Abstract
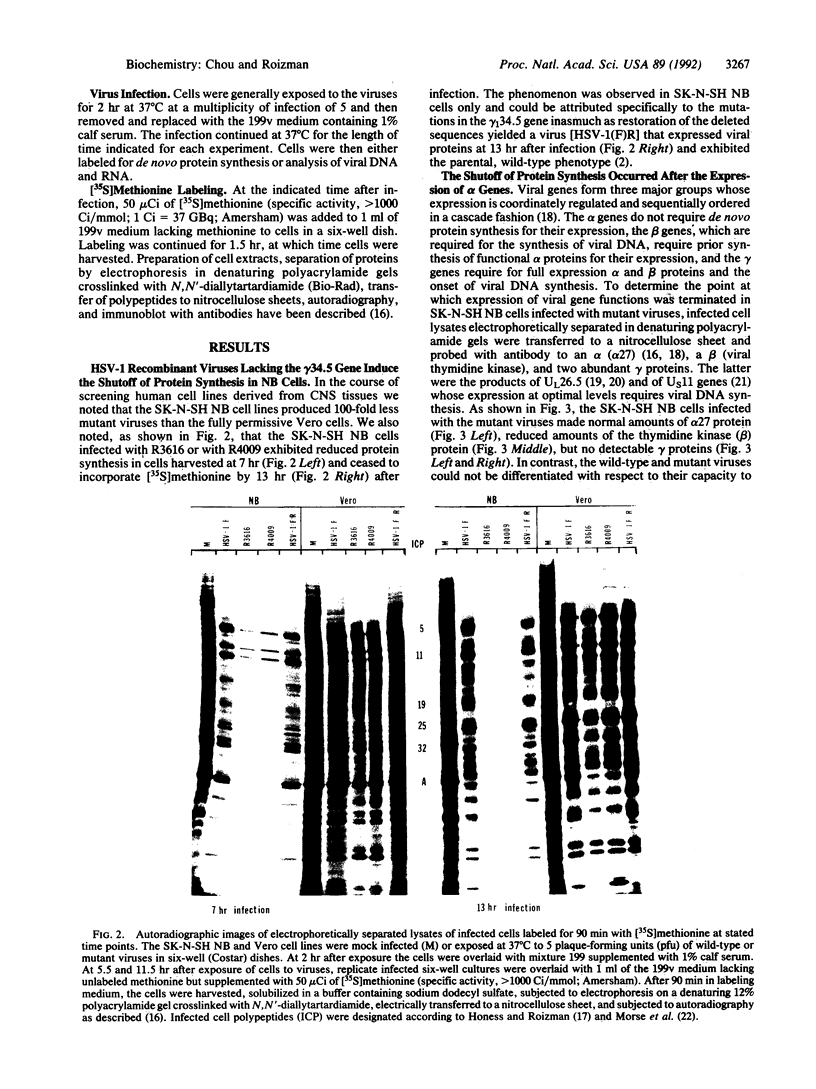
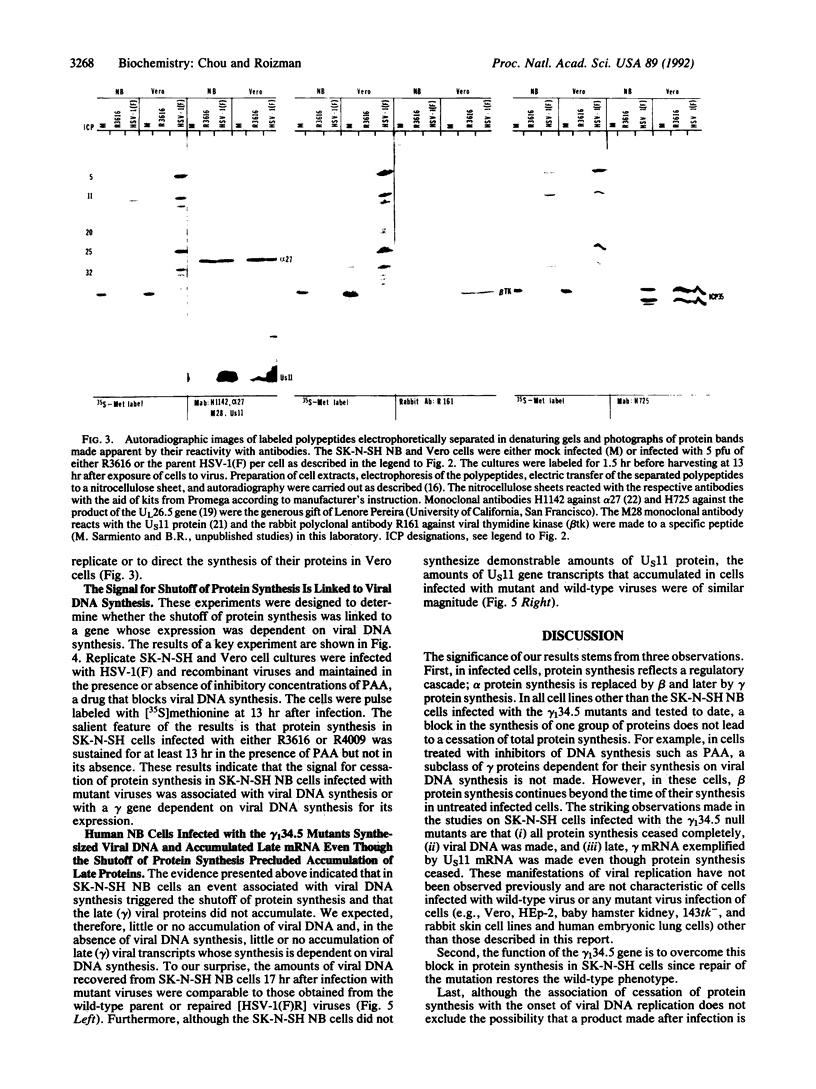
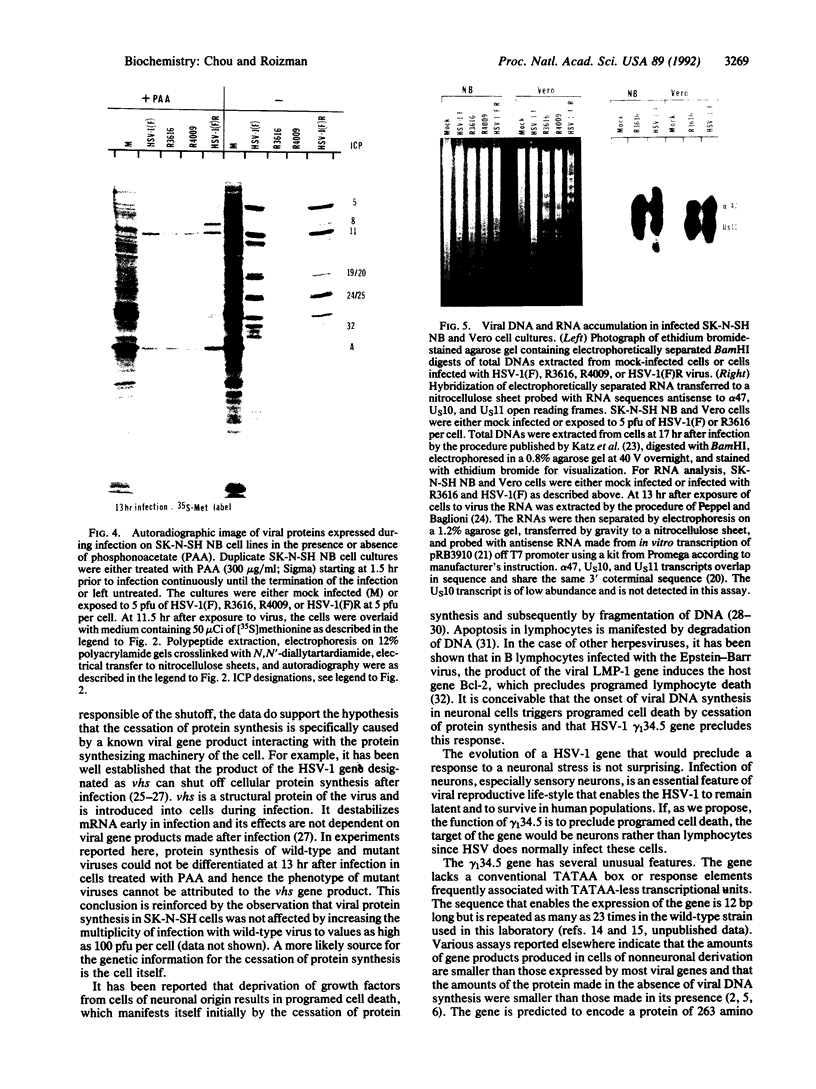
The gamma 1(34.5) gene of herpes simplex virus 1 was previously shown to play a role in viral virulence since deletion of the gene reduced by a factor of approximately 100,000 the capacity of the virus to replicate in the central nervous system and cause mortality in the mouse. Here we show that in the human neuroblastoma cell line SK-N-SH of neuronal origin gamma 1(34.5) null mutants expressed early proteins, viral DNA, and mRNA of late genes. However, the onset of viral DNA synthesis triggered complete cessation of incorporation of radioactive precursors into proteins. The mutant and wild-type viruses replicated and could not be differentiated in cell lines or cell strains of nonneuronal origin. The results indicate that in the absence of the gamma 1(34.5) gene the SK-N-SH neuroblastoma cells triggered a response similar to the programed cell death of neuronal cells induced by metabolic stress. The gamma 1(34.5) protein precludes this cell response possibly in order to enable the protein synthesis necessary for viral replication.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackermann M., Braun D. K., Pereira L., Roizman B. Characterization of herpes simplex virus 1 alpha proteins 0, 4, and 27 with monoclonal antibodies. J Virol. 1984 Oct;52(1):108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M., Chou J., Sarmiento M., Lerner R. A., Roizman B. Identification by antibody to a synthetic peptide of a protein specified by a diploid gene located in the terminal repeats of the L component of herpes simplex virus genome. J Virol. 1986 Jun;58(3):843–850. doi: 10.1128/jvi.58.3.843-850.