Abstract
Phosphoinositide-specific-phospholipase Cβ (PLCβ) is the main effector of Gαq stimulation which is coupled to receptors that bind acetylcholine, bradykinin, dopamine, angiotensin II as well as other hormones and neurotransmitters. Using a yeast two-hybrid and other approaches, we have recently found that the same region of PLCβ that binds Gαq also interacts with Component 3 Promoter of RNA induced silencing complex (RISC) (C3PO), which is required for efficient activity of the RNA-induced silencing complex. In purified form, C3PO competes with Gαq for PLCβ binding and at high concentration can quench PLCβ activation. Additionally, we have found that the binding of PLCβ to C3PO inhibits its nuclease activity leading to reversal of RNA-induced silencing of specific genes. In cells, we found that PLCβ distributes between the plasma membrane where it localizes with Gαq, and in the cytosol where it localizes with C3PO. When cells are actively processing small interfering RNAs the interaction between PLCβ and C3PO gets stronger and leads to changes in the cellular distribution of PLCβ. The magnitude of attenuation is specific for different silencing RNAs. Our studies imply a direct link between calcium responses mediated through Gαq and post-transcriptional gene regulation through PLCβ.
Keywords: phospholipase Cβ, G protein signalling, RNA silencing, calcium signalling, component 3 promoter of RNA silencing
INTRODUCTION
Overview of the Phospholipase Cβ -G protein signaling
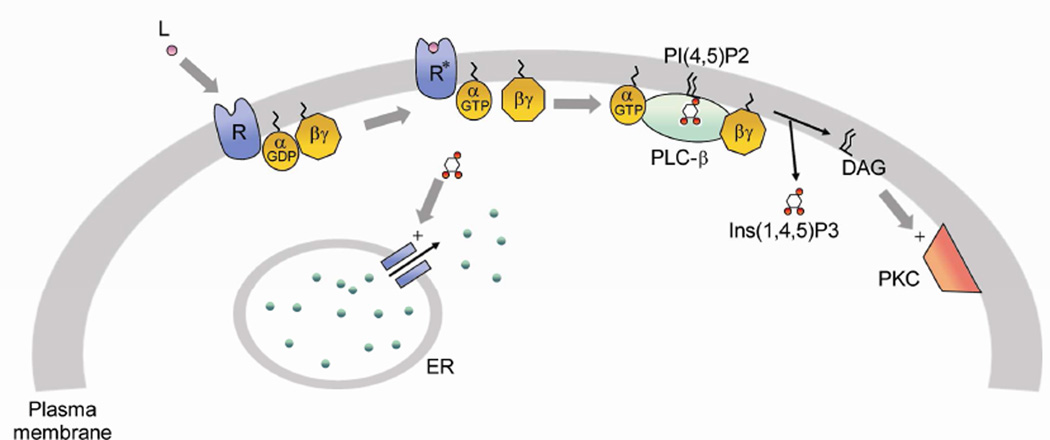
PLCβs are the main effectors of the Gαq family of heterotrimeric G proteins. Gαq subunits are coupled to receptors for acetylcholine, dopamine, angiotensin II, bradykinin, and others (see (Rebecchi and Pentylana, 2000, Suh et al., 2008)). This pathway is initiated by the binding of a ligand to its specific G-protein coupled receptor (GPCR) (Fig. 1). The ligand-bound receptor catalyzes the exchange of GTP for GDP on Gαq subunits. The GTP-bound Gαq has a much lower affinity for Gβγ, but a much higher affinity for its primary effector, PLCβ (Runnels and Scarlata, 1999). The association of Gαq(GTP) to PLCβ greatly increases PLCβ’s ability to hydrolyze its major substrate phosphatidylinositol 4,5 bisphosphate (PIP2) into two second messengers: inositol 1,4,5- trisphosphate (IP3) which promotes the release of intracellular Ca2+ from the endoplasmic reticulum resulting in activation of a host of Ca2+-sensitive proteins, and diacylglycerol (DAG) which helps to activate protein kinase C. PLCβ (β1–4) enzymes vary in their tissue distribution and their ability to be activated by Gαq. Most of the work described here utilized PLCβ1 since it is strongly activated by Gαq, is highly expressed in neural tissue (see (Aisiku et al., 2011, Berstein et al., 1992, Ciruela et al., 2000, Cocco et al., 2002)), and PLCβ3 which has a more ubiquitous distribution.
Fig. 1.
The PLCβ-G protein signaling pathway.
