Abstract
The RAD51 family is integral for homologous recombination (HR) mediated DNA repair and maintaining chromosome integrity. RAD51D, the fourth member of the family, is a known ovarian cancer susceptibility gene and required for the repair of interstrand crosslink DNA damage and preserving chromosomal stability. In this report, we describe the RNF138 E3 ubiquitin ligase that interacts with and ubiquitinates the RAD51D HR protein. RNF138 is a member of an E3 ligase family that contains an amino-terminal RING finger domain and a putative carboxyl-terminal ubiquitin interaction motif. In mammalian cells, depletion of RNF138 increased the stability of the RAD51D protein, suggesting that RNF138 governs ubiquitin-proteasome-mediated degradation of RAD51D. However, RNF138 depletion conferred sensitivity to DNA damaging agents, reduced RAD51 focus formation, and increased chromosomal instability. Site-specific mutagenesis of the RNF138 RING finger domain demonstrated that it was necessary for RAD51D ubiquitination. Presence of RNF138 also enhanced the interaction between RAD51D and a known interacting RAD51 family member XRCC2 in a yeast three-hybrid assay. Therefore, RNF138 is a newly identified regulatory component of the HR mediated DNA repair pathway that has implications toward understanding how ubiquitination modifies the functions of the RAD51 paralog protein complex.
Keywords: Homologous recombination, DNA double-stranded breaks, DNA interstrand crosslinks, RAD51D, RNF138, E3 ligase, Chromosome integrity, DNA repair, Ubiquitination
1. Introduction
When DNA damage affects both strands, such as interstrand crosslinks (ICLs) or double-strand breaks (DSBs), cells become more vulnerable to chromosomal deletions and rearrangements. Homologous recombination (HR) is a major DNA repair pathway that resolves these lesions. HR is a dynamic process involving protein complexes that are tightly regulated to guide DNA damage signaling, lesion processing, and invasion of a damaged DNA strand onto a homologous template [1,2]. Decreased HR leads to the accumulation of mutations and genome instability associated with carcinogenesis, whereas increased HR levels may lead to hyper-recombination phenotypes that contribute to radiation treatment and chemotherapy drug resistance [3].
The seven mammalian RAD51 family members are crucial HR components [4]. Even though they are involved in multiple HR steps, the regulatory mechanisms are still being investigated. Ubiquitin-specific post-translational modifications (PTMs) act as key orchestrators of the HR pathway, which include polyubiquitin landscapes and mobilization of histones that surround the damaged chromatin, ubiquitin associated recruitment and signaling, and ubiquitin-mediated protein modifications [5–11]. RAD51 is the only family member known to be ubiquitinated and is subject to proteasome-mediated degradation following exposure to ionizing radiation [12–14]. In the absence of RAD51C, the RAD51 protein is ubiquitinated independently of DSB formation [12]. Therefore, RAD51C appears to be involved in the ubiquitin transition of RAD51 in response to DNA damage and targeting for proteasomal degradation. The E3 ubiquitin ligase RAD18 acts as an adapter between RAD51C and RNF8/UBC13 catalyzed polyubiquitin chains that surround damaged chromatin [5]. In addition, RAP80 has an analogous role by recruiting the BRCA1 complex to RNF8/RNF168 synthesized polyubiquitin chains at DSB [15,16].
RAD51D is the fourth member of the RAD51 family and is a known ovarian cancer and possibly a breast cancer susceptibility gene [17–23]. Mutations in RAD51D confer extensive chromosomal instability and sensitivity to DNA damage, primarily crosslinks [24,25]. RAD51D is also necessary for the recruitment of RAD51 to DNA damage sites and facilitates homologous pairing when associated with XRCC2 [25,26]. Identification of post-translational modification mechanisms of RAD51D could make it possible to more efficiently diagnose HR-deficient ovarian cancers and to develop personalized treatment strategies [27]. Therefore, to identify RAD51D interacting proteins, we performed yeast two-hybrid screens against RAD51D and identified the RNF138 E3 ubiquitin ligase as a candidate that may be involved in ubiquitin modification of RAD51D.
RNF138 was initially reported as part of the Wnt signaling pathway involved in secondary axis formation in Xenopus embryos [28]. It was later linked to DNA damage response as a phosphorylated ATM target on Serine 124 following ionizing radiation [29]. Given the emerging role of ubiquitin signaling cascades in governing HR, we hypothesized that RNF138 E3 ligase activity regulates RAD51D function. In this manuscript, we demonstrate that RNF138 directly interacts with RAD51D and is required for ubiquitination of the RAD51D protein. Consistent with a role during HR, depletion of RNF138 increased sensitivity to DNA damaging agents, reduced RAD51 foci formation, and increased levels of chromosomal aberrations. The data presented here suggest that RNF138-dependent ubiquitination of RAD51D is an essential step during HR DNA repair and offers a potential explanation regarding the selection for increased RNF138 expression levels during carcinogenesis [30–32].
2. Materials and Methods
2.1. Cell culture and transfections
Mouse embryonic fibroblast (MEF) and HeLa cell lines were maintained at 37°C with 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM; HyClone) supplemented with 10% newborn calf serum (Atlanta Biologicals), 1% penicillin/streptomycin, and 1% glutamine. The MEF cell lines MEFC20 (Rad51d+/+Trp53−/−), MEF258 (Rad51d−/−Trp53−/−), M7 (Rad51d+/+Trp53+/+), and MEF172AG (Rad51d−/− Trp53−/− HARad51d) were described previously [25]. Plasmid constructs were transfected using Lipofectamine Reagents (Invitrogen) or Mirus TransIT-LT1 according to manufacturer’s instructions. Ten micrograms per milliliter cycloheximide (Sigma) was used for protein stability experiments.
2.2. Plasmids
RNF138 (MmRnf138 NM_207623.1) expression vectors were generated by PCR amplification of mouse liver cDNA with Platinum Taq DNA Polymerase High Fidelity (Invitrogen) using MmRnf138-specific primers RNF138s1 (5′-CTTGGTACCTCCGAGGAACTTTCGGCGG-3′) and RNF138as1 (5′-CTTGGATCCTGTAGGTTGCAAGGAGGCAG-3′) followed by cloning of the amplification products into the KpnI/BamHI sites of pcDNA3.1/Hygro+ (Invitrogen) with either HA or Myc epitope tags. Mutagenesis of the RNF138 RING finger domain (H36A,C39S) was accomplished by subcloning MmRnf138 cDNA into pUC19 for conventional PCR based site-directed mutagenesis with primers: RNF138mut1 (5’-GGCCTGTCAGGCCGTTTTCTCTAGAAAATGTTT CCTGACTG-3’) and RNF138mut2 (5’ CAGTCAGGAAACATTTTCTAGAGAAAA CGGCCTGACAGGCC 3’). The underlined portion denotes sequence targeted for mutagenesis. All PCR derived expression constructs were confirmed by sequencing both strands. The Myc-ubiquitin expression plasmid was kindly provided by Dr. Xiongbin Lu (Department of Cancer Biology, MD Anderson) and the HA-ubiquitin plasmid purchased from Addgene. The RAD51D Walker A ATPase mutant plasmid constructs were described previously [33], and RAD51D deletion constructs containing residues 4–77 and residues 77–329 were a gift from Dr. Joanna Albala (Lawrence Livermore National Laboratory) [34]. The yeast expression vector pVT100u was a generous gift from Dr. David Schild (Lawrence Berkley National Laboratory, Berkley). MmRnf138 cDNA was subcloned into the HindIII/BamHI sites of pVT100u for use in yeast three-hybrid experiments. Splice variant constructs, MmRnf138-Δ7 and MmRnf138-Δ5, were cloned into the BamHI and EcoRI sites of pGADT7 and pGBKT7 for use in yeast two-hybrid experiments.
2.3. Yeast two- and three-hybrid
For yeast two-hybrid screening, a Mus musculus pre-transformed normalized universal cDNA library (Clontech) was screened using mouse full-length RAD51D. A total of 3.3 × 107 clones were assayed (cfu/ml of diploids × resuspension volume). Liquid β-galactosidase assays were performed using ortho-nitrophenyl-β-galactopyranoside (ONPG: Sigma) [35]. Yeast two-hybrid expression vectors pGADT7 and pGBKT7 (Clontech) were co-transformed into Y187 haploids using the EZ Transformation Kit (Zymo). Yeast three-hybrid experiments were performed using Y190 haploids transformed with pGADT7, pGBKT7, and pVT100u expression constructs [36].
2.4. Immunoprecipitations
For co-immunoprecipitations, vectors encoding HA-tagged and Myc-tagged proteins were co-transfected into HeLa cells. Whole cell extracts were prepared after 24 h using mammalian protein extraction reagent (M-PER; Thermo-Scientific) or 1X Cell Lysis Buffer (20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, 1% TritonX-100) containing a protease inhibitor cocktail (Complete Mini; Roche Life Sciences). Three to five hundred micrograms of whole cell extract was incubated with anti-HA agarose beads (3F10; Roche) or anti-Myc magnetic beads (9E10; Thermo-Scientific) for 16 h at 4°C with gentle rocking in incubation buffer (20 mM Tris, 100 mM NaCl, 100 mM EDTA) or 1X Cell Lysis Buffer. Precipitated proteins were washed 3 times with 1× PBST or 1X Cell Lysis Buffer, eluted by boiling in Laemmli buffer for 10 minutes, and resolved on 12% SDS-PAGE or 4 – 20% SDS-PAGE (Bio-Rad). For in-vivo ubiquitination assays, cells were treated with 25 µM MG132 (Sigma) 4 h prior to preparation of whole cell extracts.
