Abstract
EF-G is a universally conserved translational GTPase that promotes the translocation of tRNA and mRNA through the ribosome. EF-G binds to the ribosome in a GTP-bound form and subsequently catalyzes GTP hydrolysis. The contribution of the ribosome-stimulated GTP hydrolysis by EF-G to tRNA/mRNA translocation remains debated. Here, we show that while EF-G•GDP does not stably bind to the ribosome and induce translocation, EFG• GDP in complex with phosphate group analogues BeF3− and AlF4− promotes the translocation of tRNA and mRNA. Furthermore, the rates of mRNA translocation induced by EF-G in the presence of GTP and a non-hydrolysable analogue of GTP, GDP•BeF3−are similar. Our results are consistent with the model suggesting that GTP hydrolysis is not directly coupled to mRNA/tRNA translocation. Hence, GTP binding is required to induce the activated, translocation-competent conformation of EF-G while GTP hydrolysis triggers EF-G release from the ribosome.
Introduction
During protein synthesis, tRNAs and their associated codons on the mRNA are translocated from the A (aminoacyl) to the P (peptidyl) and the E (exit) sites of the ribosome. This process is induced by the binding of a universally conserved elongation factor G (EF-G) in bacteria and elongation factor 2 (EF-2) in eukaryotes. EF-G accelerates translocation by ~50,000 fold[1, 2]. Domain I of EF-G (Fig. 1B) comprises the G’ and G subdomains; the latter hydrolyzes GTP and is structurally similar to the G-domains in other G-proteins[3, 4]. EF-G binds to the ribosome with high affinity only in the GTP-bound form[5, 6]. EF-G has low intrinsic GTPase activity, which is enhanced via the interaction of the G domain of EF-G with the universally conserved sarcin-ricin loop (SRL) of the 23S rRNA of the large ribosomal subunit[7–9]. GTP hydrolysis and the subsequent release of inorganic phosphate (Pi) trigger EF-G dissociation from the ribosome[6, 10].
Figure 1.
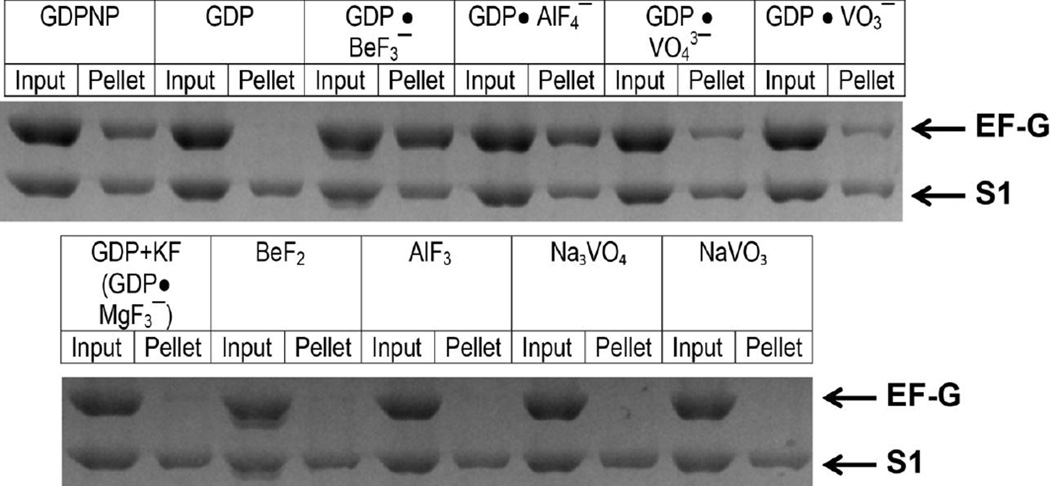
Binding of EF-G to vacant ribosomes in the presence of various nucleotides and phosphate analogues (as indicated) measured by the pelleting assay. EF-G was incubated with vacant ribosomes. Half of each sample was pelleted through a sucrose cushion (pellet); the other half was used as a loading control (input). Protein content of ribosome pellets was analyzed using SDS-PAGE. The band corresponding to EF-G and the largest ribosomal protein S1 are indicated by arrows.
The role of GTP hydrolysis in the translocation of tRNAs and mRNA is still debated. Early experiments demonstrated that GTP hydrolysis is not required for translocation because the replacement of GTP with non-hydrolysable analogues of GTP blocked EF-G release from the ribosome but allowed for a single round of translocation[5, 6]. However, more recent kinetic experiments suggested that GTP is hydrolyzed by EF-G at rates significantly faster than the rate of translocation; hence, GTP hydrolysis precedes translocation[1]. Furthermore, translocation in the presence of GTP was shown to be up to 50-fold faster than translocation in the presence of non-hydrolysable analogues of GTP, thus, suggesting that GTP hydrolysis contributes to the acceleration of translocation[1]. Additional kinetic experiments revealed that the release of inorganic phosphate following GTP hydrolysis occurs at rates similar to the rate of tRNA/mRNA translocation[10]. Therefore, the release of inorganic phosphate may be coupled to translocation[10]. It was hypothesized that GTP hydrolysis and the following inorganic phosphate release may trigger interdomain rearrangements in EF-G; namely, the movement of domain IV of EF-G relative to the rest of EF-G[10–14] that induces the translocation of tRNAs. Indeed, domain IV of EF-G was shown to be critical for the catalysis of translocation[1, 15] and is thought to displace the peptidyl-tRNA from the A site because domain IV is bound to the A site of the small subunit in the posttranslocation state of the ribosome[9, 16].
Although the model that directly links GTP hydrolysis and translocation is popular in the field, there are a number of experimental observations that are not consistent with this model. Several independent reports suggested that substituting GTP with non-hydrolysable analogues of GTP only moderately (by 2–3 fold) slow down the rate of mRNA translocation[17, 18]. Amino acid substitutions in the G domain of EF-G that inhibited the GTPase activity of EF-G reduced the rate of mRNA/tRNA translocation by only 7 to 30 fold[12, 19–21]. Furthermore, a number of antibiotics, such as viomycin or hygromycin B, strongly inhibit translocation by binding to the ribosome without impeding the binding of EF-G, GTP hydrolysis or Pi release[1, 22, 23]. Hence, at least in the presence of inhibitors of translocation, GTP hydrolysis and translocation may be completely decoupled. Consistent with this model, single-molecule Förster resonance energy transfer (smFRET) experiments show that translocation on average requires multiple EF-G binding events[20, 24, 25] suggesting that EF-G dissociation triggered by Pi release sometimes occurs before the tRNA/mRNA are translocated. These results suggest the possibility that Pi release and tRNA/mRNA translocation evolved to occur concurrently but they are not mechanistically coupled.
It was suggested that one potential source of discrepancies between the different reports on the role of GTP hydrolysis in translocation was the possible contamination of commercially available synthetic non-hydrolysable GTP analogues with GTP[12]. Although we are not aware of publications that unequivocally demonstrated the presence of GTP contamination in nonhydrolysable GTP analogues, we aimed to overcome this conceivable caveat to further examine the role of GTP hydrolysis in translocation.
