Abstract
Pseudoknots are a fundamental RNA tertiary structure with important roles in regulation of mRNA translation. Molecular force spectroscopic approaches such as optical tweezers can track the pseudoknot’s unfolding intermediate states by pulling the RNA chain from both ends, but the kinetic unfolding pathway induced by this method may be different from that in vivo, which occurs during translation and proceeds from the 5′ to 3′ end. Here we developed a ribosome-mimicking, nanopore pulling assay for dissecting the vectorial unfolding mechanism of pseudoknots. The pseudoknot unfolding pathway in the nanopore, either from the 5′ to 3′ end or in the reverse direction, can be controlled by a DNA leader that is attached to the pseudoknot at the 5′ or 3′ ends. The different nanopore conductance between DNA and RNA translocation serves as a marker for the position and structure of the unfolding RNA in the pore. With this design, we provided evidence that the pseudoknot unfolding is a two-step, multistate, metal ion-regulated process depending on the pulling direction. Most notably, unfolding in both directions is rate-limited by the unzipping of the first helix domain (first step), which is Helix-1 in the 5′ → 3′ direction and Helix-2 in the 3′ → 5′ direction, suggesting that the initial unfolding step in either pulling direction needs to overcome an energy barrier contributed by the noncanonical triplex base-pairs and coaxial stacking interactions for the tertiary structure stabilization. These findings provide new insights into RNA vectorial unfolding mechanisms, which play an important role in biological functions including frameshifting.
Graphical abstract
INTRODUCTION
Gene expression can be regulated through folding and unfolding mRNA tertiary structures.1–6 Pseudoknots are a fundamental structure found in almost all classes of RNAs7–15 such as human telomerase RNA,6 self-splicing introns of ribozymes16,17 and many noncoding RNAs.18,19 The typical H-type pseudoknot is formed by base-pairing between the loop of a hairpin and a complementary fragment outside the stem in the RNA chain. Such a structure, containing multiple stemloops, can be stabilized through noncanonical triplex base pairs and coaxial stacking interactions.6,20–22 Remarkably, in the frameshifting mechanism for translational control of viral protein synthesis (e.g., HIV, SARS and HCV),7,9,11,14 the ribosome needs to disrupt a downstream pseudoknot in a retroviral RNA to continue the translation with a shifted open reading frame (ORF). This RNA unfolding-regulated mechanism enables expression of different proteins from the same viral RNA genome for virus replication and proliferation. Pseudoknots are also the basic structures of riboswitches (e.g., PreQ-I), a class of regulatory fragments in mRNAs that can change conformation upon binding a small metabolic molecule for protein expression modulation (for a review see ref 23 and references therein). Therefore, understanding the mechanism by which pseudoknots unfold enable the development of new antiviral drugs, which reduce or eliminate virus production24–26 by modulating the stability of RNA structures.27–31
Many studies using thermodynamic analysis6,12,32–34 and computer simulations20–22,35–37 have produced a large amount of information linking the kinetics and thermodynamics of pseudoknots to their functional properties.38,39 Recently, the molecular force spectroscopes, e.g., optical tweezers and AFM, have been successfully applied to the study of pseudoknot unfolding kinetics.40–45 By pulling the pseudoknot from both ends, this approach can identify the intermediate states along the folding pathway.46–48 However, current methods also have limitations. Heating or pulling from both ends of molecules may disrupt a RNA structure starting from the most unstable regions, and the relative stabilities of different regions determine their unfolding order.21,49 This unfolding pathway could differ from that in vivo, in which the pseudoknot in mRNA undergoes a vectorial unfolding process,50 i.e., directional unfolding along the RNA backbone caused by the ribosome in the 5′ → 3′ direction.
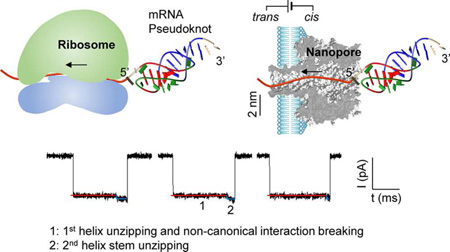
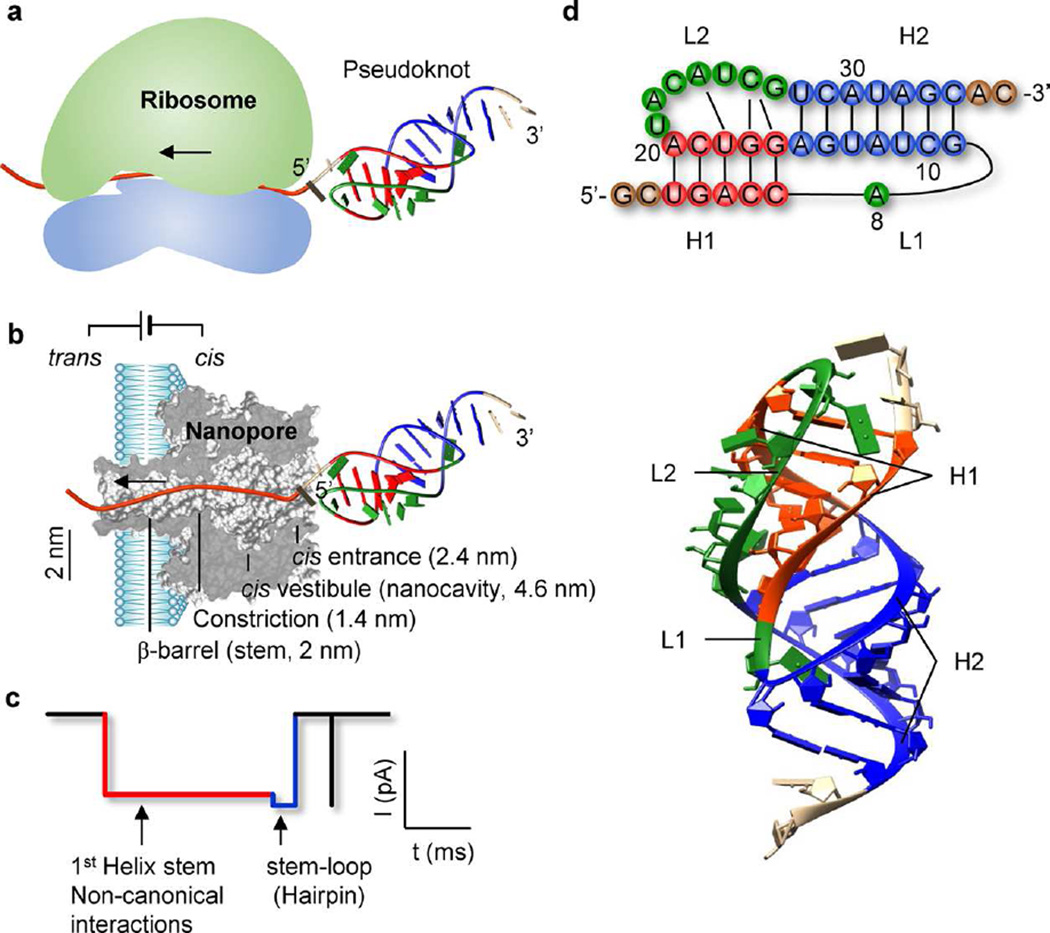
In this report, we investigated a ribosome-mimicking, protein nanopore-based pulling assay for dissecting pseudoknot’s vectorial unfolding mechanism (Figure 1). In cells, the ribosome forms a channel to embrace mRNA for translation from 5′ to 3′ end (Figure 1a). Similarly, the α-hemolysin protein pore is 1.4 nm wide in the constrictive region51 (Figure 1b). Such a pore size only allows translocation of single-stranded nucleic acids,52 but folded nucleic acids must unfold to pass through the pore.53–55 In the past two decades, the nanopore translocation of various DNAs and RNAs,52,56–60 including long RNAs (under denaturing conditions)61 and tRNAs,62 have been extensively studied, driven by the goal of the next generation sequencing.63,64 Furthermore, the nanopore has been developed as a force spectroscopy for exploring protein-nucleic acid interactions,65 unfolding of proteins66,67 and unzipping of nucleic acid structures such as hairpins,68,69 G-quadruplex,53–55,70 and microRNAs.71,72 Parallel to these efforts, the methodology of nanopore force spectroscopy (voltage-ramp) has been established for nucleic acids kinetic analysis.41,73–75 These prior studies provide a solid basis for elucidating nanopore unfolding of RNA tertiary structures. Recently, the braked translocation of tRNAs in the nanopore has been reported62 for the purpose of detecting tRNA population, but the RNA unfolding mechanisms have not been elucidated.
Figure 1.
Mimicking ribosomal unfolding of RNA pseudoknot in a protein nanopore. (a,b) Cartoons showing the unfolding of a downstream pseudoknot as ribosome slides on mRNA from the 5′ to 3′ end for protein expression (a), and vectorial unfolding of a tagged-pseudoknot in the α-hemolysin protein channel (b). (c) Current signature in response to the unfolding of a pseudoknot in the nanopore, with distinct blocking levels revealing the unfolding sequence of different parts in the pseudoknot. (d) 2D and 3D structures of the RNA pseudoknot located at the 5′ end of the gene 32 mRNA of bacteriophage T2 (PDB id: 2tpk), which contains two helices, H1 (red, U3-C7:G16-A20) and H2 (blue, G9-A15:U28-C34),76 and two loops (green), L1 (A8) and L2 (U21-G27). The base triple interactions between loops and helices (identified by RNAview110) in magenta and the coaxial stacking between two helices are illustrated.
