Abstract
The success in recent clinical trials using T cell receptor (TCR)-genetically engineered T cells to treat melanoma has encouraged the use of this approach toward other malignancies and viral infections. Although hepatitis C virus (HCV) infection is being treated with a new set of successful direct anti-viral agents, potential for virologic breakthrough or relapse by immune escape variants remains. Additionally, many HCV+ patients have HCV-associated disease, including hepatocellular carcinoma (HCC), which does not respond to these novel drugs. Further exploration of other approaches to address HCV infection and its associated disease are highly warranted. Here, we demonstrate the therapeutic potential of PBL-derived T cells genetically engineered with a high-affinity, HLA-A2-restricted, HCV NS3:1406-1415-reactive TCR. HCV1406 TCR-transduced T cells can recognize naturally processed antigen and elicit CD8-independent recognition of both peptide-loaded targets and HCV+ human HCC cell lines. Furthermore, these cells can mediate regression of established HCV+ HCC in vivo. Our results suggest that HCV TCR-engineered antigen-reactive T cells may be a plausible immunotherapy option to treat HCV-associated malignancies, such as HCC.
Electronic supplementary material
The online version of this article (doi:10.1007/s00262-016-1800-2) contains supplementary material, which is available to authorized users.
Keywords: T cell, T cell receptor, Cancer, Adoptive cell transfer, Hepatitis C virus, Hepatocellular carcinoma
Introduction
We and others have reported that retroviral vectors containing T cell receptor (TCR) genes can be used to redirect the reactivity of peripheral blood-derived lymphocyte (PBL)-derived T cells to recognize tumor cells [1–8]. Recent clinical success with TCR gene-modified T cells to treat malignancies such as melanoma [9–11] encourages the investigation of using this approach to treat other malignancies and viral infections. The available technology to genetically engineer T cells reactive to an antigen of choice allows for the possibility to generate autologous T cells with new anti-viral and anti-tumor immunity to treat any patient, so long as an effective TCR against a viral/tumor antigen has been identified and target cells can be recognized.
Hepatitis C virus (HCV) is a model target for exploring the potential use of such adoptive transfer techniques to treat virally infected cells and tumor cells. HCV infects approximately 130–150 million people globally [12], and chronic infection can lead to associated liver diseases including cirrhosis and hepatocellular carcinoma (HCC). These diseases are a leading cause of liver transplantation in the USA and Europe [13, 14]. Although standard combined therapy of pegylated-IFNα and ribavirin (RBV) has had some success, there has been much greater clinical response when treating HCV+ patients with newly FDA-approved NS3/4A protease inhibitors boceprevir, telaprevir, and simeprevir [15–17]. Despite this recent success, the rapidly mutating HCV genome can generate drug-resistant variants, which might lead to virologic breakthrough or relapse [18–20]. Moreover, many patients who may be cured of HCV infection by these novel drugs may have already developed associated disease or malignancies that cannot be treated effectively by these anti-viral agents. These issues combined with hindered preventative and therapeutic vaccine development [21, 22] warrant exploration of other novel methods to treat HCV infection and associated disease such as HCC.
We have previously identified an HLA-A2-restricted, HCV NS3:1406-1415-reactive TCR isolated from a T cell clone found in an HLA-A2− patient who received an HLA-A2+ liver allograft [23]. We have also demonstrated our ability to genetically engineer Jurkat cells with this high-affinity receptor to recognize HCV+ targets [1]. In this study, we demonstrate that transduced PBL-derived T cells can recognize naturally processed and presented HCV NS3 protein in hepatocellular carcinoma cell lines in vitro and can inhibit growth of established HCV+ tumors in vivo. These results indicate that HCV TCR-engineered antigen-reactive T cells may be a plausible immunotherapy option to treat HCV-associated malignancies, such as HCC.
Materials and methods
Cell lines, media, and reagents
293GP, PG13, T2, COS (monkey kidney tumor, HLA-A2−), HepG2 (human hepatocellular carcinoma, HLA-A2+), and Huh-7 (human hepatocellular carcinoma, HLA-A2−) cell lines were obtained from the American Type Culture Collection (Rockford, MD, USA). All medium were obtained from Corning Life Sciences (Corning, NY, USA) unless otherwise noted. T2 cell line was maintained in complete medium consisting of RPMI 1640 medium supplemented with 10 % fetal bovine serum (FBS; Tissue Culture Biologics, Long Beach, CA, USA). 293GP, COS, COS/A2, HepG2, and Huh-7 cell lines were maintained in DMEM supplemented with 10 % FBS. PG13 cells were maintained in Iscove’s DMEM supplemented with 10 % FBS.
We generated two types of HCV+ tumor lines. First, we engineered COS, COS/A2, and HepG2 cells to express full-length HCV NS3 protein. Due to observed downregulation of NS3-GFP expression in vivo, we also generated HepG2 and Huh-7 cell lines expressing HCV NS3:1406-1415 epitope as a minigene for in vitro and in vivo use. COS and COS/A2 cells were transiently transfected to express the full-length HCV NS3 protein using a pcDNAIII vector encoding HCV NS3 linked to GFP by the self-cleaving viral sequence P2A. Cells were plated in a 24-well tissue culture plate to yield 70–80 % confluency and were transfected with 3 µg DNA and 6 µl of Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) over 48 h. Because HepG2 cells were resistant to lipid-based transfection, a modified SAMEN retroviral vector encoding HCV NS3-P2A-GFP was used to transduce HepG2 cells. Flow cytometry was used to confirm expression of full-length HCV NS3 by measuring intracellular GFP levels.
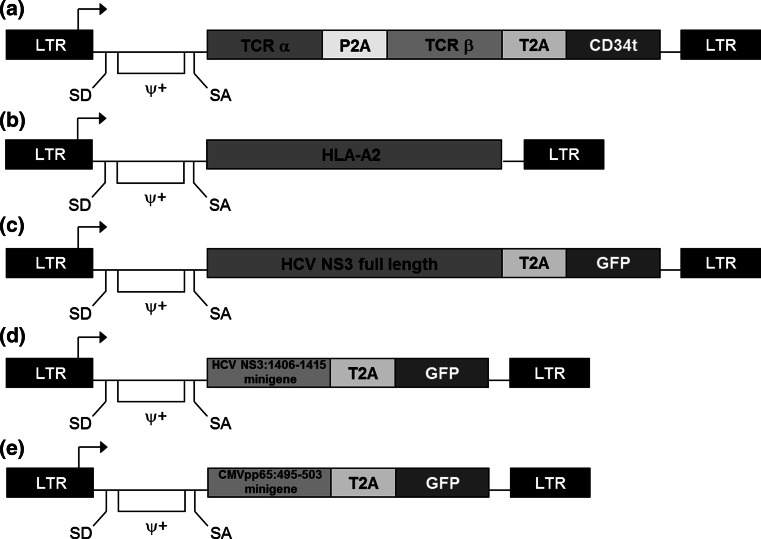
To generate cell lines expressing HCV NS3:1406-1415 or CMVpp65:495-503 epitopes as minigenes, pMFG retroviral vectors containing the respective epitope linked to eGFP by P2A and containing neo r gene were used to transduce HepG2 and Huh-7 cell lines. A modified SAMEN retroviral vector containing HLA-A2 was used to transduce Huh-7 and COS cell lines. Flow cytometry was used to confirm expression of HCV:1406-1415 (GFP), CMVpp65:495-503 (GFP), or HLA-A2 (anti-HLA-A2-APC mAb (Biolegend, San Diego, CA)). Positive cells were sorted for high and uniform expression of GFP or HLA-A2, and the resulting cell lines were maintained in DMEM/10 % FBS. HCV+ and CMV+ cell lines were supplemented with 500 µg/ml G418 (Research Products International, Mount Prospect, IL, USA). Schematics of the described retroviral vectors are provided in Fig. 1.