Complexes of GDP with phosphate analogues vanadate, beryllium and aluminum fluorides are known to mimic GTP or the transition state intermediate of GTP hydrolysis[26, 27]. Previous structural studies of a number of GTPases and ATPases suggested that GDP (or ADP)•BeF3− bound to the active site of an enzyme mimics the pre-hydrolysis state of the hydrolysis reaction while the geometry of GDP (ADP)•AlF4− and GDP(ADP)•VO43− resemble the transition or posthydrolysis state of the reaction[27–31]. Furthermore, it was previously reported that vanadate inhibits the ability of EF-G to induce RRF (ribosome recycling factor)-assisted ribosome dissociation into subunits after the termination of protein synthesis[32]. Since GTP hydrolysis was previously shown to be essential for the ribosome recycling[33, 34], inhibition of EFG/ RRF-assisted subunit dissociation by vanadate suggests that vanadate can bind to EF-G•GDP and act as a GTP analogue.
Here we test whether BeF3−, AlF4−, and VO43− can act on EF-G as phosphate analogues and support ribosomal translocation. We show that EF-G in the presence of beryllium fluoride and GDP that was purified from GTP contamination induces translocation at rates similar to the rates observed in the presence of GTP. Our results indicate that EF-G induces fast and efficient translocation in the GTP-bound pre-hydrolysis state. Thus, our results are consistent with the model suggesting that GTP hydrolysis by EF-G is not directly coupled to the translocation of mRNA and tRNA.
Results
EF-G stably binds to the ribosome in the presence of GDP and phosphate analogues
Because EF-G binds stably to the ribosome only in the GTP-bound form, we first aimed to test whether BeF3−, AlF4−, and VO43− stabilize the binding of EF-G to the ribosome in the presence of GDP. Consistent with previous reports, we found that commercially available GDP contains significant amounts of GTP[17, 35, 36]. Nevertheless, GDP and GTP could be easily separated using anion exchange chromatography[35]. Purified GDP was used in all experiments presented in this work.
To examine EF-G binding to the ribosome in the presence of various phosphate analogues we used a non-equilibrium ribosome pelleting assay[12, 37]. Vacant ribosomes from E. coli were incubated with EF-G in the presence of GDP or a non-hydrolysable analogue of GTP, β,γ -imidoguanosine 5’-triphosphate(GDPNP). Then the amount of EF-G bound to the ribosome after pelleting the solution through a sucrose cushion was determined using SDS-PAGE. Consistent with published reports[5, 6], EF-G was stably bound to the ribosome in the presence of GDPNP but was absent in the ribosome pellet in the presence of GDP. However, the addition of beryllium fluoride or aluminum fluoride to GDP resulted in the stable binding of EF-G to the ribosome (Fig. 1, Supplementary Fig. 1) suggesting that GDP•BeF3− and GDP•AlF4− can bind to EF-G and act as GTP analogues. Addition of either sodium orthovanadate (NaVO3) or metavanadate (Na3VO4) led to a sub-stoichiometric EF-G binding indicating that vanadate-GDP is less potent in stabilizing the GTP-bound conformation of EF-G (Fig.1). It has been reported that Mg2+, which is present in the polyamine buffer used for the pelleting experiments, can make a complex with fluoride and GDP (GDP•MgF3−)[27] in the active sites of GTPases, thus making another GTP analogue. When we incubated EF-G with ribosomes in the presence of potassium fluoride and GDP, no EF-G binding was observed (Fig. 1). Hence, the stabilization of EF-G binding observed in the presence of GDP, beryllium fluoride and aluminum fluoride in Mg-containing buffer is not due to the formation of an EF-G•GDP•MgF3− complex. Finally, vanadate, beryllium and aluminum fluorides alone (i.e. in the absence of GDP) did not stabilize EF-G binding (Fig. 1). Taken together, these pelleting experiments indicate that beryllium and aluminum fluorides (and, to a lesser extent, vanadate) can bind to EF-G•GDP and act as GTP analogues by trapping EF-G on the ribosome.
EF-G induces tRNA translocation in the presence of GDP combined with beryllium or aluminum fluoride
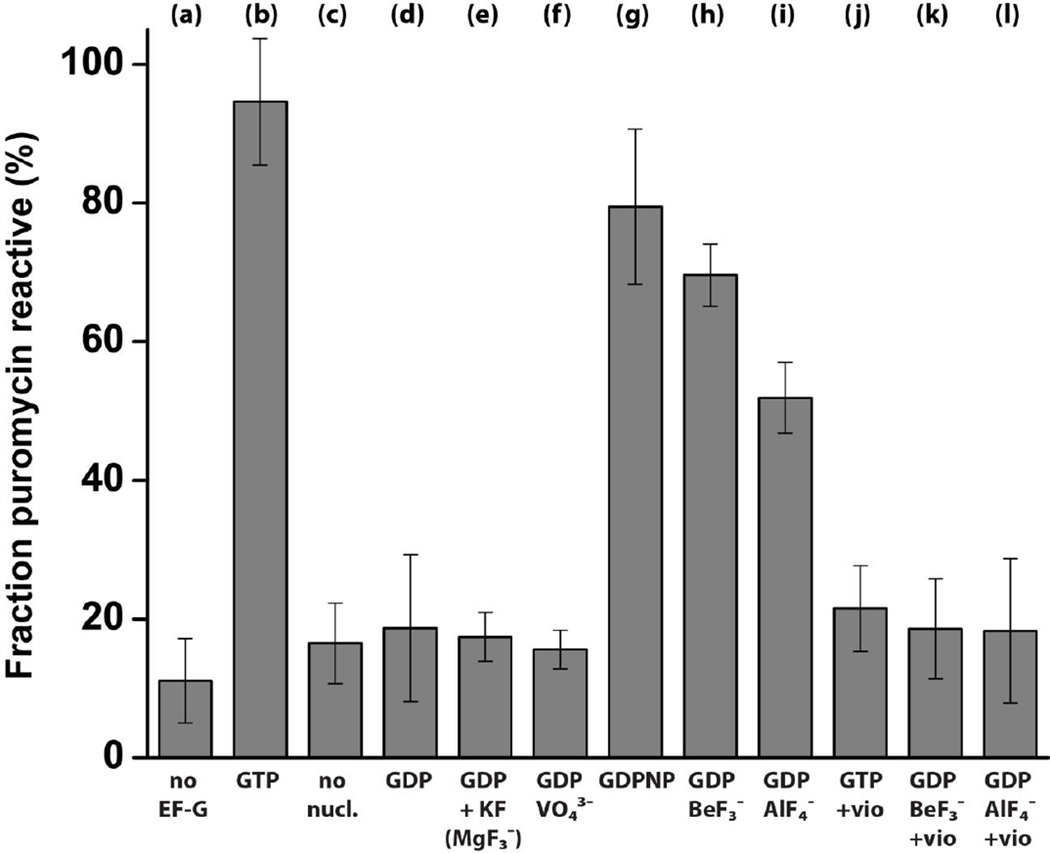
We next tested whether phosphate analogues can activate EF-G and support translocation. Pretranslocation ribosome complexes were assembled via the non-enzymatic binding of deacylated elongator tRNAMet to the P site followed by the binding of N-acetyl-[3H]Tyr-tRNATyr to the A site of the ribosome in the presence of a defined mRNA. Pretranslocation ribosomes were then incubated with EF-G and various nucleotide/phosphate analogues and the extent of translocation was measured by the reactivity of N-acetyl-[3H]Tyr-tRNATyr toward the A-site substrate of the peptidyl-transferase reaction, the antibiotic puromycin. In agreement with previous reports[36, 38], in the absence of EF-G, less than 15% of N-acetyl-[3H]Tyr-tRNATyr bound to the ribosome reacted with puromycin indicating that N-acetyl-[3H]Tyr-tRNATyr remains bound to the A site (Fig. 2a). EF-G•GTP induced efficient translocation as over 95% N-acetyl-[3H]Tyr-tRNATyr became puromycin-reactive (Fig. 2b), indicating the movement of N-acetyl-[3H]Tyr-tRNATyr into the P site of the ribosome. EF-G did not show significant translocation activity in the absence of nucleotides (Fig. 2c). Likewise, no translocation was observed in the presence of GDP, GDP with potassium fluoride or GDP with sodium vanadate (Fig. 2d–f). In contrast, 80%, 70% and 52% of ribosome-bound N-acetyl-[3H]Tyr-tRNATyr was puromycinreactive after incubation of pretranslocation ribosomes with EF-G•GDPNP, EF-G•GDP•BeF3− and EF-G•GDP•AlF4−, respectively (Fig. 2g–i). Although we tested a wide range of GDP, beryllium and aluminum fluoride concentrations, EF-G•GDP•BeF3− and EF-G•GDP•AlF4− induced translocation of N-acetyl-[3H]Tyr-tRNATyr into the P site only in a fraction of the ribosomes. Possible reasons for the incompleteness of translocation are considered in the Discussion. These results indicate that GDP•BeF3− and GDP•AlF4− act similarly to GDPNP by mimicking GTP and inducing an EF-G conformation which is active in translocation.
