Abstract
High-quality antibodies (Abs) are critical to neuroscience research, as they remain the primary affinity proteomics reagent used to label and capture endogenously expressed protein targets in the nervous system. As in other fields, neuroscientists are frequently confronted with inaccurate and irreproducible Ab-based results and/or reporting. The UC Davis/NIH NeuroMab Facility was created with the mission of addressing the unmet need for high-quality Abs in neuroscience research by applying a unique approach to generate and validate mouse monoclonal antibodies (mAbs) optimized for use against mammalian brain (i.e., NeuroMabs). Here we describe our methodology of multi-step mAb screening focused on identifying mAbs exhibiting efficacy and specificity in labeling mammalian brain samples. We provide examples from NeuroMab screens, and from the subsequent specialized validation of those selected as NeuroMabs. We highlight the particular challenges and considerations of determining specificity for brain immunolabeling. We also describe why our emphasis on extensive validation of large numbers of candidates by immunoblotting and immunohistochemistry against brain samples is essential for identifying those that exhibit efficacy and specificity in those applications to become NeuroMabs. We describe the special attention given to candidates with less common non-IgG1 IgG subclasses that can facilitate simultaneous multiplex labeling with subclass-specific secondary antibodies. We detail our recent use of recombinant cloning of NeuroMabs as a method to archive all NeuroMabs, to unambiguously define NeuroMabs at the DNA sequence level, and to re-engineer IgG1 NeuroMabs to less common IgG subclasses to facilitate their use in multiplex labeling. Finally, we provide suggestions to facilitate Ab development and use, as to design, execution and interpretation of Ab-based neuroscience experiments. Reproducibility in neuroscience research will improve with enhanced Ab validation, unambiguous identification of Abs used in published experiments, and end user proficiency in Ab-based assays.
Keywords: Brain, immunoblot, immunofluorescence, immunohistochemistry, neuroscience, validation
Introduction
Antibodies (Abs) are valuable and essential reagents for many proteomic level applications that are key to the effective pursuit of molecular and cellular neuroscience research. While Abs are only one class of the diverse binder types used in neuroscience research, they remain the primary tool for the labeling and capture of molecular targets in cells and tissues from the nervous system. They provide neuroscientists with familiar, stable and high-affinity reagents for which there are a wide array of readily available reagents for their subsequent detection and/or capture. Antibodies can be developed and validated for a broad range of labeling and capture applications, and compared to many other classes of binders, can generally be used under a broad range of assay conditions (although, as discussed below, sample preparation and assay conditions can fundamentally impact the nature of Ab-antigen interaction). High-quality and well-characterized polyclonal or monoclonal Abs (pAbs or mAbs, respectively) provide enormous benefits to neuroscience research due to their wide availability, familiarity and ease of use, such that the same Ab can be used globally across many independent laboratories. Antibodies have been crucial to the expansion of knowledge regarding the expression, localization, structure, function, and molecular interactions of a wide variety of proteins expressed in the nervous system, leading to significant advances in the field. However, in spite of the importance of Abs in neuroscience research, and the large number of commercial and public suppliers of Abs, neuroscientists are often faced with challenges and frustrations when applying Abs in their experiments [1–3]. Lack of reliable results using Abs, and their reproducibility, whether due to the poor quality of the Abs themselves, or their application in experiments under conditions for which the Ab was not validated for use, or insufficient reporting of the details of the Abs themselves, has caused considerable frustration among researchers in many fields of research [4]. In the neuroscience arena this has resulted in documentation of numerous incorrect and irreproducible results using Abs, for examples see [5–11]. This is coupled with frustration with the high cost of many commercial Abs (francesscientist.wordpress.com/2010/10/12/scamming-the-antibody/). The estimates of the resulting cost of “bad” Abs to global research budgets in the hundreds of millions of US dollars annually [12].
The UC Davis/NIH NeuroMab Facility was created in 2005, in large part as a response to inquiries from neuroscientists interested in having the same systematic approach to mAb generation and characterization we were using to generate high quality anti-ion channel mAbs in our research laboratory, for examples see [13–16], applied more broadly to targets of general importance to the neuroscience research community. This approach is similar to that used in many other laboratories for stringent in-house validation of Abs, and similar to that described in a prominent publication on Ab quality and validation [17], but with a specific focus on validation in brain samples. NeuroMab aims to provide mAbs (i.e., NeuroMabs), validated for the critical applications of immunoblotting (IB) and immunohistochemistry (IHC) against mammalian brain samples, and to distribute them on a non-profit low cost basis. Over the last ten years, the NeuroMab Facility has established a strong track record towards this mission by both following a proven neuroscience-based approach to screening that is briefly described here, and by its subsequent involvement in controlling quality of the final mAb product distributed to end users. Here we describe the approaches used at the NeuroMab Facility to generate and validate mAbs for neuroscience research applications, provide examples of validation data from such efforts, and use this as a platform to discuss some aspects of the general state of Abs for neuroscience research.
NeuroMab generation, characterization and validation
NeuroMab projects begin with input from the neuroscience community regarding potential targets that are important for neuroscience research and for which no reliable, widely available, and/or renewable binder currently exists. A Scientific Advisory Board and NIH program official prioritize target suggestions based on information regarding the scientific justification and biological importance of the target, and the availability and suitability of existing Abs or other binders. The suggesting lab is also asked to provide a list of available reagents for the project, including plasmids for expression of full-length recombinant target protein in mammalian cells, and samples from knockout (KO) mice if available. Suggesting labs are also asked to provide any other information relevant to the target that may facilitate the screening and validation (e.g., the spatial and temporal expression pattern of the target in brain, any unusual aspects of the target protein as far as instability during sample preparation, behavior on SDS gels, etc.). Through this process, the NeuroMab Facility has taken on projects for a broad array of targets, ranging from novel proteins for which little information and few reagents were available, to well-characterized proteins for which the quality of, or accessibility to, existing Abs was not reliable, or for which there is a dwindling supply of a non-renewable reagent (e.g., a pAb). The array of targets we have pursued include cytoskeletal proteins, enzymes, receptors, ion channels, transcription factors and other proteins of diverse and/or unknown function, as well as specific epigenetic marks on DNA and proteins, specific phosphorylation sites, splice junctions of alternatively spliced proteins, newly generated ends of proteolytic cleavage products, disease-associated mutations, adducts arising from bioterrorism/chemical warfare agents, and many other targets for which no reliable or renewable reagent existed.
Immunogen design and production
A critical step in Ab production is the design and production of an appropriate immunogen. While many sophisticated approaches can be used in immunogen design, guided by intensive efforts in the area of vaccine development, for example see [18], our proximate goal is to generate research reagents for labeling and capturing targets from brain tissue, so our immunogen design is guided by only a few simple considerations. The design of our immunogens is similar to that used by the Human Protein Atlas project except we do not limit the length of our immunogens to 150 amino acids [19]. One key feature is that the immunogen be hydrophilic and, in a best-case scenario, have extensive representation of charged residues to increase the probability that these regions will be surface-accessible in aqueous solutions. The immunogen should also contain extensive regions not present in other proteins, including other highly related members of the same family. It should also provide a reagent that is likely to yield mAbs that recognize the specific target across species (i.e., mAbs that recognize mammalian orthologs, but not paralogs within a given species). For complex polypeptide targets such as ion channel subunits and other integral membrane proteins, we have commonly used fragments of intracellular or extracellular loops, or N- or C-terminal domains. For smaller targets, we have used full-length protein immunogens to successfully generate target-specific NeuroMabs. We prefer that our immunogens are at least 5 kDa in size, and do not have any known requirements for establishing or maintaining proper folding. In cases where the only attractive options for immunogens were a few separate short stretches of less than 50 amino acids each, we generated fusions of these fragments. We have a focus on human targets, as many of our neuroscience end users study human samples and the mechanisms underlying human neurological disease. However, much of this research also involves animal models, especially rodents, such that specific immunoreactivity against both human and rodent orthologs is desirable. Selecting human sequences that share substantial identity with rodent sequences allows us to use rat and mouse brain tissue in screens, which offers advantages as discussed below.
Our most common immunogens are recombinant protein fragments, which typically generate a more robust and varied immune response, and yield a high number of mAbs that work well in different applications. Smaller synthetic peptides are used almost exclusively for highly focused projects such as disease-associated mutations, posttranslational modifications (PTMs), splice junctions, etc. Because of their size and complexity, designing recombinant protein immunogens that produce maximal target specificity and minimal cross-reactivity can be challenging. Expression, solubility, and affinity purification of recombinant mammalian proteins in E coli can be difficult compared with the ease of outsourcing chemical synthesis of a peptide immunogen. However, in our experience, the time, effort and cost put towards generating or obtaining recombinant protein immunogens has been worthwhile. From a retrospective analysis of 414 NeuroMab projects, we find a much higher project success rate for generating at least one validated NeuroMab when using recombinant protein immunogens (211 out of 290 projects or a 73% success rate) than when using synthetic peptide immunogens (47 out of 124 projects or a 38% success rate). One step we have incorporated into our workflow is to validate each affinity-purified immunogen by tandem mass spectrometry (MS) analysis, which has substantially reduced the number of failed projects due to an incorrect immunogen.
