Gloury et al. reveal an essential role for the Id3–E-protein axis in the transcriptional regulation of humoral immunity.
Abstract
The generation of high-affinity antibodies requires germinal center (GC) development and differentiation of long-lived plasma cells in a multilayered process that is tightly controlled by the activity of multiple transcription factors. Here, we reveal a new layer of complexity by demonstrating that dynamic changes in Id3 and E-protein activity govern both GC and plasma cell differentiation. We show that down-regulation of Id3 in B cells is essential for releasing E2A and E2-2, which in a redundant manner are required for antigen-induced B cell differentiation. We demonstrate that this pathway controls the expression of multiple key factors, including Blimp1, Xbp1, and CXCR4, and is therefore critical for establishing the transcriptional network that controls GC B cell and plasma cell differentiation.
In response to their cognate antigen, activated B cells either differentiate into antibody-secreting plasma blasts or are recruited into a germinal center (GC), where they undergo multiple rounds of proliferation, class-switch recombination (CSR), and affinity maturation. GC B cells that have successfully been selected and expanded in the GC can then give rise to long-lived plasma cells or memory B cells (Nutt et al., 2015). Thus, humoral immunity is generated in a multilayered process that includes the generation of short-lived, low-affinity plasma blasts, as well as affinity-matured, long-lived plasma cells and memory B cells. During this differentiation process, B cells undergo a dramatic remodeling of their transcriptional profile (Shi et al., 2015), which is tightly controlled by a network of regulators that ensure effective humoral immunity (Nutt et al., 2015).
When B cells adopt a GC fate, they induce expression of Bcl6, which is essential for the efficient proliferation and survival of GC B cells during CSR and somatic hypermutation that underpins antigen-receptor affinity maturation (Fukuda et al., 1997; Basso and Dalla-Favera, 2012). Plasma cell differentiation on the other hand is driven by the transcriptional regulator Blimp1, which is essential for the generation of antibody-secreting plasma cells (Shapiro-Shelef et al., 2003; Kallies et al., 2004, 2007), and Xbp1, required for efficient antibody-secretion (Reimold et al., 2001; Shaffer et al., 2004; Taubenheim et al., 2012). Importantly, in addition to these master regulators of fate determination in the B cell lineage, other transcriptional regulators operate that are critical for the initiation, timing and efficiency of these differentiation processes. For example, Bach2 is required for the generation of GC B cells as a result of its role in repressing Blimp1 expression and preventing premature commitment to plasma cell differentiation (Muto et al., 2004, 2010). Similarly, a transcriptional module consisting of IRF8 and PU.1 acts to limit plasma cell differentiation (Carotta et al., 2014).
Although plasma cell development is dependent on Blimp1, we have shown previously that plasma cell differentiation is initiated independently of this transcriptional regulator. Loss of activity of Pax5, a transcription factor required for B cell lineage commitment (Nutt et al., 1999), defines a preplasmablast stage that precedes plasma cell differentiation (Kallies et al., 2007). A major regulator of the early steps of plasma cell differentiation is the transcription factor IRF4, which is required for both Blimp1 expression and the differentiation of plasma cells (Klein et al., 2006; Sciammas et al., 2006). However, the role of IRF4 in antigen-induced B cell differentiation is much broader, as it is also required for the early stages of the GC response and CSR (Ochiai et al., 2013; Willis et al., 2014). Indeed, recent data suggest that IRF4 is involved in regulating fundamental processes of cellular metabolism and survival (Man et al., 2013), thus its role in B cell differentiation may be of a more pleiotropic nature. In line with this notion, IRF4 is up-regulated early during B cell activation, preceding commitment to both the GC and plasma cell fates (Ochiai et al., 2013).
Inhibitors of DNA-binding/differentiation (Id) proteins, in particular Id2 and Id3, have been implicated in the differentiation of several immune cell subsets including B cells (Kee et al., 2001). Id proteins heterodimerize with basic helix-loop-helix transcription factors such as E-proteins and prevent their binding to DNA (Kee, 2009). E-proteins function by forming dimers that can bind to E-boxes within promoter/enhancer regions of target genes to facilitate transcription. Because Id proteins lack a DNA-binding domain, an Id/E-protein heterodimer cannot bind DNA. Thus, Id proteins negatively regulate E-protein activity (Murre, 2005). Id proteins, particularly Id2, have been implicated in humoral immunity. For example, Id2 was shown to negatively regulate CSR to IgE, and enforced expression of Id2 resulted in down-regulation of activation induced cytidine deaminase (AID) expression and a block in CSR (Gonda et al., 2003; Sugai et al., 2003; Kishida et al., 2007). Interestingly, Id2 was proposed to act not only via inhibition of E-protein family members, but also by blocking the activity of Pax5 (Gonda et al., 2003). Id3 is expressed in B cells and is involved in CSR and proliferation in response to B cell receptor cross-linking (Pan et al., 1999; Shaffer et al., 2002; Hayakawa et al., 2007).
In B cells, E2A (with two isoforms, E12 and E47) is the dominant E-protein and of singular importance as it is required for B cell commitment and early B cell development (Bain et al., 1994; Zhuang et al., 1994; Lin et al., 2010). Although expressed throughout the B cell lineage, E2A is dispensable for mature B cell survival and for plasma cell differentiation. It does, however, play a critical role in AID expression and is important for maintaining GC (Sayegh et al., 2003; Kwon et al., 2008). E2-2, another E-protein expressed in B cells, appears to play only minor roles in B cell development and differentiation (Wikström et al., 2006). However, despite the critical roles that E and Id proteins play during early B cell lineage commitment and their continuous expression throughout the B cell lineage, their function in mature B cells is only partially understood.
In this study, we uncover an essential role for the Id3–E-protein axis in the transcriptional regulation of humoral immunity. We show that down-regulation of Id3 expression in B cells is essential for releasing E2A and E2-2 activity, which is necessary for both GC B cell and plasma cell differentiation. We demonstrate that this pathway controls the expression of multiple key factors required for antigen-induced B cell differentiation, including Blimp1, Xbp1, Obf1, Mef2b, and CXCR4, and is therefore critical for establishing the transcriptional network that controls GC B cell and plasma cell differentiation. This work reveals a novel and unexpected layer of complexity in the transcriptional network that governs peripheral B cell differentiation in response to antigen.
RESULTS
Id3 is a negative regulator of plasma cell differentiation
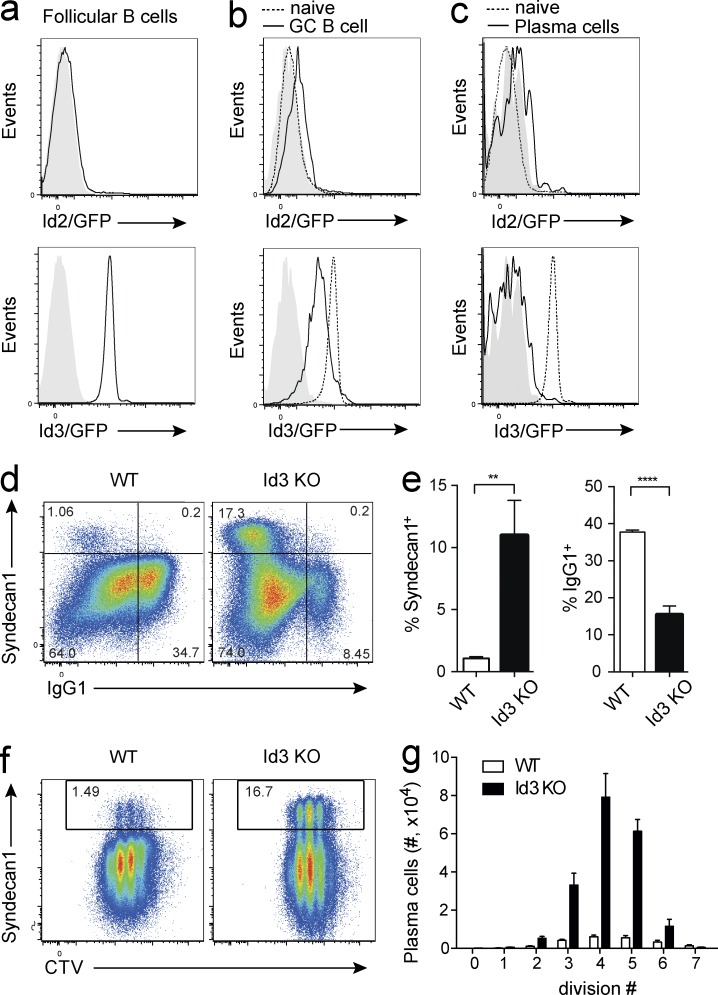
To examine the expression of Id proteins in B lineage cells, we made use of Id2 and Id3 reporter mice. Id3GFP mice carry a GFP expression cassette that was inserted into exon 1 of the Id3 locus. This insertion results in a nonfunctional protein. Thus, heterozygote mice can be used as Id3 reporters, whereas homozygote mice are Id3 deficient (Miyazaki et al., 2011). In Id2GFP mice, an IRES-GFP cassette is inserted into the 3′-untranslated region of the Id2 gene, without disrupting protein expression or function (Jackson et al., 2011). Id2 was not expressed in mature follicular B cells, but in contrast, we observed high amounts of Id3 in this population (Fig. 1 a). To examine the expression of Id2 and Id3 in activated B cells and during B cell differentiation in vivo, we immunized reporter mice with the T cell–dependent antigen nitrophenol coupled to keyhole-limpet hemocyanin (NP-KLH). Whereas Id2 was expressed at very low amounts in GC B cells, Id3 was clearly detectable, albeit at lower amounts compared with the high expression in follicular B cells (Fig. 1 b). Neither Id2 nor Id3 were expressed in mature plasma cells in the bone marrow (Fig. 1 c). Thus, the predominant Id protein in the B cell lineage is Id3, which is expressed at uniformly high amounts in follicular B cells, whereas it is dynamically down-regulated in GC B cells and extinguished in plasma cells.
Figure 1.
Id3 is a negative regulator of plasma cell differentiation. (a–c) Expression of Id2 (top, black line) and Id3 (bottom, black line) in splenic follicular B cells (a; CD19+CD23hiCD21lo) and GC B cells (b; B220+NP+Fas+) at day 7 after immunization with NP-KLH in alum, and plasma cells of the bone marrow (c; B220loSyndecan-1+), identified by flow cytometry using Id2GFP/GFP and Id3GFP/+ reporter mice, respectively. WT is indicated in filled gray histograms. Dotted line in b and c shows expression in follicular B cells for comparison. (d–g) Small resting B cells were isolated from WT and Id3 KO mice and cultured for 4 d with CD40L and IL-4. Proliferation was measured by cell division tracker Cell Trace Violet (CTV). (d) Expression of the plasma cell marker Syndecan-1 and surface IgG1 as determined by flow cytometry. (e) Percentage of plasma cells and IgG1+ cells in cultures. **, P = 0.0023; ****, P < 0.0001. (f) Cell division history and expression of plasma cell marker Syndecan-1. (g) Plasma cell numbers per division, as indicated. Histograms in a–c are representative of three to five individual mice analyzed per genotype over four independent experiments. Data in d–g are representative (d and f) or cumulative (e and g) of four to six individual mice per genotype over two independent experiments each. Graphs show mean ± SEM; statistical significance (e) was determined by Student’s t test.
