinovirus gB homologs with the conservation of the two critical arginines in every sequence except for PtrRV1 which had a lysine (Fig. 3, aa823-828).
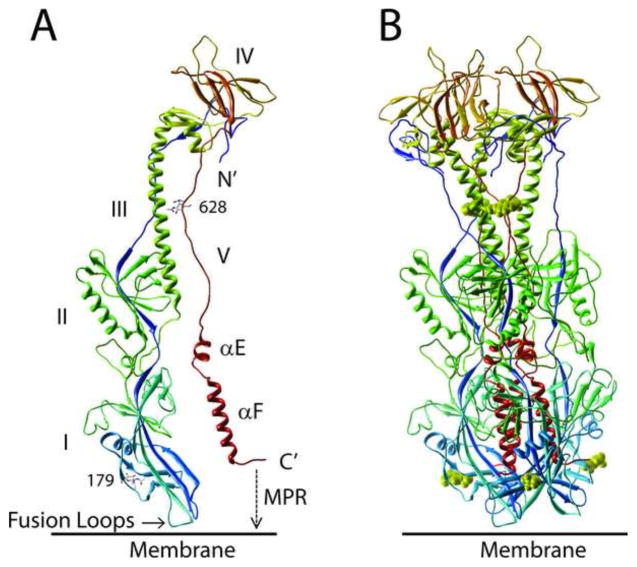
A structural model of KSHV gB revealed strong similarities to the structures of the HSV-1 and EBV gB homologs. As shown in Figure 3, the rhadinovirus gB homologs show strong sequence homology to the EBV gB, including the conservation of 10 cysteines forming 5 critical disulphide-linkages. The crystal structure of EBV gB ectodomain (3fvc) has been determined, and was found to have a similar architecture to the gB of herpes simplex virus (Heldwein et al., 2006) and the G protein of vesicular stomatitus virus (Roche et al., 2006), which have been classified as class III viral fusion proteins. We generated top-scoring homology model structures of KSHV gB from the crystal structure of EBV gB (Backovic et al., 2009) using MODELLER, RAMP and ROSETTA programs, as described in Methods. This model was estimated to be of high quality <4.1 ± 0.9 Angstroms all atom RMSD with a probability of 5.479e-3 using the ModFOLD4 server (Buenavista et al., 2012) that the model was correct. The model was deposited in the “Model Archive” (accession – ma-ay03r). The EBV and HSV1 gB homologs form trimer structures with three subunits wrapping around each other through multiple contact surfaces (Backovic et al., 2009; Heldwein et al., 2006). The proposed model structures of the KSHV gB ectodomain monomer and trimer are shown in Figure 7A and B.
Figure 7.
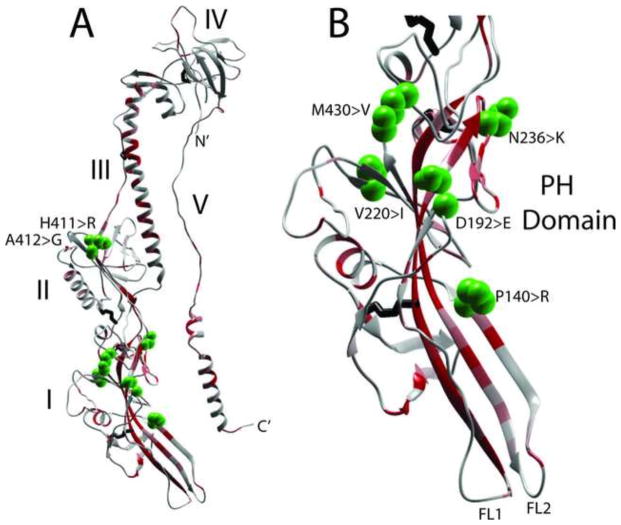
Structural model of KSHV gB ectodomain. A homology model of KSHV gB ectodomain (aa51-685) was determined from the crystal structure of the post-fusion EBV gB(Backovic et al., 2009), using MODELLER, RAMP and ROSETTA programs. A) monomer and B) trimer structure. The gB subunits are colored in blue (N-terminus) to red (C-terminus). Numbers indicate conserved Asn residues that are glycosylated in EBV gB (see Fig. 3); a single NAG molecule is modeled. The domain structures are numbered I–V, as defined in the EBV and HSV1 gB structures (Backovic et al., 2009; Heldwein et al., 2006). The ectodomain extends C-terminal of the two alpha helical domains (αE, αF) into the amphipathic membrane proximal region (MPR) and the membrane-spanning transmembrane domain (not shown), and the hydrophobic fusion loops are indicated.
A strong structural homology to the EBV gB was evident for KSHV gB, showing an elongated rod-like molecule with conservation of five domains that contain β-sheet and α-helical secondary structures (Fig. 7). The structure only shows the KSHV gB ectodomain (aa51-685) as it terminates prior to the membrane proximal regions (MPR) discussed in Figure 5 and the transmembrane helix (TMH) (see Figure 3). The structure lacks the N-terminus of KSHV gB due to the disorder in this region in EBV gB, and thus does not contain the RGD domain. The position of the two N-linked glycosylation sites of KSHV (Asn179, Asn628) that are conserved with known glycosylation sites in EBV gB are shown (Figs. 3, 4 and 7). The C-terminus of the ectodomain is located at the same end of the molecule as the putative fusion loops, predicted to function in fusion of the virion and cellular membranes during virus entry (Backovic et al., 2007). The putative KSHV fusion loop 1 (FL1) within domain I contains hydrophobic residues Trp209 and Phe210, which are completely conserved in the seven rhadinovirus gB homologs, with the exception of PtrRV2 in which Phe210 (KSHV numbering) is replaced with Leu (Figs. 3 and 4). These hydrophobic residues correspond to Trp193 and Leu194 within the EBV FL1 (Fig. 3), which are critical for fusion activity (Backovic et al., 2007). Whereas EBV FL1 contains two adjacent hydrophobic residues Ile195 and Trp196, the rhadinoviruses have a completely conserved “Pro-Gly” dipeptide (Pro211 and Gly212 in KSHV) at this position, indicating a sharp delineation of the hydrophobic structure of FL1 (Fig. 3). The KSHV fusion loop 2 (FL2) contains Leu128 and Thr129 corresponding to Trp112 and Tyr113 within EBV FL2 (Fig. 3). While the Thr residue is conserved in all of the rhadinovirus sequences, the Leu residue is only present in the FL2 of KSHV and the chimpanzee PtrRV1. The macaque RFHVMn and RFHVMm contain a Met in place of Leu at this position, while the chimpanzee PtrRV2 and macaque RRV and MneRV2 contain a Trp residue, similar to EBV.