Fig. 1.
Structures of retroviral vectors used for gene transfer. A modified SAMEN retroviral backbone was used for transferring TCR, HLA-A2, and HCV NS3 genes to alternate effectors. pMFG retroviral vectors were used to transduce HCV NS3:1406-1415 and CMVpp65:495-503 minigenes into tumor cell lines. a TCR retroviral vector containing the HCV1406 TCR α and β chain genes fused by a P2A self-cleaving peptide linker. A truncated version of the CD34 molecule (CD34t), which serves as a marker for transduction, was fused to the 3′ end of the TCR β chain via the T2A self-cleaving peptide. b HLA-A2 encoding retroviral vector used to transduce Huh-7 and COS cell lines. HCV antigen vectors containing either c full-length HCV NS3 gene in SAMEN or d only the 1406-1415 epitope minigene in pMFG both fused to GFP by a T2A self-cleaving peptide linker. A pMFG retroviral vector encoding e CMVpp65:495-503 minigene fused to GFP by T2A was also used as a negative control. LTR long terminal repeat, ψ+ packaging signal, SD splice donor, SA splice acceptor
T cells
All peripheral blood mononuclear cells (PBMCs) used in this study came from apheresis products purchased from Key Biologics (Memphis, TN, USA). Normal PBL-derived T cells were isolated from the PBMCs of normal healthy donors using Ficoll–Hypaque (Sigma-Aldrich, St. Louis, MO, USA) density gradient centrifugation. All T cells were maintained in complete medium consisting of AIM-V medium (Life Technologies, Carlsbad, CA, USA) supplemented with 5 % heat-inactivated pooled human AB serum (hAB; Valley Biomedical, Inc., Winchester, VA, USA), 300 IU/mL recombinant human IL-2 (rhIL-2; Novartis Pharmaceuticals Corporation, East Hanover, NJ, USA), and 100 ng/mL recombinant human IL-15 (rhIL-15; National Institutes of Health, Biological Resources Branch, Bethesda, MD, USA) at 37 °C in a humidified 5 % CO2 incubator.
Retroviral transduction
Retroviral supernatants were prepared using a stable retroviral producer cell line PG13 expressing HCV1406 TCR in a modified SAMEN retroviral vector containing the TCR alpha chain, P2A self-cleaving linker, TCR beta chain, T2A self-cleaving-linker, and truncated CD34 molecule (CD34t) as a transgene expression marker (Fig. 1). The original SAMEN retroviral vector described by Treisman and colleagues [24] has been modified from its original components in stepwise fashion to include TCR chain genes [25] and later a CD34t selection marker [26] for our TCR cloning purposes. Of note, our CD34t cassette differs from a truncated CD34 molecule described by Rettig and colleagues [27] in that it is not a hybrid molecule and serves only as a selection/monitoring marker not associated with a suicide switch. This modified SAMEN backbone was also used for transfer of other genes into tumor lines discussed above. Generation of a stable producer cell line was accomplished as follows: Three million 293GP cells seeded in a 10-cm poly-d-Lysine-coated tissue culture plate (Thermo Scientific, Bannockburn, IL, USA) were cotransfected with 20 µg retroviral vector DNA and 5 µg of a plasmid containing the vesicular stomatitis virus envelope gene in 50 µl Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). Media was replaced 6 h post-transfection, and viral supernatant was collected and 0.45 µm filtered after a 4-h incubation at 37 °C in 5 % CO2−. Two million PG13 cells seeded in a 10-cm tissue culture plate were transduced over 72 h using this fresh viral supernatant at 37 °C in 5 % CO2. Transduction efficiency was analyzed by measuring CD34 expression using flow cytometry after immunofluorescence staining with anti-CD34-PE mAb (Biolegend, San Diego, CA, USA). CD34-positive cells were sorted for high and uniform expression using a FACSAria IIIu cell sorter (BD BioSciences, San Jose, CA, USA), and the resulting high-tighter producer cell line was maintained in Iscove’s DMEM/10 % FBS.
Preparation of retrovirus for T cell transductions was accomplished by treating the HCV1406 TCR -expressing PG13 stable producer cell line (described above) seeded overnight at 8 × 106 cells/T-175 flask with sodium butyrate media (Iscove’s DMEM supplemented with 10 % FBS, 1 mM sodium butyrate (Sigma-Aldrich, St. Louis, MO, USA) and 10 mM HEPES (Corning Life Sciences, Corning, NY, USA) for 8–10 h. Media was replaced with Iscove’s DMEM/10 % FBS, and fresh viral supernatants were harvested and 0.45 µm filtered the next day. Virus was used fresh or stored at −20 °C for later use.
T cells were transduced by spinoculation as previously described [2]. Mono-specific TCR-redirected T cells derived from healthy donors used in experimental data presented in Figs. 2, 3, 4, 5 and 6 were activated for 3 days prior to spinoculation using 50 ng/mL anti-CD3 monoclonal antibody (Miltenyi Biotec, Bergisch Gladbach, Germany) in complete medium. Bi-specific (CMV-reactive/TCR-redirected) T cells used in experiments presented in Supplementary Figure S1 were stimulated with 5 µg/mL CMV pp65:495-503 peptide in complete medium 3 days prior to spinoculation. 24-well flat-bottom non-tissue culture-treated plates (Thermo Fisher Scientific, Bannockburn, IL, USA) were treated with 0.5 mL/well 30 µg/mL retronectin (Takara Bio, Otsu, Shiga, Japan) overnight at 4 °C. Plates were blocked with 0.5 mL/well 2 % PBSA [bovine serum albumin (BSA; Fisher Scientific, Fair Lawn, NJ, USA) in phosphate-buffered saline (PBS; Corning Life Sciences, Corning, NY, USA)] for 30 min at room temperature (RT) and washed with 2 mL/well PBS. Two milliliters of fresh or frozen retrovirus supernatant was added to each well, and the plates were spun for 2 h at 2,000×g at 32 °C and aspirated. Two million activated T cells were gently added to the viral-coated plates in 1 mL of AIM-V/5 % hAb, 600 IU/mL rhIL-2, and 200 ng/mL rhIL-15 and mixed with 1 mL filtered viral supernatant. The plates were spun again for 2 h at 2,000×g at 32 °C. After 24 h, the transduced T cells were transferred to tissue-culture-treated flasks. Three days later, transduction efficiency was determined by FACS analysis using anti-CD34 mAb, and T cells were sorted for TCR-transduced T cells by positive selection with magnetic beads labeled with anti-CD34 (Miltenyi Biotec, Bergisch Gladbach, Germany) and maintained in complete medium. T cells may also have been selectively sorted for CD4+ or CD8+ transduced T cells using respective immunomagnetic beads (Miltenyi Biotec). Purity of selection was confirmed by immunofluorescence.
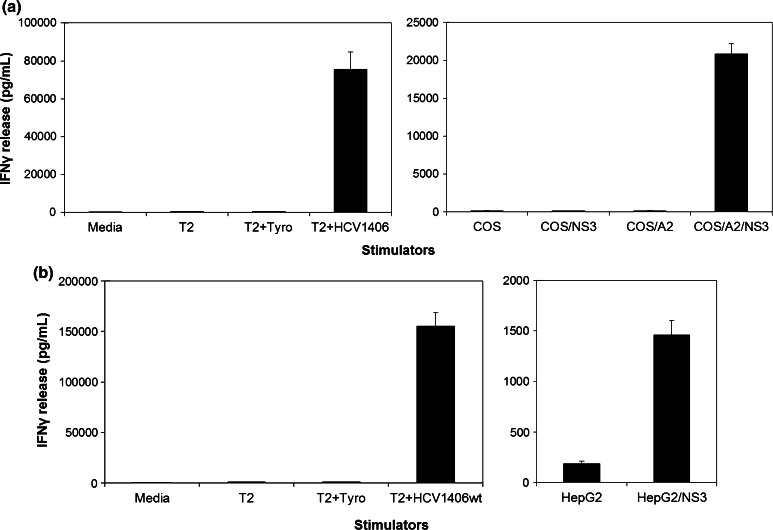
Fig. 2.