In the current study, the RNA pseudoknot was forced to unfold either from the 5′ to 3′ end (the same as ribosome-induced unfolding) or in the reverse direction, controlled by a DNA leader covalently attached to the 5′ or 3′ end of pseudoknot. Because of the different nanopore conductance for DNA and RNA translocation, we were able to monitor the position of unfolded RNA in the pore, therefore determining the pseudoknot’s stability, tracking its stepwise unfolding and translocation, and identifying intermediate states along the unfolding pathway (Figure 1c). Importantly, the different unfolding kinetics observed between two pulling directions allows us to identify noncanonical interactions involved in pseudoknot’s stability.
Our target is the gene 32 mRNA pseudoknot of bacteriophage T276 (Figure 1d). This short RNA contains all the structural elements and interactions needed for pseudoknot assembly, therefore providing a good model for studying the RNA unfolding. T2 pseudoknot’s structure is highly similar to other pseudoknots, including those involved in viral frameshifting (e.g., HIV),39 making it possible to study nanopore-based unfolding of these critical RNA structures and their mechanisms for controlling gene regulation.
RESULTS AND DISCUSSION
Construction of DNA-Tagged Pseudoknots and Nanopore Signatures
The 36-nt T2 pseudoknot (Figure 1d) comprises two short helical stems, H1 at the 5′ end (5-bp) and H2 at the 3′ end (7-bp).76 The inner end of H1 and the outer (3′-terminal) end of H2 are connected via a single-nucleotide loop L1 (A8), which makes a sharp turn associated with a strong structural tension. The outer (5′-terminal) end of H1 and the inner end of H2 are linked via another loop L2 (7 nts). L2 spans across the shallow groove of H1 to form tertiary interactions with H1 through noncanonical base pairing, making H1 virtually a stable triplex. Such a structure allows coaxial stacking of H1 and H2 on each other, forming a long quasi-continuous helix with surprisingly high stability.76
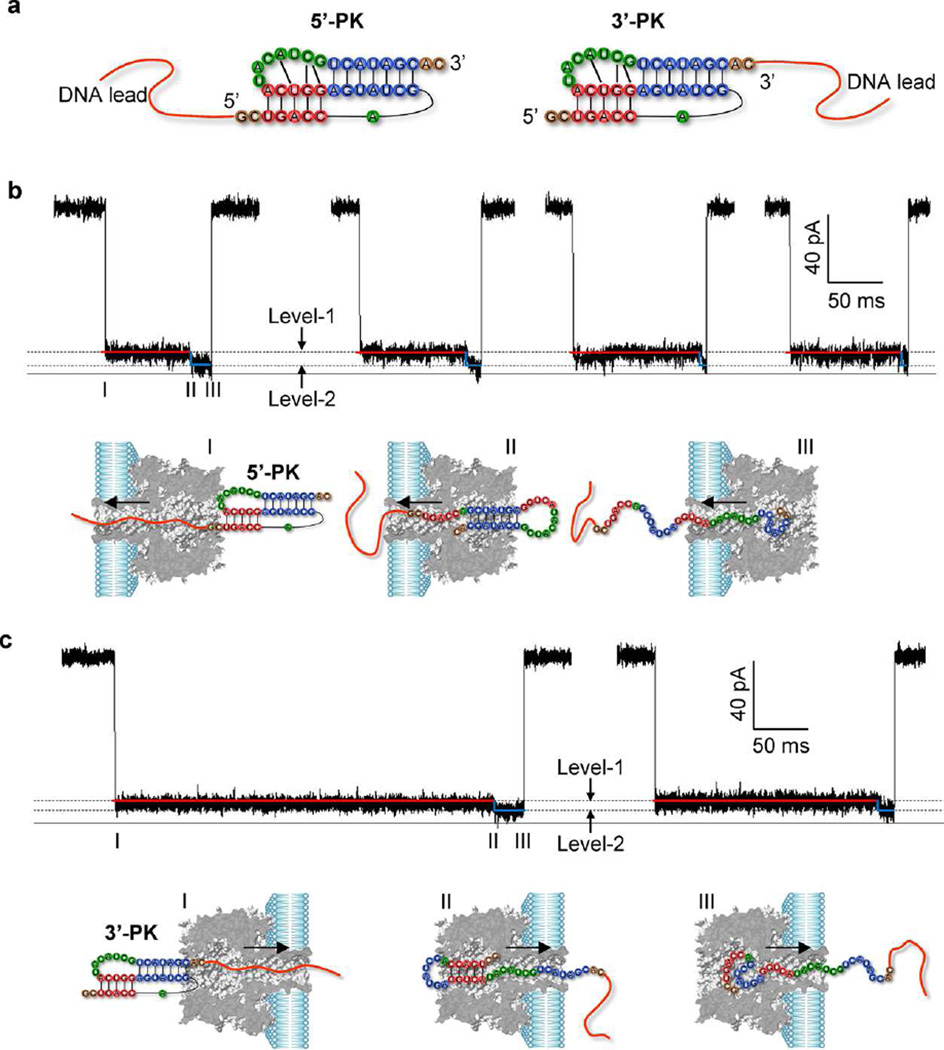
We attached a Poly(CAT)10 leader to the 5′ end or 3′ end of the pseudoknot RNA to form DNA-RNA chimeras 5′-PK and 3′-PK (Table 1, Figure 2a). Under a transmembrane voltage, the DNA leader is captured in the nanopore and drives the unfolding of the downstream pseudoknot from the 5′ to 3′ end for 5′-PK, which mimics the ribosomal unfolding direction, and from the 3′ to 5′ end for 3′-PK. The pulling force on DNA/RNA in the pore is ~10 pN at 120 mV (Figure S1 for force evaluation), which is consistent with the force in prior nanopore pulling experiment,77 and similar to the force for pseudoknot unfolding in the force spectroscopy experiment.46
Table 1.
Sequences of Nucleic Acids Used in This Study
| Nucleic acids | Sequence |
|---|---|
| 5′-PK | |
| 3′-PK | |
| T2-PK | |
| 5′-PK-mut | |
| 3′-PK-mut | |
| 2H | |
| Poly(CAT)10 | |
| miR-155 |
Figure 2.
Nanopore signatures revealing vectorial unfolding procedure of T2 pseudoknot. (a) Construction of DNA-tagged pseudoknot chimeras. A poly(CAT)10 ssDNA is attached to the 5′ end (5′-PK) and 3′ end (3′-PK) of T2 pseudoknot for trapping the molecule into the pore and drive its unfolding. (b,c) Representative two-level signatures (Level-1 and Level-2) for stepwise unfolding of 5′-PK from H1 at 5′ end to H2 at 3′ end (b), and 3′-PK from H2 at 3′ end to H1 at 5′ end (c). Models below the current traces illustrate the suggested molecular configurations of 5′-PK (b) and 3′-PK (c) for each unfolding step. The nanopore currents were recorded at +120 mV in 1 M NaCl and 10 mM MgCl2 buffered with 25 mM MOPS (pH 7.4).
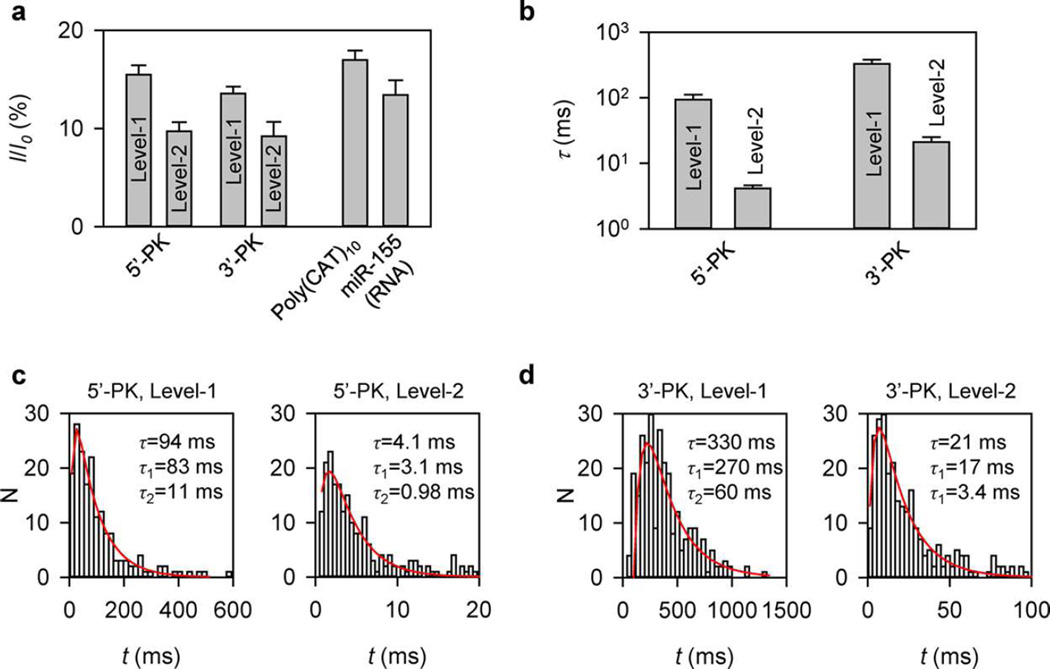
Monitored at +120 mV in 1 M NaCl and 10 mM MgCl2, both 5′-PK and 3′-PK presented on cis side of the nanopore generated a two-level blocking event (Figure 2b and c). Its conductance always started at Level-1, and terminated at Level-2, which was slightly lower than Level-1. The Level-1 stage dominated the entire block duration. The relative conductance (I/I0) of Level-1 and Level-2 was 15 ± 1% and 9.7 ± 0.9% for 5′-PK, and 13 ± 1% and 9.2 ± 1.5% for 3′-PK (Figure 3a and Figure S2). This block pattern was not observed for either the DNA leader alone or a short linear form RNA without a tertiary structure (Figure S3), and was distinct from the signal for nanopore unzipping of a hairpin68 or a double-stranded oligonucleotide.72,77,78 We hypothesize that this unique current pattern is the signature of the chimeras that form a pseudoknot, and reveals the trapping, unfolding and translocation of the pseudoknot in the nanopore.