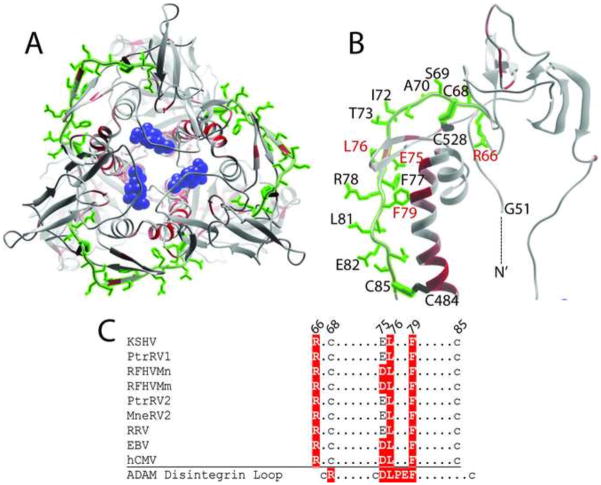
The disintegrin-like domain (DLD) of KSHV gB is highly conserved and maps to an exposed loop connecting domains III and IV. Disintegrin domains have been identified in proteins that bind to a number of different integrins, including integrin α9β1. A conserved disintegin motif “RX6DLPEF” was identified within members of the ADAMs family of metalloproteases, which binds to α9β1 integrin (Eto et al., 2002). The gB homologs of KSHV and hCMV contain disintegrin-like domains (DLD), which have weak similarity to the disintegrin motif in the ADAMs family, and bind to α9β1 integrin (Feire et al., 2004; Walker et al., 2014). To map the DLD motif onto the model structure of KSHV gB, a protein meta-functional signature (MFS) analysis of gB was performed, which combines sequence, structure, evolution and amino acid property information to provide a quantitative measurement of the functional importance of each amino acid in a protein (Horst and Samudrala, 2010; Wang et al., 2008). MFS scores were determined and visualized as a temperature gradient in Chimera and mapped onto the KSHV gB structure. The DLD motif of KSHV gB maps to the N-terminal region between Arg66 and Cys85, at the juncture of domains III and IV (see Fig. 3). We examined the position of the DLD motif on the model structure of KSHV gB, coloring the amino acid side chains green. The DLD motif was positioned on the exposed surface of the gB ectodomain connecting the N-terminus to domains III and IV through disulphide bonds linking Cys68 and Cys85 to Cys528 and Cys484, respectively (Fig. 8A and B). While the consensus disintegrin binding motif in the ADAMs family contained a highly conserved “DLPEF” sequence that was critical for binding to α9β1, this motif was only minimally conserved in the gB homologs of hCMV and KSHV (Fig. 8C). A comparison of the gB homologs of the RV1 and RV2 rhadinovirus lineages showed that, like EBV and HCMV, the motif “DLxxF” was conserved in the RFHVMn and RFHVMm gB sequences (Figs. 3 and 8C). The other rhadinovirus sequences, including KSHV contained the sequence “ELxxF” at this position, with a conserved substitution of Glu for Asp. hCMV, EBV and all of the rhadinovirus gB homologs contained a conserved Arg upstream of the “E/DLxxF motif, similar to the upstream Arg in the ADAMs consensus disintegrin motif. The model structure of KSHV gB demonstrates the exposed position of most of the amino acid side chains from Cys68 to Cys95 in the disintegrin-like domain (Fig. 8A and B).
Figure 8.
Structural model and conservation of a disintegrin-like domain (DLD) in KSHV gB. A conserved DLD motif (aa66-85) identified previously in KSHV gB (Walker et al., 2014) was mapped onto the KSHV gB trimer (A) and monomer (B) model structures. The amino acid side chains in the motif were colored green and are shown in (B). The functional importance of each amino acid was determined by meta-functional signature (MFS) analysis and the scores were mapped onto the KSHV gB structure as a temperature gradient. Predicted disulfide linkages between C68/C528 and C85/C484 are shown. The N-terminal G51 of the ectodomain is indicated. C) Conservation of the DLD motif across the RV1 and RV2 rhadinovirus gB sequences compared to the homologous regions in EBV and hCMV gB. Residues conserved with the disintegrin loop of the ADAMS family, which bind to α9β1 integrin (Feire et al., 2004), are colored red (B) and highlighted in red (C). The NAG molecule bound to N628 is colored blue in B.
KSHV gB domain I contains a highly conserved pleckstrin homology domain. As in EBV and HSV1 gB (Backovic et al., 2009; Heldwein et al., 2006), the KSHV gB domain I contains a fold resembling a pleckstrin-homology (PH) domain (Fig. 9). PH domains are characterized by a β-sandwich composed of two nearly orthogonal β-sheets of four and three strands, respectively (Blomberg et al., 1999). The PH domain fold is believed to be a stable structural fold that is able to display ligand-binding domains that can be utilized for various functions (Lemmon et al., 1996). In many cases, PH domains contain a ligand binding pocket (PHBP) formed by variable length loops between the beta strands, which has an overall positive charge and binds acidic phospholipids. The KSHV gB PH domain contains the two cysteine residues, Cys52 and Cys58, not found in EBV gB. These cysteine residues are predicted to form a disulfide bridge within a loop of the PH domain (Figs. 3, 9). The KSHV gB PH domain is adjacent to a long loop containing the furin protease cleavage site after Arg439 (Fig. 9). The β-sheet structure of the KSHV gB PH-like domain is shown in Fig. 9 using the strand numbering used for the EBV gB structure (Backovic et al., 2009). Strands β4 and β11 are the central strands of the curving 4-strand β-sheet within the PH domain, containing 3 long strands and one short strand (Fig. 9). The convex side of this subdomain, containing a putative PH binding face (PHBF) discussed below, is covered by a short 2-strand β-sheet, an α-helix and a β-hairpin turn (Fig. 9). The C-terminal region of the β11 strand is a critical component of one of the β-sheets of the PH domain, while the N-terminus is proximal to the hydrophobic residues of FL1. The N-terminal region of the β4 strand interacts with the C-terminal region of β11 within the PH domain, while the C-terminus is proximal to the hydrophobic residues of FL2. Opposite the PHBF, on the concave side of this structure, the β-strands of the PH domain open with loop structures forming a possible PH binding pocket (PHBP) similar to the binding sites for phosphoinositide (Lemmon et al., 1996). The protein meta-functional signature (MFS) analysis (Horst and Samudrala, 2010; Wang et al., 2008) revealed a high concentration of functional sites (red) in domain I of the KSHV gB, especially within the PH domain and associated β-sheet structures of the fusion loops (Fig. 9).
Figure 9.
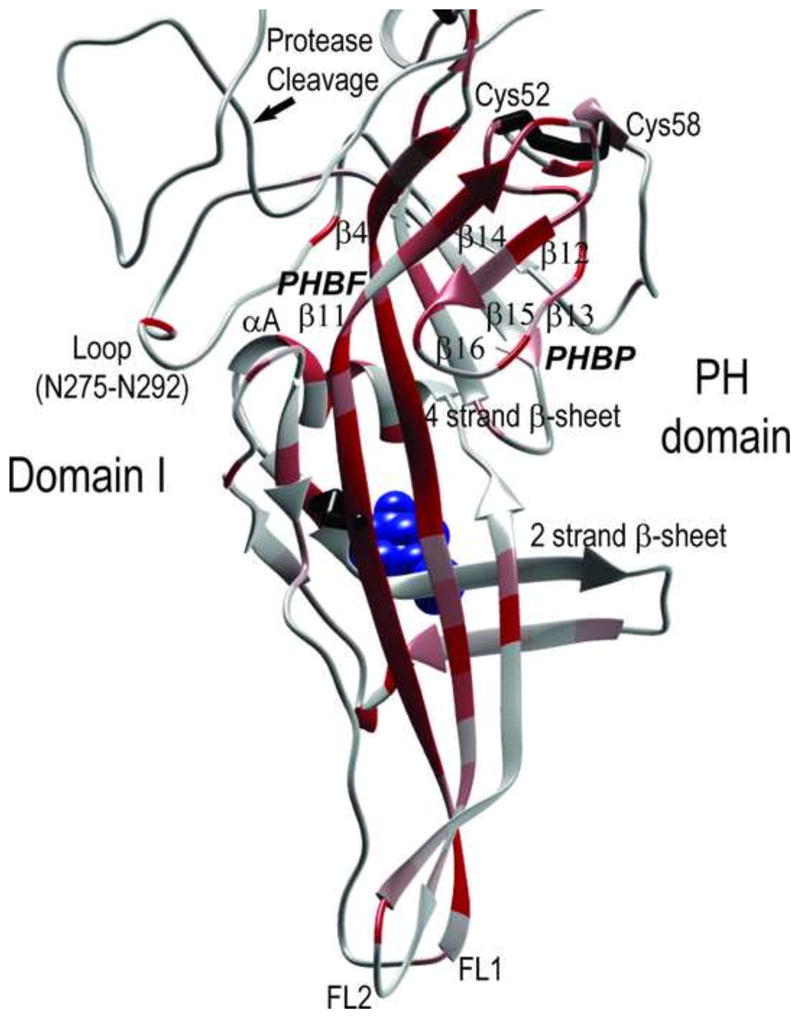
Model structure of KSHV gB domain I containing a pleckstrin homology (PH) domain and associated hydrophobic fusion loops. The homology model of domain I of KSHV gB is shown with the functional importance of each amino acid indicated by a temperature gradient of MFS scores. The high scoring pleckstrin homology (PH) domain containing orthogonal β-sheets of four and three strands is shown, and the proposed convex binding face (PHBF) and concave binding pocket (PHBP) are indicated. The β-strands are numbered by homology to the EBV gB structure, and the proposed rhadinovirus-specific disulfide bridge between C52 and C58 in the PH domain is shown. The loop between N275 and N292 and the alpha helix (αA) that cover the PHBF are denoted. The positions of the furin protease cleavage site and the NAG molecule (blue) bound to N179 are indicated. The extensions of strands β11 and β4 within the PH domain form the hydrophobic fusion loops FL1 and FL2, respectively.
