Abstract
Accumulating evidence suggests that epigenetic alterations play an important role in chemically-induced carcinogenesis. Although the epigenome and genome may be equally important in carcinogenicity, the genotoxicity of chemical agents and exposure-related transcriptomic responses have been more thoroughly studied and characterized. To better understand the evidence for epigenetic alterations of human carcinogens, and the potential association with genotoxic endpoints, we conducted a systematic review of published studies of genotoxic carcinogens that reported epigenetic endpoints. Specifically, we searched for publications reporting epigenetic effects for the 28 agents and occupations included in Monograph Volume 100F of the International Agency for the Research on Cancer (IARC) that were classified as “carcinogenic to humans” (Group 1) with strong evidence of genotoxic mechanisms of carcinogenesis. We identified a total of 158 studies that evaluated epigenetic alterations for 12 of these 28 carcinogenic agents and occupations (1,3-butadiene, 4-aminobiphenyl, aflatoxins, benzene, benzidine, benzo[a]pyrene, coke production, formaldehyde, occupational exposure as a painter, sulfur mustard, and vinyl chloride). Aberrant DNA methylation was most commonly studied, followed by altered expression of non-coding RNAs and histone changes (totaling 85, 59 and 25 studies, respectively). For 3 carcinogens (aflatoxins, benzene and benzo[a]pyrene), 10 or more studies reported epigenetic effects. However, epigenetic studies were sparse for the remaining 9 carcinogens; for 4 agents, only 1 or 2 published reports were identified. While further research is needed to better identify carcinogenesis-associated epigenetic perturbations for many potential carcinogens, published reports on specific epigenetic endpoints can be systematically identified and increasingly incorporated in cancer hazard assessments.
Keywords: epigenetics, toxicology, cancer, genotoxicity, hazard assessment
1. Introduction
Epigenetic alterations represent non-genotoxic mechanisms of carcinogenesis that may occur independently or concomitantly with genotoxic aberrations. Further, the epigenomic landscape may directly influence the genotoxic potential of a chemical; for example, several studies have indicated preferential binding of reactive chemicals to regions of DNA that harbor specific histone modification marks and/or DNA methylation patterns [1–6].
There are several major types of epigenetic and epigenomic alterations: DNA methylation, histones/chromatin structure, nucleosome positioning, and expression of non-coding RNAs, all of which can alter gene activity without change to the DNA sequence. A wealth of data demonstrates that changes in these epigenetic marks may occur as a consequence of exposure to environmental chemicals [7, 8], and may play a role in the etiology of various human diseases, including cancer [9]. It has been demonstrated that chemically-induced epigenetic alterations occur early during exposure and may also have significance as biomarkers of carcinogen exposure.
To enable incorporation of epigenetic endpoints in chemical safety assessments, further characterization of the role of epigenetic alterations induced by chemical exposure is necessary [10]. Specifically, additional studies are needed to characterize the relationship between epigenetic alterations and toxicity phenotypes, and the epigenetic-specific dose-response [11]. Several recent publications [9, 12] reviewed the current state of knowledge of epigenetics and cancer, and the application of epigenetic endpoints in cancer hazard assessments, including for chemical carcinogens. Despite the fact that the utilization of epigenetic assays in the evaluation of carcinogens is still in the very early stages, the recent surge in reports of epigenetic marks, and the advances in the technology used to detect them, has yielded better understanding of epigenetics mechanisms of carcinogenesis. Appropriately, “Epigenetic Alterations” were recently listed as one of 10 “key characteristics of human carcinogens” [13]. Beginning with Volume 112 in 2015, the International Agency for the Research on Cancer (IARC) Monographs Programme incorporates a formal search for studies on epigenetic effects in all evaluations. However, it is recognized that most carcinogens were evaluated by IARC before new data on their epigenetic effects became available [9]. Additionally, the US Environmental Protection Agency has held workshops and evaluations regarding environmental chemicals and epigenetics (http://cfpub.epa.gov/ncea/cfm/recordisplay.cfm?deid=308271), but no standard yet exists regarding how regulators are to incorporate these endpoints into chemical assessments.
To enable a comprehensive analysis of the level of evidence that exists on the epigenetic effects of human carcinogens that also act by a genotoxic mechanism, we conducted a systematic literature review. This information will allow for improved understanding of the amount of available data on epigenetic alterations associated with chemical carcinogens.
2. Methodology
We conducted a systematic review of scientific studies indexed in PubMed that investigated epigenetic alterations caused by human carcinogens that have strong evidence of a genotoxic mechanism of carcinogenesis. We used the Health Assessment Workspace Collaborative (HAWC), a publically available online tool (https://hawcproject.org) for curating published literature for the assessment of chemicals or groups of chemicals. The review focused on human carcinogens as classified by the IARC [14]. As of 2015, there are 118 agents classified as “carcinogenic to humans” (Group 1) by IARC (http://monographs.iarc.fr/ENG/Classification/index.php). These include chemical agents and related occupations, personal habits and indoor combustions, radiation, arsenic, metals, fibers, and dusts, biological agents, and pharmaceuticals. For the purpose of this systematic review, we focused on environmental and occupational hazards; specifically, the agents and occupations listed in the IARC monographs volume 100F, one of six monographs comprising volume 100, which was a re-evaluation of data on Group 1 carcinogens conducted by IARC Monographs Working Groups in 2012. Of the 31 chemicals and associated occupations included in the volume 100F, 28 were included in this review because they were (1) classified as carcinogenic to humans, and (2) the IARC Monographs working group concluded that there was strong evidence for genotoxicity as a mechanism of carcinogenesis. We identified published report of epigenetic alterations that occurred as a consequence of exposure for 12 of these 28 chemicals and occupations (Table 1).
Table 1.
Chemicals and associated occupations in IARC Monographs Volume 100F that were included in the systematic literature review.
| Chemical or associated occupation | Original volume No. |
Evidence of genotoxicity: animal* |
Evidence of genotoxicity: human* |
Epigenetics publications |
|---|---|---|---|---|
| 1,3-Butadiene | 97 | yes | yes | yes |
| 2-Naphthylamine | 99 | yes | yes | no |
| 4,4'-Methylenebis(2-chlorobenzenamine) (MOCA) | 99 | yes | yes | yes |
| 4-Aminobiphenyl | 99 | yes | yes | yes |
| Aflatoxins (naturally occurring mixtures) | 82 | yes | yes | yes |
| Benzene | supp. 7 | yes | yes | yes |
| Benzidine | 99 | yes | yes | yes |
| Benzo[a]pyrene | 92 | yes | yes | yes |
| Bis(chloromethyl)ether and chlormethyl methyl ether |
supp. 7 | no | moderate -to- strong |
no |
| Coal gasification | 92 | yes† | no | no |
| Coal-tar pitch | 92 | yes‡ | Moderate | no |
| Coke production, occupational exposures | 92 | yes§ | yes# | yes |
| Dyes metabolized to benzidine | ||||
| Ethylene oxide | 97 | yes | yes | no |
| Formaldehyde | 88 | yes | yes | yes |
| Isopropyl alcohol manufacture by the strong- acid process |
supp. 7 | no | plausible | no |
| Mineral oils, untreated or mildly treated | supp. 7 | no | weak | no |
| Mists from strong inorganic acids | 54 | no | plausible | no |
| Occupational exposure as a painter | 98 | no | yes# | yes |
| Occupational exposure during aluminium production |
92 | no | weak-to- moderate# |
no |
| Occupational exposures during coal-tar distillation |
92 | yes§ | yes | no |
| Occupational exposures during iron and steel founding |
supp. 7 | no | yes# | no |
| Occupational exposures in the rubber manufacturing industry |
supp. 7 | no | yes# | no |
| Ortho-toluidine | 99 | yes | moderate | no |
| Shale oils | supp. 7 | yes | no | no |
| Soot, as found in occupational exposure of chimney-sweeps |
92 | no | moderate # | no |
| Sulfur mustard | supp. 7 | yes | yes | yes |
| Vinyl chloride | 97 | yes | yes | yes |
as summarized in IARC monographs volume 100F
animal mechanistic data based on treatment with coke-oven tar
animal mechanistic data based on treatment with coal-tars or manufactured gas plant residues
animal mechanistic data based on treatment with coal-tars
genotoxicity is attributable to the presence of known genotoxic chemicals in the exposure scenario
Using HAWC, we queried available literature in the PubMed database using search terms for both epigenetic alterations and the chemicals listed in Table 1. The full list of search terms that were used is provided in Supplemental Table 1. The inclusion criteria for the assessment are summarized in Table 2.
Table 2.
Systematic review inclusion criteria.
1. The study evaluated a chemical that:
|
| 2. The study evaluated epigenetic alterations that occurred as an apparent consequence of exposure to the chemical of interest. |
| 3. The publication included original data. |
| 4. The study was published in English. |
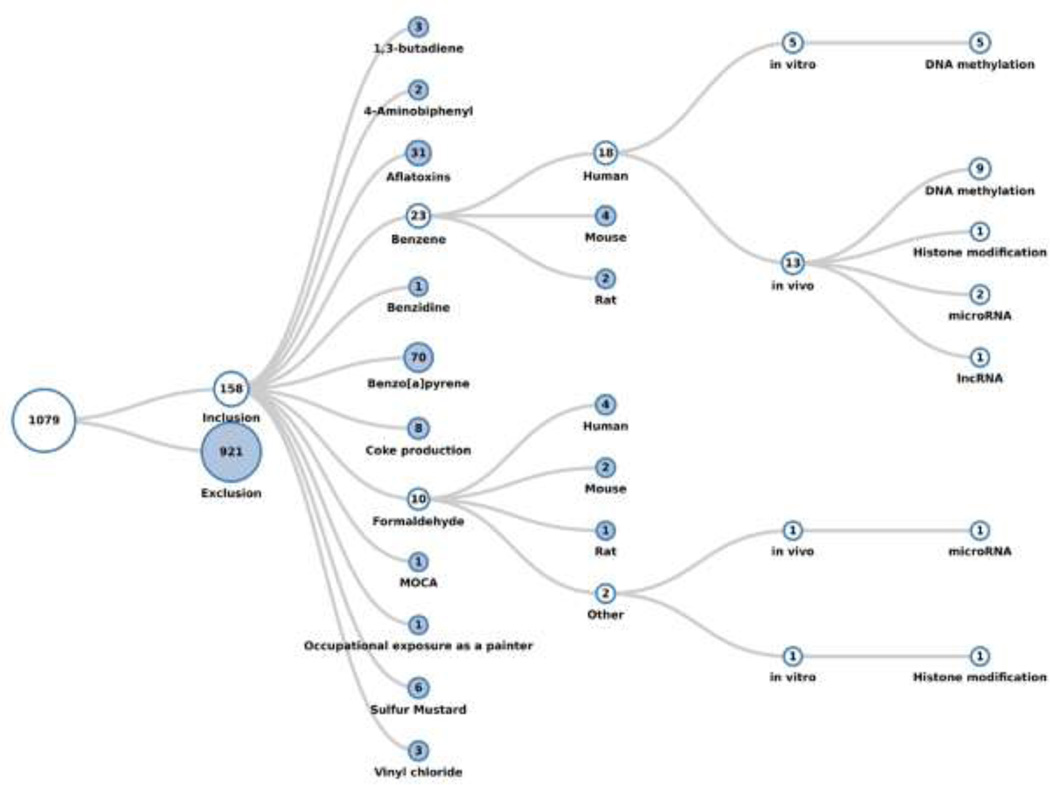
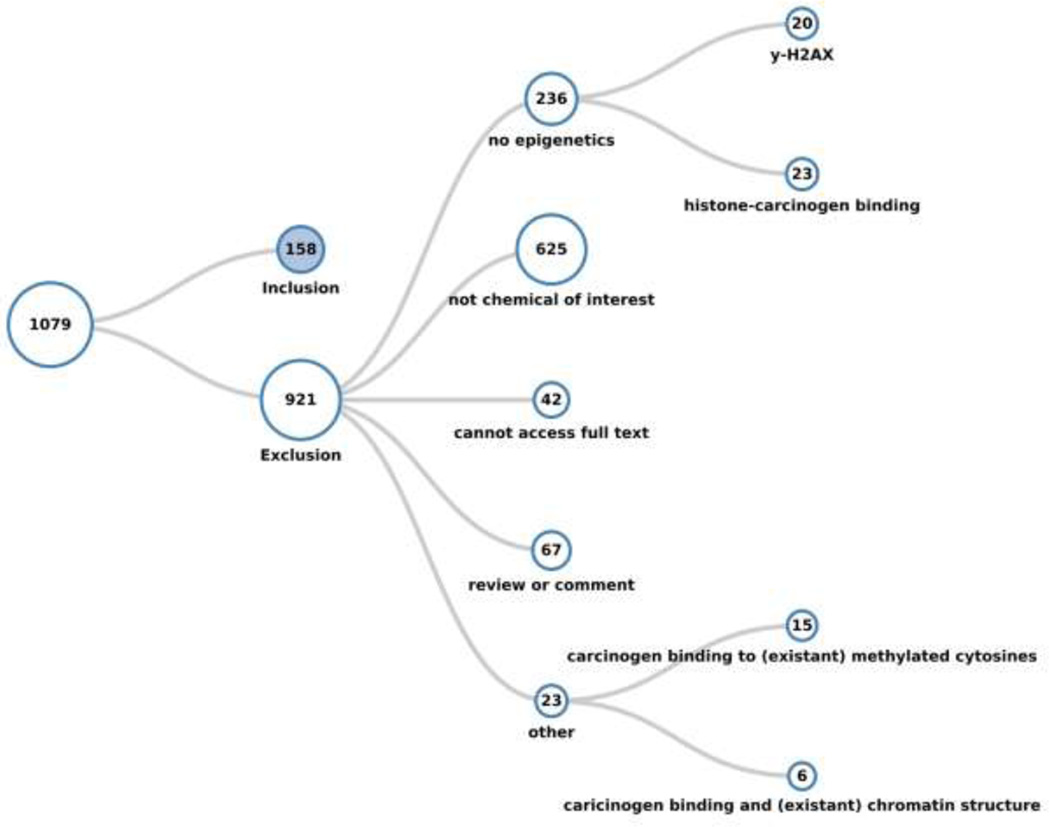
A total of 1,079 references were returned as of the search date 12/06/2015. Of these, 158 met the inclusion criteria and 921 were excluded. Using HAWC, the included studies were classified first by chemical, and then organized into species (human, mouse, rat, or other), type of study (in vitro or in vivo), and finally the category of epigenetic endpoint(s) studied (DNA methylation, histone modification, or non-coding RNA). All exclusions were documented and most (85%) publications were excluded because they did not report epigenetic effects, concern any of the chemicals or occupations of interest, or report primary data, or because were unavailable in full text in English. Studies that reported γ-H2AX, which is highly correlative with double strand breaks and is commonly used as a sensitive marker of such DNA damage [15], were sub-categorized within the studies that did not report epigenetic data, as were studies that reported binding of a carcinogen to histone proteins. Studies tagged within the “other” exclusion category included those studies that described changes in the expression of histone modification genes but not histone modifications themselves, as well as studies that discussed associations between carcinogen exposure and epigenetic features (cytosine methylation or chromatin structure/nucleosome positioning), but did not describe epigenetic alterations that appeared to be caused by the exposure to the agent or occupation of interest. Because this review focused on epigenetic alterations that were attributed to exposure to a carcinogen, these studies were excluded; however, we consider these studies pertinent to the subject at hand and, thus, appropriated tagged them. Visualizations of the literature review that demonstrate the organization of the inclusion and exclusion categories are shown in Figures 1 and 2, respectively. The publicly available systematic literature review with literature tags is freely accessible at the following link: https://hawcproject.org/lit/assessment/185/.
Figure 1.
Literature tree of the 1,079 studies returned by the search after assignment to appropriate categories. Each chemical expands into several sub-categories: first by species, second by type of study (in vitro or in vivo), and finally by the type of epigenetic modification studied. The numbers in each circle indicate how many studies exist within that category. The “branches” for benzene studies in humans, and for formaldehyde studies conducted in a system other than human, mouse, or rat are expanded. There is evident variation in the number of reported studies of epigenetic alterations across the chemicals.
Figure 2.
Literature tree of the studies excluded from the systematic review. The studies that did not meet the inclusion criteria were tagged into several categories as shown that may be of general or future interest, although they were deemed irrelevant to the present review.
3. Categories of epigenetic alterations induced by chemicals and associated occupations included in the systematic review
DNA methylation
DNA methylation, the addition of methyl groups from the universal donor S-adenosyl-L-methionine (SAM) to DNA cytosine residues, is the most extensively studied epigenetic mechanism. Methylation of DNA is a dynamic and well-balanced process of DNA methylation and DNA demethylation reactions. Methylation of DNA is initiated and established by members of the family of de novo DNA methyltransferases DNMT3 (DNMT3A and DNMT3B), and is maintained during DNA replication by the maintenance DNA methyltransferase DNMT1. DNA demethylation is achieved through two different mechanisms: (i) a “passive” replication-dependent mechanism during cell division, and (ii) an “active” replication-independent mechanism. During active DNA demethylation, a family of ten-eleven-translocation (TET) proteins sequentially oxidizes 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxycytosine (5caC), which are later removed and replaced by cytosine via base excision DNA repair mechanisms.
