Abstract
Why do some host-parasite interactions become less antagonistic over evolutionary time? Vertical transmission can select for reduced antagonism. Vertical transmission also promotes coevolution between hosts and parasites. Therefore, we hypothesized that coevolution itself may underlie transitions to reduced antagonism. To test the coevolution hypothesis, we selected for reduced antagonism between the host Caenorhabditis elegans and its parasite Serratia marcescens. This parasite is horizontally transmitted, which allowed us to study coevolution independently of vertical transmission. After 20 generations, we observed a response to selection when coevolution was possible: reduced antagonism evolved in the co-passaged treatment. Reduced antagonism, however, did not evolve when hosts or parasites were independently selected, without coevolution. In addition, we found strong local adaptation for reduced antagonism between replicate host/parasite lines in the co-passaged treatment. Taken together, these results strongly suggest that coevolution was critical to the rapid evolution of reduced antagonism.
Keywords: Evolution of virulence, experimental evolution, experimental coevolution, Caenorhabditis elegans, Serratia marcescens
Introduction
Species interactions vary enormously, from highly antagonistic (e.g. host-parasite and predator-prey) to mutualistic. Moreover, the nature of a given interaction is not always evolutionarily stable: interspecific interactions can shift readily between parasitism and mutualism (e.g. Clay 1990; Herre 1993; Noda et al. 1997; Nishiguchi and Nair 2003; Sawada et al. 2003; Thompson 2005; Fenn and Blaxter 2006; Petersen and Tisa 2013) (reviewed in Thompson 1994). The manner in which two partners interact may even vary within and between populations of the same species (Smith 1968; Thompson 1988; Burdon et al. 1999; Kraaijeveld and Godfray 1999; Thompson and Cunningham 2002; Weeks et al. 2007). Of particular interest are those cases in which mutualisms seem to have arisen from parasitic interactions (e.g. Jeon 1972; Carroll 1988; Bandi et al. 1999; Hentschel et al. 2000; Dedeine et al. 2001; Dale et al. 2002; Weeks et al. 2007; Degnan et al. 2009; Hosokawa et al. 2010). These cases raise a puzzling, though pressing question: why, from an evolutionary standpoint, do transitions towards reduced antagonism occur?
Studies of parasite transmission mode have demonstrated that vertical transmission, from parent to offspring, can select for reduced antagonism (Bull et al. 1991; Herre 1993; Clayton and Tompkins 1994; Lipsitch et al. 1996; Turner et al. 1998; Messenger et al. 1999; Stewart et al. 2005; Sachs and Wilcox 2006). According to theory, the alignment of host and parasite fitness selects for reduced antagonism. Fitness alignment refers to a positive covariance of host and parasite fitness. Under vertical transmission, parasite fitness is contingent upon host survival and reproduction, and this positive fitness covariance favors reduced antagonism. Under horizontal transmission, the covariance of host and parasite fitness can be negative: selection for increased parasite transmission between hosts may select for increased within-host reproduction and thereby increased antagonism (Anderson and May 1982; Ewald 1987; Bull 1994; Frank 1996; Wade 2007). The direction of selection on antagonism thus varies with transmission mode.
The opportunity for coevolution also varies with transmission mode. Vertical transmission provides a unique opportunity for strong coevolution, because host and parasite lineages are paired over multiple generations. Conversely, horizontal transmission impedes tight coevolution between a single host and parasite lineage, because the parasite lineage is continually transmitted between different host lineages. Therefore, in contrasting vertical with horizontal transmission, prior studies have not only compared experimental conditions with selection for and against antagonism. They have also inadvertently compared experimental conditions with high and low potential for coevolution, respectively.
To build upon this prior work, we tested the hypothesis that coevolution is fundamental to the evolution of reduced antagonism. We tested the role of coevolution, independently of vertical transmission, by using a horizontally transmitted parasite. This enabled us to impose selection for reduced antagonism directly, rather than indirectly through transmission mode. Accordingly, we did not manipulate transmission mode. Rather, we manipulated only the potential for coevolution in order to compare the degree of reduced antagonism achieved when coevolution was possible vs. that achieved when coevolution was prevented.
We tested this coevolution hypothesis through experimental evolution of the interaction between a nematode host, Caenorhabditis elegans, and a virulent bacterial parasite, Serratia marcescens. These experimental host and parasite populations were previously under selection for increased antagonism (Morran et al. 2011). In the present study, we reversed this selection. We favored reduced antagonism by selecting for hosts and parasites that were able to persistently interact, such that hosts survived to reproduction without clearing the parasites. In the co-passaged treatment, we allowed coevolution by selecting for reduced antagonism simultaneously in both partners. We contrasted this with the singly passaged treatments in which we prevented coevolution by selecting on one partner while holding the other constant. In all treatments, parasites were transmitted horizontally, not vertically, in order to investigate coevolution independently of transmission mode.
If coevolution contributes to the reduction in antagonism between hosts and parasites, we predicted that the response to selection for reduced antagonism would be greater under co-passaging than under singly passaging. Our results support this prediction: reduced antagonism, in the form of a diminished fecundity cost of infection in hosts, evolved only in the pairing of co-passaged hosts and parasites. Moreover, shifts in host or parasite phenotypes alone could not explain the reduced antagonism of the co-passaged pairing. Rather, the interaction of host and parasite lineages was a significant factor in the reduction in antagonism. In addition, we found strong local adaptation between co-passaged host and parasite lineages. Our results argue that coevolution underlies the observed reduction in antagonism.
Methods
Host and parasite populations
Caenorhabditis elegans is a model for the study of host-parasite interactions (Kurz and Ewbank 2000), and there is reason to believe that C. elegans and Serratia marcescens interact in nature (Schulenburg et al. 2004; Schulenburg and Ewbank 2004; Pradel et al. 2007). This interaction is fascinating with respect to transitions in antagonism: S. marcescens is a virulent parasite not only of nematodes, but also of insects, corals, and humans (nosocomial). Yet some nematode species form mutualistic associations with S. marcescens and closely-related species (Petersen and Tisa 2013). The diversity of interactions between nematodes and Serratia argues that transitions in antagonism are common. Our experimental evolution harnesses the evolutionary lability of this association to conduct a general test of the role that coevolution plays in transitions to reduced antagonism between naturally interacting species.