Recognition of naturally processed HCV NS3:14016-1415 by HCV1406 TCR-transduced T cells. PBL from a normal donor was transduced with the HCV1406 TCR retroviral vector. Stimulators included T2 cells loaded with HCV NS3:1406-1415 or tyrosinase:368-376 peptides and a COS±HLA-A2 cells or b HepG2 cells engineered to express full-length HCV NS3 protein. The amount of IFNγ release (average ± standard deviation of triplicate wells) was measured by ELISA
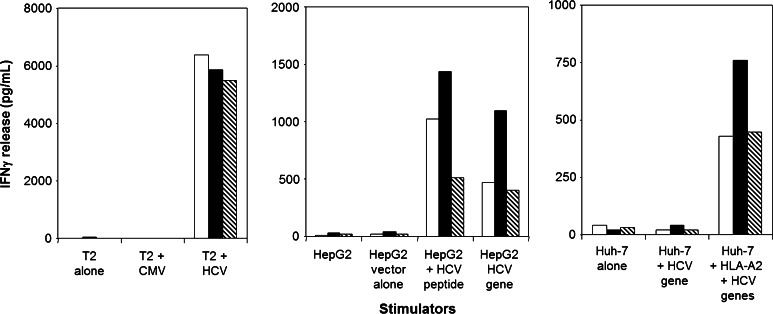
Fig. 3.
HCV1406 TCR-transduced T cell recognition of HCV+ hepatocellular carcinoma cells. HCV1406 TCR-transduced T cells from three representative normal donors [Donor A (white bars), Donor B (black bars), Donor C (striped bars)] were cocultured with various stimulators. (left panel) T2 cells alone or loaded with NS3:1406-1415 peptide or control CMVpp65 peptide; (middle panel) HepG2 cells (HLA-A2+) alone, pulsed with HCV peptide or expressing the HCV minigene; (right panel) Huh-7 cells (HLA-A2−) alone or expressing the HCV minigene ± HLA-A2. IFNγ secretion was assessed as a single-point ELISA
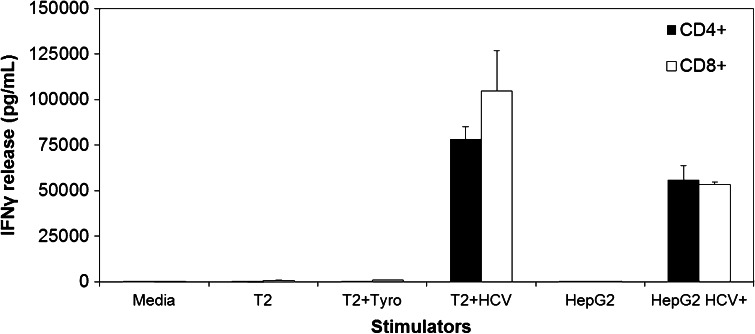
Fig. 4.
Comparison of antigen recognition by HCV1406 TCR-transduced CD4+ versus CD8+ PBL. HCV1406 TCR-transduced PBL were immunomagnetically purified into CD4+ (black bars) or CD8+ (white bars) populations. Stimulators included T2 cells pulsed with 10 µg/mL of HCV NS3:1406-1415 peptide or control tyrosinase:368-376 peptide as well as HepG2 engineered to express the HCV NS3:1406-1415 minigene. The amount of IFNγ release (average ± standard deviation of triplicate wells) was measured by ELISA
Fig. 5.
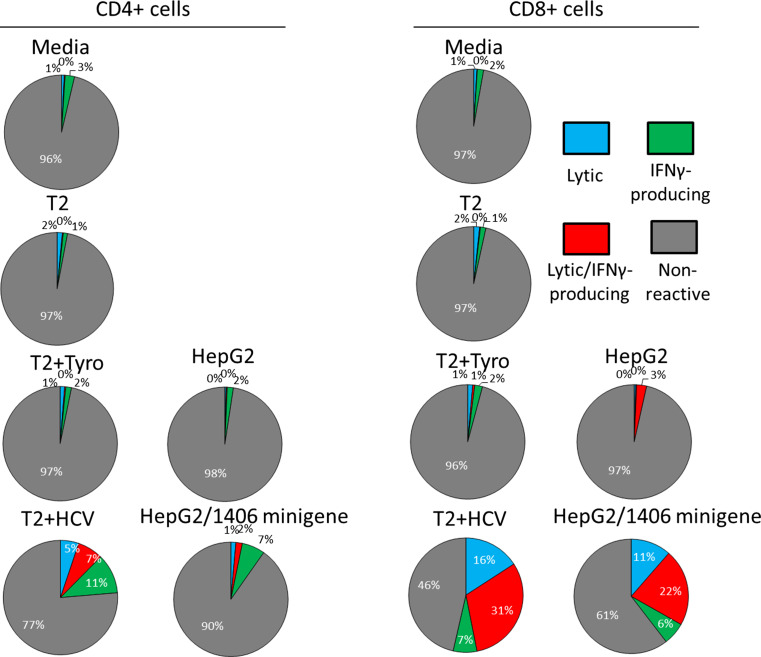
Lytic and IFNγ-producing ability of HCV1406 TCR-transduced T cells reactive against HCV NS3:1406-1415. HCV+ HepG2 cells and T2 cells pulsed with HCV NS3:1406-1415 or tyrosinase:368-376 were used as targets and cocultured with transduced PBL. Pie chart percentages are represented as CD107a+IFNγ− (lytic only = blue), CD107a+IFNγ+ (lytic and cytokine-secreting = red), CD107a−IFNγ+ (cytokine-secreting only = green), or CD107a−IFNγ− (non-reactive = gray)
Fig. 6.