To determine the role of Id3 in mature B cells upon activation, we cultured WT and Id3-deficient B cells in the presence of CD40L and IL-4, conditions that mimic T cell help and promote B cell proliferation. Although proliferation and survival were unaffected, plasma cell differentiation was strongly enhanced and CSR to IgG1 was impaired (Fig. 1, d–g). In these cultures, compared with WT, larger numbers of Id3-deficient B cells differentiated into plasma cells per division (Fig. 1, f and g). Consistent with the low expression of Id2, B cells lacking Id2 responded to stimulation similar to WT cells (unpublished data). Therefore, Id3 is a potent negative regulator of plasma cell differentiation.
Down-regulation of Id3 is required for plasma cell differentiation
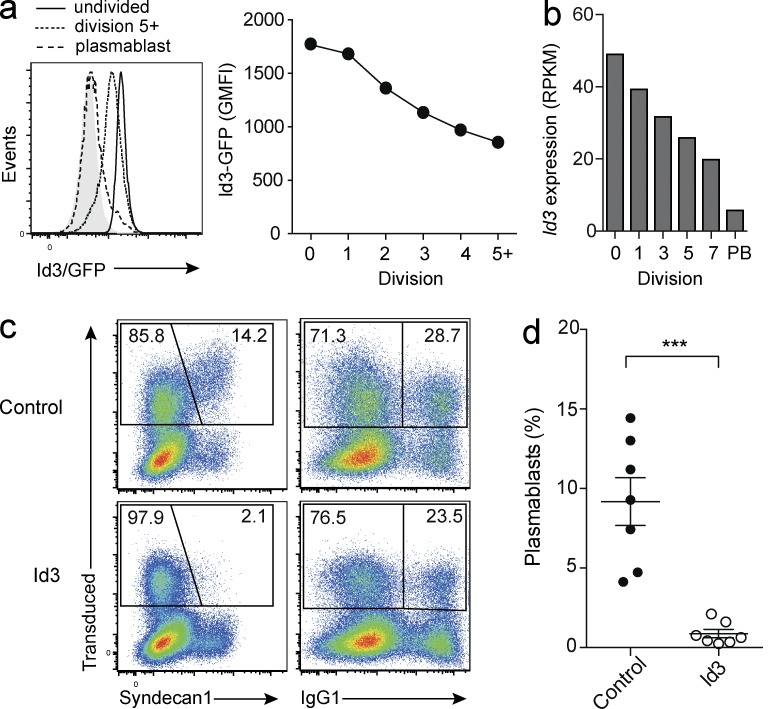
To examine more closely the dynamics of Id3 expression upon B cell activation, we labeled Id3 reporter B cells with the cell division tracker dye CTV and cultured them in vitro. Flow cytometric analysis revealed that Id3 expression was successively down-regulated with each cell division (Fig. 2 a). RNA sequencing (RNAseq) of B cells sorted according to division history (Shi et al., 2015) supported this observation and showed that Id3 transcripts decreased with each division, and were at their lowest levels in plasmablasts (Fig. 2 b). These results, together with the increased plasma cell differentiation observed in Id3-deficient B cells, indicated that Id3 acts as a negative regulator whose expression needs to be down-regulated to allow initiation of plasma cell differentiation. In line with this notion, retroviral Id3 overexpression completely blocked the formation of Syndecan-1+ cells (Fig. 2, c and d), confirming the role of Id3 as a negative regulator of plasma cell differentiation.
Figure 2.
Division-linked Id3 down-regulation is required for plasma cell differentiation. (a) Id3 expression in B cells isolated from Id3GFP/+ reporter mice gated according to their division history or plasma blast phenotype after 4 d in culture with LPS and IL-4 (left), geometric means of fluorescence intensity (GMFI) per division (right). (b) Id3 transcript amounts identified by RNAseq across cell divisions for WT B cells (division 0–7) and in vitro differentiated plasmablasts (PB). (c and d) B cells were transduced with either a control vector or a vector that forces overexpression of Id3. Representative flow cytometric analysis (c) after culture in LPS and IL-4 for 4 d. Frequency of plasmablasts (Syndecan-1+) and switched (IgG1+) cells within the retrovirally transduced population identified by Cherry fluorescence, gated on divided cells. Quantification of plasma blasts as percentage of B cells ***, P = 0.0002 (d). Flow cytometric analyses in a and c are representative of three to seven independent experiments with a total of three to seven biological replicates. Data in d are cumulative of seven independent experiments. Graphs show mean ± SEM; statistical significance (d) was determined by Student’s t test.
E-proteins are required for plasma cell differentiation
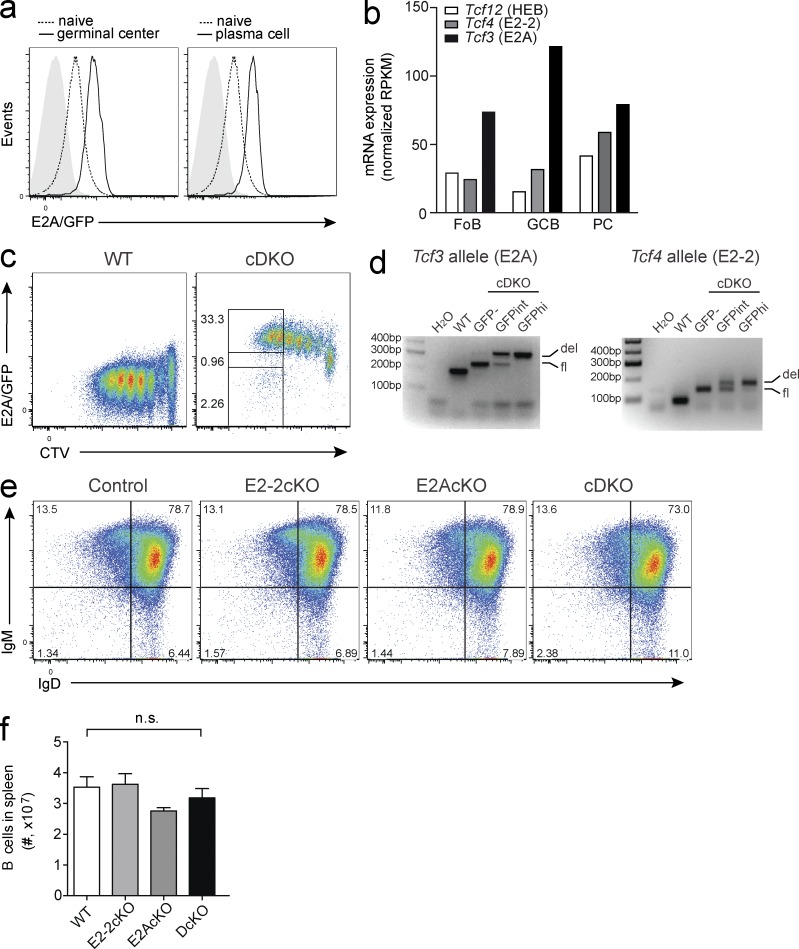
Our results revealed Id3 to be a potent inhibitor of plasma cell differentiation, suggesting that the transcriptional activity of E-protein transcription factors is required for this process. E2A is abundantly expressed in the B cell lineage and has important functions during B cell development (Kwon et al., 2008). However, despite its high expression in resting and activated B cells (Fig. 3 a), E2A was found to be dispensable for mature B cell maintenance and plasma cell differentiation (Kwon et al., 2008). Mice lacking E2A in B cells, however, showed reduced GC B cell development in comparison to controls (Kwon et al., 2008). RNAseq analysis of sorted follicular B cells, GC B cells, and plasma cells (Shi et al., 2015) confirmed high expression of Tcf3 (E2A) in B lineage cells. Furthermore, it revealed that Tcf4 (E2-2) was also abundant, particularly in GC B cells and plasma cells (Fig. 3 b). Based on these data, we hypothesized that E2-2 may compensate for the loss of E2A in mature B cells. To test this, we generated mice that lacked expression of E2A and/or E2-2 in mature B cells specifically using Cd23Cre transgenic mice (Kwon et al., 2008) that were crossed to mice carrying floxed Tcf4 alleles (cE2-2KO; Bergqvist et al., 2000), floxed Tcf3 alleles (cE2AKO; Kwon et al., 2008), or both (cDKO). Deletion frequency in mature B cells was >96% as monitored by using the GFP reporter function of the deleted Tcf3 allele and genomic PCR (Fig. 3, c and d). Importantly, neither the single nor the double conditional knock-out mice showed any gross alterations in the mature B cell compartment and maintained normal expression of surface IgD and IgM (Fig. 3, e and f).
Figure 3.
Combined deletion of E2A and E2-2 leaves mature B cells intact. (a) Expression of E2A in follicular B cells (dashed line), NP+ GC B cells (solid line, left), or NP+ plasma cells (solid line, right) analyzed by flow cytometry using E2A/GFP reporter mice 7 d after immunization with NP-KLH in alum. Filled gray histograms indicate controls lacking the GFP reporter. Data are representative of two biological replicates per genotype. (b) Expression of transcripts encoding E-proteins in different B cell populations as identified by RNAseq. Data are the combined results from two samples (Shi et al., 2015). (c and d) WT and B cells with a CD23Cre-mediated deletion of Tcf4 and Tcf3 (cDKO) were labeled with the division tracker dye CTV and cultured in CD40 and IL-4. cDKO B cells were sorted according to the indicated gates (c), and genomic PCR testing the deletion of Tcf3 (E2A; left) and Tcf4 (E2-2; right), was performed (d). FACS plots are representative of four independent experiments. (e) Splenic CD19+ B cells in mice with a CD23Cre-mediated B cell specific deletion of Tcf4 (E2-2cKO), Tcf3 (E2AcKO), or both (cDKO), or control mice (Cd23CreT/+) were analyzed for expression of mature B cell markers, surface IgM, and IgD. Data are representative of three to five biological replicates per genotype, over five separate experiments. (f) Total numbers of B cells in the spleen, pooled results from five to eight mice per genotype over two independent experiments. Graph shows mean ± SEM; statistical significance was determined by Student’s t test.