2.5. Immunoblotting
Western blot analysis was performed using mouse monoclonal anti-HA (3F10; Roche), mouse monoclonal anti-Myc (9E10; Santa Cruz Biotechnology), rabbit monoclonal anti-Myc (ab9106; Abcam), or rabbit polyclonal anti-RAD51 (H-92; Santa Cruz Biotechnology). Primary antibody incubations were followed by incubation with the appropriate species-specific HRP-conjugated or IRDye 800CW secondary antibody (Licor) secondary antibody (Santa Cruz Biotechnology). Detection was performed by ECL (West-Pico chemiluminescent substrate; Thermo-Scientific) or the Licor Odyssey Sa Imaging System. Quantitative analysis of band intensity was performed using NIH ImageJ.
2.6. RNA interference and quantitative real-time PCR
RNAi-mediated knockdowns were conducted using Mission siRNA oligos (Sigma Proligo, TX, USA). Oligos directed to the mouse Rnf138 gene corresponded to nucleotides 462–482 corresponding to exon 2 (1_Rnf138_Mm and 1_Rnf138_Mm_as duplex; siRNA1) and 1234–1254 corresponding to the 3’ UTR (2_Rnf138_Mm and 2_Rnf138_Mm_as duplex; siRNA2). As negative controls, scrambled siRNA (Sigma) or Mission siRNA oligos for the mouse Gapdh gene corresponding to nucleotides 1090–1108 (NM_008084|1 and NM_008084|1 AS dup) were used where indicated. MEFs were seeded at a concentration of 2.5 × 104 cells per well in a 24-well plate for transfection with 30nM of siRNA oligos using the N-TER nanoparticle siRNA transfection system (Sigma). Knockdown of gene expression was confirmed by quantitative real-time PCR. Total RNA was isolated from MEFs using the Aurum Total RNA isolation kit (Bio-Rad), and first-strand cDNA was generated by reverse transcription using 500ng total RNA (iScript cDNA synthesis kit; Bio-Rad). Gene specific primers for MmRnf138 full length and MmRnf138 from exons 2 to 8 were designed by Beacon Designer version 7.51 (Biosoft International): Rnf138rt1 (5’-CGTCCTACACCGAAGATG-3’) and Rnf138rt2 (5’-CTCCGCTTTCCCTCATTG-3’), RNF138s1 (5’-CTTGGTACCTCCGAGGAACTTTCGGCGG-3’) and RNF138as1 (5’-CTTGGATCCTGTAGGTTGCAAGGAGGCAG-3’). PCR reactions were performed in triplicate (iQ5 Real Time PCR detection system; Bio-Rad) using IQ SYBR Green Supermix (Bio-Rad) under the following conditions: 95°C for 3 minutes, proceeded by 40 cycles at 95°C for 10 seconds and 55°C for 30 seconds. Primer pair integrity was verified by melt-curve analysis. MmRnf138 expression levels were calculated by the comparative ddCt method and normalized to both β-Actin and Gapdh mRNA levels using the iQ5 system software (Bio-Rad).
2.7. Cell treatment
Cells were seeded at 3 × 103 cells per well in a 96-well plate and were transfected with 30nM Rnf138 siRNA1 as described above. Twenty-four hours following siRNA transfection, cells were challenged with DNA damaging agents. For MTT assays, drug treatments were as follows: cisplatin and MMS treatments were 1 h in duration, camptothecin (CPT) and HU treatments were for 24 h, while MMC treatment was carried out for 72 hours. For all conditions, media was replenished 72 h after the initiation of treatment. Cell viability was assayed seven days after the start of treatment as described [37]. For colony forming assays (CFAs), cells were treated with MMC for 24 h and expanded onto two 100-mm tissue culture dishes. Giemsa (Sigma) stained colonies were counted after seven days. Colonies containing ≥ 50 cells were scored positive.
2.8. Chromosome analysis
Giemsa stained metaphase spreads were prepared from MEFs 48 h following MMC treatment (50ng/mL) or 72 h following siRNA transfection for untreated cells [25]. The total number of specific chromosome aberrations (chromatid breaks/gaps, chromosome breaks, and chromosome radials) was recorded from at least 50 metaphase spreads (>3600 chromosomes) for each sample from two independent experiments [38]. The anaphase bridge index was calculated as described [25]. For each condition, at least one hundred cells in anaphase from two independent experiments were scored (total n ≥ 200).
2.9. Immunofluorescence
For analysis of RAD51 foci formation, 1 × 104 cells were seeded onto glass coverslips for transfection with Rnf138 siRNA. Twenty-four hours following siRNA transfection, cells were challenged with MMC (200ng/mL) for 16 hours. Immunofluoresence detection of RAD51 foci was carried out as described [25]. Cells containing five or more distinct foci were scored as positive. For each condition, a minimum of 250 cells, from at least three independent experiments, were analyzed (n ≥ 750).
2.10. Statistical analysis
Calculations of the mean, standard deviation, and standard error were performed using Microsoft Excel. Statistical analysis for comparison of experimental means was performed using Graphpad Instat 3.0 (Graphpad Software Inc.). Significance of variance was determined by ANOVA and post-tests performed when the variance was significant (P<0.05). EC50 values were calculated using non-linear regression curve analysis with Graphpad Prism 4.0.
3. Results
3.1. Identification of RAD51D and RNF138 interaction
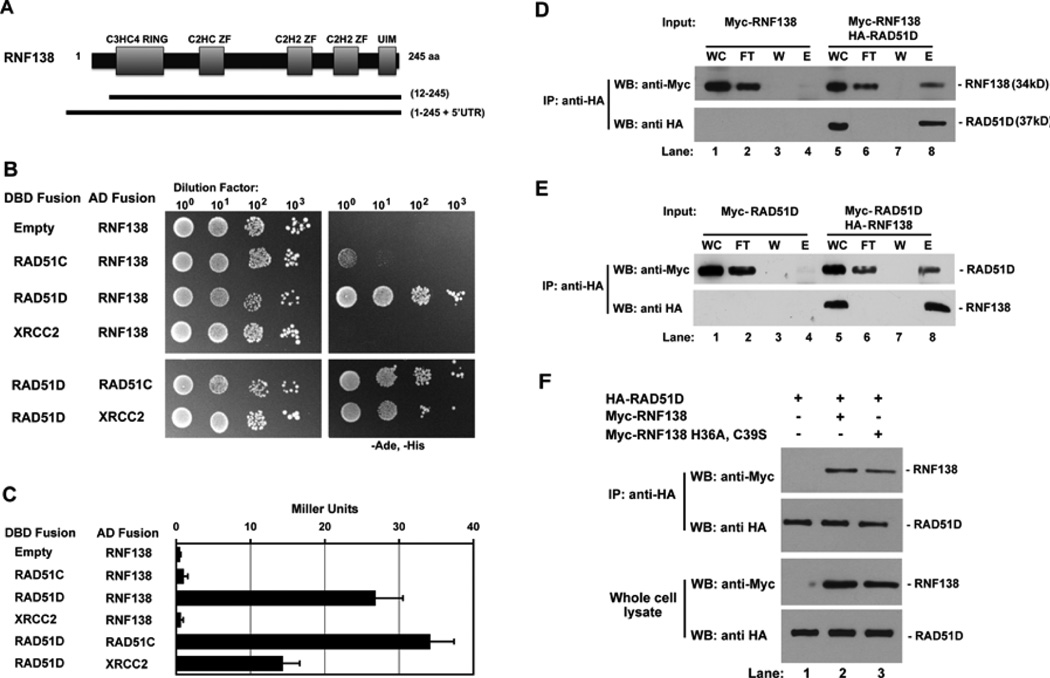
Full-length MmRAD51D was used in a yeast two-hybrid screen to identify two different cDNA clones containing MmRNF138 (Figure 1A). RNF138 belongs to the class of E3-ubiquitin ligases that have an amino-terminal C3HC4 RING finger domain, three internal zinc finger domains, and a putative carboxy-terminal ubiquitin interaction motif (UIM) [39]. One cDNA clone encoded amino-acid residues 12–245 containing the five predicted RNF138 functional domains. The second contained the full-length sequence plus an additional MmRnf138 5’UTR predicted to encode thirty amino acids.
Fig. 1.
RNF138 interacts with RAD51D. (A) RNF138 clones isolated from yeast two-hybrid screening with RAD51D. (B) Yeast two-hybrid analysis of interactions between RNF138 and RAD51 paralogs. AH109 haploids were co-transformed with the indicated GAL4 DBD and AD fusion constructs and serially diluted on to non-selective and selective growth medium. (C) Protein interaction was quantified by measuring β-galactosidase activity. Data represent mean +/− SEM from three independent experiments conducted in triplicate. (D) Lysates from cells transfected with the indicated expression constructs were subject to anti-HA immunoprecipitation followed by immunoblot analysis with anti-Myc (RNF138, top panel) and anti-HA (RAD51D, bottom panel) antibodies. (E) A reciprocal immunoprecipitation experiment was performed for HA-RNF138 and Myc-RAD51D. (F) Cells were transfected with vectors for either wild-type (WT) or RING finger domain mutant and Myc-RNF138 for anti-HA immunoprecipitation. Abbreviations: DBD Fusion- GAL4 DNA binding domain fusion, AD Fusion- GAL4 activation domain fusion, IP- immunoprecipitate, WC- whole cell lysate, FT- immunoprecipitation flow through, W- immunoprecipitation wash, E- first eluate from immunoprecipitation.
