Abstract
The comparative reaction efficiencies of currently used nucleophilic and electrophilic probes towards cysteine sulfenic acid have been thoroughly evaluated in two different settings – (i) a small molecule dipeptide based model and, (ii) a recombinant protein model. We further evaluated the stability of corresponding thioether and sulfoxide adducts under reducing conditions which are commonly encountered during proteomic protocols and in cell analysis. Powered by the development of new cyclic and linear C-nucleophiles, the unsurpassed efficiency in the capture of sulfenic acid under competitive conditions is achieved and thus holds great promise as highly potent tools for activity-based sulfenome profiling.
Graphical Abstract
Introduction
Owing to the intrinsic nucleophilicity and oxidation susceptibility, cysteine thiols (Cys-SH) are prime targets for the redox-based modulation of protein activity. Oxidation of a protein cysteine thiol (Cys-SH) to sulfenic acid (Cys-SOH) by reactive oxygen species (ROS) (e.g. H2O2), termed S-sulfenylation, is a reversible post-translational modification that plays a crucial role in regulating several protein functions.1–6 With an estimated half-life in minutes, Cys-SOH is considered a transient species whose ultimate fate depends upon factors such as presence of neighbouring thiol, level/duration of ROS and protein microenvironment.5 Essentially, absence of proximal thiols capable of forming intramolecular disulfide bond, limited solvent access and proximal hydrogen bond acceptors contribute towards Cys-SOH stabilization. High/chronic oxidative stress may cause –SOH to undergo further oxidation to irreversible sulfinic (Cys-SO2H) or sulfonic acid (Cys-SO3H). Due to the abundance of biological thiols (mM levels), an important and facile biological reaction of Cys-SOH is the disulfide formation. The nascent disulfide may undergo thiol-disulfide exchange to regenerate initial Cys-SH. Cys-SOH may undergo intramolecular reaction with adjacent amide nitrogen resulting in the formation of a cyclic species known as cyclic sulfenamide which may be reduced back to thiol via disulfide formation. Due to the central role of Cys-SOH in reversible/irreversible pathway, it serves as an important hub within the redox milieu.5 Indeed, over the past decade several groups have reported the role of Cys-SOH in regulation of several proteins such as transcription factors, kinases, phosphatases, ion channels, peroxidases and cysteine proteases, sirtuins, human serum albumin and many others.3, 6–21 Moreover, aberrant Cys-SOH formation has been shown to correlate with disease state.12, 22 The aforementioned examples and many other reports establish protein S-sulfenylation as a global signaling mechanism akin to phosphorylation and a potential drug target.5, 6, 15, 23–27
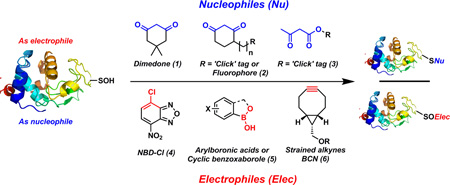
Due to the electrophilic as well as nucleophilic character of sulfur atom in Cys-SOH, the detection methods exploiting both have been reported (Fig. 1).5, 10–13, 19, 20, 25, 26, 28–43 Briefly, due to higher electrophilicity (than nucleophilicity) of Cys-SOH, vast majority of probes are nucleophilic in nature and based on cyclic 1,3-carbonyl scaffold such as dimedone (1). These nucleophilic probes reacts with Cys-SOH to give thioether adducts. Such nucleophilic probes are extensively employed to study qualitative and quantitative capture of Cys-SOH.1, 5, 9–11, 13–16, 18–22, 29–32, 34–39, 42–51 Commonly used repertoire of C-nucleophilic probes consist of commercially available 1,3-dicarbonyl-based ‘clickable’ probes such as DYn-2 (2a),15 DAz-2 (2b),10 Alk-β-KE (3),46 BP1 (7),44 and under development 6-hydroxy derivatives of phenalene-1,3-dione (8) (Fig. 2). Enrichment-ready/imaging-ready reagents such as DCP-Bio1 (2c) and DCP-Rho1 (2d) have also been made commercially available and used extensively.30 Although these nucleophilic probes are selective for protein sulfenic acid under aqueous physiological conditions, they suffer from poor reaction kinetics (kobs = 0.008 M−1s−1 – 0.05 M−1s−1) when compared to other biological reactions of Cys-SOH (such as disulfide formation or over-oxidation to sulfinic/sulfonic acid).5 To overcome this limited reactivity problem, we recently developed and reported a facile mass-spectrometery-based assay and a repurposed dipeptide-based sulfenic acid model using which we screened a library of cyclic and linear C-nucleophiles for reactivity towards sulfenic acid under aqueous conditions. We obtained the reaction rate constants of more than 150 C-nucleophiles and from this collection we identified novel classes of compounds with more than 200-fold enhanced reactivity towards Cys-SOH.25, 26
Fig. 1.
Dual chemical reactivity of sulfenic acid.
Fig. 2.
Nucleophilic and electrophilic probes reported for detecting sulfenic acid.