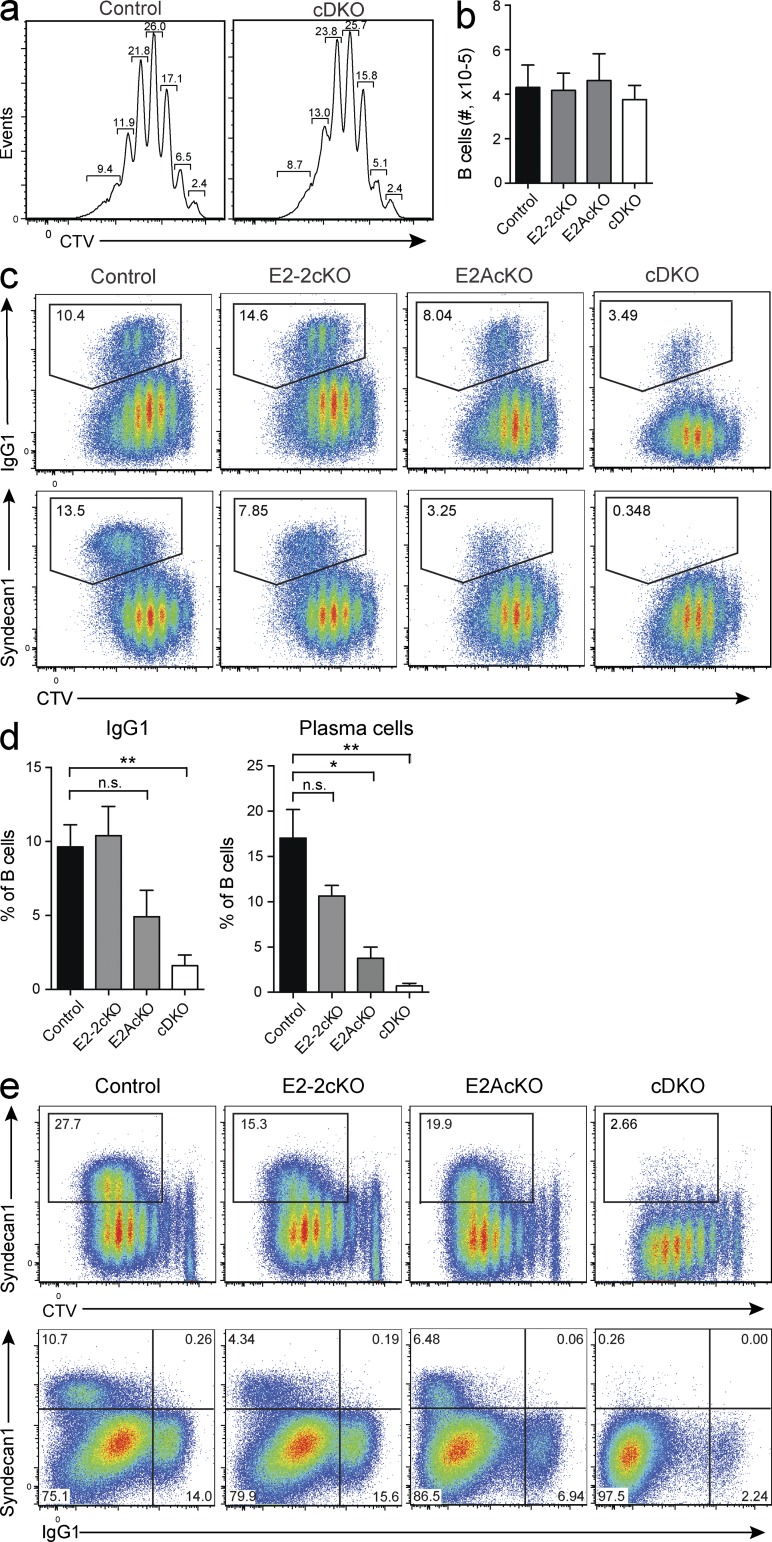
To investigate the impact of E-proteins on plasma cell differentiation, we isolated B cells from cE2-2, cE2A, and cDKO mice and from mice containing only the Cd23Cre transgene (control) and cultured them in the presence of CD40L, IL-4, and IL-5, conditions that support proliferation and plasma cell differentiation. Cell proliferation and survival were similar in all genotypes (Fig. 4, a and b), whereas CSR to IgG1 was impaired, most notably in B cells deficient for both E2-2 and E2A (Fig. 4, c and d). Strikingly, differentiation of plasma cells was completely abrogated in cDKO B cell cultures, whereas it was reduced in E2-2 and E2A single-deficient B cell cultures (Fig. 4, c and d). Similar results were observed when the cells were cultured with different cell stimuli, including LPS (Fig. 4 e). Together, these results indicate that E-proteins are required for plasma cell differentiation.
Figure 4.
The combined activity of E2A and E2-2 is required for plasma cell differentiation and immunoglobulin CSR in vitro. (a and b) Proliferation history as determined by cell division tracker CTV profile (a) and cell numbers (mean ± SEM; b) of B cells from mice with a CD23Cre-mediated deletion of Tcf4 (E2-2cKO), Tcf3 (E2AcKO), or both (cDKO), or control mice (Cd23CreT/+) after 4 d in culture with CD40L, IL-4, and IL-5. (c and d) CSR determined by IgG1 staining (c, top), and plasmablast differentiation as determined by Syndecan-1 staining (c, bottom) for B cells of the indicated genotypes cultured as in a (d) Combined data from four individual experiments. **, P = 0.0014 (left, %IgG1+); *, P < 0.0108; **, P = 0.0011 (right, %plasma cells). (e) Plasmablast differentiation and division history as determined by Syndecan-1 and CTV staining (top), and plasmablast differentiation and switching to IgG1 (bottom) in B cells of the indicated genotypes cultured in LPS (top) or LPS and IL-4 (bottom). Flow cytometry plots are representative of and bar graphs are cumulative of 4–6 biological replicates per genotype, over six independent experiments. Graphs show mean ± SEM; statistical significance was determined by Student’s t test.
E-proteins are required for GC formation and plasma cell differentiation in vivo
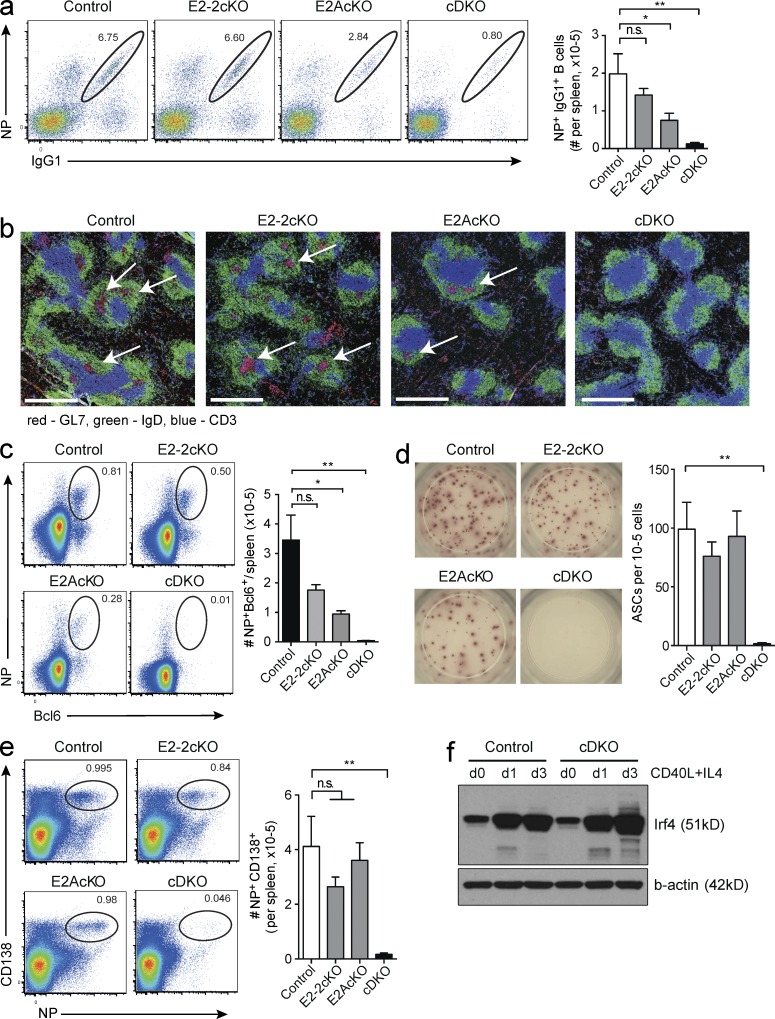
To test the function of E-proteins in vivo, we immunized mutant and control mice with NP-KLH with the adjuvant alum and analyzed them at day 7. Antigen-specific IgG1+ B cell numbers were unimpaired or mildly reduced in the cE2-2KO and cE2AKO, respectively. In contrast, they were reduced by approximately ten-fold in the double-deficient mice (Fig. 5 a). Strikingly, and in contrast to mice with B cells lacking either E2-2 or E2A, mice lacking both E2-2 and E2A in B cells did not form GCs (Figs. 5 b) and showed a dramatic reduction in Bcl6+ antigen-specific B cells (Fig. 5 c).
Figure 5.
The activity of E2A and E2-2 is required for GC formation and plasma cell differentiation. Mice with a CD23Cre-mediated B cell–specific deletion of Tcf4 (E2-2cKO), Tcf3 (E2AcKO), or both (cDKO), or control mice (Cd23CreT/+) were analyzed 7 d after immunization with NP-KLH in alum. (a) Flow cytometric analysis of antigen-specific IgG1+ cells as a percentage of switched B cells (Gr1−IgM−IgD−CD19+) in the spleen (left), and number of antigen-specific B cells per spleen (right). *, P = 0.0435; **, P = 0.0040. (b) Immunohistochemistry of spleen sections showing GC (GL7, red) indicated by arrows, B cell follicles (IgD, green), and T cell zones (CD3, blue). Bars, 0.5 mm. (c) Flow cytometric analysis of intracellular staining of Bcl6 and NP specificity, gated on total CD19+ B cells (left) and total number of NP+Bcl6+CD19+ cells per spleen (right). *,P = 0.0101; **, P = 0.0017. (d) ELISpot analysis detecting NP-specific, IgG1-secreting plasma cells. Representative results from wells loaded with 105 total splenic cells (left) and quantification (right). ASCs, antibody secreting cells; **, P = 0.0011. (e) NP-specific plasma cells as percentage of total B and plasma cells (left), and total number of NP-specific plasma cells per spleen (right) as determined by flow cytometry 7 d after NP-KLH immunization. **, P = 0.0033. (f) Expression of IRF4 determined by Western blotting in WT and E2A/E2-2 double-deficient (cDKO) B cells activated with CD40L and IL-4 for 0–3 d, as indicated. Actin is shown as a loading control. Data in a–e are from five to six mice per genotype over two separate experiments. Data in a and c–e are the mean ± SEM; statistical significance was determined by Student’s t test. Data in f is representative of two experiments.
Consistent with our in vitro data, B cells deficient in either E2-2 or E2A were able to give rise to antigen-specific plasma cells. In contrast, plasma cell differentiation was abrogated in mice with E2-2/E2A double-deficient B cells, as determined by ELISpot and flow cytometry (Fig. 5, d and e). The defective GC B cell and plasma cell differentiation in E-protein deficient B cells was reminiscent of the defect in both processes observed in the absence of the transcriptional regulator IRF4 (Klein et al., 2006; Willis et al., 2014). However, Western blotting revealed that upon stimulation, IRF4 was up-regulated in the absence of E-proteins (Fig. 5 f), clearly indicating that IRF4 expression is independent of E-protein activity.
In summary, these results show that although E2-2 and E2A are individually redundant in GC development and plasma cell differentiation, E-protein activity is essential for both processes.