Several years ago, we began to follow the association between fluorescent-tagged PLCβ and Gαq in living cells (Dowal et al., 2006). Not surprisingly, we found the majority of PLCβ localizes on the plasma membrane where it is associated with Gαq. However, we also found that a significant population of endogenous and over-produced enzyme localizes in the cytosol. While we first postulated that this cytosolic fraction acts as a reservoir during signaling, we found that this population remains in the cytosol even during Gαq activation (Dowal, Provitera, 2006) (we note that Gαq remains on the plasma membrane throughout the signaling process (Dowal, Provitera, 2006, Hughes et al., 2001)). We also considered that the cytsolic PLCβ population may modulate the PIP2 levels in internal membranes. However, this idea is unlikely given the very low basal activity of PLCβ, and noting that we have only been able to detect Gαq on the plasma membrane and not in internal locations. It is notable that PLC β2, like PLC β1 has a significant cytosolic population (Guo et al., 2010). To better understand the potential function of PLCβ in the cytoplasm, we searched for novel cytosolic binding partners using an unbiased, yeast two-hybrid approach.
PLCβ enzymes are distinguished from other PLCs by a long (~400 aa) C-terminal tail which contains the primary binding site of Gαq, 2 phosphorylation sites, a calpain cleavage site, and a nuclear localization signal (see (Rebecchi and Pentylana, 2000)). Using this region as bait, we identified the cytosolic / nuclear binding partner translin-associated factor X (TRAX).
TOPICS
PLCβ binds the TRAX subunits of the promotor of RNA silencing, C3PO
TRAX is an endoribonuclease that targets ssDNA, dsRNA and RNA (see (Jaendling and McFarlane, 2010)). Together with its partner translin, which is an RNA binding protein, TRAX has been implicated in neuronal development, cell proliferation, chromosomal translocations, spermatogenesis, and dendritic RNA trafficking in neurons (see (Li et al., 2008)). The translin /TRAX complex is an assymetric octamer with the stoichiometry 2TRAX:6translin. The shape of the octamer is spherical with the oligonuceotide binding region running through the center. Several structures of the TRAX:translin octamer from different organisms are currently available (Liu et al., 2009, Parizotto et al., 2013, Ye et al., 2011).
Using purified proteins, we found that PLCβ binds TRAX with high affinity. Removal of PLCβ’s C-terminal tail greatly diminishes binding. Pull-down, immunofluorescence and FRET studies suggest that the two proteins interact in cells (Aisiku et al., 2010). Although TRAX does not affect the enzymatic activity of PLCβ, it competes with Gαq for PLCβ binding, and in cells, over-expression of TRAX ablates Ca+2 signals mediated through Gαq- PLCβ (Aisiku, Runnels, 2010, Philip, Guo, 2012).
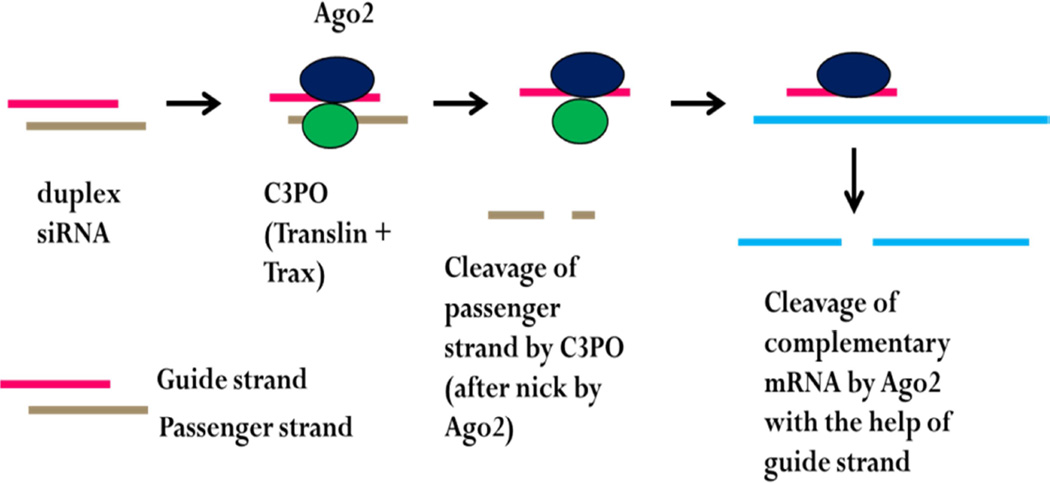
A few years ago, it was reported that a complex identified as C3PO, was needed for RNA-induced silencing (Liu, Ye, 2009, Tian et al., 2011, Ye, Huang, 2011). RNA-induced silencing begins with duplex RNAs generated in the nucleus and transported to the cytosol (Fig. 2). The double-stranded microRNA (miR) or silencing RNA (siRNA) consists of a ‘guide’ strand and a complementary passenger strand. Upon binding to RISC, the nuclease component Ago2 nicks the passenger strand. C3PO then degrades the passenger strand allowing the guide strand to hybridize to its target mRNA that is subsequently hydrolyzed by Ago2 resulting in gene silencing.
Fig. 2.