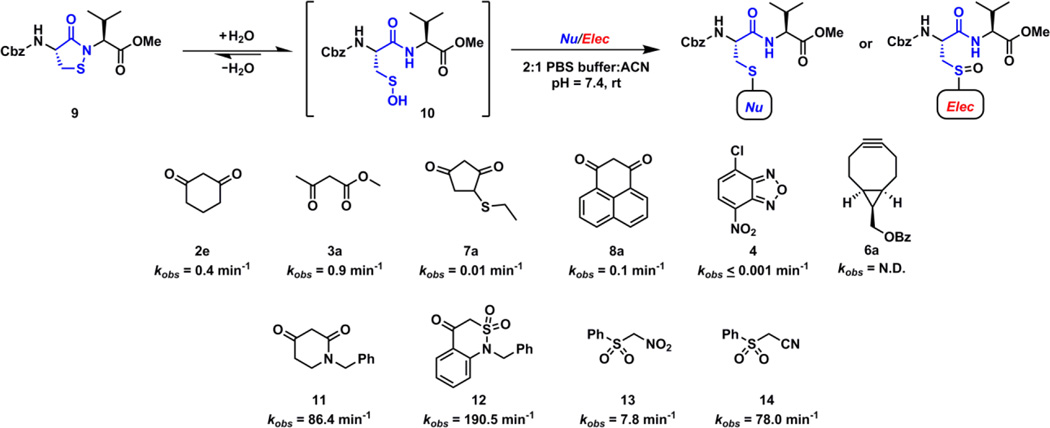
Sulfenic acid can also act as a nucleophile and react with electrophiles to produce a sulfoxide adduct (Fig. 1). Given the nucleophilic properties of thiols and several other biological species, it is generally considered that the selectivity towards Cys-SOH maybe difficult to achieve with electrophilic detection tools.47 Currently reported examples of electrophilic probes are commercially available compounds such as (i) low reactive NBD-Cl (4) which also lack the chemoselectivity and react with several biological nucleophiles including sulfenic acid (but the resulting adducts are distinguishable due to difference in their spectroscopic properties);5, 28, 38 (ii) arylboronates (5) which react with Cys-SOH in a reversible manner and thus have limited utility in proteomic-based sulfenic acid detection52 and; (iii) strained cycloalkynes (6) which were recently reported as new, highly reactive sulfenic acid traps (kobs = 13.3 M−1s−1 – 16.7 M−1s−1) with > 100-fold higher reactivity than 1,3-dicarbonyls.40 Not surprisingly, over the years strained cycloalkynes have been shown to react with other sulfur species such as protein- and small molecule thiols (present in mM concentration inside the cells) and persulfides.53–55 Moreover, the product of the reaction of Cys-SOH and strained cycloalkyne 6 is an activated vinylic sulfoxide which is a Michael-acceptor and may readily react with biological thiols as well.56 As a consequence of the promiscuous reactivity of strained cycloalkynes and the resultant sulfoxide adduct, an additional thiol-blocking step is required to increase the selectivity for sulfenic acid. As mentioned above, even though over the past decade several nucleophilic and electrophilic probes for sulfenic acid detection have been developed and commercialized, the issue of modest reactivity of such probes is well documented.5, 26, 40 Several subsequent publications reported probes with superior reactivity compared to dimedone-based probes at lower or higher pHs.44, 46, 47 However, such reports generally compared the reactivities of these probes towards Cys-SOH in specific model protein systems where the reactivity could be biased by the protein microenvironment surrounding Cys-SOH. For this reason a direct comparison of the reactivity of various nucleophilic/electrophilic probes towards Cys-SOH and the stability of resulting thioether/sulfoxide adduct under exact same conditions is needed for the better understanding of the reactivity profile of each probe. In order to do so, we chose to utilize our peptide-based sulfenic acid model with subsequent verification in a protein sulfenic acid model. The peptide-based sulfenic acid model exists in the masked form as a cyclic sulfenamide (9) which rearranges to dipeptide sulfenic acid (10) under aqueous conditions (Fig. 3).26 Since this model is not influenced by a complex protein microenvironment, the observed reaction rates are independent of factors such as steric hindrance and stabilization/destabilization of sulfenic acid.
Fig. 3.
Nucleophiles and electrophiles tested for reactivity with sulfenic acid 10.
Results and Discussion
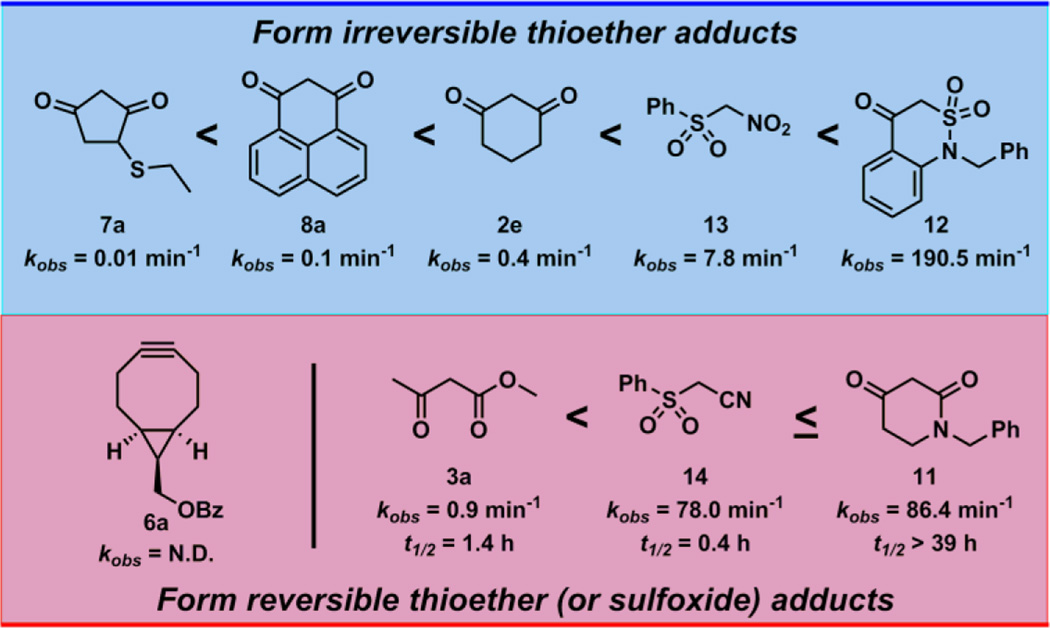
Using the peptide-based model and the previously developed LC/MS assay, we initially obtained the pseudo 1st order rate constants ([Nu/Elec] = 1 mM, [10] = 100 µM) of three nucleophilic (2e, 3a, 7a) and two electrophilic (4, 6a) compounds (Fig. 3). These compounds constitute the reactive component of prevalent and commercially available Cys-SOH probes such as 2a–d, 3, 7, 4 and 6 (Fig. 2). Nucleophilic cyclohexane-1,3-dione (2e, kobs = 0.4 min−1), methyl acetoacetate (3a, kobs = 0.9 min−1) and 4-(ethylthio)cyclopentane-1,3-dione (7a, kobs = 0.01 min−1) successfully reacted with dipeptide-SOH 10 to form corresponding thioether adduct. While 2e and 3a had similar reactivity under physiological conditions, it is noteworthy that 7a was 40-folds less reactive compared to 2e towards sulfenic acid 10. Electrophilic 4-chloro-7-nitrobenzo[c][1,2,5]oxadiazole (4, kobs < 0.001 min−1) was extremely slow to react and only a small amount of sulfoxide adduct could be detected even after 48 h (ESI, Fig. S4). Conversely, while the benzoyl derivative of strained cycloalkyne ((1R,8S,9s)-bicyclo[6.1.0]non-4-yn-9-yl)methyl benzoate (6a, kobs = N.D.) was highly reactive, the corresponding sulfoxide adduct was unstable and quickly rearranged to disulfide (Cbz-Cys-Val-OMe)2 (ESI, Fig. S5). Next, we obtained the pseudo 1st order rate constants of 1H–phenalene-1,3(2H)-dione (8a, kobs = 0.1 min−1), recently developed cyclic C-nucleophiles 1-benzylpiperidine-2,4-dione (11, kobs = 86.4 min−1), 1-benzyl-1H–benzo[c][1,2]thiazin-4(3H)-one 2,2-dioxide (12, kobs = 190.5 min−1), linear C-nucleophiles ((nitromethyl)sulfonyl)benzene (13, kobs = 7.8 min−1), and 2-(phenylsulfonyl)acetonitrile (14, kobs = 78.0 min−1) for the reaction with dipeptide-SOH 10. Due to the opposing effect of resonance stabilization and electron donating naphthyl group, 8a showed a reduced rate of reaction towards sulfenic acid 10. Cyclic and linear C-nucleophiles 11–14 showed elevated reactivity towards 10. In brief, the reactions of various nucleophiles and electrophiles towards dipeptide-SOH 10 clearly demonstrated the substantially enhanced reactivity of 11–14 compared to currently prevalent probes (Fig. 3).