Figure 2.
Puromycin reactivity of pretranslocation ribosomes incubated with EF-G in the presence of various nucleotides and phosphate analogues. Deacylated tRNAMet was bound to the P site, and N-acetyl-[3H]Tyr-tRNATyr was bound to the A site of the ribosome. Bar graphs indicate the fraction of ribosome-bound N-acetyl-[3H]Tyr-tRNATyr, measured by a filter-binding assay, that is puromycin reactive in pretranslocation ribosomes in the absence of EF-G (a) or pretranslocation ribosomes incubated with EF-G in the presence of various nucleotides and phosphate analogues (as indicated) (b–l). EF-G was added with GTP (b, j), in the absence of any nucleotides (c), with GDP (d), with GDP and KF (e), with GDP and sodium meta vanadate (f), with GDPNP (g), with GDP and beryllium fluoride (h, k) or with GDP and aluminum fluoride (i, l). In (j–l) pretranslocation ribosomes were pre-incubated with viomycin (0.5 mM). Error bars show standard deviations calculated from four to six independent measurements.
When pretranslocation ribosomes were preincubated with an inhibitor of translocation, the antibiotic viomycin, before the addition of EF-G•GTP, EF-G•GDP•BeF3− or EF-G•GDP•AlF4−, less than 20–25% of ribosome-bound N-acetyl-[3H]Tyr-tRNATyr was puromycin-reactive (Fig. 2 j–l). Viomycin is known to block translocation without hampering EF-G binding to the ribosome or GTP hydrolysis[23, 39, 40]. Hence, the increase in puromycin reactivity of ribosome-bound N-acetyl-[3H]Tyr-tRNATyr induced by EF-G•GDP•BeF3− or EF-G•GDP•AlF4− in the absence of viomycin (Fig. 2h–i) was due to the translocation of N-acetyl-[3H]Tyr-tRNATyr into the P site and not simply a result of EF-G binding to the pretranslocation ribosome.
Rates of EF-G-induced translocation in the presence of GTP and GDP·BeF3− are similar
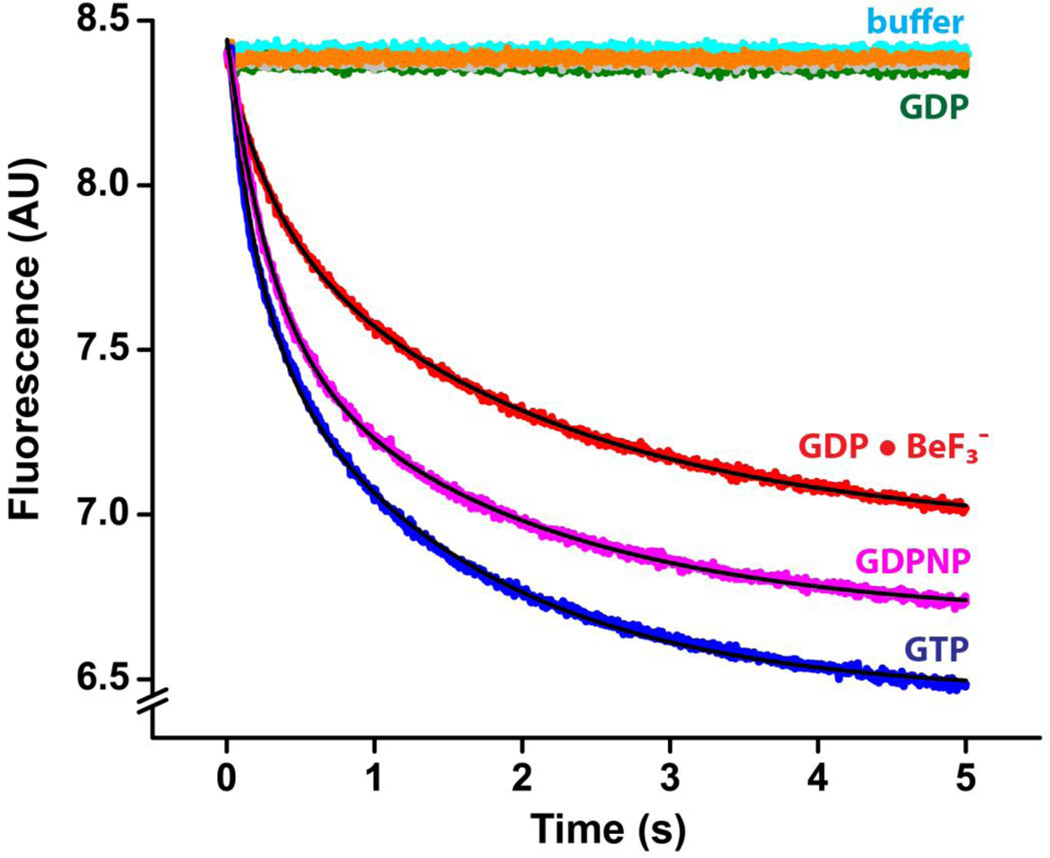
We next measured the kinetics of mRNA translocation in the presence of EF-G•GDP•BeF3− and EF-G•GDP•AlF4−. The kinetics of mRNA translocation were followed by the fluorescence quenching of a fluorescein dye attached to the 3’ end of an mRNA as it moves within the ribosome[41, 42]. Pretranslocation complexes were assembled with fluorescein-labeled mRNA, deacylated tRNAMet, N-acetyl-Tyr-tRNATyr and 70S ribosomes. When these pretranslocation ribosomes were mixed with EF-G•GTP using a stopped-flow apparatus, rapid quenching of fluorescein fluorescence was observed, indicative of mRNA translocation (Fig. 3). As has been reported previously[18, 23, 43–45], the kinetics of mRNA translocation are clearly biphasic and are best fitted by the sum of two exponentials, corresponding to the apparent rate constants k1 and k2 (Table 1); the faster rate constant k1 accounts for 40–50% of the amplitude of the change in fluorescence (Table 1). Although the biphasic manner of fluorescence changes associated with mRNA translocation is well documented[18, 23, 43–45], the physical basis of this phenomenon remains unclear and will be discussed below. We have previously used a single-exponential approximation of the bi-phasic kinetics by estimating kav, the weighted average rate constant calculated as the sum of k1 and k2 normalized to their respective contributions to the total amplitude of the fluorescence change [kav = (k1*A1 + k2*A2)/(A1 + A2)][18].