Because RAD51D is known to directly interact with RAD51C and XRCC2 [36], RNF138 containing clones were examined for possible interactions with both RAD51C and XRCC2 (Figure 1B and 1C). Whereas RNF138 and RAD51D had 79% the level of the DBD-RAD51D:AD-RAD51C positive control, β-galactosidase activity failed to increase in yeast expressing RNF138 with either RAD51C or XRCC2. Thus, among these RAD51 family members, RNF138 binding appears to be limited to RAD51D.
To verify RNF138 and RAD51D interaction, HeLa cells expressing HA-RAD51D and Myc-RNF138 were subject to anti-HA immunoprecipitation (Figure 1D and E). Myc-RNF138 was only detected when co-expressed with HA-RAD51D (Figure 1D, lane 8). A reciprocal immunoprecipitation with HA-RNF138 and Myc-RAD51 D proteins are consistent with these results (Figure 1E, lane 8). To determine whether the RNF138 RING domain mediates this interaction, two conserved RING domain amino acid residues that coordinate zinc binding were replaced (RNF138 H36A, C39S). Immunoprecipitation experiments demonstrated that the RING finger mutant also bound RAD51D (Figure 1F, lanes 2 and 3). This interaction between RNF138 H36A, C39S and RAD51D was also detected by yeast two-hybrid analysis (data not shown). Therefore, a conserved RING finger domain is not essential for RNF138 interaction with RAD51D.
3.2 RNF138 modifies interaction between RAD51D and XRCC2
Our previous work demonstrated that the RAD51D K113R and K113A highly conserved Walker A ATPase motif mutations prevented interaction with RAD51C but not XRCC2 [33]. Therefore, the RAD51D ATPase mutants were co-transformed with RNF138 and displayed less than 15% of the β-galactosidase levels compared to wild-type RAD51D (Supplemental Figure 1). These data suggest that RNF138 interaction with RAD51D requires ATP binding and hydrolysis [33,40].
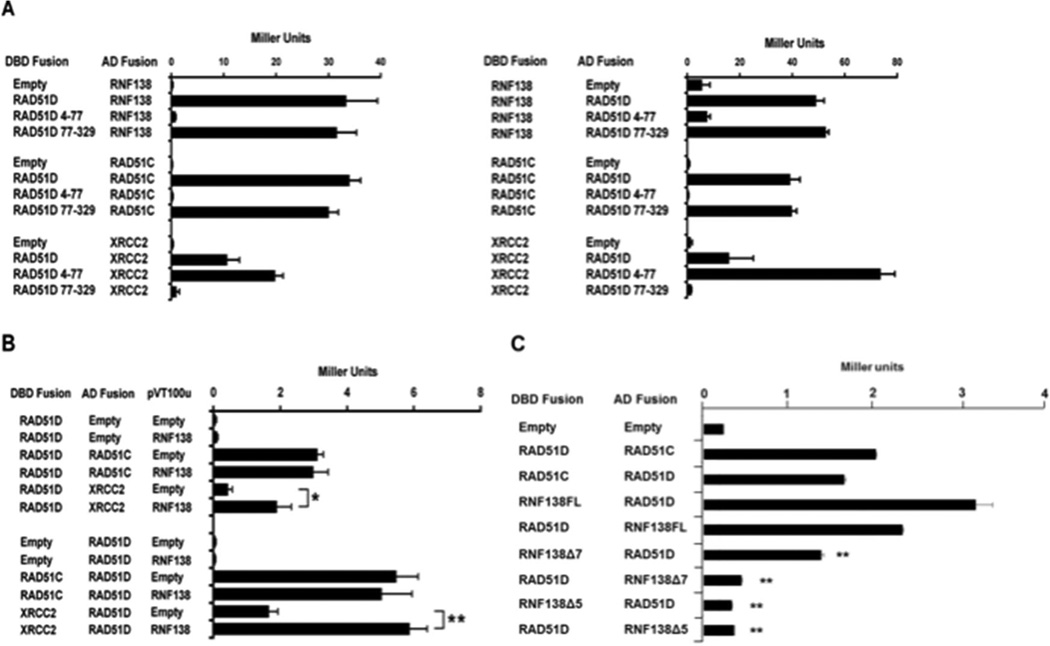
Y2H analysis was also performed using the RAD51D amino-terminal (residues 4–77) and core domain (residues 77–329) expression constructs (Figure 2A) [34,35]. It was shown previously that the RAD51D amino-terminal domain, specifically the linker region (residues 50–77), mediates binding with XRCC2, while the core domain is necessary for interaction with RAD51C. As demonstrated by ONPG analysis, the RAD51D core domain displayed a level of interaction with RNF138 similar to full-length RAD51D. Association was not detected between RAD51D 4–77 and RNF138 suggesting that RNF138 binds the RAD51D core domain, similar to RAD51C. To further narrow the region of interaction with RAD51D, the previously identified RAD51D splice variants were tested with RNF138 [35,41]. Of the seven variants, only Rad51d-Δ7b that lacks the final five amino acids of exon 7 (residues 219–223) induced β-galactosidase activity (data not shown).
Fig. 2.
RNF138 modifies interaction with RAD51C and XRCC2. (A) ONPG analysis of Y187 haploids transformed with the indicated RNF138 and RAD51D expression constructs. (B) Yeast three-hybrid analysis of Y190 haploids containing the indicated two-hybrid plasmids with either pVT100u–RNF138 or empty vector. (C) Yeast two-hybrid analysis of AH109 haploids transformed with the RNF138 splice variants and full-length RAD51D as indicated. Data represent mean +/− SEM from three independent experiments performed in triplicate, and * indicates P < 0.05, ** indicates P < 0.01.
To identify regions of RNF138 interaction, Y2H was performed using RNF138 splice variants (described below) with a deletion of exon 5 (Rnf138Δ5) or exon 7 (Rnf138Δ7) (Figure 2B). Rnf138Δ5 displayed an 8-fold decrease in the level of interaction with RAD51D compared to full-length RNF138. Rnf138Δ7 displayed a 3-fold decrease in the level of interaction. The decreased interaction was observed in both orientations and suggests the splice variants may affect RNF138 affinity for RAD51D.
To determine the potential influence of RNF138 on RAD51D interaction with RAD51C and XRCC2, a yeast three-hybrid analysis was performed (Figure 2C). Yeast were co-transformed with the yeast two-hybrid constructs for the RAD51 paralogs followed by RNF138 expression from a third vector (pVT100u). The results demonstrated that RNF138 expression had no effect on RAD51C and RAD51D interaction (Figure 2C). However, RNF138 overexpression enhanced RAD51D and XRCC2 interaction up to 3.6-fold (P < 0.01) (Figure 2C). The increased affinity between RAD51D and XRCC2 resulting from RNF138 expression, observed in both orientations, suggests RNF138 may influence the function of the BCDX2 HR complex.
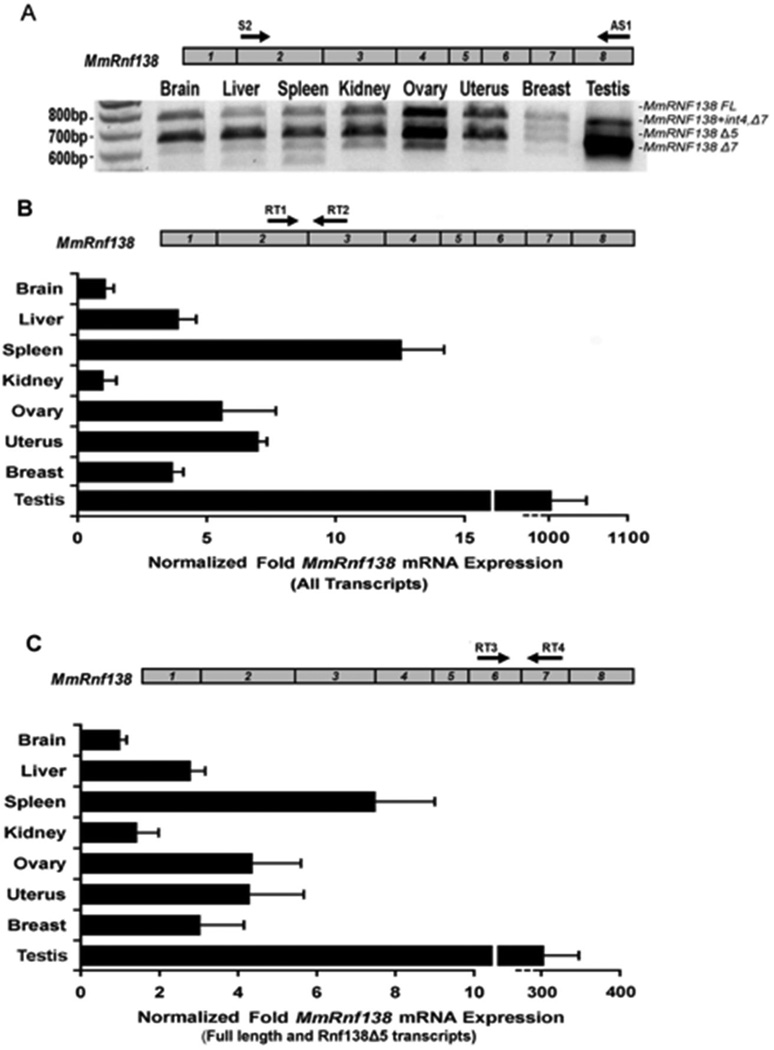
3.3 Identification of Mus musculus Rnf138 splice variants
Expression analysis of Rnf138 in eight mouse tissues using primers directed to exons 2 and 8 detected four definitive amplification products (Figure 3A). Two displayed similar expression levels in all tissues with the exception of testis, where a smaller product was predominant. RT-PCR products from brain and testis were directly cloned and sequenced to identify the four transcripts: full-length, a deletion of exon 5 (Rnf138Δ5), a deletion of exon 7 (Rnf138Δ7), and a variant from testis retaining intron 4 and a deletion of exon 7 (Rnf138+int4,Δ7). Rnf138Δ5 is predicted to remain in-frame and encode a 226 amino acid product containing all five known motifs. Rnf138Δ7 is predicted to encode a product lacking the second C2H2 zinc finger. Rnf138+int4,Δ7 is predicted to encode a truncated protein that is 149 residues long, containing a novel stretch of 18 amino acids and missing both C2H2 zinc fingers as well as the UIM (Supplemental Figure 2).