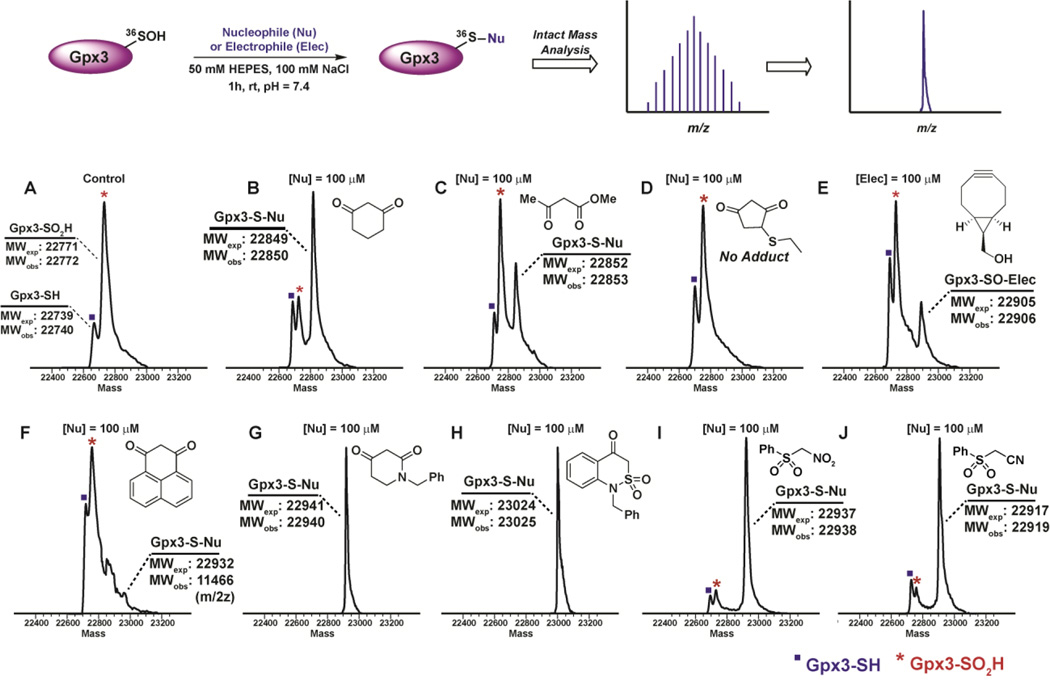
Subsequently, we examined the reactivity of above-tested nucleophiles and electrophiles towards a protein Cys-SOH. For these studies, we used our well-established Cys64Ser Cys82Ser variant of the recombinant thiol peroxidase, Gpx3.11 Incubation of nucleophiles and electrophiles at 100 µM concentration with Gpx3-SOH led to the covalent modification of Gpx3-SOH. Of note, while trapping with 2e was not quantitative, it still captured ~ 50% Gpx3-SOH (Fig. 4B). Interestingly, even though β-ketoester 3a showed a higher rate of reaction in the dipeptide-SOH model, the amount of Gpx3-SOH captured was lower than 2e, highlighting the influence of protein microenvironment on reactivity as well as the tendency of the thioether adduct of 3a to undergo loss of a methylketo (−COMe) group (Fig. 4C).25, 57 Owing to poor reaction rate, 7a failed to capture Gpx3-SOH, a finding that was consistent with data obtained in the dipeptide-SOH model (Fig. 4D). In contrast to data reported using the C165A AhpC-SOH,40 the hydroxyl version of strained cycloalkyne 6 captured only a small amount of Gpx3-SOH (Fig. 4E). This result correlates well with the observation that the sulfoxide adduct of 6a with dipeptide-SOH 10 quickly degrades (ESI, Fig. S5). Phenalene-1,3-dione 8a also showed extremely poor reactivity towards Gpx3-SOH, also consistent with results obtained in the dipeptide–SOH model (Fig. 4F). Finally, akin to their superior reactivity towards the dipeptide-SOH, cyclic C-nucleophiles 11 and 12 quantitatively trapped Gpx3-SOH (Fig. 4G–H). Linear C-nucleophiles 13 and 14 also tagged the majority of Gpx3-SOH (Figure 4I–J). These results gratifyingly confirmed the order of reactivity observed in our LC/MS assay using the dipeptide-SOH model system.
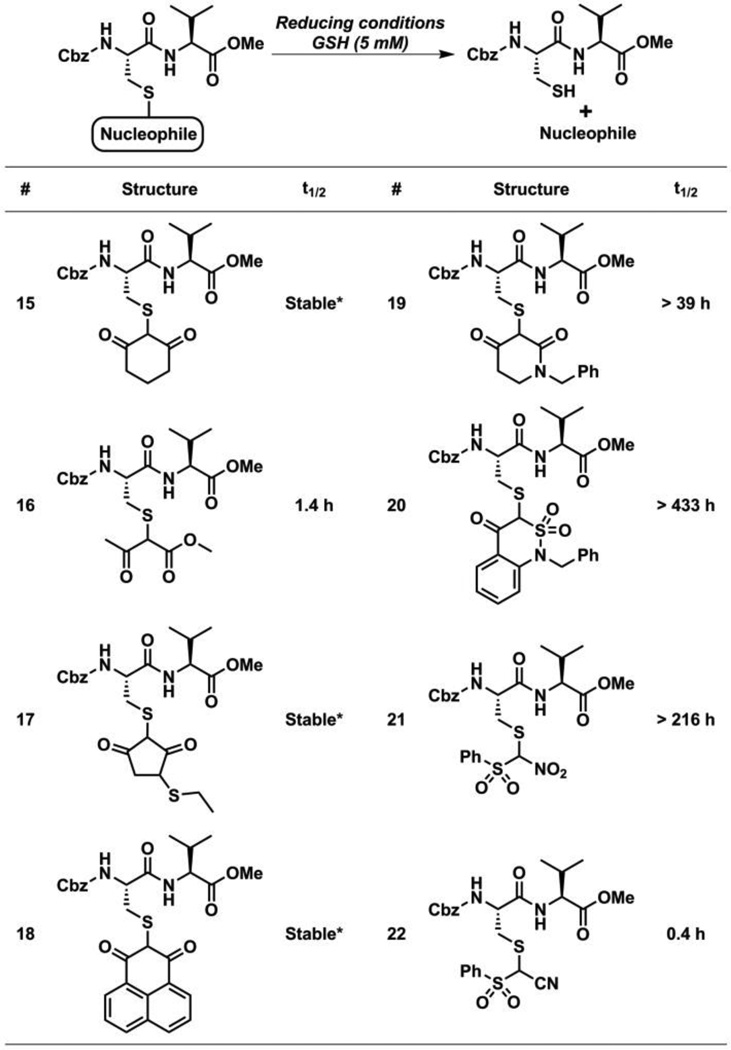
Fig. 4.