Replicate parasite populations were derived from S. marcescens strain Sm2170, which is highly virulent towards C. elegans (Schulenburg and Ewbank 2004). Caenorhabditis elegans hosts were derived from the strain PX382, an inbred line of CB4856. Five replicate populations were independently mutagenized with ethyl methanesulfonate to introduce genetic variation. These host populations were then co-passaged with populations of Sm2170 for 30 generations as part of a prior experiment (Morran et al. 2011). Assays of this experiment demonstrated that these host replicate lines adapted to resist their co-passaged parasite lines: co-passaged hosts showed significantly lower mortality rates than ancestral hosts when exposed to co-passaged parasites (Morran et al. 2011: mixed mating coevolved lines). Moreover, significant local adaptation of co-passaged parasite lines to kill sympatric co-passaged host lines strongly suggested genetic divergence between host lines under antagonistic coevolution (Morran et al. 2014: mixed mating coevolved lines).
We used these five divergent, co-passaged host populations as the ancestral replicate lines for our experimental evolution. We did so first because they provided an antagonistic starting point from which to select for reduced antagonism. Secondly, we observed a “reduced antagonism” phenotype at relatively high frequency in these host lines. Serratia marcescens typically colonizes the intestine of C. elegans hosts, resulting in loss of fecundity (Schulenburg and Ewbank 2004; Morran et al. 2011) and rapid host mortality (Mallo et al. 2002). In these lines, we observed hosts with light infections of S. marcescens in their upper intestines. Through a stereoscope, the infection is readily evident as a bright red band or cluster of colonies just below the pharynx (Fig. S1). Preliminary observations indicated that hosts carrying S. marcescens in this region survive and reproduce without clearing the infection. Because host and parasite coexist for an extended period without a total loss of fitness for host (i.e. no death or sterilization) or parasite (i.e. no host recovery), we chose this “ruby-throated” phenotype as a model of reduced antagonism.
Prior to commencing experimental evolution, we quantified the frequency of the ruby-throated phenotype in a naïve nematode line vs. one with 30 generations of prior exposure to S. marcescens. 750 L3-L4 nematodes were added to a lawn of Sm2170 (as in Serratia selection plates, described below). Sixty hours later, we counted the number of ruby-throated hermaphrodites on the plates. We assayed ten plates for each of the two host lines. To compare ruby-throated frequency between lines, we performed a Student's t-test in R v3.0.2 (R Core Team 2013)
Experimental evolution of reduced antagonism
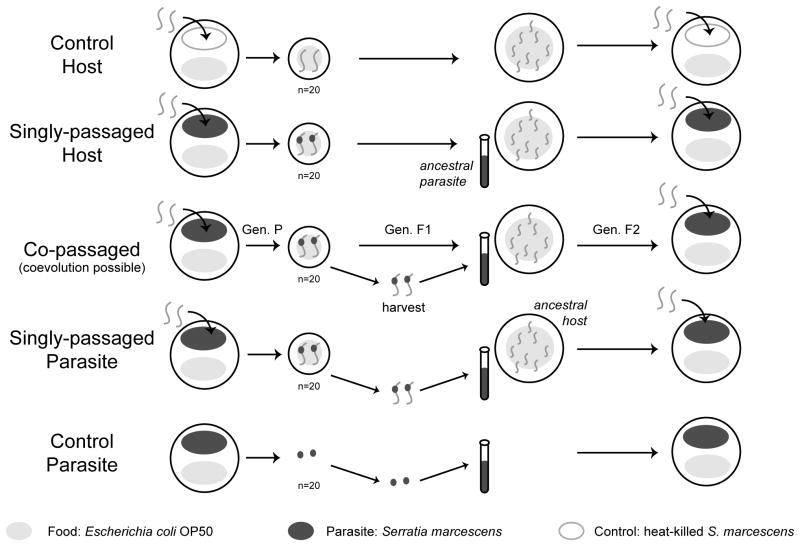
We devised experimental evolution treatments in order to select for 1. Ruby-throated hosts, which survive and reproduce with a persistent infection and/or 2. Ruby-throated parasites, which establish persistent infections without rapidly killing or sterilizing the host. Treatments selected for the evolution of reduced antagonism under conditions that did or did not permit coevolution (Fig. 1). In the “co-passaged” treatment, coevolution of host and parasite was possible: hosts and parasites were simultaneously passaged. Two “singly-passaged” treatments did not permit coevolution: selection for reduced antagonism was conducted separately on host and parasite. Each generation of selection, ruby-throated hosts and parasites were paired with static ancestral parasites and hosts, respectively. Two control treatments prevented coevolution and symbiosis: control hosts and parasites were selected at random and passaged in the absence of live parasites and hosts, respectively. Control treatments served to account for genetic drift and non-focal selection pressures. Each of these five treatments had five replicate host populations; these were the five divergent lineages resulting from a prior experimental evolution project, as discussed above. We conducted 20 generations of selection.
Figure 1. Experimental evolution design.
Five experimental evolution treatments were established to evaluate the role of coevolution in the evolution of host-parasite antagonism. Selection for the ruby-throated phenotype was performed on Serratia Selection Plates (SSPs). Twenty ruby-throated adults were then transferred to new plates to reproduce. In the co-passaged treatment, in which coevolution was possible (center row), both the F1 ruby-throated hosts and parasites were propagated for an additional generation. F2 hosts and parasites were then combined on a new SSP for the next generation of selection. In the singly-passaged treatments, ruby-throated hosts (2nd row) and parasites (4th row) were passaged with static ancestral parasite andhost populations, respectively. In the control treatments, healthy hosts (1st row) and free-living parasites (5th row) were passaged in the absence of live parasites or hosts, respectively.
Transfer and Selection on SSPs
We describe here the details of the co-passaged treatment, followed by specific modifications for the four remaining treatments (Fig. 1). Selection was performed on Serratia selection plates (SSPs), which we constructed as in Morran et al. (2011). We added 1500 L3-L4 nematodes to a lawn of S. marcescens to force hosts and parasites to interact. Hosts migrated towards the opposite half of the plate, seeded with a lawn of OP50 (food source). After 60 hours on the plates, we transferred approximately 20 ruby-throated hosts per replicate to a new plate, seeded with only OP50. Ruby-throated hosts were then allowed to reproduce. After 48 hours, parental ruby-throated hosts were separated from their F1 offspring by size and phenotype. F1 host offspring were washed with M9 buffer and transferred to a plate seeded with OP50 to reproduce for 65 hours. Ruby-throated parasites were extracted from the ∼20 parental ruby-throated hosts by washing hosts repeatedly with M9 buffer to remove external bacteria and then crushing them to release the ruby-throated bacteria. The isolated bacteria were grown on a plate for 24 hours at 28°C. Colonies were then randomly selected for growth in Luria Broth (LB) overnight at 28°C (n=10). This culture was used to seed a new SSP. 1500 L3-L4 F2 hosts were added to this plate for the next generation of selection.