Graded E-protein activity controls gene expression in activated B cells
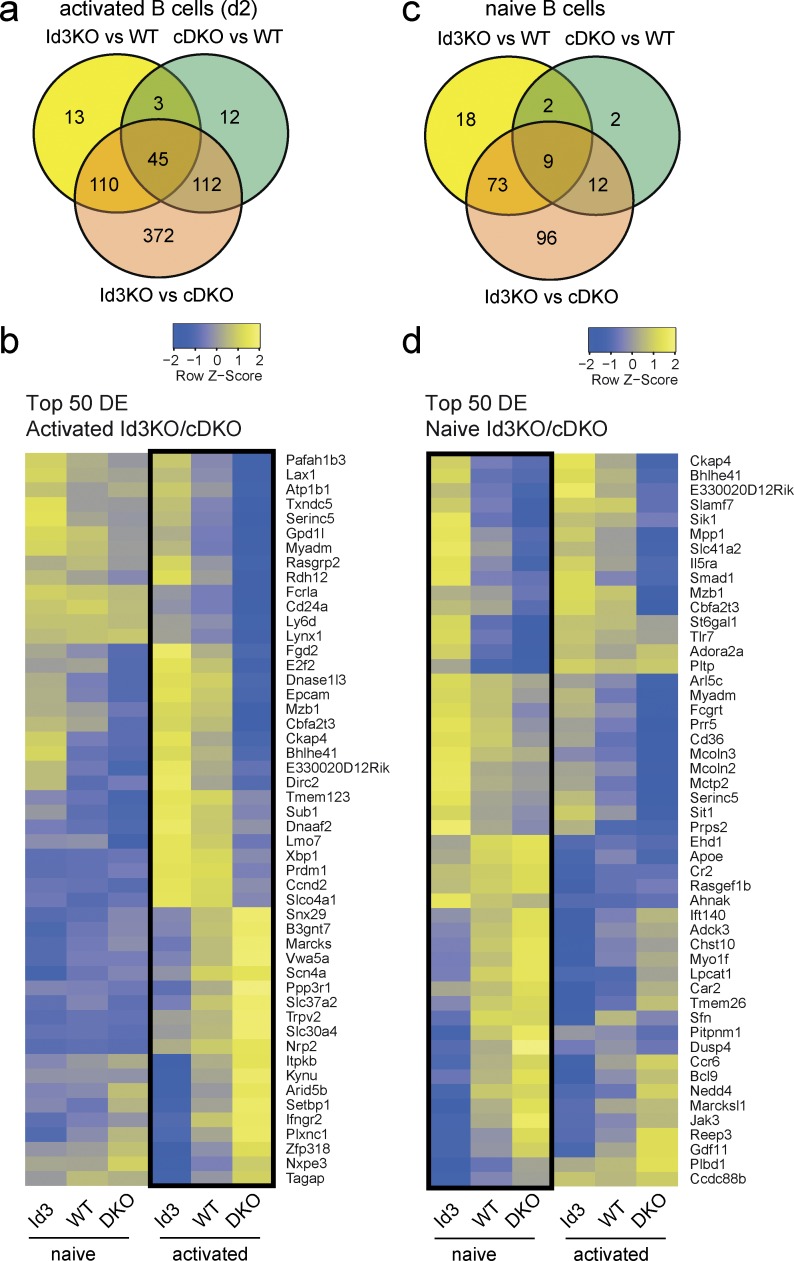
To gain mechanistic insight into the activities of E-proteins during antigen-induced B cell differentiation, we purified WT, Id3-KO, and E2-2/E2A cDKO B cells and stimulated them with CD40L plus IL-4 for 2 d. At this time point, the B cells had undergone only one or two rounds of division and had not formed plasma cells in either genotype (unpublished data). Transcriptional profiling of these activated B cells by RNAseq revealed 171 and 172 genes, respectively, that were significantly differentially expressed (DE) between WT and Id3-KO or WT and E2-2/E2A cDKO activated B cells (FDR, 0.05; >2-fold; Fig. 6, a and b). Remarkably, the number of DE genes increased to 639 when comparing Id3-deficient and E-protein double-deficient B cells, which represent cells with the highest (Id3-KO) or lowest (E2-2/E2A cDKO) E-protein activity (Fig. 6, a and b). In contrast, only 190 DE genes were found when comparing Id3-KO and E2-2/E2A cDKO naive B cells (Fig. 6, c and d). These data demonstrate that the transcriptional profile of B cells is susceptible to quantitative changes in E-protein activity, in particular after activation.
Figure 6.
Graded E-protein activity controls gene expression in activated B cells. RNAseq analysis of WT, Id3-deficient (Id3-KO), and E2A/E2-2-double-deficient (cDKO) B cells. (a–d) Venn diagrams (a and c) showing the numbers and overlap of DE genes in the comparisons as indicated, and heat maps (b and d) showing relative expression levels (row z values) of the top 50 DE genes in B cells activated for 2 d with CD40L, IL-4, and IL-5 (a and b) and freshly isolated naive B cells (c and d). Data are from the combined analysis of two separate experiments.
E2A and E2-2 regulate multiple GC B cell–associated genes
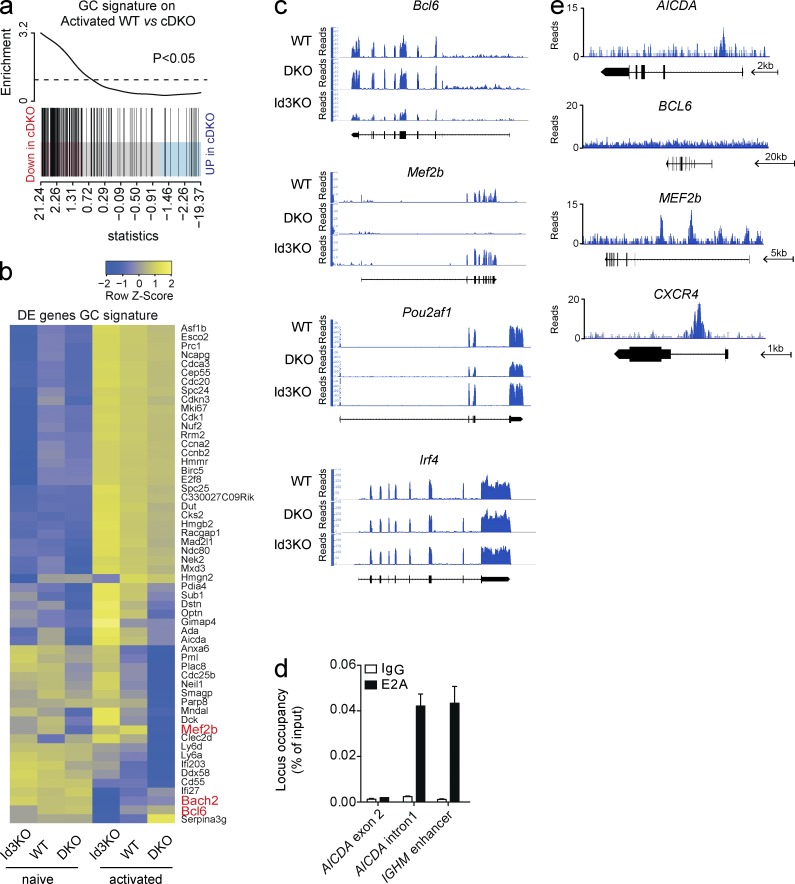
To understand how E-proteins impact on GC B cell differentiation, we generated a GC B cell transcriptional signature derived from our recent transcriptional profiling of multiple B cell subsets (Shi et al., 2015). This signature comprises 241 genes specifically up- or down-regulated in GC B cells compared with B or plasma cells. Remarkably, GC signature genes were significantly enriched among the genes that were down-regulated in the absence of E2-2 and E2A (Fig. 7 a), suggesting that E-proteins play a critical role in establishing the GC-specific transcriptional program. In total, 55 of the GC signature genes (24.6%) were deregulated in the combined absence of E2A and E2-2 (Fig. 7 b). Genes including Bcl6, Bach2, Pou2af1 (encoding Obf1), and Cxcr4, all of which are required for GC development or organization (Kim et al., 1996; Schubart et al., 1996; Allen et al., 2004; Muto et al., 2004; Caron et al., 2009; Basso and Dalla-Favera, 2010; Bannard et al., 2013; Huang et al., 2014), were DE between activated Id3-KO and E2-2/E2A cDKO B cells, but not Irf4, which was expressed at similar amounts independent of the genotype (Fig. 7, b and c).
Figure 7.
E-protein activity is required for establishing the GC B cell transcriptional signature. RNAseq analysis of WT, cDKO, and Id3KO B cells after 2 d of stimulation in vitro with CD40L, IL-4, and IL-5, and ChIPseq analysis after pull-down with an E2A antibody in the Ramos cell line. (a) Gene set enrichment plot for genes specifically up-regulated in GC B cells in the comparison between activated WT and E2A/E2-2 double-deficient (cDKO) B cells. The differential gene expression between WT and cDKO is shown as a shaded rectangle with genes horizontally ranked by moderated t statistic. Genes down-regulated in cDKO are shown in pink (t > 1) and up-regulated genes are shown in blue (t < −1). Black vertical lines represent GC B cell signature genes. (b) Heat map showing relative expression levels (row z values) of genes specifically up- or down-regulated in GC B cells that are DE between activated Id3-deficient (Id3-KO) and cDKO and WT B cells. Rows are scaled to have a mean of 0 and a standard deviation of 1. (c) RNAseq tracks of selected genes in activated B cells. (d and e) ChIP performed using a B lymphoma cell line (Ramos) and E2A-antibodies. (d) Quantitative PCR on ChIPed DNA using primers specific for previously identified E2A-binding sites in regulatory regions within the AICDA and IGHM genes (Sayegh et al., 2003; Zhang et al., 2008). IgG was used as a specificity control. Data are the mean ± SEM. (e) ChIPseq tracks showing binding of E2A in the indicated gene loci. Arrows indicate a scale and direction of transcription. Data in a and b is pooled from two separate experiments; data in c–e is representative of two individual experiments.
To determine whether these genes were direct targets of the E-proteins, we performed chromatin immunoprecipitation (ChIP) using an E2A antibody and a human B lymphoma cell line (Ramos). We validated our ChIP protocol using primers directed to known E2A-binding sites within the AICDA and immunoglobulin loci (Sayegh et al., 2003; Zhang et al., 2008; Fig. 7 d). Using ChIP sequencing (ChIPseq), we detected specific binding of E2A to 2,746 genes. Of the 639 genes detected as DE in our transcriptional profiling of Id3 and E-protein–deficient activated B cells, 162 (25.4%) were bound by E2A, suggesting that they were directly regulated by E-protein activity. This included CXCR4, IRF8, POU2AF1, and BACH2, but not BCL6 (Fig. 7 e and not depicted). However, we found multiple binding sites in MEF2b (Fig. 7 e), a transcriptional activator of BCL6 expression (Ying et al., 2013), suggesting that direct regulation of Mef2b expression by E-proteins may contribute to the failure to up-regulate Bcl6 in vivo.