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centifanto-Fitzgerald Y. M., Yamaguchi T., Kaufman H. E., Tognon M., Roizman B. Ocular disease pattern induced by herpes simplex virus is genetically determined by a specific region of viral DNA. J Exp Med. 1982 Feb 1;155(2):475–489. doi: 10.1084/jem.155.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Kern E. R., Whitley R. J., Roizman B. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science. 1990 Nov 30;250(4985):1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- Chou J., Roizman B. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+. J Virol. 1990 Mar;64(3):1014–1020. doi: 10.1128/jvi.64.3.1014-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. The terminal a sequence of the herpes simplex virus genome contains the promoter of a gene located in the repeat sequences of the L component. J Virol. 1986 Feb;57(2):629–637. doi: 10.1128/jvi.57.2.629-637.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke R. C., Cohen J. J. IL-2 addiction: withdrawal of growth factor activates a suicide program in dependent T cells. Lymphokine Res. 1986 Fall;5(4):289–299. [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Henderson S., Rowe M., Gregory C., Croom-Carter D., Wang F., Longnecker R., Kieff E., Rickinson A. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell. 1991 Jun 28;65(7):1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Roizman B. Proteins specified by herpes simplex virus. XI. Identification and relative molar rates of synthesis of structural and nonstructural herpes virus polypeptides in the infected cell. J Virol. 1973 Dec;12(6):1347–1365. doi: 10.1128/jvi.12.6.1347-1365.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javier R. T., Thompson R. L., Stevens J. G. Genetic and biological analyses of a herpes simplex virus intertypic recombinant reduced specifically for neurovirulence. J Virol. 