In the singly-passaged host treatment, ruby-throated hosts were passaged as in the co-passaged treatment. However, in place of the ruby-throated parasites, we used ancestral Sm2170 for seeding of the subsequent SSP. In the singly-passaged parasite treatment, ruby-throated parasites were passaged as in the co-passaged treatment. However, each generation, in place of the ruby-throated hosts, we obtained offspring from ancestral host populations maintained at 15°C. In the control host treatment, SSPs were constructed with heat-killed Sm2170 in place of live parasites. Twenty hosts were selected at random from SSPs and allowed to reproduce. In the control parasite treatment, SSPs were constructed as in the co-passaged treatment, but no hosts were added. Free-living parasites were passaged by randomly selecting 20 colonies from the Sm2170 lawn in order to roughly mimic the number of parasites passaged when transferring 20 ruby-throated hosts. Host and parasite lines were stored at -80°C after 20 generations of selection (Supplemental Methods).
Assays of Frequency, Fecundity and Virulence
Both an increase in frequency of the ruby-throated phenotype and an increase in fecundity of ruby-throated hosts are consistent with a response to selection for reduced antagonism. We therefore compared ruby-throated frequency and fecundity in various combinations of host and parasite lines in order to 1. Test if reduced antagonism evolved primarily when coevolution was possible (co-passaged treatment) (Table 1A,B) and 2. Evaluate the relative contributions of host vs. parasite evolution in the transition to reduced antagonism (Table 1C,D). We reduced variation resulting from sex-specific differences in the ruby-throated phenotype by selecting 20 unmated hermaphrodites to establish low-male subpopulations. Assays were then performed so as to replicate the conditions of experimental evolution.
Table 1. Pairings of host and parasite populations for assays of ruby-throated frequency and fecundity.
| A. Interaction: experimental host and parasite populations combined1 | B. Interaction: host and parasite populations paired as in experimental selection2 | ||
|---|---|---|---|
| Host | Parasite | Host | Parasite |
|
| |||
| ancestor | ancestor (Sm2170) | singly-passaged | ancestor |
| control | control | ancestor | singly-passaged |
| singly-passaged | singly-passaged | co-passaged | co-passaged |
| co-passaged | co-passaged | ||
| C. Host alone: experimental host populations with ancestral parasites3 | D. Parasite alone: experimental parasite populations with ancestral hosts4 | ||
| Host | Parasite | Host | Parasite |
|
| |||
| control | ancestor | ancestor | control |
| singly-passaged | ancestor | ancestor | singly-passaged |
| co-passaged | ancestor | ancestor | co-passaged |
Compares host and parasite lines paired according to shared evolutionary history. If coevolution contributes to reduced antagonism, the response to selection for reduced antagonism will be greatest in the co-passaged pairing.
Compares pairings that were selected upon during experimental evolution to further test the prediction that the response to selection for reduced antagonism will be greatest in the co-passaged pairing.
Compares changes in passaged host lines independent of the parasite to test if the reduced antagonism observed in the co-passaged pairing can be attributed to evolution of co-passaged host populations alone.
Compares changes in passaged parasite lines independent of the host to test if the reduced antagonism observed in the co-passaged pairing can be attributed to evolution of co-passaged parasite populations alone.
We first quantified changes from the ancestor in the frequency of formation of the ruby-throated phenotype. Two hundred L3-L4 nematodes from low-male lines were added to SSPs, constructed as outlined above. After 65 hours, we counted the total numbers of adult hermaphrodites and adult ruby-throated hermaphrodites on the OP50 halves of the SSPs. We assayed three replicates for each combination of host and parasite line (combinations given in Table 1A, C-D). For all assays, host lines were paired with sympatric parasite lines except in the local adaptation assays below. For each comparison, a separate ANOVA was performed in SPSS v21 (IBM) to test the effect of treatment, line, replicate, and the treatment by line interaction on the frequency of ruby-throated hosts. Replicate was excluded when insignificant. In all statistical analyses, treatment was a fixed effect, while line and replicate were treated as random. We additionally performed a linear contrast test of ruby-throated frequency in control, singly-passaged, and co-passaged pairings vs. ancestral pairings.
Fecundity assays were an extension of the frequency assays above. After measuring ruby-throated frequency, we selected ten ruby-throated hermaphrodites from each replicate and transferred them to individual plates. In total, we isolated 30 ruby-throated hermaphrodites (10 × 3 replicates) per combination of host and parasite line. After 48 hours, we counted the number of offspring per hermaphrodite. For each comparison (Table 1A-D), a separate ANOVA was performed on ruby-throated fecundity, as described above. We similarly measured the fecundity of uninfected hosts from all treatments in order to evaluate the contribution of host evolution alone to observed reductions in antagonism (Supplemental Methods).
We also quantified the change in parasite virulence during experimental evolution in order to evaluate the contribution of parasite evolution alone to observed reductions in antagonism. Parasite virulence was assessed through mortality assays, as described in Morran et al. (2011). We measured the mortality rate as the proportion of dead/morbid hosts after 24 hours of exposure of standard host lines to ancestral, control, singly-passaged, and co-passaged parasite lines (Supplemental Methods). An ANOVA was performed as described above.
Local adaptation
To determine if co-passaged hosts and parasites coevolved during experimental evolution, we tested the degree of local adaptation for reduced antagonism. We performed fully reciprocal cross-infections between our five co-passaged host and parasite lines (25 combinations). Fecundity was measured as described above, excepting that for each combination, 15 adult ruby-throated hermaphrodites were isolated from a single SSP. Analyses of local adaptation were based upon Morran et al. (2014). We first performed an ANOVA to test for a significant interaction effect of host and parasite line on ruby-throated fecundity. We then evaluated overall local adaptation by performing a linear contrast test of the fecundity of ruby-throated hosts in all sympatric pairings (n=5) versus all allopatric pairings (n=20) (Blanquart et al. 2013).