Mice are immunized using a short (30 day) but immunogen-intensive immunization protocol [13] that is a modified version of protocols developed to generate a strong IgG response in a much shorter time period than conventional immunization protocols [20; 21]. For each project, two naïve female BALB/c mice (6–8 weeks old) are immunized intraperitoneally using immunogen mixed with Sigma Adjuvant System adjuvant weekly for four weeks. Antiserum from the mice is analyzed for immunoreactivity against the immunogen by enzyme-linked immunosorbent assay (ELISA), by immunocytochemistry (ICC) against the target protein heterologously overexpressed in mammalian COS-1 cells, and by immunoblot against the target protein endogenously expressed in brain tissue. For three days preceding splenocyte isolation and fusion, mice receive intravenous tail vein injections of immunogen in physiological saline. Splenocytes from both mice are then harvested and pooled to maximize the population of mAb specificities, and used in a fusion with Sp2/0-Ag14 (ATCC CRL-1581) B-lymphocyte hybridomas. A small fraction (<1% or 5 × 104 cells) of pooled splenocytes is frozen as archival material and for potential future recombinant Ab generation efforts. The remainder of the splenocytes (typically 4 × 108) is used for hybridoma generation via electrofusion with a Nepa Gene ECFG21 apparatus, which replaced polyethylene glycol (PEG)-based chemical fusion after an in-house comparison revealed higher levels of long term survival of hybridoma colonies (Table 1), as well as increased stability of mAb production. These results are typical of what other investigators have found, reviewed in [22]. Following electrofusion, nascent hybridomas are plated (32 × 96-well plates for a total of 2,944 wells) and incubated for one week, after which the mAb-conditioned media (i.e., tissue culture supernatants or TCS) is harvested for use in primary screens.
Table 1.
Fusion efficiency with electrofusion (EF) and polyethylene glycol (PEG)
| % of wells with hybridomas @ 1 × 107 splenocytes/plate | % of wells with hybridomas @ 1 × 106 splenocytes/plate | % of wells with hybridomas @ 1 × 105 splenocytes/plate | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Project | PEG | EF | Fold Δ | PEG | EF | Fold Δ | PEG | EF | Fold Δ |
| N372A, B | 75% | 100% | >1 | 16% | 94% | 5.9 | 0% | 18% | NA |
| N378 | 54% | 100% | >1 | 15% | 83% | 5.5 | 2% | 40% | 20.0 |
| L93 | 32% | 86% | 2.7 | 2% | 46% | 23.0 | ND | ND | ND |
| N379 | 51% | 66% | 1.3 | 8% | 24% | 3.0 | 1% | 3% | 3.0 |
| N380 | 96% | 100% | >1 | 14% | 100% | 7.1 | 2% | 66% | 33.0 |
| N376A* | 92% | 100% | >1 | 32% | 74% | 2.3 | 2% | 9% | 4.5 |
NA = not applicable; ND = not done
Using larger capacity chamber: 100%, 100% and 50% of wells with hybridomas @ 1 × 107, 1 × 106 and 1 × 105 splenocytes/plate, resulting in >1, >3 and 25.0 fold increases, respectively.
Primary ELISA screens
The primary screen entails parallel ELISAs in two different 96-well plate formats, one employing plates coated with recombinant protein or synthetic peptide immunogen, and the other with plates containing transfected COS-1 cells. Both types of ELISAs are performed using a balanced cocktail of IgG1, IgG2a and IgG2b subclass-specific anti-mouse secondary Abs to avoid the bias inherent in ‘generic’ anti-mouse IgG heavy-and-light-chain (H+L) specific secondary Abs that we identified in a previous study [23]. For ELISAs against immunogens, recombinant protein fragments are expressed in and purified from E. coli bacteria, typically fused to a molecular tag to allow for affinity purification, and in some cases (e.g., glutathione S-transferase (GST) and maltose binding protein (MBP) tags) [24] to enhance expression and solubility. However, if used directly as an immunogen this can result in tag-specific mAbs that can interfere with effective identification of target-specific mAbs. Many of our recent projects have utilized immunogens generated by a modified version [25] of the chitin-intein system [26], which eliminates the tag during the purification process, resulting in the use of predominantly untagged immunogens and a low incidence of tag-specific mAbs. For projects employing GST-tagged proteins as immunogens, we preincubate the TCS from each of the 2,944 wells with soluble GST, which binds to and competes away anti-GST mAbs. This has yielded an effective suppression of the interfering signal from anti-GST mAbs and a subsequent increase in our ability to identify target-specific mAbs. Shorter tags (e.g., 6x His) have proven less problematic. A set of tag-specific mAbs effective for detecting tagged proteins by IB, ICC and IHC, generated as a byproduct of NeuroMab projects against tagged immunogens, have been added to the NeuroMab catalog for those end users in need of such a reagent. For ELISAs against synthetic peptide immunogens, we coat plates with bovine serum albumin (BSA)-conjugated peptides, while keyhole limpet hemocyanin (KLH)-conjugated peptides are used for immunizations. This means that only the peptide itself is in common between immunogen and ELISA target, avoiding selection of KLH-specific mAbs. The chemical properties of BSA generally predominate within the peptide-BSA conjugate, allowing for ease and predictability of use, including reliable binding to microtiter plates that may not be typical of all peptides, given their varying amino acid compositions.
In parallel, ELISA screens against transiently transfected COS-1 cells expressing full-length target protein are performed to identify mAbs that recognize the full-length target protein in a cellular context, in which it may exhibit folding conformations, binding partners, and PTMs not present in the bacterially expressed immunogen. Cells are transiently transfected, plated onto 96-well plates, and cultured for two days. Cells are fixed under standard ICC conditions, using a freshly prepared solution of 4% formaldehyde, with 0.1% Triton X-100 for permeabilization. These conditions are also similar to those used for IHC, thereby enhancing the predictive value of this ELISA screen for that critical downstream application. Another distinction between immunogen- and cell-based ELISAs is their mode of detection. While both use horseradish peroxidase (HRP)-conjugated secondary Abs, the former uses colorimetric detection, and the latter a more sensitive chemiluminescence detection [27]. ELISA screens are done in a semiautomated manner using a customized Agilent BioCel 1800 platform with plate- and liquid-handling skills.
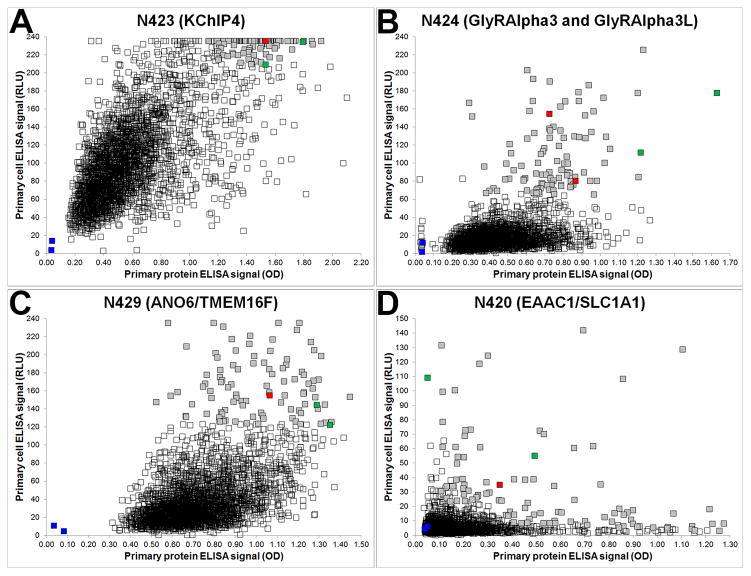
Examples of primary ELISA screen data for four NeuroMab projects are shown in Fig. 1, where the signal from the immunogen protein/peptide ELISA (x axis) is plotted against the signal from the cell-based ELISA (y axis) on a scatter plot. For each of the four projects shown, the squares represent the signal from each well of the 2,944 candidate wells, as well as signals from the positive (green squares) and negative (blue squares) serum and media controls, which are presented as an average of their 32 replicates (one from each fusion plate). The color of the candidate mAb symbols depicts those not selected for further consideration (unfilled squares), and of the 96 that were selected, those that ultimately became a NeuroMab (red squares), and those that did not (grey squares). In some cases, such as the N423, N424 and N429 projects shown in the panels in Fig. 1A–C, it is rather simple to select 96 candidates for further consideration from among those with high signals in both assays (i.e., those trending towards the upper right quadrant). However, for other projects, such as the N420 project shown in Fig. 1D, there is a wide distribution of data points, from which we may also select from wells with “low cell, high protein” signals (upper left quadrant) and from those with “high cell, low protein” signals (lower right quadrant). One important theme that has emerged from these screens, evident from the location of the plotted red squares, is the absolute magnitude of the signal strength on primary ELISAs has not been predictive of which candidates ultimately become NeuroMabs. This reinforces our need to take as many candidates as is manageable, from a cell culture and screening standpoint, forward into the downstream screens for NeuroMab-critical applications. As soon as ELISA results are available, the selected 96 candidate hybridoma cultures are expanded into 24-well culture plates.
Figure 1. Representative ELISA primary screen data.
Scatter plots show the relative distribution of ELISA data from NeuroMab projects N423 (A), N424 (B), N429 (C) and N420 (D), where protein and cell ELISA data are plotted on the x- and y-axis, respectively. Each project began with 2,944 hybridoma-containing wells and each graph shows 2,948 data points (squares) that together comprise positive (green) and negative (blue) control wells (each the average of 32 replicates), unselected wells (unfilled squares) and selected wells (grey squares), with red squares denoting the wells with candidates that ultimately became NeuroMabs.