Mechanism of RNA silencing by C3PO
PLCβ binds to C3PO in different cell lines to reverse RNA silencing
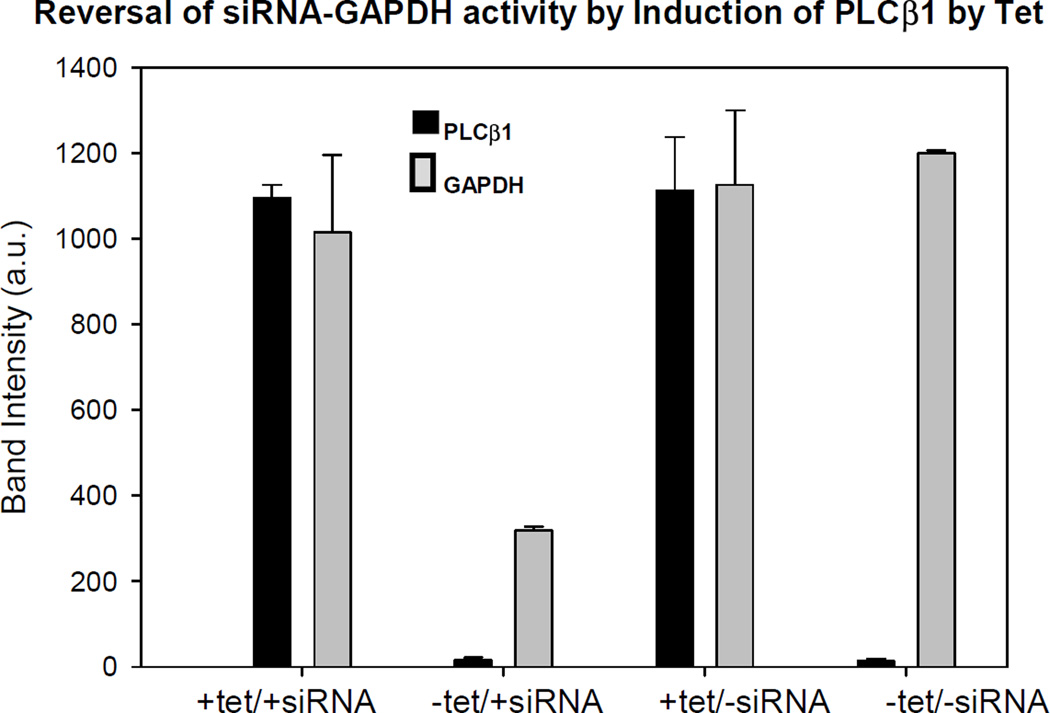
Based on the novel role of C3PO in promoting RNA silencing, we wondered whether PLCβ could interfere with this function through its interaction with TRAX. Using HEK293 cells that can be induced to over-produce PLCβ1 by treatment with tetracycline (tet), we made the surprising discovery that PLCβ can reverse siRNA down-regulation of the housekeeping enzyme, GAPDH (see Fig. 3), and this effect is also seen in other cell lines (Philip, Guo, 2012). Additionally, we found that TRAX over-expression reverses PLCβ’s reversal of RNA silencing, and PLCβ down-regulation enhances the down-regulation of GAPDH by siRNA.
Fig. 3.
From (Philip et al., 2012) -Changes in protein levels of GAPDH and PLCβ1 in HEK293 cells induced to over-express PLCβ1 by tet treatment and with siRNA (GAPDH).
When we extended this study to other genes (cyclophilin A and Hsp90) we found that expression of PLCβ did not affect siRNA down-regulation, suggesting that PLCβ only affects the knock-down of certain genes (Philip, Guo, 2012). In trying to understand why PLCβ reversed down-regulation of siRNA of GAPDH but not Hsp90 or cyclophilin A, we found that GAPDH, along with LDH, is part of complex is required for the synthesis of histone H2B (Zheng et al., 2003). Histone H2B synthesis promotes synthesis of the other histones as well as DNA allowing cells to move from the Go/G1 to the S phase and this can only occur when cells are at the proper redox state (Dai et al., 2008). We found that down-regulating GAPDH or LDH eliminates H2B production that can result in cell death. However, over-expressing PLCβ1 not only restores GAPDH and LDH levels but also histone H2B (Philip, Guo, 2012) resulting in viable cells.
Why does PLCβ1 only reverse the silencing of certain genes?