We also performed fine-scale local adaptation tests for each host and parasite pairing (Morran et al. 2014). We did so using a linear contrast test of the sympatric host-parasite pairing against all allopatric pairings: we compared the fecundity of the sympatric pairing (e.g. co-passaged host line 1 with co-passaged parasite line 1) with that of the host population plus allopatric parasite populations (e.g. co-passaged host line 1 with co-passaged parasite lines 2-5) and that of the parasite population plus allopatric host populations (e.g. co-passaged parasite line 1 with co-passaged host lines 2-5). One-tailed linear contrast tests were performed independently for lines 1-5 to evaluate the hypothesis that the fine-scale tests of local adaptation reflected the overall test of local adaptation. We report the results of tests for which equal variances were not assumed, as variances were significantly different for three of the six tests performed.
Results
Reduced antagonism is only observed under co-passaging
After 20 generations of selection, we quantified the response to selection for reduced antagonism by measuring the frequency and fecundity of ruby-throated hosts. We compared these traits in the following pairings: ancestral hosts and parasites; control hosts and parasites; singly-passaged hosts and parasites; co-passaged hosts and parasites (Table 1A). If coevolution contributes to reduced antagonism, we predicted that the co-passaged pairing would show the greatest response to selection for reduced antagonism.
Increased frequency of the ruby-throated phenotype is consistent with reduced antagonism, because host and parasite engage more frequently in persistent, non-lethal interactions. The frequency of the ruby-throated phenotype did not differ between host-parasite combinations (Table S1A: F(3,12)=0.938, p=0.453). Specifically, the frequency did not increase in evolved relative to ancestral pairings (linear contrast: p=0.254, one-tailed)(Fig. S2A, bar a≈b,c,d). Thus experimental selection did not alter rates of persistent association between host and parasite.
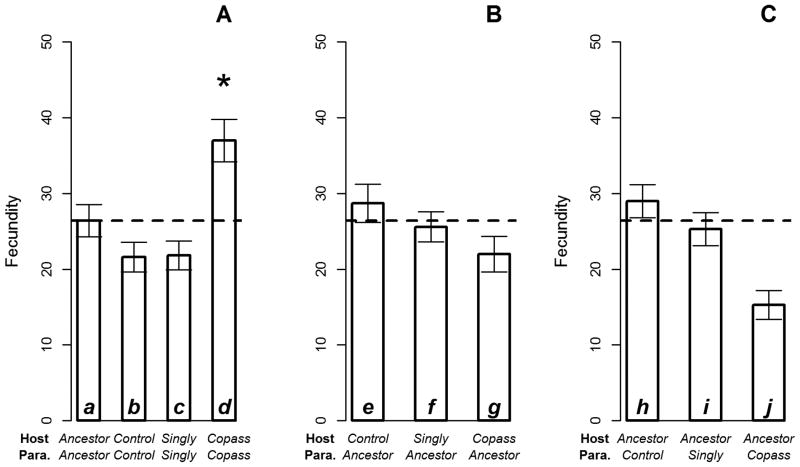
Reduced antagonism was observed when measured as changes in fecundity over the course of the experiment. Increased fecundity of ruby-throated hosts is consistent with reduced antagonism, because the fitness cost of an active infection is reduced. The fecundity of ruby-throated hosts differed between host-parasite combinations (Table S2A: F(3,12)=4.643, p=0.022)(Fig. 2A): ruby-throated hosts from co-passaged pairings displayed significantly elevated fecundity relative to ancestral (p=0.015, bar d>a), control (p<0.001, d>b), and singly-passaged (p<0.001, d>c) pairings. Control and singly-passaged pairings did not differ from the ancestral pairing (p=0.481, a≈b; p=0.521, a≈c, respectively), nor from one another (p=1.00, b≈c). Thus, among these pairings, a response to selection for reduced antagonism only occurred in the host-parasite pairs that had the potential for coevolution.
Figure 2. Fecundity of ruby-throated hosts.
The fecundity of ruby-throated hosts across different pairings of host and parasite populations. (A) Host and parasite pairings according to shared evolution history. The fecundity of co-passaged pairings was significantly elevated relative to ancestral, control, and singly-passaged pairings. (B) Hosts paired with ancestral parasites. Ruby-throated fecundity did not differ when host populations were paired with the same (ancestral) parasite population. (C) Parasites paired with ancestral hosts. Ruby-throated fecundity differed marginally when parasite populations were paired with the same (ancestral) host populations. The dashed line marks the starting point of experimental evolution: mean ruby-throated fecundity resulting from pairing ancestral hosts with ancestral parasites. Each bar is an average of fecundity counts obtained from 150 ruby-throated hermaphrodites (10 hermaphrodites per replicate assay plate, three replicates per each of five lines), and error bars give the standard error of mean fecundity counts. Individual bars are referred to by letter in the text.
However, singly-passaged hosts and parasites may have evolved reduced antagonism specifically in combination with the parasite and host populations, respectively, that they encountered during the experiment. Therefore, we also compared ruby-throated fecundity in the pairings under selection in experimental evolution: singly-passaged hosts with ancestral parasites, ancestral hosts with singly-passaged parasites, and co-passaged hosts with co-passaged parasites (Table 1B). The fecundity of ruby-throated hosts again differed significantly (Table S2B: F(2,8)=5.073, p=0.038), with the co-passaged pairing having significantly higher fecundity than either singly-passaged pairing (host: p=0.014, bar d>f; parasite: p=0.013, d>i)(Fig. 2A-C). Furthermore, singly-passaged host and parasite populations did not differ in their response to selection: ruby-throated fecundity of singly-passaged hosts with ancestral parasites did not differ from singly-passaged parasites with ancestral hosts (p=0.999, f≈i). Therefore, reduced antagonism evolved only when coevolution was possible.