Figure 3.
Pre-steady-state kinetics of translocation in the presence of phosphate analogues at pH 7.5. mRNA translocation was induced by mixing pretranslocation ribosomes (35 nM after mixing) with EF-G (1 µM after mixing) preincubated with GTP (blue), GDPNP (magenta), GDP (dark green), GDP and KF (grey), GDP and VO43− (orange) or GDP and BeF3− (red). Experiments were performed in polyamine buffer at pH 7.5. mRNA translocation was detected by the quenching of fluorescein attached to the 3’ end of mRNA using a stopped-flow apparatus. Pretranslocation ribosomes were also mixed with buffer only (cyan) to account for the rates of the photobleaching of fluorescein and spontaneous translocation. Double-exponential fits for fluorescence quenching (black lines) are reported in Table 1.
Table 1.
Rates of mRNA translocation induced by EF-G in the presence of various nucleotides and phosphate analogues.
| Nucleotide | k1, s−1 | k2, s−1 | A1/(A1+A2) | kav, s−1 |
|---|---|---|---|---|
| GTP, pH 7.5 | 4.5±0.3 | 0.7±0.02 | 0.44±0.04 | 2.3±0.2 |
| GDP•BeF3−, pH 7.5 | 3.2±0.4 | 0.5±0.02 | 0.42±0.11 | 1.8±0.4 |
| GDPNP, pH 7.5 | 3.3±0.6 | 0.6±0.06 | 0.48±0.03 | 1.9±0.4 |
| GTP, pH 6.0 | 6.9±0.5 | 0.5±0.02 | 0.39±0.03 | 3.0±0.3 |
| GDP•BeF3−, pH 6.0 | 1.8±0.4 | 0.3±0.05 | 0.32±0.01 | 0.8±0.2 |
| GDP•AlF4−, pH 6.0 | 0.30±0.01 | 0.019±0.001 | 0.17±0.01 | 0.068±0.003 |
Rates of translocation induced by EF-G in the presence of GTP or GTP analogues were measured in pre-steady-state stopped-flow kinetic experiments. EF-G and ribosome concentrations after mixing were 1 µM and 35 nM, respectively. Experiments were performed in polyamine buffer at pH 7.5 and 6.0, as indicated. k1 and k2 are the rate constants of double-exponential fits of the mRNA translocation data; A1/(A1+A2) is the relative contribution of the faster phase to the total amplitude of fluorescein quenching. About ten time traces were acquired for each experiment. Rate constants averaged from two to five experiments and respective standard deviations are presented in the table. Weighted average values (kav) [18] for mRNA translocation rates were calculated by combining the rate constants derived from the twoexponential fits: kav = (k1A1 + k2A2)/(A1 + A2).
When pretranslocation ribosomes were mixed with buffer in the absence of EF-G and GTP, no fluorescence change was observed indicating that photobleaching of the fluorophore and spontaneous translocation are negligible in the 5 s time scale (Fig. 3). Likewise, no fluorescence change was observed when pretranslocation ribosomes were mixed with EF-G preincubated with GDP demonstrating that, consistent with the results of the puromycin translocation assay (Fig.2) and published reports[17, 18, 35], EF-G•GDP does not induce rapid translocation. Consistent with previous reports [17, 18], EF-G preincubated with GDPNP induced mRNA translocation at a rate that was similar to the rate of translocation measured in the presence of EF-G and GTP (Fig.3, Table 1). Interestingly, translocation in the presence of two other synthetic nonhydrolysable analogues of GTP, β,γ -methyleneguanosine 5’-triphosphate (GDPCP) and guanosine 5’-O-(gamma-thio) triphosphate (GTPγS) was less efficient than translocation in the presence of GDPNP as indicated by the significant decrease in both amplitude and the rate of fluorescence change (Supplementary Fig. 2, Supplementary Table 1). This observation suggests that GDPNP more authentically mimics GTP in the GTP binding pocket of EF-G than GDPCP and GTPγS.
Next, we measured the mRNA translocation rate in the presence of various phosphate analogues and GDP. EF-G preincubated with either GDP and metavanadate or GDP and potassium fluoride (which could form an EF-G•GDP•MgF3− complex with Mg2+ ions in the polyamine buffer used for kinetic measurements) did not induce a measurable mRNA translocation (Fig. 3). By contrast, EF-G preincubated with GDP and beryllium fluoride induced rapid fluorescence change corresponding to mRNA translocation. Remarkably, the measured rate of translocation with EF-G•GDP•BeF3− was similar to the rate of translocation measured in the presence of EF-G and GTP (Table 1). The amplitude (but not the rate) of the fluorescence change was notably smaller in experiments performed with EF-G•GDP•BeF3− than in experiments with EF-G and GTP (Fig. 3). This observation, which is consistent with the results of the puromycin translocation assay (Fig. 2), suggests that EF-G•GDP•BeF3− induces translocation in a majority, but not in all ribosomes.
Measurement of the translocation kinetics in the presence of GDP and aluminum fluoride using a fluorescence quenching assay was complicated by the contribution of light scattering to the measured fluorescent signal. The light scattering likely results from the formation of insoluble aluminum hydroxide to which aluminum fluoride converts to over time in the polyamine buffer (pH 7.5) that was used for the kinetic, puromycin translocation assay and ribosome pelleting experiments. Indeed, a white pellet of aluminum hydroxide could be detected in aluminum fluoride containing samples by centrifugation. Hence, we repeated the kinetic experiments at pH 6.0, at which no detectable precipitation of aluminum hydroxide was observed.
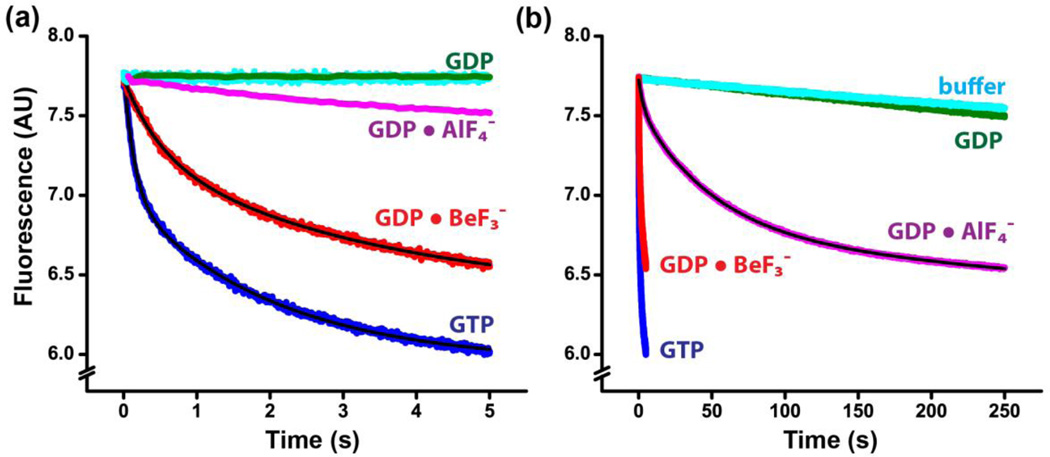
Decreasing the pH from 7.5 to 6.0 did not significantly affect either the rate or amplitude of translocation catalyzed by EF-G and GTP (Fig. 4a and Table 1). The amplitude of the fluorescence change corresponding to the translocation induced by EF-G•GDP•BeF3− did not change upon lowering the pH to 6.0, while the rate of translocation in the presence of EFG• GDP•BeF3− was about three times slower than translocation in the presence of EF-G and GTP (Fig. 4a, Table 1).