E2A and E2-2 are required for GC organization
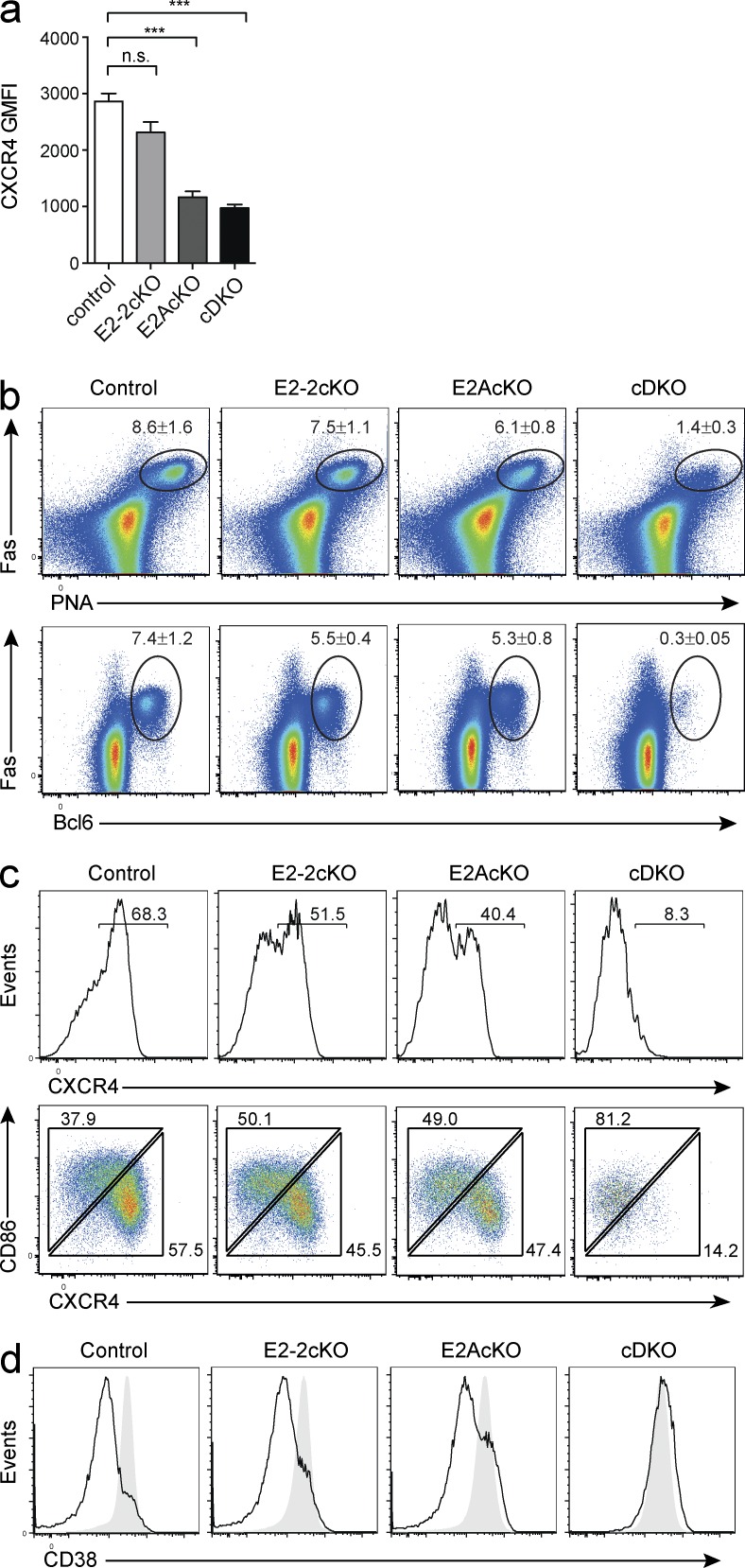
Our RNAseq and ChIPseq results revealed that E2A could bind the CXCR4 locus and affect its transcription. Indeed, when we cultured single- and double-deficient B cells in vitro and analyzed them by flow cytometry, CXCR4 expression was reduced depending on the availability of E-proteins (Fig. 8 a). To investigate this defect more thoroughly in vivo, we infected mice with influenza virus, which induces a strong and sustained GC response. Similar to immunization with NP-KLH, infection with influenza virus resulted in a severely impaired GC response in the B cells deficient in E2A and E2-2, despite the development of some Fas+PNA+ GC-like B cells (Fig. 8 b). In control mice, the GC population contained a high frequency of CXCR4+ cells, representing centroblasts of the dark zone. Consistent with a requirement for E-proteins for the expression of CXCR4, the proportion of CXCR4+ GC B cells was reduced in the E2-2 and E2A single KOs and was almost completely absent in the E-protein cDKO (Fig. 8 c, top). In parallel, expression of CD86, which marks light zone GC B cells, was impaired in cDKO B cells (Fig. 8 c, bottom). In line with this observation, cDKO Fas+PNA+ B cells did not show down-regulation of CD38, a feature of GC maturation (Fig. 8 d). Thus, E-protein activity is required for multiple aspects of GC development, including the induction of transcriptional regulators of GC B cell differentiation, as well as regulation of genes important for GC organization.
Figure 8.
E-protein activity is required for GC organization. (a) Expression of CXCR4 on B cells of the indicated genotypes after 4 d in culture with CD40L, IL-4, and IL-5. ***, P = 0.0003 (E2AcKO); ***, P = 0.0006 (cDKO). Data are the mean ± SEM of three independent cultures from one mouse per genotype and is representative of two individual experiments; statistical significance was determined by Student’s t test. (b–d) Mice with a CD23Cre-mediated B cell specific deletion of Tcf4 (E2-2cKO), Tcf3 (E2AcKO), or both (cDKO), or control mice (Cd23CreT/+) were infected with HKx31 influenza virus and analyzed 10 d later. Flow cytometric analysis of CD19+ mediastinal lymph node B cells. (b) Fas+PNA+ cells (top) and Fas+Bcl6+ cells (bottom) as percentage of total B cells. Numbers indicate the mean ± SEM. (c and d) Flow cytometric analysis of B cells expressing the GC markers Fas and PNA. Plots show expression of CXCR4 (c, top), and CXCR4 and CD86 (c, bottom), and CD38 (d, naive B cells indicated by filled gray histogram). Plots in b–d are representative of two to three individual experiments with a total of five to seven mice per genotype.
E2A regulates plasma cell differentiation and function via Blimp1 and Xbp1
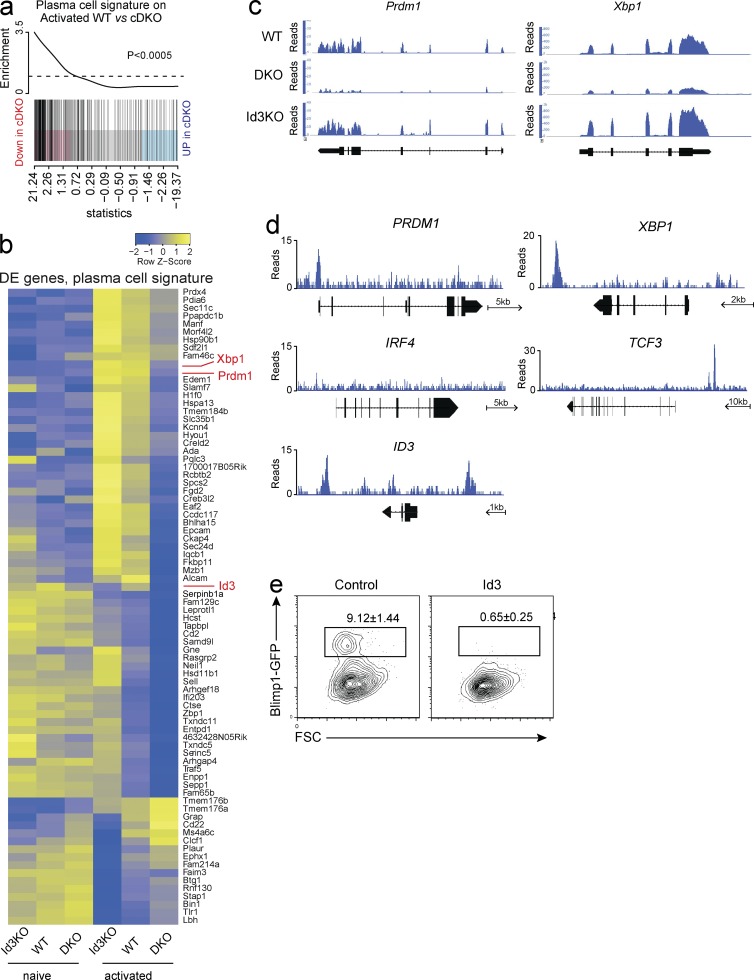
To understand the requirement for E-proteins during plasma cell differentiation, we made use of a set of 301 genes that we recently identified as specifically induced during plasma cell differentiation (plasma cell signature; Shi et al., 2015). Similar to our analysis of GC B cell–associated genes, plasma cell signature genes were significantly enriched among the genes that were down-regulated in the absence of E-protein activity, suggesting that E-proteins play a critical role in establishing the plasma-cell specific transcriptional program (Fig. 9 a). Of note and as outlined above, as plasma cells fail to develop in the combined absence of E2A and E2-2, RNAseq was performed on activated B cells that had not differentiated into plasma cells in either genotype. Thus, our data relate to the initiation phase of plasma cell differentiation. In total 64 of the plasma cell signature genes (21.3%) were not induced appropriately in the combined absence of E2A and E2-2, suggesting that the induction of the plasma cell differentiation program depends on E-protein activity (Fig. 9 b). This included Prdm1 (encoding Blimp1) a key regulator of plasma cell differentiation, expression of which was not detected in E2-2/E2A cDKO B cells but was detectable in WT and Id3-KO B cells (Fig. 9, b and c). Similarly, E-protein deficient B cells expressed much lower amounts of Xbp1, important for efficient antibody secretion of plasma cells (Fig. 9, b and c). In agreement with a direct role of E-proteins in the regulation of Blimp1 and Xbp1 expression, ChIPseq identified E2A binding sites within the promoter of PRDM1 and within the XBP1 locus (Fig. 9 d). Interestingly, E2A also bound to its own gene locus, TCF3, as well as to the ID3 gene locus, suggesting an autoregulatory loop (Fig. 9 d). In line with this idea, Id3 expression was impaired in the absence of E-proteins (Fig. 9 b). Importantly, E-box motifs (CANNTG) that were associated with the E2A-ChIPseq peaks in PRDM1 and XBP1 were conserved between human and mouse as were the binding sites identified in CXCR4, POU2AF1, MEF2B, TCF3, and ID3, suggesting that E-protein–mediated transcriptional regulation of these genes is evolutionarily conserved (unpublished data).
Figure 9.
E-protein activity is required for initiation of plasma cell differentiation. (a) Gene set enrichment plot for genes specifically up-regulated in plasma cells as identified previously (Shi et al., 2015) in the comparison between activated WT and E2A/E2-2 double-deficient (cDKO) B cells. Black vertical lines represent plasma cell signature genes. (b) Heat map showing relative expression levels (row z values) of genes specifically up- or down-regulated in plasma cells and DE between activated Id3-deficient (Id3-KO), cDKO, and WT B cells. Rows are scaled to have a mean of 0 and a standard deviation of 1. (c) RNAseq tracks of selected DE genes in activated B cells of the indicated genotype. (d) ChIPseq tracks showing binding of E2A in the indicated gene loci. Arrows indicate a scale and direction of transcription. (e) Representative flow cytometric analysis of B cells isolated from Blimp1GFP reporter mice and transduced with either a control vector or an Id3 overexpressing vector and cultured for 4 d in LPS and IL-4. Transduced cells were identified by Cherry fluorescence. Frequency (mean ± SEM) of Blimp1+ cells within the retrovirally transduced population. Data in a and b are pooled from two separate experiments; data in c and d are representative of two individual experiments; data in e are from five independent experiments.