Host evolution alone cannot explain reduced antagonism
We only observed reduced antagonism in the co-passaged pairing, arguing for a role for coevolution. However, the case for coevolution would be significantly weakened if traits in the co-passaged host or parasite populations alone could explain the reduction in antagonism. We first tested if co-passaged host populations alone could recapitulate the increase in ruby-throated fecundity of the co-passaged pairing. To do so, we asked if the fecundity of co-passaged hosts exceeded that of control and singly-passaged hosts when all were paired with ancestral parasites (Table 1C; Fig. 2B, bar g vs. e,f). The fecundity of ruby-throated hosts did not differ between any pairings with ancestral parasites (Table S2C: F(3,12.31)=0.142, p=0.933). Therefore evolution of co-passaged hosts alone cannot explain reduced antagonism in the co-passaged pairing.
Moreover, a general increase in the fecundity of healthy co-passaged hosts cannot explain the elevated ruby-throated fecundity. We observed no significant difference in the fecundity of uninfected hosts from the ancestral, control, singly-passaged, and co-passaged populations (Table S3: F(2,8)=3.160,p=0.097)(Fig. S3).
Parasite evolution alone cannot explain reduced antagonism
We then tested if the co-passaged parasite populations alone could recapitulate the increase in ruby-throated fecundity of the co-passaged pair. To do so, we asked if the fecundity of ancestral hosts infected with co-passaged parasites exceeded that of ancestral hosts infected with control and singly-passaged parasites (Table 1D; Fig. 2C, bar j vs. h,i). The difference in fecundity of ruby-throated hosts from these pairings was marginally significant (Table S2D: F(3,12)=2.784, p=0.086) due to the relatively depressed fecundity of co-passaged parasites with ancestral hosts. This result strongly argues that the evolution of co-passaged parasites alone cannot explain reduced antagonism in the co-passaged pairing.
Moreover, decreased virulence of the co-passaged parasite populations cannot explain the elevated ruby-throated fecundity. We measured the mortality rate of ancestral hosts exposed to ancestral, control, singly-passaged, and co-passaged parasites. Mortality rate differed significantly between parasite treatment (Table S4: F(3,12)=14.370, p<0.001)(Fig. S4): ancestral parasites induced a significantly higher mortality rate than all other parasites (p<0.001 for each). Mortality rate with co-passaged parasites was low but equivalent to that of control (p=0.407) and singly-passaged (p=0.878) parasites. Parasite virulence declined uniformly across all experimental treatments and thus cannot explain reduced antagonism in the co-passaged pair.
Strong local adaptation is evidence for coevolution
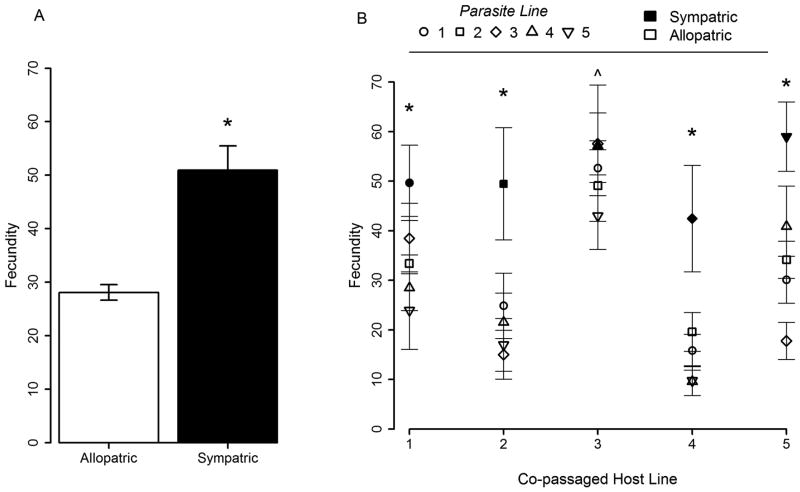
Our results indicate that coevolution was critical to the response to selection for reduced antagonism. However, the degree to which co-passaged hosts and parasites coevolved was uncertain. Tests of local adaptation are commonly used to detect reciprocal adaptation. We measured ruby-throated fecundity in fully reciprocal cross-infections among our five co-passaged host and parasite populations. Consistent with local adaptation, we found a significant interaction effect of co-passaged host and parasite population (line) on ruby-throated fecundity (Table S5A: F(16,298)=3.560, p<0.001)(Fig. 3).
Figure 3. Local adaptation of co-passaged populations.
Reciprocal cross-infections of co-passaged host and parasite lines were performed to test for local adaptation. (A) Sympatric co-passaged pairings displayed significantly higher fecundity than allopatric pairings. The allopatric bar is an average of fecundity counts obtained from 300 hermaphrodites (15 hermaphrodites from each of 20 allopatric combinations), and the sympatric bar is an average of 75 hermaphrodites (5 sympatric combinations). (B) Each co-passaged host-parasite pairing supports the results of the overall analysis: ruby-throated fecundity was significantly higher in sympatric relative to allopatric pairings for lines 1,2,4, and 5 (*) and marginally significantly for line 3 (^). Each point is an average of fecundity counts obtained from 15 hermaphrodites, and all error bars give the standard error of mean fecundity counts.
Given this significant interaction, we tested the hypothesis that ruby-throated fecundity in sympatric pairings exceeds that in allopatric pairings. We first made the comparison across all co-passaged host and parasite lines. Consistent with local adaptation, we found that the ruby-throated fecundity of sympatric pairings significantly exceeded that of allopatric pairings (Table S5B: p<0.001)(Fig. 3A).
We then made the comparison for each replicate population within the co-passaged treatment by contrasting each sympatric host-parasite pairing with its eight possible allopatric pairings (i.e. the focal host population paired with the four allopatric parasite populations and the focal parasite population paired with the four allopatric host populations). Consistent with local adaptation, the fecundity of the sympatric pairing exceeded that of allopatric pairings to a significant degree for host and parasite lines 1,2,4 and 5 and marginally for host and parasite lines 3 (Table S5B: 1: p=0.017; 2: p=0.027; 3: p=0.059; 4: p=0.046; 5: p<0.001; one-tailed tests)(Fig. 3B). These results suggest rapid divergence among replicate populations in the co-passaged treatment, consistent with coevolution of co-passaged host and parasite populations.
Discussion
In this study, we tested the hypothesis that coevolution can contribute to the evolution of reduced antagonism between hosts and parasites. We selected directly on a phenotypic indicator of reduced antagonism under conditions that either did or did not permit coevolution. Our results strongly support the coevolution hypothesis: after 20 generations of selection, reduced antagonism only evolved in response to selection when coevolution was possible (Fig. 2A).