Figure 3.
Characteristic blocking levels and durations of different steps in signatures of pseudoknots and control oligonucleotides. (a) Relative conductance (I/I0, I and I0 are current amplitudes of the block and empty pore) of Level-1 and Level-2 blocks in the 5′-PK and 3′PK signatures shown in Figure 2b and c, and blocks generated by DNA Poly(CAT)10 and RNA miR-155 translocation through the pore. Blocks of Poly(CAT)10 and miR-155 were analyzed in Figure S3. (b) Durations of Level-1 and Level-2 stages in the 5′-PK and 3′-PK signatures. (c,d) Representative histograms of Level-1 (left) and Level-2 (right) durations for 5′-PK (N = 245) (c) and 3′-PK (N = 272) (d). The duration distributions in histograms were fitted with N(t) = A/(τ1 − τ2)[e−1t/τ1−e−t/τ2], based on a two-step kinetic pathway111 (see Figure S10 for the fitting method). The duration of each level τ was the sum of fitted τ1 and τ2.
First, the Level-1 conductance for 5′-PK (I/I0 = 15 ± 1%) and 3′-PK (13 ± 1%) was similar to the blocking level of DNA translocation (I/I0 = 17 ± 1%) (Figure 3a), suggesting that Level-1 originates from the entrapment of the DNA leader in the constrictive β-barrel51 (Figure S1). Notably, 3′-PK had a slightly lower Level-1 conductance compared to 5′-PK. This is consistent with a previous observation that the 3′ → 5′ DNA translocation reduced more conductance than the 5′ → 3′ translocation.79 Furthermore, as the voltage increased from +120 mV to +180 mV, the signature duration (τ) was consistently shortened by 10-fold from 98 ± 9 ms to 8.3 ± 2.1 ms for 5′-PK, and 30-fold from 350 ± 50 ms to 12 ± 2 ms for 3′-PK (Figure S4), indicating that the folded chimera is destabilized by the voltage applied and unfolds under the pulling. As the unfolded RNA enters the β-barrel in place of DNA, the nanopore current is transitioned to Level-2 with lower conductance, because RNA translocation produced deeper blocking level than DNA72 (Figure 3a and Figure S3). Overall, Level-1 and Level-2 in the current pattern are signals for sequential translocation of the DNA leader and unfolded RNA in the nanopore stem.
In addition to pseudoknot signatures, the chimeras also produced different nonpseudoknot block patterns (Figure S6). We found that 56% of the 5′-PK signals were pseudoknot signatures, and 11% were a type of short blocks (18 ± 3 ms) similar to the Level-2 current in the pseudoknot signature, supporting that these blocks are generated by partially folded hairpins that unzip in the nanopore. The remaining 27% spikes (0.3 ± 0.2 ms) and 6% spikes with a shoulder (1.7 ± 0.2 ms) should correspond to 1- and 2-step translocation of nucleic acids in extended conformations.80 Similar structural and block pattern diversity was also identified for 3′-PK (Figure S6).
To better understand the chimera configurations in the nanopore, we investigated the T2 pseudoknot without tags (T2-PK, Table 1). Unlike tagged pseudoknot, T2-PK only generated nonpseudoknot short blocks (I/I0 = 10 ± 2%, ~ 0.1–10 ms, Figure S5 and Figure S6), suggesting that the pseudoknot structure itself cannot be trapped into the cis vestibule. This is in contrast to the DNA G-quadruplex, which can be trapped in the nanocavity and partially reduce the pore current (I/I0 ~ 50%).54 Therefore, it was expected that when 5′- or 3′-PK is captured by the nanopore, its pseudoknot domain should be anchored at the cis entrance; and the chimera cannot be trapped in the opposite orientation with pseudoknot heading into the pore. By analyzing the block frequency, we estimated that 40% of T2-PK molecules fold into pseudoknot (Figure S6). This population is similar to the 5′- and 3′-PK pseudoknots, suggesting that the tag in chimeras does not significantly affect pseudoknot formation.
Vectorial Unfolding Pathways of T2 Pseudoknot
Pulled by the DNA leader, 5′-PK is expected to unfold in the 5′ → 3′ direction, with H1, which is directly connected to the DNA leader, unfolds first during the Level-1 stage (94 ± 6 ms, Figure 2b). The unzipping of H1 releases a hairpin that consists of the H2 stem and an enlarged loop comprising L2 and the G16-A20 domain dissociated from H1. The unzipping of H1 also releases the 5′ end G1-A8 domain, which acts as an overhang of H2 to enter the β-barrel, generating the Level-2 conductance. Pulled by the overhang, the stem of such a “fishhook” hairpin would be trapped into the cis vestibule, according to the literature.81 Therefore, the Level-1/Level-2 transition marks the occurrence of pseudoknot unfolding to a partially folded hairpin structure. The Level-2 stage (4.1 ± 0.3 ms) terminates upon unzipping of the H2 hairpin. The time scale for unzipping this 7-bp RNA hairpin is consistent with unzipping a 7–10-bp hairpin in previous report (~10 ms).68 In contrast to 5′-PK, 3′-PK is expected to unfold in the 3′ → 5′ direction. The 3′ end H2 that connects to the DNA leader unfolds first during the Level-1 stage (330 ± 40 ms), resulting in a hairpin with H1. As the unfolded RNA domain is pulled into the β-barrel, the conductance is transitioned to Level-2 for unzipping H1 of the hairpin (21 ± 3 ms). In summary, 5′-PK and 3′-PK are unfolded along different pathways. For the unfolding in the 5′ → 3′ direction (5′-PK), Level-1 is for unzipping H1 and Level-2 for H2; for the opposite 3′ → 5′ direction (3′-PK), Level-1 is for unzipping H2 and Level-2 for H1.
The duration of each blocking level further reveals intermediate unfolding states. For both 5′-PK and 3′-PK, the durations of Level-1 and Level-2 stages can be better fitted with a two components (Figure 3c and d), rather than single component exponential distribution. Such a duration distribution suggests that there exists at least one intermediate state in both Level-1 and Level-2 stages. The intermediate state in the Level-1 stage probably corresponds to a partially unzipped state of H1 for 5′-PK or H2 for 3′-PK. The partially unfolded state of H1 has been identified using molecular dynamic simulation.37 At a temperature of 350 K, H1 first unfolded in the outer region (U3-A20 pair), and underwent a reversible disruption–reformation process. The final unfolding of H1 started from its inner region close to the loop and proceeded from the inner region outward. The intermediate state identified in Level-2 stage occurs during the unzipping of the remaining (second) stem, H2 in 5′-PK and H1 in 3′-PK. The intermediate state during hairpin unzipping in the nanopore has not been reported previously. We interpret that when the hairpin is squeezed into the pore, the large loop may adjust its configuration. This process may involve new base-pair formation within the loop to facilitate the unzipping of the hairpin (Simulation result below).
Tertiary Interactions for Pseudoknot Stabilization
The block patterns reveal that, in both unfolding directions, the Level-1 stage is much longer than the Level-2 stage (Figure 2 and 3). For 5′-PK, Level-1 was 24-fold longer than Level-2, and for 3′-PK, Level-1 was 16-fold longer than Level-2, indicating that the unfolding in both directions is rate-limited by the first step. The disruption of the first helix determines the barrier of the overall unfolding process. As shown in Figure 1d, both helices involve rich tertiary interactions, such as the loop-helix base triples and helix–helix coaxial stacking interactions. Besides the stability of the helix alone, the disruption of a helix also involves breaking tertiary interactions. This finding suggests that there are more factors that could influence the unfolding time than the helix stability alone.21 One potential factor could be the asymmetry of tertiary interactions, including loop-helix base triples and coaxial stacking between the two helices. Here we utilized three mutant RNAs to demonstrate this effect.