Secondary screens
Three days after expansion, TCS is collected from each well and tested in parallel secondary screens that include the NeuroMab-critical applications of IB and IHC against mammalian brain samples. However, prior to use in these labor-intensive assays, a confirmation fluorescence-linked ELISA (FLISA) versus the immunogen is performed with subclass-specific secondary Abs conjugated to fluorophores (e.g., Alexa 555-conjugated anti-IgG1, Alexa 488-conjugated anti-IgG2a, and Alexa 647-conjugated anti-IgG2b). This assay serves to confirm that TCS contain sufficient immunoreactivity to justify subsequent testing, and to determine the IgG subclass of the target-positive mAbs in these likely polyclonal TCS samples. Determining IgG subclass early in the screening process can enhance focus on candidates containing the less common IgG2a and IgG2b mAbs, which have increased value for multiplex labeling as most available mAbs are of the IgG1 subclass (e.g., the NeuroMab and Millipore mAb collections in 2012 were 70% IgG1, 20% IgG2a and 10% IgG2b) [23]. Positives from FLISA (typically most if not all of the 96 candidates) are then subjected to three parallel downstream assays. These comprise the two NeuroMab-critical applications of IB versus crude subcellular fractions from adult rat brain, and IHC on sagittal adult rat brain sections, as well as ICC on transfected cells, performed in a critical time window of a few days, due to cell culture and cryopreservation considerations detailed below. For IB analyses of brain samples, the benchmarks relate to the established or predicted molecular characteristics of the target protein (relative electrophoretic mobility and microheterogeneity of the target protein population due to alternative splicing of mRNA, PTMs, etc.) on sodium dodecyl sulfate (SDS) gels.
For IHC analyses on brain sections, the established or predicted anatomical features of endogenous protein expression at the regional, cellular and/or subcellular level need to be established a priori in order to gauge the specificity of immunolabeling for each candidate. Some NeuroMab projects target well-characterized proteins for which some validated immunolabeling data exists, and where the intent is to recapitulate this in a NeuroMab. In other cases the target is a novel protein that has never been characterized, in which case we need to develop diagnostic predictions on molecular characteristics (e.g., from UniProt and other databases) and expression patterns in brain (e.g., from Allen Brain Atlas (ABA) and other databases of mRNA expression from in situ hybridization).
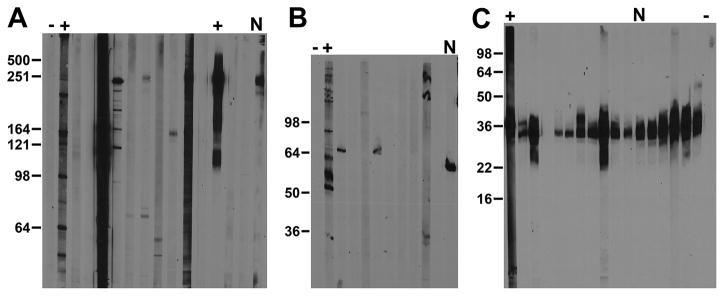
Immunoblot screening is performed on crude subcellular fractions of adult rat brain prepared from homogenates by differential centrifugation, and typically constituting a nuclear/cytoskeletal fraction, or a crude membrane fraction, the so-called P1 and P2 fractions, respectively, of Gray and Whittaker [28]. The choice of brain fraction is determined by the predicted subcellular location of the target protein, or empirically in pilot studies using available Abs or immune serum collected from the NeuroMab project mice. Samples (3 mg total protein) are size-fractionated on large format (14 cm wide × 15 cm long) SDS gels, with the resulting nitrocellulose blots cut into 30 × 0.4 cm wide vertical strips, each containing 100 μg of brain protein. The 30 strips from each gel allow for analysis of TCS from 26 candidates and a total of four positive and negative controls. Fig. 2 shows a subset of candidate TCS from three different NeuroMab projects, together with interspersed positive and negative controls (plus and minus symbols, respectively) for IB analyses of a crude membrane or P2 fraction (Fig. 2A and 2C), used for membrane protein projects (N419: voltage-gated sodium channel principal subunits; N423: an A-type potassium channel auxiliary subunit), and a low-speed pellet or P1 fraction (Fig. 2B) used for nuclear/cytoskeleton projects (N431: a GABA(A) receptor subunit). In these examples, the expected relative electrophoretic mobility of the target proteins (all are somewhat microheterogeneous due to PTMs) are 250 kDa for the N419 candidates (Fig. 2A), 50 kDa for the N431 candidates (Fig. 2B) and 30 kDa for the N423 candidates (Fig. 2C). The immunoblot analyses of N419 and N431 yielded a wide variety of banding patterns, with a minority of candidates selected as being as target-specific, driving the eventual selection of these NeuroMabs (labeled with “N”s above the lanes): N419/40 (Fig. 2A) and N431/64 (Fig. 2B) as NeuroMabs. In contrast, the N423 project yielded numerous candidates with varying IgG concentrations that exhibited different band intensities but also monospecific immunoreactivity to a band with the expected molecular characteristics, one of which, N423/75 (“N”-labeled lane in Fig. 2C) was selected as a NeuroMab based on results of other assays.
Figure 2. Representative immunoblot secondary screen data.
Images show immunoblot data from three different NeuroMab projects. (A) Anti-pan Nav1 channel N419 NeuroMab project. (B) Anti-GABA(A) receptor π subunit N431 NeuroMab project. (C) Anti-KChIP4 N423 NeuroMab project. Numbers to the left of each panel show the mobility of molecular weight standards in kDa. Adult rat brain protein samples analyzed were a P2 crude membrane fraction or RBM (A and C) or a P1 low-speed pellet or RBLSP (B). Expected relative electrophoretic mobilities for target bands were 250 kDa (A), 50 kDa (B), and 30 kDa (C). The subset of strips shown depicts multiple candidates. (A) N419/28 to N419/40. (B) N431/55 to N431/64. (C) N423/28 to N423/37. Each set includes respective positive and negative controls (plus and minus symbols, respectively). The “N” denotes candidates that ultimately became a NeuroMab. (A) N419/40. (B) N431/64. (C) N423/75.
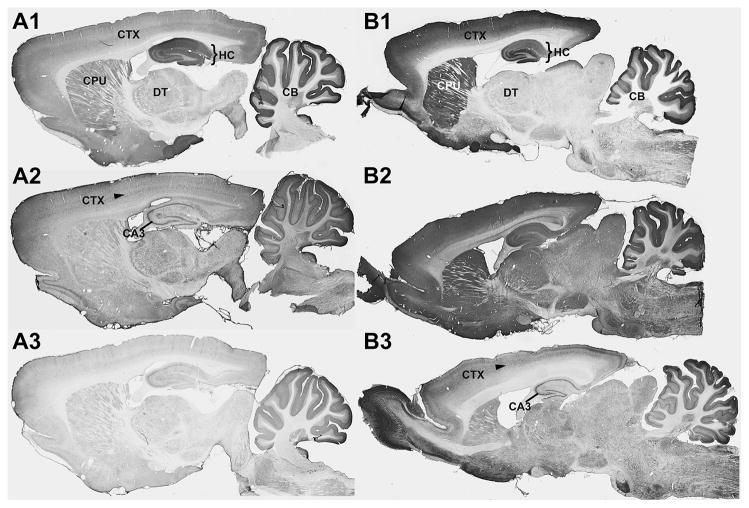
The IHC screens are typically performed on sagittal sections prepared from a formaldehyde-perfused and sucrose-cryoprotected adult female rat brain, which yields sufficient 40 μm thick sections to test all 96 candidates, as well as all appropriate controls. Figure 3 shows examples of immunolabeling from typical NeuroMab IHC screens, in which immunolabeling is detected using a protocol employing avidin-biotin complex amplification, and a nickel-enhanced immunoperoxidase/diaminobenzidine (NiDAB) reaction [29]. Each section is digitally imaged at high resolution using a high-speed automated slide scanner (Aperio ScanScope CS, Leica). Based on available in situ hybridization analyses, and previously published IHC data, immunolabeling for the GluA1/GluR1 glutamate receptor (Fig. 3A) was expected to be high in the hippocampus (bracketed region in Fig. 3) and cerebellum relative to other brain structures (e.g. thalamus), and within these regions the immunolabeling was expected to be localized to fields of neuropil that are rich in neuronal synapses, as opposed to neuronal cell bodies or other non-neuronal cell types. Among the examples shown, immunolabeling in panel A1 fits these criteria, such that N355/1, which ultimately became a NeuroMab, was judged as target-specific.
Figure 3. Representative images from IHC secondary screens.
Photomicrographs show DAB/NAS immunolabeling of sagittal sections of adult rat brain from NeuroMab projects N355 (A1, A2 and A3) and N399 (B1, B2 and B3). There were 96 candidates screened for each project and each column shown here depicts images collected from one candidate judged to meet the criteria set out for specific labeling (N355/1 in A1 and N399/19 in B1) and two other candidates from these projects that failed to meet these criteria (N355/2 in A2, N355/3 in A3, N399/17 in B2 and N399/18 in B3). The bracketed region in A1 and B1 is the hippocampus, and the arrow in A2 and B3 identifies neocortex layer 5. CA3, hippocampal cornu ammonis area 3, CB, cerebellum, CPU, caudoputamen, CTX, neocortex DT, dorsal thalamus, HC, hippocampus.
Immunolabeling for the GABA(A) receptor α2 subunit (Fig. 3B) was expected to be higher in neocortex, caudoputamen and hippocampus relative to other brain regions, and was also expected to give a synaptic neuropil-enriched pattern of labeling. Among these examples, N399/19 (panel B1) was judged target-specific, and ultimately became a NeuroMab. Other panels show candidates with labeling that did not satisfy the criteria for correct positive labeling (Fig. 3A2, A3, B2, B3). Note that panels A2 and B3 show examples of candidate TCS from unrelated projects that exhibit similar off-target labeling across layer 5 of cortex, and in the principal cells of hippocampus, and especially in CA3. Candidates with this pattern can typically be eliminated, as this is a commonly observed pattern of non-specific immunolabeling in adult rat brain [2].