Fig. 3.
RT-PCR analysis of Mus musculus Rnf138 alternative transcripts. (A) RT-PCR for Rnf138 was performed from eight tissues. A schematic of the Rnf138 gene is shown (top) with numbered boxes representing exons. (B and C) Quantitative Real-time PCR was performed for Rnf138 from the same eight tissues. Black arrows in Rnf138 exons indicate the locations of forward and reverse PCR primers: Rnf138 exons 2 and 3 (B) or exons 6 and 7 (C). Note that Rnf138 alternative transcripts lacking exon 7 predominant in testis are not amplified in (C). Rnf138 expression was normalized to Gapdh mRNA levels. Error bars indicate standard deviation from two independent experiments performed in duplicate.
To assess the tissue-expression profile, quantitative real-time PCR analysis was performed. Primers were directed to Rnf138 exons 2 and 3 for detection of Rnf138 full-length and alternative transcripts. Rnf138 transcripts were expressed at extremely high levels in testis, and a comparison of ct values demonstrated that testis Rnf138 mRNA levels were even above Gapdh. Rnf138 was also expressed at high levels in spleen, ovary, and uterus, and at lower levels in brain and kidney (Figure 3B). Rnf138Δ7 expression in testis was determined to make up a substantial proportion of the total Rnf138 expression profile, and expression of Rnf138 full length and Rnf138Δ5 combined was approximately 3-fold lower in the testis (Figure 3C).
Increased expression of RNF138 family members is associated with tumor metastasis and chemotherapy resistance [42,43]. Consistent with these reports, RNF138 mRNA expression was approximately 4-fold higher in RKO, WiDR, and SW48 human colon cancer cell lines and 6- and 12-fold higher in T47D and HCC38 breast carcinoma cells respectively (Supplemental Figure 3).
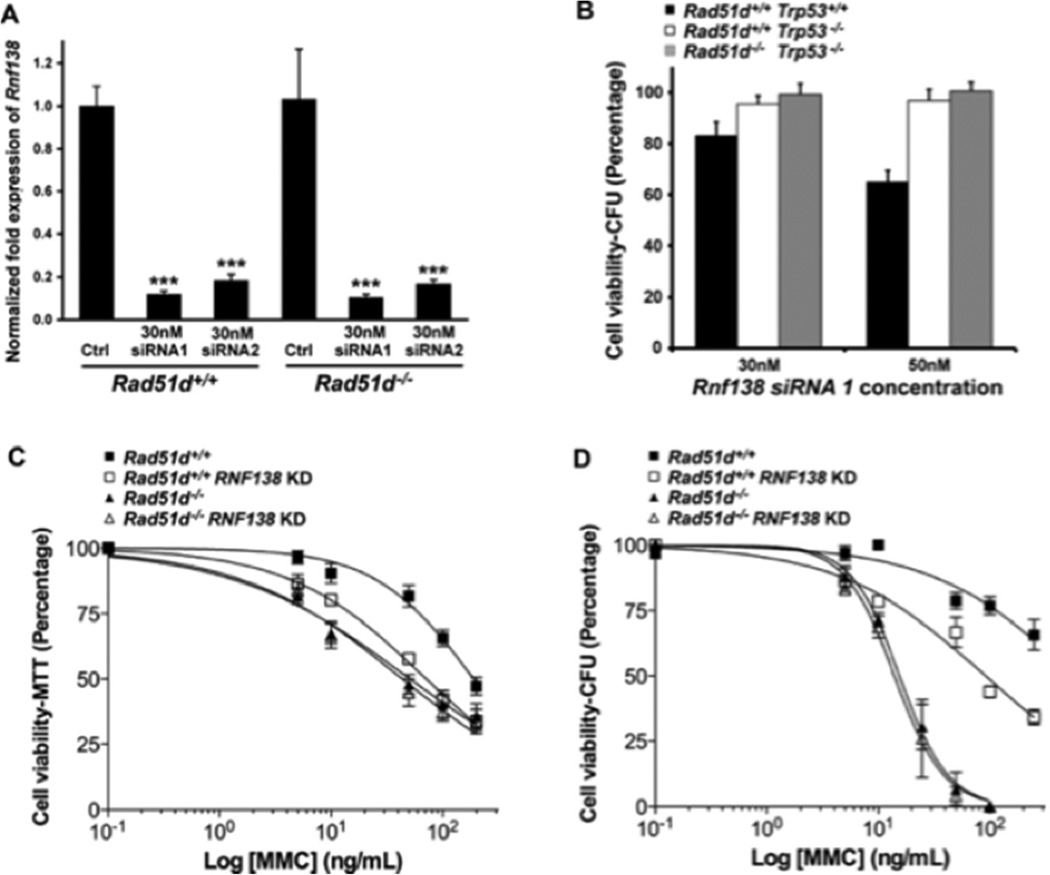
3.4. RNF138 is required for repairing DNA damage
Using siRNA, RNF138 expression was reduced in MEFs by more than 80 percent (Figure 4A), which did not alter the colony forming ability in Rad51d+/+ nor Rad51d−/− cell lines (Figure 4B). However, reduced RNF138 expression conferred increased sensitivity to DNA-crosslinking (MMC and cisplatin), DNA alkylating (MMS), and DSB generating (CPT) agents (Figure 4C, Supplemental Figure 4, and Table 1). Notably, Rad51d+/+ MEFs with reduced RNF138 expression were 2.6-fold more sensitive to MMC treatment, analogous to the 3.7-fold increase observed in Rad51d−/− cells. The EC50 values from MTT analyses were similar to Rad51d−/− cells (Table 1). In addition, transfection of Rad51d-proficient MEFs with an independent Rnf138 siRNA duplex (siRNA2) resulted in similar sensitivity to MMC (Supplemental Figure 4). Consistent with these findings, RNF138 knockdowns did not significantly increase sensitivity of Rad51d−/− cells to the same agents (Figure 4C, Table 1); Rad51d−/− MEFs were 3.7 and 1.8-fold more sensitive to MMC and cisplatin treatments compared to 4.3 and 1.9-fold for Rad51d−/− cells with reduced RNF138 levels. Furthermore, as measured by the colony forming assay, decreasing RNF138 expression conferred a 6.2-fold increased sensitivity to MMC in wild-type cells and had no further effect in Rad51d-deficient cells (Figure 4D). Thus, reduced RNF138 expression in the absence of RAD51D did not result in synthetic lethal nor synergistic effects.
Fig. 4.
RNF138 deficiency confers increased sensitivity to Mitomycin C. (A) Rnf138 expression in Rad51d+/+ (Rad51d+/+, Trp53−/−) and Rad51d−/− (Rad51d−/−, Trp53−/−) MEFs was analyzed by qPCR 24 h following siRNA transfection and normalized to both β-actin and Gapdh. Data represent mean +/− STD from a representative experiment performed in triplicate, and *** indicates P < 0.001. (B) Cell viability relative to scrambled siRNA control was measured by colony forming assay (CFA). Error bars represent mean +/− SEM from three independent experiments performed in duplicate. (C and D) MEFs were treated with the indicated amounts of Mitomycin C and cell viability assessed by (C) MTT or (D) CFA. Error bars represent +/− SEM from four independent experiments performed in triplicate for MTT assays and three independent experiments performed in duplicates for CFA using siRNA1. Note that Trp53 encodes the p53 protein and that RNF138 KD represents the RNF138 siRNA knockdown.
Table 1.