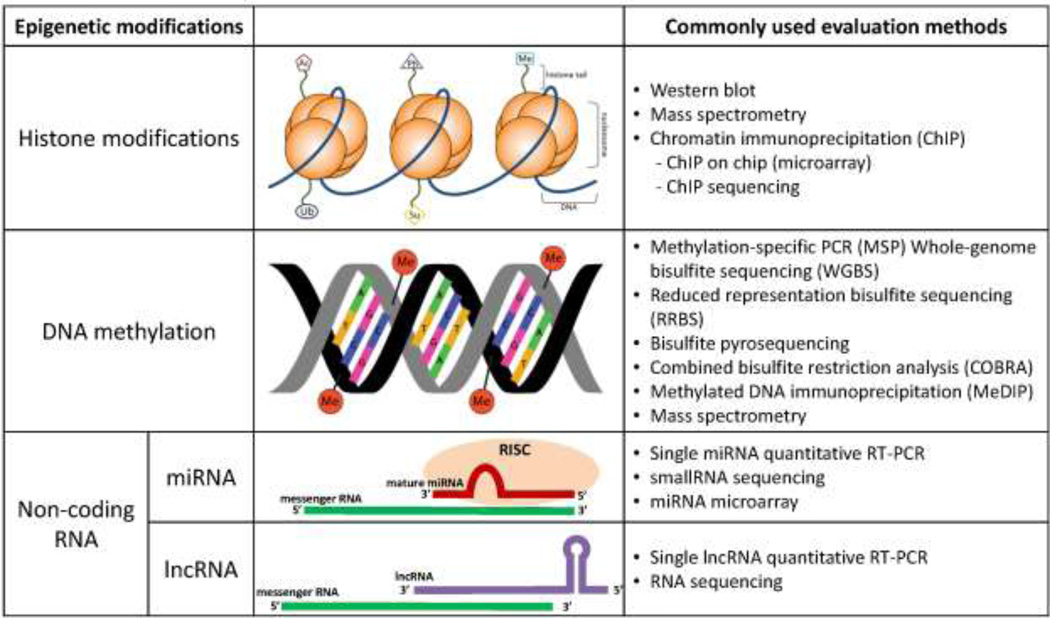
DNA methylation regulates and determines transcription, chromatin structure, chromosome integrity, and genomic imprinting [16]. While the relative effects of each of the above-mentioned nucleotide variants is not clearly understood, it has been shown that 5mC generally has a silencing effect, while 5-hmC, 5-fC, and 5-caC are intermediate variants of 5mC, although there is some evidence that these cytosine variants can interact with binding proteins and may affect transcription [17]. Importantly, aberrant DNA methylation can lead to disruption of any or all of these processes and may contribute to carcinogenesis. Generally, although not exclusively, hypermethylation of CpG island-containing promoter regions of genes (regions rich in CpG dinucleotides) is associated with their respective silencing, whereas promoter gene-specific CpG island hypomethylation is linked to gene activation, and global hypomethylation is associated with genomic instability [18, 19]. Recently, the complexities surrounding the functional importance (or lack thereof) of global or site-specific effects on DNA methylation has been highlighted [20]. Measurement of DNA methylation can be achieved by several methods, including methylation-specific PCR (MSP), combined bisulfite restriction analysis (COBRA) for gene-specific DNA methylation, whole-genome bisulfite treatment with sequencing (WGBS), methylated DNA immunoprecipitation (MeDIP), and mass spectrometry for global levels of DNA methylation [21] (Figure 3).
Figure 3.
Overview of epigenetic alterations. Simplified diagrams provide a generalized view of common epigenetic alterations, and common methods to evaluate such epigenetic marks are listed. Abbreviations: Ac – acetylation; Ph – phosphorylation; Me – methylation; Ub – ubiquitination; Su – sumoylation; PCR – polymerase chain reaction; miRNA - micro RNA; RISC – RNA induced silencing complex; lncRNA - long non-coding RNA.
Histone modifications
Histone modifications occur post-transcriptionally and can affect the accessibility of DNA to transcription factors or DNA damaging agents, thus leading to changes in transcription, as well as influencing DNA damage and repair. There are several types of histone modifications, including methylation, acetylation, phosphorylation, sumoylation, and ubiquitination of specific amino acid residues on the histone tails [22]. Such histone modifications are important regulators of chromatin state, and are highly influential of transcription when they occur at active or poised enhancer and promoter DNA elements, as well as insulators or silencers, either distally or in cis to genes [23, 24]. Histone marks are dynamically altered by “writer and eraser” enzymes (e.g. histone methyltransferases, histone acetyltransferases, histone deacetylases, and histone phosphorylases, among others) and that introduce or remove the histone modifications [25]. Histone marks are then recognized by epigenetic “reader” protein domains (e.g. bromodomains and chromodomains), which is a prerequisite for protein-histone associations that are involved in chromatin remodeling [26]. The histone modifications that have been most commonly reported in chemical exposures and associated deleterious phenotypes are methylation and acetylation of lysine residues, with the mechanistic features of these alterations dependent on the nature of the change (gain or loss) and the site of the histone mark [27]. Generally, acetylation neutralizes the positive charge of lysine residues, which weakens the interaction between the histone and DNA, causing a relaxation of the chromatin, which is generally associated with transcriptional activation. A similar effect is observed with phosphorylation of serine residues, although there are far fewer sites of phosphorylation on histone tails compared to acetylation [27]. Methylation only occurs on lysine or arginine residues, while the most commonly observed addition of methyl groups are on lysine on histone tails H3 and H4. Unlike any other histone modification, methylation has additional complexity in that residues on the histone tails may be mono-, di-, or tri-methylated. Specific methylation marks recruit proteins that are involved in the activation (or inactivation) of chromatin (and, thus, transcription).
While all of these histone modifications occur during normal cellular development and processes, dysregulation of the balance of appropriate histone modifications can lead to disease [27]. Histone modifications are of particular interest in this review because histone dynamics play a role in the toxic potential of the chemicals by influencing both transcriptional activity [28] and DNA repair mechanisms [29, 30]. Histone modifications are commonly measured by antibody-based assays, such as chromatin immunoprecipitation (ChIP) followed by microarray, PCR, or sequencing to identify gene-specific enrichment of specific histone marks, or western blotting and mass spectrometry for global levels of histone modifications (Figure 3).
Non-coding RNAs
It is estimated that over 60% of the genome is transcribed into non-coding RNAs [31], which include any RNA molecule that is not translated into a protein. Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), two types of non-coding RNAs, have various mechanisms of post-transcriptional regulation, including direct binding to RNA, recruitment of chromatin modifying enzymes to target genes, and bringing together proteins to form ribonucleoprotein complexes. A vast majority (48/54, 89%) of the studies of non-coding RNAs included in this review reported alterations in miRNAs, a pattern that is largely representative of the studies of non-coding RNAs in general. MiRNAs are short (19–25 nts) non-coding RNAs that regulate gene expression by binding to the 3’ untranslated region of the gene and either inducing RNA degradation or blocking translation of the gene [32]. The regulatory action of miRNAs has been implicated in many human cancers, and changes in miRNA expression have also been shown to be altered by exposure to environmental chemicals [33]. The most commonly used methods for measurement of non-coding RNAs are quantitative RT-PCR and microarrays for the targeted evaluation of miRNAs or lncRNAs, and sequencing to assess all of the small RNAs in a sample (sequencing also enables the discovery of new microRNAs) (Figure 3).
4. Epigenetic effects associated with carcinogenic chemicals and associated occupations
Each of the 12 human carcinogens that met the inclusion criteria are described below, briefly detailing the common routes of exposure, associated cancers, and previously reported evidence of genotoxicity, followed by a discussion of the epigenetics findings of the studies reviewed. For only 3 of these agents or occupations, 10 or more studies reporting epigenetic endpoints were identified (Table 3).
Table 3.
Number of publications concerning epigenetics for each chemical or related occupation included in the systematic literature review.
| DNA methylation | Histone modification | Non-coding RNA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human (Homo sapiens) |
Mouse (Mus musculus) |
Rat (Rattus norvegicus) |
Other (see footnotes) |
Human (Homo sapiens) |
Mouse (Mus musculus) |
Rat (Rattus norvegicus) |
Other (see footnotes) |
Human (Homo sapiens) |
Mouse (Mus musculus) |
Rat (Rattus norvegicus) |
Other (see footnotes) |
Total | ||
| Benzo[a]pyrene |
in vitro |
11 | 6 | 3*, 1† | 6 | 2 | 1‡ | 16 | 3 | 48 | ||||
|
in vivo |
5 | 4 | 3§, 2#, | 1 | 1 | 1 | 7 | 1 | 25 | |||||
| Aflatoxins (naturally occurring mixtures) |
in vitro |
1 | 1*, 1& | 1& | 7 | 1 | 12 | |||||||
|
in vivo |
10 | 2 | 1 | 1¶ | 1 | 1¶ | 3 | 2 | 21 | |||||
| Benzene |
in vitro |
5 | 1 | 1 | 1 | 8 | ||||||||
|
in vivo |
9 | 1 | 1 | 1 | 1 | 3 | 2 | 18 | ||||||
| Formaldehyde |
in vitro |
1 | 2 | 1© | 1 | 5 | ||||||||
|
in vivo |
2 | 1 | 1$ | 4 | ||||||||||
| Coke production, occupational exposures |
in vitro |
0 | ||||||||||||
|
in vivo |
6 | 2 | 8 | |||||||||||
| 1,3-Butadiene |
in vitro |
0 | ||||||||||||
|
in vivo |
3 | 3 | 6 | |||||||||||
| Sulfur mustard |
in vitro |
1 | 1 | 2 | ||||||||||
|
in vivo |
3 | 1 | 4 | |||||||||||
| Vinyl chloride |
in vitro |
0 | ||||||||||||
|
in vivo |
3 | 3 | ||||||||||||
| 4-Aminobiphenyl |
in vitro |
1 | 1 | 2 | ||||||||||
|
in vivo |
0 | |||||||||||||
| Benzidine |
in vitro |
0 | ||||||||||||
|
in vivo |
1 | 1 | ||||||||||||
| 4,4'-Methylenebis(2- chlorobenzenamine) |
in vitro |
1 | 1 | |||||||||||
|
in vivo |
0 | |||||||||||||
| Occupational exposure as a painter |
in vitro |
0 | ||||||||||||
|
in vivo |
1 | 1 | ||||||||||||
Oligodeoxynucleotides
Hamster (Mesocricetus auratus)
Frog (Xenopus borealis)
Zebrafish (Danio rerio)
Mummichog fish (Fundulus heteroclitus)
Pig (Sus domesticus)
Rainbow trout (Oncorhynchus mykiss)
Synthetic peptide
Cynomolgus macaques (Macaca fascicularis)
4.1 Benzo[a]pyrene
Routes of exposure, associated cancers, and genotoxicity
Benzo[a]pyrene is one of many polycyclic aromatic hydrocarbons (PAHs) that are products of incomplete combustion [34]. BaP is a ubiquitous environmental contaminant, with major sources including tobacco smoke, automobile exhaust, and residential and commercial heating with coal or wood. Occupational exposures occur in coke production, coal gasification and liquefaction, roofing and paving involving coal-tar pitch, aluminum production, chimney sweeping, and working in power plants. BaP can be metabolized to four different diolepoxides, all of which are DNA-reactive. Chromosomal aberrations, DNA damage (by comet assay), sister chromatid exchange, DNA adducts, micronuclei and mutations have all been reported in rodents and/or humans exposed to BaP [35–38]. Most of the mechanistic data for BaP has been conducted in experimental mammals, showing that it is a multi-tissue carcinogen that primarily induces carcinomas of the lung, skin, liver, forestomach and mammary gland. Human cancers associated with BaP exposure include: lung, skin, bladder, and various oral and esophageal carcinomas specifically associated with tobacco smoking [39].
DNA methylation
Studies of DNA methylation and BaP comprise the largest (34 studies) epigenetic category/chemical combination in our review, with the majority of the studies conducted in vitro in both human and rodent cells. BaP has been shown to decrease global DNA methylation levels in a dose-dependent manner in various in vitro systems [40–44]. This may be explained by the inhibition of enzyme-catalyzed transfer of methyl groups from S-adenosyl-L-methionine to cytosines, which is potentially caused by BaP-DNA adducts [44]. In contrast, a lack of change in global DNA methylation has also been reported in a number of in vitro experiments as well [43, 45–48]. However, sequence-specific hypo- and hyper-methylation was observed in p53-positive and p53-negative human breast cancer cell lines, primarily hypomethylation at DNA repetitive elements, in the absence of global DNA methylation changes [47].
Other examples of gene-specific aberrant methylation have been reported: human bronchial epithelial cells that were exposed to BPDE displayed an increase in DNA methyltransferase proteins relative to controls, in tandem with decreased the expression of the CDH13 gene, which is commonly down-regulated in lung cancer, among others [49]. Promoter hypermethylation and reduced expression of the IFNγ gene (another gene commonly down-regulated in various cancers) was observed in Jurkat cells and two human adenocarcinoma cell lines exposed to low, non-cytotoxic doses (0.1 and 1 nM) of BaP, as well as in cord white blood cells of women who were exposed to PAHs during pregnancy [50]. Hypermethylation of CpG islands within the p16INK4α tumor suppressor gene, as well as down-regulation of expression of the gene, was observed in human bronchial epithelial cells exposed to BaP [51]. The same trend was observed in the peripheral blood of PAH-exposed workers, relative to that of non-exposed control subjects, and the degree of methylation was associated with the internal exposure and the level of DNA damage). HeLa cells challenged with BaP displayed DNMT1-mediated promoter hypomethylation, which was associated with activation of the long interspersed nuclear element 1 (LINE-1) repetitive elements [52]. Promoter hypermethylation and reduced expression of DUSP22, a gene that interacts with cancer-associated map kinases, was observed in human Jurkat T lymphocyte cells and normal human prostrate cells exposed to BaP, as well as in blood from both new and experienced firefighters [53]. In the same study, IFNγ methylation was not altered, in contrast to the above-mentioned study [50]. Hypermethylation of the promoter region of p16 was evident in BaP-induced primary immortalized Syrian hamster dermal fibroblasts, accompanied by an overexpression of the gene [54].
In a study of newborns with potential in utero exposure to BaP, among other PAHs, the cord blood samples with detectable BaP-DNA adducts had higher levels of genomic methylation relative to the samples without adducts [55]. This hypermethylation may increase BaP-induced DNA damage, because reactive metabolic intermediates have been shown to preferentially bind to methylated CpG dinucleotides [4], with several studies demonstrating this feature in the p53 tumor suppressor gene [2, 56–58]. Relatedly, the methylation of cytosines that are flanked by BaP-guanine DNA adducts is inhibited [59, 60]
In mice administered BaP, cytosine methylation was reduced in the Igf-II gene in lung tumors [61], and methylation of the Rassf1a gene was observed in 30% of skin tumors [62]. In another study of mice, several cancer-related and aberrantly methylated genes were down-regulated (Wnt4, Fzd3, Mapk3 (Erk1), Mapk11, Foxd3, and Nanog) [63] in the BaP-treated group. Further, the DNA methyltransferase-encoding genes Dnmt3a and Dnmt3b were down-regulated in BaP-treated mice, which may have contributed to the gene-specific aberrant methylation.
Histone modifications
In the same study mentioned above that reported promoter hypomethylation and activation of LINE1 repetitive elements in BaP-exposed HeLa cells, H3K4me3 and H3K9ac, both marks of transcriptional activation, were also increased [52]. Together, these findings are suggestive of a cascade of epigenetic events that lead to activation of the LINE-1 retrotransposons, which may result in retrotrasposition and genome instability.
Exposure of MCF7 breast cancer cells to BaP resulted in a global increase in acetylation of H3K9, and a positive correlation was identified between gene expression and gene-specific H3K9 hyperacetylation [64]. Additionally, genes involved in the organization and remodeling of chromatin were identified among genetic pathways that were responsive to the BaP treatment. H3K4me2 was decreased in the promoter region of the estrogen receptor α gene (ER) in both a human breast cancer cell line exposed to BaP, as well as in liver tissue from mice exposed to BaP [65]. This histone modification, which is likely mediated by depletion of the orphan nuclear receptor NR2E3, causes down-regulation of ER, which was associated with increased BaP-induced oxidative injury.
An increase in acetylation of H3K9 and H3K14 and trimethylation of H3K4, all marks of transcriptional activation, was observed in the promoter region of Cyp1a1, an aryl hydrocarbon hydroxylase that is highly involved in drug and xenobiotic metabolism, along with up-regulation of Cyp1a1 mRNA in BaP-exposed mouse hepatoma Hepa-1 cells [66, 67].
In a study of neonatal rats administered BaP, the extent of acetylation of H3K14 and mRNA expression of StAR were both decreased, in correlation with a decrease in sperm count and serum testosterone levels, all changes that persisted into adulthood [68].
It has also been shown that BPDE-damaged DNA has more stable nucleosomes, which may interfere with nucleotide excision repair and lead to an increase in mutation rate [69].