Reactivity of nucleophiles and electrophiles with Gpx3-SOH.
We previously showed that due to the difference in reactivity, pKa, and carbanion stability of C-nucleophiles, the resulting thioether adducts also display a range of stability under reducing conditions.25, 26 As cell contains millimolar concentrations of glutathione (GSH) we tested the ability of this low-molecular weight thiol to reduce the nascent covalent thioether bond formed between 10 and nucleophiles. Initially thioether adducts of linear C-nucleophiles were shown to be reversible under reducing conditions – a view strengthened by the stability of thioether adducts 15, 17 and 18 and unstable nature of thioether adducts 16 (t1/2 = 1.4 h) and 22 (t1/2 = 0.4 h) (Table 1, ESI Fig. S6 – S21 and Fig. S34 – S37).25, 26 However, the instability of thioether adduct 19 (t1/2 > 39 h) formed by cyclic C-nucleophile 11 (ESI, Fig. S22 – S25), and the stability of thioether adduct 21 (t1/2 > 9 days) of linear C-nucleophile 14 (ESI, Fig. S30 – S33), indicates that the cyclic/linear nature of nucleophiles is not the determining factor with respect to the stability/instability of corresponding adducts (Table 1). Interestingly, enhanced reactivity of a nucleophile is also not the sole criterion for reversibility, as illustrated by the stability of thioether adduct 20 (t1/2 > 18 days) which results from trapping by the highly reactive cyclic C-nucleophile 12 (Table 1, ESI Fig. S26 – S29). Thioether adducts showing t1/2 < 48 h were then tested for reversibility in the Gpx3 model. Gpx3-SOH was first labeled with nucleophiles 3a, 11 and 14. Subsequently, each Gpx3-Nu adduct was subjected to mM glutathione (GSH), dithiothreitol (DTT) or the phosphine-based reductant, TCEP. Correlating with the reversibility trends observed in the reduction of dipeptide-based model, Gpx3-S-Nu adducts formed by β-ketoester 3a and phenylsulfonylacetonitrile 14 were almost completely reversible while the adduct corresponding to 11 showed some reversibility under reducing conditions (ESI Fig. S50).
Table 1.
Reversibility of dipeptide-S-Nu adducts under reducing conditions.
Stable adduct formation is defined as an adduct in which no detectable cleavage has occurred within 48 h.
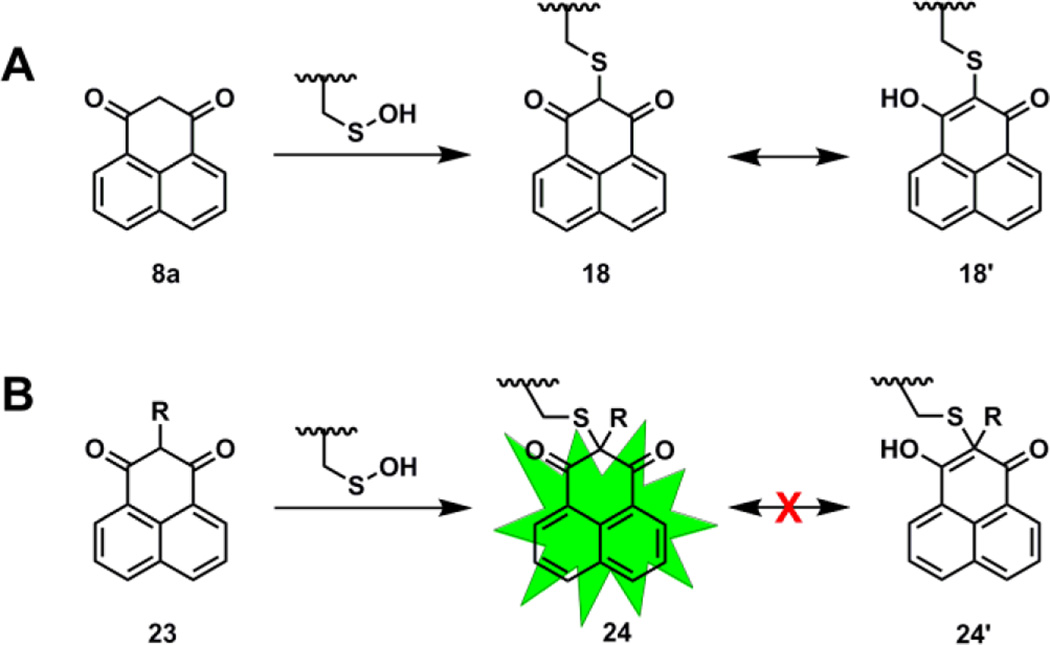
In light of the aforementioned stability data, we subsequently explored the effect of C-2 substitution on the reactivity of nucleophiles towards sulfenic acid. In this regard, one key issue is that a tertiary carbanion is less stable than a secondary carbanion. Moreover, upon reaction with sulfenic acid, a C-2 substituted nucleophile would be ‘locked’ in the keto tautomer. Such a conformational lock could potentially result in unique spectroscopic properties (i.e., UV/Vis or fluorescence) of the nucleophile before and after reaction with sulfenic acid. Such difference in UV/Vis absorption or fluorescence may be exploited to develop a ‘turn-on/turn-off switch’ based sulfenic acid detection strategy. A representative example of a nucleophile undergoing such change in spectroscopic properties is 2-substituted derivatives of phenalene-1,3-dione (8a). Keto-enol tautomerism is facile for compound 8a readily and, upon reaction with Cys-SOH, keto product 18 can easily rearrange to give enol 18’ (Fig. 5A), whereas the thioether derivative of 2-substituted- phenalene-1,3-dione (23) would be locked in the keto form 24 (Fig. 5B) resulting in turn-on fluorescence.58 Although new spectral properties of the resulting thioether adduct is an attractive possibility, the effect of C-2 substitution by an electron-donating group (EDG) or electron-withdrawing group (EWG) on nucleophile reactivity has not been studied.