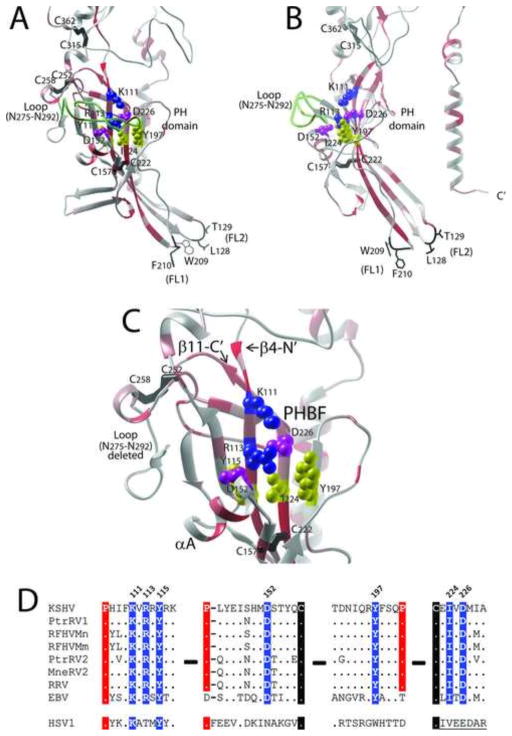
A putative heparin binding motif identified previously in KSHV gB is located within the PH domain and shows little structural similarity to known heparin binding domains. A positively charged linear amino acid motif (HIFKVRRYRKI; aa108-118) within the KSHV gB was previously identified as a heparan sulfate binding domain, since it matched the consensus heparin binding sequence “BHHBHBBHBB (H=hydrophobic; B=basic) (Akula et al., 2001). The core of this motif (KVRRYRK) is identical in all seven rhadinovirus gB sequences (Fig. 3). This motif mapped to the C-terminal end of strand β4 within the PH domain and scored highly in the MFS analysis (Fig. 10A, B, and C). While the positively charged side chains of K111 and R113 within this motif were directed to the convex side of the curved PH subdomain forming a possible binding face (PHBF), these residues were immediately flanked by the negatively charged side chains of D226 and D153 on strand β11 and the short α-helix (αA) covering the β-sheet, respectively. In addition, loop (N275-N292) blocked access to the positive charged residues (Figs. 9, 10A, B, and C). Examination of the sequence conservation between the rhadinovirus, EBV and HSV1 gB sequences revealed a complete conservation of Lys and Tyr residues corresponding to K111 and Y115 within the putative heparin binding motif of KSHV gB (Fig. 10D). However, the other positively charged residues in the KSHV sequence that matched with the consensus heparin binding sequence (ie. R114, R116) were not conserved in EBV and HSV1 gB. It is interesting to note that this subdomain contains a number of residues with side chains directed from the convex face of the β-sheet, including Y115, D152, Y197, I224 and D226 forming a possible PH-binding face (PHBF) (Fig. 10C). These residues are completely conserved in EBV gB and have semi-conservative substitutions in HSV1 gB (Fig. 10D). All of these binding face residues scored highly in the MFS analysis.
Figure 10.
The model structure of a putative heparan binding domain in KSHV gB has little similarity to known heparan binding domains. A) A previously identified positively charged heparan sulfate binding motif “HIFKVRRYRKI” (aa108-118) of KSHV gB (Akula et al., 2001) was mapped to the N-terminal region of strand β4 within the PH domain of KSHV gB. B) The side chains of basic residues K111 and R113 (blue) and polar residue Y115 (yellow) extend from the convex PH binding face of the four β-stranded sheet covered by Loop N275-N292. C) The highly conserved structure of the PH binding face positions negative residues D152 (αA helix) and D226 (β-11) (purple) and polar residue Y197 (yellow) adjacent to K111 and R113, disrupting the positive charge on this binding face. (Loop N275-N292 is deleted for visualization purposes). The proposed rhadinovirus-specific disulfide bond between C252 and C258 is shown. D) The conserved discontiguous residues, which form the convex PH binding face (PHBF) of KSHV gB are shown (highlighted in blue), with conserved prolines and cysteines highlighted in red and black, respectively.
Genetic variation in KSHV gB differs from that seen in the gB homologs of the macaque rhadinoviruses. To examine the genetic variation in gB between closely related rhadinovirus strains, the positions of non-synonymous sequence variants were mapped onto the linear gB structure. For KSHV, we compared the gB sequence of the GK18 strain (NC_009333) from a patient with classical (non-HIV) KS with the VG1 strain (ADB08179), an African B-subtype. Additional KSHV gB sequences from 16 Zambian KSHV strains (Olp et al., 2015) and strain JSC-1 (ACY00399) were also compared. The only other KSHV gB sequences available were essentially identical to GK18. A total of 19/2538 bp were different between GK18 and VG1, of which only 5 (26.3%) changed the amino acid sequence. Five additional amino acid variants were detected in the Zambian KSHV strains. These 10 amino acid differences were widely distributed across the gB sequence, with one variant F19>L occurring within the signal peptide, four variants P140>R, D192>E, N236>K and V270>I occurring within the PH subdomain of domain I, two variants H412>R and A413>G occurring in domain II, one variant M431>V adjacent to the furin protease site, and three variants L759>M, L808>M and T843>A occuring within the cytoplasmic domain (Figs. 11A and 12). Surprisingly, no amino acid differences were detected in domains III, IV or V (Fig. 12).
Figure 11.
Genetic variants in the human and macaque rhadinovirus gB homologs.
The non-synonymous sequence variants between A) KSHV strains (GK18, VG1, JSC1 and 16 Zambian KSHV variants), B) two RFHVMn strains (M78814 and 442N), C) two RRV strains (17577 and H2695) and two MneRV2 strains (J97167 and 98126) were mapped onto the linear gB sequence. Each vertical line represents a variant, and the amino acid changes from the GK18 reference sequence are shown for KSHV: F19>L (ZM95), P140>R (VG1, ZM4, 27), D192>E (ZM95, 102, 116, 117, 118), N236>K (ZM91), V270>I (VG1, ZM4, 27), H412>R (VG1, ZM4, 27), A413>G (ZM106), M431>V (VG1, ZM4, 27, 114, 128, 130), L759>M (JSC-1). The positions of the signal peptide (SP), RGD motifs, fusion loops (FL2, FL1), furin protease cleavage site (P), membrane proximal amphipathic domains (MPR), hydrophobic transmembrane domains (TMH) and cysteines and disulphide bonding pattern are indicated. Numbering is from the KSHV sequence (Fig. 3).
Figure 12.
KSHV strain variants mapped to gB structure.
The non-synonymous sequence variants of KSHV strains described in Figure 11 were mapped to the KSHV gB ectodomain model structure (aa51-685), shown with the temperature gradient of protein meta-functional signature (MFS) scores showing a quantitative measurement of the functional importance of each amino acid in a protein, as described in the text. A) complete monomer showing the five gB domains and B) enlargement of domain I containing the PH domain and the fusion loops. The amino acid side chains of the variable amino acids are shown as green spheres and the amino acid changes are shown.