Non-coding RNA
miR-29a was identified to have tumor-suppressor activity in human cells exposed to BPDE by targeting Cdc7 kinase and sensitizing cells to BPDE, thus presumably diminishing the accumulation of cells with DNA damage [70]. miR-29b-3p was also identified among BaP-mediated alterations and subsequent miRNA-mRNA interactions in mouse primary hepatocytes (miR-29b-3p and Col4a2, miR-24-3p and Flna), which were also found to be involved in cell cycle arrest and the impairment of repair mechanisms of DNA damage [71]. miR-181a-1-3p, was overexpressed in HEPG cells treated with BaP, and one of its targets, MGMT, a gene that encodes an enzyme involved in DNA damage repair, was decreased at the mRNA level [72]. In a study of human liver HepRG cells, treatment with BaP caused over-expression of miR-410 [73]. miR-892a was down-regulated in human breast adenocarcinoma MCF7 cells. miR-892a targets CYP1A1, a gene that is induced by BaP, and repression of CYP1A1 by miR-892a mediated the loss of cell viability caused by BaP exposure [74]. In a study using the human multiple myeloma cell line MM1.s, BaP exposure resulted in the up-regulation of 27 miRNAs, 7 of which (miR-25, miR-15a, miR-16, miR-92, miR-125b, miR-141, and miR-200a) have been reported to repress the p53 tumor suppressor gene [75]. miR-34c, which is also associated with p53 expression by a positive feedback loop, was increased in a correlated manner with phosphorylated p53 in human bronchial epithelial cells treated with BaP, and the up-regulation of miR-34c prevented BaP-induced malignant transformation [76]. miR-622 and miR-506 displayed tumor-suppressor properties in anti-benzo[a]pyrene-trans-7,8-diol-9,10-epoxide transformed human bronchial epithelial cells by suppressing K-ras (protein) and N-ras (both protein and mRNA) expression [77, 78]. In contrast, miRs-106a, -638, -494 and -22 were all increased and were all identified as having oncogenic properties in another experiment using anti-benzo[a]pyrene-trans-7,8-diol-9,10-epoxide transformed human bronchial epithelial cells. Both miR-22 and miR-494 target the tumor suppressor gene PTEN, and the increase in the abundance of these two miRNAs was accompanied by a decrease in PTEN protein level (with no effect on PTEN mRNA) in the transformed cells [79, 80]. miR-106a inhibited the suppression of cell proliferation and cell cycle arrest and apoptosis, and promoted tumor growth in transfected nude mice [81]. miR-638 was increased both in human bronchial epithelial cells and in peripheral lymphocytes collected from 86 workers who were exposed to PAHs, and the overexpression of this miRNA aggravated BaPinduced cell DNA damage, likely due to suppression of BRCA1 [81].
Silencing of lncRNA-DQ786227 expression in BaP-treated human bronchial BEAS-2B cells inhibited cell proliferation and colony formation, and increased apoptosis. These findings were corroborated by the dramatic promotion of the ability of BEAS-2B-T cells to form colonies in vitro, and of tumor development in nude mice induced by expression of lncRNA-DQ786227 [82]. Similarly, silencing of the lncRNAs AF118081 and LOC728228 inhibited cell growth and tumor invasion in BaP-treated human 16HBE cells, and downregulation of AF118081 clearly suppressed tumor growth in nude mice [83, 84].
4.2 Aflatoxins (naturally occurring mixtures)
Routes of exposure, associated cancers, and genotoxicity
Aflatoxins are naturally occurring potent hepatocarcinogens produced by Aspergillus flavus and Aspergillus parasiticus, with aflatoxin B1 (AFB1) considered to be the most toxic type. Human exposure to AFB1 primarily occurs by consumption of contaminated food sources, most commonly stored grains, but occupational exposure also occurs during processing and handling of contaminated grains (inhalation and dermal). The carcinogenicity is attributed to the metabolic activation of AFB1 to a genotoxic epoxide, with a high prevalence of point mutations in the p53 gene [85, 86]. AFB1 exposure causes sister chromatid exchange, micronuclei, chromosomal alterations, and DNA and protein adducts [87].
DNA methylation
Many of the studies of AFB1 investigated gene-specific DNA methylation changes. In two studies, Zhang et al. demonstrated inactivation of the tumor-suppressor RASSF1, MGMT, and p16 genes by promoter hypermethylation in the promoter region of tumor DNA in human hepatocellular carcinoma (HCC) patients who were exposed to AFB1 [88, 89]. Further, the methylation status of the promoter regions of all three of these genes was significantly positively associated with the level of AFB1-DNA adducts in the tumor tissues, and methylation of MGMT was associated with mutations in the tumor suppressor TP53 gene. Feng, et al also showed that hypermethylation of the RASSF1 gene was associated with AFB1-DNA adducts in human HCC tumor tissue [90]. A significant association was observed between promoter hypermethylation of the glutathione S-transferase pi (GSTP1) gene and the level of AFB1-DNA adducts in human HCC tumor tissue, and a marginally significant association was found for adjacent non-tumor tissue [91]. The level of GSTP1 mRNA was inversely associated with promoter hypermethylation in a majority of the tumor samples, and a loss of this detoxifying enzyme that is involved in xenobiotic metabolism may be related to the associated DNA damage also observed in the tumors in this study. In addition to the site-specific gene methylation, hypomethylation of repetitive DNA elements, a characteristic indicative of genomic instability, has also been reported as a result of AFB1 exposure in both HCC and cancer-free patients with confirmed AFB1 exposure [92, 93].
AFB1 exposure in pregnant women was found to be associated with aberrant DNA methylation in blood collected from their infants at 2-8 years of age [94]. AFB1-associated differential methylation was observed in growth factor genes, including FGF12 and IGF1, and immune-related genes, including CCL28, TLR2 and TGFB1I, exemplifying pathologically important epigenetic alterations induced by exposure to a genotoxic chemical at a critical developmental stage.
In mice, at least partial methylation of CpG sites was observed in 43 of 49 (88%) of lung tumors analyzed for p19Arf promoter hypermethylation, and methylation of transcription factor binding sites or consensus sequences was confirmed in 21 tumors [95]. There was a general increase in DNA methylation levels in oocytes collected from high dose mycotoxin-fed mice, as well as in a study of porcine oocytes exposed in vitro to AFB1, [96, 97], which may be causative of decreased developmental competence of oocytes in mice that ingest AFB1. A study of rat AFB1-induced liver tumors demonstrated that the gamma-glutamyl transpeptidase (GGT) gene was hypermethylated in hepatic tumors, but the correlation between GGT activity and methylation was not clear, and the regulatory mechanism of methylation of GGT differs from that of fetal liver development [98].
In vitro studies demonstrated a role of cytosine methylation on mutation spectrum, with increased methylation of CpG sites associated with increased mutation frequency, particularly for TP53 and in a codon-specific manner [2, 99].
Histone modifications
The levels of H3K9me3 and H4K20me3 (marks of transcriptional activation) were increased in oocytes from mycotoxin-fed mice, while H3K27me3 and H4K20me2 (marks of transcriptional repression and activation, respectively) were decreased. These alterations were observed along with increased global DNA hypermethylation, and may play a role in decreased developmental competence of oocytes in mice that ingest AFB1, although the mechanisms are not clear [96]. Similarly, in a study of porcine oocytes exposed in vitro to AFB1, the levels H3K27me3 (transcription repressive mark) and H3K4me2 (transcription activator) decreased, whereas the level of the transcription repressor mark H3K9me3 increased [97].
Non-coding RNA
In a comprehensive study of the miRNome in mouse primary hepatocytes exposed to a panel of both genotoxic and non-genotoxic chemicals, miRNA-mRNA interactions were identified for AFB1 (miR-301b-3p and Papss2), which were also found to be involved in cell cycle arrest and the impairment of repair mechanisms of DNA damage [71]. Liu et al [100] observed dysregulation of several miRNAs in the liver of rats that were exposed to AFB1 for four weeks, with upregulated miR-34a-5p facilitating p53-mediated DNA damage repair. The level of miR34a-5p was increased in the circulating blood of the rats, preceding any significant increase in alanine transaminase activity; thus, miR-34a-5p may represent a sensitive biomarker of AFB1-induced DNA damage in the liver. Another study of rats with AFB1-induced liver cancer also reported up-regulation of miR-34a, as well a loss of members of the miR-17-92 family, of which members play a tumor-suppressor role, in tumors [101].
Up-regulation of miR-429, which inhibits apoptosis and induces progression of tumor cell growth, was observed in human liver tumors in HCC patients that were confirmed to be exposed to AFB1, and was significantly correlated with high levels of AFB1-DNA adducts [102]. The same research group found that miR-24, which has been reported to be an “oncomir” [103], was upregulated in liver tumors from patients who resided in regions with high AFB1 exposure [104]. Further, in a large case-control hospital study in China that investigated polymorphisms in pre-miRNAs as potential risk and prognostic biomarkers of AFB1-related HCC, rs28599926 in miR-1268a was identified as one such candidate [105].
The majority of studies that reported alterations to miRNA expression caused by aflatoxins utilized in vivo data. One study, however, investigated the changes in miRNA expression in human liver HepRG cells, and observed a dose- and time-dependent down-regulation of miR-122 [73]. This AFB1-induced loss of miR-122 was attributed to inhibition of the HNF4A/miR-122 regulatory pathway.
Additionally, the H19 gene, which encodes a lncRNA, was up-regulated in human HepG2 cells treated with AFB1 [106]. This overexpression promoted cell cycle progression in an E2F1-dependent manner.
4.3 Benzene
Routes of exposure, associated cancers, and genotoxicity
Benzene is a solvent that has historically been used in printing inks, gasoline, and chemical and drug production. Currently, the main use of benzene is in the manufacture of organic chemicals, and it is an intermediate in the production of several products that are used in drugs, insecticides, plastics, and dyes. Exposure to benzene is typically dermal or by inhalation in occupational settings, but it is present in the atmosphere, particularly in proximity to gas stations and in areas of high vehicular traffic. Benzene is leukemogenic, with excess cases of various types of leukemia (primarily acute myelogenous leukemia) reported in workers exposed to benzene [107, 108]. Limited studies have also associated benzene exposure with increased risk of lung and kidney cancer. The carcinogenicity of benzene is contingent on metabolic activation, with benzoquinones in the bone marrow implicated in the ultimate toxicity. Benzene leads to genotoxic effects at the hematopoietic stem cell level; specifically, DNA double strand breaks and chromosomal aberrations that are known to be causative of hematopoietic cancers occur in benzene-exposed human patients [109–111].
DNA Methylation
Benzene induced global DNA hypomethylationin human lymphoblastoid TK6 cells at concentrations of 1, 10, and 100 µM [112]. However, no significant global DNA methylation changes were observed in a study using normal hepatic L02 cells or human myeloid HL-60 cells that were incubated with benzene for 48 hours and which displayed changes in gene expression levels [113, 114], although the exposure concentrations tested were similar or higher than those used in the Tabish et al. [112] study.
While the in vitro studies of benzene-induced changes in DNA methylation are conflicting, global and repetitive element DNA hypomethylation has been reported in humans exposed to low levels of benzene (as confirmed by personal air samplers) [115, 116]. Further, hypermethylation of the p15 promoter, which likely contributes to deregulation of cell proliferation and is associated with acute myelogenous leukemia, was observed in benzene-exposed individuals. Gene-specific DNA methylation has also been reported in individuals exposed to benzene. Three hypermethylated genes with concurrent mRNA down-regulation (PRKG1, PARD3, and EPHA8) and two hypomethylated genes with increased mRNA level (STAT3, IFNGR1) were identified in benzene poisoning patients [117]. Subsequent pathway analysis identified STAT3 as a central player in several enriched carcinogenesis-relevant genesets and pathways, including acute myeloid leukemia and the JAK-STAT cascade. Promoter DNA hypermethylation of the tumor suppressor genes p15 and p16 was observed in benzene-exposed workers, along with a decrease in the mRNA level [118]. A study of pregnant mice revealed that benzene exposure induced global hypomethylation, but that p15 promoter methylation was unchanged in both fetal livers and maternal bone marrow cells [119], indicating that this epigenetic response to benzene exposure may be species-specific. In a study using rat bone marrow cells [120], genes that control apoptosis (the primary mechanism of cytotoxicity induced by benzene) were investigated. Addition of a DNA methyltransferase inhibitor to the benzene-exposed cells increased the mRNA levels of Bax and Cas3 (apoptosis inhibitors), and decreased the level of cell death in benzene-exposed rat bone marrow cells. This indicates that benzene-induced cytotoxicity is modulated by epigenetic regulation of apoptosis-inhibiting genes. A decrease in the expression of Pten, a tumor suppressor gene, and a significant increase of Pten methylation level was observed in rats exposed to benzene and in F32 human lymphoblast cells incubated with benzene [121]. Both the decrease in mRNA and the increase in promoter methylation were observed in a dose-dependent manner. Expression of the repair gene PARP-1 was decreased in tandem with promoter hypermethylation in human lymphoblasoid F32 cells treated with 10mM benzene [122].
Histone modifications
Reduced histone H4 and H3 acetylation and H3K4 methylation, and increased H3K9 methylation, were observed in the promoter region of topoisomerase IIα (Topo IIα) in patients with benzene exposure [123], accompanied by decreased Topo IIα activity, expression, and mRNA level. These findings demonstrate the involvement of histone modifications in the decrease of Topo IIα, a mechanism that is implicated in benzene-induced hematotoxicity. In the same study mentioned above (section B.3.a) that exposed rat bone marrow cells to benzene [120], the inhibition of histone deacetylation and apoptosis was also investigated. Inhibition of histone deacetylation increased the mRNA level of Bcl-2, an apoptosis inhibitor, in benzene-exposed rat BMCs, indicating that histone modification is also a mechanism of benzene-induced cytotoxicity. In contrast, no changes in the acetylation of histones H3, H4, and H3K56, nor methylation of histones H3K9 and H3K27 were observed in a study of pregnant mice dosed with 200 mg/kg benzene on gestational days 8, 10, 12, and 14 relative to control mice, in either maternal bone marrow cells or fetal livers [119].
Non-coding RNA
A total of 6 miRNAs were up-regulated (miR-34a, miR-205, miR-10b, let-7d, miR-185 and miR-423-2-5p) and 7 down-regulated (miR-133a, miR-543, miR-130a, miR-27b, miR-223, miR-142-5p and miR-320b) in the blood of individuals with chronic benzene poisoning compared to healthy controls [124]. An association between benzene and aberrant miRNA expression was also reported in a non-occupational setting: miR-223 expression in pregnant women and indoor dwelling concentrations of benzene and toluene (smoking-related volatile organic compounds) were positively associated and appeared to decrease the number of regulatory T-cells in maternal and cord blood [125]. Mice that were injected with benzene for 4 weeks showed significant hematotoxicity, as well as changes in expression of several miRNAs in the bone marrow cells of exposed mice: 5 miRNAs were over-expressed and 45 miRNAs were downregulated [126]. The over-expressed miRNAs were miR-34a-5p, miR-129b-5p, miR-451a, miR-144-5p and miR-129b-3p, and the most highly down-regulated miRNAs were miR-33-5p, miR-128-1-5p, miR-188-5p, miR-211-5p, miR-224-5p, miR-504-5p, miR-5107-3p, miR-5120, and let-7i-3p.
Additionally, in a study of benzene-exposed workers, the expression of two lncRNAs (NR_045623 and NR_028291) was higher in the blood of exposed workers relative to controls [127]. These lncRNAs and their associated mRNAs are involved in immune response, hematopoiesis, B cell receptor signaling and chronic myeloid leukemia gene networks, suggesting their association with benzene-induced hemotoxicity and leukemogenesis.
4.4 Formaldehyde
Routes of exposure, associated cancers, and genotoxicity
Formaldehyde is used in the production of binders (wood production, pulp/paper) as well as in plastics, coatings, and textile finishing, and is also commonly used as a preservative. Exposure to formaldehyde occurs both environmentally and occupationally; formaldehyde is also a natural product in most living systems, including fruits and other foods, and is endogenously formed as a byproduct of oxidative metabolism in mammals (including humans). Occupational exposure occurs in the production of formaldehyde or in any of the above-mentioned industrial uses, while non-occupational exogenous sources of formaldehyde include tobacco smoke and automobile exhaust. Formaldehyde is associated with nasopharyngeal cancer and leukemia in humans, and nasal cavity, lung, leukemia and hematopoietic cancers in laboratory animals [128]. Formaldehyde can react directly with DNA, and increased frequency of micronuclei, DNA-protein crosslinks, DNA strand breaks, and sister chromatid exchange have been observed in the blood and/or nasal mucosal cells of exposed workers [129–131], as well as in various human and rodent in vitro systems [128, 132, 133].
DNA methylation
A study of DNA methylation and formaldehyde reported a time-related decrease in global DNA methylation in human 16HBE cells treated with formaldehyde for 24 hr once per week for 24 weeks. Formaldehyde exposure also resulted in down-regulation of expression of the DNA methyltransferase genes DNMT3a and DNMT3b, and up-regulation of DNMT1 and MBD2, all at both the mRNA and protein level. These results indicate that loss of global DNA methylation, an epigenetic alteration associated with genomic instability, after long-term exposure to a low dose of formaldehyde may be one of the possible underlying carcinogenic mechanisms of formaldehyde [134].
Histone modifications
In a study of human pulmonary epithelial cells, histone H3 was more highly phosphorylated at serine 10 and 28 (H3S10 and H3S28) after exposure to formaldehyde compared with normal human lung fibroblasts [135, 136], particularly within the promoter region of the proto-oncogenes FOS and JUN, indicating a relationship between formaldehyde-inducted tumorigenesis and H3S10 and H3S28 phosphorylation. Another study demonstrated that binding of formaldehyde to lysine residues on histone 4 only occurred in the absence of post-translational modifications of histone 4, indicating that the balance between histone acetylation and deacetylation could be disturbed by the attachment of formaldehyde on lysine residues [137].