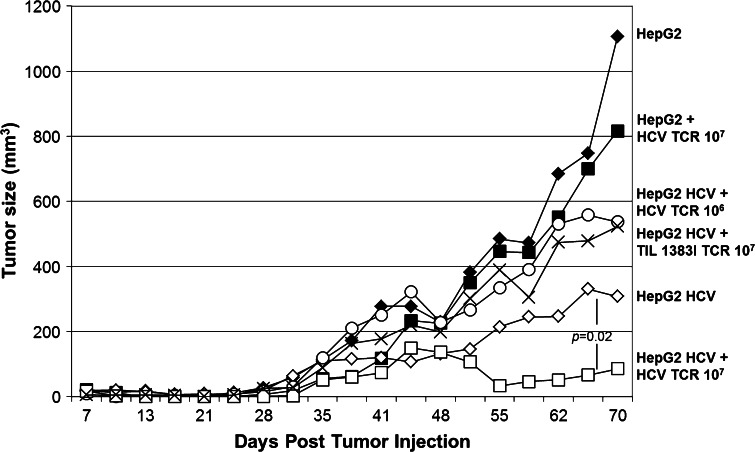
Growth inhibition of established hepatocellular carcinoma tumors by HCV1406 TCR-transduced T cells. Parental HepG2 (black) or HCV+ HepG2 (white) tumors were established in scid/beige mice (n = 5 per treatment group). Mice were given no T cells (diamonds), 107 TIL 1383I TCR-transduced T cells (circles), or 106 (×’s) or 107 (squares) HCV1406 TCR-transduced T cells on day 7 post-tumor challenge. Statistically significant differences in tumor growth over 70 days was determined using the Wilcoxon rank sum test, p = 0.02
Immunofluorescence
Cell surface expression of the TCR and other T cell markers was measured via immunofluorescence staining and quantified via flow cytometry. mAbs used to characterize transduced T cell cultures in these experiments included: anti-CD3-APC/Cy7, anti-CD34-PerCP/Cy5.5, anti-CD4-FITC, anti-CD8-Alexa Fluor 700, anti-CD25-Brilliant Violet (BV) 711, anti-CD69-PE/Cy7, anti-PD-1-BV 421, anti-TIM-3-BV 605 (all Biolegend, San Diego, CA, USA). mAbs used in bifunctional reactivity assays included the following: anti-CD3-APC/Cy7, anti-CD4-PE-Cy7, anti-CD8-PerCP/Cy5.5, anti-CD34-APC, anti-IFNγ-BV 421, and anti-CD107a-BV 510 (all Biolegend). PE-labeled HLA-A*0201 tetramer folded around CMVpp65:495-503 (Beckman Coulter, Marseille, France) and PE-labeled HLA-A*0201 dextramer folded around HCV NS3:1406-1415 (Immudex, Copenhagen, Denmark) were also used. Flow cytometry was performed using a LSRFortessa flow cytometer (BD Biosciences, San Jose, CA), and data were analyzed with FlowJoX software (Treestar, Ashland, OR, USA).
Peptides
HCV NS3:1406-1415 (KLVALGINAV), CMVpp65:495-503 (NLVPMVATV), and tyrosinase:368-376 (TMDGTMSQV) were purchased at 95 % purity from Synthetic Biomolecules (San Diego, CA, USA). Peptides were stored in 100 % dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO, USA) at −80 °C in 5 µg/µL aliquots.
Cytokine release assay
Antigen reactivity by HCV1406 TCR-transduced T cells was measured in cytokine release assays as described [3]. Briefly, T2 or tumor targets (where indicated) were loaded with 10 µg/mL peptide for 2 h prior to coculture. 1 × 105 responder and 1 × 105 stimulator cells were cocultured in triplicate in a 1:1 ratio in 96-well U-bottom tissue culture plates in 200 µL complete medium. Of note, data portrayed in Fig. 3 are a representative of multiple single-point ELISA experiments, each showing similar results. Cocultures were incubated at 37 °C for 20 h, and supernatants were harvested. The amount of IFNγ released was measured via ELISA (R&D Systems, Minneapolis, MN, USA).
Bifunctional reactivity assay
Percentages of cytokine producing and/or lytic HCV1406 TCR-transduced T cells were measured in an intracellular cytokine/CD107a-detection assay. 3 × 105 responder and 3 × 105 stimulator cells were cocultured in a 1:1 ratio in 96-well U-bottom tissue culture plates in 200 µL complete medium. 5 µL anti-CD107a mAb, 5.0 ng/mL brefeldin-A, and 2.0 nM monensin (all Biolegend, San Diego, CA, USA) were added at the start of coculture. Cocultures were incubated at 37 °C for 5 h, and cells were stained for immunofluorescence against cell surface antigens for 20 min at RT. Subsequently, cells were fixed in Fixation Buffer (Biolegend) for 20 min, washed 3 times in permeabilization and wash buffer (Biolegend), and immunofluorescence stained for intracellular IFNγ for 20 min at RT. Data were acquired using an LSRFortessa flow cytometer (BD Biosciences, San Jose, CA, USA). CD3+CD34+ events (transduced T cells) were gated into CD4+CD8− or CD4−CD8+ populations. Percentages of CD107a+IFNγ− (lytic only), CD107a+IFNγ+ (lytic and cytokine-secreting), CD107a−IFNγ+ (cytokine-secreting only), or CD107a−IFNγ− (non-reactive) cells were calculated for both CD3+CD34+CD4+CD8− and CD3+CD34+CD4+CD8− cell populations and converted into pie charts.
In vivo xenograft model
Prior to tumor challenge, scid/beige mice (n = 5 per treatment group) were given 2 Gy total body irradiation. 107 HCV+ or HCV− HepG2 cells were injected subcutaneously in 0.1 cc saline. Generally, palpable tumors formed within 7 days after tumor challenge. Once palpable tumors formed, mice were adoptively transferred to 106 or 107 HCV1406 TCR-transduced T cells, 107 tyrosinase-reactive TIL 1383I TCR-transduced T cells, or no T cells. Tumor size was measured every three days through day 70 post-tumor challenge. A Wilcoxon rank sum test was used to determine significant difference in tumor burden between treatment groups.
Ethics statement
All applicable international, national, and/or institutional guidelines for the care and use animals were followed. All animal studies were conducted under approved Medical University of South Carolina (MUSC) Institutional Animal Care and Use Committee (IACUC) protocols. All recombinant DNA and retroviral transductions work was done under approved MUSC and Loyola Institutional Biosafety Committee (IBC) protocols. This article does not contain any studies with human participants performed by any of the authors. All human materials used were either established and de-identified tumor cell lines or PBMC purchased from commercial sources.
Results
Recognition of naturally processed HCV NS3 by HCV TCR-transduced T cells
Although we have shown that TCR-transduced Jurkat cells can recognize HCV NS3:1406-1415 peptide-loaded T2 cells, it is critical to establish that TCR gene-modified primary T cells can recognize both peptide-loaded targets and HCV+ tumors to validate its potential use in adoptive cell transfer (ACT). We first examined our ability to generate properly functioning transduced primary T cells by simultaneously testing specificity of both the parental and introduced TCR. We first generated a population of single antigen-reactive T cells by stimulating PBMC with the CMVpp65:495-503 peptide. We then transduced these CMV-reactive T cells with the retroviral vector encoding HCV1406 TCR described in Fig. 1. When stimulated with HCV+ tumor cells, IFNγ was produced by cells that also bound HLA-A2/CMVpp65:495-503 tetramer (Supplementary figure S1). This demonstrates our ability to redirect specificity of T cells while maintaining proper endogenous signaling and parental TCR specificity.
The best system to assess the ability of TCR-transduced T cells to recognize HCV antigen would be to use HCV-infected primary liver cells, but human liver containing HCC cells infected with HCV was not available for our experiments. As an alternative, COS and COS/A2 cells were transiently transfected with a pcDNAIII vector encoding the full-length HCV protein NS3 linked to GFP, allowing us to test HLA-A2-restricted recognition of naturally processed and presented peptide from its full-length cognate protein. Typical transfections yielded 40–60 % GFP expression (data not shown). Anti-CD3-activated PBL-derived T cells transduced with HCV1406 TCR for in vitro experiments were anti-CD34 immunomagnetically enriched for TCR transgene expression. Populations are routinely ≥95 % CD34+ post-magnetic sort. Representative immunofluorescence analysis for experimental HCV1406 TCR-transduced T cells is shown in Supplementary figure S2. Both CD4+ and CD8+ transduced populations (CD3+CD34+) are highly activated (CD25+CD69+) with low levels of exhaustion markers PD-1 and TIM-3. Due to poor staining with available Vβ11 antibodies, dextramer binding is shown to measure expression of the introduced TCR. These HCV TCR-transduced T cells were capable of recognizing the naturally processed HCV NS3 in an HLA-A2-dependent manner (Fig. 2a). Robust IFNγ secretion was observed when stimulated with COS/A2/NS3 but not any other cell lines. Additionally, HepG2 cells transduced to express NS3 also stimulated large IFNγ secretion by these transduced T cells (Fig. 2b). These cells have also been observed to secrete IL-2, TNF-α and others by intracellular cytokine staining, but for simplicity, only IFNγ production is shown.