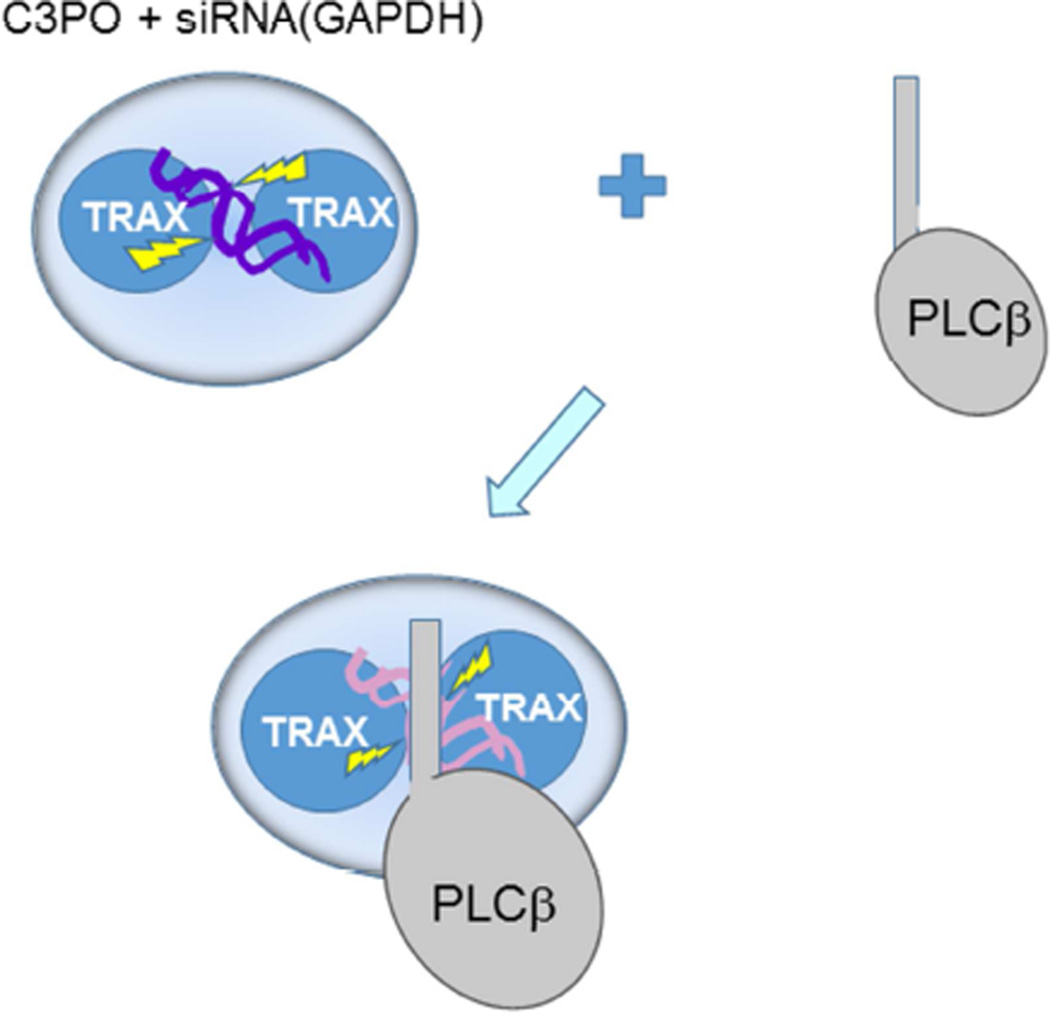
The ability of PLCβ to reverse down-regulation of GAPDH and LDH but not Hsp90 or cyclophilin A was intriguing. One possible reason may be that cells contain different RISC complexes which have varying affinities to PLCβ, or that RISC processes RNAs differently depending on their structure / sequence which are differentially affected by PLCβ. We initially addressed this question by characterizing the biophysical properties of the C3PO-PLCβ complex (Sahu et al., 2014). C3PO crystallizes as an asymmetric octamer with two TRAX subunits and 6 translin subunits (Liu, Ye, 2009, Tian, Simanshu, 2011). Using a combination of native gel electrophoresis and fluorescence-based measurements, our studies suggested that a single PLCβ molecule binds to an external site either on one TRAX subunits of C3PO or between the two TRAX molecules (see Fig. 4).
Fig. 4.
Cartoon depicting the location of the two TRAX subunits of C3PO with bound dsRNA where the lightening bolts depict hydrolytic interactions. Binding of the C-terminus of PLCβ, as depicted by a long stem, inhibits productive C3PO-oligonucleotide interactions.
Our cell studies suggested that PLCβ affects the ability of C3PO to process different siRNAs. We attached a fluorescent probe / quencher pair to the ends of different siRNA molecules to monitor hydrolysis of different sequences by C3PO in real time. Our studies found that C3PO hydrolyzes siRNA (GAPDH) at a much faster rate than siRNA (Hsp90) (Sahu, Philip, 2014). However, when PLCβ is bound to C3PO, the hydrolysis rate for siRNA (GAPDH) is significantly reduced while the rate for siRNA (Hsp90) is unchanged (see Fig. 5, from (Sahu, Philip, 2014)).
Fig. 5.
Cleavage rates of FAM-BH labeled siRNAs by C3PO in the presence and absence of PLCβ
We then determined whether differences in hydrolysis rates of the different siRNAs were caused by changes in binding of these siRNA to C3PO in the presence of PLCβ. We measured the change in binding affinity of C3PO to small (18nt) DNAs in the presence and absence of PLCβ. We find that the presence of PLCβ reduces oligonucleotide binding by an order of magnitude.