Non-coding RNA
miRNAs have been demonstrated to be dysregulated upon in vitro exposure to formaldehyde in human lung epithelial A549 cells [138], and in the olfactory bulb [139] of mice, nasal epithelium cells of non-human primates (macaques) [140], and in the nose and WBCs of rats [141] exposed in vivo to formaldehyde by inhalation. The five most differentially expressed miRNAs in the human lung cells were miR-33, miR-450, miR-330, miR-181a, and miR-10b (all down-regulated), the predicted mRNA targets of which are associated with inflammatory response pathways; specifically, the IL-8 pathway. An up-regulation of cytokine release in formaldehyde-exposed cells confirmed the involvement of miRNA expression on formaldehyde-induced inflammatory response [138]. Differentially expressed miRNAs were postulated to be related to increased expression of inflammatory response genes in formaldehyde-exposed rats as well [141]. In regard to cross-species formaldehyde-induced changes in miRNA expression, let-7a, let-7c, let-7f, miR-10b, miR-126, miR-21, and miR-23a were all significantly decreased in both the study using human lung cells and the study of the nose of rats exposed to formaldehyde. The expression of 13 miRNAs was significantly dysregulated in the nasal epithelium cells of macaques exposed to formaldehyde, with miR-125b and miR-152 being the most increased in expression and miR-145 and miR-142-3p being the most decreased. An up-regulation in the expression of integrin-linked kinase-associated genes that are targets of miR-142-3p was observed, as was a down-regulation of apoptosis-related gene targets of miR-125b, demonstrating the mechanistic involvement of these miRNAs in the formaldehyde-induced cellular disease state [140]. In the olfactory bulb of mice exposed to formaldehyde by inhalation, the alterations in miRNA expression was more profound after 1 day of exposure for 6 hours relative to 7 days of 6 hours/day of exposure [139]. Functional annotation analysis of the predicted targets of the 18 miRNAs that were differentially expressed after exposure to formaldehyde for 1 day demonstrated enrichment for cancer and transcriptional regulation pathways, suggesting the involvement of dysregulation of microRNAs in formaldehyde-induced carcinogenesis.
4.5 Coke production, occupational exposures
Routes of exposure, associated cancers, and genotoxicity
Coke is produced by coal carbonization and is used as a fuel in iron-making blast furnaces and other metal-smelting processes. Coke oven workers are primarily exposed to PAHs, and may be exposed to a large number of other compounds, such as asbestos, silica, amines, metals, sulfur dioxide and sulfuric acid. An increased risk of lung cancer has been reported in coke oven workers, and cohort studies of bladder or skin cancer among coke oven workers have been conducted, although the data are inadequate for evaluation of the association with occupational exposures during coke production. The genotoxic effects of coke oven emissions are largely attributed to the presence of PAHs, several of which have been shown to be individually genotoxic in both in vitro and in vivo systems (benzo[a]pyrene, benzo[c]phenanthrene, benzo[b]fluouranthrene) [142]. An increased frequency of sister chromatid exchange, DNA strand breaks [143], micronuclei frequency [144], and benzo[a]pyrene diol epoxide (BPDE)-DNA adducts [145] have been reported in peripheral blood lymphocytes from coke oven workers in comparison to age-matched controls.
DNA methylation
Studies have shown an association between aberrant DNA methylation patterns and exposure to PAHs among coke oven workers [146]. Promoter methylation of the tumor suppressor genes p14ARK and p16INK4 was increased in peripheral blood mononuclear cells, along with increased urinary levels of 1-hydroxypyrene (an indicator of exposure to PAHs) in coke oven workers relative to water pump workers [147]. DNA damage, as evaluated by a comet assay, was also significantly higher in the coke oven workers. Studies have reported increased methylation of LINE-1 and Alu repetitive DNA elements, and gene-specific hypomethylation of the tumor suppressor genes p53 and HIC1 in peripheral blood [148, 149]. The changes in DNA methylation of repetitive elements were positively correlated with urinary biomarkers of PAH exposure and with BPDE-DNA adducts in the blood, while p53 promoter hypomethylation was significantly correlated with micronuclei formation. LINE-1 hypomethylation, as well as hypomethylation and suppression of the DNA methyltransferase gene MGMT, was observed in both the blood of coke-oven workers, as well as in human bronchial epithelial cells (16HBE) treated with coke oven emissions [150]. The LINE-1 methylation was inversely associated with comet tail length and micronucleus frequency (indicators of DNA damage) in the coke oven workers, and with BPDE-DNA adducts in the in vitro assay.
Non-coding RNA
miRNA profiling was conducted in coke oven workers [151], and the association between differentially expressed miRNAs and PAH exposure was evaluated. Five significantly differentially expressed miRNAs that were associated with heightened levels of urinary PAHs and/or plasma benzo[a]pyrene-r-7,t-8,c-10-tetrahydrotetrol-albumin were identified: miR-24-3p, miR-27a-3p, and miR-142-5p, miR-28-5p (all down-regulated), and miR-150-5p (up-regulated). The dysregulation of all 5 of these miRNAs was associated with increased micronuclei frequency, an indicator of DNA damage and a common marker of genotoxicity.
4.6 1,3-butadiene
Routes of exposure, associated cancers, and genotoxicity
1,3-Butadiene is a gas monomer used in the production of synthetic rubber. Exposure typically occurs in occupational settings in the production of 1,3-butadiene itself, as well as in production of rubber and plastics and petroleum refining and distribution. 1,3-Butadiene is also widely detected in ambient air, albeit at much lower levels than in occupational settings, from sources such as vehicle exhaust, cigarette smoke, and wood fires. An excess of hematopoietic cancers has been reported among workers occupationally exposed to 1,3-butadiene, and, to a lesser extent, lung and bladder cancers. Further, 1,3-butadiene is a multi-site carcinogen in experimental animals [152]. The carcinogenicity of 1,3-butadiene is contingent upon the metabolism of 1,3-butadiene to reactive epoxides, which can bind with DNA and proteins [153, 154]. 1,3-Butadiene-DNA adducts have been observed in occupationally-exposed humans and experimental animals, and have been associated with mutations in cancer-related genes [155–159].
DNA methylation
In studies in mice, loss of global DNA methylation was observed in the liver following relatively high exposure (625 ppm) to 1,3-butadiene by inhalation for 6 hours per day, 5 days per week for two weeks [160]. The extent of global DNA hypomethylation was strain-specific and also varied across target and non-target tissues of 1,3-butadiene-induced carcinogenesis [161, 162]. For example, a loss of methylation within repetitive DNA elements was observed in the lung and liver (target tissues of carcinogenesis), but not in the kidney (non-target tissue of carcinogenesis) in C57BL/6J mice. These results indicate that aberration of normal DNA structure is likely associated with the carcinogenic mechanisms of 1,3-butadiene, and that this epigenetic mechanism may be driven by underlying genetic differences.
Histone modifications
A loss of trimethylation at histone H3 lysine 9 (H3K9me3), histone H3 lysine 27 (H3K27me3), and histone H4 lysine 20 (H4K20me3) was observed in a dose-dependent manner in the liver of mice exposed to 1,3-butadiene for 6 hours a day, 5 days a week for 2 weeks [160]. Loss of these histone modifications is known to impair the maintenance of proper chromatin structure, diminish cellular maintenance and regulation of the cell cycle, disrupt the balance between cell proliferation and differentiation, and severely reduce cell viability [163, 164]. These histone modifications in the liver have also been shown to vary across several inbred mouse strains, as well as in target and non-target tissues of carcinogenesis [161, 162]. Interestingly, an increase in the repressive histone marks H3K9me3, H3K27me3, and H4K20me3 was observed in the kidney, a non-target tissue of carcinogenesis, in C57BL/6J mice that were subjected to short-term exposure to 1,3-butadiene. The same pattern was observed in the liver of 1,3-butadiene-exposed CAST/EiJ mice, which had the lowest abundance of DNA adducts among 7 mouse strains, suggesting a possible protective effect conferred by this histone mark. In contrast, H3K27 acetylation, an indicator of transcriptionally active (i.e., relaxed) chromatin [165], was significantly increased in the liver of 1,3-butadiene-exposed mice.
4.7 Sulfur mustard
Routes of exposure, associated cancers, and genotoxicity
Mustard gas is a chemical warfare agent. Exposure to sulfur mustard occurs either in production of the chemical, or in contaminated areas where mustard gas was released. Exposure to sulfur mustard causes respiratory cancers in humans and experimental animals [166–169], and the carcinogenicity of sulfur mustard is attributed to its genotoxicity. Exposure to sulfur mustard has been shown to cause guanine-guanine DNA crosslinks, sister chromatid exchange, micronuclei, and mutations in humans and rodents [170–173].
DNA Methylation
Global DNA methylation was evaluated in sulfur mustard-exposed early endothelial cells, as well as in human skin samples obtained from a patient 1 year after an accidental exposure to pure sulfur mustard. A global increase of DNA methylation was observed in both the in vitro study and in human skin samples [174].
Non-coding RNA
An in vitro study of miRNA expression using normal human epidermal keratinocytes exposed to sulfur mustard reported dysregulation of metabolic activity, proliferation and keratin-1 expression as a result of up-regulation of miR-203 and miR-210 [175]. These two miRNAs were selected for study based on their impact on proliferation and differentiation in epidermal cells (miR-203) and involvement in the control of cell proliferation and induction by oxygen depletion in keratinocytes (miR-210). In mouse early endothelial cells incubated with various sub-lethal concentrations of sulfur mustard, a total of 66 miRNAs were significantly differentially expressed compared to control cultures [176]. Of those, up-regulation of miR-92a of the miR-17-92 cluster (oncomir-1), which plays a central role in carcinogenesis [177], had the strongest correlation with sulfur mustard concentration.
4.8 Vinyl chloride
Routes of exposure, associated cancers, and genotoxicity
Vinyl chloride is primarily used in polyvinyl chloride (PVC) production, and inhalational exposure is the main route of exposure. Non-occupational exposures are very low, but are higher in populations living in relatively close proximity to industrial emission sources. In humans, vinyl chloride exposure is associated with angiosarcoma of the liver, hepatocellular carcionoma (HCC), lung cancer, and malignant neoplasms of connective and soft tissues. The reactive metabolites of vinyl chloride, chloroethylene oxide and chloroacetaldehyde, are reactive with both DNA and protein [178–180]. Vinyl chloride induces an increased frequency of sister chromatid exchange, micronuclei formation, and chromosomal aberrations [181]. Mutations in cancer-related genes have been reported in both humans and rats exposed to vinyl chloride [181, 182].
DNA methylation
In a study of angiosarcoma patients, the majority of whom had confirmed chronic occupational exposure to vinyl chloride, promoter methylation of p14ARF was confirmed in 5 of 19 cases (26%), p16INKa showed aberrant promoter methylation in 12 of 19 cases (63%), and methylation of the promoter region of both of these tumor suppressor genes was observed in 3 (16%) cases. Increased promoter methylation correlated with transcriptional down-regulation. The aberrant p14ARF methylation occurred independently of p53 mutation, which was detected in 6 of 19 (32%) cases [183]. However, p16INKa promoter hypermethylation was associated with KRAS mutations in HCC patients who were occupationally exposed to vinyl chloride [184].
4.9 4-Aminobiphenyl
Routes of exposure, associated cancers, and genotoxicity
4-Aminobiphenyl is an aromatic amine used as a dye intermediate and as a rubber antioxidant, and human exposure predominantly occurs in occupational settings. Industrial production of 4-aminobiphenyl was ceased in 1955, and current exposures are due to contamination or metabolic release from benzidine. 4-Aminobiphenyl is also a byproduct of tobacco combustion, and has been detected in fumes from cooking oils. Bladder carcinoma is the primary cancer associated with exposure to 4-aminobiphenyl, observed in human chemical plant workers and in experimental animal models. Multiple metabolic pathways activate aromatic amines, including 4-aminobiphenyl, to DNA-reactive intermediates, which are known to result in mutations. 4-Aminobiphenyl-DNA adducts have been detected in human bladder, lung, and breast tissue of exposed humans [185–187]. Mutations in the HPRT locus and in the H-ras gene have been detected in human and mouse tissues, respectively, after exposure to 4-aminobiphenyl [87].
Histone modifications
Histone H3K4 mono-methylation, a mark whose function is not well understood, was not altered by BaP treatment alone (5µM for 48 hours) in normal human mammary epithelial cells, but was decreased when cells were treated with 4-aminobiphenyl alone (also 5µM for 48 hours), suggesting that this histone modification is carcinogen-specific [188].
Non-coding RNA
In an in vitro study using human HepG2 cells exposed to 4-aminobiphenyl as a model of DNA damage, the expression of 27 miRNAs was at least 3-fold higher in the 4-ABP-treated cells relative to the control group [189]. Additionally, 16 DNA repair-related genes were down-regulation in 4-aminobiphenyl-treated cells. miRNA-513a-5p and miRNA-630 were predicted to be implicated in the deregulation of FANCG and RAD18 genes, respectively, which are both involved in DNA damage repair. Overexpression and knockdown of miRNA-513a-5p and miRNA-630 reduced and increased the expression of FANCG and RAD18 proteins, respectively. The authors concluded that miRNA-513a-5p and miRNA-630 may have an inhibitory effect on DNA repair genes, ultimately leading to DNA damage.
4.10 Benzidine
Routes of exposure, associated cancers, and genotoxicity
Benzidine is, and has primarily been used as, the base for various types of dyes used in fabrics, as well as for visual detection of blood cells in laboratory settings. Benzidine is only allowed to be used in closed systems, and limited amounts are released into the environment. Bladder carcinoma is the primary cancer that has been associated with occupational exposure to benzidine, and is a multi-target carcinogen in experimental animals (primarily a hepatocarcinogen when administered by injection or ingestion). Like 4-aminobiphenyl, benzidine is an aromatic amine and can be metabolized to DNA-reactive intermediates that can lead to chromosomal aberrations, DNA strand breaks, formation of micronuclei, DNA adducts, and mutations in oncogenes [190–192].
There was only report of epigenetic response after exposure to benzidine, in which the H-ras oncogene was hypomethylated (entire gene) in benzidine-induced liver tumors relative that of non-tumor tissue in B6C3F1 mice, and an increase in the expression of the gene was also detected. The K-ras oncogene was also hypomethylated in half of the mice. These results suggest that hypomethylation of oncogenes may provide an epigenetic mechanism for facilitating their aberrant expression. The lack of a DNA methylated sites observed in the H-ras oncogene in the liver of B6C3F1 mice may indicate an increased potential for its expression, which could account for the high propensity for hepatoma development in this strain [193].
4.11 4,4’-Methylenebis(2-chlorobenzenamine)
Routes of exposure, associated cancers, and genotoxicity
4,4’-Methylenebis(2-chlorobenzenamine), also referred to as MOCA, is a curing agent used in the manufacture of urethane rubber products. The majority of human exposures occur in occupational settings, with non-occupational exposures in areas contaminated with MOCA or consumption of foods that were grown in contaminated soil. Limited human data is suggestive of an association between MOCA exposure and bladder cancer, and MOCA caused lung, liver, and bladder cancer in experimental animals [194]. The assignment of MOCA as a “Group 1 carcinogen” by IARC was largely based on the strong evidence of genotoxic mechanisms of action, involving metabolism of the aromatic amine to DNA-reactive intermediates, which lead to DNA adducts, mutations, sister chromatid exchange, and increased micronuclei [195–197].
Only one study investigated epigenetic alterations caused by 4,4'-methylene-bis(2-chloroaniline) (MOCA). The authors found that rat spleen cells incubated with 10 mM MOCA increased phosphorylation in the histone fraction of the cells after 4 hours of exposure [198].
4.12 Occupational exposure as a painter
Routes of exposure, associated cancers, and genotoxicity
Paint products are composed of up to thousands of chemical compounds for various purposes (pigment, driers, binders, and corrosion inhibitors, among others), some of which are volatile and/or hazardous. In recent years, many hazardous chemicals, such as benzene, phthalates, chromium, and lead, have been reduced or removed from paint. Associations have been reported between bladder cancer and occupational exposure as a painter, and childhood leukemia and maternal exposure during painting. Chromosomal aberrations, increased micronuclei and sister chromatid exchange have all been reported in occupational painters [199– 201]; however, the genotoxic mechanisms associated with occupational exposure as a painter are attributed to the genotoxic effects of the individual constituents of paints, e.g. benzene, toluene, styrene, and PAHs.
A study conducted in 150 non-smoking car painters from several workshops in the southwest of Colombia found a significant increase in DNA methylation in the promoter region of GSTP1 and p16INK4a in exfoliated urothelial cells of exposed workers compared to references, and these gene-specific alterations were associated with an increase in micronuclei frequency [202], an indicator of genotoxicity. Because the exact chemical composition of the exposure is not reported here, the molecular findings can only be associated to the general category “occupational exposure as a painter,” which is one of the occupational exposures included in the IARC monograph volume 100F.
5. Summary
While the number of studies devoted to understanding the epigenetic alterations caused by exposure to chemical carcinogens is rapidly increasing, there remains a dearth of well-designed comprehensive studies that identify epigenetic alterations that are associated with the carcinogenic process. Importantly, there is neither a “gold standard” for the evaluation of epigenetic or epigenomic signatures in toxicology studies (i.e., what types of epigenetic effects shall be evaluated to draw inferences about epigenetic effects of chemicals), nor guidelines for the types of assays to be used so that this information can be used in cancer hazard evaluations.