Fold-sensitivity of Rnf138 and Rad51d-deficient MEFs to DNA damaging agents.
| Agent | RNF138 KD | Rad51d−/− |
Rad51d−/− RNF138 KD |
|---|---|---|---|
| MMC | 2.64 | 3.68 | 4.31 |
| Cisplatin | 1.62 | 1.84 | 1.93 |
| MMS | 1.48 | 2.32 | 2.84 |
| Camptothecin | 1.55 | 2.27 | 2.99 |
| Hydroxyurea | 1.22 | 0.97 | 0.97 |
Fold sensitivities were determined using the EC50 values obtained from the MTT assays. RNF138 siRNA1 was used for these experiments, and EC50 values were calculated from semi-log plots fitted to non-linear regression analysis.
3.5. RNF138 is localized to the nucleus and necessary for RAD51 foci formation
DNA repair proteins predictably display nuclear localization, and RNF138 does contain a putative “RRER” NLS adjacent to the RING domain [44,45]. EGFP-RNF138 localization, as expected, was predominantly nuclear and overlapped with the DAPI nuclear stain (data not shown). EGFP-RAD51D displayed nuclear localization but was also distributed throughout the nuclear and cytoplasmic compartments, consistent with our previous findings [35].
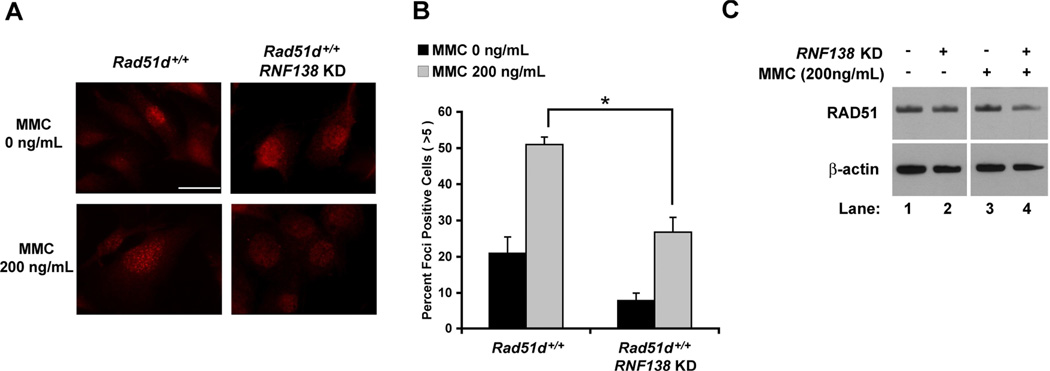
MMC treatment (200ng/mL) increased the percentage of wild-type MEFs containing 5 or more distinct RAD51 foci from 21 percent for untreated control cells to 51 percent following a 16 h exposure (Figure 5A and B) [46]. Reduced RNF138 expression resulted in a 1.9-fold decrease in RAD51 foci formation following MMC treatment (p < 0.05). In addition, the number of cells containing RAD51 foci was less in untreated RNF138-deficient versus RNF138-proficient MEFs. Although, a notable increase in RAD51 foci is still detected upon MMC treatment. Western analysis demonstrated that depletion of RNF138 had no substantial effect on RAD51 protein levels (Figure 5C). RAD51 foci formation in RNF138 KD MEFs, using siRNA2, following treatment with neocarzinostatin (200ng/ml; 2 h) was consistent with these findings (data not shown). These data indicate that RNF138 is needed for RAD51 foci formation and HR repair of DNA damage.
Fig. 5.
RNF138 is necessary for increased RAD51 foci formation in response to DNA damage. (A) Immunofluorescence visualization of RAD51 foci in Rad51d+/+ (Rad51d+/+, Trp53−/−) MEFs. (B) Quantification of RAD51 foci formation. Data represent mean +/− SEM from three independent experiments using RNF138 siRNA2 and * indicates P < 0.05. (C) Western analysis of RAD51 protein levels. β-actin was used as a loading control.
3.6 RNF138 is required to maintain chromosome integrity
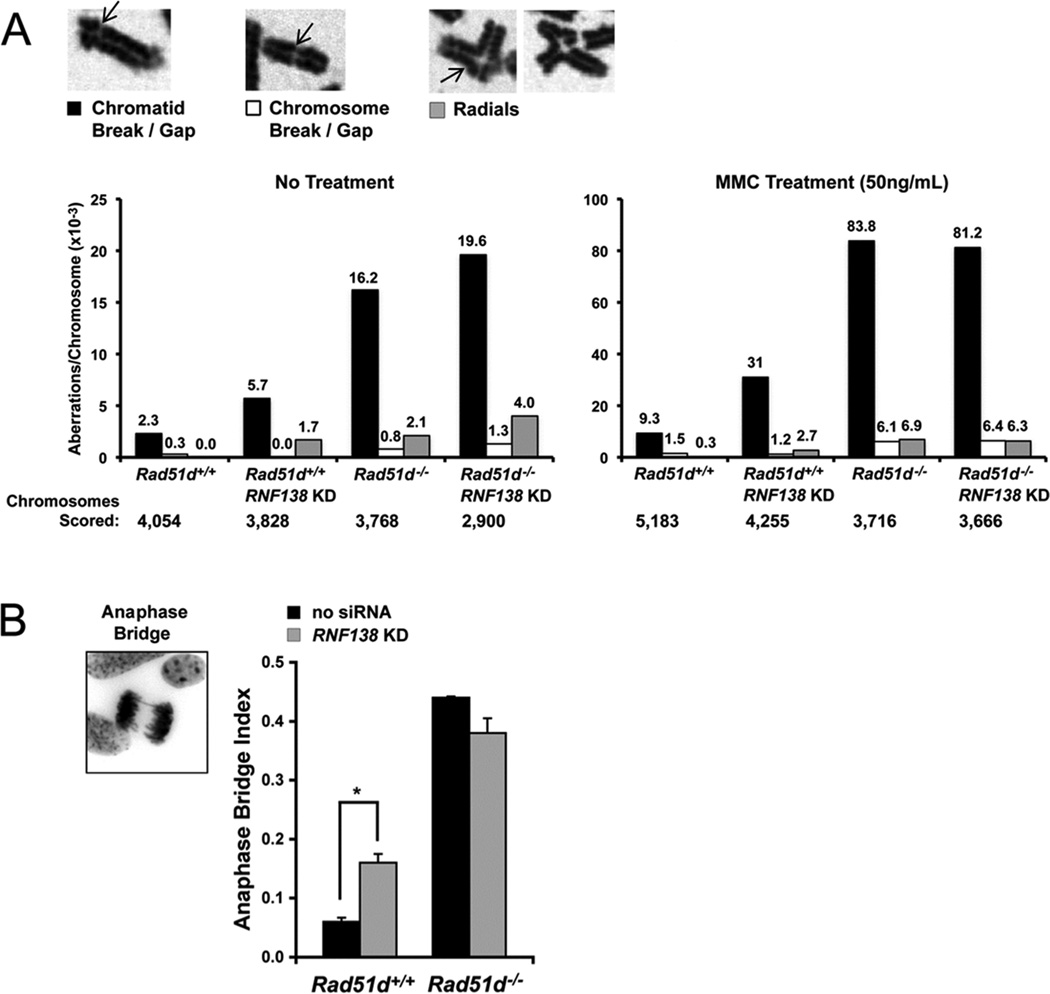
Cells with HR defects exhibit genome instability as characterized by chromosome aberrations and gross chromosome rearrangements. Consistent with a role during HR, reduced RNF138 expression resulted in a 3-fold increase in chromosomal instability (Figure 6A, left panel). Similar to our previous reports, Rad51d−/− MEFs had a 7.6-fold increase in the total number of spontaneous chromosome aberrations [25]. Chromatid breaks accounted for the majority of the aberrations (black bars), and cells deficient for both RAD51D and RNF138 had a slight increase in chromosome aberrations (Figure 6A). Following treatment with MMC (50 ng/ml), RNF138-deficient cells exhibited a 3-fold increase in total chromosome aberrations (Figure 6A, right panel). In comparison, chromatid and chromosome breaks/gaps and chromosome radials were increased 8.7-fold in Rad51d−/− MEFs following MMC treatment. Analogous to untreated cells, chromosomal aberrations in double-deficient MEFs were not higher than those in Rad51d−/− MEFs. In addition, the frequency of anaphase bridge formation was increased by 2.5-fold 72hr following transfection with Rnf138 siRNA (Figure 6B). Taken together, these data suggest RNF138 participates with RAD51D at a common step to repair DNA damage and maintain chromosome integrity.
Fig. 6.
RNF138 maintains chromosome integrity. (A) Metaphase spreads were prepared from Rad51d+/+ (Rad51d+/+, Trp53−/−) and Rad51d−/− (Rad51d−/−, Trp53−/−) MEFs following mock transfection or transfection with 30 nM Rnf138 siRNA1. Cells were either untreated (left panel) or treated with 50 ng/mL MMC (right panel). Spreads were scored for chromatid breaks/gaps, chromosome breaks/gaps, and chromosome radials, represented by arrows. The frequency of each aberration per chromosome is displayed above the bar (10−3). Note the change in scale between untreated and treated MEFs. (B) Giemsa stained cells were scored for the presence of anaphase bridges. Error bars represent mean +/− STD from two independent experiments and * indicates P < 0.05.