To test if E2A and E2-2 activity was indeed required for induction of Blimp1 expression in mouse B cells, we inhibited E-protein activity by retroviral overexpression of Id3 in Blimp1GFP-reporter B cells. Consistent with the idea that E2A/E2-2 directly activated the Blimp1 locus, Id3 overexpression completely abrogated LPS induced Blimp1/GFP expression (Fig. 9 e). Thus, E-protein transcriptional activity is required for the expression of key mediators of GC development and plasma cell differentiation.
DISCUSSION
Understanding the molecular requirements of plasma cell and GC B cell differentiation is central to improving vaccination strategies, targeting immune deficiencies, or treating B cell–derived malignancies. Despite significant progress, however, many questions remain, particularly concerning the partitioning of GC and plasma cell fate. Here, we identify a new transcriptional circuit critical for both GC B cell and plasma cell differentiation. We show that the activity of the E-protein transcription factors E2A and E2-2 is essential for humoral immune responses and is tightly controlled by the expression of Id3. This sheds new light on the initiating events of GC and plasma cell development and allows for a better understanding of how differentiation in response to antigen is orchestrated in the B cell lineage.
During an infection it is critical to promote rapid plasma cell differentiation to generate large amounts of antibody that may prevent or slow down the spreading of the pathogen. Concurrently, some antigen-specific B cells need to be retained to allow clonal population expansion, CSR, and affinity maturation in the GC. This is not only essential for the generation of long-lived plasma cells, producing high-affinity antibody but also for retention of antigen-specificity in the form of memory B cells (Tarlinton and Good-Jacobson, 2013). Although multiple molecular regulators of antigen-induced B cell differentiation have been identified, there is uncertainty as to how terminal differentiation into plasma cells and retention of antigen-specific B cells as self-renewing GC and memory B cells is partitioned and timed. Our results demonstrate that the Id3-E2A–E2-2 axis is a critical component of the transcriptional network that governs this process. Although deletion of either E2A or E2-2 alone did not have a major impact on GC or plasma cell differentiation, the combined absence of these E-proteins resulted in a complete block in antigen-induced B cell differentiation. Our data demonstrate that E-proteins play a major role in initiating and orchestrating the transcriptional networks associated with GC and plasma cell differentiation. Expression of genes critical for plasma cell differentiation including Prdm1 (encoding Blimp1) and Xbp1 failed to be induced in the absence of E-protein activity. In addition to these essential factors, numerous other components of the plasma cell transcriptional signature were not induced appropriately, suggesting that E-proteins target multiple pathways that underpin plasma cell differentiation. This may be similar during GC development, as we found multiple critical targets of E-proteins that are either involved in the transcriptional regulation of GC B cell development or the local organization of productive GC structures. This included Mef2b, an upstream regulator of Bcl6, and Cxcr4, a chemokine receptor involved in GC dark and light zone organization (Victora and Nussenzweig, 2012; Ying et al., 2013). Consequently, multiple components of the transcriptional signature associated with GC B cells were deregulated in the absence of E2A and E2-2. Thus, E-proteins are major regulators of both GC B cell and plasma cell differentiation.
Importantly, we found that Id3, but not Id2, is a critical regulator of the E2A/E2-2 transcriptional module. Id3 is highly expressed in mature B cells and therefore limits activity of E-proteins in resting B cells. After B cell activation, Id3 is down-regulated, thereby releasing E-protein activity and allowing for GC B cell and plasma cell differentiation. Although our results demonstrate that E2A and E2-2 are required for GC B cell development, it remains unclear how E-protein activity is regulated in the GC, where tight control of E-protein activity appears to be important. This is supported by the observation that Id3 expression is maintained in GC B cells, albeit at lower levels compared with naive B cells, suggesting that a titration of E-protein activity is critical for the segregation of GC B cell and plasma cell differentiation. Indeed, deletion of E2A alone in B cells results in a reduction in GC B cells as reported previously (Kwon et al., 2008). The notion that E-protein activity and Id protein expression in B cells need to be tightly controlled is also supported by several studies that report high frequencies of mutations in Id3 or aberrant expression of Id2 in Hodgkin's and Burkitt's lymphoma (Renné et al., 2006; Love et al., 2012; Richter et al., 2012; Schmitz et al., 2014). Although some of these mutations impact on cell proliferation and survival, our data suggest that certain mutations may also block antigen-induced B cell differentiation and could thus contribute to lymphoma development.
Several transcriptional circuits have been identified as being required to coordinate B cell differentiation in response to antigen. Most prominently, this includes the IRF4-Bcl6-Blimp1 transcriptional network that is central to GC B cell and plasma cell differentiation. Interestingly, the way in which the Id3-E2A–E2-2 axis controls antigen-induced differentiation in the B cell lineage appears to be distinct from other transcriptional circuits guiding B cell differentiation. Whereas transcription factors such as Blimp1, Bcl6, or IRF4, are induced only after B cell activation, E2A and E2-2 are abundantly expressed throughout both development and maintenance of mature B cells and do not need to be produced de novo. They are kept in check by Id3, which too is highly expressed in naive B cells and appears to neutralize the activity of both E-proteins. As activated B cells down-regulate Id3 expression in a cell division dependent manner, E-protein activity is released, thereby allowing for the initiation of plasma cell differentiation. The evolution of such a brake and release mechanism may be important to ensure a rapid and quantitatively appropriate response to antigenic challenges. Such a mechanism may also play a role in restraining premature plasma cell differentiation. This is supported by the observation that B cell survival in the periphery requires tonic antigen-receptor signaling (Zhang et al., 2008), which however, does not lead to substantial GC or plasma cell differentiation. Thus, Id3 may be one of the factors that renders B cells relatively inert to weak stimuli and thereby prevents unnecessary differentiation with concomitant depletion of the mature B cell pool.
Importantly, Id3 expression is compatible with recruitment of B cells into the GC, thereby allowing extensive proliferation and affinity maturation. This is important, as plasma cell differentiation is a proliferation-dependent process, with cells undergoing more divisions being more likely to differentiate into plasma cells (Hasbold et al., 2004; Nutt et al., 2015). Although it has been proposed that B cell fate is allocated in a stochastic fashion during this process (Duffy et al., 2012), it is clear that transcriptional regulators play critical roles in determining the likelihood of differentiation versus self-renewal. Id3 may fulfill such a role, by limiting the probability of undergoing differentiation. Thus, Id3-controlled progression into plasma cell differentiation could be envisaged after two not necessarily exclusive models. Proliferation-dependent down-regulation of Id3 may serve to delay plasma cell differentiation, thereby creating a window of opportunity during which CSR and affinity maturation can occur before differentiation. Interestingly, such a switch in GC output has been proposed before (Shlomchik and Weisel, 2012). Alternatively, B cell receptor signaling strength may directly determine the extent of Id3 down-regulation in the GC, meaning affinity maturation itself could define the timing of plasma cell differentiation, a long held model of GC B cell differentiation (Tarlinton and Smith, 2000). However, data showing diminished BCR signaling in GC B cells argue against this latter possibility (Khalil et al., 2012). The work reported here further unravels the tightly controlled molecular network that governs B cell differentiation in response to antigen and may allow better understanding of the relative contribution of stochastic and deterministic modes to this process.
MATERIAL AND METHODS
Mice
Id2GFPand Blimp1GFP mice were described previously (Kallies et al., 2004; Jackson et al., 2011). Id3GFP MICE were supplied by C. Murre (University of California, San Diego, La Jolla, CA), Tcf3fl/fl, Tcf4fl/fl, and CD23Cre were supplied by M. Busslinger (Institute for Molecular Pathology, Vienna, Austria) and were described previously (Bergqvist et al., 2000; Kwon et al., 2008; Miyazaki et al., 2011). Id3GFP/+ mice were used as Id3 reporter mice, and Id3GFP/GFP were used as Id3-deficient (Id3-KO) mice (Miyazaki et al., 2011). As deletion of the floxed Tcf3 allele results in the expression of a GFP reporter, Tcf3fl/+/Cd23CreT/+ mice were used as Tcf3 (E2A) reporter mice (Kwon et al., 2008). To unambiguously identify B cells that had deleted Tcf3, flow-cytometric analysis of Tcf3fl/fl/Cd23CreT/+ (E2AcKO) and Tcf3fl/fl/Tcf4fl/fl cDKO mice was performed by gating on GFP+ B cells reporting deletion of the Tcf3fl/fl allele as previously described (Kwon et al., 2008). Cd23CreT/+ mice were used as control mice. All mice were maintained on a C57BL/6 background and animal experiments were undertaken according to Animal Experimental Ethics Committee guidelines and approval by The Walter and Eliza Hall Institute of Medical Research (Melbourne, Australia).
Antibodies and flow cytometry
Monoclonal antibodies to the following mouse E-proteins were used in multiparameter flow cytometric analysis: B220/CD45R (RA3-6B2), CD19 (eBio1D3), CD86 (GL1), CXCR4/CD184 (2B11), and GL7 (GL7) from eBioscience; CD138/Syndecan-1 (281-2), Fas/CD95 (Jo2), and IgG1 (X56) from BD; and peanut agglutinin (PNA) from Vector Laboratories. Antibodies to the following proteins were generated and conjugated in-house: Bcl6 (7D1-10), CD21 (7G6), CD23 (B3B4), CD38 (NIMR5), CD138/Syndecan-1 (7/11-8B8), Gr1/Ly6c (RB6-8C5), IgD (11-26C), and IgM (331.12). All antibodies were titrated for optimum concentration before use, with dilutions ranging from 1/100 to 1/1,600. FcγRII/III (24G2; supernatant) was used for blocking. Biotinylated monoclonal antibodies were detected with streptavidin conjugated to PECy7 (BD). NP was conjugated in-house. Viable cells were identified by propidium iodide or SytoxBlue (Invitrogen) exclusion. For intracellular staining, cells were fixed, permeabilized, and stained using the reagents and protocol in the Foxp3 Staining Buffer Set (eBioscience). Cells were analyzed on FACSCanto II or LSR Fortessa cytometers (BD). Cells were sorted on a FACS Aria cytometer (BD). Data were processed using FlowJo software.
Western blotting
Total protein extracts were produced from equivalent numbers of cells with DISC buffer (20 mM Tris-Cl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 10% glycerol, and complete protease inhibitor [Roche]). Rat monoclonal anti-IRF4 (3E4) was generated in-house and detected with an anti–rat Ig conjugated to HRP (Santa Cruz Biotechnology, Inc.). Equal loading of lanes with proteins was confirmed by probing with HRP–conjugated anti–β-actin (I-19; Santa Cruz Biotechnology, Inc.). Antibodies were used within the dilution range of 1/500 to 1/3,000.
Cell isolation and culture
Small resting B cells were isolated from spleens by lysis of red blood cells, followed by Percoll gradient and purification via negative selection using magnetic beads (B cell isolation kit; Miltenyi Biotec). Cells were sometimes labeled with CTV (Invitrogen), by resuspending at 107 cells/ml in PBS, adding CTV to a final concentration of 10 µM and incubating at 37°C for 10 minutes before quenching with ice-cold media. Cells were washed and cultured in CD40L (100 ng/ml; R&D Systems) with recombinant mouse IL-4 (10 ng/ml; R&D Systems) with or without recombinant mouse IL-5 (5 ng/ml; R&D Systems) or LPS from E. coli, (20 µg/ml; Sigma-Aldrich) with or without recombinant mouse IL-4. Cultures stimulated with CD40L were plated at a cell density of 2 × 105 cells/ml, and those that were stimulated with LPS were plated at a cell density of 5 × 105 cells/ml. Cultures were incubated at 37°C and analyzed by flow cytometry 4 d later. Cell numbers were assessed by adding 10,000 rainbow calibration particles (BD) to wells before collection and staining.