The complete gB sequences have been determined for two strains of RFHVMn, strain M78814 (AGY30687) and strain 442N (AF204166). The RFHVMn nucleotide sequences varied between the two strains at 137 positions, with 22 non-synonymous changes resulting in amino acid differences (16.1%). The majority of these changes flanked the furin protease site (Fig. 11B). A number of other changes were in the N-terminal domain downstream of the RGD motif or at the junction between domains I and II adjacent to aa300 (KSHV numbering). Other variants were scattered in domain II upstream of the furin cleavage site. In contrast to KSHV gB variants, no sequence differences were detected in the PH subdomain nor in the cytoplasmic domain. We also examined the sequence variation of the gB homologs of two different strains of the rhesus macaque RRV, strains 17577 (NP_570749) and 26–95 (AF210726), and two different strains of the pig-tailed macaque MneRV2, strains J97167 (AJE29647) and 98126 (Auerbach et al., 2000). For the rhesus RRV strains, 71 nucleotide changes were observed with 19 (26.3%) non-synonymous changes altering amino acid sequences. While the nucleotide sequence of the pig-tail MneRV2 strain 98126 was not published, there were 19 non-synonymous changes between the J97167 and 98126 strains of MneRV2, which altered amino acid sequences. In both the RRV and MneRV2 cases, the amino acid changes mapped to the same regions as the changes in the RFHVMn strains. The majority of the variants mapped near the furin protease cleavage site, with additional variants mapping at the junction of domains I and II (Fig. 11C). No variants were detected in the PH domain, fusion loops or in the cytoplasmic domain.
SUMMARY
In this report, we have analyzed, characterized and compared the virion-associated gB of KSHV with gB homologs of other Old World primate rhadinoviruses of the RV1 and RV2 lineages. Complete sequences were available for the gB homologs of rhadinoviruses infecting humans: KSHV (RV1 lineage), rhesus macaques: RFHVMm (RV1 lineage) and RRV (RV2 lineage), and pig-tailed macaques: RFHMn (RV1 lineage) and MneRV2/PMRV (RV2 lineage), respectively. The gB sequences of two distinct strains/isolates were available for each rhadinovirus, except for RFHVMm. To enlarge the evolutionary comparison, we have cloned and determined the complete gB sequence of the rhadinoviruses infecting chimpanzee: PtrRV1 (RV1 lineage) and PtrRV2 (RV2 lineage). Phylogenetic analysis of the gB sequences confirmed earlier studies with DNA polymerase sequences demonstrating the existence of the two distinct lineages of KSHV-like herpesviruses (Schultz et al., 2000), the RV1 group consisting of KSHV, PtrRV1, RFHVMn and RFHVMm, and the RV2 group consisting of PtrRV2, RRV and MneRV2/PMRV.
Other examples of closely related herpesvirus lineages co-infecting a single host species include human HSV-1 and HSV-2 and human herpesvirus-6A, -6B, and -7 (HHV-6A, HHV-6B, HHV-7). Based on sequence comparisons of the gB homologs, the RV1 and RV2 rhadinoviruses were more closely related to each other (59–64% amino acid identity) than HHV-6 and HHV-7 (56% amino acid identity), but more distantly related than HSV-1 and HSV-2 (86% amino acid identity). Recently, HHV-6A and HHV-6B have been designated as distinct viruses and their gB homologs were 96.1% identical. The emergence of such closely-related pairs of viruses is not thought to be due to host speciation events, but probably represents the development of novel host cell tropism and variations in viral life cycle, as has been proposed for the emergence of the three major subfamilies of the Herpesviridae (McGeoch et al., 1995). Although the biological differences between the two lineages of Old World primate rhadinoviruses have not yet been studied in detail, there are apparent differences in virus infectivity and transmission. The macaque members of the RV2 lineage of rhadinoviruses, including RRV and MneRV2, are easily isolated and grown lytically to high titer in cell culture in vitro (Bruce et al., 2009; Desrosiers et al., 1997). In contrast, the RV1 rhadinoviruses, including KSHV and RFHV, establish a more latent type of infection and are difficult to culture lytically in vitro (Moore and Chang, 2001)(our unpublished results). We have shown that RV2 rhadinoviruses establish lytic infections in Vero and HEK293 cells, whereas KSHV infections are latent in the same cell types (DeMaster and Rose, 2014). While the RV1 rhadinoviruses, KSHV and RFHV, appear to be latent in infected lymphoma and/or KS-like tumor cells (Antman and Chang, 2000; Bruce et al., 2006), the RV2 rhadinoviruses, RRV and MneRV2 appear to be lytically replicating in infected lymphoma cells (Bruce et al., 2012).
We previously determined that the KSHV gB contains an arginine-glycine-aspartic acid (RGD) motif that binds to the cell surface receptor, integrin αVβ3, and showed that the gB RGD domain mediates infection and cell adhesion through specific interactions with αVβ3 (Garrigues et al., 2008). Sequence analysis revealed additional RGD motifs in the N-terminus of the gB homologs of the chimpanzee and macaque RV1 rhadinoviruses. A strong conservation of the amino acid sequences flanking the RGD motifs with a consensus sequence of “S/TRGDTF/P” was observed. The RGD domain of RFHVMn most closely resembles the RGD domain of vitronectin, the integrin αVβ3 receptor, suggesting convergent evolution to optimize RGD binding with integrin αVβ3.
Our previous studies showed that KSHV virions bind to specific cell surface microdomains containing αVβ3, and that KSHV attachment and entry are dependent on αVβ3 integrin (Garrigues et al., 2014a; Garrigues et al., 2014b). Thus, the interaction of the gB RGD motif and αVβ3 is a major determinant of cell tropism for KSHV and most likely other RV1 rhadinoviruses. In contrast, no RGD motifs were detected in the gB homologs of the RV2 rhadinoviruses, including macaque RRV and MneRV2, or chimpanzee PtrRV2. Furthermore, we have shown that the RRV virion does not interact with purified integrins αVβ3 or α3β1 (Garrigues et al., 2008). Thus, members of the RV1 rhadinovirus lineage could share a tropism for cells expressing integrin αVβ3, which is not shared with members of the RV2 rhadinovirus lineage or other herpesvirus lineages. The gain of function to interact with αVβ3 integrin expressing cells may have played a critical role in the emergence of the RV1 rhadinovirus lineage.
We have shown that the gB homologs of KSHV and the macaque and chimpanzee rhadinoviruses contain eight conserved potential N-linked glycosylation sites (NXS/T). Analysis of the model KSHV gB structure reveals that all of these potential glycosylation sites are exposed on the surface of the protein trimer structure, except for the site at Asn628 in the core of Domain III. Four of the surface exposed glycosylation sites are located within the intrachain loop structures, Domain I: Cys158-Cys222, Cys252-Cys258; Domain II: Cys315-Cys362, and Domain IV: Cys550-Cys558. The putative glycosylation site at Asn628 in KSHV gB is conserved in EBV gB (Asn629), and this site has been determined to be glycosylated in EBV gB (Backovic et al., 2009). This site is conserved in HSV1 gB (Asn 674), and is flanked by Ile671 and Phe683 on the C-terminal arm of the HSV1 gB ectodomain. Mutational studies of HSV-1 gB indicate that Asn674 is not a site of glycosylation (Cai et al., 1988). However, Ile671 and Phe683 of HSV1 gB C-terminal arm are conserved with Ile625 and Phe631 of KSHV gB, flanking the Asn628 glycosylation site. These HSV1 residues have been shown to induce packing of the C-terminal arm against the coiled-coil structure of Domain III, providing the driving force for gB refolding from a prefusion to a postfusion conformation (Connolly and Longnecker, 2012). Thus, glycosylation of the KSHV (Asn628)/EBV(Asn629) may play a role in the conformational change of gB during virus entry of gammaherpesviruses.
Our sequence analysis is compatible with the hypothesis that the gB homologs of KSHV and the macaque and chimpanzee rhadinoviruses, like the homologs of other herpesviruses, are integral membrane proteins. A highly hydrophobic transmembrane helix domain from Leu733 to Leu752 in KSHV gB would anchor the protein in the membrane. This hydrophobic region is highly conserved in the macaque and chimpanzee RV1 and RV2 gB sequences. Two amphipathic domains (MPRa and MPRb) with strong hydrophobic character were identified immediately upstream and proximal to the transmembrane domain. The MPR regions were highly conserved across the rhadinovirus gB homologs. Similar hydrophobic membrane proximal regions have been identified in other herpesvirus gB homologs, where they have been implicated in membrane association and fusion events during virus entry (Shelly et al., 2012). Analysis of the amphipathic structure of the MPR regions revealed strong homology between the KSHV, HSV1 and EBV gB sequences, suggesting common functions.