Figure 4.
Pre-steady-state kinetics of translocation in the presence of phosphate analogues at pH 6.0. mRNA translocation was induced by mixing pretranslocation ribosomes (35 nM after mixing) EF-G (1 µM after mixing) preincubated with GTP (blue), GDP (dark green), GDP and BeF3− (red) or GDP and AlF4− (magenta). Pretranslocation ribosomes were also mixed with buffer only (cyan) to account for the rates of the photobleaching of fluorescein and spontaneous translocation. Experiments were performed in polyamine buffer at pH 6.0. Double-exponential fits for fluorescence quenching (black lines) are reported in Table 1. Same kinetic traces are shown in the two time scales set to 5 s (panel a) and 250 s (panel b).
At pH 6.0, a slow decrease in fluorescence was observed when pretranslocation ribosomes were mixed with EF-G pre-incubated with GDP•AlF4− (Fig. 4). The amplitude of fluorescence change was about half the amplitude observed in the presence of EF-G and GTP suggesting that EF-G•GDP•AlF4− induced translocation only in a fraction of the ribosomes. In the time scale of slow translocation brought by EF-G•GDP•AlF4− (250 s) photobleaching of fluorescein and/or spontaneous translocation make a measurable contribution to the observed fluorescence change as evident from the mixing of pretranslocation ribosomes with buffer not containing EF-G and GTP (Fig. 4b). The decrease in fluorescence corresponding to photobleaching and/or spontaneous translocation can be fit by a linear function giving the rate constant of ~0.001 s−1. Hence, we fit the decrease in fluorescence brought by EF-G•GDP•AlF4− by the sum of two exponentials and a linear component corresponding to photobleaching and/or spontaneous translocation. The rate of translocation determined in the presence of EF-G•GDP•AlF4− was about 40 times slower than the rate of translocation observed in the presence of GTP (Table 1). Hence, EF-G•GDP•AlF4−, which likely resembles the geometry of the transition state of the GTP hydrolysis reaction[27], is significantly less active in promoting of tRNA/mRNA translocation than EF-G•GTP and EF-G•GDP•BeF3−.
Discussion
GDP·BeF3−and GDP·AlF4− act as analogues of GTP
Our binding and translocation experiments unambiguously show that, consistent with previous reports, EF-G•GDP does not stably bind to the ribosome and induce tRNA/mRNA translocation. By contrast, EF-G•GDP•BeF3− and EF-G•GDP•AlF4− trap EF-G on the ribosome (Fig.1) and promote tRNA/mRNA translocation (Fig. 2–4). Hence, GDP•BeF3− and GDP•AlF4− act as non-hydrolysable analogues of GTP. Importantly, since no translocation was observed in the presence of EF-G•GDP, EF-G with AlF3 (without GDP) or EF-G with BeF2 (without GDP), translocation observed in the presence of EF-G•GDP•BeF3− and EF-G·GDP•AlF4− cannot be due to GTP contamination in any of the reaction components.
Interestingly, phosphate analogues dramatically differ in their ability to support EF-G-induced translocation. GDP•VO43− weakly stabilized EF-G binding but did not support translocation. GDP•AlF4− strongly stabilizes EF-G binding to the ribosome (Fig. 1), however, translocation with EF-G•GDP•AlF4− was almost two orders of magnitude slower than translocation catalyzed by EF-G•GTP. Finally, EF-G•GDP•BeF3− induces translocation at a similar rate (at pH 7.5) as EF-G•GTP. Previous studies of ATPases and GTPases, whose structure were determined in the presence of ADP (or GDP) and phosphate analogues, suggested that GDP•BeF3− bound to the active site on the enzyme resembles the geometry of a prehydrolysis state of the reaction while GDP•AlF4− and GDP•VO43− mimic the transition or posthydrolysis state of the reaction[27–31]. Hence, our data suggest that EF-G is the most potent in promoting tRNA/mRNA translocation when it is bound to GTP in the pre-hydrolysis state of the hydrolysis reaction.
Notably, translocation in the presence of EF-G•GDP•AlF4− and EF-G•GDP•BeF3− occurred only in a fraction of ribosomes in both puromycin and fluorescence quenching translocation assays even with extended observation time (Fig. 3–4). Kinetic and puromycin assay experiments were done at saturating concentrations of EF-G, GDP and phosphate analogues, i.e. a further increase in the concentrations of EF-G, GDP and phosphate analogues did not accelerate the rate of translocation nor its extent in the ribosome population. Translocation in the presence of EFG• GTP occurs in ~95% of ribosomes ruling out a possibility that incomplete translocation observed with EF-G•GDP•AlF4− and EF-G•GDP•BeF3− is due to the heterogeneity in the population of pretranslocation ribosomes. Hence, it appears that a fraction of EF-G•GDP•AlF4− and EF-G•GDP•BeF3− bind to pretranslocation ribosomes in a conformation incapable to induce translocation. However, these inactive EF-G molecules may remain bound to the ribosome and prevent translocation to occur upon binding of EF-G in an active conformation. The idea that EFG can be stably bound to the ribosome in a translocation-incompetent conformation is supported by the observation that GDP•VO43− can trap EF-G on the ribosome in the ribosome pelleting assay (Fig. 1) but is completely incapable of supporting translocation (Fig. 2–3). A possible reason for the conformational heterogeneity in EF-G is that aluminum and beryllium fluoride can form multiple structurally different species that are in equilibrium with each other. For instance, aluminum fluoride can form AlF3, AlF4−, AlF3(OH)−, AlF2(OH)2− and AlF(OH)3− species while beryllium fluoride can interchange between BeF3−, BeF42− and BeF2 species[46, 47]. Further investigation is required to elucidate which of these species of aluminum and beryllium fluoride stabilize the translocation-competent conformation of ribosome-bound EF-G.
Why is fluorescence quenching corresponding to mRNA translocation biphasic?
As it was previously documented, the interpretation of mRNA translocation kinetic experiments that are based on the quenching of a fluorophore (pyrene or fluorescein) is complicated by the biphasic character of the fluorescence change. No commonly accepted explanation of this biphasic behavior has so far emerged. Only the fast phase of mRNA translocation kinetics is often considered as the signal corresponding to mRNA translocation [23, 43, 44, 48], while others fit the data to a single exponential[10, 41, 49, 50]. However, conditions that slow down the rate of translocation, such as the addition of antibiotics, decrease the rates of both phases[18, 23]. In addition, when translocation is inhibited by antibiotics, the contribution of the fast phase to the total amplitude of fluorescence change was reported to decrease while the contribution of the slow phase showed the reciprocal increase [18, 23] thus implicating both the fast and slow phases in the translocation of mRNA. It is noteworthy that the rate of fluorophore photobleaching/spontaneous translocation is ~ 3 orders of magnitude slower than the rate of the slow phase of fluorophore quenching induced by EF-G•GTP (Fig. 3 and 4) thus ruling out photobleaching/spontaneous translocation as possible processes responsible for the appearance of the slow kinetic phase. Nevertheless, one cannot exclude the possibility that the slow phase of fluorophore quenching corresponds to a structural rearrangement of the ribosome that is not concurrent, but instead subsequent to mRNA translocation.