Notably, one third (4 out of 12) of the chemicals and occupational hazards included in this review had a maximum of only two published reports of epigenetic alterations, and there were only 3 for which at least 10 studies were identified to have reported epigenetic endpoints (Table 3). Among the chemical agents for which there were between 2 and 10 reports, nearly half were human in vivo studies (14/32) of DNA methylation or non-coding RNAs, demonstrating strength in the human relevance of epigenetic alterations for these chemicals, while highlighting the need for additional animal and/or in vitro studies for further understanding of the mechanistic role of such marks in carcinogenesis. Overall, studies of aberrant DNA methylation represented the most commonly studied epigenetic feature, followed by changes in the expression of noncoding RNAs, and finally histone modifications (87, 60, and 25, respectively). While a wealth of in vitro data may be especially important for a better understanding of the mechanistic role of epigenetic alterations, (e.g. as seen among the studies of benzo[a]pyrene), substantial evidence of epigenetic alterations in humans may provide relatively stronger confidence of the relevance of such alterations in human cancers (e.g., human in vivo data represented the majority of studies found for benzene, coke production, and sulfur mustard). Future studies may increase confidence of the role of specific epigenetic alterations by prioritizing those for which the most mechanistic data has been reported, and applying them to human samples, where possible. Alternately, those epigenetic alterations that have been most frequently reported in human cancers may be prioritized in in vitro epigenetic mechanistic studies.
As emphasized by Herceg et al [9], epigenetic mechanisms represent an essential tool for cancer hazard identification, particularly for non-genotoxic and non-receptor-mediated carcinogens, or chemicals and hazards for which human carcinogenicity data are inconclusive. However, most chemicals that were classified as known or suspected carcinogens by IARC were reviewed before epigenetics data were available, and there remains a need for additional studies of epigenetic alterations and cancer hazards. Importantly, delineation between normal epigenetic processes in cells and the epigenetic alterations that have a causal relationship to cancer is needed for all epigenetic marks, not only for DNA methylation [9], to effectively identify cancer-relevant epigenetic marks. Many of the studies included in this review demonstrated an association between epigenetic alterations and changes in expression of cancer-relevant genes, or reported epigenomic signatures that may be generally linked to genomic instability, e.g., global DNA and repetitive element demethylation. However, many studies lack the report of a definitive causal association between an epigenetic even and carcinogenesis. To gain a more comprehensive understanding of adverse health outcomes that may be causally linked to epigenetic changes, studies that include apical end-points, functional metrics, and full dose-response characterization are needed [11, 203].
6. Future Research Needs
Due to the increasing recognition that epigenetic events are involved in the pathogenesis of numerous pathological states, the interest in monitoring epigenetic perturbations in various models of disease phenotypes is rising. Epigenetic marks represent a class of biomarkers with great potential in the identification of exposure status, damage response, and/or disease state. However, to effectively utilize epigenetic endpoints as markers of exposure and/or damage, a better understanding is needed to decide which epigenetic alterations are most informative of specific types of damage or disease, as well as how these marks compare to currently used markers of carcinogenicity (e.g. DNA adducts or chromosomal aberrations). Because it is well-established that epigenetic alterations are as equally important in the carcinogenic process as genetic changes, it is essential that the focus of future research is devoted to discovery of cancer-specific carcinogen exposure-related epigenetic changes, as well as the investigation of the extended evolution of any given epigenetic alteration with respect to carcinogenesis. To fully understand the importance of epigenetic and epigenomic responses to environmental stressors, studies that investigate and compare both epigenetic data with functional measures (such as gene and protein expression) within the same study and controlled exposure scenario are needed.
When the target tissue of oncogenesis is an internal organ, biomarkers in accessible surrogate tissues are required for the evaluation of molecular changes in humans. For example, because miRNAs are detectable in biological fluids, including blood and urine, they represent a potentially easily accessible and informative class of biomarkers of both exposure as well as effect [204]. An additional initiative in the study of epigenetics in cancer hazard assessment is to determine whether changes in the epigenome and transcriptome, as well as the level of DNA damage, of easily accessible tissues (e.g. peripheral blood, skin) can inform the same responses of target tissues.
Another necessity is the incorporation of several time points in exposure studies, which would facilitate a better understanding of the evolution and persistence of epigenetic and transcriptional alterations during and after cessation of exposure to environmental toxicants. Epigenetic marks potentially represent early markers of carcinogenesis, because many epigenetic alterations have been shown to occur before other molecular events that are associated with cancer. Previously reported short time-course epigenetics studies primarily demonstrated reversibility of most effects after cessation of an exogenous stimulus, while a few have shown that some epigenetic marks (particularly histone lysine acetylation) persist for weeks and even months [205]. However, the plasticity of the epigenome presents a challenge in understanding the relevance of specific epigenetic alterations to cancer, and additional studies that incorporate time point studies, and/or cell type-specific analyses of epigenetic marks, are essential.
Additionally, a major unresolved issue is the lack of specificity of any given carcinogen-induced epigenetic alteration. This can be illustrated by the fact that a loss of DNA methylation has been reported after exposure to a broad range of chemicals. Similarly, the role of non-coding RNAs, the “youngest” of the types of epigenetic alterations presented herein in terms of discovery and characterization, in cancer remains poorly understood. Although studies of changes in microRNA expression represent the fastest-growing category of epigenetic alteration reported in toxicity studies, there are few studies confirming the involvement of a particular miRNA in carcinogenesis associated with exposure to a specific chemical. It is notable that miR-10b, miR-24, 125a, 125b, miR-92, and miR-142 as some of the top-most differentially expressed microRNAs after exposure to at least 3 different chemicals included in this review, strengthening their characterization as cancer- and chemical exposure-relevant miRNAs.
Finally, future studies are needed to address the mechanistic relationship between specific epigenetic alterations and DNA damage, and how this relationship is associated with carcinogenesis. For example, in vitro assays are needed to evaluate the supposed potentiation effect(s) of certain epigenetic alterations in tandem with, or preceding, genotoxicity. Further, genomic analysis of site-specific DNA damage in association with epigenetic marks of condensed or relaxed chromatin will offer insight into the relationship between chromatin dynamics and genotoxicity.
7. Conclusions
A major challenge in the application of these epigenetic findings in regulatory science is the question of “how” to effectively include the findings. Epigenetic endpoints are currently being increasingly used in cancer hazard assessments; for example, “Epigenetic alterations” has recently been included as one of ten key characteristics of human carcinogens [13]. However, while there is extensive information about the fundamental role of epigenetic alterations in cancer development and progression, the understanding of the mechanistic significance and specificity of carcinogen-induced epigenetic abnormalities in the carcinogenic process is insufficient. For example, several studies have demonstrated a mechanistic link between DNA hypomethylation (the most highly reported, and thus assumed best-characterized, epigenetic alteration among the studies included in this review) and genetic changes, and established the role of this epigenetic alteration in carcinogenesis [206–208]. In contrast, there is not a single study among an extensive list of observational reports on carcinogen-induced DNA hypomethylation that demonstrated a mechanistic link between loss of DNA methylation and cancer development.
Moving forward, the currently best-characterized epigenetic mechanisms of carcinogenesis should be increasingly incorporated in cancer hazard assessments, while ongoing research must continue to investigate and characterize epigenetic alterations in models of chemical exposure and disease for which data is lacking. Together, these initiatives will enable the efficacious inclusion and application of epigenetics data in the assessment of potential and known chemical carcinogens, a practice that is essential for the comprehensive understanding of chemical carcinogenesis.
Supplementary Material
Acknowledgments
This work was funded, in part, by a grant from the National Institutes of health (R01 ES023195). The authors thank Andrew Shapiro for his assistance with HAWC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclaimer: The views expressed in this manuscript do not necessarily represent those of the U.S. Food and Drug Administration.
References
- 1.Hu W, Feng Z, Tang MS. Preferential carcinogen-DNA adduct formation at codons 12 and 14 in the human K-ras gene and their possible mechanisms. Biochemistry. 2003;42:10012–10023. doi: 10.1021/bi034631s. [DOI] [PubMed] [Google Scholar]
- 2.Chen JX, Zheng Y, West M, Tang MS. Carcinogens preferentially bind at methylated CpG in the p53 mutational hot spots. Cancer Res. 1998;58:2070–2075. [PubMed] [Google Scholar]
- 3.Yoon JH, Smith LE, Feng Z, Tang M, Lee CS, Pfeifer GP. Methylated CpG dinucleotides are the preferential targets for G-to-T transversion mutations induced by benzo[a]pyrene diol epoxide in mammalian cells: similarities with the p53 mutation spectrum in smoking-associated lung cancers. Cancer Res. 2001;61:7110–7117. [PubMed] [Google Scholar]
- 4.Tretyakova N, Guza R, Matter B. Endogenous cytosine methylation and the formation of carcinogen carcinogen-DNA adducts. Nucleic Acids Symp Ser (Oxf) 2008:49–50. doi: 10.1093/nass/nrn025. [DOI] [PubMed] [Google Scholar]
- 5.Denissenko MF, Chen JX, Tang MS, Pfeifer GP. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc Natl Acad Sci U S A. 1997;94:3893–3898. doi: 10.1073/pnas.94.8.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, Casero RA, Sears CL, Baylin SB. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marlowe J, Teo SS, Chibout SD, Pognan F, Moggs J. Mapping the epigenome--impact for toxicology. EXS. 2009;99:259–288. doi: 10.1007/978-3-7643-8336-7_10. [DOI] [PubMed] [Google Scholar]
- 8.Stein RA. Epigenetics and environmental exposures. J Epidemiol Community Health. 2012;66:8–13. doi: 10.1136/jech.2010.130690. [DOI] [PubMed] [Google Scholar]
- 9.Herceg Z, Lambert MP, van Veldhoven K, Demetriou C, Vineis P, Smith MT, Straif K, Wild CP. Towards incorporating epigenetic mechanisms into carcinogen identification and evaluation. Carcinogenesis. 2013;34:1955–1967. doi: 10.1093/carcin/bgt212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koturbash I, Beland FA, Pogribny IP. Role of epigenetic events in chemical carcinogenesis--a justification for incorporating epigenetic evaluations in cancer risk assessment. Toxicol Mech Methods. 2011;21:289–297. doi: 10.3109/15376516.2011.557881. [DOI] [PubMed] [Google Scholar]
- 11.Alyea RA, Moore NP, LeBaron MJ, Gollapudi BB, Rasoulpour RJ. Is the current product safety assessment paradigm protective for epigenetic mechanisms? J Pharmacol Toxicol Methods. 2012;66:207–214. doi: 10.1016/j.vascn.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Pogribny IP, Rusyn I. Environmental toxicants, epigenetics, and cancer. Adv Exp Med Biol. 2013;754:215–232. doi: 10.1007/978-1-4419-9967-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith MT, Guyton KZ, Gibbons CF, Fritz JM, Portier CJ, Rusyn I, DeMarini DM, Caldwell JC, Kavlock RJ, Lambert P, Hecht SS, Bucher JR, Stewart BW, Baan R, Cogliano VJ, Straif K. Key Characteristics of Carcinogens as a Basis for Organizing Data on Mechanisms of Carcinogenesis. Environ Health Perspect. 2015 doi: 10.1289/ehp.1509912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearce N, Blair A, Vineis P, Ahrens W, Andersen A, Anto JM, Armstrong BK, Baccarelli AA, Beland FA, Berrington A, Bertazzi PA, Birnbaum LS, Brownson RC, Bucher JR, Cantor KP, Cardis E, Cherrie JW, Christiani DC, Cocco P, Coggon D, Comba P, Demers PA, Dement JM, Douwes J, Eisen EA, Engel LS, Fenske RA, Fleming LE, Fletcher T, Fontham E, Forastiere F, Frentzel-Beyme R, Fritschi L, Gerin M, Goldberg M, Grandjean P, Grimsrud TK, Gustavsson P, Haines A, Hartge P, Hansen J, Hauptmann M, Heederik D, Hemminki K, Hemon D, Hertz-Picciotto I, Hoppin JA, Huff J, Jarvholm B, Kang D, Karagas MR, Kjaerheim K, Kjuus H, Kogevinas M, Kriebel D, Kristensen P, Kromhout H, Laden F, Lebailly P, LeMasters G, Lubin JH, Lynch CF, Lynge E, t Mannetje A, McMichael AJ, McLaughlin JR, Marrett L, Martuzzi M, Merchant JA, Merler E, Merletti F, Miller A, Mirer FE, Monson R, Nordby KC, Olshan AF, Parent ME, Perera FP, Perry MJ, Pesatori AC, Pirastu R, Porta M, Pukkala E, Rice C, Richardson DB, Ritter L, Ritz B, Ronckers CM, Rushton L, Rusiecki JA, Rusyn I, Samet JM, Sandler DP, de Sanjose S, Schernhammer E, Costantini AS, Seixas N, Shy C, Siemiatycki J, Silverman DT, Simonato L, Smith AH, Smith MT, Spinelli JJ, Spitz MR, Stallones L, Stayner LT, Steenland K, Stenzel M, Stewart BW, Stewart PA, Symanski E, Terracini B, Tolbert PE, Vainio H, Vena J, Vermeulen R, Victora CG, Ward EM, Weinberg CR, Weisenburger D, Wesseling C, Weiderpass E, Zahm SH. IARC monographs: 40 years of evaluating carcinogenic hazards to humans. Environ Health Perspect. 2015;123:507–514. doi: 10.1289/ehp.1409149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma A, Singh K, Almasan A. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol Biol. 2012;920:613–626. doi: 10.1007/978-1-61779-998-3_40. [DOI] [PubMed] [Google Scholar]
- 16.Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi Y, Cao L. DNA methylation, its mediators and genome integrity. Int J Biol Sci. 2015;11:604–617. doi: 10.7150/ijbs.11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song CX, Yi C, He C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol. 2012;30:1107–1116. doi: 10.1038/nbt.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe Y, Maekawa M. Methylation of DNA in cancer. Adv Clin Chem. 2010;52:145–167. doi: 10.1016/s0065-2423(10)52006-7. [DOI] [PubMed] [Google Scholar]
- 20.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 21.Shen L, Waterland RA. Methods of DNA methylation analysis. Curr Opin Clin Nutr Metab Care. 2007;10:576–581. doi: 10.1097/MCO.0b013e3282bf6f43. [DOI] [PubMed] [Google Scholar]