Fig. 5.
Effect of C-2 substitution on the UV-vis and fluorescence properties of the nucleophiles and corresponding thioether adducts.
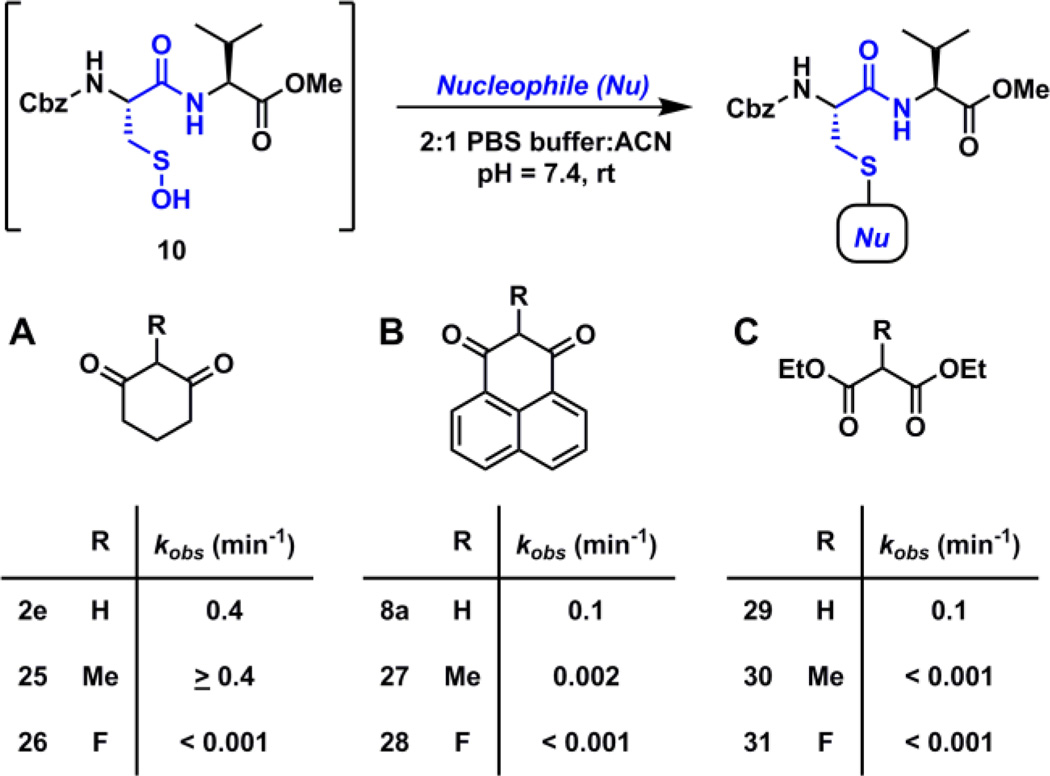
To examine the issue of C-2 substitution, we studied the reactivity of dipeptide sulfenic acid 10 with C-2 methylated (EDG) and C-2 fluorinated (EWG) derivatives of three nucleophiles – (a) cyclohexane-1,3-dione (2e), (b) 1H–phenalene-1,3(2H)-dione (8a), and (c) diethylmalonate (29). The analogs were prepared following literature reported procedures.59 The kinetic studies data are summarized in Table 2. Briefly, while 2-methylcyclohexane-1,3-dione (25, kobs > 0.4 min−1) showed slightly higher reactivity than 2e (kobs = 0.4 min−1), 2-fluorocyclohexane-1,3-dione (26, kobs < 0.001 min−1) was very slow to react (Table 2A). Conversely, both 2-methyl-1H–phenalene-1,3(2H)-dione (27, kobs = 0.002 min−1) and 2-fluoro-1H–phenalene-1,3(2H)-dione (28, kobs < 0.001 min−1) showed reduced reactivity, as compared to 8a (kobs = 0.1 min−1) (Table 2B). Diethylmalonate (29, kobs = 0.1 min−1) exhibited similar reactivity to 8a and its C-2 substituted derivatives diethyl 2-methylmalonate (29, kobs < 0.001 min−1) and diethyl 2-fluoromalonate (30, kobs < 0.001 min−1) were extremely slow to react as well (Table 2C).
Table 2.
Effect of C-2 substitution on the reactivity of nucleophiles towards dipeptide-SOH.
The poor reactivity of C-2 fluorinated analogs 26, 28 and 31 may be attributed to the strongly electron-withdrawing effect resulting in hydration of carbonyl group. However, the reactivity of C-2 methylated analogs towards dipeptide-SOH 10 is more complex to explain. While 25 showed slightly enhanced reactivity compared to 2e the resulting thioether adduct quickly underwent carbonyl hydration due to the electron-withdrawing effect of -SR group (ESI Fig. S51 – S52). Similar carbonyl hydration was observed in case of thioether adduct of 27 (ESI Fig. S53). Another consequence of hydration is that the diketo nucleophile becomes a poorer leaving group and the corresponding thioether adducts are stable under reducing conditions (ESI Fig. S38 – S41). The hydration effect is not observed in case of the thioether adduct 15 of 2e or 18 of 8a because under aqueous conditions they readily undergo keto-enol tautomerism and favor the enol form. Diethylmalonate 29 has a comparatively high pKa value of 13 and its C-2 methyl derivative 30 is expected to have an even higher pKa, which would result in reduced reactivity towards sulfenic acid 10, as was observed. In general, C-2 substitution is detrimental for the reactivity of C-nucleophiles towards sulfenic acid and reduced the reaction rates by 100-folds or more.
Like many other critical post-translational modifications, such as phosphorylation and methylation, protein sulfenylation is reversible with an estimated half-life occurring on the timescale of minutes.5 In the case of sulfenic acid, its transient nature is often attributed to its reaction with GSH and protein thiol group and these reactions compete with chemical probes. We illustrate this issue by calculating the percent of Human Serum Albumin sulfenic acid (HSA-SOH) and oxidized MAP Kinase Phosphatase 3 (oxMKP3) trapped by various nucleophiles in the presence of mM GSH (ESI, Fig. S54). It is clear from the calculations that in both cases, the majority of C-nucleophiles on which current probes are based would capture only a fraction of sulfenic acid (Table 3, entries 2e, 3a, 7a and 8a). By contrast, the same calculations with more recently developed cyclic and linear C-nucleophiles predict near quantitative capture of both HSA-SOH and oxMKP3, even at low nucleophile concentration ([Nu] = 100 µM) (Table 3, entries 11–14). Collectively, these data highlight key differences among chemical probes for detecting sulfenic acid in the face of competing biological reactions.