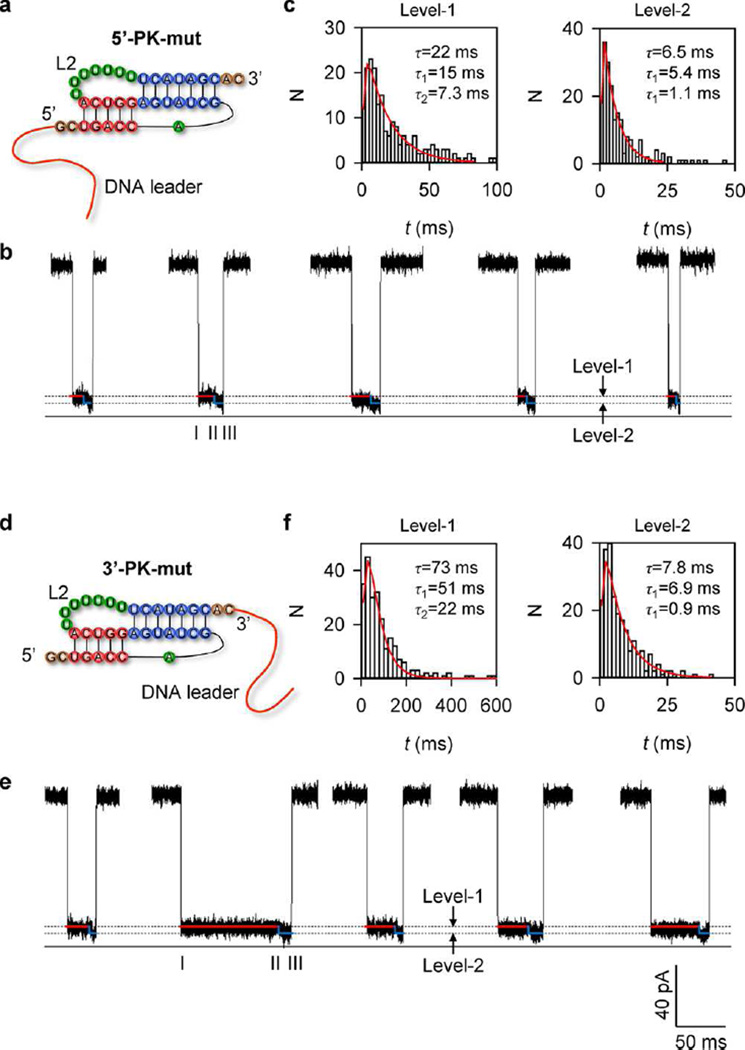
Mutations that disrupt loop-stem tertiary contacts have been found to dramatically reduce biological activity, often due to the decreasing thermodynamic stability of the native pseudoknot.13,33,82–88 To investigate the effect of the loop-stem tertiary contacts on the unfolding kinetics of the T2 pseudoknot, we substituted L2 with a poly(U) loop. Elastic property studies have shown that a single-stranded RNA poly(U) chain tends to be more disordered with weaker loopstem tertiary interactions.89,90 Similar to 5′- and 3′-PK, we attached a DNA leader to either end of the mutant pseudoknot to form the chimeras 5′-PK-mut and 3′-PK-mut (Figure 4a and d). Both mutant chimeras generated the two-level unfolding signatures (Figure 4b and e). However, the substitution of L2 with the poly(U) loop weakens loop-stem tertiary interactions, causing reduction in the dominating Level-1 duration (prior to the PK→ HP transition) compared to their wild-type (WT) counterparts, with reduction ratios of 4.3 (94 ± 6 ms/22 ± 4 ms) and 4.5 (330 ± 40 ms/73 ± 10 ms) for the 5′- and 3′-unfolding, respectively (Figure 4c and f), This result supports the possible correlation between the loop-helix tertiary interaction and the unfolding kinetic barrier. The barrier difference with respect to the corresponding WT chimeras was 0.84 kcal·mol−1 for unfolding in the 5′ → 3′ direction (5′-PK vs 5′-PK-mut), and 0.86 kcal·mol−1 in the 3′ → 5′ direction (3′-PK vs 3′-PK-mut) (Table 2), suggesting that the unfolding of T2 pseudoknot from either direction needs to overcome a similar initial barrier to break the loop-stem tertiary interaction.
Figure 4.
Unfolding of 5′- and 3′-PK-mut in the nanopore. (a) 2D structure of 5′-PK-mut, with L2 changed to poly(U). (b) Representative two-level signatures (Level-1 and Level-2) for stepwise unfolding of 5′-PK-mut from H1 at 5′ end to H2 at 3′ end. (c) Representative histograms of Level-1 (left) and level-2 (right) durations (N = 252), which were both fitted with a two-step exponential distribution as 5′-PK in Figure 3. (d) 2D structure of 3′-PK-mut. (e) Representative two-level signatures (Level-1 and Level-2) for stepwise unfolding of 3′-PK-mut from H2 at 3′ end to H1 at 5′ end. (f) Representative histograms of Level-1 (left) and Level-2 (right) durations (N = 239) and the two-step exponential fitting as 3′-PK in Figure 3.
Table 2.
Relative Stability of T2 Pseudoknot and Its Variants Revealed by Their Barrier Energy Differences
| ΔG (kcal·mol−1) | |
|---|---|
| 5′-PK vs 5′-PK-mut | −0.86a |
| 3′-PK vs 3′-PK-mut | −0.84a |
| 5′-PK vs 2H | −1.3b |
| 3′-PK vs 2H | −2.0b |
The unfolding barrier difference with respect to the unfolding of wild-type (WT) pseudoknot. ΔG = −RT ln(τWT/τmut). τWT and τmut were Level-1 block duration of WT and mutant pseudoknots.
The unfolding barrier difference with respect to the unfolding of 2H. ΔG = -RT ln(τ/τ2H), where τ2H is the block duration of 2H and τ is the block duration of 5′- and 3′-PK.
The poly(U) loop substitution also influences the Level-2 duration (prior to the HP → coil transition). For the 5′ pulling, poly(U) is located within the 12-nt hairpin loop, which prevents the proper base pairing and the formation of the intermediate structure. This large loop may prolong the Level-2 duration (6.5 ± 1.2 ms for 5′-PK-mut compared with 4.1 ± 0.3 ms for 5′-PK) due to the larger entropic barrier for loop compaction. For the 3′ pulling, even though the substitution of the L2 loop with poly(U) does not change the pathway of the HP → coil transition, the Level-2 duration decreased (7.8 ± 1.1 ms for 3′-PK-mut compared with 21 ± 3 ms for 3′-PK), probably because the 3′-poly(U) tail of the helix is highly flexible, causing an adjustable stretching of the chain and hence an efficient pulling. In another explanation, after the initial H2 unzipping (Level-1), the H1-L2 tertiary interactions may not be fully broken while the unfolded RNA domain has already entered the β-barrel (Level-2). Therefore, the Level-2 stage may involve the breaking of H1-L2 interactions in addition to H1 unzipping.
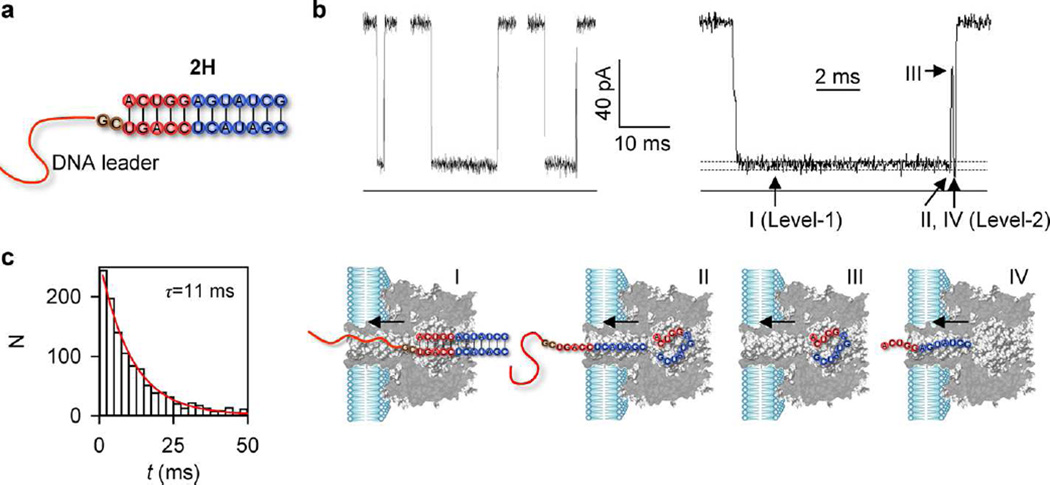
To eliminate the loop-helix tertiary interactions, we constructed a truncated RNA duplex 2H without loops (Table 1 and Figure 5a). 2H contains all 5 bps from H1 and 7 bps from H2. Since the two stems in the pseudoknot are coaxially stacked on each other to form a long (12 bps) quasi-continuous helical 3D structure, 2H should keep the stability of the (coaxially stacked) helical part but lack loop-helix tertiary interactions. This RNA duplex can be unzipped upon pulling by the attached DNA leader. The resulting signature reveals a multistep unzipping and translocation process (Figure 5b and Figure S7), which has been dissected in our previous studies.72 The 2H signature (11 ± 2 ms, Figure 5c) was 8.9- and 31-fold shorter than 5′- and 3′-PK signatures, and 2.6- and 8-fold shorter than 5′- and 3′-PK-mut signatures. Therefore, the unfolding of 5′-PK and 3′-PK, which contains coaxially stacking and strong loop-helix interactions, is harder than the unfolding of the 5′-PK-mut and 3′-PK-mut, which may involve weak loop-helix interactions, and much harder than unfolding of 2H, which has no loop-involved tertiary interactions. With respect to 2H, an additional energy barrier of 1.3 and 2.0 kcal·mol−1 are needed for 5′- and 3′-PK (Table 2). This additional energy cost may correspond to the disruption of loop-stem tertiary interactions, as suggested by previous simulation results.20 Notably, 3′-PK requires a longer time (330 ms) than 5′-PK (94 ms) to unfold in the rate-limiting Level-1 stage. This difference may be primarily caused by the different stabilities of the two helices. Breaking the 7-bp H2 (as the first step for unfolding 3′-PK) is slower than breaking the less stable 5-bp H1 (as the first step for unfolding 5′-PK).