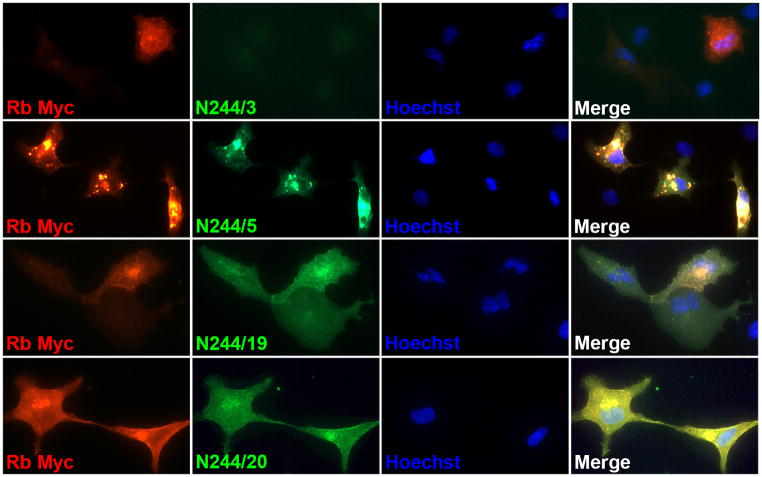
While not a NeuroMab-critical application, each of the 96 candidate TCS is subjected to multiplex immunofluorescence (IF) ICC (IF-ICC) versus heterologous cells expressing full-length target protein. This is done as additional confirmation of target specificity. Transient transfection yields target-positive (transfected) cells in the same field of view as target-negative (untransfected) cells, allowing for a simple, straightforward analysis of concordance (or lack thereof) of candidate TCS when independent tags are utilized in the transfection. Figure 4 shows examples from such a screen, in this case from the N244 anti-SynCAM4 project, in which Myc-tagged SynCAM4 was expressed in COS-1 cells. Concordance of Myc (red) and mAb candidate (green) immunolabeling signals can be seen for N244/5 and N244/20, and to a lesser extent, N244/19, yielding a yellow signal in the merged image, while N244/3 exhibits discord in the signals, in this case due to a negative candidate, although in some cases it can be due to the candidate TCS labeling all cells.
Figure 4. Representative images from IF-ICC secondary screens.
Photomicrographs show IF immunolabeling of COS-1 cells transiently transfected with a Myc-tagged rat SynCAM4 mammalian expression construct using a rabbit anti-Myc pAb (red), candidate mouse mAbs (green) and Hoechst nuclear stain (blue). Each row depicts a set of images collected from a different candidates, one presented as typical of a negative candidate (N244/3), and three represented as positive candidates (N244/5, N244/19 and N244/20), of which N244/5 ultimately became a NeuroMab.
In the 3–4 day period during which these secondary screens are being completed, the hybridoma cells have been further expanded into 6 cm dishes, and are sufficiently dense to cryopreserve as two seed vials in liquid nitrogen. The goal is to have the results of the secondary screens available by this time, so that a small subset (e.g., 5) of the top candidates can be subcloned by limiting dilution. This is critical as these cultures have substantial polyclonality at this stage, and it is advantageous to separate the desired hybridomas from the others prior to subjecting the polyclonal cultures to a freeze/thaw cycle, which empirical evidence suggests imposes a negative selection on the most valuable hybridomas. Harvesting for cryopreservation yields another 12 mL of TCS that can be used for any tertiary screens.
Tertiary screens
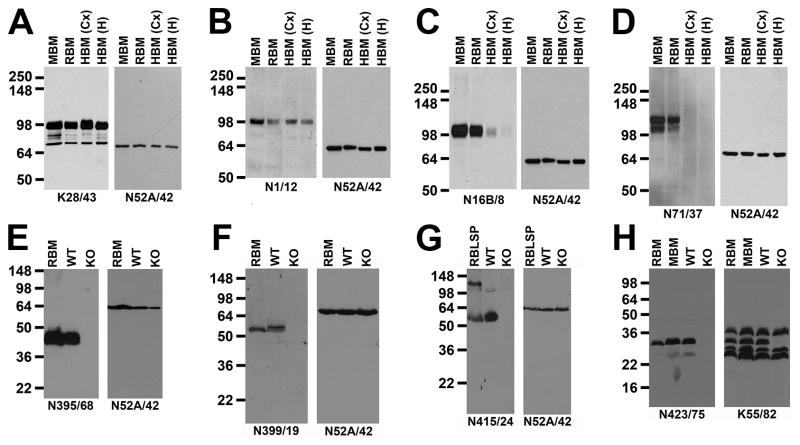
A variety of tertiary screens, the nature of which in part depends on the availability of samples, are performed on selected TCS, and/or on TCS from subcloned candidates, after their confirmation by ELISA. IB-positive mAbs are assayed against blots containing human, mouse, and rat brain (a positive control) samples. Due to the large size of the human brain it is not practical to make whole brain samples, and the region from which the sample is derived needs to be selected based on information regarding the regional pattern of target expression to avoid a false negative signal from bona fide human-positive mAbs. Human samples can have variable postmortem intervals prior to preservation during which the target proteins can be degraded, again resulting in a false positive. However, the translational value of assaying for anti-human brain efficacy is sufficiently high to pursue these assays with these caveats in mind. Fig. 5 shows examples from such IB assays, employing the anti-Mortalin/GRP75 NeuroMab N52A/42 as a loading control, showing two NeuroMabs (anti-PSD-95 NeuroMab K28/43 and anti-KCC2 NeuroMab N1/12) that exhibit comparable immunoreactivity across rodent and human brain samples, and two (anti-Kv3.1b NeuroMab N16B/8 and anti-HCN2 NeuroMab N71/37) that exhibit preferential immunoreactivity against rodent samples. We also perform IB analyses against samples from KO or transgenic mice. Such validation is crucial in order to determine Ab specificity in native tissue such as brain [2]. Examples of such analyses are shown in Fig. 5E–H, again using N52A/42 as a loading control. The same comparison of KO versus WT brain samples can be used to define IHC labeling specificity (Fig. 6C,D), as detailed below. When justified by the project, we perform additional rounds of IF-ICC against transfected cells assaying for cross-reactivity against related members of the target protein family. While we typically aim to generate monospecific mAbs, recombinant immunogens often contain both target-unique regions, and regions conserved across family members, and can yield “pan-specific” mAbs, or mAbs that recognizes all members of a protein family. Such “pan-specific” mAbs can be useful for labeling [15] or capturing [30] all members of a protein family.
Figure 5. Tertiary screening by IB against samples from human brain, and from KO mouse brain.
Panels (A–D) depict results from NeuroMab TCS screening against mouse, rat and human brain P2 crude membrane fraction samples (MBM: adult mouse brain; RBM: adult rat brain; HBM (adult human brain, from Cx: neocortex; Hi: hippocampus). (A) Anti-PSD-95 NeuroMab K28/43. (B) Anti-KCC2 NeuroMab N1/12. (C) Anti-Kv3.1b NeuroMab N16B/8. (D) Anti-HCN2 NeuroMab N71/37. Panels (E–H) depict results from NeuroMab TCS screening against brain samples from WT mice, and from the KO mouse corresponding to the NeuroMab target. Samples in panels (E, F) and (H) are P2 crude membrane fractions from adult rat brain (RBM), or adult mouse brains from WT and KO mice. Panel (G) has P1 low-speed pellet fractions from adult rat brain (RBLSP), or from adult WT and KO mouse brains. (E) Anti-Navβ2 subunit NeuroMab N395/68. (F) Anti-GABA(A) receptor α2 subunit NeuroMab N399/19. (G) Anti-GABA(A) receptor α5 subunit NeuroMab N415/24. (H) Anti-KChIP4 Kv channel subunit NeuroMab N423/75. In all cases duplicate immunoblots were probed the anti-Mortalin/GRP75 NeuroMab N52A/42 as a control to show comparable loading of all samples, with the exception of (H) in which the anti-pan-KChIP NeuroMab K55/82 was used to show the selective elimination of the KChIP4 band within the brain KChIP population in the KChIP4 KO sample. Numbers to the left of each panel show the mobility of molecular weight standards in kDa.
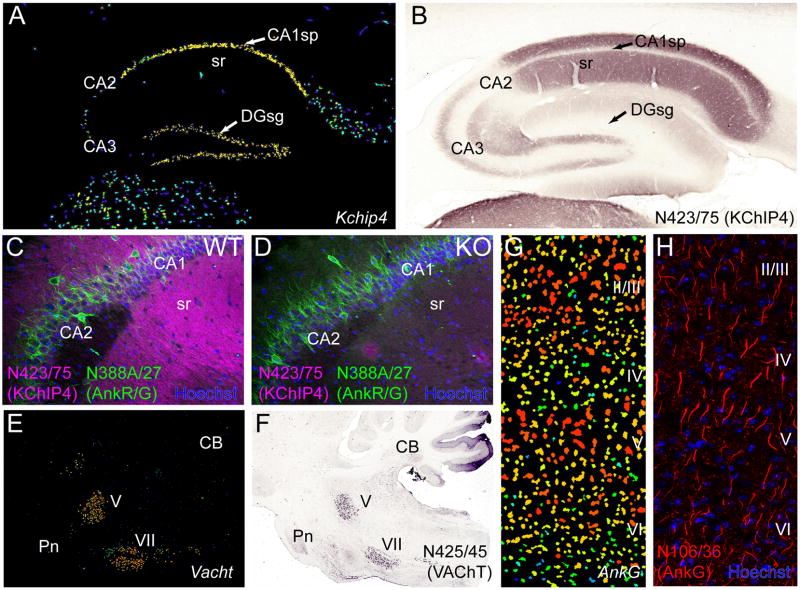
Figure 6. Tertiary screening by IHC.