An alternative explanation of biphasic kinetics is the existence of two populations of ribosomes that translocate at different rates[23, 51]. Pretranslocation ribosomes are known to spontaneously fluctuate between the classical, non-rotated and hybrid-state, rotated conformations while EF-G is thought to transiently stabilize the rotated, hybrid state conformation during translocation[18, 52–55]. Hence, it has been hypothesized that EF-G binding to rotated ribosomes results in fast translocation while EF-G binding to non-rotated ribosomes also leads to translocation but at the slower rate[51]. This hypothesis, however, is rendered doubtful by the lack of correlation between variations in the distribution between rotated, hybrid and non-rotated, classical conformations of the ribosome and the amplitudes of the fast and slow phases of the mRNA kinetic assay[18, 45, 54]. Furthermore, previous FRET studies revealed that, unlike the kinetics of mRNA translocation, the kinetics of the reverse intersubunit rotation and swivel-like motion of the head domain of the small ribosomal subunit, which are thought to be coupled to mRNA translocation, are monophasic [18, 48]. These observations further challenge the idea of translocation heterogeneity within the ribosome population that, nevertheless, cannot be ruled out. To analyze the rate of translocation in a potentially heterogeneous ribosome population, we calculated a pseudo-first-order rate constant (kav) as a weighted average of the fast and slow rate phases (Table 1).
Finally, the deviation from single exponential behavior in our kinetic traces may indicate that mRNA translocation process itself is heterogeneous, i.e. translocation occurs through more than one pathway. It is has been shown that mRNA translocation is accompanied by the reverse rotation of the small ribosomal subunit relative to the large subunit into the nonrotated conformation of the ribosome [18] and the back-swiveling motion of the head domain of the small ribosomal subunit into the “non-swiveled” conformation [48]. However, it is not quite clear whether these two rearrangements happen concurrently or sequentially. It is also possible that mRNA translocation may be accompanied by either intersubunit rotation or the 30S head swivel, i.e. through a couple of alternative pathways. The degree of the 30S swivel/back-swivel and intersubunit rotation that accompany translocation may also vary. Pathway heterogeneity, indicated by the appearance of nonexponential kinetic traces, was previously observed for a number of processes ranging from fluorophore quenching [56, 57] to protein folding [58] whose kinetics were described by the stretched exponential function y=y0 +A*exp(−kstretched*t)β where kstretched is the stretched exponential rate constant; β is the stretched exponential parameter (0<β≤1)[56–58]. Our kinetic data are fit equally well by the sum of two exponentials and the stretched exponential function (Fig. 5, Supplementary Table 2) suggesting that translocation pathway heterogeneity may cause a deviation in mRNA translocation kinetics from single exponential behavior.
Figure 5.
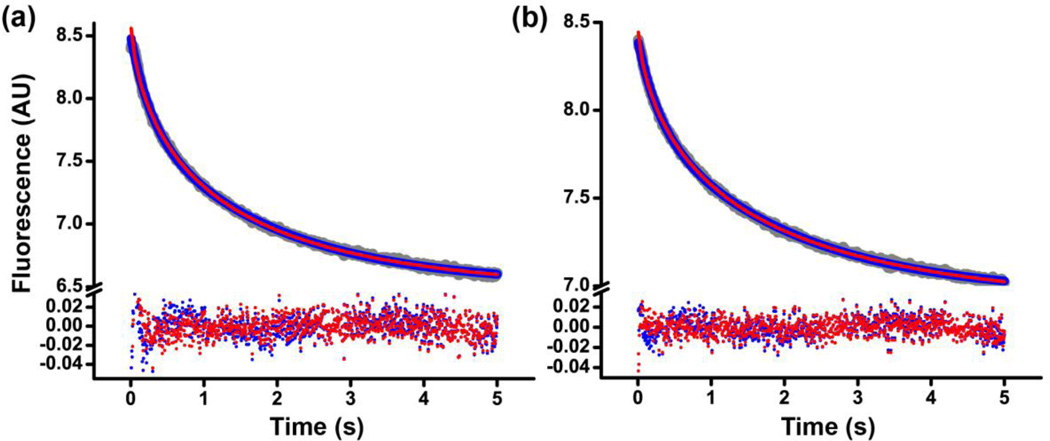
Comparison of double-exponential and stretched exponential fits of kinetics of mRNA translocation induced by EF-G•GTP (a) or EF-G•GDP•BeF3− (b). Fluorescein quenching is shown in grey, double-exponential and stretched exponential fits in blue and red, respectively. The corresponding residuals obtained by subtracting the fitted curves from the raw data are shown at the bottom. The reduced chi-squared and the coefficient of determination R2 for both double-exponential and stretched exponentials fits in (a) were 2*10−4 and 0.999, respectively; in (b) − 1*10−4 and 0.999, respectively. Experiments were performed in polyamine buffer at pH 7.5.
At this point, it seems difficult to unambiguously determine what the reason for the apparent biphasic character of mRNA translocation kinetics is. Nevertheless, whether (i) only the rates of the fast phase are considered or (ii) the weighted average of the fast and slow phases are compared or (iii) the stretch exponential rate constants are examined, the rates of translocation induced by EF-G•GTP and EF-G•GDP•BeF3− appear similar.
The role of GTP hydrolysis in translocation
Macromolecules and macromolecular complexes move in a unidirectional manner by converting the energy of a chemical reaction into mechanical movement. There are two fundamental mechanisms of such conversion. In the first mechanism, called the power stroke, chemical change occurs either concurrently with the movement or precedes it [59]. In the second mechanism, called the Brownian ratchet, the movement occurs spontaneously and precedes the chemical change[59]. In the Brownian ratchet mechanism, the chemical change traps the macromolecule in the post-movement state thus acting like a pawl that rectifies the movement of the wheel of a mechanical ratchet. The model suggesting that GTP hydrolysis precedes translocation and triggers the large movement of domain IV of EF-G, which promotes the translocation of tRNA/mRNA[1, 12, 14, 24], is consistent with the power stroke mechanism.
By contrast, our data suggest that the rate of translocation induced by EF-G bound with the non-hydrolysable analogue of GTP, GDP•BeF3− is similar to the rate of translocation induced by EF-G•GTP and, thus, argue against the direct involvement of GTP hydrolysis in translocation. Our results are also consistent with previously published data showing that replacement of GTP with the synthetic non-hydrolysable analogue of GTP, GDPNP, only moderately slows down translocation[17, 18]. Nevertheless, consistent with previous reports[5, 6, 35, 36], our results suggest that GTP binding is required to induce the translocation-competent conformation of EFG. Therefore, translocation is likely promoted by the energy stored in EF-G•GTP, although GTP hydrolysis itself is not directly coupled to translocation. These mechanistic features are consistent with the Brownian ratchet model, in which by binding to the A site of the small subunit [9, 16], domain IV of EF-G acts as the pawl of the Brownian ratchet mechanism of translocation. The Brownian ratchet and the power stroke mechanisms are idealized models; ribosomal translocation may combine features of both models [60]. Nevertheless, recent optical tweezers measurements of ribosomal translocation against an applied force also suggested that the ribosome likely translocates by the Brownian ratchet mechanism [60].
Taken together, our data provide new insights into the role of GTP hydrolysis in translocation and raise a possibility that phosphate analogues beryllium and aluminum fluorides can be used in structural studies of the catalytic mechanism of GTP hydrolysis by the EF-G-ribosome complex.