Figure 5.
Unzipping of the RNA duplex 2H in the nanopore. (a) Structure of 2H, a poly(CAT)10-tagged, truncated pseudoknot that only retains the 5-bp H1 (red) and 7-bp H2 (blue) but without loops. (b) Representative multilevel blocking signatures for 2H (left) and an expanded signature (right) showing RNA unzipping and translocation as in the model below the traces: I, trapping the DNA leader in the pore (Level-1, I/I0 = 14.9 ± 1.2%); II, unzipping of 2H at the end of Level-1 stage followed by RNA translocation (Level-2, I/I0 = 9.7 ± 1.1%); III, intermediate partial block for the complementary RNA strand residing in the nanocavity (I/I0 = 52 ± 1.9%); and IV, the terminal Level-2 resistive pulse (I/I0 = 9.6 ± 1.2%) for translocation of the complementary RNA strand. Level-2 was lower than Level-1, in consistence with the translocation of DNA followed by RNA in the pseudoknot signatures (Figure 2b and c). (c) Duration histograms for the 2H signatures, fitted with a single-exponential distribution.
Kinetics Simulation
To better understand the mechanism of RNA pseudoknot unfolding through a nanopore, we used kinetic Monte Carlo (KMC) method (ref 91 and therein for algorithm) to simulate the unfolding process. The KMC method is powerful to provide detailed transition pathways and the distribution of unfolding time.91 We used the first passage time (FPT), i.e., the time for the first formation of a conformation from the initial conformations to characterize the unfolding time. For a specific target conformation, FPT is a random variable, but the mean FPT can be directly compared with the mean dwell time obtained by experiments. In the pseudoknot unfolding process, electric force on the DNA leader (Poly(CAT)10) in the nanopore exerts a pulling force on the RNA pseudoknot. To simulate the effect of the electric pulling force on the kinetic rates, we introduced a voltage V-dependent free energy change g(V) generated by the pulling force. The rate constant for the opening and closing of a base stack91,92 was calculated based on the Metropolis rule:93 k = min (k0, k0e−ΔG(V)/kBT), where ΔG (V) = ΔG (0) + g (V) and ΔG(0) is the force-free free energy difference between the closed and open states of a base stack. The kinetic barrier was modified by the additional change g(V) due to the external electric field V. The prefactor k0 is the attempt frequency, kB is the Boltzmann constant, and T is the temperature. To model the helix zipping, we followed Cocco et al.94 and restricted the initialization of the unzipping at the helix end where the force is applied. Therefore, a typical conformation in the unfolding process of an N-bp pseudoknot has the first n base pairs opened along the pulling direction and the rest N–n base pairs closed. After the disruption of the first helix, the hairpin loop of the remaining helix is enlarged. Depending on the sequence, the enlarged hairpin loop may allow new base pairs and mismatches to be formed within the loop, resulting in a stable stem-loop structure with smaller loop size. The enthalpies and entropies for such conformations were predicted using the V-fold model95 (Table S1).
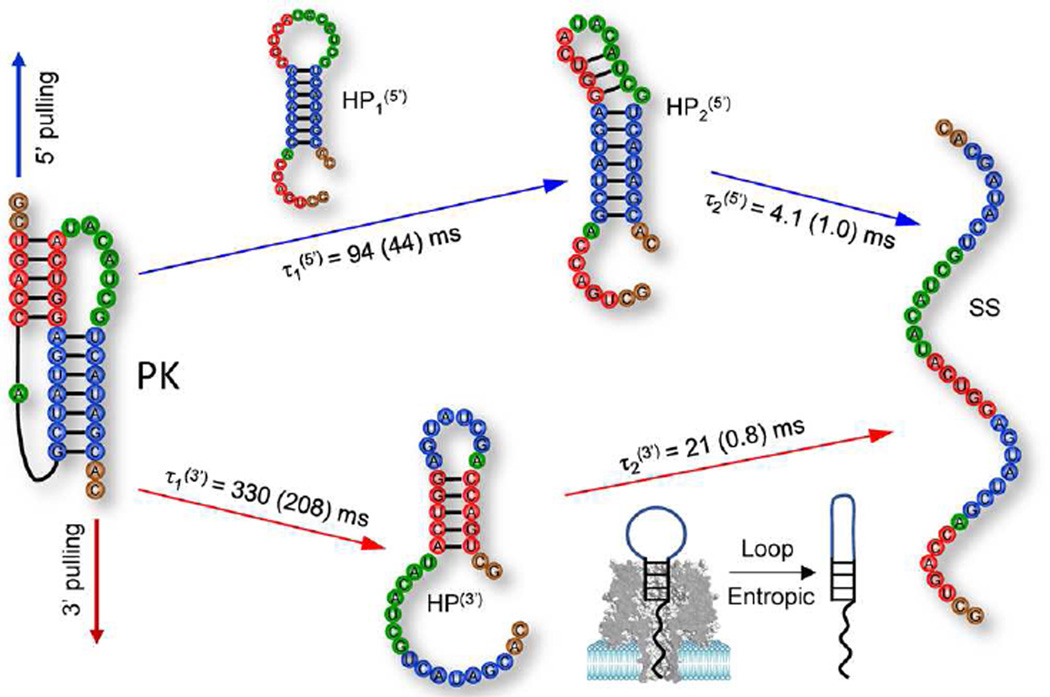
As shown in Figure 6, the pseudoknot unfolded through different pathways for different pulling directions. For the pulling from the 5′ end, the 5′ H1 (shown in red) was first disrupted, causing the conformational transition from the pseudoknot to a 12-nt hairpin HP1(5′). Because of the formation of the base pairs within its loop, HP1(5′) quickly folded into a compact stem-loop structure HP2(5′), which contains a 1-nt bulge loop and a 5-nt hairpin loop. Finally further pulling from the 5′ side caused the disruption of the base pairs in HP2(5′). Pulling from the 3′ end led to a different intermediate hairpin HP(3′). In contrast to HP1(5′), the sequence of the 8-nt large loop in HP(3′) does not allow the formation of stable intraloop base pairs.
Figure 6.
Predicted pathways of the nanopore unfolding of the T2 pseudoknot for the pulling from the 5′ end (blue) and from the 3′ end (red), respectively. Experimental dwell times and the mean first passage times (FPT) from KMC simulations (in parentheses) for the different steps are shown for comparison. The large difference of τ2(3′) (11 ms vs 0.8 ms) may be accounted for by the pausing time for the 8-nt loop in hairpin HP(3′) to overcome (mainly entropic) kinetic barrier to form compact conformations so HP(3′) can pass through the cis opening of the nanopore. In our model, we used g(V) = 1.8 kcal mol−1. Assuming g(V) = -Qeff·V as the electric potential-induced reduction in the energy barrier with V = 120 mV for the potential drop across the nanopore in the experimental setup, we found the effective net charge Qeff ≈ 0.7 e for the chain inside the pore.68 Noted that Qeff is sensitive to the molecular types and the solution conditions, such as ion concentrations and pH value.
Furthermore, the predicted unfolding times from our KMC simulations were consistent with the experimental data, except for the time for the unfolding of HP(3′) (τ2(3′)) due to the reason explained in the following. The large size of the 8-nt loop in HP(3′) may cause the structure to pause at the cis opening of the nanopore. Translocation would occur only after the loop forms a more compact conformation (Figure 6). The time duration required for loop-compaction is more pronounced for HP(3′) than for HP2(5′) due to the larger loop size (8 nts) and hence a larger entropic barrier. Without considering the HP(3′) pause, the KMC simulation predicted a dwell time of 0.8 ms. The pause of HP(3′) due to the large loop size may account for the difference between the experimental τ2(3′) result (21 ms) and the KMC result (0.8 ms).