- 22.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014;28:672–682. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18:R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nature Reviews Genetics. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 25.Torres IO, Fujimori DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol. 2015;35:68–75. doi: 10.1016/j.sbi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 29.Waters R, van Eijk P, Reed S. Histone modification and chromatin remodeling during NER. DNA Repair (Amst) 2015;36:105–113. doi: 10.1016/j.dnarep.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 30.O'Hagan HM. Chromatin modifications during repair of environmental exposure-induced DNA damage: a potential mechanism for stable epigenetic alterations. Environ Mol Mutagen. 2014;55:278–291. doi: 10.1002/em.21830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meseure D, Drak Alsibai K, Nicolas A, Bieche I, Morillon A. Long Noncoding RNAs as New Architects in Cancer Epigenetics, Prognostic Biomarkers, and Potential Therapeutic Targets. Biomed Res Int. 2015;2015:320214. doi: 10.1155/2015/320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 33.Pogribny IP, Beland FA, Rusyn I. The role of microRNAs in the development and progression of chemical-associated cancers. Toxicol Appl Pharmacol. 2015 doi: 10.1016/j.taap.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 34.Tong WM, Lee MK, Galendo D, Wang ZQ, Sabapathy K. Aflatoxin-B exposure does not lead to p53 mutations but results in enhanced liver cancer of Hupki (human p53 knock-in) mice. Int J Cancer. 2006;119:745–749. doi: 10.1002/ijc.21890. [DOI] [PubMed] [Google Scholar]
- 35.Kalina I, Brezani P, Gajdosova D, Binkova B, Salagovic J, Habalova V, Mrackova G, Dobias L, Sram RJ. Cytogenetic monitoring in coke oven workers. Mutat Res. 1998;417:9–17. doi: 10.1016/s1383-5718(98)00089-8. [DOI] [PubMed] [Google Scholar]
- 36.Brauze D, Wielgosz SM, Pawlak AL, Baer-Dubowska W. Effect of the route of benzo[a]pyrene administration on sister chromatid exchange and DNA binding in bone marrow of mice differing with respect to cytochrome P450 1A1 induction. Toxicol Lett. 1997;91:211–217. doi: 10.1016/s0378-4274(97)00024-6. [DOI] [PubMed] [Google Scholar]
- 37.Kliesch U, Roupova I, Adler ID. Induction of chromosome damage in mouse bone marrow by benzo[a]pyrene. Mutat Res. 1982;102:265–273. doi: 10.1016/0165-1218(82)90136-7. [DOI] [PubMed] [Google Scholar]
- 38.DeMarini DM, Landi S, Tian D, Hanley NM, Li X, Hu F, Roop BC, Mass MJ, Keohavong P, Gao W, Olivier M, Hainaut P, Mumford JL. Lung tumor KRAS and TP53 mutations in nonsmokers reflect exposure to PAH-rich coal combustion emissions. Cancer Res. 2001;61:6679–6681. [PubMed] [Google Scholar]
- 39.IARC. Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. IARC Monogr Eval Carcinog Risks Hum. 2010;92:1–853. [PMC free article] [PubMed] [Google Scholar]
- 40.Huang H, Hu G, Cai J, Xia B, Liu J, Li X, Gao W, Zhang J, Liu Y, Zhuang Z. Role of poly(ADP-ribose) glycohydrolase silencing in DNA hypomethylation induced by benzo(a)pyrene. Biochem Biophys Res Commun. 2014;452:708–714. doi: 10.1016/j.bbrc.2014.08.146. [DOI] [PubMed] [Google Scholar]
- 41.Minero AS, Lukashevich OV, Cherepanova NA, Kolbanovskiy A, Geacintov NE, Gromova ES. Probing murine methyltransfease Dnmt3a interactions with benzo[a]pyrene-modified DNA by fluorescence methods. FEBS J. 2012;279:3965–3980. doi: 10.1111/j.1742-4658.2012.08756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson VL, Jones PA. Chemical carcinogen-mediated decreases in DNA 5-methylcytosine content of BALB/3T3 cells. Carcinogenesis. 1984;5:1027–1031. doi: 10.1093/carcin/5.8.1027. [DOI] [PubMed] [Google Scholar]
- 43.Wilson VL, Jones PA. Inhibition of DNA methylation by chemical carcinogens in vitro. Cell. 1983;32:239–246. doi: 10.1016/0092-8674(83)90514-7. [DOI] [PubMed] [Google Scholar]
- 44.Wojciechowski MF, Meehan T. Inhibition of DNA methyltransferases in vitro by benzo[a]pyrene diol epoxide-modified substrates. J Biol Chem. 1984;259:9711–9716. [PubMed] [Google Scholar]
- 45.Diala ES, Hoffman RM. DNA methylation levels in normal and chemically-transformed mouse 3T3 cells. Biochem Biophys Res Commun. 1982;104:1489–1494. doi: 10.1016/0006-291x(82)91419-x. [DOI] [PubMed] [Google Scholar]
- 46.Yauk CL, Polyzos A, Rowan-Carroll A, Kortubash I, Williams A, Kovalchuk O. Tandem repeat mutation, global DNA methylation, and regulation of DNA methyltransferases in cultured mouse embryonic fibroblast cells chronically exposed to chemicals with different modes of action. Environ Mol Mutagen. 2008;49:26–35. doi: 10.1002/em.20359. [DOI] [PubMed] [Google Scholar]
- 47.Sadikovic B, Rodenhiser DI. Benzopyrene exposure disrupts DNA methylation and growth dynamics in breast cancer cells. Toxicol Appl Pharmacol. 2006;216:458–468. doi: 10.1016/j.taap.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 48.Tommasi S, Kim SI, Zhong X, Wu X, Pfeifer GP, Besaratinia A. Investigating the epigenetic effects of a prototype smoke-derived carcinogen in human cells. PLoS One. 2010;5:e10594. doi: 10.1371/journal.pone.0010594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Damiani LA, Yingling CM, Leng S, Romo PE, Nakamura J, Belinsky SA. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res. 2008;68:9005–9014. doi: 10.1158/0008-5472.CAN-08-1276. [DOI] [PubMed] [Google Scholar]
- 50.Tang WY, Levin L, Talaska G, Cheung YY, Herbstman J, Tang D, Miller RL, Perera F, Ho SM. Maternal exposure to polycyclic aromatic hydrocarbons and 5'-CpG methylation of interferon-gamma in cord white blood cells. Environ Health Perspect. 2012;120:1195–1200. doi: 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang P, Ma J, Zhang B, Duan H, He Z, Zeng J, Zeng X, Li D, Wang Q, Xiao Y, Liu C, Xiao Q, Chen L, Zhu X, Xing X, Li Z, Zhang S, Zhang Z, Ma L, Wang E, Zhuang Z, Zheng Y, Chen W. CpG site-specific hypermethylation of p16INK4alpha in peripheral blood lymphocytes of PAH-exposed workers. Cancer Epidemiol Biomarkers Prev. 2012;21:182–190. doi: 10.1158/1055-9965.EPI-11-0784. [DOI] [PubMed] [Google Scholar]
- 52.Teneng I, Montoya-Durango DE, Quertermous JL, Lacy ME, Ramos KS. Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics. 2011;6:355–367. doi: 10.4161/epi.6.3.14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ouyang B, Baxter CS, Lam HM, Yeramaneni S, Levin L, Haynes E, Ho SM. Hypomethylation of dual specificity phosphatase 22 promoter correlates with duration of service in firefighters and is inducible by low-dose benzo[a]pyrene. J Occup Environ Med. 2012;54:774–780. doi: 10.1097/JOM.0b013e31825296bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yasaei H, Gilham E, Pickles JC, Roberts TP, O'Donovan M, Newbold RF. Carcinogen-specific mutational and epigenetic alterations in INK4A, INK4B and p53 tumour-suppressor genes drive induced senescence bypass in normal diploid mammalian cells. Oncogene. 2013;32:171–179. doi: 10.1038/onc.2012.45. [DOI] [PubMed] [Google Scholar]
- 55.Herbstman JB, Tang D, Zhu D, Qu L, Sjodin A, Li Z, Camann D, Perera FP. Prenatal exposure to polycyclic aromatic hydrocarbons, benzo[a]pyrene-DNA adducts, and genomic DNA methylation in cord blood. Environ Health Perspect. 2012;120:733–738. doi: 10.1289/ehp.1104056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Satterwhite JE, Trumbo CM, Danell AS, Hvastkovs EG. Electrochemical study on the effects of epigenetic cytosine methylation on anti-benzo[a]pyrene diol epoxide damage at TP53 oligomers. Anal Chem. 2013;85:1183–1191. doi: 10.1021/ac303077h. [DOI] [PubMed] [Google Scholar]
- 57.Dong H, Bonala RR, Suzuki N, Johnson F, Grollman AP, Shibutani S. Mutagenic potential of benzo[a]pyrene-derived DNA adducts positioned in codon 273 of the human P53 gene. Biochemistry. 2004;43:15922–15928. doi: 10.1021/bi0482194. [DOI] [PubMed] [Google Scholar]
- 58.Weisenberger DJ, Romano LJ. Cytosine methylation in a CpG sequence leads to enhanced reactivity with Benzo[a]pyrene diol epoxide that correlates with a conformational change. J Biol Chem. 1999;274:23948–23955. doi: 10.1074/jbc.274.34.23948. [DOI] [PubMed] [Google Scholar]
- 59.Subach OM, Baskunov VB, Darii MV, Maltseva DV, Alexandrov DA, Kirsanova OV, Kolbanovskiy A, Kolbanovskiy M, Johnson F, Bonala R, Geacintov NE, Gromova ES. Impact of benzo[a]pyrene-2'-deoxyguanosine lesions on methylation of DNA by SssI and HhaI DNA methyltransferases. Biochemistry. 2006;45:6142–6159. doi: 10.1021/bi0511639. [DOI] [PubMed] [Google Scholar]
- 60.Subach OM, Maltseva DV, Shastry A, Kolbanovskiy A, Klimasauskas S, Geacintov NE, Gromova ES. The stereochemistry of benzo[a]pyrene-2'-deoxyguanosine adducts affects DNA methylation by SssI and HhaI DNA methyltransferases. FEBS J. 2007;274:2121–2134. doi: 10.1111/j.1742-4658.2007.05754.x. [DOI] [PubMed] [Google Scholar]
- 61.Tao L, Li Y, Wang W, Kramer PM, Gunning WT, Lubet RA, Steele VE, Pereira MA. Effect of budesonide on the methylation and mRNA expression of the insulin-like growth factor 2 and c-myc genes in mouse lung tumors. Mol Carcinog. 2002;35:93–102. doi: 10.1002/mc.10078. [DOI] [PubMed] [Google Scholar]
- 62.Tommasi S, Dammann R, Zhang Z, Wang Y, Liu L, Tsark WM, Wilczynski SP, Li J, You M, Pfeifer GP. Tumor susceptibility of Rassf1a knockout mice. Cancer Res. 2005;65:92–98. [PubMed] [Google Scholar]
- 63.Tommasi S, Zheng A, Yoon JI, Besaratinia A. Epigenetic targeting of the Nanog pathway and signaling networks during chemical carcinogenesis. Carcinogenesis. 2014;35:1726–1736. doi: 10.1093/carcin/bgu026. [DOI] [PubMed] [Google Scholar]
- 64.Sadikovic B, Andrews J, Carter D, Robinson J, Rodenhiser DI. Genome-wide H3K9 histone acetylation profiles are altered in benzopyrene-treated MCF7 breast cancer cells. J Biol Chem. 2008;283:4051–4060. doi: 10.1074/jbc.M707506200. [DOI] [PubMed] [Google Scholar]
- 65.Khanal T, Kim D, Johnson A, Choubey D, Kim K. Deregulation of NR2E3, an orphan nuclear receptor, by benzo(a)pyrene-induced oxidative stress is associated with histone modification status change of the estrogen receptor gene promoter. Toxicol Lett. 2015;237:228–236. doi: 10.1016/j.toxlet.2015.06.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schnekenburger M, Peng L, Puga A. HDAC1 bound to the Cyp1a1 promoter blocks histone acetylation associated with Ah receptor-mediated trans-activation. Biochim Biophys Acta. 2007;1769:569–578. doi: 10.1016/j.bbaexp.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ovesen JL, Schnekenburger M, Puga A. Aryl hydrocarbon receptor ligands of widely different toxic equivalency factors induce similar histone marks in target gene chromatin. Toxicol Sci. 2011;121:123–131. doi: 10.1093/toxsci/kfr032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang J, Zhu H, Li C, Ding Y, Zhou Z, Wu Q. Neonatal exposure to benzo[a]pyrene decreases the levels of serum testosterone and histone H3K14 acetylation of the StAR promoter in the testes of SD rats. Toxicology. 2012;302:285–291. doi: 10.1016/j.tox.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 69.Mann DB, Springer DL, Smerdon MJ. DNA damage can alter the stability of nucleosomes: effects are dependent on damage type. Proc Natl Acad Sci U S A. 1997;94:2215–2220. doi: 10.1073/pnas.94.6.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barkley LR, Santocanale C. MicroRNA-29a regulates the benzo[a]pyrene dihydrodiol epoxide-induced DNA damage response through Cdc7 kinase in lung cancer cells. Oncogenesis. 2013;2:e57. doi: 10.1038/oncsis.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rieswijk L, Brauers KJ, Coonen ML, van Breda SG, Jennen DG, Kleinjans JC. Evaluating microRNA profiles reveals discriminative responses following genotoxic or non-genotoxic carcinogen exposure in primary mouse hepatocytes. Mutagenesis. 2015 doi: 10.1093/mutage/gev036. [DOI] [PubMed] [Google Scholar]
- 72.Caiment F, Gaj S, Claessen S, Kleinjans J. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic Acids Res. 2015;43:2525–2534. doi: 10.1093/nar/gkv115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marrone AK, Tryndyak V, Beland FA, Pogribny IP. MicroRNA Responses to the Genotoxic Carcinogens Aflatoxin B1 and Benzo[a]pyrene in Human HepaRG Cells. Toxicol Sci. 2016;149:496–502. doi: 10.1093/toxsci/kfv253. [DOI] [PubMed] [Google Scholar]
- 74.Choi YM, An S, Lee EM, Kim K, Choi SJ, Kim JS, Jang HH, An IS, Bae S. CYP1A1 is a target of miR-892a-mediated post-transcriptional repression. Int J Oncol. 2012;41:331–336. doi: 10.3892/ijo.2012.1418. [DOI] [PubMed] [Google Scholar]
- 75.Gordon MW, Yan F, Zhong X, Mazumder PB, Xu-Monette ZY, Zou D, Young KH, Ramos KS, Li Y. Regulation of p53-targeting microRNAs by polycyclic aromatic hydrocarbons: Implications in the etiology of multiple myeloma. Mol Carcinog. 2015;54:1060–1069. doi: 10.1002/mc.22175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han Z, Zhang Y, Xu Y, Ji J, Xu W, Zhao Y, Luo F, Wang B, Bian Q, Liu Q. Cell cycle changes mediated by the p53/miR-34c axis are involved in the malignant transformation of human bronchial epithelial cells by benzo[a]pyrene. Toxicol Lett. 2014;225:275–284. doi: 10.1016/j.toxlet.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 77.Han Z, Yang Q, Liu B, Wu J, Li Y, Yang C, Jiang Y. MicroRNA-622 functions as a tumor suppressor by targeting K-Ras and enhancing the anticarcinogenic effect of resveratrol. Carcinogenesis. 2012;33:131–139. doi: 10.1093/carcin/bgr226. [DOI] [PubMed] [Google Scholar]
- 78.Zhao Y, Liu H, Li Y, Wu J, Greenlee AR, Yang C, Jiang Y. The role of miR-506 in transformed 16HBE cells induced by anti-benzo[a]pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Toxicol Lett. 2011;205:320–326. doi: 10.1016/j.toxlet.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 79.Liu L, Jiang Y, Zhang H, Greenlee AR, Yu R, Yang Q. miR-22 functions as a micro-oncogene in transformed human bronchial epithelial cells induced by anti-benzo[a]pyrene-7,8-diol-9,10-epoxide. Toxicol In Vitro. 2010;24:1168–1175. doi: 10.1016/j.tiv.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 80.Liu L, Jiang Y, Zhang H, Greenlee AR, Han Z. Overexpressed miR-494 down-regulates PTEN gene expression in cells transformed by anti-benzo(a)pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Life Sci. 2010;86:192–198. doi: 10.1016/j.lfs.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 81.Li D, Wang Q, Liu C, Duan H, Zeng X, Zhang B, Li X, Zhao J, Tang S, Li Z, Xing X, Yang P, Chen L, Zeng J, Zhu X, Zhang S, Zhang Z, Ma L, He Z, Wang E, Xiao Y, Zheng Y, Chen W. Aberrant expression of miR-638 contributes to benzo(a)pyrene-induced human cell transformation. Toxicol Sci. 2012;125:382–391. doi: 10.1093/toxsci/kfr299. [DOI] [PubMed] [Google Scholar]
- 82.Gao L, Mai A, Li X, Lai Y, Zheng J, Yang Q, Wu J, Nan A, Ye S, Jiang Y. LncRNA-DQ786227-mediated cell malignant transformation induced by benzo(a)pyrene. Toxicol Lett. 2013;223:205–210. doi: 10.1016/j.toxlet.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 83.Yang Q, Zhang S, Liu H, Wu J, Xu E, Peng B, Jiang Y. Oncogenic role of long noncoding RNA AF118081 in anti-benzo[a]pyrene-trans-7,8-dihydrodiol-9,10-epoxide-transformed 16HBE cells. Toxicol Lett. 2014;229:430–439. doi: 10.1016/j.toxlet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 84.Hu G, Yang T, Zheng J, Dai J, Nan A, Lai Y, Zhang Y, Yang C, Jiang Y. Functional role and mechanism of lncRNA LOC728228 in malignant 16HBE cells transformed by anti-benzopyrene-trans-7,8-dihydrodiol-9,10-epoxide. Mol Carcinog. 2015;54(Suppl 1):E192–E204. doi: 10.1002/mc.22314. [DOI] [PubMed] [Google Scholar]
- 85.Gomaa AI, Khan SA, Toledano MB, Waked I, Taylor-Robinson SD. Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World J Gastroenterol. 2008;14:4300–4308. doi: 10.3748/wjg.14.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tam AS, Foley JF, Devereux TR, Maronpot RR, Massey TE. High frequency and heterogeneous distribution of p53 mutations in aflatoxin B1-induced mouse lung tumors. Cancer Res. 1999;59:3634–3640. [PubMed] [Google Scholar]