Materials and Methods
Materials and sample preparation
All chemicals and reagents were purchased from Sigma with the exception of puromycin (Acros Organics). tRNAMet, and tRNATyr were purchased from Chemblock. The mRNAs were synthesized by IDT. Ribosomes, 6-histidine-tagged EF-G and aminoacylated tRNAs were prepared as previously described[61, 62]. To prepare stock solutions of phosphate analogues, Na3VO4 and NaVO3 were dissolved in water at 40 °C to the final concentration of 5 mM; AlF3 was dissolved in 5 mM K-acetate, pH 5.3 at 50 °C (final concentration 60 mM); BeF2 was diluted in water to the final concentration of 50 mM. GDP was purified using a 6 ml BioRad Qcolumn as previously described[62].
Ribosome pelleting assay
Ribosome pelleting assay was performed as previously described [12, 37] with few modifications. The ribosome-EF-G complexes were assembled in polyamine buffer (30 mM Hepes-KOH, pH 7.5, 6 mM MgCl2, 150 mM NH4Cl, 2 mM spermidine, 0.1 mM spermine, 6 mM β-mercaptoethanol) by incubating 2.0 µM 70S ribosomes with 2.0 µM EF-G at 37°C for 10 minutes; the concentrations of nucleotides and salt analogs were 0.5 mM for GDP, GDPNP, BeF2 and KF, 15 mM for AlF3, and 2 mM for Na3VO4 and NaVO3 as indicated in Fig. 1. Then, 10 µl of each sample were saved as the loading control for the subsequent SDS-PAGE analysis while 10 µl were layered onto a 400 µl sucrose cushion (1.1 M sucrose in polyamine buffer with concentration of MgCl2 increased to 20 mM) and centrifuged at 120000 rpm for 90 minutes, at 4°C using Beckman TLA 120.1 rotor. Pellets were resuspended in SDS-PAGE sample buffer; all samples were run on a 10% Laemmli SDS-PAGE and visualized by coomassie blue staining.
Filter binding and puromycin assay
Pre-translocation ribosomal complexes were assembled in polyamine buffer, pH 7.5 as follows: deacylated elongator tRNAMet was bound to the P site by incubating 600 nM 70S ribosomes with 1.2 µM mRNA (5’ GGC AAG GAG GUA AAA AUG UAC AAA GUA UAA 3’; the Shine-Dalgarno sequence and start codon are underlined) and 1.2 µM tRNAMet at 37 °C for 20 minutes. Radiolabeled tRNA was then bound to the A site by adding N-acetyl-[3H]Tyr-tRNATyr (final concentration 1.1 µM) and incubating at 37 °C for 30 minutes. The translocation reaction was carried out in polyamine buffer by incubating 300 nM pre-translocation ribosomes with EF-G (final concentration 2.0 µM) at 37 °C for 10 minutes; concentrations of nucleotides and salt analogs were 0.5 mM for GDP, GDPNP, GTP, and BeF2; 15 mM for AlF3; and 2 mM for Na3VO4. Subsequently, 5 µl of the translocation reaction mix were added to a nitrocellulose membrane filter, which was then washed three times with 3 ml of ice-cold polyamine buffer containing 20 mM MgCl2. Filters were then dried at 70 °C for 15 minutes, immersed in scintillation fluid, and radioactivity was quantified. The puromycin reactivity of ribosomal complexes was measured as follows: 5 µl of the translocation reaction mix were incubated with puromycin (final concentration 1 mM) in polyamine buffer at 37°C for 10 minutes. The puromycin reaction was stopped by diluting the reaction mix to 80 µl by MgSO4-saturated 0.15 M sodium acetate, pH 5.3, and the N-acetyl-[3H]Tyr-puromycin product was extracted by adding 1 ml ethyl acetate. Radioactivity was measured as 800 µl of the ethyl acetate phase was mixed with scintillation fluid.
Stopped-flow measurements of pre-steady-state translocation kinetics
Kinetics of mRNA translocation were measured as previously described with minor modifications [18, 41, 42]. Pretranslocation complexes were constructed by the incubation of 70S ribosomes (1 µM) with fluorescein-labeled mRNA (5’-GGC AAG GAG GUA AAA AUG UAC AAA-3’- fluorescein, synthesized by IDT, 0.85 µM) and deacylated tRNAMet (2 µM) in polyamine buffer (30 mM HEPES•KOH, pH 7.5, 150 mM NH4Cl, 6 mM MgCl2, 2 mM spermidine, 0.1 mM spermine, or 30 mM MES•KOH, pH 6.0, 150 mM NH4Cl, 6 mM MgCl2, 2 mM spermidine, 0.1 mM spermine) for 15 minutes at 37 °C, followed by an incubation with N-acetyl-Tyr-tRNATyr (1.5 µM) for 30 minutes at 37 °C. Pretranslocation ribosomes were mixed with EF-G and GTP (or GTP analogues) using an Applied Photophysics stopped-flow fluorometer. Final concentrations after mixing were: 35 nM ribosomes, 1 µM EF-G, 0.5 mM GTP, 0.5 mM GDPNP (GDPCP or GTPγS), 0.5 mM GDP, 0.5 mM BeF2, 15 mM AlF3, 0.5 mM KF, 0.5 mM Na3VO4. Fluorescein was excited at 490 nm and fluorescence emission was detected using a 515 nm long-pass filter. All stopped-flow experiments were done at 23°C; monochromator slits were adjusted to 9.3 nm. Translocation of the mRNA resulted in a partial quenching of fluorescein coupled to the 3’ end of the mRNA (8). Time traces were analyzed using Origin. As reported previously (9, 11, 12), the kinetics of mRNA translocation are clearly biphasic and are best fitted to the sum of two exponentials (y=y0+A1*exp(−k1*t)+A2*exp(−k2*t)), corresponding to the apparent rate constants k1 and k2. The rate of translocation was defined as the weighted average rate constant kav (Table 1), calculated as the sum of k1 and k2 normalized to their respective contributions to the total amplitude of fluorescence change [kav = (k1*A1 + k2*A2)/(A1 + A2)] (9). Fluorescence change corresponding to translocation in the presence of EFG, GDP and aluminum fluoride was fit the sum of two exponentials and a linear component (0.001*t) corresponding to photobleaching and/or spontaneous translocation (y=y0+A1*exp(−k1*t)+A2*exp(−k2*t)−0.001*t).
Supplementary Material
RESEARCH HIGLIGHTS.
GDP•BeF3− and GDP•AlF4− trap EF-G bound to the ribosome.
EF-G•GDP•BeF3− and EF-G•GDP•AlF4− promote mRNA translocation.
The rates of mRNA translocation induced by EF-G•GTP and EFG• GDP•BeF3− are similar.
GTP hydrolysis is not directly involved in promoting mRNA/tRNA translocation.