Panels (A, B) show that discordance between regional localization of transcript and protein in mammalian brain can confound simple Ab evaluation. (A) Pseudocolor image of a sagittal section from mouse brain processed for in situ hybridization to mRNA for Kchip4 obtained from the ABA (image 70546039_100) showing prominent transcript expression in the principal cell layers of the hippocampus, where cell bodies reside. (B) A sagittal section from rat brain immunolabeled for KChIP4 using NeuroMab N423/75 TCS by nickel enhanced diaminobenzidine (NiDAB) histochemistry showing a lack of labeling in the principal cellular layers of the hippocampus, but prominent labeling in the molecular layers, particularly stratum radiatum (sr) which is rich in dendrites. (C, D) Antibody specificity confirmed by KO validation. (C) IF labeling for KChIP4 using NeuroMab N423/75 TCS (magenta) in a sagittal section from a WT mouse brain shows the same prominent immunolabeling pattern in the dendrite rich molecular layers of hippocampus observed in rat (B). (D) Immunolabeling for KChIP4 is absent in the KChIP4 KO brain. In contrast, immunolabeling with the anti-ankyrin-R/G NeuroMab N388A/27 TCS (green) does not differ between WT and KO. Dye staining with Hoechst 33342 (blue) labels cellular nuclei. (E, F) Cellular specificity of transcript expression can guide Ab evaluation. (E) Pseudocolor image of in situ hybridization to VAChT mRNA in a sagittal mouse section from the ABA (image 79762433_86) showing specific transcript expression in the cranial trigeminal motor nucleus (V) and facial nucleus (VII) of the brainstem. (F) Immunolabeling by NiDAB histochemistry using NeuroMab N425/45 TCS in a sagittal section through the brainstem of a rat shows prominent signal for VAChT in the same cell nuclei (V, VII) as mRNA. (G,H) Subcellular target localization can guide NeuroMab evaluation. (G) Pseudocolor image of in situ hybridization of Ankyrin-G mRNA from the ABA (image 68196979_98) showing intense cellular expression throughout all cortical laminae (II–VI). (H) IF labeling for Ankyrin-G (magenta) with NeuroMab N106/36 TCS and staining against cellular nuclei with the dye Hoechst 33342 (blue). In contrast to mRNA, which is localized to cell bodies and resembles the nuclear dye stain, protein expression is restricted to axon initial segments. CA1–3, hippocampal cornu ammonis areas 1–3, CB, cerebellum, DG, hippocampal dentate gyrus, Pn, pons, sg, stratum granulosum (granule cell body layer), sp, stratum pyramidale (pyramidal cell body layer), sr, stratum radiatum (pyramidal cell molecular layer).
Value of a multistep, real time approach to screening
Taking a large set of candidate mAbs through the multistep screening process described above, including the NeuroMab-critical screens of IB and IHC on mammalian brain samples, typically yields a set of mAbs (i.e., NeuroMabs) extensively validated for these specific applications. Our experience suggests that taking such large numbers (96) of ELISA-positive candidates through these screens is important to identifying at least one mAb that is effective and specific for these applications. Conversely, we posit that performing mAb projects in which a more limited number of ELISA-positive candidates (e.g., 5–10) enter the downstream validation pipeline, such as typically offered by commercial custom mAb vendors, is not likely to yield such mAbs. While we spend a great deal of time and effort on screening this large number of candidates for these NeuroMab-critical applications, there are levels of characterization that are routine at other facilities, and that we do not perform. One prominent example is determination of absolute binding affinity against a recombinant protein or synthetic peptide. We have focused instead on identifying mAbs that recognize endogenous target in native tissue, in which mAb binding may be impacted by PTMs, protein-protein interactions, tissue characteristics or other aspects of the biological target protein, and that may alter binding of certain mAbs and not others in a manner unrelated to binding affinities against “naked” recombinant or synthetic preparations.
The candidate mAbs that pass all of the tests mentioned above are added to the NeuroMab catalog for distribution to end users in the research community. However, we retain frozen hybridoma seeds and TCS samples for all ELISA-positive candidates. We select NeuroMabs based on the two NeuroMab-critical applications and we cannot predict how the selected NeuroMabs will perform in other applications or even in the same applications performed under sample preparation and/or binding conditions distinct from those under which they were validated. Should a given NeuroMab not work for an application that is not part of our screening paradigm (e.g., immunoelectron microscopy), it is possible to return to the original candidate sample set, rescreen for these applications, and provide them to end users with those specialized research needs.
By definition Ab binding is dependent on the conditions under which the sample is prepared, and the binding is performed [3; 31–33]. For IB analyses, this includes extraction and denaturation of the sample prior to SDS-PAGE [34], and for ICC and IHC, the nature of fixation (which fixative, concentration, pH, temperature and/or time of fixation). Fundamentally results can be obtained with changes in fixation (e.g., changing from 4% formaldehyde, pH 7.4 fixation to 2% formaldehyde, pH 6.0 fixation) [33]. Making more extensive changes, for example adding glutaraldehyde as is typical for sample preparation for immunoelectron microscopy, presents even additional challenges. In these cases, Ab validation needs to be redone under these specific conditions. There are also numerous protocols for retrieving antigenicity from fixed tissue samples in which fixation has altered access to or the nature of the epitope [35–38], and preparing samples in this manner requires revalidation. Tissues from KO animals are invaluable in these assays [2; 33]. As to the binding reaction itself, the pH, buffer composition, salt concentration, blocking agent, detergents, temperature and time are among factors that can profoundly influence the interaction of Ab and target. Given that the details of sample preparation and binding can profoundly influence efficacy and specificity, it is likely that discrepancies in these procedures, due to a variety of factors including lack of transparency in reporting, and/or inattention to detail by experimentalists, are the basis for at least some of the issues underlying the irreproducibility of Ab-based results [4]. Protocols used for validation at NeuroMab are posted on our website, so that end users can replicate these exact conditions in their own laboratories, or are at least aware when they are using conditions distinct from those used for mAb validation.
Challenges and considerations in determining Ab specificity in brain IHC
The mammalian brain is the most heterogeneous tissue in the body, and presents special challenges for generation and validation of specific Abs for use therein. Target and off-target proteins could potentially be localized in a variety of cell types including those related to vasculature (endothelial cells, smooth muscle), multiple types of glia, or any of the myriad classes of neurons present in specific brain regions, networks and laminae [39–42]. Neuroscientists, therefore, must be exceptionally cognizant of the potential for off-target binding to molecular entities. However, the defined architecture of the brain and its cells at the regional, network, cellular and subcellular level can provide a wealth of information that can be used in Ab validation. Open access databases of mRNA expression patterns in mammalian brain, such as the ABA, can provide valuable information on regional and cellular patterns. Regional differences in protein expression can be extremely useful in determining specificity of Ab labeling, as these can be quite dramatic. In the case of the NeuroMab target RGS14, a priori knowledge that within the brain, RGS14 was primarily expressed in hippocampal region CA2, led to the relatively straightforward identification of the N133/21 NeuroMab, that has subsequently found use in defining CA2 circuitry [43]. In some cases, while gene expression can be extremely informative, one must also take into consideration the subcellular localization of the encoded protein. Having knowledge of brain architecture and the network of interconnectivity between regions and cells within regions is crucial to evaluating the specificity of target binding when proteins are trafficked to distinct compartments which may lie at some distance from the site of the mRNA in the cell body, see discussion in [44]. As one example of a target for which there exists partial overlap of mRNA and protein, the VAChT mRNA is specifically localized to brainstem cranial nerve motor neurons and immunolabeling of VAChT protein with NeuroMab N425/45 shows the same expression pattern (Fig. 6E, F). However, there is also additional dense neuropil labeling throughout the caudoputamen, the target of dopaminergic neurons in the substantia nigra. Striatal labeling of this kind was therefore one of the diagnostic criteria for identifying a successful candidate. In some cases there is a strong discordance between transcript and protein expression for proteins localized to axonal and dendritic neuronal processes, which may lie far from the cell soma where mRNA is typically localized (Fig. 6). The potassium channel interacting protein KChIP4 is a good example of such discordance (Fig. 6A,B). While the mRNA is localized to the principal cell layers of the hippocampus, protein labeling with NeuroMab N423/75 is found overlying the molecular layers that are primarily comprised of the apical dendrites of these cells, with little labeling in the principal cell layers themselves. The scaffolding protein Ankyrin-G provides another example, as its localization to axon initial segments and nodes of Ranvier yields a specific immunolabeling pattern quite distinct from its mRNA that is present in cell somata (Fig. 6G, H). While such considerations can be confounding, they can also be used to advantage by a trained neuroanatomist who is able to correlate the sites of immunolabeling to the in situ hybridization signal in the cell bodies of origin. While the cellular heterogeneity of the brain, and the complexity of the cells therein, present challenges to Ab validation, they also offer distinct opportunities for determining Ab specificity.