Of note, HepG2 cells expressing full-length NS3-GFP downregulated antigen expression over time in vivo, suggesting that this cell line would not be of optimal use for adoptive transfer experiments after tumor challenge. To address this, we generated HCV+ human HCC cell lines expressing HCV NS3:1406-1415 as a minigene, and these cells did not downregulate antigen expression or presentation in vivo. Subsequently, these cell lines were used for both cytokine release assays and in vivo tumor regression experiments. T cells expressing HCV1406 TCR secreted large amounts of IFNγ when stimulated by HepG2 cells (HLA-A2+) either pulsed with HCV NS3:1406-1415 peptide or expressing the HCV minigene (Fig. 3). Similarly, these transduced T cells recognized Huh-7 cells (HLA-A2−) only when transfected to express the HCV minigene and HLA-A2. These data suggest that we can engineer TCR gene-modified T cells to recognize naturally processed and presented HCV NS3 antigen.
CD8-indpendent HCV NS3:1406-1415 antigen recognition by HCV TCR-transduced PBL
To verify CD8-independent recognition of both peptide and tumor, we purified transduced CD4+ and CD8+ by immunomagnetic separation and cocultured with peptide-loaded and tumor targets. Figure 4 demonstrates that CD8-independent recognition is conserved against peptide and HCV+ tumor as both purified CD4+ and CD8+ transduced T cells secreted robust amounts of IFNγ when stimulated by HCV-loaded T2 cells and HCV+ HepG2 cells. Moreover, our HCV TCR-transduced T cells displayed lytic behavior as measured by CD107a surface expression against peptide-pulsed targets and HCV+ HepG2 cells (Fig. 5). Counterstaining for intracellular IFNγ also revealed that reactive T cells can be lytic, cytokine-secreting, or both, and that CD4+ and CD8+ exhibited distinct heterogeneous polyfunctional phenotypes. Therefore, we are able to generate both CD8+ and CD4+ effectors and MHC class I-restricted helper function against this HCV antigen.
HCV TCR-transduced T cell-mediated regression of established HCV+ HCC in vivo
While our in vitro data firmly demonstrate that both CD4+ and CD8+ transduced T cells can mediate polyfunctional behavior, it is necessary to validate in vivo recognition in a xenograft mouse model to establish potential preclinical benefit. We established HCC tumors in scid/beige mice (n = 5 per treatment group) by subcutaneous injection of HCV+ or HCV− HepG2 cells. Once the tumors were palpable (day 7 post-injection), transduced T cells were adoptively transferred. HCV+ tumor-bearing mice exhibited reduced tumor growth after adoptive transfer of 107 HCV TCR-transduced T cells but not when treated with tyrosinase-reactive TIL 1383I TCR-transduced T cells or a lower dose (106) of HCV TCR-transduced T cells (Fig. 6). It is important to note that in the experiment presented HCV+ HepG2 tumors in untreated mice grew at a slower rate after day 30 than in other HCV+ tumor-bearing mice groups. While we attribute this to experimental variation, other experiments demonstrated more uniform HCV+ tumor growth in other treatment groups (data not shown). Regardless, the change in tumor burden at day 70 between HCV TCR-transduced T cell-treated HCV+ and HCV− HepG2-tumor-bearing mice exhibited significance with a p value of 0.02 as determined by Wilcoxon rank sum test. In at least one instance, HCV+ tumors harvested from 107 HCV TCR-transduced T cells exhibited antigen loss, which could explain why tumors were not completely eliminated even though displaying a statistically significant reduction in tumor burden. In our experience, TCR-transduced human T cells engrafted into mice are generally not detected beyond 5–7 days after infusion, even with IL-2 being administered (data not shown). Thus, we were not able to measure frequency or phenotype of T cells post-transfer. Taken together, this evidence further supports the HCV+ HCC recognition capability and potential for our HCV TCR-transduced T cells to be used in ACT.
Discussion
For any adoptive immunotherapy to be a viable treatment option, it must be clearly evident that autologous antigen-reactive T cells are capable of being generated. Additionally, physiologically relevant targets need to be recognized by functional T cells. While other studies have examined NY-ESO-1 [28–32] or Glypican 3 [31, 33–38] as biomarkers or therapeutic targets for HCC, we have chosen to investigate HCV for its non-self nature, its immunogenicity, and the fact that HCV is a major risk factor for the development of HCC. While the role of HCV proteins in hepatocarcinogenesis is not well described, HCV is mainly thought to cause HCC via indirect pathways including chronic inflammation, cell death, proliferation, and cirrhosis [39–42]. That being said, it is possible that HCV TCR-transduced T cells would target both normal and malignant hepatocytes, potentially lead to severe adverse events, as noted by another group investigating HCV TCR-redirected T cells [43]. This accompanied with cirrhosis and liver dysfunction in many HCC patients would cause concern for systemic infusion of HCV TCR-transduced T cells.
Thus, designed clinical trials for this immunotherapy are planned to involve CT or ultrasound guided intratumoral injection in a manner similar to transarterial chemoembolization (TACE). A similar intratumoral technique was used successfully in one of our previous clinical trials [44]. Such carefully thought approaches would help preclude significant exposure of normal hepatocytes to the HCV TCR-transduced T cells and minimize adverse events. In this study, we have shown that PBL-derived T cells engineered to express a TCR reactive to HCV NS3:1406-1415 are capable of recognizing naturally processed and presented HCV NS3 protein in hepatocellular carcinoma cell lines in vitro and can mediate statistically significant regression of established HCV+ tumors in vivo.
To date, only a limited number of naturally occurring CD8-independent TCRs have been cloned and characterized. The ability of this receptor to transfer reactivity to both effector and helper T cells is advantageous for an effective immunotherapy candidate. Both CD4+ and CD8+ transduced PBL-derived T cells can recognize both peptide-loaded targets and a variety of HCV+ tumor cells lines. Our in vivo tumor regression model further supports our in vitro data that this TCR is capable of recognizing HCV+ tumors. Further analysis of differential adoptive transfer of CD8+ T cells with and without CD4+ T cells may better distinguish the importance of generating CD4+-reactive T cells for eliminating tumor, which may influence treatment modality in patients. Since one of the fundamental problems typical of chronic or recurrent HCV infections is a weak or absent HCV-specific CD4+ T cell response [45–48], the ability to engineer CD4+ T cells capable of secreting cytokines and exhibiting cytolytic activity is extremely beneficial for an effective treatment.
Of interest is the phenotypic heterogeneity observed in transduced T cells reactive against peptide-loaded targets and naturally processed HCV+ tumors. While both CD4+ and CD8+ T cells are capable of recognizing these targets, they differentially expressed IFNγ and CD107a in different proportions. While most CD4+ T cells secreted IFNγ in response to these targets, some cells exhibited cytolytic activity as indicated by CD107a expression. Those that did express CD107a were less likely to also secrete cytokine. Conversely, a greater number of CD8+ T cells exhibited a lytic phenotype; those that secreted IFNγ were more likely to simultaneously express CD107a as well. These data support other reports that CD4+ T cells can exhibit cytolytic activity [49–52] and provide insight into the heterogenous behavior of T cells expressing the same TCR.