3.7. RNF138-dependent ubiquitination of RAD51D and proteasome-mediated degradation
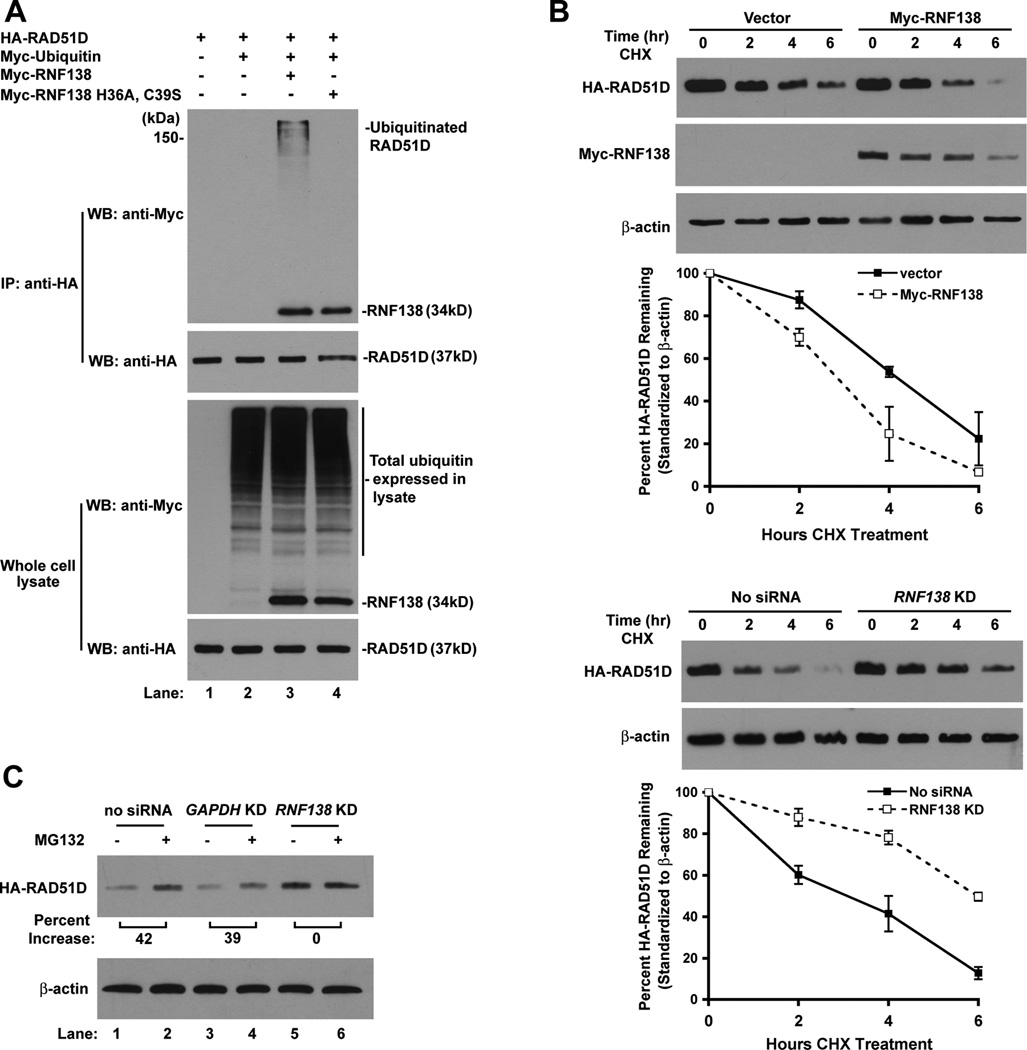
The direct interaction between RNF138 and RAD51D suggests RNF138 may target RAD51D for ubiquitination. Therefore, HA-RAD51D, Myc-RNF138, and Myc-Ubiquitin constructs were transfected into mammalian cells followed by anti-HA immunoprecipitation. HA-RAD51D migrated as high molecular weight bands, indicating RAD51D was ubiquitinated at multiple sites or ubiquitin branching occurred (Figure 7A). In addition, RAD51D ubiquitination failed to be detected when the RNF138 E3 ligase null mutant (H36A, C39S) was expressed (Figure 7A, lane 4), despite retaining its ability to interact with RAD51D (Figure 1F).
Fig. 7.
RNF138 mediates degradation of RAD51D through the ubiquitin-proteasome pathway. (A) Analysis of RNF138 facilitated RAD51D ubiquitination in vivo. (B) Rad51d−/− Trp53−/− HARad51d MEFs were transfected with Myc-RNF138 (upper panels) or 30nM Rnf138 siRNA1 (lower panels). HA-RAD51D protein levels were assessed 2, 4, and 6 hours following initiation of CHX block. HA-RAD51D band intensity was normalized to β-actin and plotted as percent protein remaining for each time point. Data represent mean +/− STD from two representative experiments. (C) MEFs were treated with CHX (100µg/mL) or CHX in combination with MG132 (20µg/mL). Cell lysates were prepared at the 4 h time point, and HA-RAD51D band intensity was normalized to β-actin.
To determine whether RNF138-directed ubiquitination modulates RAD51D protein stability, the half-life was examined in cells stably expressing HA-RAD51D [25]. Increased RNF138 expression decreased HA-RAD51D half-life from 4.2 hours to 3.0 hours (Figure 7B, top panel). Depletion of endogenous RNF138 by siRNA resulted in the accumulation of HA-RAD51D and extended the half-life to 6.6 hours (Figure 7B, bottom panel). These data indicate that RNF138-mediated ubiquitination negatively regulates RAD51D.
To examine whether decreased HA-RAD51D half-life can be attributed to proteasome degradation, protein levels were assessed following treatment with the MG132 proteasome inhibitor. HA-RAD51D was elevated up to 42% in MG132 treated samples (Figure 7C, lanes 1 – 4), indicating RAD51D is at least in part regulated by the proteasome. Intriguingly, proteasome inhibition did not further increase HA-RAD51D protein stability following knockdown of RNF138 expression (Figure 7C, lanes 5 – 6). To further demonstrate ubiquitination of RAD51D, Myc-RAD51D was demonstrated to be ubiquitinated upon addition of MG132 with no manipulation of RNF138 expression (Supplemental Figure 5A). In addition, the ubiquitin signal intensity of Myc-RAD51D was nearly 3-fold higher than Myc-RAD51C ubiquitin signal intensity (Supplemental Figure 5B). These results imply that in the absence of RNF138, RAD51D ubiquitination is diminished and no longer targeted for degradation by the proteasome.
4. Discussion
Maintaining genomic integrity is of fundamental biological and clinical importance, and RAD51D is a critical component of the homologous recombination DNA repair system. Defects within the RAD51D gene confer chromosome instability that lead to cell death unless cell cycle checkpoint mechanisms have been compromised. We demonstrated previously that Rad51d–deficient mouse embryos survived until mid-gestation, and cells derived from the embryos were not able to proliferate in culture [47]. However, removing the p53 checkpoint partially bypassed the embryo lethal phenotype, and Rad51d−/− Trp53−/− cells were able to proliferate even though there were high levels of chromosome instability and shortened telomeres [25,48]. Given the importance of RAD51D, it will be critical to identify RAD51D interacting factors and determine their functions as well as post-translational modifications.
Through a yeast two-hybrid screen, we discovered that the RNF138 E3 ubiquitin ligase is a RAD51D interacting partner. This was an intriguing connection because ubiquitin signaling cascades have emerged as essential components of HR DNA damage response [7]. RNF138 is a member of a small family of proteins distinguished by having both a RING finger domain and an ubiquitin interacting motif [39]. RING domains are found in known HR proteins such as RAD18, BRCA1, RNF8, and RNF168. Interestingly, RNF138 has also been identified as a potential target for ATM-mediated phosphorylation in response to ionizing radiation [29].
Consistent with a role during HR, reduced RNF138 expression increased the hallmarks of chromosome instability and conferred sensitivity to mitomycin C and moderate sensitivity to cisplatin and methyl methanesulfonate treatments, similar to Rad51d-deficient MEFs [25,49]. Reduced RNF138 expression in Rad51d-deficient MEFs did not confer additional sensitivity to DNA damaging agents or a more severe chromosome instability phenotype, suggesting that RNF138 and RAD51D have an epistatic relationship and act at the same HR step. We also demonstrated that RNF138 facilitates ubiquitination and proteasome degradation of RAD51D. Therefore, RNF138 may regulate HR by removing RAD51D from locations along the chromosome that encompass DSB sites to facilitate accessibility and recruitment of downstream HR factors such as RAD51.
RNF138 was first identified as part of the Wnt-p-catenin signaling pathway [28], cooperating with the E2–25K to induce the ubiquitination of T cell factor/lymphoid enhancer factor (TCF/LEF). Ubiquitinated TCF/LEF was targeted for proteasome degradation leading to decreased expression of Wnt-induced genes. Using comparative genomic hybridization analysis, two previous investigations discovered that the genomic region encoding RNF138 was significantly amplified in breast cancers [31,32]. Consistent with these findings, our q-RT-PCR analysis revealed that RNF138 expression was 2 to 12 fold higher in breast carcinoma cell lines relative to the non-tumorigenic MCF10 cells. Cells with increased RNF138 expression are expected to have a corresponding reduction in RAD51D levels, possibly contributing to genomic instability during carcinogenesis. In addition, sequence information provided by expressed sequence tag (EST) databases and our RT-PCR results suggested the mouse Rnf138 message is alternatively spliced into at least three variants: one lacking exon 5 (RNF138Δ5), one retaining intron 4 and missing exon 7 (RNF138+int4, Δ7), and a third lacking exon 7 (RNF138Δ7). Each is expressed in mouse tissues at similar levels except testis, where the RNF138Δ7 transcript is predominant. Therefore, RNF138 expression levels or altered splicing patterns may potentially be used as predictors for sensitivity to DNA crosslinking agents [50].