Acknowledgments
This work was supported by grant from the US National Institute of Health no. GM-099719 (to D.N.E.). The authors thank George Makhatadze and Joseph Wedekind for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 2.Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- 3.AEvarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, et al. Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J. 1994;13:3669–3677. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czworkowski J, Wang J, Steitz TA, Moore PB. The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inoue-Yokosawa N, Ishikawa C, Kaziro Y. The role of guanosine triphosphate in translocation reaction catalyzed by elongation factor G. J Biol Chem. 1974;249:4321–4323. [PubMed] [Google Scholar]
- 6.Belitsina NV, Glukhova MA, Spirin AS. Translocation in ribosomes by attachment-detachment of elongation factor G without GTP cleavage: evidence from a column-bound ribosome system. FEBS Lett. 1975;54:35–38. doi: 10.1016/0014-5793(75)81062-3. [DOI] [PubMed] [Google Scholar]
- 7.Parmeggiani A, Sander G. Properties and regulation of the GTPase activities of elongation factors Tu and G, and of initiation factor 2. Mol Cell Biochem. 1981;35:129–158. doi: 10.1007/BF02357085. [DOI] [PubMed] [Google Scholar]
- 8.Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 9.Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 11.Peske F, Matassova NB, Savelsbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell. 2000;6:501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 12.Cunha CE, Belardinelli R, Peske F, Holtkamp W, Wintermeyer W, Rodnina MV. Dual use of GTP hydrolysis by elongation factor G on the ribosome. Translation. 2013;1:e24315-e-11. doi: 10.4161/trla.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holtkamp W, Cunha CE, Peske F, Konevega AL, Wintermeyer W, Rodnina MV. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014;33:1073–1085. doi: 10.1002/embj.201387465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Gagnon MG, Bulkley D, Steitz TA. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell. 2015;160:219–227. doi: 10.1016/j.cell.2014.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martemyanov KA, Gudkov AT. Domain IV of elongation factor G from Thermus thermophilus is strictly required for translocation. FEBS Lett. 1999;452:155–159. doi: 10.1016/s0014-5793(99)00635-3. [DOI] [PubMed] [Google Scholar]
- 16.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 17.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohr D, Wintermeyer W, Rodnina MV. Arginines 29 and 59 of elongation factor G are important for GTP hydrolysis or translocation on the ribosome. EMBO J. 2000;19:3458–3464. doi: 10.1093/emboj/19.13.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salsi E, Farah E, Dann J, Ermolenko DN. Following movement of domain IV of elongation factor G during ribosomal translocation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15060–15065. doi: 10.1073/pnas.1410873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koripella RK, Holm M, Dourado D, Mandava CS, Flores S, Sanyal S. A conserved histidine in switch-II of EF-G moderates release of inorganic phosphate. Sci Rep. 2015;5:12970. doi: 10.1038/srep12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wintermeyer W, Savelsbergh A, Semenkov YP, Katunin VI, Rodnina MV. Mechanism of elongation factor G function in tRNA translocation on the ribosome. Cold Spring Harb Symp Quant Biol. 2001;66:449–458. doi: 10.1101/sqb.2001.66.449. [DOI] [PubMed] [Google Scholar]
- 23.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Petrov A, Tsai A, O'Leary SE, Puglisi JD. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol. 2013;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010;17:1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lassila JK, Zalatan JG, Herschlag D. Biological phosphoryl-transfer reactions: understanding mechanism and catalysis. Annu Rev Biochem. 2011;80:669–702. doi: 10.1146/annurev-biochem-060409-092741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korhonen HJ, Conway LP, Hodgson DR. Phosphate analogues in the dissection of mechanism. Current opinion in chemical biology. 2014;21:63–72. doi: 10.1016/j.cbpa.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Fisher AJ, Smith CA, Thoden JB, Smith R, Sutoh K, Holden HM, et al. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 29.Kagawa R, Montgomery MG, Braig K, Leslie AG, Walker JE. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004;23:2734–2744. doi: 10.1038/sj.emboj.7600293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen B, Doucleff M, Wemmer DE, De Carlo S, Huang HH, Nogales E, et al. ATP ground-and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, sigma54. Structure. 2007;15:429–440. doi: 10.1016/j.str.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavriljuk K, Gazdag EM, Itzen A, Kotting C, Goody RS, Gerwert K. Catalytic mechanism of a mammalian Rab. RabGAP complex in atomic detail. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21348–21353. doi: 10.1073/pnas.1214431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA. 2009;15:772–780. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peske F, Rodnina MV, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Zavialov AV, Hauryliuk VV, Ehrenberg M. Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol Cell. 2005;18:675–686. doi: 10.1016/j.molcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 35.Zavialov AV, Hauryliuk VV, Ehrenberg M. Guanine-nucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs. J Biol. 2005;4:9. doi: 10.1186/jbiol24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. Rna. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koutmou KS, McDonald ME, Brunelle JL, Green R. RF3:GTP promotes rapid dissociation of the class 1 termination factor. RNA. 2014;20:609–620. doi: 10.1261/rna.042523.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 39.Modolell J, Vazquez The inhibition of ribosomal translocation by viomycin. Eur J Biochem. 1977;81:491–497. doi: 10.1111/j.1432-1033.1977.tb11974.x. [DOI] [PubMed] [Google Scholar]
- 40.Holm M, Borg A, Ehrenberg M, Sanyal S. Molecular mechanism of viomycin inhibition of peptide elongation in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:978–983. doi: 10.1073/pnas.1517541113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 42.Svidritskiy E, Ling C, Ermolenko DN, Korostelev AA. Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12283–12288. doi: 10.1073/pnas.1304922110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munro JB, Altman RB, Tung CS, Cate JH, Sanbonmatsu KY, Blanchard SC. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi X, Chiu K, Ghosh S, Joseph S. Bases in 16S rRNA important for subunit association, tRNA binding, and translocation. Biochemistry. 2009;48:6772–6782. doi: 10.1021/bi900472a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antonny B, Chabre M. Characterization of the aluminum and beryllium fluoride species which activate transducin. Analysis of the binding and dissociation kinetics. J Biol Chem. 1992;267:6710–6718. [PubMed] [Google Scholar]
- 47.Baxter NJ, Blackburn GM, Marston JP, Hounslow AM, Cliff MJ, Bermel W, et al. Anionic charge is prioritized over geometry in aluminum and magnesium fluoride transition state analogs of phosphoryl transfer enzymes. J Am Chem Soc. 2008;130:3952–3958. doi: 10.1021/ja078000n. [DOI] [PubMed] [Google Scholar]
- 48.Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–241. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ticu C, Nechifor R, Nguyen B, Desrosiers M, Wilson KS. Conformational changes in switch I of EF-G drive its directional cycling on and off the ribosome. EMBO J. 2009;28:2053–2065. doi: 10.1038/emboj.2009.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie P. An explanation of biphasic characters of mRNA translocation in the ribosome. Biosystems. 2014;118:1–7. doi: 10.1016/j.biosystems.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munro JB, Altman RB, O'Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 56.Wong AL, Harris JM, Marshall DB. Measurements of Energy Dispersion at Liquid Solid Interfaces - Fluorescence Quenching of Pyrene Bound to Fumed Silica. Can J Phys. 1990;68:1027–1034. [Google Scholar]
- 57.Berberan-Santos MN, Bodunov EN, Valeur B. Mathematical functions for the analysis of luminescence decays with underlying distributions 1. Kohlrausch decay function (stretched exponential) Chem Phys. 2005;315:171–182. [Google Scholar]
- 58.Sabelko J, Ervin J, Gruebele M. Observation of strange kinetics in protein folding. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6031–6036. doi: 10.1073/pnas.96.11.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howard J. Protein power strokes. Curr Biol. 2006;16:R517–R519. doi: 10.1016/j.cub.2006.06.045. [DOI] [PubMed] [Google Scholar]
- 60.Liu T, Kaplan A, Alexander L, Yan S, Wen JD, Lancaster L, et al. Direct measurement of the mechanical work during translocation by the ribosome. Elife. 2014;3:e03406. doi: 10.7554/eLife.03406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 62.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of Intersubunit Movement of the Ribosome in Solution Using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.