We also have established a proof of principle in generating “bifunctional” T cells by selecting for CMV-reactive T cells in PBL and subsequently transducing them with our HCV TCR. This approach in generating dual-reactive T cells capable of recognizing two distinct antigens might be an effective tool to combat the instability of the HCV genome. Several groups have reported that mutations in the HCV genome can lead to HCV antigen escape variants [53–58]. In this way, it is speculated that HCV can evade the immune response, resulting in disease progression. Although it is not clear to what extent antigen loss actually leads to disease progression, our TCR gene transfer methodology offers the opportunity to treat patients with HCV antigen loss variants. We have previously identified a second high-affinity HCV NS3:1073-1081 TCR and demonstrated its ability to transfer reactivity to Jurkat and primary T cells against peptide and HCV+ tumor [59]. Engineering T cells to simultaneously express both TCRs may be effective against HCV immune escape variants for treatment of either infection or HCV-associated malignancies, but this approach has yet to be evaluated for its capacity to target two defined antigens in vivo. Taken together, these data suggest that this TCR is a useful reagent to engineer both CD4+ and CD8+ T cells capable of recognizing and eliminating HCV+ tumor targets and may be a useful tool to treat patients with HCV-associated diseases, such as HCC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to acknowledge their funding sources kindly provided by the National Cancer Institute (NCI) of the National Institutes of Health (NIH). These include grants P01 CA154778 (Nishimura), R01 CA104947 (Nishimura), R01 CA104947-S1 (Nishimura), R01 CA90873 (Nishimura), R01 CA102280 (Nishimura), R21 CA153789 (Nishimura), and F30 CA180731 (Spear).
Abbreviations
- CD34t
Truncated CD34 molecule
- FDA
Food and Drug Administration
- HCC
Hepatocellular carcinoma
- IACUC
Institutional Animal Care and Use Committee
- IBC
Institutional Biosafety Committee
- MUSC
Medical University of South Carolina
- NS3
Non-structural protein 3
- NS4A
Non-structural protein 4A
- PBSA
Phosphate-buffered saline with bovine serum albumin
- pMHC
Peptide–major histocompatibility complex
- RBV
Ribavirin
- rhIL-2
Recombinant human interleukin-2
- rhIL-15
Recombinant human interleukin-15
Compliances with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Callender GG, Rosen HR, Roszkowski JJ, Lyons GE, Li M, Moore T, Brasic N, McKee MD, Nishimura MI. Identification of a hepatitis C virus-reactive T cell receptor that does not require CD8 for target cell recognition. Hepatology. 2006;43(5):973–981. doi: 10.1002/hep.21157. [DOI] [PubMed] [Google Scholar]
- 2.Clay TM, Custer MC, Sachs J, Hwu P, Rosenberg SA, Nishimura MI. Efficient transfer of a tumor antigen-reactive TCR to human peripheral blood lymphocytes confers anti-tumor reactivity. J Immunol. 1999;163(1):507–513. [PubMed] [Google Scholar]
- 3.Cole DJ, Weil DP, Shilyansky J, Custer M, Kawakami Y, Rosenberg SA, Nishimura MI. Characterization of the functional specificity of a cloned T-cell receptor heterodimer recognizing the MART-1 melanoma antigen. Cancer Res. 1995;55(4):748–752. [PubMed] [Google Scholar]
- 4.Kuball J, Schmitz FW, Voss RH, Ferreira EA, Engel R, Guillaume P, Strand S, Romero P, Huber C, Sherman LA, Theobald M. Cooperation of human tumor-reactive CD4+ and CD8+ T cells after redirection of their specificity by a high-affinity p53A2.1-specific TCR. Immunity. 2005;22(1):117–129. doi: 10.1016/j.immuni.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Rosati SF, Parkhurst MR, Hong Y, Zheng Z, Feldman SA, Rao M, Abate-Daga D, Beard RE, Xu H, Black MA, Robbins PF, Schrump DA, Rosenberg SA, Morgan RA. A novel murine T-cell receptor targeting NY-ESO-1. J Immunother. 2014;37(3):135–146. doi: 10.1097/CJI.0000000000000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roszkowski JJ, Lyons GE, Kast WM, Yee C, Van Besien K, Nishimura MI. Simultaneous generation of CD8+ and CD4+ melanoma-reactive T cells by retroviral-mediated transfer of a single T-cell receptor. Cancer Res. 2005;65(4):1570–1576. doi: 10.1158/0008-5472.CAN-04-2076. [DOI] [PubMed] [Google Scholar]
- 7.Shilyansky J, Nishimura MI, Yannelli JR, Kawakami Y, Jacknin LS, Charmley P, Rosenberg SA. T-cell receptor usage by melanoma-specific clonal and highly oligoclonal tumor-infiltrating lymphocyte lines. Proc Natl Acad Sci USA. 1994;91(7):2829–2833. doi: 10.1073/pnas.91.7.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanislawski T, Voss RH, Lotz C, Sadovnikova E, Willemsen RA, Kuball J, Ruppert T, Bolhuis RL, Melief CJ, Huber C, Stauss HJ, Theobald M. Circumventing tolerance to a human MDM2-derived tumor antigen by TCR gene transfer. Nat Immunol. 2001;2(10):962–970. doi: 10.1038/ni1001-962. [DOI] [PubMed] [Google Scholar]
- 9.Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, Wunderlich JR, Nahvi AV, Helman LJ, Mackall CL, Kammula US, Hughes MS, Restifo NP, Raffeld M, Lee CC, Levy CL, Li YF, El-Gamil M, Schwarz SL, Laurencot C, Rosenberg SA. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29(7):917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, Dudley ME, Feldman SA, Yang JC, Sherry RM, Phan GQ, Hughes MS, Kammula US, Miller AD, Hessman CJ, Stewart AA, Restifo NP, Quezado MM, Alimchandani M, Rosenberg AZ, Nath A, Wang T, Bielekova B, Wuest SC, Akula N, McMahon FJ, Wilde S, Mosetter B, Schendel DJ, Laurencot CM, Rosenberg SA. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother. 2013;36(2):133–151. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chodon T, Comin-Anduix B, Chmielowski B, Koya RC, Wu Z, Auerbach M, Ng C, Avramis E, Seja E, Villanueva A, McCannel TA, Ishiyama A, Czernin J, Radu CG, Wang X, Gjertson DW, Cochran AJ, Cornetta K, Wong DJ, Kaplan-Lefko P, Hamid O, Samlowski W, Cohen PA, Daniels GA, Mukherji B, Yang L, Zack JA, Kohn DB, Heath JR, Glaspy JA, Witte ON, Baltimore D, Economou JS, Ribas A. Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma. Clin Cancer Res. 2014;20(9):2457–2465. doi: 10.1158/1078-0432.CCR-13-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hepatitis C fact sheet (2015) World Health Organization. http://www.who.int/mediacentre/factsheets/fs164/en/. Accessed July 2015
- 13.Hepatitis C FAQs (2015) National Centers for Disease Control and Prevention. http://www.cdc.gov/hepatitis/hcv/hcvfaq.htm. Accessed July 2015
- 14.Campos-Varela I, Lai JC, Verna EC, O’Leary JG, Todd Stravitz R, Forman LM, Trotter JF, Brown RS, Terrault NA. Hepatitis C genotype influences post-liver transplant outcomes. Transplantation. 2015;99(4):835–840. doi: 10.1097/TP.0000000000000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hézode C, Forestier N, Dusheiko G, Ferenci P, Pol S, Goeser T, Bronowicki J-P, Bourlière M, Gharakhanian S, Bengtsson L, McNair L, George S, Kieffer T, Kwong A, Kauffman RS, Alam J, Pawlotsky J-M, Zeuzem S. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N Engl J Med. 2009;360(18):1839–1850. doi: 10.1056/NEJMoa0807650. [DOI] [PubMed] [Google Scholar]