We found no support for the idea that reduced antagonism arose from the evolution of host or parasite traits alone (Fig. 2B,C). Co-passaged host or parasite lineages, when paired with ancestral parasites or hosts, respectively, did not show reduced antagonism. We particularly note that declines in parasite virulence alone were insufficient for reduced antagonism. The virulence of the co-passaged parasite lineages towards ancestral hosts was no lower than that of singly-passaged and control parasite lineages: virulence declined in all experimental and control parasite lineages (Fig. S4). A possible explanation for this is that the free-living generations during experimental evolution of the parasite lines (growth in LB or on agar plates) exerted negative or relaxed selection on virulence or correlated traits (Caraco and Wang 2008; Friman et al. 2009; Mikonranta et al. 2012; Wasik et al. 2015). Regardless of the mechanism, differential evolution of virulence in our experimental lines cannot explain the reduced antagonism observed in the co-passaged pairing.
Our results argue that the evolution of reduced antagonism required coevolution, which may result in local adaptation (Parker 1985; Lively 1989). We found strong evidence of local adaptation of co-passaged host and parasite lineages for reduced antagonism (Fig. 3), further demonstrating that co-passaged host and parasite lineages did indeed coevolve. These results also demonstrate that adaptation of co-passaged host and parasite replicate lines occurred rapidly, within 20 generations of selection. This is particularly noteworthy given that genetic drift likely reduced the fixation probability of beneficial mutations in our experiment (Gillespie 1998). Thus, our results may have been stronger with larger effective population sizes. Finally, local adaptation indicates that replicate pairs diverged rapidly, arriving at reduced antagonism via distinct evolutionary routes. We did use genetically distinct ancestral populations to establish our five replicate host lines, which may explain why we see such a strong signal of divergence between our pairings. However, the use of distinct ancestral host lineages makes the repeated evolution of reduced antagonism in the co-passaged treatment even more striking.
Taken together, our findings argue that reduced antagonism arises from the interaction of reciprocally adapting host and parasite lineages. We therefore propose that coevolution may contribute to the evolution of reduced antagonism when selection is imposed by fitness alignment under vertical transmission (positive fitness covariance) (Bull et al. 1991; Herre 1993; Clayton and Tompkins 1994; Lipsitch et al. 1996; Turner et al. 1998; Messenger et al. 1999; Stewart et al. 2005; Sachs and Wilcox 2006). Our findings also show that reduced antagonism can evolve without vertical transmission if selection is directly imposed and coevolution is present. It is unlikely that we imposed any indirect selection via fitness alignment, because the parasite is horizontally transmitted. Moreover, parasite fitness also depended on reproduction during the “free-living” phase: multiple free-living generations alternated with selection events during infection. Similar transmission modes are common in nature, including in mutualisms of Vibrio bacteria with squid and rhizobia with legumes (as reviewed in Bright and Bulgheresi 2010).
In addition, fitness alignment should correspond to an increase in the frequency of the ruby-throated phenotype, which we did not observe. When a parasite genotype establishes the ruby-throated phenotype with minimal cost to its host, that host genotype will comprise a larger proportion of the next host generation. Assuming genetic specificity of ruby-throated host and parasite, those parasite offspring will then have more opportunities for host establishment. The frequency of the ruby-throated phenotype should accordingly increase. The fact that we failed to observe any increase in frequency argues against fitness alignment in our study. The lack of response in ruby-throated frequency in our study may alternately be due to limited additive genetic variance for this trait following prior evolution of these host populations (Morran et al. 2011).
Previous studies of fitness alignment have demonstrated that reduced antagonism arises from an interaction of host and parasite genotypes, which hints at the significance of coevolution (Traub 1939; Bull et al. 1991; Bull and Molineux 1992). After selection under vertical transmission of bacteriophage f1 with Escherichia coli, Bull and Molineux (1992) observed increases in the growth rate of infected host populations and in phage-mediated protection against infection by alternate phages. For most experimental lines, growth rate and protection were greater when phage were paired with their co-passaged hosts than with the ancestor. Several studies, however, present contradictory results: evolution of host (Helling et al. 1981; Bouma and Lenski 1988) or parasite traits alone (Jeon 1972; Sachs and Wilcox 2006; Weeks et al. 2007; Jansen et al. 2015) could explain the observed reduction in antagonism. Importantly, these studies all examined the endpoint of long-term selection under vertical transmission, rather than directly testing the role of coevolution.
Here, we directly tested the contribution of coevolution under horizontal transmission. Our experimental coevolution design is particularly powerful in contrasting coevolution with independent evolution (Brockhurst and Koskella 2013) and could be applied to many other experimental symbiosis models in which partners can be dissociated (Denison et al. 2003; e.g. Stewart et al. 2005; Sachs and Wilcox 2006; Hillesland and Stahl 2010; Jansen et al. 2015). Overall, we find that coevolution is critical to the evolution of reduced antagonism. Similar investigations of natural symbioses are required to determine if coevolution is generally a factor in evolutionary transitions towards reduced antagonism. Further support for the significance of host-parasite coevolution would argue for its inclusion in models of virulence evolution, which primarily focus upon parasite evolution alone (though see van Baalen 1998; Restif et al. 2001; Gandon et al. 2002; Day and Burns 2003; Restif and Koella 2003; Little et al. 2010).
Supplementary Material
Acknowledgments
We are grateful to L Sloan, A Bhattacharya, R Parrish, S Klosak, T O'Sullivan, S Slowinski, J Xu, and J Hite for contributions to this study. We thank E Ragsdale, J Powers, and the IU Light Microscopy Imaging Center for assistance. Funding was provided by the National Institutes of Health (1F32GM096482-01 to LTM; IU Common Themes in Reproductive Diversity training grant to AKG), the National Science Foundation (Graduate Research Fellowship to AKG), and the IU Hutton Honors College (Research Grant to KSS).