Immunization
Immunizations were performed with the hapten 4(hydroxy-3-nitrophenyl)acetyl coupled to the protein Keyhole Limpet Haemocyanin (NP-KLH) at a molar ratio of ∼16:1. 100 µg of antigen was precipitated on alum and delivered by i.p. injection.
Immunohistochemistry
Tissue samples were frozen in OCT (Tissue-Tek; Sakura). 7-µm sections were cut using a microtome (Leica) at −20°C, and then stained with rat anti–mouse GL7 followed by goat anti–rat Alexa Fluor 633 (Invitrogen). Slides were then blocked with normal rat serum and further stained with biotinylated rat anti–mouse IgD (Southern Biotech) and rabbit anti–mouse CD3 (Neo Markers) concurrently, followed by streptavidin Alexa Fluor 488 (Invitrogen) and goat anti–rabbit Alexa Fluor 555 (Invitrogen). Coverslips were mounted with ProLong Gold antifade reagent (Life Technologies) and sealed with clear nail polish. Slides were imaged on an LSM 780 C4.47 laser scanning confocal microscope (Zeiss) at 10× magnification.
ELISpot
Splenocytes were added to a 96-well cellulose ester membrane plate (MAHAS4510; Millipore) coated with 20 µg/ml of NP-BSA and incubated for at least 20 h at 37°C and 5–10% CO2. Anti-NP IgG1 was shown using goat anti–mouse IgG1 conjugated to HRP (1070–05; SouthernBiotech) and visualized with substrate 3-amino-9-ethylCarbazole (AEC; Sigma-Aldrich). Spots were counted using an automated reader (AID ELISpot Reader System; software version 4).
Retroviral overexpression of Id3
Retroviral supernatant was produced from HEK293 human embryonic kidney cells transfected with retroviral expression plasmid (pMSCV) containing either a mCherry or an Id3-IRES-mCherry expression cassette. For retroviral transduction, small resting B cells were isolated from the spleens of WT mice and were activated for approximately 40 h with LPS + IL-4. The cells were then cultured for a further 48 h on plates that had previously been coated with retronectin and retrovirus. Coated plates were generated by incubating non–tissue-culture plates with retronectin (in-house) at 4°C overnight, after which retroviral supernatant was spun onto the coated plate at 3800 rpm at 4°C for 1 h.
ChIP and ChIPseq
2 × 107 cells from the human Ramos cell line were cross-linked by the addition of 1% formaldehyde at room temperature for 10 min and quenched with 0.25 M glycine, followed by sonication and immunoprecipitation with 10 µg of anti-E2A (sc-349; Santa Cruz Biotechnology, Inc.) and the corresponding rabbit polyclonal IgG control antibody (sc-2027; Santa Cruz Biotechnology, Inc.). DNA fragments were amplified by PCR primers designed to target the AICDA UTR and IGHM enhancer as positive controls and AICDA exon 2 as a negative control.
The following primers were used: AICDA exon 2, forward, 5′-TGAACCGGAGGAAGTTTCTTTA-3′, and reverse, 5′-AACCAAAGTCCAGTGAAAAGGA-3′; AICDA UTR, forward, 5′-ACTTTACCGAGGGAGTGTTTCA-3′, and reverse 5′-TAGTGATGGCAAAATGACTTGG-3′; IGHM enhancer, forward, 5′-CACCTCTTCACAACCAGAAGTG-3′, and reverse 5′-ATTAATTGAGCGAAGCTGGAAG-3′.
For sequencing, DNA fragments were blunt-end ligated to the Illumina adaptors, amplified, and sequenced with an Illumina HiSeq Sequencer. 43 million 50-bp single-end reads were generated for the sample (ChIP-seq data available from Gene Expression Omnibus under accession no. GSE72832).
Genome alignment/conservation analysis
Sequences within the human genome that corresponded to a peak via E2A ChIP-Seq were blasted against the mouse genome using a basic local alignment search tool (BLAST), the results of which were then aligned to the human input sequence using ClustalW.
Influenza infection
Mice were anaesthetized with methoxyfluorane, and then inoculated intranasally with 1 × 104 PFU of influenza virus strain HKx31 (H3N2).
RNAseq
RNAseq was performed following an adapted protocol. Small resting B cells were purified as described above and either subjected to RNA extraction or cultured in CD40L and IL-4. After 2 d, cells had undergone one to two divisions and mature plasma cells had not developed in either of the genotypes. RNA purification was performed following the manufacturer’s protocol using the RNeasy Plus Mini kit (QIAGEN). RNAseq was carried out on an Illumina HiSeq sequencer at Beijing Genome Institute (Shenzhen, China). Approximately 7 million pairs of 90-bp reads were generated per library. Two or more biological replicates were generated and sequenced for each sample (RNA-seq data available from Gene Expression Omnibus under accession no. GSE72832).
Bioinformatic analysis
Sequence reads were aligned to the GRCm38/mm10 build of the Mus musculus genome using the Subread aligner (Liao et al., 2013). Only uniquely mapped read pairs (fragments) were retained. Genewise counts were obtained using featureCounts (Liao et al., 2014). Fragments overlapping exons in annotation build 38.1 of the National Center of Biotechnology Information (NCBI) RefSeq database were included. Genes were filtered from downstream analysis if they failed to achieve a CPM (counts per million mapped fragments) value of ≥0.5 in at least two libraries. Counts were converted to log2 counts per million, quantile normalized, and precision weighted with the voom function of the limma package (Law et al., 2014; Ritchie et al., 2015). A linear model was fitted to each gene, and empirical Bayes moderated t statistics were used to assess differences in expression (Smyth, 2004). Genes were called DE if they achieved a false discovery rate of ≤0.05 and had an expression change of two folds or greater. The so-called DE genes also had to have at least 8 FPKMs (fragments per kilobase of exon length per million mapped fragments) in one or both of the two cell types being compared. The gene set enrichment plots were generated with the barcodeplot function in limma. Gene set enrichment analysis was carried out using the roast method in limma, with 999 rotations (Wu et al., 2010). One-sided p-values are reported.
To generate a GC B cell signature, we made use of our RNAseq dataset that includes expression data for GC B cells, follicular B cells, marginal zone B cells, B1 cells, and plasma cells (Shi et al., 2015). To be included in the GC signature, genes had to have at least threefold higher (GC up) or lower (GC down) expression in GC B cells compared to all other populations. A FDR of 0.05 had to be achieved in any comparison. Each GC up signature gene had to have an expression abundance of ≥32 FPKMs in GC B cells, and each GC down signature gene had to have an expression abundance of ≥32 FPKMs in all populations, except GC B cells.
For ChIPseq data analysis, sequence reads were mapped to human genome hg19 using the Subread aligner. Only uniquely mapped reads were retained. E2A binding peaks were identified using MACS2 v2.1.0 program (Zhang et al., 2008) with a p-value cutoff of 10−5. Genes with binding peaks falling within between 20 kb upstream and 5 kb downstream of the gene body were considered to be E2A-binding targets. NCBI RefSeq human annotation build 37.2 was used to obtain chromosomal coordinates of genes. Binding peaks were assigned to genes using the featureCounts program. Gene symbols were used to match genes in ChIPseq data with those in RNAseq data.
Statistical analyses
If not stated otherwise, unpaired Student's t tests were performed to test for statistical significance.
ACKNOWLEDGEMENTS
We would like to thank Cornelius Murre and Meinrad Busslinger for mice, and Roger Sciammas (University of Chicago, Chicago, IL) for discussion. We also wish to thank Lauren Wilkins, Tania Camilleri, and Liana Mackiewicz for technical support.
This work was supported by grants and fellowships from the National Health and Medical Research Council of Australia (NHMRC; project grant 1023454 to W. Shi; project grant 1049416 to A. Kallies and D. Zotos; and program grant 1054925 to S.L. Nutt, D.M. Tarlinton, P.D. Hodgkin, L.M. Corcoran, and G.T. Belz), the Sylvia and Charles Viertel Foundation (A. Kallies and G.T. Belz), the Australian Research Council (S.L. Nutt, G.T. Belz, and A. Kallie), and the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC Independent Research Institute Infrastructure Support scheme.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- ChIPseq
- ChIP sequencing
- CSR
- class-switch recombination
- CTV
- Cell Trace Violet
- DE
- differentially expressed
- GC
- germinal center
- NP-KLH
- nitro-phenol coupled to keyhole-limpet haemocyanin
- RNAseq
- RNA sequencing
References
- Allen C.D., Ansel K.M., Low C., Lesley R., Tamamura H., Fujii N., and Cyster J.G.. 2004. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol. 5:943–952. 10.1038/ni1100 [DOI] [PubMed] [Google Scholar]
- Bain G., Maandag E.C., Izon D.J., Amsen D., Kruisbeek A.M., Weintraub B.C., Krop I., Schlissel M.S., Feeney A.J., van Roon M., et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 79:885–892. 10.1016/0092-8674(94)90077-9 [DOI] [PubMed] [Google Scholar]
- Bannard O., Horton R.M., Allen C.D., An J., Nagasawa T., and Cyster J.G.. 2013. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 39:912–924. 10.1016/j.immuni.2013.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso K., and Dalla-Favera R.. 2010. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 105:193–210. 10.1016/S0065-2776(10)05007-8 [DOI] [PubMed] [Google Scholar]
- Basso K., and Dalla-Favera R.. 2012. Roles of BCL6 in normal and transformed germinal center B cells. Immunol. Rev. 247:172–183. 10.1111/j.1600-065X.2012.01112.x [DOI] [PubMed] [Google Scholar]
- Bergqvist I., Eriksson M., Saarikettu J., Eriksson B., Corneliussen B., Grundström T., and Holmberg D.. 2000. The basic helix-loop-helix transcription factor E2-2 is involved in T lymphocyte development. Eur. J. Immunol. 30:2857–2863. [DOI] [PubMed] [Google Scholar]
- Caron G., Le Gallou S., Lamy T., Tarte K., and Fest T.. 2009. CXCR4 expression functionally discriminates centroblasts versus centrocytes within human germinal center B cells. J. Immunol. 182:7595–7602. 10.4049/jimmunol.0804272 [DOI] [PubMed] [Google Scholar]
- Carotta S., Willis S.N., Hasbold J., Inouye M., Pang S.H., Emslie D., Light A., Chopin M., Shi W., Wang H., et al. 2014. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J. Exp. Med. 211:2169–2181. 10.1084/jem.20140425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy K.R., Wellard C.J., Markham J.F., Zhou J.H., Holmberg R., Hawkins E.D., Hasbold J., Dowling M.R., and Hodgkin P.D.. 2012. Activation-induced B cell fates are selected by intracellular stochastic competition. Science. 335:338–341. 10.1126/science.1213230 [DOI] [PubMed] [Google Scholar]