TRAX expression affects PLCβ-mediated Ca2+ signals
Our previous work showed that when TRAX is over-expressed, Gαq-PLCβ mediated Ca2+ signals are ablated (Philip, Guo, 2012) suggesting that excess TRAX sequesters PLCβ in the cytoplasm and away from Gαq. To test this idea, we followed the changes in colocalization between PLCβ and TRAX when cells are stimulated by carbachol to activate Gαq, we find that the colocalization between TRAX and PLCβ is reduced by 50%. Alternately, when unstimulated cells are treated with siRNA (GAPDH) or siRNA(Hsp90), the colocalization between TRAX and PLCβ increases over 60% suggesting a dynamic equilibrium between the two proteins (Philip, Guo, 2012).
Taken together, our studies show that PLCβ binds to C3PO and that this binding reduces the ability fo C3PO to bind and process different oligonucleotides. Since our report, changes in the association between PLCβ and C3PO have been found to initiate angiogenesis triggered by neighboring cancer cells (Cheng et al., 2014) suggesting that PLCβ – C3PO association may impact the state of the cells.
Role of PLCβ and TRAX in neuronal differentiation
As mentioned above, we found that PLCβ may be linked to cell cycle progression indirectly through histone H2B (Philip, Guo, 2012). We have also found that PLCβ increases the expression of a large cluster genes that code for H2B subtypes (Philip et al., 2013), as well as for proteins involved in maintaining the redox state and positively regulating cell death (unpublished). We also surveyed changes in microRNAs (miRNAs) with PLCβ expression. MiRNAs that are significantly down-regulated are closely linked to various leukemias and lymphomas, and PLCβ1 over-expression is directly linked to these diseases (Follo et al., 2009). Interestingly, we found that the level of miR221 changed by several orders of magnitude with PLCβ over-expression. MiR221 has been reported to mediate PC12 differentiation (Hamada et al., 2012). Based on these observations, we then determined whether PLCβ might impact PC12 differientiation through its interaction with TRAX.
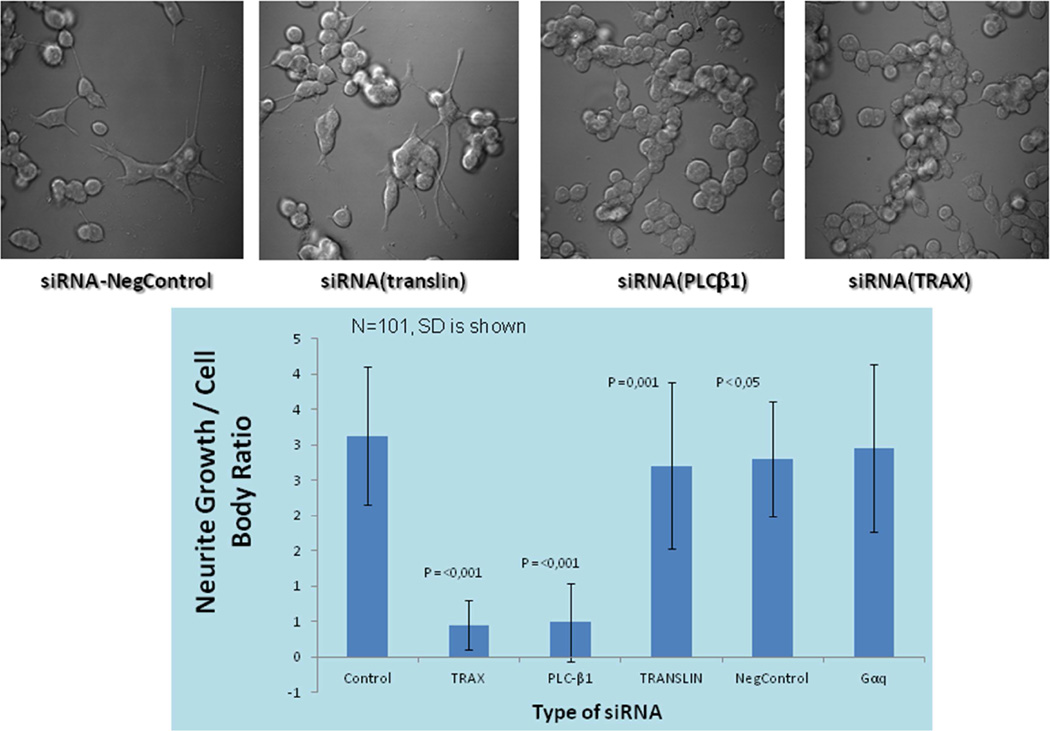
In their undifferentiated form, PC12 cells are fairly circular and divide rapidly. Upon treatment with nerve growth factor (NGF) (see (Bradshaw et al., 2015) for review), the cells stop dividing and sprout neurites (Drubin et al., 1985). Differentiation of these cells is generally considered complete when the length of the neurites is 3–4 times the length of the cell body. We found that PC12 cells differentiate normally after siRNA-induced down-regulation. However, down-regulating TRAX or PLCβ by 73% and 78%, respectively, prevented differentiation and preserved proliferation (Fig. 6 right images and graph). Thus, it is possible that the absence of PLCβ allows for rapid processing of miRs by C3PO that preserve the undifferentiated state. While it has been noted that over-expression of PLCβ1 does not impact PC12 growth or differentiation (Bortul et al., 2001), our studies show that a certain level of cytosolic PLCβ1 is required for differentiation.