To further explore the biological role of KSHV gB, we have generated homology model structures for the gB homolog of KSHV from the crystal structure of EBV gB (Backovic et al., 2009). This model was of high quality and closely matched the EBV structure, with conservation of sequence and domain structures. We performed a protein meta-functional signature (MFS) analysis of gB, which combines sequence, structure, evolution and amino acid property information to provide a quantitative measurement of the functional importance of each amino acid in a protein (Horst and Samudrala, 2010; Wang et al., 2008). The MFS scores were determined and visualized as a temperature gradient in Chimera and mapped onto the KSHV gB structure. Using this structure, we mapped the position of two motifs previously implicated in gB-mediated virus entry, including a disintegrin-like binding domain (Walker et al., 2014) and a heparan sulfate binding domain (Akula et al., 2001). We could not map the N-terminal RGD domain of KSHV gB, since the N-terminus of EBV gB was not resolved in the crystal structure. Our analysis revealed that the disintegrin-like domain was highly conserved in the gB homologs of KSHV and the macaque and chimpanzee RV1 and RV2 rhadinovirus homologs. Furthermore, this domain was positioned on the surface of the gB ectodomain connecting the N-terminus to domains III and IV, with exposure of the amino acid side chains, making it highly available for protein-protein interactions. A similar disintegrin-like domain was originally detected in human CMV, which had homology to the disintegrin motif in the ADAMS family of metalloproteases (Feire et al., 2004). A conserved motif of “RX8E/DLX2F in the rhadinovirus, EBV and CMV gB sequences weakly matched the ADAMS family “RX6DLPEF” motif, which mediates binding to integrin α9β1(Feire et al., 2004).
Our analysis revealed that a putative heparan sulfate binding motif “108HIFKVRRYRK117” of KSHV gB was highly conserved in the gB homologs of the macaque and chimpanzee RV1 and RV2 rhadinoviruses. This motif was originally identified due to a match with one of the heparin binding consensus sequences, and in the context of an isolated peptide was found to bind heparan sulfate (Akula et al., 2001). This motif mapped to strand β4 within the pleckstrin homology (PH) domain which was highly conserved in the KSHV, EBV and HSV1 structures. Examination of the putative KSHV gB heparin binding motif within the context of the conserved β-sheet structure revealed few similarities to typical heparin-binding domains. Such domains are accessible to the solvent and consist of flat surfaces or shallow grooves lined by 4–16 positively charged lysines and arginines where side chains would interact with the negatively charged heparan sulfate (Murphy et al., 2007; Schlessinger et al., 2000; Sue et al., 2004). Analysis of the 3-dimensional structure of the KSHV gB PH domain showed that K111, R113 and Y115 within the putative heparin binding peptide formed a possible binding site on the convex face of the PH domain. However, the adjacent side chains of the negatively charged D152 and D226 are predicted to inhibit interaction with negatively charged heparan sulfate moieties, and the N275-N292 loop structure would occlude the binding site. The side chains of the other positively charged residues R114 and R116 matching the heparin binding motif consensus are positioned on the opposite side of the conserved beta sheet structure, limiting their availability for binding heparan sulfate chains. This analysis indicates that the structure of this domain is not consistent with the ability to bind heparan sulfate chains, as previously proposed (Akula et al., 2001). Although a positively charged heparin binding motif “KPKKNRKPK” was identified in HSV1 gB, this motif mapped to a completely different region of the protein at aa67-75 within the N-terminal region of HSV-1 gB, in a position analogous to the RGD motif in KSHV gB (Laquerre et al., 1998). No similarly charged sequence was present in the KSHV gB N-terminal region. Thus, the role of heparin binding in gB-mediated virus entry should be further investigated.
Within the KSHV PH domain, the β4 strand containing the putative heparin binding domain and the β11 strand both scored highly in the metafunctional sequence analysis with the vast majority of the amino acid residues attaining the maximal score. To determine a possible role for this domain, we have examined the ability of the central peptide sequence “HIFKVRRYRKIATS” on strand β4 to alter KSHV virion binding and infection. We found that the peptide could significantly block binding of KSHV virions to cell surfaces, and moreover, could significantly block KSHV infection (data not shown). While this inhibitory activity could be due to the ability of the isolated peptide to bind heparan sulfate, as shown previously (Akula et al., 2001), it could also be due to interactions with other cellular or viral proteins implicated in KSHV binding and entry. Since the PH domain and its extended β-strands, including β4 and β11, form the hydrophobic loops implicated in fusion of the virion and cellular membranes, they may play critical roles in KSHV binding and entry.
Finally, we compared the gB sequences from closely related strains of KSHV, RFHVMn, RRV and MneRV2, and mapped the non-synonymous mutations altering amino acid sequences onto the gB structure. Eleven amino acid differences were detected between the available KSHV gB sequences. Four of these variations occurred within domain I, in the vicinity of the PH domain and associated fusion loops, suggestive of a concentrated adaptive evolution targeting this critical region of gB. Additional variations flanked the protease cleavage site or occurred within the intracellular C-terminal domain. In contrast, the two RRV strains, the two MneRV2 strains and the two RFHMn strains showed significantly more differences between the strain pairs, with the most sequence variation surrounding the internal furin protease cleavage site in Domains I and II. No changes were detected in the fusion loop, PH domain or intracellular domains. The differences in the adaptive gB variations of the KSHV strains and those seen in the different strains of the macaque rhadinoviruses suggests fundamental differences in the host-virus interaction.
MATERIALS AND METHODS
Tissue sources
Saliva samples were obtained from two chimpanzees (Austin and Cordova) at M.D. Anderson (provided by S. Shapiro), which were naturally infected with PtrRV1 and PtrRV2 rhadinoviruses.
DNA isolation
Frozen tissue samples were quickly thawed in the presence of a standard proteinase K extraction buffer containing 0.1% SDS by homogenization in a disposable homogenizer. Samples were digested at 55° C for several hours and the DNA was purified by standard phenol/chloroform extraction and ethanol precipitation.
Oligonucleotide primers
Consensus-degenerate hybrid oligonucleotide primers (CODEHOPs) were derived from the conserved motifs in the gammaherpesvirus ORF 7 (Transport Protein (TP)) (“NYSKA” primer) and ORF 9 (DNA polymerase) (“CVNVB” primer), essentially as described in (Rose, 2005). Gene specific oligonucleotide primers were designed from the sequences of the different PCR fragments obtained using the CODEHOPS PCR strategy.
Cloning and sequence analysis of chimpanzee rhadinovirus gB genes
CODEHOP PCR primers were used with various combinations of other CODEHOP and gene specific primers to obtain overlapping PCR fragments, essentially as described (Schultz et al., 2000). The correct-size PCR products were purified from excess primers using a Qiagen spin column and sequenced directly with CODEHOP or gene-specific primers. Additional internal gene-specific primers were used to obtain overlapping sequences within the larger PCR products. Multiple PCR products and clones were sequenced in both orientations. Sequence assembly was done using Sequencher 4.0.5b10 (GeneCodes). The PtrRV1 (Austin) and PtrRV2 (Cordova) sequences were deposited at Genbank, with accession numbers KU984977 and KU984976, respectively.
Sequence and phylogenetic analysis
Pairwise nucleotide and encoded amino acid alignments were performed using GenePro software (Riverside Scientific, Bainbridge Island, WA). Multiple sequence alignment was done using ClustalW (EMBL, Heidelberg, Germany). Phylogenetic analysis of amino acid sequences was done by protein maximum likelihood as implemented at LIRMM.fr (Dereeper et al., 2008). Neural network predictions of mucin type GalNAc O-glycosylation sites was performed using the NetOGlyc 2.0 Prediction Server at http://www.cbs.dtu.dk/services/NetOGlyc/ (Hansen et al., 1998). Signal sequence prediction was performed using SignalP v1.1 at Center for Biological Sequence Analysis (CBS) at http://www.cbs.dtu.dk/services/SignalP/. Transmembrane helices predictions were performed using TMHMM v.2.0 at the CBS (http://www.cbs.dtu.dk/services/TMHMM/)(Krogh et al., 2001), TMpred at EMBNET http://www.ch.embnet.org/software/TMPRED_form.html, PSI-PRED at the PSIPRED v2.1 server http://bioinf.cs.ucl.ac.uk/psiform.html (McGuffin et al., 2000), TM-Finder at HSC Bioinformatics Centre http://www.bioinformatics-canada.org/TM/ (Deber et al., 2001).