Table 3.
Amount of protein-SOH captured by nucleophiles in the presence of GSH at steady state.
| # | Nucleophiles |
k2 (M−1s−1) |
% HSA-SOH trapped by Nu |
%oxMKP3 trapped by Nu |
# | Nucleophiles |
k2 (M−1s−1) |
% HSA-SOH trapped by Nu |
% OXMKP3 trapped by Nu |
|---|---|---|---|---|---|---|---|---|---|
| 2e | 6.7 | 10% | 14% | 11 | 1442.8 | 96% | 97% | ||
| 3a | 15.0 | 20% | 27% | 12 | 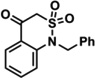 |
3175.6 | 98% | 99% | |
| 7a | 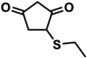 |
0.17 | 0.3% | 0.4% | 13 | 130.0 | 69% | 76% | |
| 8a | 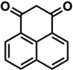 |
1.7 | 3% | 4% | 14 | 1300 | 96% | 97% |
The impact of the slow reaction rates of currently used probes in the detection and quantitative analysis of protein-SOH is countered by using large amount of starting biological material (> 30 mg), higher probe concentrations (5 mM – 10 mM) and longer incubation periods.19, 41 It is speculated that higher probe concentrations and longer incubation times increase the rate of false positive identification.21 Thus, an ideal probe for proteomic studies would have the following characteristics: (i) sufficiently fast rate of reaction with protein-SOH at low probe concentrations; (ii) form stable adducts with protein-SOH; and (iii) retain chemoselectivity for sulfenic acid. Many of the commercially available probes do not satisfy one or more of these criterions. For example, most commonly used probes such as DYn-2 (2a), DAz-2 (2b), DCP-Bio1 (2c), DCP-Rho1 (2d) and many more are based on cyclohexane1,3-dione (2e) core, which has modest reactivity towards SOH (kobs = 0.4 min−1). Factors such as the poor reactivity of cyclopentane-1,3-dione (kobs = 0.01 min−1), strongly anionic nature and presence of a biotin group (poor cell permeability and increased complexity for peptide identification) make BP1 (7) less attractive for proteomic analysis of protein-SOH. On the other hand, while β-ketoester-based Alk-β-KE (3) gives reaction rates (kobs = 0.9 min−1) similar to 2e, the corresponding thioether adduct is susceptible to reduction as well as deacetylation. Recently reported strained cycloalkyne BCN-Bio (6) was highly reactive towards small molecule SOH (10), but the corresponding sulfoxide adduct was highly unstable under the conditions of LC/MS analysis. Moreover, BCN-bio (6) showed poor reactivity towards Gpx3-SOH and is not chemoselective for sulfenic acid.53–55 The concept of exploiting 2-substituted-phenalene-1,3(2H)-diones (8, 27, 28) as fluorescent turn-on probes using conformational-lock is attractive, however such compounds suffered from poor reactivity towards dipeptide-SOH 10 (kobs < 0.001 min−1) as well as Gpx3-SOH.
In an effort to address above limitations and develop “next-generation” sulfenic acid probes, we performed side-by-side comparisons with recently developed cyclic (11, 12) and linear C-nucleophiles (13, 14). With kobs ranging from 7.8 min−1 to 190.5 min−1, C-nucleophiles 11–14 showed 20- to 500-fold rate enhancement, compared to 2e. Moreover, these nucleophiles quantitatively trapped Gpx3-SOH at low concentrations (100 µM). Thioether adducts resulting from reaction of cyclic 11 and linear 14 with sulfenic acid 10 (i.e., 19 and 22) exhibited varying stability under reducing conditions. With a t1/2 = 39 h, thioether 19 was substantially more stable than 22 (t1/2 = 0.4 h), indicating that cyclic C-nucleophile 11 may be suitable for future probe development. Thioether adducts 20 and 21 showed robust stability under reducing conditions, suggesting that C-nucleophiles 12 and 13 are excellent candidates for “next-generation” sulfenic acid probes.
Conclusion
In conclusion, we have compared the reactivity of commonly used/commercially available nucleophilic and electrophilic probes with recently developed cyclic and linear C-nucleophiles with sulfenic acid. Using the rate constants obtained from our dipeptide-SOH model, and comparing them with known rates of glutathionylation for protein-SOH, our data indicate that while current probes are sufficient for qualitative analysis, they are not capable of quantitative trapping and may not be well-suited to detect more short-lived sulfenic acid modifications. By extension, “next-generation” probes with superior reaction rates and sufficient stability will enable more comprehensive analysis of the sulfenylome. Another important consideration is the effect of probe/protein structure on chemical reactivity. Along these lines, to obtain deep coverage of the sulfenylome, it may be valuable to explore multiple probes with varying reactivity and distinct structural features. Further, we have established the reversibility of the thioether adduct resulting from β-ketoester-based Alk-β-KE (3) and other nucleophiles, such as 14, under reducing conditions encountered during trapping or in common use during proteomic sample preparation. Such reversibility can result in false negative results and, thus, it is critical to evaluate the stability of thioether (or sulfoxide) adducts formed by the reaction of nucleophilic (or electrophilic) probes (Fig. 6). Finally, we highlight the utility of dipeptide sulfenic acid 10 as model for screening and a facile way to compare the reactivity of nucleophiles and electrophiles with sulfenic acid. Moreover, dipeptide-SOH 10 provides a straightforward way to prepare the corresponding thioether (or sulfoxide) adducts in order to study their stability under various conditions.
Fig. 6.
Ratification of various nucleophiles and electrophiles on the basis of sulfenic acid reactivity and stability/reversibility of corresponding thioether (or sulfoxide) adduct.
Supplementary Material
Acknowledgments
We thank Dr. Brent Martin (University of Michigan) for conversations pertaining to 2-substituted-phenalene-1,3(2H)-diones. This work is supported by the National Institutes of Health under awards No. R01 GM102187 and R01 CA174864 to K.S.C.
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: Experimental procedures, 1H NMR, 13C NMR, and LC-MS data including stability studies.
The authors declare no competing financial interest.