Fig. 6.
Down-regulation of PLCβ1 or TRAX ablates PC12 cell Differentition
We repeated the studies of Hamada and coworkers, 2012, described above that show a very large and significant increase in miR221 with PC12 differentiation. However, after 4 independent attempts, we could not detect any increase in miR221. In fact, the levels of miR221 are extremely low levels in undifferentiated PC12 cells and these levels remained very low throughout the differentiation process. Unless the affinity of miR221 for the C3PO-RISC machinery was extremely strong, it is unclear whether it could impact cellular levels of any protein. Therefore, other miRs must be involved in PC12 cell differentiation. Studies are now underway to characterize these miRs.
PLCβ1 and TRAX are closely involved in neuronal disease and development
PLCβ1 is highly expressed in the brain (Gerfen et al., 1988) and PLCβ1 deficiencies have been found in patients with neurological disorders such as epileptic encephalopathies which are reproduced in knock-out mice (see (Albert et al., 1997)). PLCβ1 is associated with neural growth and its expression correlates with synaptic plasticity in rat cortex (Baxter et al., 1995, Hannan et al., 1998). PLCβ1 mutagenesis and deficiencies underlie a large number of neurological and psychiatric problems including memory loss, schizophrenia and hot/cold sensitivity (see (García del Caño et al., 2014)).
TRAX and translin are present at high levels in neurons. Mutagenesis studies show that these proteins play key roles in function and development in cultured cells of neuronal linage and in the neural system of whole animals, although the underlying basis is unknown (Finkenstadt et al., 2000, Kobayashi et al., 1998, Stein et al., 2006, Wu et al., 1997). The ability of TRAX in the context of C3PO to regulate these processes through generation of specific miRs is now being investigated.
Alternate mechanisms?
We have extensive molecular and cellular evidence that PLCβ directly binds to the TRAX subunits of C3PO and reduces the rate of hydrolysis of specific oligonucleotides, such as siRNA (GAPDH) (Sahu, Philip, 2014) to reverse RNA silencing (Philip, Guo, 2012) leading to changes in the genetic composition of the cell (Philip, Sahu, 2013) which may lead to changes in cell function such as differentiation. There is a possibility that other pathways induced by PLCβ play a role, although these potential pathways are not apparent at the moment. While it is possible that changes in mRNA can be induced via IRBIT at high cellular levels of IP3 (Mikoshiba, 2015), we know that both active and inactive PLCβ reverses siRNA (GAPDH) showing that IP3 production is not involved (Philip, Guo, 2012) and which negates the many Ca2+ mediated signaling pathways that could be involved. We found that receptor tyrosine kinase ligands do not appear to be involved (Philip, Guo, 2012). Additionally, we find that siRNA treatment affects Gαq/ PLCβ – mediated Ca2+ release almost immediately after treatment arguing against the contribution of longer term translational processes. We also find that agonists directed at non-Gαq families do not show effects (Philip, Guo, 2012). Taken together, these observations support direct PLCβ –C3PO effects.
Conclusions
In summary, we have shown that PLCβ binds to C3PO leading to changes in the ability of C3PO to promote RNA-induced silencing. The impact of PLCβ on C3PO function are certain to have direct impact on the cellular content of miRs and in turn, on the protein products.
Acknowlegements
The authors would like to thank Barbara Rosatiand David McKinnon for their help with the microRNA studies. This work was suppored by NIH GM116178.