Structural modeling
Comparative modeling techniques were applied to model the monomeric structure of KSHV gB based on the alignment of the KSHV sequence to the EBV crystal structure (3fvc) (see Fig. 2), using the RAMP (Samudrala and Moult, 1998), Rosetta (Das and Baker, 2008) and MODELLER (Sali and Blundell, 1993) software suites. Oligomerization was modeled by superimposing the top 100 scoring monomer models from each modeling suite onto the three chains of 3fvc with the UCSF Chimera tool “Match Maker.” The monomer model which produced the fewest clashes was selected for further optimization using fragment-guided molecular dynamics (Zhang et al., 2011). The KSHV gB model was deposited in the “Model Archive” (modelarchive.org) under accession record “ma-ayo3r”. Protein meta-functional signatures (MFS) were generated using the MFS web server (http://protinfo.compbio.washington.edu/mfs) (Wang et al., 2008) and applied to the new structure file with the temperature factor field replaced by the MFS scores to enable visual inspection of functionally important regions using Chimera.
Highlights.
The sequences of the glycoprotein B genes of the chimpanzee PtrRV1 and PtrRV2 rhadinoviruses were determined
The evolutionary conservation of sequence and structural motifs was determined for the Kaposi’s sarcoma-associated herpesvirus (KSHV) and related Old World primate RV1 and RV2 rhadinoviruses
An RGD motif, shown to be involved in KSHV binding and entry through the integrin aVb3, was conserved within the RV1 rhadinovirus lineage but not within the RV2 rhadinovirus lineage
A structural model of the KSHV gB was determined and functional and sequence variants were mapped to the model structure.
Acknowledgments
We acknowledge S. Shapiro, University of Texas M.D. Anderson Cancer Center for his generous gift of chimpanzee saliva. This work was supported by the National Center for Research Resources (NCRR) and the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health (NIH) through grant RR023343 (to T. Rose) and by the National Institute for Dental and Craniofacial Research (NIDCR) through grants DE018927, DE021954 and (to T. Rose). The content is solely the responsibility of the authors and does not necessarily represent official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adair BD, Xiong JP, Maddock C, Goodman SL, Arnaout MA, Yeager M. Three-dimensional EM structure of the ectodomain of integrin {alpha}V{beta}3 in a complex with fibronectin. J Cell Biol. 2005;168:1109–1118. doi: 10.1083/jcb.200410068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akula SM, Pramod NP, Wang FZ, Chandran B. Human herpesvirus 8 envelope-associated glycoprotein B interacts with heparan sulfate-like moieties. Virology. 2001;284:235–249. doi: 10.1006/viro.2001.0921. [DOI] [PubMed] [Google Scholar]
- Alexander L, Denekamp L, Knapp A, Auerbach MR, Damania B, Desrosiers RC. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi’s sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol. 2000;74:3388–3398. doi: 10.1128/jvi.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antman K, Chang Y. Kaposi’s sarcoma. The New England journal of medicine. 2000;342:1027–1038. doi: 10.1056/NEJM200004063421407. [DOI] [PubMed] [Google Scholar]
- Auerbach MR, Czajak SC, Johnson WE, Desrosiers RC, Alexander L. Species specificity of macaque rhadinovirus glycoprotein B sequences. J Virol. 2000;74:584–590. doi: 10.1128/jvi.74.1.584-590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backovic M, Jardetzky TS, Longnecker R. Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity. J Virol. 2007;81:9596–9600. doi: 10.1128/JVI.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backovic M, Longnecker R, Jardetzky TS. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci U S A. 2009;106:2880–2885. doi: 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghian A, Luftig M, Black JB, Meng YX, Pau CP, Voss T, Pellett PE, Kousoulas KG. Glycoprotein B of human herpesvirus 8 is a component of the virion in a cleaved form composed of amino- and carboxyl-terminal fragments. Virology. 2000;269:18–25. doi: 10.1006/viro.2000.0198. [DOI] [PubMed] [Google Scholar]
- Blomberg N, Baraldi E, Nilges M, Saraste M. The PH superfold: a structural scaffold for multiple functions. Trends Biochem Sci. 1999;24:441–445. doi: 10.1016/s0968-0004(99)01472-3. [DOI] [PubMed] [Google Scholar]
- Britt WJ. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology. 1984;135:369–378. doi: 10.1016/0042-6822(84)90193-4. [DOI] [PubMed] [Google Scholar]
- Britt WJ, Vugler LG. Processing of the gp55-116 envelope glycoprotein complex (gB) of human cytomegalovirus. J Virol. 1989;63:403–410. doi: 10.1128/jvi.63.1.403-410.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt WJ, Vugler LG. Oligomerization of the human cytomegalovirus major envelope glycoprotein complex gB (gp55-116) J Virol. 1992;66:6747–6754. doi: 10.1128/jvi.66.11.6747-6754.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AG, Bakke AM, Bielefeldt-Ohmann H, Ryan JT, Thouless ME, Tsai CC, Rose TM. High levels of retroperitoneal fibromatosis (RF)-associated herpesvirus in RF lesions in macaques are associated with ORF73 LANA expression in spindleoid tumour cells. The Journal of general virology. 2006;87:3529–3538. doi: 10.1099/vir.0.82339-0. [DOI] [PubMed] [Google Scholar]
- Bruce AG, Bakke AM, Gravett CA, DeMaster LK, Bielefeldt-Ohmann H, Burnside KL, Rose TM. The ORF59 DNA polymerase processivity factor homologs of Old World primate RV2 rhadinoviruses are highly conserved nuclear antigens expressed in differentiated epithelium in infected macaques. Virology journal. 2009;6:205. doi: 10.1186/1743-422X-6-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AG, Bielefeldt-Ohmann H, Barcy S, Bakke AM, Lewis P, Tsai CC, Murnane RD, Rose TM. Macaque homologs of EBV and KSHV show uniquely different associations with simian AIDS-related lymphomas. PLoS pathogens. 2012;8:e1002962. doi: 10.1371/journal.ppat.1002962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AG, Ryan JT, Thomas MJ, Peng X, Grundhoff A, Tsai CC, Rose TM. Next-Generation Sequence Analysis of the Genome of RFHVMn, the Macaque Homolog of Kaposi’s Sarcoma (KS)-Associated Herpesvirus, from a KS-Like Tumor of a Pig-Tailed Macaque. J Virol. 2013;87:13676–13693. doi: 10.1128/JVI.02331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AG, Thouless ME, Haines AS, Pallen MJ, Grundhoff A, Rose TM. Complete genome sequence of Pig-tailed macaque rhadinovirus 2 and its evolutionary relationship with rhesus macaque rhadinovirus and human herpesvirus 8/Kaposi’s sarcoma-associated herpesvirus. J Virol. 2015;89:3888–3909. doi: 10.1128/JVI.03597-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenavista MT, Roche DB, McGuffin LJ. Improvement of 3D protein models using multiple templates guided by single-template model quality assessment. Bioinformatics. 2012;28:1851–1857. doi: 10.1093/bioinformatics/bts292. [DOI] [PubMed] [Google Scholar]
- Burke HG, Heldwein EE. Crystal Structure of the Human Cytomegalovirus Glycoprotein B. PLoS pathogens. 2015;11:e1005227. doi: 10.1371/journal.ppat.1005227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai WZ, Person S, DebRoy C, Gu BH. Functional regions and structural features of the gB glycoprotein of herpes simplex virus type 1. An analysis of linker insertion mutants. J Mol Biol. 1988;201:575–588. doi: 10.1016/0022-2836(88)90639-0. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma [see comments] Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Claesson-Welsh L, Spear PG. Amino-terminal sequence, synthesis, and membrane insertion of glycoprotein B of herpes simplex virus type 1. J Virol. 1987;61:1–7. doi: 10.1128/jvi.61.1.1-7.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly SA, Longnecker R. Residues within the C-terminal arm of the herpes simplex virus 1 glycoprotein B ectodomain contribute to its refolding during the fusion step of virus entry. J Virol. 2012;86:6386–6393. doi: 10.1128/JVI.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Deber CM, Wang C, Liu LP, Prior AS, Agrawal S, Muskat BL, Cuticchia AJ. TM Finder: a prediction program for transmembrane protein segments using a combination of hydrophobicity and nonpolar phase helicity scales. Protein Sci. 2001;10:212–219. doi: 10.1110/ps.30301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaster LK, Rose TM. A critical Sp1 element in the rhesus rhadinovirus (RRV) Rta promoter confers high-level activity that correlates with cellular permissivity for viral replication. Virology. 2014;448:196–209. doi: 10.1016/j.virol.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers RC, Sasseville VG, Czajak SC, Zhang X, Mansfield KG, Kaur A, Johnson RP, Lackner AA, Jung JU. A herpesvirus of rhesus monkeys related to the human Kaposi’s sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto K, Huet C, Tarui T, Kupriyanov S, Liu HZ, Puzon-McLaughlin W, Zhang XP, Sheppard D, Engvall E, Takada Y. Functional classification of ADAMs based on a conserved motif for binding to integrin alpha 9beta 1: implications for sperm-egg binding and other cell interactions. J Biol Chem. 2002;277:17804–17810. doi: 10.1074/jbc.M200086200. [DOI] [PubMed] [Google Scholar]