List of Works Cited
- Anderson RM, May RM. Coevolution of hosts and parasites. Parasitology. 1982;85:411–426. doi: 10.1017/s0031182000055360. [DOI] [PubMed] [Google Scholar]
- Bandi C, McCall JW, Genchi C, Corona S, Venco L, Sacchi L. Effects of tetracycline on the filarial worms Brugia pahangi and Dirofilaria immitis and their bacterial endosymbionts Wolbachia. Int J Parasitol. 1999;29:357–364. doi: 10.1016/s0020-7519(98)00200-8. [DOI] [PubMed] [Google Scholar]
- Blanquart F, Kaltz O, Nuismer SL, Gandon S. A practical guide to measuring local adaptation. Ecol Lett. 2013;16:1195–1205. doi: 10.1111/ele.12150. [DOI] [PubMed] [Google Scholar]
- Bouma JE, Lenski RE. Evolution of a bacteria/plasmid association. Nature. 1988;335:351–352. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nature. 2010;8:218–230. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhurst MA, Koskella B. Experimental coevolution of species interactions. Trends Ecol Evol. 2013;28:367–375. doi: 10.1016/j.tree.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Bull J. Perspective: virulence. Evolution. 1994:1423–1437. doi: 10.1111/j.1558-5646.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Molineux IJ. Molecular genetics of adaptation in an experimental model of cooperation. Evolution. 1992;46:882–895. doi: 10.1111/j.1558-5646.1992.tb00606.x. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Molineux IJ, Rice WR. Selection of benevolence in a host-parasite system. Evolution. 1991;45:875–882. doi: 10.1111/j.1558-5646.1991.tb04356.x. [DOI] [PubMed] [Google Scholar]
- Burdon J, Gibson A, Searle SD, Woods M, Brockwell J. Variation in the effectiveness of symbiotic associations between native rhizobia and temperate Australian Acacia: within-species interactions. J Appl Ecol. 1999;36:398–408. [Google Scholar]
- Caraco T, Wang IN. Free-living pathogens: Life-history constraints and strain competition. J Theor Biol. 2008;250:569–579. doi: 10.1016/j.jtbi.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll G. Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecology. 1988:2–9. [Google Scholar]
- Clay K. Comparative demography of three graminoids infected by systemic, clavicipitaceous fungi. Ecology. 1990:558–570. [Google Scholar]
- Clayton DH, Tompkins DM. Ectoparasite virulence is linked to mode of transmission. Proc Roy Soc B. 1994;256:211–217. doi: 10.1098/rspb.1994.0072. [DOI] [PubMed] [Google Scholar]
- Dale C, Plague GR, Wang B, Ochman H, Moran NA. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Nat Acad Sci USA. 2002;99:12397–12402. doi: 10.1073/pnas.182213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T, Burns J. A consideration of patterns of virulence arising from host-parasite coevolution. Evolution. 2003;57:671–676. doi: 10.1111/j.0014-3820.2003.tb01558.x. [DOI] [PubMed] [Google Scholar]
- Dedeine F, Vavre F, Fleury F, Loppin B, Hochberg ME, Boulétreau M. Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp Proc Nat Acad Sci USA. 2001;98:6247–6252. doi: 10.1073/pnas.101304298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Nat Acad Sci USA. 2009;106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison R, Kiers E, West S. Darwinian agriculture: when can humans find solutions beyond the reach of natural selection? Quarterly Review of Biology. 2003;78:145–168. doi: 10.1086/374951. [DOI] [PubMed] [Google Scholar]
- Ewald PW. Transmission modes and evolution of the parasitism-mutualism continuum. Ann NY Acad Sci. 1987;503:295–306. doi: 10.1111/j.1749-6632.1987.tb40616.x. [DOI] [PubMed] [Google Scholar]
- Fenn K, Blaxter M. Wolbachia genomes: revealing the biology of parasitism and mutualism. Trends Parasitol. 2006;22:60–65. doi: 10.1016/j.pt.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Frank SA. Models of parasite virulence. Quart Rev Biol. 1996;71:37–78. doi: 10.1086/419267. [DOI] [PubMed] [Google Scholar]
- Friman VP, Lindstedt C, Hiltunen T, Laakso J, Mappes J. Predation on multiple trophic levels shapes the evolution of pathogen virulence. PloS One. 2009;4:1–6. doi: 10.1371/journal.pone.0006761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandon S, van Baalen M, Jansen VAA. The evolution of parasite virulence, superinfection, and host resistance. Am Nat. 2002;159:658–669. doi: 10.1086/339993. [DOI] [PubMed] [Google Scholar]
- Gillespie JH. Population Genetics: A Concise Guide. John Hopkins University Press; Baltimore: 1998. [Google Scholar]
- Helling R, Kinney T, Adams J. The maintenance of plasmid-containing organisms in populations of Escherichia coli. J Gen Microbiol. 1981;123:129–141. doi: 10.1099/00221287-123-1-129. [DOI] [PubMed] [Google Scholar]
- Hentschel U, Steinert M, Hacker J. Common molecular mechanisms of symbiosis and pathogenesis. Trends Microbiol. 2000;8:226–231. doi: 10.1016/s0966-842x(00)01758-3. [DOI] [PubMed] [Google Scholar]
- Herre EA. Population structure and the evolution of virulence in nematode parasites of fig wasps. Science. 1993;259:1442–1445. doi: 10.1126/science.259.5100.1442. [DOI] [PubMed] [Google Scholar]
- Hillesland KL, Stahl DA. Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Nat Acad Sci USA. 2010;107:2124–2129. doi: 10.1073/pnas.0908456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Nat Acad Sci USA. 2010;107:769–774. doi: 10.1073/pnas.0911476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen G, Crummenerl LL, Gilbert F, Mohr T, Pfefferkorn R, Thänert R, Rosenstiel P, Schulenburg H. Evolutionary transition from pathogenicity to commensalism: global regulator mutations mediate fitness gains through virulence attenuation. Mol Biol Evol. 2015 doi: 10.1093/molbev/msv160. msv160v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon K. Development of cellular dependence on infective organisms: micrurgical studies in amoebas. Science. 1972;176:1122–1123. doi: 10.1126/science.176.4039.1122. [DOI] [PubMed] [Google Scholar]
- Kraaijeveld A, Godfray H. Geographic patterns in the evolution of resistance and virulence in Drosophila and its parasitoids. Am Nat. 1999;153:S61–S74. doi: 10.1086/303212. [DOI] [PubMed] [Google Scholar]