The suggestion from the data that degrading RAD51D is necessary for its function creates an interesting conundrum. Absence of RNF138 is expected to increase intracellular RAD51D levels but rather confers an HR-deficient phenotype. Recent work suggests that ubiquitin-regulated transcription factors contain degrons essential for both activation and proteasomal destruction [51]. These unknown interrelated mechanisms that make use of proteolytic and nonproteolytic activities of the ubiquitin-proteasome system may also be involved at the RAD51D steps during HR. Such a system would explain how absence of RNF138, which leads to increased accumulation of RAD51D, is critical for the repair of DNA damage. Work here does not exclude the possibility that multiple ubiquitin linkages form along RAD51D that are necessary for both RAD51D function as well as degradation [52].
The ubiquitin landscape along DNA break sites is complex with the proteasome being implicated during HR mediated repair [53–55]. Notably, inhibition of proteasome activity by MG132 treatment specifically suppressed HR repair of ionizing radiation-induced DNA damage and prevented RAD51 and BRCA1 foci formation [56]. In addition, RAD51 is subject to ubiquitin-mediated degradation following exposure to ionizing radiation [12–14]. However, it is unknown whether these processes are linked to the RNF8/RNF168 ubiquitin signaling cascade. The work described here suggests that RNF138 functions during HR via ubiquitination of RAD51D and also potentially could serve as an adapter between RAD51D and RAD18/RNF8 synthesized polyubiquitin chains found at sites of DNA damage [5].
While this manuscript was in revision, RNF138 was demonstrated to accumulate at DSBs to promote end resection and homologous recombination [57,58]. Processing of the breaks by the Mre11-Rad50-Nbs1 (MRN) complex was shown to provide RNF138 binding sites prior to CtIP binding activity, which was dependent upon the RNF138 ZNF domains. Consistent with these findings, loss of RNF138 activity stimulated DSB processing by the NHEJ pathway. In fact, DNA damage sensitivity and decreased formation of RAD51 foci may be influenced by the loss of RAD51D modification by RNF138 in addition to the end resection defect in RNF138-depleted cells. How RNF138 acts at these early stages and potentially regulates RAD51D activity now needs to be resolved as well as to determine whether RNF138 mechanisms differ depending upon the type of lesion encountered. It will also be useful to determine if UBE2D, the identified RNF138 E2 ubiquitination mediator, is required for the interplay of these DNA repair pathways during DNA crosslink repair.
In summary, identifying RAD51D post-translational modifications may uncover mechanisms of carcinogenesis pathways as well as potential cancer drug targets. The work presented here strongly suggests that ubiquitination of the RAD51D HR protein by the RNF138 E3 ligase plays an important role during the repair of DNA interstrand crosslink damage. The details of this ubiquitination need discerned to better understand the significance of this interaction. Sites along RAD51D that are ubiquitinated will need to be identified as well as the ubiquitin linkage arrangements required for function. This will provide insight into the mechanisms necessary for recruiting RAD51 and other downstream proteins to the sites of DNA damage.
Supplementary Material
Highlights.
RNF138 is required to repair DNA damage, primarily interstrand crosslinks, and to maintain chromosome stability as well as RAD51 focus formation.
The RNF138 E3 ligase interacts with and mediates RAD51D stability.
The RNF138 RING finger domain is necessary for ubiquitination of the RAD51D complex.
RNF138 and RAD51D act at the same step during HR mediated repair.
Acknowledgments
The authors wish to thank Dr. Changanamkandath Rajesh, Dr. Preeti Rajesh, and Mr. Cedric Johnson for technical assistance. We would also like to express appreciation to Drs. Joanna Albala, David Schild, and Xiongbin Lu for providing plasmid constructs. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R15GM110615. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Additional support was provided to DLP by the American Cancer Society (RSG-030158-01-GMC) and to NMR from a SPARC Graduate Research Grant from the Office of the Vice President for Research at the University of South Carolina.
Abbreviations
- HR
homologous recombination
- CFA
colony forming assay
- NT
N-terminus
- ssDNA
single-stranded DNA
- ICL
interstrand crosslink
- DSB
double-strand break
- MEF
mouse embryo fibroblasts
- MMC
mitomycin C
- MMS
methyl methanesulfonate
- PTM
post-translational modification
- UIM
ubiquitin interaction motif
- Y2H
yeast two-hybrid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Polo SE. Reshaping Chromatin after DNA Damage: The Choreography of Histone Proteins. J Mol Biol. 2014;427:626–636. doi: 10.1016/j.jmb.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao H, Zhu M, Dou G, Zhao H, Zhu B, Li J, Liao J, Xu X. BCL10 regulates RNF8/RNF168-mediated ubiquitination in the DNA damage response. Cell Cycle. 2014;13:1777–1787. doi: 10.4161/cc.28707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: roles in DNA damage signalling recombinational repair and tumorigenesis. Semin Cell Dev Biol. 2011;22:898–905. doi: 10.1016/j.semcdb.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 7.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, Shkedy D, Smorodinsky NI, van Vliet N, Kuster B, Mann M, Ciechanover A, Dahm-Daphi J, Kanaar R, Hu MC, Chen DJ, Oren M, Shiloh Y. Requirement of ATM-Dependent Monoubiquitylation of Histone H2B for Timely Repair of DNA Double-Strand Breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, Zou L, Komatsu K. Regulation of Homologous Recombination by RNF20-Dependent H2B Ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett BT, Knight KL. Cellular localization of human Rad51C and regulation of ubiquitin-mediated proteolysis of Rad51. J Cell Biochem. 2005;96:1095–1109. doi: 10.1002/jcb.20640. [DOI] [PubMed] [Google Scholar]
- 13.Yamamori T, Meike S, Nagane M, Yasui H, Inanami O. ER stress suppresses DNA double-strand break repair and sensitizes tumor cells to ionizing radiation by stimulating proteasomal degradation of Rad51. FEBS Lett. 2013;587:3348–3353. doi: 10.1016/j.febslet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J, Zhou L, Wu G, Konig H, Lin X, Li G, Qiu XL, Chen CF, Hu CM, Goldblatt E, Bhatia R, Chamberlin AR, Chen PL, Lee WH. A novel small molecule RAD51 inactivator overcomes imatinib-resistance in chronic myeloid leukaemia. EMBO Mol Med. 2013;5:353–365. doi: 10.1002/emmm.201201760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 16.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 17.Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, Bowden G, Kalmyrzaev B, Warren-Perry M, Snape K, Adlard JW, Barwell J, Berg J, Brady AF, Brewer C, Brice G, Chapman C, Cook J, Davidson R, Donaldson A, Douglas F, Greenhalgh L, Henderson A, Izatt L, Kumar A, Lalloo F, Miedzybrodzka Z, Morrison PJ, Paterson J, Porteous M, Rogers MT, Shanley S, Walker L, Eccles D, Evans DG, Renwick A, Seal S, Lord CJ, Ashworth A, Reis-Filho JS, Antoniou AC, Rahman N. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osher DJ, De Leeneer K, Michils G, Hamel N, Tomiak E, Poppe B, Leunen K, Legius E, Shuen A, Smith E, Arseneau J, Tonin P, Matthijs G, Claes K, Tischkowitz MD, Foulkes WD. Mutation analysis of RAD51D in non-BRCA1/2 ovarian and breast cancer families. Br J Cancer. 2012;106:1460–1463. doi: 10.1038/bjc.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelttari LM, Kiiski J, Nurminen R, Kallioniemi A, Schleutker J, Gylfe A, Aaltonen LA, Leminen A, Heikkila P, Blomqvist C, Butzow R, Aittomaki K, Nevanlinna H. A Finnish founder mutation in RAD51D: analysis in breast ovarian, prostate and colorectal cancer. J Med Genet. 2012;49:429–432. doi: 10.1136/jmedgenet-2012-100852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson ER, Rowley SM, Sawyer S, Confab K, Eccles DM, Trainer AH, Mitchell G, James PA, Campbell IG. Analysis of RAD51D in ovarian cancer patients and families with a history of ovarian or breast cancer. PLoS One. 2013;8:e54772. doi: 10.1371/journal.pone.0054772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weissman SM, Weiss SM, Newlin AC. Genetic testing by cancer site: ovary. Cancer J. 2012;18:320–327. doi: 10.1097/PPO.0b013e31826246c2. [DOI] [PubMed] [Google Scholar]