- Fan Z, Grantham ML, Smith MS, Anderson ES, Cardelli JA, Muggeridge MI. Truncation of herpes simplex virus type 2 glycoprotein B increases its cell surface expression and activity in cell-cell fusion, but these properties are unrelated. J Virol. 2002;76:9271–9283. doi: 10.1128/JVI.76.18.9271-9283.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feire AL, Koss H, Compton T. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc Natl Acad Sci U S A. 2004;101:15470–15475. doi: 10.1073/pnas.0406821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues HJ, DeMaster LK, Rubinchikova YE, Rose TM. KSHV attachment and entry are dependent on alphaVbeta3 integrin localized to specific cell surface microdomains and do not correlate with the presence of heparan sulfate. Virology. 2014a;464–465:118–133. doi: 10.1016/j.virol.2014.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues HJ, Rubinchikova YE, Dipersio CM, Rose TM. Integrin alphaVbeta3 Binds to the RGD motif of glycoprotein B of Kaposi’s sarcoma-associated herpesvirus and functions as an RGD-dependent entry receptor. J Virol. 2008;82:1570–1580. doi: 10.1128/JVI.01673-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues HJ, Rubinchikova YE, Rose TM. KSHV cell attachment sites revealed by ultra sensitive tyramide signal amplification (TSA) localize to membrane microdomains that are up-regulated on mitotic cells. Virology. 2014b;452–453:75–85. doi: 10.1016/j.virol.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greensill J, Schulz TF. Rhadinoviruses (gamma2-herpesviruses) of Old World primates: models for KSHV/HHV8-associated disease? Aids. 2000;14:S11–19. [PubMed] [Google Scholar]
- Greensill J, Sheldon JA, Murthy KK, Bessonette JS, Beer BE, Schulz TF. A chimpanzee rhadinovirus sequence related to Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8: increased detection after HIV-1 infection in the absence of disease. Aids. 2000a;14:F129–135. doi: 10.1097/00002030-200012010-00001. [DOI] [PubMed] [Google Scholar]
- Greensill J, Sheldon JA, Renwick NM, Beer BE, Norley S, Goudsmit J, Schulz TF. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among old world primates? J Virol. 2000b;74:1572–1577. doi: 10.1128/jvi.74.3.1572-1577.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gretch DR, Gehrz RC, Stinski MF. Characterization of a human cytomegalovirus glycoprotein complex (gcI) The Journal of general virology. 1988;69:1205–1215. doi: 10.1099/0022-1317-69-6-1205. [DOI] [PubMed] [Google Scholar]
- Haffey ML, Spear PG. Alterations in glycoprotein gB specified by mutants and their partial revertants in herpes simplex virus type 1 and relationship to other mutant phenotypes. J Virol. 1980;35:114–128. doi: 10.1128/jvi.35.1.114-128.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J Virol. 2007;81:4858–4865. doi: 10.1128/JVI.02755-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JE, Lund O, Tolstrup N, Gooley AA, Williams KL, Brunak S. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj J. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- Heineman TC, Hall SL. VZV gB endocytosis and Golgi localization are mediated by YXXphi motifs in its cytoplasmic domain. Virology. 2001;285:42–49. doi: 10.1006/viro.2001.0930. [DOI] [PubMed] [Google Scholar]
- Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Crystal structure of glycoprotein B from herpes simplex virus 1. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- Hensler HR, Tomaszewski MJ, Rappocciolo G, Rinaldo CR, Jenkins FJ. Human herpesvirus 8 glycoprotein B binds the entry receptor DC-SIGN. Virus research. 2014;190:97–103. doi: 10.1016/j.virusres.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst JA, Samudrala R. A protein sequence meta-functional signature for calcium binding residue prediction. Pattern recognition letters. 2010;31:2103–2112. doi: 10.1016/j.patrec.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka M, Nagahama M, Kim WS, Watanabe T, Hatsuzawa K, Ikemizu J, Murakami K, Nakayama K. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J Biol Chem. 1991;266:12127–12130. [PubMed] [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A. 2004;101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Gessain A. KSHV-like herpesviruses in chimps and gorillas. Nature. 2000a;407:151–152. doi: 10.1038/35025145. [DOI] [PubMed] [Google Scholar]
- Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Gessain A. A novel gamma 2-herpesvirus of the Rhadinovirus 2 lineage in chimpanzees. Genome Res. 2001;11:1511–1519. doi: 10.1101/gr.158601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste V, Mauclere P, Dubreuil G, Lewis J, Georges-Courbot MC, Rigoulet J, Petit T, Gessain A. Simian Homologues of Human Gamma-2 and Betaherpesviruses in Mandrill and Drill Monkeys. J Virol. 2000b;74:11993–11999. doi: 10.1128/jvi.74.24.11993-11999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laquerre S, Argnani R, Anderson DB, Zucchini S, Manservigi R, Glorioso JC. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J Virol. 1998;72:6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Schlessinger J. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- Mansfield KG, Westmoreland SV, DeBakker CD, Czajak S, Lackner AA, Desrosiers RC. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J Virol. 1999;73:10320–10328. doi: 10.1128/jvi.73.12.10320-10328.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ, Cook S, Dolan A, Jamieson FE, Telford EA. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Meyer GA, Radsak KD. Identification of a novel signal sequence that targets transmembrane proteins to the nuclear envelope inner membrane. J Biol Chem. 2000;275:3857–3866. doi: 10.1074/jbc.275.6.3857. [DOI] [PubMed] [Google Scholar]
- Moore PS, Chang Y. Molecular virology of Kaposi’s sarcoma-associated herpesvirus. Philos Trans R Soc Lond B Biol Sci. 2001;356:499–516. doi: 10.1098/rstb.2000.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JW, Cho Y, Sachpatzidis A, Fan C, Hodsdon ME, Lolis E. Structural and functional basis of CXCL12 (stromal cell-derived factor-1 alpha) binding to heparin. J Biol Chem. 2007;282:10018–10027. doi: 10.1074/jbc.M608796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff S, Sa-Carvalho D, Rieder E, Mason PW, Blystone SD, Brown EJ, Baxt B. Foot-and-mouth disease virus virulent for cattle utilizes the integrin alpha(v)beta3 as its receptor. J Virol. 1998;72:3587–3594. doi: 10.1128/jvi.72.5.3587-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norais N, Tang D, Kaur S, Chamberlain SH, Masiarz FR, Burke RL, Marcus F. Disulfide bonds of herpes simplex virus type 2 glycoprotein gB. J Virol. 1996;70:7379–7387. doi: 10.1128/jvi.70.11.7379-7387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olp LN, Jeanniard A, Marimo C, West JT, Wood C. Whole-Genome Sequencing of Kaposi’s Sarcoma-Associated Herpesvirus from Zambian Kaposi’s Sarcoma Biopsy Specimens Reveals Unique Viral Diversity. J Virol. 2015;89:12299–12308. doi: 10.1128/JVI.01712-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett PE, Biggin MD, Barrell B, Roizman B. Epstein-Barr virus genome may encode a protein showing significant amino acid and predicted secondary structure homology with glycoprotein B of herpes simplex virus 1. J Virol. 1985a;56:807–813. doi: 10.1128/jvi.56.3.807-813.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett PE, Kousoulas KG, Pereira L, Roizman B. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J Virol. 1985b;53:243–253. doi: 10.1128/jvi.53.1.243-253.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L. Function of glycoprotein B homologues of the family herpesviridae. Infect Agents Dis. 1994;3:9–28. [PubMed] [Google Scholar]