Distinct mouse IgG subclasses as a benefit to multiplex labeling
Mice have four subclasses of IgG (in order of typical abundance): IgG1, IgG2a, IgG2b, and IgG3. We recently found that ‘generic’ anti-mouse IgG H+L secondary Abs have a bias towards recognizing IgG1 mAbs that confounds equivalent detection of bound primary mouse Abs of different IgG subclasses [23]. As such, we now perform all early stage assays using balanced cocktails of anti-IgG1, anti-IgG2a and anti-IgG2b subclass-specific anti-mouse secondary Abs to avoid detection bias when the IgG subclasses of candidate mAbs are not yet known; we do not include anti-IgG3 secondary Abs as the characteristics of these mAbs can complicate purification and stability in storage [45]. The availability of mouse IgG mAbs that are by definition a single IgG subclass, along with specific and effective subclass-specific secondary Abs conjugated to different fluorophores, provides to mouse mAbs an important asset not present in rabbit mAbs, that allows for combining multiple Abs in simultaneous multiplex IF labeling [23]. As the vast majority ( 70%) of commercially available mouse mAbs are of the IgG1 subclass, identifying and isolating IgG2a and IgG2b mouse mAbs has become a high priority, as this allows our end users to have additional experimental versatility. From a given project we may select more than one validated NeuroMab specifically to include representatives of different IgG subclasses.
Recombinant NeuroMabs as a route to archiving and engineering
As detailed above, we use natural as opposed to recombinant approaches to generate NeuroMabs. However, we are now generating recombinant NeuroMabs by post facto cloning from hybridomas, using a protocol developed by Wright and colleagues [46; 47]. This archiving of NeuroMabs is independent of cryopreserved hybridomas and is a route to enhancing their unambiguous identification by sequence-based methods such as MS-based sequencing of the native or recombinant mAb preparation [12]. We express and validate each recombinant NeuroMab in COS-1 cells in a side-by-side comparison with the native mAb to ensure that the native characteristics are faithfully represented in the recombinant form. Cloning of NeuroMabs also allows for their engineering into alternate forms that may facilitate their use in specific applications, our initial focus being re-engineering the constant region to alter the IgG subclass, especially for the extremely common IgG1s, to generate collections of otherwise identical mAbs of different IgG subclasses that enhances the flexibility of their use in simultaneous multiplex labeling experiments [23].
NeuroMab and the outlook for the future of Ab quality
In general, neuroscientists have been at the forefront in advocating for enhanced reliability and reproducibility of Ab-based research, promoting both stringent validation of Abs [2; 33; 48], and transparent, unambiguous description of those used in published research [49]. Neuroscience journals with high publication standards such as the Journal of Comparative Neurology (JCN) have implemented strict measures for manuscripts reporting Ab-based research [1; 50]. This is in part due to numerous instances of variable Ab characterization and data quality, some of which was revealed by the availability of KO mice, and in which the reported Ab-based localization was found to be unchanged in the KO samples [1; 50]. These mandatory standards include a requirement for detailed descriptions of Ab validation, regardless of whether the Ab originated from a commercial vendor, came from another academic laboratory, or was generated de novo by the authors for the study. Once an Ab meets these standards, it is added to a database of JCN-Abs (onlinelibrary.wiley.com/journal/10.1002/(ISSN)1096–9861/homepage/jcn_antibody_database.htm).
The neuroscience community has also been instrumental in promoting the use of a standardized reporting system for research reagents, including and especially Abs, through the Neuroscience Information Framework (www.neuinfo.org), which serves as a prominent portal to the Antibody Registry (www.antibodyregistry.org), a database of over 2.4 million Abs, each of which has a unique identifier (e.g., “AB_1234567”), to ensure the unambiguous description of any particular Ab. This unique identifier is then built into the larger Research Resource Identification Initiative (www.scicrunch.org/resources) or RRID (Table 2) [51] such that each Antibody Registry unique identifier becomes a corresponding RRID identifier (e.g., “AB_1234567” becomes “RRID:AB_1234567”). Certain neuroscience journals, including JCN, have embraced this effort, requiring an Antibody Registry/RRID number for all Abs used, while other journals have been making slower progress in this regard [51]. The full implementation of the RRID:AB system should go a long way towards clearly defining which Ab was used for which experiment, as it is fairly common that references to Abs are incomplete, insufficiently clear, or in some cases completely absent, a problem exacerbated by journals with strict space limitations that have decided to dispense with experimental details [52]. As both a service to our end users, and as a reporting tool, the NeuroMab Facility strives to track publications employing the use of NeuroMabs (neuromab.ucdavis.edu/publications.cfm). However, these efforts are often confounded by the lack of unambiguous detail of exactly which NeuroMab was used (e.g., the NeuroMab catalog contains four anti-Ankyrin-G mAbs, which vary in their efficacy in IB versus and IHC, and in IgG subclass). As such, stating that an experiment employed an anti-Ankyrin G mAb from NeuroMab would not allow for reproducibility of the experiment. This is even more common for large Ab distributors, who may distribute several Abs against a single target. We have long encouraged our end users to provide the specific NeuroMab clone number (e.g., N106/36, N106/65, etc.) as an unambiguous transparent identifier in all publications, a practice that should be even clearer now that a unique “RRID:AB_” identifier has been assigned to each NeuroMab (and extended to the level of TCS versus pure IgG preparations). This systematic registry of research resources is a major step towards increasing the reproducibility of experiments involving Abs [53] and is consistent with calls for greater transparency in research [49], and research reproducibility [54], an emerging focus of the research community.
Table 2.
NeuroMabs highlighted in current manuscript and their associated Research Resource Identifiers (RRIDs)
| Target | Accession # | Clone | IgG Subclass | RRID TCS | RRID Pure |
|---|---|---|---|---|---|
| Ankyrin-G (staining) | E9PE32 | N106/36 | IgG2a | RRID:AB_10697718 | RRID:AB_10673030 |
| Ankyrin-R | P16157 | N388A/10 | IgG2b | RRID:AB_2336901 | RRID:AB_2491109 |
| GABA(A)R, Alpha2 | P23576 | N399/19 | IgG1 | RRID:AB_2336904 | NA* |
| GABA(A)R, Alpha5 | P31644 | N415/24 | IgG1 | RRID:AB_2491075 | RRID:AB_2491187 |
| GABA(A)R, Pi | O00591 | N431/64 | IgG2a | RRID:AB_2493092 | RRID:AB_2532051 |
| GluR1 glutamate receptor | P19490 | N355/1 | IgG1 | RRID:AB_2315839 | RRID:AB_2315840 |
| HCN2 | Q9JKA9 | N71/37 | IgG1 | RRID:AB_10672304 | RRID:AB_2279449 |
| KCC2 | NP_599190 | N1/12 | IgG2a | RRID:AB_10697875 | RRID:AB_10672851 |
| KChIP4 K+ channel | Q3YAB7 | N423/75 | IgG2a | RRID:AB_2491080 | RRID:AB_2493100 |
| Mortalin/GRP75 | P38647 | N52A/42 | IgG1 | RRID:AB_10674108 | RRID:AB_2120479 |
| Navbeta2 Na+ channel | Q56A07 | N395/68 | IgG2b | RRID:AB_2315895 | RRID:AB_2315896 |
| Navbeta4 Na+ channel | Q7M730 | N168/6 | IgG1 | RRID:AB_10673578 | RRID:AB_2301402 |
| Pan-KChIP K+ channel | - | K55/82 | IgG2a | RRID:AB_2132595 | RRID:AB_10673406 |
| Pan-MAGUK | - | K28/86 | IgG1 | RRID:AB_10698179 | RRID:AB_10673115 |
| Pan-Nav1 Na+ channel | - | N419/40 | IgG2a | RRID:AB_2491079 | RRID:AB_2491098 |
| RGS14 | O08773 | N133/21 | IgG2a | RRID:AB_10698026 | RRID:AB_2179931 |
| SynCAM4 | Q8R464 | N244/5 | IgG1 | RRID:AB_10673109 | RRID:AB_10676101 |
| VAChT | Q62666 | N425/45 | IgG1 | RRID:AB_2532047 | NA* |
Currently not available
NeuroMab: an emphasis on “back-end” validation for neuroscience research
Any process for developing and screening Abs or other binders can be thought of in terms of “front-end” and “back-end” stages, using terminology associated with the production of semiconductors. The front-end comprises the fairly generic phases of immunogen design, production and purification; immunizations and Ab generation (or in the case of some binders, library generation); and primary screening and Ab selection. The back-end includes screens for efficacy and specificity in “real world” applications, as most end users want to use Abs for more than ELISAs. Almost all front-end tasks in Ab production conclude with a high-throughput binding assay to identify or select a manageable number of candidates, in our case ELISAs. This is similar to what is offered to clients at Ab core facilities and custom Ab producers, and presumably what is standard in commercial Ab generation. It is the back-end processes that can differ dramatically in their extent and nature, and what defines NeuroMab as distinct. Most core facilities and custom Ab producers, and presumably most commercial Ab developers, deem this to be the task of the client or end user, understandable as the diverse array of applications, nuances of the target protein behavior and the myriad sample preparation and binding conditions a client might require are more than what generic Ab production systems can handle. As is clear from the process detailed above, the NeuroMab Facility spends much time and effort on back-end validation, with over two-thirds of our effort and project costs devoted to taking the 96 candidate TCS through the NeuroMab-critical validation steps, which we have tailored to the most standard applications and associated methodologies of our end user community. A substantial portion of the front-end costs would also be eliminated by reducing the number of ELISA positives carried forward through expansion and cryopreservation, as typical for many core facilities and custom suppliers who provide clients with 5–10 ELISA positive candidates for further consideration. However, our experience, and as reinforced by the data shown in Fig. 1, is that the top 5–10 ELISA positives are not necessarily those that ultimately prove to be effective and specific in real world applications against endogenous proteins in tissue samples.