Abbreviations
- PLCβ
phospholipase Cβ
- C3PO
component 3 promoter of RNA silencing
- GAPDH
glyceraldehyde-3-phosphate dehyrodrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aisiku O, Dowal L, Scarlata S. Protein kinase C phosphorylation of PLC[beta]1 regulates its cellular localization. Archives of Biochemistry and Biophysics. 2011;509:186–190. doi: 10.1016/j.abb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aisiku OR, Runnels LW, Scarlata S. Identification of a Novel Binding Partner of Phospholipase Cβ1: Translin-Associated Factor X. PLoS ONE. 2010;5:e15001. doi: 10.1371/journal.pone.0015001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert JL, Boyle JP, Roberts JA, Challiss RA, Gubby SE, Boarder MR. Regulation of brain capillary endothelial cells by P2Y receptors coupled to Ca2+, phospholipase C and mitogen-activated protein kinase. Br J Pharmacol. 1997;122:935–941. doi: 10.1038/sj.bjp.0701453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter RM, Cohen P, Obermeier A, Ullrich A, Downes CP, Doza YN. Phosphotyrosine residues in the nerve-growth-factor receptor (Trk-A). Their role in the activation of inositolphospholipid metabolism and protein kinase cascades in phaeochromocytoma (PC12) cells. European Journal of Biochemistry. 1995;234:84–91. doi: 10.1111/j.1432-1033.1995.084_c.x. [DOI] [PubMed] [Google Scholar]
- Berstein G, Blank JL, Jhon D-Y, Exton JH, Rhee SG, Ross EM. Phospholipase C-β1 is a GTPase-Activating Protein for Gq/11, Its Physiologic Regulator. Cell. 1992;70:411–418. doi: 10.1016/0092-8674(92)90165-9. [DOI] [PubMed] [Google Scholar]
- Bortul R, Aluigi M, Tazzari PL, Tabellini G, Baldini G, Bareggi R, et al. Phosphoinositide-specific phospholipase C beta 1 expression is not linked to nerve growth factor-induced differentiation, cell survival or cell cycle control in PC12 rat pheocromocytoma cells. J Cell Biochem. 2001;84:56–67. doi: 10.1002/jcb.1266. [DOI] [PubMed] [Google Scholar]
- Bradshaw RA, Pundavela J, Biarc J, Chalkley RJ, Burlingame AL, Hondermarck H. NGF and ProNGF: Regulation of neuronal and neoplastic responses through receptor signaling. Advances in Biological Regulation. 2015;58:16–27. doi: 10.1016/j.jbior.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J-Y, Wang S-H, Lin J, Tsai Y-C, Yu J, Wu J-C, et al. Globo-H Ceramide Shed from Cancer Cells Triggers Translin-Associated Factor X-Dependent Angiogenesis. Cancer Research. 2014;74:6856–6866. doi: 10.1158/0008-5472.CAN-14-1651. [DOI] [PubMed] [Google Scholar]
- Ciruela A, Hinchliffe KA, Divecha N, Irvine RF. Nuclear targeting of the beta isoform of type II phosphatidylinositol phosphate kinase (phosphatidylinositol 5-phosphate 4-kinase) by its alpha-helix 7. Biochem J. 2000;346(Pt 3):587–591. [PMC free article] [PubMed] [Google Scholar]
- Cocco L, Capitini S, Maraldi N, Mazzotti G, Barnabei O, Rizzoli R, et al. Inositides in the nucleus: regulation of PI-PLCβ1. Adv Enzyme Regul. 2002;42:181–193. doi: 10.1016/s0065-2571(01)00030-9. [DOI] [PubMed] [Google Scholar]
- Dai R-P, Yu F-X, Goh S-R, Chng H-W, Tan Y-L, Fu J-L, et al. Histone 2B (H2B) Expression Is Confined to a Proper NAD+/NADH Redox Status. Journal of Biological Chemistry. 2008;283:26894–26901. doi: 10.1074/jbc.M804307200. [DOI] [PubMed] [Google Scholar]
- Dowal L, Provitera P, Scarlata S. Stable association between G alpha(q) and phospholipase C beta 1 in living cells. J Biol Chem. 2006;281:23999–24014. doi: 10.1074/jbc.M512330200. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Feinstein SC, Shooter EM, Kirschner MW. Nerve growth factor-induced neurite outgrowth in PC12 cells involves the coordinate induction of microtubule assembly and assembly-promoting factors. The Journal of Cell Biology. 1985;101:1799–1807. doi: 10.1083/jcb.101.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkenstadt PM, Kang W-S, Jeon M, Taira E, Tang W, Baraban JM. Somatodendritic Localization of Translin, a Component of the Translin/Trax RNA Binding Complex. Journal of Neurochemistry. 2000;75:1754–1762. doi: 10.1046/j.1471-4159.2000.0751754.x. [DOI] [PubMed] [Google Scholar]
- Follo MY, Finelli C, Clissa C, Mongiorgi S, Bosi C, Martinelli G, et al. Phosphoinositide-Phospholipase C β1 Mono-Allelic Deletion Is Associated With Myelodysplastic Syndromes Evolution Into Acute Myeloid Leukemia. Journal of Clinical Oncology. 2009;27:782–790. doi: 10.1200/JCO.2008.19.3748. [DOI] [PubMed] [Google Scholar]