- Pereira L, Hoffman M, Gallo D, Cremer N. Monoclonal antibodies to human cytomegalovirus: three surface membrane proteins with unique immunological and electrophoretic properties specify cross-reactive determinants. Infect Immun. 1982;36:924–932. doi: 10.1128/iai.36.3.924-932.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Hoffman M, Tatsuno M, Dondero D. Polymorphism of human cytomegalovirus glycoproteins characterized by monoclonal antibodies. Virology. 1984;139:73–86. doi: 10.1016/0042-6822(84)90331-3. [DOI] [PubMed] [Google Scholar]
- Person S, Kousoulas KG, Knowles RW, Read GS, Holland TC, Keller PM, Warner SC. Glycoprotein processing in mutants of HSV-1 that induce cell fusion. Virology. 1982;117:293–306. doi: 10.1016/0042-6822(82)90470-6. [DOI] [PubMed] [Google Scholar]
- Pertel PE, Spear PG, Longnecker R. Human herpesvirus-8 glycoprotein B interacts with Epstein-Barr virus (EBV) glycoprotein 110 but fails to complement the infectivity of EBV mutants. Virology. 1998;251:402–413. doi: 10.1006/viro.1998.9412. [DOI] [PubMed] [Google Scholar]
- Rasile L, Ghosh K, Raviprakash K, Ghosh HP. Effects of deletions in the carboxy-terminal hydrophobic region of herpes simplex virus glycoprotein gB on intracellular transport and membrane anchoring. J Virol. 1993;67:4856–4866. doi: 10.1128/jvi.67.8.4856-4866.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen L, Mullenax J, Nelson R, Merigan TC. Viral polypeptides detected by a complement-dependent neutralizing murine monoclonal antibody to human cytomegalovirus. J Virol. 1985;55:274–280. doi: 10.1128/jvi.55.2.274-280.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- Rose TM. CODEHOP-mediated PCR - a powerful technique for the identification and characterization of viral genomes. Virology journal. 2005;2:20. doi: 10.1186/1743-422X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TM, Bosch ML, Strand K. Glycoprotein B of the RFHV/KSHV subfamily of herpes viruses. #6,022,542 United States Patent. 2000
- Rose TM, Ryan JT, Schultz ER, Raden BW, Tsai CC. Analysis of 4.3 kilobases of divergent locus B of macaque retroperitoneal fibromatosis-associated herpesvirus reveals a close similarity in gene sequence and genome organization to Kaposi’s sarcoma-associated herpesvirus. J Virol. 2003;77:5084–5097. doi: 10.1128/JVI.77.9.5084-5097.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TM, Schultz ER, Henikoff JG, Pietrokovski S, McCallum CM, Henikoff S. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic acids research. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TM, Strand KB, Schultz ER, Schaefer G, Rankin GW, Jr, Thouless ME, Tsai CC, Bosch ML. Identification of two homologs of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TM, Strand KB, Bosch ML. Glycoprotein B of the RFHV/KSHV subfamily of herpesviruses. #6,015,565 United States Patent. 1999
- Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–497. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Samudrala R, Moult J. An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. J Mol Biol. 1998;275:895–916. doi: 10.1006/jmbi.1997.1479. [DOI] [PubMed] [Google Scholar]
- Sarmiento M, Spear PG. Membrane proteins specified by herpes simplex viruses. IV. Conformation of the virion glycoprotein designated VP7(B2) J Virol. 1979;29:1159–1167. doi: 10.1128/jvi.29.3.1159-1167.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- Schultz ER, Rankin GW, Jr, Blanc MP, Raden BW, Tsai CC, Rose TM. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi’s sarcoma-associated herpesvirus. J Virol. 2000;74:4919–4928. doi: 10.1128/jvi.74.10.4919-4928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles RP, Bergquam EP, Axthelm MK, Wong SW. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cahill N, Prout ME, Miller G. A functionally distinct TATA box required for late progression through the Epstein-Barr virus life cycle. J Virol. 1998;72:8338–8343. doi: 10.1128/jvi.72.10.8338-8343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly SS, Cairns TM, Whitbeck JC, Lou H, Krummenacher C, Cohen GH, Eisenberg RJ. The membrane-proximal region (MPR) of herpes simplex virus gB regulates association of the fusion loops with lipid membranes. mBio. 2012;3 doi: 10.1128/mBio.00429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri ME, Sundqvist VA. An investigation into the heparin-binding properties of a synthetic peptide deduced from the antigenic domain 2 of human cytomegalovirus glycoprotein B. Scandinavian journal of immunology. 2001;53:282–289. doi: 10.1046/j.1365-3083.2001.00878.x. [DOI] [PubMed] [Google Scholar]
- Spaete RR, Thayer RM, Probert WS, Masiarz FR, Chamberlain SH, Rasmussen L, Merigan TC, Pachl C. Human cytomegalovirus strain Towne glycoprotein B is processed by proteolytic cleavage. Virology. 1988;167:207–225. doi: 10.1016/0042-6822(88)90071-2. [DOI] [PubMed] [Google Scholar]
- Spear PG. Membrane proteins specified by herpes simplex viruses. I. Identification of four glycoprotein precursors and their products in type 1-infected cells. J Virol. 1976;17:991–1008. doi: 10.1128/jvi.17.3.991-1008.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear PG, Longnecker R. Herpesvirus entry: an update. J Virol. 2003;77:10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand K, Harper E, Thormahlen S, Thouless ME, Tsai C, Rose T, Bosch ML. Two distinct lineages of macaque gamma herpesviruses related to the Kaposi’s sarcoma associated herpesvirus. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2000;16:253–269. doi: 10.1016/s1386-6532(99)00080-3. [DOI] [PubMed] [Google Scholar]
- Sue SC, Chen JY, Lee SC, Wu WG, Huang TH. Solution structure and heparin interaction of human hepatoma-derived growth factor. J Mol Biol. 2004;343:1365–1377. doi: 10.1016/j.jmb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Triantafilou K, Triantafilou M, Takada Y, Fernandez N. Human parechovirus 1 utilizes integrins alphavbeta3 and alphavbeta1 as receptors. J Virol. 2000;74:5856–5862. doi: 10.1128/jvi.74.13.5856-5862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugizov S, Maidji E, Xiao J, Pereira L. An acidic cluster in the cytosolic domain of human cytomegalovirus glycoprotein B is a signal for endocytosis from the plasma membrane. J Virol. 1999;73:8677–8688. doi: 10.1128/jvi.73.10.8677-8688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LR, Hussein HA, Akula SM. Disintegrin-like domain of glycoprotein B regulates Kaposi’s sarcoma-associated herpesvirus infection of cells. J Gen Virol. 2014;95:1770–1782. doi: 10.1099/vir.0.066829-0. [DOI] [PubMed] [Google Scholar]
- Wang K, Horst JA, Cheng G, Nickle DC, Samudrala R. Protein meta-functional signatures from combining sequence, structure, evolution, and amino acid property information. PLoS computational biology. 2008;4:e1000181. doi: 10.1371/journal.pcbi.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenske EA, Bratton MW, Courtney RJ. Endo-beta-N-acetylglucosaminidase H sensitivity of precursors to herpes simplex virus type 1 glycoproteins gB and gC. J Virol. 1982;44:241–248. doi: 10.1128/jvi.44.1.241-248.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- Wong-Ho E, Wu TT, Davis ZH, Zhang B, Huang J, Gong H, Deng H, Liu F, Glaunsinger B, Sun R. Unconventional sequence requirement for viral late gene core promoters of murine gammaherpesvirus 68. J Virol. 2014;88:3411–3422. doi: 10.1128/JVI.01374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liang Y, Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19:1784–1795. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]