- 16.Kwo PY, Lawitz EJ, McCone J, Schiff ER, Vierling JM, Pound D, Davis MN, Galati JS, Gordon SC, Ravendhran N, Rossaro L, Anderson FH, Jacobson IM, Rubin R, Koury K, Pedicone LD, Brass CA, Chaudhri E, Albrecht JK. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet. 2010;376(9742):705–716. doi: 10.1016/S0140-6736(10)60934-8. [DOI] [PubMed] [Google Scholar]
- 17.Manns M, Marcellin P, Poordad F, de Araujo ES, Buti M, Horsmans Y, Janczewska E, Villamil F, Scott J, Peeters M, Lenz O, Ouwerkerk-Mahadevan S, De La Rosa G, Kalmeijer R, Sinha R, Beumont-Mauviel M. Simeprevir with pegylated interferon alfa 2a or 2b plus ribavirin in treatment-naive patients with chronic hepatitis C virus genotype 1 infection (QUEST-2): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet. 2014;384(9941):414–426. doi: 10.1016/S0140-6736(14)60538-9. [DOI] [PubMed] [Google Scholar]
- 18.Berger KL, Scherer J, Ranga M, Sha N, Stern JO, Quinson AM, Kukolj G. Baseline polymorphisms and emergence of drug resistance in the NS3/4A protease of HCV genotype-1 following TREATMENT with faldaprevir plus pegylated interferon Alfa-2a and ribavirin in phase 2 and phase 3 studies. Antimicrob Agents Chemother. 2015;59(10):6017–6025. doi: 10.1128/AAC.00932-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Luca A, Di Giambenedetto S, Lo Presti A, Sierra S, Prosperi M, Cella E, Giovanetti M, Torti C, Caudai C, Vicenti I, Saladini F, Almi P, Grima P, Blanc P, Fabbiani M, Rossetti B, Gagliardini R, Kaiser R, Ciccozzi M, Zazzi M. Two distinct hepatitis C virus genotype 1a clades have different geographical distribution and association with natural resistance to NS3 protease inhibitors. Open Forum Infect Dis. 2015;2(2):ofv43. doi: 10.1093/ofid/ofv043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagpal N, Goyal S, Wahi D, Jain R, Jamal S, Singh A, Rana P, Grover A. Molecular principles behind Boceprevir resistance due to mutations in hepatitis C NS3/4A protease. Gene. 2015;570(1):115–121. doi: 10.1016/j.gene.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Lechmann M, Liang TJ. Vaccine development for hepatitis C. Semin Liv Dis. 2000;20(2):211–226. doi: 10.1055/s-2000-9947. [DOI] [PubMed] [Google Scholar]
- 22.Trujillo-Murillo Kdel C, Garza-Rodriguez Mdel L, Martinez-Rodriguez HG, Barrera-Saldana HA, Bosques-Padilla F, Ramos-Jimenez J, Rivas-Estilla AM. Experimental models for hepatitis C virus (HCV): new opportunities for combating hepatitis C. Ann Hepatol. 2004;3(2):54–62. [PubMed] [Google Scholar]
- 23.Rosen HR, Hinrichs DJ, Leistikow RL, Callender G, Wertheimer AM, Nishimura MI, Lewinsohn DM. Cutting edge: identification of hepatitis C virus-specific CD8+ T cells restricted by donor HLA alleles following liver transplantation. J Immunol. 2004;173(9):5355–5359. doi: 10.4049/jimmunol.173.9.5355. [DOI] [PubMed] [Google Scholar]
- 24.Treisman J, Hwu P, Minamoto S, Shafer GE, Cowherd R, Morgan RA, Rosenberg SA. Interleukin-2-transduced lymphocytes grow in an autocrine fashion and remain responsive to antigen. Blood. 1995;85(1):139–145. [PubMed] [Google Scholar]
- 25.Roszkowski JJ, Yu DC, Rubinstein MP, McKee MD, Cole DJ, Nishimura MI. CD8-independent tumor cell recognition is a property of the T cell receptor and not the T cell. J Immunol. 2003;170(5):2582–2589. doi: 10.4049/jimmunol.170.5.2582. [DOI] [PubMed] [Google Scholar]
- 26.Norell H, Zhang Y, McCracken J, Martins da Palma T, Lesher A, Liu Y, Roszkowski JJ, Temple A, Callender GG, Clay T, Orentas R, Guevara-Patino J, Nishimura MI. CD34-based enrichment of genetically engineered human T cells for clinical use results in dramatically enhanced tumor targeting. Cancer Immunol Immunother. 2010;59(6):851–862. doi: 10.1007/s00262-009-0810-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rettig MP, Ritchey JK, Meyerrose TE, Haug JS, DiPersio JF. Transduction and selection of human T cells with novel CD34/thymidine kinase chimeric suicide genes for the treatment of graft-versus-host disease. Mol Ther. 2003;8(1):29–41. doi: 10.1016/S1525-0016(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 28.Flecken T, Schmidt N, Hild S, Gostick E, Drognitz O, Zeiser R, Schemmer P, Bruns H, Eiermann T, Price DA, Blum HE, Neumann-Haefelin C, Thimme R. Immunodominance and functional alterations of tumor-associated antigen-specific CD8+ T-cell responses in hepatocellular carcinoma. Hepatology. 2014;59(4):1415–1426. doi: 10.1002/hep.26731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt N, Flecken T, Thimme R. Tumor-associated antigen specific CD8 T cells in hepatocellular carcinoma—a promising target for immunotherapy. Oncoimmunology. 2014;3(9):e954919. doi: 10.4161/21624011.2014.954919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang XY, Chen HS, Zhang HG, Pang XW, Qiao H, Peng JR, Qin LL, Fei R, Mei MH, Leng XS, Gnjatic S, Ritter G, Simpson AJ, Old LJ, Chen WF. The spontaneous CD8+ T-cell response to HLA-A2-restricted NY-ESO-1b peptide in hepatocellular carcinoma patients. Clin Cancer Res. 2004;10(20):6946–6955. doi: 10.1158/1078-0432.CCR-04-0502. [DOI] [PubMed] [Google Scholar]
- 31.Sideras K, Bots SJ, Biermann K, Sprengers D, Polak WG, Jn IJ, de Man RA, Pan Q, Sleijfer S, Bruno MJ, Kwekkeboom J. Tumour antigen expression in hepatocellular carcinoma in a low-endemic western area. Br J Cancer. 2015;112(12):1911–1920. doi: 10.1038/bjc.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu H, Gu N, Liu ZB, Zheng M, Xiong F, Wang SY, Li N, Lu J. NY-ESO-1 expression in hepatocellular carcinoma: a potential new marker for early recurrence after surgery. Oncol Lett. 2012;3(1):39–44. doi: 10.3892/ol.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao W, Kim H, Ho M. Human monoclonal antibody targeting the heparan sulfate chains of glypican-3 inhibits HGF-mediated migration and motility of hepatocellular carcinoma cells. PLoS One. 2015;10(9):e0137664. doi: 10.1371/journal.pone.0137664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geramizadeh B, Seirfar N. Diagnostic value of arginase-1 and glypican-3 in differential diagnosis of hepatocellular carcinoma, cholangiocarcinoma and metastatic carcinoma of liver. Hepat Mon. 2015;15(7):e30336. doi: 10.5812/hepatmon30336v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanaoka H, Nagaya T, Sato K, Nakamura Y, Watanabe R, Harada T, Gao W, Feng M, Phung Y, Kim I, Paik CH, Choyke PL, Ho M, Kobayashi H. Glypican-3 targeted human heavy chain antibody as a drug carrier for hepatocellular carcinoma therapy. Mol Pharm. 2015;12(6):2151–2157. doi: 10.1021/acs.molpharmaceut.5b00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, Yao M, Pan LH, Qian Q, Yao DF. Glypican-3 is a biomarker and a therapeutic target of hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2015;14(4):361–366. doi: 10.1016/S1499-3872(15)60396-4. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y, Liu H, Weng H, Zhang X, Li P, Fan CL, Li B, Dong PL, Li L, Dooley S, Ding HG. Glypican-3 promotes epithelial-mesenchymal transition of hepatocellular carcinoma cells through ERK signaling pathway. Int J Oncol. 2015;46(3):1275–1285. doi: 10.3892/ijo.2015.2827. [DOI] [PubMed] [Google Scholar]