- García del Caño G, Montaña M, Aretxabala X, González-Burguera I, López de Jesús M, Barrondo S, et al. Nuclear phospholipase C-β1 and diacylglycerol LIPASE-α in brain cortical neurons. Advances in Biological Regulation. 2014;54:12–23. doi: 10.1016/j.jbior.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Choi WC, Suh PG, Rhee SG. Phospholipase C I and II brain isoenzymes: immunohistochemical localization in neuronal systems in rat brain. Proc Natl Acad Sci USA. 1988;85:3208–3212. doi: 10.1073/pnas.85.9.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Golebiewska U, D'Amico S, Scarlata S. The Small G Protein Rac1 Activates Phospholipase Cδ1 through Phospholipase Cβ2. Journal of Biological Chemistry. 2010;285:24999–25008. doi: 10.1074/jbc.M110.132654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N, Fujita Y, Kojima T, Kitamoto A, Akao Y, Nozawa Y, et al. MicroRNA expression profiling of NGF-treated PC12 cells revealed a critical role for miR-221 in neuronal differentiation. Neurochemistry International. 2012;60:743–750. doi: 10.1016/j.neuint.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Hannan AJ, Kind PC, Blakemore C. Phospholipase C-β1 expression correlates with neuronal differentiation and synaptic plasticity in rat somatosensory cortex. Neuropharmacology. 1998;37:593–605. doi: 10.1016/s0028-3908(98)00056-2. [DOI] [PubMed] [Google Scholar]
- Hughes TE, Zhang H, Logothetis D, Berlot CH. Visualization of a functional Gαq-green fluorescent protein fusion in living cells. J Biol Chem. 2001;276:4227–4235. doi: 10.1074/jbc.M007608200. [DOI] [PubMed] [Google Scholar]
- Jaendling A, McFarlane RJ. Biological roles of translin and translin-associated factor-X: RNA metabolism comes to the fore. Biochemical Journal. 2010;429:225–234. doi: 10.1042/BJ20100273. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Takashima A, Anzai K. The Dendritic Translocation of Translin Protein in the Form of BC1 RNA Protein Particles in Developing Rat Hippocampal Neurons in Primary Culture. Biochemical and Biophysical Research Communications. 1998;253:448–453. doi: 10.1006/bbrc.1998.9704. [DOI] [PubMed] [Google Scholar]
- Li Z, Wu Y, Baraban JM. The Translin/Trax RNA binding complex: Clues to function in the nervous system. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2008;1779:479–485. doi: 10.1016/j.bbagrm.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, et al. C3PO, an Endoribonuclease That Promotes RNAi by Facilitating RISC Activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikoshiba K. Role of IP3 receptor signaling in cell functions and diseases. Advances in Biological Regulation. 2015;57:217–227. doi: 10.1016/j.jbior.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Parizotto EA, Lowe ED, Parker JS. Structural basis for duplex RNA recognition and cleavage by Archaeoglobus fulgidus C3PO. Nat Struct Mol Biol. 2013;20:380–386. doi: 10.1038/nsmb.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip F, Guo Y, Aisiku O, Scarlata S. Phospholipase Cβ1 is linked to RNA interference of specific genes through translin-associated factor X. The FASEB Journal. 2012;26:4903–4913. doi: 10.1096/fj.12-213934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip F, Sahu S, Caso G, Scarlata S. Role of phospholipase C-beta in RNA interference. Adv Biol Regul. 2013;53:319–330. doi: 10.1016/j.jbior.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecchi M, Pentylana S. Structure, function and control of phosphoinositide-specific phospholipase C. Physiological Reviews. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- Runnels LW, Scarlata S. Determination of the affinities between heterotrimeric G protein subunits and their phospholipase C- β effectors. Biochemistry. 1999;38:1488–1496. doi: 10.1021/bi9821519. [DOI] [PubMed] [Google Scholar]
- Sahu S, Philip F, Scarlata S. Hydrolysis Rates of Different Small Interfering RNAs (siRNAs) by the RNA Silencing Promoter Complex, C3PO, Determines Their Regulation by Phospholipase Cβ. Journal of Biological Chemistry. 2014;289:5134–5144. doi: 10.1074/jbc.M113.531467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JM, Bergman W, Fang Y, Davison L, Brensinger C, Robinson MB, et al. Behavioral and Neurochemical Alterations in Mice Lacking the RNA-Binding Protein Translin. The Journal of Neuroscience. 2006;26:2184–2196. doi: 10.1523/JNEUROSCI.4437-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh P, Park J, Manzoli L, Cocco L, Peak J, Katan M, et al. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB reports. 2008;41:415–434. doi: 10.5483/bmbrep.2008.41.6.415. [DOI] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ascano M, Diaz-Avalos R, Park AY, Juranek SA, et al. Multimeric assembly and biochemical characterization of the Trax–translin endonuclease complex. Nat Struct Mol Biol. 2011;18:658–664. doi: 10.1038/nsmb.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X-Q, Gu W, Meng X, Hecht NB. The RNA-binding protein, TB-RBP, is the mouse homologue of translin, a recombination protein associated with chromosomal translocations. Proceedings of the National Academy of Sciences. 1997;94:5640–5645. doi: 10.1073/pnas.94.11.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, et al. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol. 2011;18:650–657. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Roeder RG, Luo Y. S Phase Activation of the Histone H2B Promoter by OCA-S, a Coactivator Complex that Contains GAPDH as a Key Component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]