Given our extensive focus on the back-end validation of NeuroMabs for critical applications in brain samples, the NeuroMab model may not be directly relevant to large-scale efforts to address what many end users feel is a rising tide of substandard Ab-based data across broad areas of biomedical research [4]. On the other hand, as a model for an intensive, rigorous and step-wise approach to Ab validation with a focus on serving end users in a particular research community, NeuroMab may be well placed to offer an example that is relevant to both academic and commercial efforts to provide reliable binders for affinity proteomics. We remain convinced that the overall time, effort and cost devoted to extensive back-end validation of large panels of candidate mAbs, performed with the neuroscientist end user in mind, is justified in providing them with a reliable reagent. Moreover, we argue that the costs associated with the back-end
NeuroMab validation steps will be recouped by the savings of avoiding having these assays done for the first time in numerous independently-funded laboratories, or even worse, should this validation not be done, by the cost to the research system at-large of having incorrect results enter the literature.
Suggestions for responsible antibody development and use
Numerous questions have been recently raised regarding the transparency and reproducibility of Ab-based research [4; 12], and to what extent each link in the chain from Ab developers to distributors, end users, peer reviewers, journal editors and funding agencies are responsible for ensuring adequate Ab validation, use and transparent reporting. Each step in this chain has practical considerations that confound what may be viewed as optimal. Even the largest Ab developers do not have the resources to validate every Ab for every imaginable end user application. However, it seems reasonable to validate Abs in a small number of standard applications (e.g., IB, IHC) under a set of standard conditions, whose details are made clear to end users, to determine whether the Ab effectively and specifically recognized the full-length target protein endogenously expressed in native tissues, and not merely show data against purified or overexpressed targets. Validation against samples from KO animals would be an added bonus that would set an Ab above most others generated against the same target (as a case in point, there are more than 400 NeuroMabs in the catalog and more than 130 of them have been KO-validated). Antibody distributors (developers and resellers) should have internal quality control systems in place to assay each new Ab production lot relative to established standards, with inferior lots rejected, and should the problem persist, the Ab pulled from the catalog. We have performed such quality control at NeuroMab for all production lots prior to their release. In the case of Ab end users, while “Caveat emptor” always applies, regardless of the reputation of the developer or distributor. However, it does seem that there should be some level of confidence that, to paraphrase Rimm and colleagues “what is on the label corresponds to what is in the tube” [17], as is the case with most other research reagents, especially those obtained at a premium price. It seems unreasonable to pay a high mark-up for a commercial Ab and then need to validate it in-house, especially since extensive Ab validation, as performed in house at the NeuroMab Facility or in certain end user labs [17], is costly in time and effort. That said, any data generated using the Ab will ultimately reflect on the end user and their research program, such that it is in their best interest to perform some basic level of validation, with appropriate positive and negative primary and secondary Ab controls, prior to embarking on key experiments. However, it remains that the optimal balance between these efforts on the part of developers, distributors and end users is not clear, and it remains the choice of scientists at each step in the chain to strive for accuracy and transparency in their efforts. Peer reviewers and journal editors play a unique role in enforcing high standards, for example by requiring transparency in experimental details, and including the use of unambiguous research Ab identifiers such as RRIDs, as well as minimal standards for reporting the extent and nature of validation for each Ab used, as recently proposed for research involving cell lines [55]. Funding agencies can contribute by supporting and/or maintaining curated lists of validated Abs that generate high-quality and reproducible data, and ensure that all funded investigators, study section members and program officials are aware of these lists. Funding agencies should continue to support efforts in the non-profit sector to provide highly validated and renewable Abs to the research community, if for no other reason than for the arguments that have been presented that this is a cost effective approach to the management of research funding [12].
Conclusions
The generation and validation of high-quality Abs, and the subsequent generation of sound and reproducible data from Ab-based experiments, will continue to play a major role in the future of rigorous and effective proteomics level biomedical research. As such, every link in the Ab chain bears a responsibility for vigilance in experimental design, execution and interpretation, during Ab generation and validation, in evaluating production lots, and when using these reagents in the research laboratory setting, so as not to diminish the potential value that Abs can provide to researchers. An increased focus at each step, and its transparent reporting, will ensure research reproducibility, and avoid many of the problems that have been recently highlighted regarding Ab-based research. The UC Davis/NIH NeuroMab Facility strives to contribute towards this aim, primarily in the neuroscience research arena, but also more broadly, with the goal of serving as a resource, not only for the NeuroMab reagents themselves, but also as a model that may benefit efforts towards generating and validating highly validated Abs and other affinity reagents for biomedical research.
Highlights.
High-quality antibodies are key to proteomic-level neuroscience research.
Irreproducible antibody data cause considerable frustration among neuroscientists.
We make mouse monoclonal antibodies validated via brain-based screens (NeuroMabs).
Determining NeuroMab specificity for staining brain tissue is a unique challenge.
We suggest ideas for responsible and reproducible antibody development and use.
Acknowledgments
This work was funded by NIH research grants U24 NS050606 and R24 NS092991 to J. S. Trimmer. We thank the current and former staff members of the UC Davis/NIH NeuroMab Facility and the Trimmer laboratory for their contributions and dedicated efforts, Ms. Randi Jenkins for valuable institutional support at UC Davis, and Dr. Randall Stewart at the National Institute of Neurological Disorders and Stroke for support and helpful advice.
Abbreviations
- ABA
Allen Brain Atlas
- Abs
antibodies
- BSA
bovine serum albumin
- GST
glutathione S-transferase
- ELISA
enzyme-linked immunosorbent assay
- FLISA
fluorescence-linked immunosorbent assay
- H+L
heavy-and-light-chain
- HRP
horseradish peroxidase
- IB
immunoblotting
- ICC
immunocytochemistry
- IF
immunofluorescence
- IHC
immunohistochemistry
- KLH
keyhole limpet hemocyanin
- KO
knockout
- mAbs
monoclonal antibodies
- MBP
maltose binding protein
- MS
mass spectrometry
- NiDAB
nickel enhanced diaminobenzidine
- pAbs
polyclonal antibodies
- PEG
polyethylene glycol
- PTM
posttranslational modification
- SDS
sodium dodecyl sulfate
- sr
stratum radiatum
- TCS
tissue culture supernatants
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saper CB. An open letter to our readers on the use of antibodies. J Comp Neurol. 2005;493:477–478. doi: 10.1002/cne.20839. [DOI] [PubMed] [Google Scholar]
- 2.Rhodes KJ, Trimmer JS. Antibodies as valuable neuroscience research tools versus reagents of mass distraction. J Neurosci. 2006;26:8017–8020. doi: 10.1523/JNEUROSCI.2728-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saper CB. A guide to the perplexed on the specificity of antibodies. J Histochem Cytochem. 2009;57:1–5. doi: 10.1369/jhc.2008.952770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015;521:274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 5.Moser N, Mechawar N, Jones I, Gochberg-Sarver A, Orr-Urtreger A, et al. Evaluating the suitability of nicotinic acetylcholine receptor antibodies for standard immunodetection procedures. J Neurochem. 2007;102:479–492. doi: 10.1111/j.1471-4159.2007.04498.x. [DOI] [PubMed] [Google Scholar]
- 6.Lu X, Bartfai T. Analyzing the validity of GalR1 and GalR2 antibodies using knockout mice. Naunyn Schmiedebergs Arch Pharmacol. 2009;379:417–420. doi: 10.1007/s00210-009-0394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herkenham M, Rathore P, Brown P, Listwak SJ. Cautionary notes on the use of NF-kappaB p65 and p50 antibodies for CNS studies. J Neuroinflammation. 2011;8:141. doi: 10.1186/1742-2094-8-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayala R, Kett LR, Leach TL, Young AB, Dunah AW, et al. Metabotropic glutamate receptor 1 (mGluR1): antibody specificity and receptor expression in cultured primary neurons. J Neurosci Methods. 2012;204:221–226. doi: 10.1016/j.jneumeth.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 9.Baek JH, Darlington CL, Smith PF, Ashton JC. Antibody testing for brain immunohistochemistry: brain immunolabeling for the cannabinoid CB(2) receptor. J Neurosci Methods. 2013;216:87–95. doi: 10.1016/j.jneumeth.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 10.Jackson TC, Bayir H, Ikonomovic MD, Janesko-Feldman K, Mi Z, et al. Detection of PHLPP1alpha/beta in human and mouse brain by different anti-PHLPP1 antibodies. Sci Rep. 2015;5:9377. doi: 10.1038/srep09377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider EH, Seifert R. The histamine H-receptor and the central and peripheral nervous system: A critical analysis of the literature. Neuropharmacology. 2015 doi: 10.1016/j.neuropharm.2015.05.004. in press. [DOI] [PubMed] [Google Scholar]