- 38.Dargel C, Bassani-Sternberg M, Hasreiter J, Zani F, Bockmann JH, Thiele F, Bohne F, Wisskirchen K, Wilde S, Sprinzl MF, Schendel DJ, Krackhardt AM, Uckert W, Wohlleber D, Schiemann M, Stemmer K, Heikenwalder M, Busch DH, Richter G, Mann M, Protzer U. T Cells engineered to express a T-cell receptor specific for glypican-3 to recognize and kill hepatoma cells in vitro and in mice. Gastroenterology. 2015;149(4):1042–1052. doi: 10.1053/j.gastro.2015.05.055. [DOI] [PubMed] [Google Scholar]
- 39.But DY, Lai CL, Yuen MF. Natural history of hepatitis-related hepatocellular carcinoma. World J Gastroenterol. 2008;14(11):1652–1656. doi: 10.3748/wjg.14.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Oliveria Andrade LJ, D’Oliveira A, Melo RC, De Souza EC, Costa Silva CA, Parana R. Association between hepatitis C and hepatocellular carcinoma. J Glob Infect Dis. 2009;1(1):33–37. doi: 10.4103/0974-777X.52979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koike K. Hepatitis C virus contributes to hepatocarcinogenesis by modulating metabolic and intracellular signaling pathways. J Gastroenterol Hepatol. 2007;22(Suppl 1):S108–S111. doi: 10.1111/j.1440-1746.2006.04669.x. [DOI] [PubMed] [Google Scholar]
- 42.Pawlotsky JM. Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol. 2004;12(2):96–102. doi: 10.1016/j.tim.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 43.Pasetto A, Frelin L, Aleman S, Holmstrom F, Brass A, Ahlen G, Brenndorfer ED, Lohmann V, Bartenschlager R, Sallberg M, Bertoletti A, Chen M. TCR-redirected human T cells inhibit hepatitis C virus replication: hepatotoxic potential is linked to antigen specificity and functional avidity. J Immunol. 2012;189(9):4510–4519. doi: 10.4049/jimmunol.1201613. [DOI] [PubMed] [Google Scholar]
- 44.Duval L, Schmidt H, Kaltoft K, Fode K, Jensen JJ, Sorensen SM, Nishimura MI, von der Maase H. Adoptive transfer of allogeneic cytotoxic T lymphocytes equipped with a HLA-A2 restricted MART-1 T-cell receptor: a phase I trial in metastatic melanoma. Clin Cancer Res. 2006;12(4):1229–1236. doi: 10.1158/1078-0432.CCR-05-1485. [DOI] [PubMed] [Google Scholar]
- 45.Gerlach JT, Diepolder HM, Jung MC, Gruener NH, Schraut WW, Zachoval R, Hoffmann R, Schirren CA, Santantonio T, Pape GR. Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C. Gastroenterology. 1999;117(4):933–941. doi: 10.1016/S0016-5085(99)70353-7. [DOI] [PubMed] [Google Scholar]
- 46.Missale G, Bertoni R, Lamonaca V, Valli A, Massari M, Mori C, Rumi MG, Houghton M, Fiaccadori F, Ferrari C. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J Clin Invest. 1996;98(3):706–714. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194(10):1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semmo N, Klenerman P. CD4+ T cell responses in hepatitis C virus infection. World J Gastroenterol. 2007;13(36):4831–4838. doi: 10.3748/wjg.v13.i36.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freeman ML, Burkum CE, Cookenham T, Roberts AD, Lanzer KG, Huston GE, Jensen MK, Sidney J, Peters B, Kohlmeier JE, Woodland DL, van Dyk LF, Sette A, Blackman MA. CD4 T cells specific for a latency-associated gamma-herpesvirus epitope are polyfunctional and cytotoxic. J Immunol. 2014;193(12):5827–5834. doi: 10.4049/jimmunol.1302060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keesen TS, Gomes JA, Fares RC, de Araujo FF, Ferreira KS, Chaves AT, Rocha MO, Correa-Oliveira R. Characterization of CD4(+) cytotoxic lymphocytes and apoptosis markers induced by Trypanossoma cruzi infection. Scand J Immunol. 2012;76(3):311–319. doi: 10.1111/j.1365-3083.2012.02730.x. [DOI] [PubMed] [Google Scholar]
- 51.Kitano S, Tsuji T, Liu C, Hirschhorn-Cymerman D, Kyi C, Mu Z, Allison JP, Gnjatic S, Yuan JD, Wolchok JD. Enhancement of tumor-reactive cytotoxic CD4+ T cell responses after ipilimumab treatment in four advanced melanoma patients. Cancer Immunol Res. 2013;1(4):235–244. doi: 10.1158/2326-6066.CIR-13-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morales O, Depil S, Mrizak D, Martin N, Ndour PA, Dufosse F, Miroux C, Coll J, de Launoit Y, Auriault C, Pancre V, Delhem N. EBV latency II-derived peptides induce a specific CD4+ cytotoxic T-cell activity and not a CD4+ regulatory T-cell response. J Immunother. 2012;35(3):254–266. doi: 10.1097/CJI.0b013e31824d72c5. [DOI] [PubMed] [Google Scholar]
- 53.Campo DS, Dimitrova Z, Yamasaki L, Skums P, Lau DT, Vaughan G, Forbi JC, Teo CG, Khudyakov Y. Next-generation sequencing reveals large connected networks of intra-host HCV variants. BMC Genom. 2014;15(Suppl 5):S4. doi: 10.1186/1471-2164-15-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cusick MF, Yang M, Gill JC, Eckels DD. Naturally occurring CD4+ T-cell epitope variants act as altered peptide ligands leading to impaired helper T-cell responses in hepatitis C virus infection. Hum Immunol. 2011;72(5):379–385. doi: 10.1016/j.humimm.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gededzha MP, Mphahlele MJ, Selabe SG. Characterization of HCV genotype 5a envelope proteins: implications for vaccine development and therapeutic entry target. Hepat Mon. 2014;14(11):e23660. doi: 10.5812/hepatmon.23660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kolls JK, Szabo G. The genetics of hepatitis C virus underlie its ability to escape humoral immunity. J Clin Invest. 2015;125(1):97–98. doi: 10.1172/JCI79424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skums P, Bunimovich L, Khudyakov Y. Antigenic cooperation among intrahost HCV variants organized into a complex network of cross-immunoreactivity. Proc Natl Acad Sci USA. 2015;112(21):6653–6658. doi: 10.1073/pnas.1422942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulsenheimer A, Paranhos-Baccala G, Komurian-Pradel F, Raziorrouh B, Kurktschiev P, Diepolder HM, Zachoval R, Spannagl M, Jung MC, Gruener NH. Lack of variant specific CD8+ T-cell response against mutant and pre-existing variants leads to outgrowth of particular clones in acute hepatitis C. Virol J. 2013;10:295. doi: 10.1186/1743-422X-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Liu Y, Moxley KM, Golden-Mason L, Hughes MG, Liu T, Heemskerk MH, Rosen HR, Nishimura MI. Transduction of human T cells with a novel T-cell receptor confers anti-HCV reactivity. PLoS Pathog. 2010;6(7):e1001018. doi: 10.1371/journal.ppat.1001018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.