References
- 1.Reddie KG, Carroll KS. Expanding the functional diversity of proteins through cysteine oxidation. Curr Opin Chem Biol. 2008;12:746–754. doi: 10.1016/j.cbpa.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 2.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo Conte M, Carroll KS. The redox biochemistry of protein sulfenylation and sulfinylation. J Biol Chem. 2013;288:26480–26488. doi: 10.1074/jbc.R113.467738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulsen CE, Carroll KS. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta V, Carroll KS. Sulfenic acid chemistry, detection and cellular lifetime. Biochim Biophys Acta. 2014;1840:847–875. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chouchani ET, Kazak L, Jedrychowski MP, Lu GZ, Erickson BK, Szpyt J, Pierce KA, Laznik-Bogoslavski D, Vetrivelan R, Clish CB, et al. Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature. 2016;532:112–116. doi: 10.1038/nature17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denu JM, Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 8.Carballal S, Radi R, Kirk MC, Barnes S, Freeman BA, Alvarez B. Sulfenic acid formation in human serum albumin by hydrogen peroxide and peroxynitrite. Biochemistry. 2003;42:9906–9914. doi: 10.1021/bi027434m. [DOI] [PubMed] [Google Scholar]
- 9.Depuydt M, Leonard SE, Vertommen D, Denoncin K, Morsomme P, Wahni K, Messens J, Carroll KS, Collet JF. A periplasmic reducing system protects single cysteine residues from oxidation. Science. 2009;326:1109–1111. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- 10.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 11.Paulsen CE, Carroll KS. Chemical dissection of an essential redox switch in yeast. Chem Biol. 2009;16:217–225. doi: 10.1016/j.chembiol.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Seo YH, Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Natl Acad Sci U S A. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leonard SE, Garcia FJ, Goodsell DS, Carroll KS. Redox-based probes for protein tyrosine phosphatases. Angew Chem Int Ed Engl. 2011;50:4423–4427. doi: 10.1002/anie.201007871. [DOI] [PubMed] [Google Scholar]
- 14.Barrett TJ, Pattison DI, Leonard SE, Carroll KS, Davies MJ, Hawkins CL. Inactivation of thiol-dependent enzymes by hypothiocyanous acid: role of sulfenyl thiocyanate and sulfenic acid intermediates. Free Radic Biol Med. 2012;52:1075–1085. doi: 10.1016/j.freeradbiomed.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol. 2012;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svoboda LK, Reddie KG, Zhang L, Vesely ED, Williams ES, Schumacher SM, O’Connell RP, Shaw R, Day SM, Anumonwo JM, et al. Redox-sensitive sulfenic acid modification regulates surface expression of the cardiovascular voltage-gated potassium channel Kv1.5. Circ Res. 2012;111:842–853. doi: 10.1161/CIRCRESAHA.111.263525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alili L, Sack M, von Montfort C, Giri S, Das S, Carroll KS, Zanger K, Seal S, Brenneisen P. Downregulation of tumor growth and invasion by redox-active nanoparticles. Antioxid Redox Signal. 2013;19:765–778. doi: 10.1089/ars.2012.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulathu Y, Garcia FJ, Mevissen TE, Busch M, Arnaudo N, Carroll KS, Barford D, Komander D. Regulation of A20 and other OTU deubiquitinases by reversible oxidation. Nat Commun. 2013;4:1569. doi: 10.1038/ncomms2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Gupta V, Carroll KS, Liebler DC. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat Commun. 2014;5:4776. doi: 10.1038/ncomms5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gould NS, Evans P, Martinez-Acedo P, Marino SM, Gladyshev VN, Carroll KS, Ischiropoulos H. Site-Specific Proteomic Mapping Identifies Selectively Modified Regulatory Cysteine Residues in Functionally Distinct Protein Networks. Chem Biol. 2015;22:965–975. doi: 10.1016/j.chembiol.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devarie-Baez NO, Silva Lopez EI, Furdui CM. Biological chemistry and functionality of protein sulfenic acids and related thiol modifications. Free Radical Research. 2016;50:172–194. doi: 10.3109/10715762.2015.1090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zito E, Hansen HG, Yeo GS, Fujii J, Ron D. Endoplasmic reticulum thiol oxidase deficiency leads to ascorbic acid depletion and noncanonical scurvy in mice. Mol Cell. 2012;48:39–51. doi: 10.1016/j.molcel.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truong TH, Carroll KS. Redox regulation of epidermal growth factor receptor signaling through cysteine oxidation. Biochemistry. 2012;51:9954–9965. doi: 10.1021/bi301441e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Truong TH, Carroll KS. Redox regulation of protein kinases. Crit Rev Biochem Mol Biol. 2013;48:332–356. doi: 10.3109/10409238.2013.790873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta V, Carroll KS. Rational design of reversible and irreversible cysteine sulfenic acid-targeted linear C-nucleophiles. Chem Commun (Camb) 2016;52:3414–3417. doi: 10.1039/c6cc00228e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta V, Carroll KS. Profiling the Reactivity of Cyclic C-Nucleophiles towards Electrophilic Sulfur in Cysteine Sulfenic Acid. Chem Sci. 2016;7:400–415. doi: 10.1039/c5sc02569a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visscher M, Arkin MR, Dansen TB. Covalent targeting of acquired cysteines in cancer. Curr Opin Chem Biol. 2016;30:61–67. doi: 10.1016/j.cbpa.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis HR, Poole LB. Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochemistry. 1997;36:15013–15018. doi: 10.1021/bi972191x. [DOI] [PubMed] [Google Scholar]
- 29.Poole LB, Zeng BB, Knaggs SA, Yakubu M, King SB. Synthesis of chemical probes to map sulfenic acid modifications on proteins. Bioconjug Chem. 2005;16:1624–1628. doi: 10.1021/bc050257s. [DOI] [PubMed] [Google Scholar]
- 30.Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW, King SB. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug Chem. 2007;18:2004–2017. doi: 10.1021/bc700257a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddie KG, Seo YH, Muse Iii WB, Leonard SE, Carroll KS. A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol Biosyst. 2008;4:521–531. doi: 10.1039/b719986d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo YH, Carroll KS. Facile synthesis and biological evaluation of a cell-permeable probe to detect redox-regulated proteins. Bioorg Med Chem Lett. 2009;19:356–359. doi: 10.1016/j.bmcl.2008.11.073. [DOI] [PubMed] [Google Scholar]