Regulation of Pseudoknot Stability by Magnesium Ions and in Near-Physiology Ionic Condition
Mg2+ ions are critical for stabilizing tertiary structures and regulating biological functions of RNA structures such as pseudoknots34 and riboswitches96 and RNA-drug complexes.97 The nanopore’s capability in revealing the pseudoknot unfolding procedure allows investigating the Mg2+ effect on each unfolding step. We found that in the absence of Mg2+, the durations of both Level-1 and Level-2 stages were considerably shortened (Table 3). For 5′-PK, the durations of Level-1 and level-2 were shortened to 37 ± 5 ms and 2.4 ± 0.6 ms without Mg2+, compared to 94 ± 6 ms and 4.1 ± 0.3 ms in 10 mM Mg2+. Similarly, both stage durations for 3′-PK were shortened to 210 ± 25 ms and 11 ± 1 ms in the absence of Mg2+ from 330 ± 40 ms and 21 ± 3 ms in 10 mM Mg2+. Therefore, Mg2+ can prolong the durations of both stages for unfolding from either direction, suggesting that Mg2+ ions interact with and stabilize both the pseudoknot tertiary structure and its partially unfolded hairpins. This finding seems to be different from the Mg2+ effect on VPK pseudoknot,98 in which only one Mg2+ ion was considered to bind in a binding pocket formed by nucleotides in loop 1 and the major groove of stem 2.
Table 3.
Pseudoknot Blocking Durations at Various Mg2+ and Salt Concentrations
| 5′-PK |
3′-PK |
||||
|---|---|---|---|---|---|
| [Na+] | [Mg2+] | Level-1 (ms) | Level-2 (ms) | Level-1 (ms) | Level-2 (ms) |
| 1Ma | 10 mM | 94 ± 6 | 4.1 ± 0.3 | 330 ± 40 | 21 ± 3 |
| 0 mM | 37 ± 5 | 2.4 ± 0.6 | 210 ± 25 | 11 ± 1 | |
| 140 mMb | 10 mM | 18 ± 3 | 0.81 ± 0.09 | 41 ± 8 | 1.8 ± 0.2 |
| 1 mM | 13 ± 2 | 0.71 ± 0.13 | 25 ± 4.8 | 1.5 ± 0.1 | |
| 0 mM | 9.5 ± 1.7 | 0.67 ± 0.19 | 8.9 ± 2.3 | 0.49 ± 0.23 | |
cis and trans solutions contained 1 M NaCl, buffered with 25 mM MOPS (pH 7.4), without or with 10 mM MgCl2.
cis (5′-PK or 3′-PK presented), 140 mM NaCl, 5 mM KCl, 1 mM CaCl2; trans, 2 M NaCl. Both cis and trans solutions were buffered with 25 mM MOPS (pH 7.4) without or with 1 mM and 10 mM MgCl2.
To detect the pseudoknot unfolding at low salt concentration in near-physiological conditions, we added 140 mM NaCl in the cis solution in which pseudoknot was presented, and added 2 M NaCl in the trans solution. Such an asymmetric solution design allows the pseudoknot to be studied in low salt concentration on one side, while the high salt concentration in trans region ensures a high pore conductance needed for distinguishing different blocking levels without influencing the properties of pseudoknot in cis. In addition, the salt gradient across the pore can enhance the capture rate of nucleic acids molecules.72,99 We found that both 5′-PK and 3′-PK in 140 mM NaCl generated the same two-level signatures as in 1 M NaCl (Figure S8). However, the durations of all blocking levels (Figure S8) in their signatures were shortened compared with high salt concentration (Table 3). For 5′-PK in 140 mM NaCl, the duration of Level-1 and Level-2 stages were shortened to 18 ± 3 ms and 0.81 ± 0.09 ms, compared with 94 ± 7 ms and 4.1 ± 0.3 ms in 1 M NaCl. Similarly, low salt concentration reduced 3′-PK’s Level-1 and Level-2 durations to 41 ± 8 ms and 1.8 ± 0.2 ms, compared with 330 ± 40 ms and 21 ± 3 ms in high salt concentration. Notably, the duration of both levels in 3′-PK and 5′-PK signatures can be fitted into a two-component exponential function as in 1 M NaCl. All these results suggest that, similar to Mg2+, low Na+ concentration can also lower the stability of pseudoknot and its intermediate structures. However, low salt concentration does not change the unfolding pathway of pseudoknot in each direction. Therefore, the unfolding pathways dissected in 1 M NaCl could be implicated in the living cell environment.
Metal ions can screen the negative charges on nucleic acids and stabilize the folded RNA structures. Divalent Mg2+ ions are more effective compared with monovalent Na+ in screening negative charges on nucleic acids. Low Mg2+ concentration may be sufficient to stabilize a compact RNA structure. It has been found that Mg2+ at the millimolar (mM) level can cause similar folding stability as monovalent ion at the molar (M) level.100–102 We identified the similar trend in the unfolding kinetics of T2 pseudoknot. Overall, the pseudoknot stability can be regulated by both divalent Mg2+ and monovalent Na+. This finding suggests a possibility to program the pseudoknot stability by the combined use of multiple metal ions, in order for understanding the ribosomal unfolding of RNA tertiary structure in vivo and RNA-based drug design.103
DISCUSSION
The nanopore’s capability in unfolding along the backbone of biopolymer, i.e., vectorial unfolding, may help to reveal different intermediate states and unfolding pathway that is different from the thermodynamic approaches. UV–Vis thermal denaturation experiments reveal essentially identical melting profiles for T2, 3′-PK, 5′-PK, 3′-PK-mut, and 5′-PK-mut (Figure S9). The melting temperatures observed are comparable to those previously reported for the related gene 32 mRNA pseudoknot of bacteriophage T4.89,90 Thus, the melting studies indicate that the presence of poly(CAT)10 extensions on the pseudoknot structures do not alter their thermal stability. This, in turn, suggests that the pseudoknot structures remain unperturbed by the DNA leader. Furthermore, we do not observe significant differences in thermal stability (melting profiles) of pseudoknots containing different nucleobases in the loop regions of L2. Interestingly, pseudoknots containing base substitutions in the loop region do produce distinct signals in the nanopore experiments. This suggests that the nanopore has a unique capacity to detect these subtle differences in pseudoknot sequence/structure. It is possible that interactions of nucleobases in the loop with protein functional groups in the vestibule of the nanopore yield the observed differences in dwell time (or residual current). Alternatively, or in addition, the vectorial unfolding induced by pulling the pseudoknot through the pore may provide a distinct and more sensitive means, compared to thermal denaturation which unfolds globally and has a different unfolding mechanism, for detecting differences in the stability of these folded nucleic acid structures
The nanopore results indicate that the unfolding of the T2 pseudoknot involves two main steps with multiple intermediate states. This observation matches the results from molecular dynamics simulations,21,22,36,37 theoretical predictions,20,35,101 and thermal melting experiments6,12,32–34,104 for other pseudoknots such as T4 RNA and the human telomerase RNA pseudoknot. In combination with theoretical analysis, the nanopore kinetics experiment provided valuable insights into the interplay between the different interactions in RNA pseudoknot unfolding. These interactions include secondary structural interactions such as base pairing and base stacking, and tertiary structural interactions such as coaxial stacking and loop-helix base triple interactions. Unfolding of the pseudoknot involves disruptions of base pairs in the helix, coaxial stacking between helices, and loop-helix base triple contacts. For the sequential unfolding under specific directional pulling in the nanopore, coaxial stacking is broken after the unfolding of a helix. Furthermore, the competition and cooperation of the different interactions led to the different pathways. For the T2 pseudoknot, although the 5-bp H1 is less stable than the 7-bp H2, breaking H1 as the first step in 5′-PK is slower than breaking H2 as the second step due to the extensive stabilizing tertiary contacts between L2 and H1. Furthermore, comparison with the helix-only duplex system suggested that the kinetic barrier of breaking H1 (in 5′-PK) corresponds to the disruption of the triples, which indicated that the rate-limiting step is the disruption of first few base pairs from the 5′-end that results in a concomitant disruption of the triples. In contrast, for 3′-PK, the unfolding of H1 (as the second step) is very fast because the unfolding of H2 (as the first step) cooperatively causes destabilization of L2 and the tertiary contacts between L2 and H1.
In addition to the pseudoknot stability and unfolding kinetics, the nanopore signatures can be used to directly identify different structures of the designed chimeras. These structures, their corresponding molecular processes in the nanopore, and their populations are summarized in Figure S6. These T2-RNAs can not only form pseudoknot, but also form partially folded stem-loop hairpins, or adopt the extended conformation in our in vitro condition. The WT pseudoknot populations (T2-PK, 5′-PK and 3-PK) were about 50%, and the mutation (5′- and 3′-PK-mut) decreased the pseudoknot populations. This population difference is consistent with their mechanical stability difference revealed by the nanopore. The partially folded structures, if verified as a common property for different pseudoknots, may play functional roles such as controlling the frameshift efficiency.28,39,105 The nanopore results suggest that pseudoknot-forming RNA molecules may not all fold into the pseudoknot structure. Folded and unfolded RNAs are in equilibrium, and for most retroviruses, probably only the folded RNA can lead to the frameshifting. Therefore, the important follow-up studies can be envisioned to investigate how the in vitro observed pseudoknot folding and unfolding efficiency is correlated with their associated in vivo physiology function such as frameshifting.