- 12.Bradbury A, Pluckthun A. Reproducibility: Standardize antibodies used in research. Nature. 2015;518:27–29. doi: 10.1038/518027a. [DOI] [PubMed] [Google Scholar]
- 13.Bekele-Arcuri Z, Matos MF, Manganas L, Strassle BW, Monaghan MM, et al. Generation and characterization of subtype-specific monoclonal antibodies to K+ channel alpha- and beta-subunit polypeptides. Neuropharmacology. 1996;35:851–865. doi: 10.1016/0028-3908(96)00128-1. [DOI] [PubMed] [Google Scholar]
- 14.Rhodes KJ, Strassle BW, Monaghan MM, Bekele-Arcuri Z, Matos MF, et al. Association and colocalization of the Kvbeta1 and Kvbeta2 beta-subunits with Kv1 alpha-subunits in mammalian brain K+ channel complexes. J Neurosci. 1997;17:8246–8258. doi: 10.1523/JNEUROSCI.17-21-08246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasband MN, Peles E, Trimmer JS, Levinson SR, Lux SE, et al. Dependence of nodal sodium channel clustering on paranodal axoglial contact in the developing CNS. J Neurosci. 1999;19:7516–7528. doi: 10.1523/JNEUROSCI.19-17-07516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhodes KJ, Carroll KI, Sung MA, Doliveira LC, Monaghan MM, et al. KChIPs and Kv4 alpha subunits as integral components of A-type potassium channels in mammalian brain. J Neurosci. 2004;24:7903–7915. doi: 10.1523/JNEUROSCI.0776-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bordeaux J, Welsh A, Agarwal S, Killiam E, Baquero M, et al. Antibody validation. BioTechniques. 2010;48:197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodrigues AF, Soares HR, Guerreiro MR, Alves PM, Coroadinha AS. Viral vaccines and their manufacturing cell substrates: New trends and designs in modern vaccinology. Biotechnol J. 2015;10:1329–1344. doi: 10.1002/biot.201400387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Baumgarten H, Schulze M, Hebell T. Methods of Immunizing Mice. Monoclonal Antibodies. In: Peters JH, Baumgarten H, editors. Monoclonal Antibodies. Springer-Verlag; 1992. pp. 50–57. [Google Scholar]
- 21.Hu JG, Yokoyama T, Kitagawa T. Studies on the optimal immunization schedule of experimental animals. IV. The optimal age and sex of mice, and the influence of booster injections. Chem Pharm Bull (Tokyo) 1990;38:448–451. doi: 10.1248/cpb.38.448. [DOI] [PubMed] [Google Scholar]
- 22.Kanduser M, Usaj M. Cell electrofusion: past and future perspectives for antibody production and cancer cell vaccines. Expert Opin Drug Deliv. 2014;11:1885–1898. doi: 10.1517/17425247.2014.938632. [DOI] [PubMed] [Google Scholar]
- 23.Manning CF, Bundros AM, Trimmer JS. Benefits and pitfalls of secondary antibodies: why choosing the right secondary is of primary importance. PLoS ONE. 2012;7:e38313. doi: 10.1371/journal.pone.0038313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell MR, Engleka MJ, Malik A, Strickler JE. To fuse or not to fuse: what is your purpose? Protein Sci. 2013;22:1466–1477. doi: 10.1002/pro.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wogulis M, Chew ER, Donohoue PD, Wilson DK. Identification of formyl kynurenine formamidase and kynurenine aminotransferase from Saccharomyces cerevisiae using crystallographic, bioinformatic and biochemical evidence. Biochemistry. 2008;47:1608–1621. doi: 10.1021/bi701172v. [DOI] [PubMed] [Google Scholar]
- 26.Chong S, Mersha FB, Comb DG, Scott ME, Landry D, et al. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 27.Lewkowich IP, Campbell JD, HayGlass KT. Comparison of chemiluminescent assays and colorimetric ELISAs for quantification of murine IL-12, human IL-4 and murine IL-4: chemiluminescent substrates provide markedly enhanced sensitivity. J Immunol Methods. 2001;247:111–118. doi: 10.1016/s0022-1759(00)00306-9. [DOI] [PubMed] [Google Scholar]
- 28.Gray EG, Whittaker VP. The isolation of nerve endings from brain: an electron-microscopic study of cell fragments derived by homogenization and centrifugation. J Anat. 1962;96:79–88. [PMC free article] [PubMed] [Google Scholar]
- 29.Rhodes KJ, Keilbaugh SA, Barrezueta NX, Lopez KL, Trimmer JS. Association and colocalization of K+ channel alpha- and beta-subunit polypeptides in rat brain. J Neurosci. 1995;15:5360–5371. doi: 10.1523/JNEUROSCI.15-07-05360.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berendt FJ, Park KS, Trimmer JS. Multisite phosphorylation of voltage-gated sodium channel alpha subunits from rat brain. J Proteome Res. 2010;9:1976–1984. doi: 10.1021/pr901171q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmseth S, Lehre KP, Danbolt NC. Specificity controls for immunocytochemistry. Anat Embryol (Berl) 2006;211:257–266. doi: 10.1007/s00429-005-0077-6. [DOI] [PubMed] [Google Scholar]
- 32.Holmseth S, Zhou Y, Follin-Arbelet VV, Lehre KP, Bergles DE, et al. Specificity controls for immunocytochemistry: the antigen preadsorption test can lead to inaccurate assessment of antibody specificity. J Histochem Cytochem. 2012;60:174–187. doi: 10.1369/0022155411434828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorincz A, Nusser Z. Specificity of immunoreactions: the importance of testing specificity in each method. J Neurosci. 2008;28:9083–9086. doi: 10.1523/JNEUROSCI.2494-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong B, Rhodes KJ, Bekele-Arcuri Z, Trimmer JS. Type I and type II Na(+) channel alpha-subunit polypeptides exhibit distinct spatial and temporal patterning, and association with auxiliary subunits in rat brain. J Comp Neurol. 1999;412:342–352. [PubMed] [Google Scholar]
- 35.Fritschy JM, Weinmann O, Wenzel A, Benke D. Synapse-specific localization of NMDA and GABA(A) receptor subunits revealed by antigen-retrieval immunohistochemistry. J Comp Neurol. 1998;390:194–210. [PubMed] [Google Scholar]
- 36.Jiao Y, Sun Z, Lee T, Fusco FR, Kimble TD, et al. A simple and sensitive antigen retrieval method for free-floating and slide-mounted tissue sections. J Neurosci Methods. 1999;93:149–162. doi: 10.1016/s0165-0270(99)00142-9. [DOI] [PubMed] [Google Scholar]
- 37.Fukaya M, Watanabe M. Improved immunohistochemical detection of postsynaptically located PSD-95/SAP90 protein family by protease section pretreatment: a study in the adult mouse brain. J Comp Neurol. 2000;426:572–586. [PubMed] [Google Scholar]
- 38.Lyck L, Dalmau I, Chemnitz J, Finsen B, Schroder HD. Immunohistochemical markers for quantitative studies of neurons and glia in human neocortex. J Histochem Cytochem. 2008;56:201–221. doi: 10.1369/jhc.7A7187.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schitine C, Nogaroli L, Costa MR, Hedin-Pereira C. Astrocyte heterogeneity in the brain: from development to disease. Front Cell Neurosci. 2015;9:76. doi: 10.3389/fncel.2015.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeFelipe J, Lopez-Cruz PL, Benavides-Piccione R, Bielza C, Larranaga P, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14:202–216. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lodato S, Shetty AS, Arlotta P. Cerebral cortex assembly: generating and reprogramming projection neuron diversity. Trends Neurosci. 2015;38:117–125. doi: 10.1016/j.tins.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menshawi K, Mohr JP, Gutierrez J. A Functional Perspective on the Embryology and Anatomy of the Cerebral Blood Supply. J Stroke. 2015;17:144–158. doi: 10.5853/jos.2015.17.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohara K, Pignatelli M, Rivest AJ, Jung HY, Kitamura T, et al. Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat Neurosci. 2014;17:269–279. doi: 10.1038/nn.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trimmer JS. Subcellular localization of K+ channels in mammalian brain neurons: remarkable precision in the midst of extraordinary complexity. Neuron. 2015;85:238–256. doi: 10.1016/j.neuron.2014.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weir DM. Handbook of Experimental Immunology. 3. Blackwell; 1978. [Google Scholar]
- 46.Crosnier C, Staudt N, Wright GJ. A rapid and scalable method for selecting recombinant mouse monoclonal antibodies. BMC Biol. 2010;8:76. doi: 10.1186/1741-7007-8-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller-Sienerth N, Crosnier C, Wright GJ, Staudt N. Cloning of recombinant monoclonal antibodies from hybridomas in a single mammalian expression plasmid. Methods Mol Biol. 2014;1131:229–240. doi: 10.1007/978-1-62703-992-5_14. [DOI] [PubMed] [Google Scholar]
- 48.Anderson CN, Grant SG. High throughput protein expression screening in the nervous system--needs and limitations. J Physiol. 2006;575:367–372. doi: 10.1113/jphysiol.2006.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landis SC, Amara SG, Asadullah K, Austin CP, Blumenstein R, et al. A call for transparent reporting to optimize the predictive value of preclinical research. Nature. 2012;490:187–191. doi: 10.1038/nature11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saper CB, Sawchenko PE. Magic peptides, magic antibodies: guidelines for appropriate controls for immunohistochemistry. J Comp Neurol. 2003;465:161–163. doi: 10.1002/cne.10858. [DOI] [PubMed] [Google Scholar]
- 51.Bandrowski A, Brush M, Grethe JS, Haendel MA, Kennedy DN, et al. The Resource Identification Initiative: A cultural shift in publishing. F1000Research. 2015;4:134. doi: 10.12688/f1000research.6555.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nature Publising Group. Journals unite for reproducibility. Nature. 2014;515:7. doi: 10.1038/515007a. [DOI] [PubMed] [Google Scholar]
- 53.Vasilevsky NA, Brush MH, Paddock H, Ponting L, Tripathy SJ, et al. On the reproducibility of science: unique identification of research resources in the biomedical literature. Peer J. 2013;1:e148. doi: 10.7717/peerj.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins FS, Tabak LA. Policy: NIH plans to enhance reproducibility. Nature. 2014;505:612–613. doi: 10.1038/505612a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lorsch JR, Collins FS, Lippincott-Schwartz J. Cell Biology. Fixing problems with cell lines. Science. 2014;346:1452–1453. doi: 10.1126/science.1259110. [DOI] [PMC free article] [PubMed] [Google Scholar]