- 33.Leonard SE, Carroll KS. Chemical ‘omics’ approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 34.Seo YH, Carroll KS. Quantification of protein sulfenic acid modifications using isotope-coded dimedone and iododimedone. Angew Chem Int Ed Engl. 2011;50:1342–1345. doi: 10.1002/anie.201007175. [DOI] [PubMed] [Google Scholar]
- 35.Truong TH, Garcia FJ, Seo YH, Carroll KS. Isotope-coded chemical reporter and acid-cleavable affinity reagents for monitoring protein sulfenic acids. Bioorg Med Chem Lett. 2011;21:5015–5020. doi: 10.1016/j.bmcl.2011.04.115. [DOI] [PubMed] [Google Scholar]
- 36.Garcia FJ, Carroll KS. Redox-based probes as tools to monitor oxidized protein tyrosine phosphatases in living cells. Eur J Med Chem. 2014;88:28–33. doi: 10.1016/j.ejmech.2014.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Acedo P, Gupta V, Carroll KS. Proteomic analysis of peptides tagged with dimedone and related probes. J Mass Spectrom. 2014;49:257–265. doi: 10.1002/jms.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan J, Carroll KS. Chemical biology approaches to study protein cysteine sulfenylation. Biopolymers. 2014;101:165–172. doi: 10.1002/bip.22255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brewer TF, Garcia FJ, Onak CS, Carroll KS, Chang CJ. Chemical approaches to discovery and study of sources and targets of hydrogen peroxide redox signaling through NADPH oxidase proteins. Annu Rev Biochem. 2015;84:765–790. doi: 10.1146/annurev-biochem-060614-034018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poole TH, Reisz JA, Zhao WL, Poole LB, Furdui CM, King SB. Strained Cycloalkynes as New Protein Sulfenic Acid Traps. J Am Chem Soc. 2014;136:6167–6170. doi: 10.1021/ja500364r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Gupta V, Tallman KA, Porter NA, Carroll KS, Liebler DC. Global, in situ, site-specific analysis of protein S-sulfenylation. Nat Protoc. 2015;10:1022–1037. doi: 10.1038/nprot.2015.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia FJ, Carroll KS. An immunochemical approach to detect oxidized protein tyrosine phosphatases using a selective C-nucleophile tag. Mol Biosyst. 2016 doi: 10.1039/c5mb00847f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, Carroll KS, Liebler DC. The Expanding Landscape of the Thiol Redox Proteome. Mol Cell Proteomics. 2016;15:1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qian J, Klomsiri C, Wright MW, King SB, Tsang AW, Poole LB, Furdui CM. Simple synthesis of 1,3-cyclopentanedione derived probes for labeling sulfenic acid proteins. Chem Commun (Camb) 2011;47:9203–9205. doi: 10.1039/c1cc12127h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wani R, Qian J, Yin L, Bechtold E, King SB, Poole LB, Paek E, Tsang AW, Furdui CM. Isoform-specific regulation of Akt by PDGF-induced reactive oxygen species. Proc Natl Acad Sci U S A. 2011;108:10550–10555. doi: 10.1073/pnas.1011665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian J, Wani R, Klomsiri C, Poole LB, Tsang AW, Furdui CM. A simple and effective strategy for labeling cysteine sulfenic acid in proteins by utilization of beta-ketoesters as cleavable probes. Chem Commun (Camb) 2012;48:4091–4093. doi: 10.1039/c2cc17868k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furdui CM, Poole LB. Chemical approaches to detect and analyze protein sulfenic acids. Mass Spectrometry Reviews. 2014;33:126–146. doi: 10.1002/mas.21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan J, Carroll KS. Light-Mediated Sulfenic Acid Generation from Photocaged Cysteine Sulfoxide. Org Lett. 2015;17:6014–6017. doi: 10.1021/acs.orglett.5b02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waszczak C, Akter S, Eeckhout D, Persiau G, Wahni K, Bodra N, Van Molle I, De Smet B, Vertommen D, Gevaert K, et al. Sulfenome mining in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2014;111:11545–11550. doi: 10.1073/pnas.1411607111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akter S, Huang J, Waszczak C, Jacques S, Gevaert K, Van Breusegem F, Messens J. Cysteines under ROS attack in plants: a proteomics view. J Exp Bot. 2015;66:2935–2944. doi: 10.1093/jxb/erv044. [DOI] [PubMed] [Google Scholar]
- 51.Akter S, Huang J, Bodra N, De Smet B, Wahni K, Rombaut D, Pauwels J, Gevaert K, Carroll K, Van Breusegem F, et al. DYn-2 Based Identification of Arabidopsis Sulfenomes. Mol Cell Proteomics. 2015;14:1183–1200. doi: 10.1074/mcp.M114.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu CT, Benkovic SJ. Capturing a sulfenic acid with arylboronic acids and benzoxaborole. J Am Chem Soc. 2013;135:14544–14547. doi: 10.1021/ja407628a. [DOI] [PubMed] [Google Scholar]
- 53.Sun Y, Ma XW, Cheng K, Wu BY, Duan JL, Chen H, Bu LH, Zhang RP, Hu XM, Deng ZX, et al. Strained Cyclooctyne as a Molecular Platform for Construction of Multimodal Imaging Probes. Angew Chem Int Edit. 2015;54:5981–5984. doi: 10.1002/anie.201500941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galardon E, Padovani D. Reactivity of persulfides toward strained bicyclo[6.1.0]nonyne derivatives: relevance to chemical tagging of proteins. Bioconjug Chem. 2015;26:1013–1016. doi: 10.1021/acs.bioconjchem.5b00243. [DOI] [PubMed] [Google Scholar]
- 55.Tian H, Sakmar TP, Huber T. A simple method for enhancing the bioorthogonality of cyclooctyne reagent. Chem Commun (Camb) 2016;52:5451–5454. doi: 10.1039/c6cc01321j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicponski DR, Marchi JM. Selectivity Reversal during Thia-Michael Additions Using Tetrabutylammonium Hydroxide: Operationally Simple and Extremely High Turnover. Synthesis. 2014;46:1725–1730. [Google Scholar]
- 57.Ruddraraju KV, Parsons ZD, Llufrio EM, Frost NL, Gates KS. Reactions of 1,3-Diketones with a Dipeptide Isothiazolidin-3-one: Toward Agents That Covalently Capture Oxidized Protein Tyrosine Phosphatase 1B. The Journal of Organic Chemistry. 2015;80:12015–12026. doi: 10.1021/acs.joc.5b01949. [DOI] [PubMed] [Google Scholar]
- 58.Wang S, Xu S, Hu G, Bai X, James TD, Wang L. A Fluorescent Chemodosimeter for Live-Cell Monitoring of Aqueous Sulfides. Analytical Chemistry. 2016;88:1434–1439. doi: 10.1021/acs.analchem.5b04194. [DOI] [PubMed] [Google Scholar]
- 59.Sloop JC, Churley M, Guzman A, Moseley S, Stalker S, Weyand J, Yi J. Synthesis and Reactivity of Fluorinated Cyclic Ketones: Initial Findings. American Journal of Organic Chemistry. 2014;4:1–10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.