CONCLUSION AND PERSPECTIVE
The nanopore single-molecule sensor combined with molecular design enables investigation of the vectorial, stepwise, multi-state unfolding of RNA pseudoknots. The nanopore data provides a direct comparison between the pseudoknot unfolding pathways under different pulling directions. The pulling direction dependent kinetics shows that in either pulling direction, the unfolding is rate-limited by the first step, namely, the disruption of the pseudoknot into a hairpin structure. It is possible that this unfolding step needs to disrupt noncanonical interactions such as triplex and coaxial stacking involved in the stabilization of the global RNA tertiary structure. As verified by the Monte Carlo kinetic simulation, the pseudoknot loop can also contribute to the unfolding rates by either forming new short helix or by adjusting its loop configurations. It could be possible to adapt the nanopore approach to study other RNA pseudoknots and riboswitches, and the unique information collected using this approach may provide new insights into in vivo unfolding mechanisms, gene regulation mechanisms such as frameshifting,106–109 and the design of new antiviral drugs.
METHODS
Chemicals and Nucleic Acids
All chemicals include NaCl, KCl, MgCl2, 3-(N-morpholino) propanesulfonic acid (MOPS), diethylpyrocarbonate (DEPC) were obtained from Sigma-Aldrich (St. Louis, MO, USA) and used as received. Lipid 1,2-diphytanoyl-sn-glycero-3-phosphocholine for bilayer formation was purchased from Avanti Polar Lipids (Alabaster, AL, USA) and used without further purification. DNAs, RNAs and DNA-RNA chimeras used in this study (Table 1) were synthesized by Integrated DNA Technologies Inc. (Coralville, IA).
Pseudoknot Formation
Synthetic DNA-RNA chimeras were first denatured in DEPC treated water at 95 °C for 5 min, immediately snap-cooled on ice, and adjusted to 50 mM NaCl, 25 mM MOPS (pH 7.4), 10 mM MgCl2 (or without MgCl2), or other composition of buffers as mentioned in the electrophysiology measurements and then either kept on ice or prewarmed at 37.5 °C for 3 h before electrophysiology measurement.
Single Protein Pore Formation, Current Recording and Event Analysis
Lipid bilayer composed of 1,2-diphytanoyl-sn-glycero-3-phosphocholine was formed on a ~150 µm orifice in a Teflon film that separates two Teflon chambers. For symmetric solution, each chamber contains 2 mL of solution containing 1 M NaCl, 10 mM MgCl2 and 25 mM MOPS (pH 7.4). For near-physiological experiments (asymmetric solutions), cis chamber was added with 140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0–10 mM MgCl2, and 25 mM MOPS (pH 7.4), and trans chamber with 2 M NaCl, 0–10 mM MgCl2, and 25 mM MOPS (pH 7.4). The concentration of all chimeras in the nanopore experiments was 100 nM. The concentration of T2-PK was 300 nM. 1–5 µL of α-hemolysin solution was added to the cis chamber to form single protein pores in the lipid bilayer membrane from the cis side. The ionic current through the pore was recorded using an Axopatch 200B amplifier (Molecular Devices Inc., Sunnyvale, CA). The amplified signal was filtered with a built-in 4-pole low-pass Bessel Filter at 5 kHz before acquired into the computer using a DigiData 1440A A/D converter (Molecular Devices) at a sampling rate of 20 kHz. The data recording and acquisition were controlled through a Clampex program (Molecular Devices) and nanopore current traces were analyzed using a Clampfit 10.4 software (Molecular Devices). Because the two-level pseudoknot unfolding signatures are distinguished from other types of blocks (Figure S6), there was no need to set cutoff for analyzing the signature duration and current amplitude. The duration of each current level (Level-1 and Level-2) in a signature was measured by two cursors in the software from the beginning to the end of the level. The duration values for all signatures were collected to construct histograms. The duration distribution was fitted with a two-exponential function (Figure S10) to obtain the durations of two intermediate states τ1 and τ2. Their sum (τ1 + τ2) yielded the mean duration τ of Level-1 or Level-2. The duration values from at least three experiments (n ≥ 3) with independent nanopores were used to obtain the mean τ, and its standard deviation (SD). The residual currents were obtained by fitting the event current amplitude histogram using Gaussian function in Clampfit. The standard deviation for each result value was based on at least three independent experiments. All nanopore pulling experiments were conducted at room temperature 22 ± 1 °C.
Melting Temperature Measurement
The melting temperatures of T2 RNA pseudoknot and its derivatives were determined by monitoring the increase in absorbance at 260 nm as a function of temperature. The temperature was increased from 22 to 95 °C at a rate of 0.5 °C/min. The melting temperature was determined using Cary WinUV-Thermal software. The first derivative of each smoothed absorbance curve with respect to temperature was calculated (∂A/∂T), and normalized by dividing the derivative value by the absorbance at 90 °C (ref 104).
Supplementary Material
Acknowledgments
We are grateful to the National Institutes of Health for support of this work through initial R01-GM079613 (L.Q.G.) and current R01-GM114204 (L.Q.G.), R01-GM063732 (S.J.C.), and ES021007 (K.S.G.).
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.5b07910.
More typical events for 5′-PK and 3′-PK and their blocking level, single strand DNA and RNA blocking levels, event duration’s voltage dependence, typical events for 2H, 5′-PK and 3′-PK events and duration in near physiological condition, various blocking events in additional to pseudoknot blocking events and their percentage, energy parameters used for the KMC simulations. (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Cruz JA, Westhof E. Cell. 2009;136:604. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Gleitsman KR, Herschlag DH. RNA. 2014;20:1732. doi: 10.1261/rna.044362.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JT. Science. 2012;338:1435. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 4.Mandal M, Breaker RR. Nat. Rev. Mol. Cell Biol. 2004;5:451. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 5.Mortimer SA, Kidwell MA, Doudna JA. Nat. Rev. Genet. 2014;15:469. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- 6.Theimer CA, Feigon J. Curr. Opin. Struct. Biol. 2006;16:307. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Belew AT, Meskauskas A, Musalgaonkar S, Advani VM, Sulima SO, Kasprzak WK, Shapiro BA, Dinman JD. Nature. 2014;512:265. doi: 10.1038/nature13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JL, Greider CW. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8080. doi: 10.1073/pnas.0502259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Nature. 2013;503:539. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narayanan R, Velmurugu Y, Kuznetsov SV, Ansari A. J. Am. Chem. Soc. 2011;133:18767. doi: 10.1021/ja205737v. [DOI] [PubMed] [Google Scholar]
- 11.Plant EP, Perez-Alvarado GC, Jacobs JL, Mukhopadhyay B, Hennig M, Dinman JD. PLoS Biol. 2005;3:e172. doi: 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staple DW, Butcher SE. PLoS Biol. 2005;3:e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theimer CA, Blois CA, Feigon J. Mol. Cell. 2005;17:671. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Tuerk C, MacDougal S, Gold L. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6988. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Bao P, Leibowitz MJ, Zhang Y. RNA. 2009;15:1986. doi: 10.1261/rna.1638609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herschlag D. RNA. 2015;21:527. doi: 10.1261/rna.050914.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serganov A, Patel DJ. Nat. Rev. Genet. 2007;8:776. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris KV, Mattick JS. Nat. Rev. Genet. 2014;15:423. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinn JL, Chang HY. Annu. Rev. Biochem. 2012;81:145. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao S, Giedroc DP, Chen SJ. RNA. 2010;16:538. doi: 10.1261/rna.1800210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho SS, Pincus DL, Thirumalai D. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17349. doi: 10.1073/pnas.0906625106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biyun S, Cho SS, Thirumalai D. J. Am. Chem. Soc. 2011;133:20634. doi: 10.1021/ja2092823. [DOI] [PubMed] [Google Scholar]
- 23.Peselis A, Serganov A. Wiley interdisciplinary reviews. RNA. 2014;5:803. doi: 10.1002/wrna.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brierley I, Pennell S, Gilbert RJ. Nat. Rev. Microbiol. 2007;5:598. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao S, Chen SJ. Phys. Biol. 2008;5:016002. doi: 10.1088/1478-3975/5/1/016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farabaugh PJ. Annu. Rev. Genet. 1996;30:507. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 27.Blount KF, Breaker RR. Nat. Biotechnol. 2006;24:1558. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 28.Brierley I, Digard P, Inglis SC. Cell. 1989;57:537. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. Cell. 2009;139:770. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 30.Tucker BJ, Breaker RR. Curr. Opin. Struct. Biol. 2005;15:342. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Verhounig A, Karcher D, Bock R. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6204. doi: 10.1073/pnas.0914423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gluick TC, Draper DE. J. Mol. Biol. 1994;241:246. doi: 10.1006/jmbi.1994.1493. [DOI] [PubMed] [Google Scholar]
- 33.Soto AM, Misra V, Draper DE. Biochemistry. 2007;46:2973. doi: 10.1021/bi0616753. [DOI] [PubMed] [Google Scholar]
- 34.Nixon PL, Giedroc DP. Biochemistry. 1998;37:16116. doi: 10.1021/bi981726z. [DOI] [PubMed] [Google Scholar]
- 35.Cao S, Chen SJ. RNA. 2009;15:696. doi: 10.1261/rna.1429009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yingling YG, Shapiro BA. J. Mol. Graphics Modell. 2006;25:261. doi: 10.1016/j.jmgm.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Zhang J, Wang W. J. Am. Chem. Soc. 2011;133:6882. doi: 10.1021/ja1109425. [DOI] [PubMed] [Google Scholar]