- 87.IARC. Chemical Agents and Related Occupations - A Review of Human Carcinogens. IARC Monogr Eval Carcinog Risks Hum. 2012;100F:1–567. [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang YJ, Ahsan H, Chen Y, Lunn RM, Wang LY, Chen SY, Lee PH, Chen CJ, Santella RM. High frequency of promoter hypermethylation of RASSF1A and p16 and its relationship to aflatoxin B1-DNA adduct levels in human hepatocellular carcinoma. Mol Carcinog. 2002;35:85–92. doi: 10.1002/mc.10076. [DOI] [PubMed] [Google Scholar]
- 89.Zhang YJ, Chen Y, Ahsan H, Lunn RM, Lee PH, Chen CJ, Santella RM. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation and its relationship to aflatoxin B1-DNA adducts and p53 mutation in hepatocellular carcinoma. Int J Cancer. 2003;103:440–444. doi: 10.1002/ijc.10852. [DOI] [PubMed] [Google Scholar]
- 90.Feng Y, Xue WJ, Li P, Sha ZY, Huang H, Rui L, Li HX, Mao QS. RASSF1A hypermethylation is associated with aflatoxin B1 and polycyclic aromatic hydrocarbon exposure in hepatocellular carcinoma. Hepatogastroenterology. 2012;59:1883–1888. doi: 10.5754/hge11731. [DOI] [PubMed] [Google Scholar]
- 91.Zhang YJ, Chen Y, Ahsan H, Lunn RM, Chen SY, Lee PH, Chen CJ, Santella RM. Silencing of glutathione S-transferase P1 by promoter hypermethylation and its relationship to environmental chemical carcinogens in hepatocellular carcinoma. Cancer Lett. 2005;221:135–143. doi: 10.1016/j.canlet.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 92.Wu HC, Wang Q, Yang HI, Tsai WY, Chen CJ, Santella RM. Global DNA methylation in a population with aflatoxin B1 exposure. Epigenetics. 2013;8:962–969. doi: 10.4161/epi.25696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang YJ, Wu HC, Yazici H, Yu MW, Lee PH, Santella RM. Global hypomethylation in hepatocellular carcinoma and its relationship to aflatoxin B(1) exposure. World J Hepatol. 2012;4:169–175. doi: 10.4254/wjh.v4.i5.169.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hernandez-Vargas H, Castelino J, Silver MJ, Dominguez-Salas P, Cros MP, Durand G, Calvez-Kelm FL, Prentice AM, Wild CP, Moore SE, Hennig BJ, Herceg Z, Gong YY, Routledge MN. Exposure to aflatoxin B1 in utero is associated with DNA methylation in white blood cells of infants in The Gambia. Int J Epidemiol. 2015;44:1238–1248. doi: 10.1093/ije/dyv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tam AS, Devereux TR, Patel AC, Foley JF, Maronpot RR, Massey TE. Perturbations of the Ink4a/Arf gene locus in aflatoxin B1-induced mouse lung tumors. Carcinogenesis. 2003;24:121–132. doi: 10.1093/carcin/24.1.121. [DOI] [PubMed] [Google Scholar]
- 96.Zhu CC, Hou YJ, Han J, Liu HL, Cui XS, Kim NH, Sun SC. Effect of mycotoxin-containing diets on epigenetic modifications of mouse oocytes by fluorescence microscopy analysis. Microsc Microanal. 2014;20:1158–1166. doi: 10.1017/S1431927614000919. [DOI] [PubMed] [Google Scholar]
- 97.Liu J, Wang QC, Han J, Xiong B, Sun SC. Aflatoxin B1 is toxic to porcine oocyte maturation. Mutagenesis. 2015;30:527–535. doi: 10.1093/mutage/gev015. [DOI] [PubMed] [Google Scholar]
- 98.Baik JH, Griffiths S, Giuili G, Manson M, Siegrist S, Guellaen G. DNA methylation patterns of the rat gamma-glutamyl transpeptidase gene in embryonic, adult and neoplastic liver. Carcinogenesis. 1991;12:1035–1039. doi: 10.1093/carcin/12.6.1035. [DOI] [PubMed] [Google Scholar]
- 99.Chan KT, Hsieh DP, Lung ML. In vitro aflatoxin B1-induced p53 mutations. Cancer Lett. 2003;199:1–7. doi: 10.1016/s0304-3835(03)00337-9. [DOI] [PubMed] [Google Scholar]
- 100.Liu C, Yu H, Zhang Y, Li D, Xing X, Chen L, Zeng X, Xu D, Fan Q, Xiao Y, Chen W, Wang Q. Upregulation of miR-34a-5p antagonizes AFB1-induced genotoxicity in F344 rat liver. Toxicon. 2015;106:46–56. doi: 10.1016/j.toxicon.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 101.Yang W, Lian J, Feng Y, Srinivas S, Guo Z, Zhong H, Zhuang Z, Wang S. Genome-wide miRNA-profiling of aflatoxin B1-induced hepatic injury using deep sequencing. Toxicol Lett. 2014;226:140–149. doi: 10.1016/j.toxlet.2014.01.021. [DOI] [PubMed] [Google Scholar]
- 102.Huang XY, Yao JG, Huang HD, Wang C, Ma Y, Xia Q, Long XD. MicroRNA-429 Modulates Hepatocellular Carcinoma Prognosis and Tumorigenesis. Gastroenterol Res Pract. 2013;2013:804128. doi: 10.1155/2013/804128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luzi E, Marini F, Giusti F, Galli G, Cavalli L, Brandi ML. The negative feedback-loop between the oncomir Mir-24-1 and menin modulates the Men1 tumorigenesis by mimicking the "Knudson's second hit". PLoS One. 2012;7:e39767. doi: 10.1371/journal.pone.0039767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu YX, Long XD, Xi ZF, Ma Y, Huang XY, Yao JG, Wang C, Xing TY, Xia Q. MicroRNA-24 modulates aflatoxin B1-related hepatocellular carcinoma prognosis and tumorigenesis. Biomed Res Int. 2014;2014:482926. doi: 10.1155/2014/482926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Long XD, Huang XY, Yao JG, Liao P, Tang YJ, Ma Y, Xia Q. Polymorphisms in the precursor microRNAs and aflatoxin B1-related hepatocellular carcinoma. Mol Carcinog. 2015 doi: 10.1002/mc.22350. [DOI] [PubMed] [Google Scholar]
- 106.Lv J, Yu YQ, Li SQ, Luo L, Wang Q. Aflatoxin B1 promotes cell growth and invasion in hepatocellular carcinoma HepG2 cells through H19 and E2F1. Asian Pac J Cancer Prev. 2014;15:2565–2570. doi: 10.7314/apjcp.2014.15.6.2565. [DOI] [PubMed] [Google Scholar]
- 107.Infante PF, Rinsky RA, Wagoner JK, Young RJ. Leukaemia in benzene workers. Lancet. 1977;2:76–78. doi: 10.1016/s0140-6736(77)90074-5. [DOI] [PubMed] [Google Scholar]
- 108.Infante PF. Benzene exposure and multiple myeloma: a detailed meta-analysis of benzene cohort studies. Ann N Y Acad Sci. 2006;1076:90–109. doi: 10.1196/annals.1371.081. [DOI] [PubMed] [Google Scholar]
- 109.Tough IM, Brown WM. Chromosome Aberrations and Exposure to Ambient Benzene. Lancet. 1965;1:684. doi: 10.1016/s0140-6736(65)91835-0. [DOI] [PubMed] [Google Scholar]
- 110.Zhang L, Rothman N, Wang Y, Hayes RB, Yin S, Titenko-Holland N, Dosemeci M, Wang YZ, Kolachana P, Lu W, Xi L, Li GL, Smith MT. Benzene increases aneuploidy in the lymphocytes of exposed workers: a comparison of data obtained by fluorescence in situ hybridization in interphase and metaphase cells. Environ Mol Mutagen. 1999;34:260–268. [PubMed] [Google Scholar]
- 111.Lau A, Belanger CL, Winn LM. In utero and acute exposure to benzene: investigation of DNA double-strand breaks and DNA recombination in mice. Mutat Res. 2009;676:74–82. doi: 10.1016/j.mrgentox.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 112.Tabish AM, Poels K, Hoet P, Godderis L. Epigenetic factors in cancer risk: effect of chemical carcinogens on global DNA methylation pattern in human TK6 cells. PLoS One. 2012;7:e34674. doi: 10.1371/journal.pone.0034674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hu J, Ma H, Zhang W, Yu Z, Sheng G, Fu J. Effects of benzene and its metabolites on global DNA methylation in human normal hepatic L02 cells. Environ Toxicol. 2014;29:108–116. doi: 10.1002/tox.20777. [DOI] [PubMed] [Google Scholar]
- 114.Nishikawa T, Izumo K, Miyahara E, Horiuchi M, Okamoto Y, Kawano Y, Takeuchi T. Benzene induces cytotoxicity without metabolic activation. J Occup Health. 2011;53:84–92. doi: 10.1539/joh.10-002-oa. [DOI] [PubMed] [Google Scholar]
- 115.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, Bertazzi PA, Yang AS. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 116.Fustinoni S, Rossella F, Polledri E, Bollati V, Campo L, Byun HM, Agnello L, Consonni D, Pesatori AC, Baccarelli A, Bertazzi PA. Global DNA methylation and low-level exposure to benzene. Med Lav. 2012;103:84–95. [PubMed] [Google Scholar]
- 117.Yang J, Bai W, Niu P, Tian L, Gao A. Aberrant hypomethylated STAT3 was identified as a biomarker of chronic benzene poisoning through integrating DNA methylation and mRNA expression data. Exp Mol Pathol. 2014;96:346–353. doi: 10.1016/j.yexmp.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 118.Xing C, Wang QF, Li B, Tian H, Ni Y, Yin S, Li G. Methylation and expression analysis of tumor suppressor genes p15 and p16 in benzene poisoning. Chem Biol Interact. 2010;184:306–309. doi: 10.1016/j.cbi.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 119.Philbrook NA, Winn LM. Investigating the effects of in utero benzene exposure on epigenetic modifications in maternal and fetal CD-1 mice. Toxicol Appl Pharmacol. 2015 doi: 10.1016/j.taap.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 120.Gao A, Zuo X, Song S, Guo W, Tian L. Epigenetic modification involved in benzene-induced apoptosis through regulating apoptosis-related genes expression. Cell Biol Int. 2011;35:391–396. doi: 10.1042/CBI20100256. [DOI] [PubMed] [Google Scholar]
- 121.Yang J, Zuo X, Bai W, Niu P, Tian L, Gao A. PTEN methylation involved in benzene-induced hematotoxicity. Exp Mol Pathol. 2014;96:300–306. doi: 10.1016/j.yexmp.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 122.Gao A, Zuo X, Liu Q, Lu X, Guo W, Tian L. Methylation of PARP-1 promoter involved in the regulation of benzene-induced decrease of PARP-1 mRNA expression. Toxicol Lett. 2010;195:114–118. doi: 10.1016/j.toxlet.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 123.Yu K, Shi YF, Yang KY, Zhuang Y, Zhu RH, Xu X, Cai G. Decreased topoisomerase IIalpha expression and altered histone and regulatory factors of topoisomerase IIalpha promoter in patients with chronic benzene poisoning. Toxicol Lett. 2011;203:111–117. doi: 10.1016/j.toxlet.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 124.Bai W, Chen Y, Yang J, Niu P, Tian L, Gao A. Aberrant miRNA profiles associated with chronic benzene poisoning. Exp Mol Pathol. 2014;96:426–430. doi: 10.1016/j.yexmp.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 125.Herberth G, Bauer M, Gasch M, Hinz D, Roder S, Olek S, Kohajda T, Rolle-Kampczyk U, von Bergen M, Sack U, Borte M, Lehmann I. Lifestyle, F. Environmental, g. Their Influence on Newborns Allergy Risk study, Maternal and cord blood miR-223 expression associates with prenatal tobacco smoke exposure and low regulatory T-cell numbers. J Allergy Clin Immunol. 2014;133:543–550. doi: 10.1016/j.jaci.2013.06.036. [DOI] [PubMed] [Google Scholar]
- 126.Wei H, Zhang J, Tan K, Sun R, Yin L, Pu Y. Benzene-Induced Aberrant miRNA Expression Profile in Hematopoietic Progenitor Cells in C57BL/6 Mice. Int J Mol Sci. 2015;16:27058–27071. doi: 10.3390/ijms161126001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bai W, Yang J, Yang G, Niu P, Tian L, Gao A. Long non-coding RNA NR_045623 and NR_028291 involved in benzene hematotoxicity in occupationally benzene-exposed workers. Exp Mol Pathol. 2014;96:354–360. doi: 10.1016/j.yexmp.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 128.IARC. Formaldehyde, 2-butoxyethanol and 1-tert-butoxypropan-2-ol. IARC Monogr Eval Carcinog Risks Hum. 2006;88:1–478. [PMC free article] [PubMed] [Google Scholar]
- 129.Costa S, Coelho P, Costa C, Silva S, Mayan O, Santos LS, Gaspar J, Teixeira JP. Genotoxic damage in pathology anatomy laboratory workers exposed to formaldehyde. Toxicology. 2008;252:40–48. doi: 10.1016/j.tox.2008.07.056. [DOI] [PubMed] [Google Scholar]
- 130.Ying CJ, Ye XL, Xie H, Yan WS, Zhao MY, Xia T, Yin SY. Lymphocyte subsets and sister-chromatid exchanges in the students exposed to formaldehyde vapor. Biomed Environ Sci. 1999;12:88–94. [PubMed] [Google Scholar]
- 131.Ye X, Yan W, Xie H, Zhao M, Ying C. Cytogenetic analysis of nasal mucosa cells and lymphocytes from high-level long-term formaldehyde exposed workers and low-level short-term exposed waiters. Mutat Res. 2005;588:22–27. doi: 10.1016/j.mrgentox.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 132.Speit G, Schutz P, Hogel J, Schmid O. Characterization of the genotoxic potential of formaldehyde in V79 cells. Mutagenesis. 2007;22:387–394. doi: 10.1093/mutage/gem031. [DOI] [PubMed] [Google Scholar]
- 133.Schmid O, Speit G. Genotoxic effects induced by formaldehyde in human blood and implications for the interpretation of biomonitoring studies. Mutagenesis. 2007;22:69–74. doi: 10.1093/mutage/gel053. [DOI] [PubMed] [Google Scholar]
- 134.Liu Q, Yang L, Gong C, Tao G, Huang H, Liu J, Zhang H, Wu D, Xia B, Hu G, Wang K, Zhuang Z. Effects of long-term low-dose formaldehyde exposure on global genomic hypomethylation in 16HBE cells. Toxicol Lett. 2011;205:235–240. doi: 10.1016/j.toxlet.2011.05.1039. [DOI] [PubMed] [Google Scholar]
- 135.Ibuki Y, Toyooka T, Zhao X, Yoshida I. Cigarette sidestream smoke induces histone H3 phosphorylation via JNK and PI3K/Akt pathways, leading to the expression of proto-oncogenes. Carcinogenesis. 2014;35:1228–1237. doi: 10.1093/carcin/bgt492. [DOI] [PubMed] [Google Scholar]
- 136.Yoshida I, Ibuki Y. Formaldehyde-induced histone H3 phosphorylation via JNK and the expression of proto-oncogenes. Mutat Res. 2014;770:9–18. doi: 10.1016/j.mrfmmm.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 137.Lu K, Boysen G, Gao L, Collins LB, Swenberg JA. Formaldehyde-induced histone modifications in vitro. Chem Res Toxicol. 2008;21:1586–1593. doi: 10.1021/tx8000576. [DOI] [PubMed] [Google Scholar]
- 138.Rager JE, Smeester L, Jaspers I, Sexton KG, Fry RC. Epigenetic changes induced by air toxics: formaldehyde exposure alters miRNA expression profiles in human lung cells. Environ Health Perspect. 2011;119:494–500. doi: 10.1289/ehp.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li G, Yang J, Ling S. Formaldehyde exposure alters miRNA expression profiles in the olfactory bulb. Inhal Toxicol. 2015;27:387–393. doi: 10.3109/08958378.2015.1062580. [DOI] [PubMed] [Google Scholar]
- 140.Rager JE, Moeller BC, Doyle-Eisele M, Kracko D, Swenberg JA, Fry RC. Formaldehyde and epigenetic alterations: microRNA changes in the nasal epithelium of nonhuman primates. Environ Health Perspect. 2013;121:339–344. doi: 10.1289/ehp.1205582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rager JE, Moeller BC, Miller SK, Kracko D, Doyle-Eisele M, Swenberg JA, Fry RC. Formaldehyde-associated changes in microRNAs: tissue and temporal specificity in the rat nose, white blood cells, and bone marrow. Toxicol Sci. 2014;138:36–46. doi: 10.1093/toxsci/kft267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.IARC. Polynuclear aromatic compounds, Part 1, Chemical, environmental and experimental data. IARC Monogr Eval Carcinog Risk Chem Hum. 1983;32:1–453. [PubMed] [Google Scholar]
- 143.Popp W, Vahrenholz C, Schell C, Grimmer G, Dettbarn G, Kraus R, Brauksiepe A, Schmeling B, Gutzeit T, von Bulow J, Norpoth K. DNA single strand breakage, DNA adducts, and sister chromatid exchange in lymphocytes and phenanthrene and pyrene metabolites in urine of coke oven workers. Occup Environ Med. 1997;54:176–183. doi: 10.1136/oem.54.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu AL, Lu WQ, Wang ZZ, Chen WH, Lu WH, Yuan J, Nan PH, Sun JY, Zou YL, Zhou LH, Zhang C, Wu TC. Elevated levels of urinary 8-hydroxy-2 -deoxyguanosine, lymphocytic micronuclei, and serum glutathione S-transferase in workers exposed to coke oven emissions. Environ Health Perspect. 2006;114:673–677. doi: 10.1289/ehp.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pavanello S, Kapka L, Siwinska E, Mielzynska D, Bolognesi C, Clonfero E. Micronuclei related to anti-B[a]PDE-DNA adduct in peripheral blood lymphocytes of heavily polycyclic aromatic hydrocarbon-exposed nonsmoking coke-oven workers and controls. Cancer Epidemiol Biomarkers Prev. 2008;17:2795–2799. doi: 10.1158/1055-9965.EPI-08-0346. [DOI] [PubMed] [Google Scholar]
- 146.Chao MR, Wang CJ, Wu MT, Pan CH, Kuo CY, Yang HJ, Chang LW, Hu CW. Repeated measurements of urinary methylated/oxidative DNA lesions, acute toxicity, and mutagenicity in coke oven workers. Cancer Epidemiol Biomarkers Prev. 2008;17:3381–3389. doi: 10.1158/1055-9965.EPI-08-0721. [DOI] [PubMed] [Google Scholar]
- 147.Zhang H, Li X, Ge L, Yang J, Sun J, Niu Q. Methylation of CpG island of p14(ARK), p15(INK4b) and p16(INK4a) genes in coke oven workers. Hum Exp Toxicol. 2015;34:191–197. doi: 10.1177/0960327114533576. [DOI] [PubMed] [Google Scholar]