- Fukuda T., Yoshida T., Okada S., Hatano M., Miki T., Ishibashi K., Okabe S., Koseki H., Hirosawa S., Taniguchi M., et al. 1997. Disruption of the Bcl6 gene results in an impaired germinal center formation. J. Exp. Med. 186:439–448. 10.1084/jem.186.3.439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda H., Sugai M., Nambu Y., Katakai T., Agata Y., Mori K.J., Yokota Y., and Shimizu A.. 2003. The balance between Pax5 and Id2 activities is the key to AID gene expression. J. Exp. Med. 198:1427–1437. 10.1084/jem.20030802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasbold J., Corcoran L.M., Tarlinton D.M., Tangye S.G., and Hodgkin P.D.. 2004. Evidence from the generation of immunoglobulin G-secreting cells that stochastic mechanisms regulate lymphocyte differentiation. Nat. Immunol. 5:55–63. 10.1038/ni1016 [DOI] [PubMed] [Google Scholar]
- Hayakawa I., Tedder T.F., and Zhuang Y.. 2007. B-lymphocyte depletion ameliorates Sjögren’s syndrome in Id3 knockout mice. Immunology. 122:73–79. 10.1111/j.1365-2567.2007.02614.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Geng H., Boss I., Wang L., and Melnick A.. 2014. Cooperative transcriptional repression by BCL6 and BACH2 in germinal center B-cell differentiation. Blood. 123:1012–1020. 10.1182/blood-2013-07-518605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.T., Hu Y., Liu R., Masson F., D’Amico A., Carotta S., Xin A., Camilleri M.J., Mount A.M., Kallies A., et al. 2011. Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages. EMBO J. 30:2690–2704. 10.1038/emboj.2011.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Hasbold J., Tarlinton D.M., Dietrich W., Corcoran L.M., Hodgkin P.D., and Nutt S.L.. 2004. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J. Exp. Med. 200:967–977. 10.1084/jem.20040973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies A., Hasbold J., Fairfax K., Pridans C., Emslie D., McKenzie B.S., Lew A.M., Corcoran L.M., Hodgkin P.D., Tarlinton D.M., and Nutt S.L.. 2007. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 26:555–566. 10.1016/j.immuni.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Kee B.L. 2009. E and ID proteins branch out. Nat. Rev. Immunol. 9:175–184. 10.1038/nri2507 [DOI] [PubMed] [Google Scholar]
- Kee B.L., Rivera R.R., and Murre C.. 2001. Id3 inhibits B lymphocyte progenitor growth and survival in response to TGF-beta. Nat. Immunol. 2:242–247. 10.1038/85303 [DOI] [PubMed] [Google Scholar]
- Khalil A.M., Cambier J.C., and Shlomchik M.J.. 2012. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science. 336:1178–1181. 10.1126/science.1213368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim U., Qin X.F., Gong S., Stevens S., Luo Y., Nussenzweig M., and Roeder R.G.. 1996. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 383:542–547. 10.1038/383542a0 [DOI] [PubMed] [Google Scholar]
- Kishida T., Hiromura Y., Shin-Ya M., Asada H., Kuriyama H., Sugai M., Shimizu A., Yokota Y., Hama T., Imanishi J., et al. 2007. IL-21 induces inhibitor of differentiation 2 and leads to complete abrogation of anaphylaxis in mice. J. Immunol. 179:8554–8561. 10.4049/jimmunol.179.12.8554 [DOI] [PubMed] [Google Scholar]
- Klein U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., and Dalla-Favera R.. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782. 10.1038/ni1357 [DOI] [PubMed] [Google Scholar]
- Kwon K., Hutter C., Sun Q., Bilic I., Cobaleda C., Malin S., and Busslinger M.. 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 28:751–762. 10.1016/j.immuni.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Law C.W., Chen Y., Shi W., and Smyth G.K.. 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., and Shi W.. 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lin Y.C., Jhunjhunwala S., Benner C., Heinz S., Welinder E., Mansson R., Sigvardsson M., Hagman J., Espinoza C.A., Dutkowski J., et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 11:635–643. 10.1038/ni.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love C., Sun Z., Jima D., Li G., Zhang J., Miles R., Richards K.L., Dunphy C.H., Choi W.W., Srivastava G., et al. 2012. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 44:1321–1325. 10.1038/ng.2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man K., Miasari M., Shi W., Xin A., Henstridge D.C., Preston S., Pellegrini M., Belz G.T., Smyth G.K., Febbraio M.A., et al. 2013. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14:1155–1165. 10.1038/ni.2710 [DOI] [PubMed] [Google Scholar]
- Miyazaki M., Rivera R.R., Miyazaki K., Lin Y.C., Agata Y., and Murre C.. 2011. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat. Immunol. 12:992–1001. 10.1038/ni.2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C. 2005. Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 6:1079–1086. 10.1038/ni1260 [DOI] [PubMed] [Google Scholar]
- Muto A., Tashiro S., Nakajima O., Hoshino H., Takahashi S., Sakoda E., Ikebe D., Yamamoto M., and Igarashi K.. 2004. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 429:566–571. 10.1038/nature02596 [DOI] [PubMed] [Google Scholar]
- Muto A., Ochiai K., Kimura Y., Itoh-Nakadai A., Calame K.L., Ikebe D., Tashiro S., and Igarashi K.. 2010. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 29:4048–4061. 10.1038/emboj.2010.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Heavey B., Rolink A.G., and Busslinger M.. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 401:556–562. 10.1038/44076 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Hodgkin P.D., Tarlinton D.M., and Corcoran L.M.. 2015. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 15:160–171. 10.1038/nri3795 [DOI] [PubMed] [Google Scholar]
- Ochiai K., Maienschein-Cline M., Simonetti G., Chen J., Rosenthal R., Brink R., Chong A.S., Klein U., Dinner A.R., Singh H., and Sciammas R.. 2013. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 38:918–929. 10.1016/j.immuni.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Sato S., Frederick J.P., Sun X.H., and Zhuang Y.. 1999. Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol. Cell. Biol. 19:5969–5980. 10.1128/MCB.19.9.5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold A.M., Iwakoshi N.N., Manis J., Vallabhajosyula P., Szomolanyi-Tsuda E., Gravallese E.M., Friend D., Grusby M.J., Alt F., and Glimcher L.H.. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 412:300–307. 10.1038/35085509 [DOI] [PubMed] [Google Scholar]
- Renné C., Martin-Subero J.I., Eickernjäger M., Hansmann M.L., Küppers R., Siebert R., and Bräuninger A.. 2006. Aberrant expression of ID2, a suppressor of B-cell-specific gene expression, in Hodgkin’s lymphoma. Am. J. Pathol. 169:655–664. 10.2353/ajpath.2006.060020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J., Schlesner M., Hoffmann S., Kreuz M., Leich E., Burkhardt B., Rosolowski M., Ammerpohl O., Wagener R., Bernhart S.H., et al. ICGC MMML-Seq Project . 2012. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat. Genet. 44:1316–1320. 10.1038/ng.2469 [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh C.E., Quong M.W., Agata Y., and Murre C.. 2003. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 4:586–593. 10.1038/ni923 [DOI] [PubMed] [Google Scholar]
- Schmitz R., Ceribelli M., Pittaluga S., Wright G., and Staudt L.M.. 2014. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb. Perspect. Med. 4:4 10.1101/cshperspect.a014282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubart D.B., Rolink A., Kosco-Vilbois M.H., Botteri F., and Matthias P.. 1996. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 383:538–542. [DOI] [PubMed] [Google Scholar]
- Sciammas R., Shaffer A.L., Schatz J.H., Zhao H., Staudt L.M., and Singh H.. 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 25:225–236. 10.1016/j.immuni.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Shaffer A.L., Lin K.I., Kuo T.C., Yu X., Hurt E.M., Rosenwald A., Giltnane J.M., Yang L., Zhao H., Calame K., and Staudt L.M.. 2002. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 17:51–62. 10.1016/S1074-7613(02)00335-7 [DOI] [PubMed] [Google Scholar]
- Shaffer A.L., Shapiro-Shelef M., Iwakoshi N.N., Lee A.H., Qian S.B., Zhao H., Yu X., Yang L., Tan B.K., Rosenwald A., et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 21:81–93. 10.1016/j.immuni.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M., Lin K.I., McHeyzer-Williams L.J., Liao J., McHeyzer-Williams M.G., and Calame K.. 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 19:607–620. 10.1016/S1074-7613(03)00267-X [DOI] [PubMed] [Google Scholar]
- Shi W., Liao Y., Willis S.N., Taubenheim N., Inouye M., Tarlinton D.M., Smyth G.K., Hodgkin P.D., Nutt S.L., and Corcoran L.M.. 2015. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 16:663–673. 10.1038/ni.3154 [DOI] [PubMed] [Google Scholar]
- Shlomchik M.J., and Weisel F.. 2012. Germinal centers. Immunol. Rev. 247:5–10. 10.1111/j.1600-065X.2012.01125.x [DOI] [PubMed] [Google Scholar]
- Smyth G.K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:e3. [DOI] [PubMed] [Google Scholar]
- Sugai M., Gonda H., Kusunoki T., Katakai T., Yokota Y., and Shimizu A.. 2003. Essential role of Id2 in negative regulation of IgE class switching. Nat. Immunol. 4:25–30. 10.1038/ni874 [DOI] [PubMed] [Google Scholar]
- Tarlinton D., and Good-Jacobson K.. 2013. Diversity among memory B cells: origin, consequences, and utility. Science. 341:1205–1211. 10.1126/science.1241146 [DOI] [PubMed] [Google Scholar]
- Tarlinton D.M., and Smith K.G.. 2000. Dissecting affinity maturation: a model explaining selection of antibody-forming cells and memory B cells in the germinal centre. Immunol. Today. 21:436–441. 10.1016/S0167-5699(00)01687-X [DOI] [PubMed] [Google Scholar]
- Taubenheim N., Tarlinton D.M., Crawford S., Corcoran L.M., Hodgkin P.D., and Nutt S.L.. 2012. High rate of antibody secretion is not integral to plasma cell differentiation as revealed by XBP-1 deficiency. J. Immunol. 189:3328–3338. 10.4049/jimmunol.1201042 [DOI] [PubMed] [Google Scholar]
- Victora G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Wikström I., Forssell J., Goncalves M., Colucci F., and Holmberg D.. 2006. E2-2 regulates the expansion of pro-B cells and follicular versus marginal zone decisions. J. Immunol. 177:6723–6729. 10.4049/jimmunol.177.10.6723 [DOI] [PubMed] [Google Scholar]
- Willis S.N., Good-Jacobson K.L., Curtis J., Light A., Tellier J., Shi W., Smyth G.K., Tarlinton D.M., Belz G.T., Corcoran L.M., et al. 2014. Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J. Immunol. 192:3200–3206. 10.4049/jimmunol.1303216 [DOI] [PubMed] [Google Scholar]
- Wu D., Lim E., Vaillant F., Asselin-Labat M.L., Visvader J.E., and Smyth G.K.. 2010. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics. 26:2176–2182. 10.1093/bioinformatics/btq401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying C.Y., Dominguez-Sola D., Fabi M., Lorenz I.C., Hussein S., Bansal M., Califano A., Pasqualucci L., Basso K., and Dalla-Favera R.. 2013. MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat. Immunol. 14:1084–1092. 10.1038/ni.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Soriano P., and Weintraub H.. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell. 79:875–884. 10.1016/0092-8674(94)90076-0 [DOI] [PubMed] [Google Scholar]