- 38.Cornish PV, Hennig M, Giedroc DP. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12694. doi: 10.1073/pnas.0506166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giedroc DP, Theimer CA, Nixon PL. J. Mol. Biol. 2000;298:167. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cecconi C, Shank EA, Bustamante C, Marqusee S. Science. 2005;309:2057. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 41.Dudko OK, Hummer G, Szabo A. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15755. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Science. 2008;319:630. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kellermayer MS, Smith SB, Granzier HL, Bustamante C. Science. 1997;276:1112. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 44.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Science. 2001;292:733. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 45.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Science. 1997;276:1109. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 46.Chen G, Wen JD, Tinoco I., Jr RNA. 2007;13:2175. doi: 10.1261/rna.676707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tinoco I, Jr, Li PT, Bustamante C. Q. Rev. Biophys. 2006;39:325. doi: 10.1017/S0033583506004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green L, Kim CH, Bustamante C, Tinoco I., Jr J. Mol. Biol. 2008;375:511. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao S, Chen SJ. J. Mol. Biol. 2007;367:909. doi: 10.1016/j.jmb.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yagawa K, Yamano K, Oguro T, Maeda M, Sato T, Momose T, Kawano S, Endo T. Protein science: a publication of the Protein Society. 2010;19:693. doi: 10.1002/pro.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Science. 1996;274:1859. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 52.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13770. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shim J, Gu LQ. Methods. 2012;57:40. doi: 10.1016/j.ymeth.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shim JW, Gu LQ. J. Phys. Chem. B. 2008;112:8354. doi: 10.1021/jp0775911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.An N, Fleming AM, Middleton EG, Burrows CJ. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14325. doi: 10.1073/pnas.1415944111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys. J. 1999;77:3227. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Butler TZ, Gundlach JH, Troll M. Biophys. J. 2007;93:3229. doi: 10.1529/biophysj.107.107003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Butler TZ, Gundlach JH, Troll MA. Biophys. J. 2006;90:190. doi: 10.1529/biophysj.105.068957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clamer M, Höfler L, Mikhailova E, Viero G, Bayley H. ACS Nano. 2014;8:1364. doi: 10.1021/nn4050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cracknell JA, Japrung D, Bayley H. Nano Lett. 2013;13:2500. doi: 10.1021/nl400560r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Japrung D, Henricus M, Li Q, Maglia G, Bayley H. Biophys. J. 2010;98:1856. doi: 10.1016/j.bpj.2009.12.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith AM, AbuShumays RL, Akeson M, Bernick DL. Front. Bioeng. Biotechnol. 2015 doi: 10.3389/fbioe.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laszlo AH, Derrington IM, Ross BC, Brinkerhoff H, Adey A, Nova IC, Craig JM, Langford KW, Samson JM, Daza R, Doering K, Shendure J, Gundlach JH. Nat. Biotechnol. 2014;32:829. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, Akeson M. Nat. Nanotechnol. 2010;5:798. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. Nat. Methods. 2007;4:315. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 66.Rodriguez-Larrea D, Bayley H. Nat. Nanotechnol. 2013;8:288. doi: 10.1038/nnano.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodriguez-Larrea D, Bayley H. Nat. Commun. 2014;5:4841. doi: 10.1038/ncomms5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathe J, Visram H, Viasnoff V, Rabin Y, Meller A. Biophys. J. 2004;87:3205. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Phys. Rev. Lett. 2003;90:238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 70.An N, Fleming AM, White HS, Burrows CJ. ACS Nano. 2015;9:4296. doi: 10.1021/acsnano.5b00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang X, Wang Y, Fricke BL, Gu LQ. ACS Nano. 2014;8:3444. doi: 10.1021/nn406339n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Zheng D, Tan Q, Wang MX, Gu LQ. Nat. Nanotechnol. 2011;6:668. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dudko OK, Mathé J, Meller A. In: Methods in Enzymology. Nils GW, editor. Vol. 475. Academic Press; 2010. p. 565. [DOI] [PubMed] [Google Scholar]
- 74.Dudko OK, Mathe J, Szabo A, Meller A, Hummer G. Biophys. J. 2007;92:4188. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin J, Fabian M, Sonenberg N, Meller A. Biophys. J. 2012;102:1427. doi: 10.1016/j.bpj.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holland JA, Hansen MR, Du Z, Hoffman DW. RNA. 1999;5:257. doi: 10.1017/s1355838299981360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y, Tian K, Hunter LL, Ritzo B, Gu LQ. Nanoscale. 2014;6:11372. doi: 10.1039/c4nr03195d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kang I, Wang Y, Reagan C, Fu Y, Wang MX, Gu LQ. Sci. Rep. 2013;3:2381. doi: 10.1038/srep02381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mathe J, Aksimentiev A, Nelson DR, Schulten K, Meller A. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12377. doi: 10.1073/pnas.0502947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maglia G, Restrepo MR, Mikhailova E, Bayley H. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19720. doi: 10.1073/pnas.0808296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ding Y, Fleming AM, White HS, Burrows CJ. J. Phys. Chem. B. 2014;118:12873. doi: 10.1021/jp5101413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19202. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Butcher SE, Pyle AM. Acc. Chem. Res. 2011;44:1302. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 84.Cash DD, Cohen-Zontag O, Kim N-K, Shefer K, Brown Y, Ulyanov NB, Tzfati Y, Feigon J. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10970. doi: 10.1073/pnas.1309590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chauhan S, Woodson SA. J. Am. Chem. Soc. 2008;130:1296. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu F, Theimer CA. J. Mol. Biol. 2012;423:719. doi: 10.1016/j.jmb.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 87.Nixon PL, Cornish PV, Suram SV, Giedroc DP. Biochemistry. 2002;41:10665. doi: 10.1021/bi025843c. [DOI] [PubMed] [Google Scholar]
- 88.Zhou Y, Kierzek E, Loo ZP, Antonio M, Yau YH, Chuah YW, Geifman-Shochat S, Kierzek R, Chen G. Nucleic Acids Res. 2013;41:6664. doi: 10.1093/nar/gkt352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Seol Y, Skinner GM, Visscher K. Phys. Rev. Lett. 2004;93:118102. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
- 90.Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Phys. Rev. Lett. 2007;98:158103. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]
- 91.Xu X, Chen S-J. J. Am. Chem. Soc. 2012;134:12499. doi: 10.1021/ja3013819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cao S, Chen S-J. Biophys. J. 2009;96:4024. doi: 10.1016/j.bpj.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. J. Chem. Phys. 1953;21:1087. [Google Scholar]
- 94.Cocco S, Marko JF, Monasson R. Eur. Phys. J. E: Soft Matter Biol. Phys. 2003;10:153. doi: 10.1140/epje/e2003-00019-8. [DOI] [PubMed] [Google Scholar]
- 95.Xu X, Zhao P, Chen S-J. PLoS One. 2014;9:e107504. doi: 10.1371/journal.pone.0107504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hayes RL, Noel JK, Mohanty U, Whitford PC, Hennelly SP, Onuchic JN, Sanbonmatsu KY. J. Am. Chem. Soc. 2012;134:12043. doi: 10.1021/ja301454u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hermann T. Angew. Chem., Int. Ed. 2000;39:1890. doi: 10.1002/1521-3773(20000602)39:11<1890::aid-anie1890>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 98.Gonzalez RL, Jr, Tinoco I., Jr J. Mol. Biol. 1999;289:1267. doi: 10.1006/jmbi.1999.2841. [DOI] [PubMed] [Google Scholar]
- 99.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Nat. Nanotechnol. 2010;5:160. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bai Y, Chu VB, Lipfert J, Pande VS, Herschlag D, Doniach S. J. Am. Chem. Soc. 2008;130:12334. doi: 10.1021/ja800854u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen SJ. Annu. Rev. Biophys. 2008;37:197. doi: 10.1146/annurev.biophys.37.032807.125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Draper DE. Biophys. J. 2008;95:5489. doi: 10.1529/biophysj.108.131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dinman JD, Ruiz-Echevarria MJ, Peltz SW. Trends Biotechnol. 1998;16:190. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qiu H, Kaluarachchi K, Du Z, Hoffman DW, Giedroc DP. Biochemistry. 1996;35:4176. doi: 10.1021/bi9527348. [DOI] [PubMed] [Google Scholar]
- 105.Draper DE. Curr. Opin. Cell Biol. 1990;2:1099. doi: 10.1016/0955-0674(90)90162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dulude D, Baril M, Brakier-Gingras L. Nucleic Acids Res. 2002;30:5094. doi: 10.1093/nar/gkf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kontos H, Napthine S, Brierley I. Molecular and cellular biology. 2001;21:8657. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Plant EP, Jacobs KL, Harger JW, Meskauskas A, Jacobs JL, Baxter JL, Petrov AN, Dinman JD. RNA. 2003;9:168. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Staple DW, Butcher SE. J. Mol. Biol. 2005;349:1011. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 110.Yang H, Jossinet F, Leontis N, Chen L, Westbrook J, Berman H, Westhof E. Nucleic Acids Res. 2003;31:3450. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xie S. Single Mol. 2001;2:229. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.