- 148.Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolognesi C, Bertazzi PA, Baccarelli A. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer. 2009;125:1692–1697. doi: 10.1002/ijc.24492. [DOI] [PubMed] [Google Scholar]
- 149.Pavanello S, Pesatori AC, Dioni L, Hoxha M, Bollati V, Siwinska E, Mielzynska D, Bolognesi C, Bertazzi PA, Baccarelli A. Shorter telomere length in peripheral blood lymphocytes of workers exposed to polycyclic aromatic hydrocarbons. Carcinogenesis. 2010;31:216–221. doi: 10.1093/carcin/bgp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Duan H, He Z, Ma J, Zhang B, Sheng Z, Bin P, Cheng J, Niu Y, Dong H, Lin H, Dai Y, Zhu B, Chen W, Xiao Y, Zheng Y. Global and MGMT promoter hypomethylation independently associated with genomic instability of lymphocytes in subjects exposed to high-dose polycyclic aromatic hydrocarbon. Archives of Toxicology. 2013;87:2013–2022. doi: 10.1007/s00204-013-1046-0. [DOI] [PubMed] [Google Scholar]
- 151.Deng Q, Huang S, Zhang X, Zhang W, Feng J, Wang T, Hu D, Guan L, Li J, Dai X, Deng H, Zhang X, Wu T. Plasma microRNA expression and micronuclei frequency in workers exposed to polycyclic aromatic hydrocarbons. Environ Health Perspect. 2014;122:719–725. doi: 10.1289/ehp.1307080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Melnick RL, Huff J, Barrett JC, Maronpot RR, Lucier G, Portier CJ. Cell proliferation and chemical carcinogenesis: Symposium overview. Environmental Health Perspectives. 1993;101:3–7. doi: 10.1289/ehp.93101s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Huff JE, Melnick RL, Solleveld HA, Haseman JK, Powers M, Miller RA. Multiple organ carcinogenicity of 1,3-butadiene in B6C3F1 mice after 60 weeks of inhalation exposure. Science. 1985;227:548–549. doi: 10.1126/science.3966163. [DOI] [PubMed] [Google Scholar]
- 154.Jackson TE, Lilly PD, Recio L, Schlosser PM, Medinsky MA. Inhibition of cytochrome P450 2E1 decreases, but does not eliminate, genotoxicity mediated by 1,3-butadiene. Toxicol Sci. 2000;55:266–273. doi: 10.1093/toxsci/55.2.266. [DOI] [PubMed] [Google Scholar]
- 155.Sills RC, Hong HL, Boorman GA, Devereux TR, Melnick RL. Point mutations of K-ras and H-ras genes in forestomach neoplasms from control B6C3F1 mice and following exposure to 1,3-butadiene, isoprene or chloroprene for up to 2-years. Chem Biol Interact. 2001;135–136:373–386. doi: 10.1016/s0009-2797(01)00179-x. [DOI] [PubMed] [Google Scholar]
- 156.Lee DH, Kim TH, Lee SY, Kim HJ, Rhee SK, Yoon B, Pfeifer GP, Lee CS. Mutations induced by 1,3-butadiene metabolites, butadiene diolepoxide, and 1,2,3,4-diepoxybutane at the Hprt locus in CHO-K1 cells. Mol Cells. 2002;14:411–419. [PubMed] [Google Scholar]
- 157.Abdel-Rahman SZ, Ammenheuser MM, Ward JB., Jr Human sensitivity to 1,3-butadiene: role of microsomal epoxide hydrolase polymorphisms. Carcinogenesis. 2001;22:415–423. doi: 10.1093/carcin/22.3.415. [DOI] [PubMed] [Google Scholar]
- 158.Abdel-Rahman SZ, El-Zein RA, Ammenheuser MM, Yang Z, Stock TH, Morandi M, Ward JB., Jr Variability in human sensitivity to 1,3-butadiene: Influence of the allelic variants of the microsomal epoxide hydrolase gene. Environ Mol Mutagen. 2003;41:140–146. doi: 10.1002/em.10142. [DOI] [PubMed] [Google Scholar]
- 159.Ton TV, Hong HH, Devereux TR, Melnick RL, Sills RC, Kim Y. Evaluation of genetic alterations in cancer-related genes in lung and brain tumors from B6C3F1 mice exposed to 1,3-butadiene or chloroprene. Chem Biol Interact. 2007;166:112–120. doi: 10.1016/j.cbi.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 160.Koturbash I, Scherhag A, Sorrentino J, Sexton K, Bodnar W, Tryndyak V, Latendresse JR, Swenberg JA, Beland FA, Pogribny IP, Rusyn I. Epigenetic alterations in liver of C57BL/6J mice after short-term inhalational exposure to 1,3-butadiene. Environ Health Perspect. 2011;119:635–640. doi: 10.1289/ehp.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Koturbash I, Scherhag A, Sorrentino J, Sexton K, Bodnar W, Swenberg JA, Beland FA, Pardo-Manuel Devillena F, Rusyn I, Pogribny IP. Epigenetic mechanisms of mouse interstrain variability in genotoxicity of the environmental toxicant 1,3-butadiene. Toxicol Sci. 2011;122:448–456. doi: 10.1093/toxsci/kfr133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chappell G, Kobets T, O'Brien B, Tretyakova N, Sangaraju D, Kosyk O, Sexton KG, Bodnar W, Pogribny IP, Rusyn I. Epigenetic events determine tissue-specific toxicity of inhalational exposure to the genotoxic chemical 1,3-butadiene in male C57BL/6J mice. Toxicol Sci. 2014;142:375–384. doi: 10.1093/toxsci/kfu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yang H, Mizzen CA. The multiple facets of histone H4-lysine 20 methylation. Biochem Cell Biol. 2009;87:151–161. doi: 10.1139/O08-131. [DOI] [PubMed] [Google Scholar]
- 164.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 165.Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, Yoon YS, Ren B, He C, Jin P. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yamakido M, Ishioka S, Hiyama K, Maeda A. Former poison gas workers and cancer: incidence and inhibition of tumor formation by treatment with biological response modifier N-CWS. Environ Health Perspect. 1996;104(Suppl 3):485–488. doi: 10.1289/ehp.96104s3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nishimoto Y, Yamakido M, Shigenobu T, Onari K, Yukutake M. Long-term observation of poison gas workers with special reference to respiratory cancers. J UOEH. 1983;5(Suppl):89–94. [PubMed] [Google Scholar]
- 168.Heston WE. Pulmonary tumors in strain A mice exposed to mustard gas. Proc Soc Exp Biol Med. 1953;82:457–460. doi: 10.3181/00379727-82-20146. [DOI] [PubMed] [Google Scholar]
- 169.Heston WE. Occurrence of tumors in mice injected subcutaneously with sulfur mustard and nitrogen mustard. J Natl Cancer Inst. 1953;14:131–140. [PubMed] [Google Scholar]
- 170.Lin P, Vaughan FL, Bernstein IA. Formation of interstrand DNA cross-links by bis-(2-chloroethyl)sulfide (BCES): a possible cytotoxic mechanism in rat keratinocytes. Biochem Biophys Res Commun. 1996;218:556–561. doi: 10.1006/bbrc.1996.0099. [DOI] [PubMed] [Google Scholar]
- 171.Shahin S, Cullinane C, Gray PJ. Mitochondrial and nuclear DNA damage induced by sulphur mustard in keratinocytes. Chem Biol Interact. 2001;138:231–245. doi: 10.1016/s0009-2797(01)00275-7. [DOI] [PubMed] [Google Scholar]
- 172.Roberts JJ, Brent TP, Crathorn AR. Evidence for the inactivation and repair of the mammalian DNA template after alkylation by mustard gas and half mustard gas. Eur J Cancer. 1971;7:515–524. doi: 10.1016/0014-2964(71)90056-9. [DOI] [PubMed] [Google Scholar]
- 173.IARC. IARC monographs on the evaluation of the carcinogenic risk of chemicals to man: some aziridines N-, S- & O-mustards and selenium. IARC Monogr Eval Carcinog Risk Chem Man. 1975;9:1–268. [PubMed] [Google Scholar]
- 174.Steinritz D, Schmidt A, Balszuweit F, Thiermann H, Simons T, Striepling E, Bolck B, Bloch W. Epigenetic modulations in early endothelial cells and DNA hypermethylation in human skin after sulfur mustard exposure. Toxicol Lett. 2015 doi: 10.1016/j.toxlet.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 175.Deppe J, Steinritz D, Santovito D, Egea V, Schmidt A, Weber C, Ries C. Upregulation of miR-203 and miR-210 affect growth and differentiation of keratinocytes after exposure to sulfur mustard in normoxia and hypoxia. Toxicol Lett. 2015 doi: 10.1016/j.toxlet.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 176.Schmidt A, Steinritz D, Thiermann H, Meineke V, Abend M. Alteration of miRNA expression in early endothelial cells after exposure with sub-lethal sulfur mustard concentrations. Toxicol Lett. 2015 doi: 10.1016/j.toxlet.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 177.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Guengerich FP, Mason PS, Stott WT, Fox TR, Watanabe PG. Roles of 2-haloethylene oxides and 2-haloacetaldehydes derived from vinyl bromide and vinyl chloride in irreversible binding to protein and DNA. Cancer Res. 1981;41:4391–4398. [PubMed] [Google Scholar]
- 179.Guengerich FP. Roles of the vinyl chloride oxidation products 1-chlorooxirane and 2-chloroacetaldehyde in the in vitro formation of etheno adducts of nucleic acid bases [corrected] Chem Res Toxicol. 1992;5:2–5. doi: 10.1021/tx00025a001. [DOI] [PubMed] [Google Scholar]
- 180.Maltoni C, Lefemine G, Chieco P, Carretti D. Vinyl chloride carcinogenesis: current results and perspectives. Med. Lav. 1974;65:421–444. [PubMed] [Google Scholar]
- 181.IARC. IARC monographs on the evaluation of carcinogenic risks to humans. Volume 97. 1,3-butadiene, ethylene oxide and vinyl halides (vinyl fluoride, vinyl chloride and vinyl bromide) IARC Monogr Eval Carcinog Risks Hum. 2008;97:3–471. [PMC free article] [PubMed] [Google Scholar]
- 182.Cheng KC, Preston BD, Cahill DS, Dosanjh MK, Singer B, Loeb LA. The vinyl chloride DNA derivative N2,3-ethenoguanine produces G----A transitions in Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:9974–9978. doi: 10.1073/pnas.88.22.9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Weihrauch M, Markwarth A, Lehnert G, Wittekind C, Wrbitzky R, Tannapfel A. Abnormalities of the ARF-p53 pathway in primary angiosarcomas of the liver. Hum Pathol. 2002;33:884–892. doi: 10.1053/hupa.2002.126880. [DOI] [PubMed] [Google Scholar]
- 184.Weihrauch M, Benicke M, Lehnert G, Wittekind C, Wrbitzky R, Tannapfel A. Frequent k- ras −2 mutations and p16(INK4A)methylation in hepatocellular carcinomas in workers exposed to vinyl chloride. Br J Cancer. 2001;84:982–989. doi: 10.1054/bjoc.2000.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Beland FA, Beranek DT, Dooley KL, Heflich RH, Kadlubar FF. Arylamine-DNA adducts in vitro and in vivo: their role in bacterial mutagenesis and urinary bladder carcinogenesis. Environ Health Perspect. 1983;49:125–134. doi: 10.1289/ehp.8349125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lin D, Lay JO, Jr, Bryant MS, Malaveille C, Friesen M, Bartsch H, Lang NP, Kadlubar FF. Analysis of 4-aminobiphenyl-DNA adducts in human urinary bladder and lung by alkaline hydrolysis and negative ion gas chromatography-mass spectrometry. Environ Health Perspect. 1994;102(Suppl 6):11–16. doi: 10.1289/ehp.94102s611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Reimann F, Erdogan G. Malformations, anomalies and variations in patients with severe iron deficiency. Blut. 1976;32:423–432. doi: 10.1007/BF01013882. [DOI] [PubMed] [Google Scholar]
- 188.Bradley C, van der Meer R, Roodi N, Yan H, Chandrasekharan MB, Sun ZW, Mernaugh RL, Parl FF. Carcinogen-induced histone alteration in normal human mammary epithelial cells. Carcinogenesis. 2007;28:2184–2192. doi: 10.1093/carcin/bgm100. [DOI] [PubMed] [Google Scholar]
- 189.Huan LC, Wu JC, Chiou BH, Chen CH, Ma N, Chang CY, Tsen YK, Chen SC. MicroRNA regulation of DNA repair gene expression in 4-aminobiphenyl-treated HepG2 cells. Toxicology. 2014;322:69–77. doi: 10.1016/j.tox.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 190.Mirkova ET, Lalchev SG. The genetic toxicity of the human carcinogens benzidine and benzidine-based dyes: chromosomal analysis in exposed workers. Prog Clin Biol Res. 1990;340C:397–405. [PubMed] [Google Scholar]
- 191.Xiang CQ, Shen CL, Wu ZR, Qin YQ, Zhang YY, Liu CZ, Chen JG, Zhang SN. Detection of mutant p53 protein in workers occupationally exposed to benzidine. J Occup Health. 2007;49:279–284. doi: 10.1539/joh.49.279. [DOI] [PubMed] [Google Scholar]
- 192.Rothman N, Bhatnagar VK, Hayes RB, Zenser TV, Kashyap SK, Butler MA, Bell DA, Lakshmi V, Jaeger M, Kashyap R, Hirvonen A, Schulte PA, Dosemeci M, Hsu F, Parikh DJ, Davis BB, Talaska G. The impact of interindividual variation in NAT2 activity on benzidine urinary metabolites and urothelial DNA adducts in exposed workers. Proc Natl Acad Sci U S A. 1996;93:5084–5089. doi: 10.1073/pnas.93.10.5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vorce RL, Goodman JI. Altered methylation of ras oncogenes in benzidine-induced B6C3F1 mouse liver tumors. Toxicol Appl Pharmacol. 1989;100:398–410. doi: 10.1016/0041-008x(89)90288-3. [DOI] [PubMed] [Google Scholar]
- 194.Stula EF, Barnes JR, Sherman H, Reinhardt CF, Zapp JA., Jr Urinary bladder tumors in dogs from 4,4'-methylene-bis (2-chloroaniline) (MOCA) J Environ Pathol Toxicol. 1978;1:31–50. [PubMed] [Google Scholar]
- 195.Kaderlik KR, Talaska G, DeBord DG, Osorio AM, Kadlubar FF. 4,4'-Methylene-bis(2-chloroaniline)-DNA adduct analysis in human exfoliated urothelial cells by 32P-postlabeling. Cancer Epidemiol Biomarkers Prev. 1993;2:63–69. [PubMed] [Google Scholar]
- 196.Murray EB, Edwards JW. Micronuclei in peripheral lymphocytes and exfoliated urothelial cells of workers exposed to 4,4'-methylenebis-(2-chloroaniline) (MOCA) Mutat Res. 1999;446:175–180. doi: 10.1016/s1383-5718(99)00180-1. [DOI] [PubMed] [Google Scholar]
- 197.Silk NA, Lay JO, Jr, Martin CN. Covalent binding of 4,4'-methylenebis-(2-chloroaniline) to rat liver DNA in vivo and of its N-hydroxylated derivative to DNA in vitro. Biochem Pharmacol. 1989;38:279–287. doi: 10.1016/0006-2952(89)90038-5. [DOI] [PubMed] [Google Scholar]
- 198.DeBord DG, Cheever KL, Booth-Jones AD, Swearengin TF, Savage RE., Jr Alterations of histone phosphorylation in rat spleen cells after treatment with the aromatic amine, 4,4'-methylene-bis(2-chloroaniline) J Biochem Toxicol. 1995;10:19–23. [PubMed] [Google Scholar]
- 199.Pinto D, Ceballos JM, Garcia G, Guzman P, Del Razo LM, Vera E, Gomez H, Garcia A, Gonsebatt ME. Increased cytogenetic damage in outdoor painters. Mutat Res. 2000;467:105–111. doi: 10.1016/s1383-5718(00)00024-3. [DOI] [PubMed] [Google Scholar]
- 200.Testa A, Festa F, Ranaldi R, Giachelia M, Tirindelli D, De Marco A, Owczarek M, Guidotti M, Cozzi R. A multi-biomarker analysis of DNA damage in automobile painters. Environ Mol Mutagen. 2005;46:182–188. doi: 10.1002/em.20147. [DOI] [PubMed] [Google Scholar]
- 201.IARC. Painting, firefighting, and shiftwork. IARC Monogr Eval Carcinog Risks Hum. 2010;98:9–764. [PMC free article] [PubMed] [Google Scholar]
- 202.Hoyos-Giraldo LS, Escobar-Hoyos LF, Saavedra-Trujillo D, Reyes-Carvajal I, Munoz A, Londono-Velasco E, Tello A, Cajas-Salazar N, Ruiz M, Carvajal S, Santella RM. Gene-specific promoter methylation is associated with micronuclei frequency in urothelial cells from individuals exposed to organic solvents and paints. J Expo Sci Environ Epidemiol. 2015 doi: 10.1038/jes.2015.28. [DOI] [PubMed] [Google Scholar]
- 203.Ray PD, Yosim A, Fry RC. Incorporating epigenetic data into the risk assessment process for the toxic metals arsenic, cadmium, chromium, lead, and mercury: strategies and challenges. Front Genet. 2014;5:201. doi: 10.3389/fgene.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Siddeek B, Inoubli L, Lakhdari N, Rachel PB, Fussell KC, Schneider S, Mauduit C, Benahmed M. MicroRNAs as potential biomarkers in diseases and toxicology. Mutat Res Genet Toxicol Environ Mutagen. 2014;764–765:46–57. doi: 10.1016/j.mrgentox.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 205.de Conti A, Kobets T, Tryndyak V, Burnett SD, Han T, Fuscoe JC, Beland FA, Doerge DR, Pogribny IP. Persistence of furan-induced epigenetic aberrations in the livers of F344 rats. Toxicol Sci. 2015;144:217–226. doi: 10.1093/toxsci/kfu313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H, Jaenisch R. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc.Natl.Acad Sci U.S.A. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 208.Chen HJ, Zhang L, Cox J, Cunningham JA, Chung FL. DNA adducts of 2,3-epoxy-4-hydroxynonanal: detection of 7-(1', 2'-dihydroxyheptyl)-3H-imidazo[2,1-i]purine and 1,N6-ethenoadenine by gas chromatography/negative ion chemical ionization/mass spectrometry. Chem Res Toxicol. 1998;11:1474–1480. doi: 10.1021/tx980107o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.