- Kurz CL, Ewbank JJ. Caenorhabditis elegans for the study of host–pathogen interactions. Trends Microbiol. 2000;8:142–144. doi: 10.1016/s0966-842x(99)01691-1. [DOI] [PubMed] [Google Scholar]
- Lipsitch M, Siller S, Nowak AM. The evolution of virulence in pathogens with vertical and horizontal transmission. Evolution. 1996;50:1729–1741. doi: 10.1111/j.1558-5646.1996.tb03560.x. [DOI] [PubMed] [Google Scholar]
- Little TJ, Shuker DM, Colegrave N, Day T, Graham AL. The coevolution of virulence: tolerance in perspective. PLoS Pathog. 2010;6:e1001006. doi: 10.1371/journal.ppat.1001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lively CM. Adaptation by a parasitic trematode to local populations of its snail host. Evolution. 1989;43:1663–1671. doi: 10.1111/j.1558-5646.1989.tb02616.x. [DOI] [PubMed] [Google Scholar]
- Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S, Kohara Y, Ewbank JJ. Inducible antibacterial defense system in C. elegans. Curr Biol. 2002;12:1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- Messenger AL, Molineux IJ, Bull JJ. Virulence evolution in a virus obeys a trade-off. Proc Roy Soc B. 1999;266:397–404. doi: 10.1098/rspb.1999.0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikonranta L, Friman VP, Laakso J, Vinatzer BA. Life history trade-offs and relaxed selection can decrease bacterial virulence in environmental reservoirs. PloS One. 2012;7:1–9. doi: 10.1371/journal.pone.0043801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran L, Schmidt O, Gelarden I, Parrish R, II, Lively CM. Running with the Red Queen: host-parasite coevolution selects for biparental sex. Science. 2011;333:216–218. doi: 10.1126/science.1206360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morran LT, Parrish R, II, Gelarden I, Allen MB, Lively CM. Experimental coevolution: rapid local adaptation by parasites depends on host mating system. Am Nat. 2014;184:S91–S100. doi: 10.1086/676930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi MK, Nair VS. Evolution of symbiosis in the Vibrionaceae: a combined approach using molecules and physiology. Int J Syst Evol Microbiol. 2003;53:2019–2026. doi: 10.1099/ijs.0.02792-0. [DOI] [PubMed] [Google Scholar]
- Noda H, Munderloh UG, Kurtti TJ. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl Environ Microbiol. 1997;63:3926–3932. doi: 10.1128/aem.63.10.3926-3932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MA. Local population differentiation for compatibility in an annual legume and its host-specific fungal pathogen. Evolution. 1985;39:713–723. doi: 10.1111/j.1558-5646.1985.tb00414.x. [DOI] [PubMed] [Google Scholar]
- Petersen LM, Tisa LS. Friend or foe? A review of the mechanisms that drive Serratia towards diverse lifestyles. Can J Microbiol. 2013;59:627–640. doi: 10.1139/cjm-2013-0343. [DOI] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Nat Acad Sci USA. 2007;104:2295–2300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. http://www.R-project.org. [Google Scholar]
- Restif O, Hochberg ME, Koella JC. Virulence and age at reproduction: new insights into host-parasite coevolution. J Evol Biol. 2001;14:967–979. [Google Scholar]
- Restif O, Koella J. Shared control of epidemiological traits in a coevolutionary model of host-parasite interactions. Am Nat. 2003;161:827–836. doi: 10.1086/375171. [DOI] [PubMed] [Google Scholar]
- Sachs JL, Wilcox TP. A shift to parasitism in the jellyfish symbiont Symbiodinium microadriaticum. Proc Roy Soc B. 2006;273:425–429. doi: 10.1098/rspb.2005.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada H, Kuykendall LD, Young JM. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J Gen Appl Microbiol. 2003;49:155–179. doi: 10.2323/jgam.49.155. [DOI] [PubMed] [Google Scholar]
- Schulenburg H, CL K, JJ E. Evolution of the innate immune system: the worm perspective. Immunol Rev. 2004;198:36–58. doi: 10.1111/j.0105-2896.2004.0125.x. [DOI] [PubMed] [Google Scholar]
- Schulenburg H, Ewbank J. Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol Biol. 2004;4:49. doi: 10.1186/1471-2148-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NG. The advantage of being parasitized. Nature. 1968;219:690–694. doi: 10.1038/219690a0. [DOI] [PubMed] [Google Scholar]
- Stewart AD, Logsdon JM, Kelley SE. An empirical study of the evolution of virulence under both horizontal and vertical transmission. Evolution. 2005;59:730–739. [PubMed] [Google Scholar]
- Thompson J. In: Extreme specialization in mutualists. Thompson J, editor. The Coevolutionary Process. University of Chicago Press; Chicago: 1994. pp. 167–185. [Google Scholar]
- Thompson JN. Variation in interspecific interactions. Ann Rev Ecol Syst. 1988:65–87. [Google Scholar]
- Thompson JN. The Geographic Mosaic of Coevolution. University of Chicago Press; Chicago, IL, USA: 2005. [Google Scholar]
- Thompson JN, Cunningham BM. Geographic structure and dynamics of coevolutionary selection. Nature. 2002;417:735–738. doi: 10.1038/nature00810. [DOI] [PubMed] [Google Scholar]
- Traub E. Epidemiology of lymphocytic choriomeningitis in a mouse stock observed for four years. J Exp Med. 1939;69:801–817. doi: 10.1084/jem.69.6.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PE, Cooper VS, Lenski RE. Tradeoff between horizontal and vertical modes of transmission in bacterial plasmids. Evolution. 1998;52 doi: 10.1111/j.1558-5646.1998.tb01634.x. [DOI] [PubMed] [Google Scholar]
- van Baalen M. Coevolution of recovery ability and virulence. Proc Roy Soc B. 1998;265:317–325. doi: 10.1098/rspb.1998.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ. The co-evolutionary genetics of ecological communities. Nature Reviews Genetics. 2007;8:185–195. doi: 10.1038/nrg2031. [DOI] [PubMed] [Google Scholar]
- Wasik BR, Bhushan A, Ogbunugafor CB, Turner PE. Delayed transmission selects for increased survival of vesicular stomatitis virus. Evolution. 2015;69:117–125. doi: 10.1111/evo.12544. [DOI] [PubMed] [Google Scholar]
- Weeks AR, Turelli M, Harcombe WR, Reynolds KT, Hoffmann AA. From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 2007;5:e114. doi: 10.1371/journal.pbio.0050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.