1987 Jun;61(6):1978–1984. doi: 10.1128/jvi.61.6.1978-1984.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. M., Jr, Chang J. Y., Koike T., Martin D. P. Why do neurons die when deprived of trophic factor? Neurobiol Aging. 1989 Sep-Oct;10(5):549–553. doi: 10.1016/0197-4580(89)90127-9. [DOI] [PubMed] [Google Scholar]
- Katz J. P., Bodin E. T., Coen D. M. Quantitative polymerase chain reaction analysis of herpes simplex virus DNA in ganglia of mice infected with replication-incompetent mutants. J Virol. 1990 Sep;64(9):4288–4295. doi: 10.1128/jvi.64.9.4288-4295.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikorian C. R., Read G. S. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J Virol. 1991 Jan;65(1):112–122. doi: 10.1128/jvi.65.1.112-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong A. D., Kruper J. A., Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988 Mar;62(3):912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. Y., Roizman B. The promoter, transcriptional unit, and coding sequence of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J Virol. 1991 Jan;65(1):206–212. doi: 10.1128/jvi.65.1.206-212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord K. A., Hoffman-Liebermann B., Liebermann D. A. Sequence of MyD116 cDNA: a novel myeloid differentiation primary response gene induced by IL6. Nucleic Acids Res. 1990 May 11;18(9):2823–2823. doi: 10.1093/nar/18.9.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Barnett B. C. Neurovirulence factor. Nature. 1991 Oct 17;353(6345):609–609. doi: 10.1038/353609b0. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Post L. E., Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980 Nov;22(1 Pt 1):243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Morse L. S., Pereira L., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus (HSV) DNA. X. Mapping of viral genes by analysis of polypeptides and functions specified by HSV-1 X HSV-2 recombinants. J Virol. 