- 22.Wickramanyake A, Bernier G, Pennil C, Casadei S, Agnew KJ, Stray SM, Mandell J, Garcia RL, Walsh T, King MC, Swisher EM. Loss of function germline mutations in RAD51D in women with ovarian carcinoma. Gynecol Oncol. 2012;127:552–555. doi: 10.1016/j.ygyno.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker JL, Schwab RB, Wallace AM, Madlensky L. Breast cancer in a RAD51D mutation carrier: case report and review of the literature. Clin Breast Cancer. 2015;15:e71–e75. doi: 10.1016/j.clbc.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Hinz JM, Tebbs RS, Wilson PF, Nham PB, Salazar EP, Nagasawa H, Urbin SS, Bedford JS, Thompson LH. Repression of mutagenesis by Rad51D–mediated homologous recombination. Nucleic Acids Res. 2006;34:1358–1368. doi: 10.1093/nar/gkl020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smiraldo PG, Gruver AM, Osborn JC, Pittman DL. Extensive chromosomal instability in Rad51d–deficient mouse cells. Cancer Res. 2005;65:2089–2096. doi: 10.1158/0008-5472.CAN-04-2079. [DOI] [PubMed] [Google Scholar]
- 26.Kurumizaka H, Ikawa S, Nakada M, Enomoto R, Kagawa W, Kinebuchi T, Yamazoe M, Yokoyama S, Shibata T. Homologous pairing and ring and filament structure formation activities of the human Xrcc2*Rad51D complex. J Biol Chem. 2002;277:14315–14320. doi: 10.1074/jbc.M105719200. [DOI] [PubMed] [Google Scholar]
- 27.Konstantinopoulos PA, Wilson AJ, Saskowski J, Wass E, Khabele D. Suberoylanilide hydroxamic acid (SAHA) enhances olaparib activity by targeting homologous recombination DNA repair in ovarian cancer. Gynecol Oncol. 2014;133:599–606. doi: 10.1016/j.ygyno.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada M, Ohnishi J, Ohkawara B, Iemura S, Satoh K, Hyodo-Miura J, Kawachi K, Natsume T, Shibuya H. NARF an nemo-like kinase (NLK)-associated ring finger protein regulates the ubiquitylation and degradation of T cell factor/lymphoid enhancer factor (TCF/LEF) J Biol Chem. 2006;281:20749–20760. doi: 10.1074/jbc.M602089200. [DOI] [PubMed] [Google Scholar]
- 29.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 30.Anderson TW, Wright C, Brooks WS. The E3 Ubiquitin Ligase NARF Promotes Colony Formation in vitro and Exhibits Enhanced Expression Levels in Glioblastoma Multiforme in vivo. ajur.uni.edu. 2010 [Google Scholar]
- 31.Han W, Jung EM, Cho J, Lee JW, Hwang KT, Yang SJ, Kang JJ, Bae JY, Jeon YK, Park IA, Nicolau M, Jeffrey SS, Noh DY. DNA copy number alterations and expression of relevant genes in triple-negative breast cancer. Genes Chromosomes Cancer. 2008;47:490–499. doi: 10.1002/gcc.20550. [DOI] [PubMed] [Google Scholar]
- 32.Klijn C, Holstege H, de Ridder J, Liu X, Reinders M, Jonkers J, Wessels L. Identification of cancer genes using a statistical framework for multiexperiment analysis of nondiscretized array CGH data. Nucleic Acids Res. 2008;36:e13. doi: 10.1093/nar/gkm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gruver AM, Miller KA, Rajesh C, Smiraldo PG, Kaliyaperumal S, Balder R, Stiles KM, Albala JS, Pittman DL. The ATPase motif in RAD51D is required for resistance to DNA interstrand crosslinking agents and interaction with RAD51C. Mutagenesis. 2005;20:433–440. doi: 10.1093/mutage/gei059. [DOI] [PubMed] [Google Scholar]
- 34.Miller KA, Sawicka D, Barsky D, Albala JS. Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res. 2004;32:169–178. doi: 10.1093/nar/gkg925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruver AM, Yard BD, McInnes C, Rajesh C, Pittman DL. Functional characterization and identification of mouse Rad51d splice variants. BMC Mol Biol. 2009;10:27. doi: 10.1186/1471-2199-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schild D, Lio Y, Collins DW, Tsomondo T, Chen DJ. Evidence for simultaneous protein interactions between human rad51 paralogs. J Biol Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- 37.Rajesh C, Baker DK, Pierce AJ, Pittman DL. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 2011;39:132–145. doi: 10.1093/nar/gkq738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Savage J. Classification and relationships of induced chromosomal structural changes. Journal of Medical Genetics. 1975;12:103–122. doi: 10.1136/jmg.13.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giannini AL, Gao Y, Bijlmakers MJ. T-cell regulator RNF125/TRAC-1 belongs to a novel family of ubiquitin ligases with zinc fingers and a ubiquitin-binding domain. Biochem J. 2008;410:101–111. doi: 10.1042/BJ20070995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forget AL, Loftus MS, McGrew DA, Bennett BT, Knight KL. The human Rad51 K133A mutant is functional for DNA double-strand break repair in human cells. Biochemistry. 2007;46:3566–3575. doi: 10.1021/bi062128k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawabata M, Akiyama K, Kawabata T. Genomic structure multiple alternative transcripts of the mouse TRAD/RAD51L3/RAD51D gene a member of the recA/RAD51 gene family. Biochim Biophys Acta. 2004;1679:107–116. doi: 10.1016/j.bbaexp.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Sasaki H, Miura K, Horii A, Kaneko N, Fujibuchi W, Kiseleva L, Gu Z, Murata Y, Karasawa H, Mizoi T, Kobayashi T, Kinouchi M, Ohnuma S, Yazaki N, Unno M, Sasaki I. Orthotopic implantation mouse model and cDNA microarray analysis indicates several genes potentially involved in lymph node metastasis of colorectal cancer. Cancer Sci. 2008;99:711–719. doi: 10.1111/j.1349-7006.2008.00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ju W, Yoo BC, Kim IJ, Kim JW, Kim SC, Lee HP. Identification of genes with differential expression in chemoresistant epithelial ovarian cancer using high-density oligonucleotide microarrays. Oncol Res. 2009;18:47–56. doi: 10.3727/096504009789954672. [DOI] [PubMed] [Google Scholar]
- 44.Kaneko H, Orii KO, Matsui E, Shimozawa N, Fukao T, Matsumoto T, Shimamoto A, Furuichi Y, Hayakawa S, Kasahara K, Kondo N. BLM (the causative gene of Bloom syndrome) protein translocation into the nucleus by a nuclear localization signal. Biochem Biophys Res Commun. 1997;240:348–353. doi: 10.1006/bbrc.1997.7648. [DOI] [PubMed] [Google Scholar]
- 45.da Costa Lima TD, Moura DM, Reis CR, Vasconcelos JR, Ellis L, Carrington M, Figueiredo RC, de Melo Neto OP. Functional characterization of three leishmania poly(a) binding protein homologues with distinct binding properties to RNA and protein partners. Eukaryot Cell. 2010;9:1484–1494. doi: 10.1128/EC.00148-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci U S A. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pittman DL, Schimenti JC. Midgestation lethality in mice deficient for the RecA-related gene Rad51d/Rad51l3. Genesis. 2000;26:167–173. doi: 10.1002/(sici)1526-968x(200003)26:3<167::aid-gene1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 48.Tarsounas M, Munoz P, Claas A, Smiraldo PG, Pittman DL, Blasco MA, West SC. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004;117:337–347. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- 49.Sasaki MS, Takata M, Sonoda E, Tachibana A, Takeda S. Recombination repair pathway in the maintenance of chromosomal integrity against DNA interstrand crosslinks. Cytogenet Genome Res. 2004;104:28–34. doi: 10.1159/000077463. [DOI] [PubMed] [Google Scholar]
- 50.Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, Salter J, Orr N, Parton M, Smith IE, Reis-Filho JS, Dowsett M, Ashworth A, Turner NC. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res. 2010;16:6159–6168. doi: 10.1158/1078-0432.CCR-10-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geng F, Wenzel S, Tansey WP. Ubiquitin and proteasomes in transcription. Annu Rev Biochem. 2012;81:177–201. doi: 10.1146/annurev-biochem-052110-120012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gudmundsdottir K, Lord CJ, Ashworth A. The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene. 2007;26:7601–7606. doi: 10.1038/sj.onc.1210579. [DOI] [PubMed] [Google Scholar]
- 54.Kristensen CN, Bystol KM, Li B, Serrano L, Brenneman MA. Depletion of DSS1 protein disables homologous recombinational repair in human cells. Mutat Res. 2010;694:60–64. doi: 10.1016/j.mrfmmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 55.Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, Greenblatt JF. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 56.Murakawa Y, Sonoda E, Barber LJ, Zeng W, Yokomori K, Kimura H, Niimi A, Lehmann A, Zhao GY, Hochegger H, Boulton SJ, Takeda S. Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res. 2007;67:8536–8543. doi: 10.1158/0008-5472.CAN-07-1166. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt CK, Galanty Y, Sczaniecka-Clift M, Coates J, Jhujh S, Demir M, Cornwell M, Beli P, Jackson SP. Systematic E2 screening reveals a UBE2D–RNF138-CtIP axis promoting DNA repair. Nat Cell Biol. 2015;17:1458–1470. doi: 10.1038/ncb3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ismail IH, Gagné JP, Genois MM, Strickfaden H, McDonald D, Xu Z, Poirier GG, Masson JY, Hendzel MJ. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat Cell Biol. 2015;17:1446–1457. doi: 10.1038/ncb3259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.