1978 May;26(2):389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppel K., Baglioni C. A simple and fast method to extract RNA from tissue culture cells. Biotechniques. 1990 Dec;9(6):711–713. [PubMed] [Google Scholar]
- Rawson C. L., Loo D. T., Duimstra J. R., Hedstrom O. R., Schmidt E. E., Barnes D. W. Death of serum-free mouse embryo cells caused by epidermal growth factor deprivation. J Cell Biol. 1991 May;113(3):671–680. doi: 10.1083/jcb.113.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read G. S., Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate early) viral polypeptides. J Virol. 1983 May;46(2):498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller R. J., Roizman B. Herpes simplex virus 1 RNA-binding protein US11 negatively regulates the accumulation of a truncated viral mRNA. J Virol. 1991 Nov;65(11):5873–5879. doi: 10.1128/jvi.65.11.5873-5879.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Taha M. Y., Clements G. B., Brown S. M. The herpes simplex virus type 2 (HG52) variant JH2604 has a 1488 bp deletion which eliminates neurovirulence in mice. J Gen Virol. 1989 Nov;70(Pt 11):3073–3078. doi: 10.1099/0022-1317-70-11-3073. [DOI] [PubMed] [Google Scholar]
- Thompson R. L., Rogers S. K., Zerhusen M. A. Herpes simplex virus neurovirulence and productive infection of neural cells is associated with a function which maps between 0.82 and 0.832 map units on the HSV genome. Virology. 1989 Oct;172(2):435–450. doi: 10.1016/0042-6822(89)90186-4. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. T. Programmed cell death: apoptosis and oncogenesis. Cell. 1991 Jun 28;65(7):1097–1098. doi: 10.1016/0092-8674(91)90002-g. [DOI] [PubMed] [Google Scholar]