Abstract
The nucleus is the repository for the eukaryotic cell’s genetic blueprint, which must be protected from harm to ensure survival. Multiple quality control (QC) pathways operate in the nucleus to maintain the integrity of the DNA, the fidelity of the DNA code during replication, its transcription into mRNA, and the functional structure of the proteins that are required for DNA maintenance, mRNA transcription, and other important nuclear processes. Although we understand a great deal about DNA and RNA QC mechanisms, we know far less about nuclear protein quality control (PQC) mechanisms despite that fact that many human diseases are causally linked to protein misfolding in the nucleus. In this review, we discuss what is known about nuclear PQC and we highlight new questions that have emerged from recent developments in nuclear PQC studies.
Introduction
The misfolding of proteins is an unavoidable problem that every eukaryotic organelle encounters. Protein misfolding occurs via numerous mechanisms: genetic mutations, errors in transcription or translation, problems during nascent peptide folding, and stressors that damage structures of normally folded proteins. The devastating issue of protein misfolding is highlighted by the human pathologies that are causally linked to misfolded protein aggregation such as Alzheimer’s, Parkinson’s, and amyotrophic lateral sclerosis (ALS) [1]. To prevent aggregation, eukaryotic cells have evolved robust and often interconnected compartmental protein quality control (PQC) pathways that manage misfolded proteins by either refolding them into functional proteins through chaperones [2], sequestering them into large inclusions via small heat shock proteins or chaperones [2], or degrading them through the ubiquitin-proteasome system [3] or autophagy [4,5] (Figure 1). From a historical view, much of what we have learned about eukaryotic PQC systems and their role in maintaining proteostasis has largely come from studies in the cytoplasm and endoplasmic reticulum (ER) [6]. By contrast, much less is known about PQC mechanisms in the nucleus. However, at least 15 human diseases are associated with misfolded protein aggregation in the nucleus and include Huntington’s, many spinal cerebellar ataxias (SCAs), and spinal bulbar muscular atrophy (SBMA) [7].
Figure 1. Overall summary of general PQC.
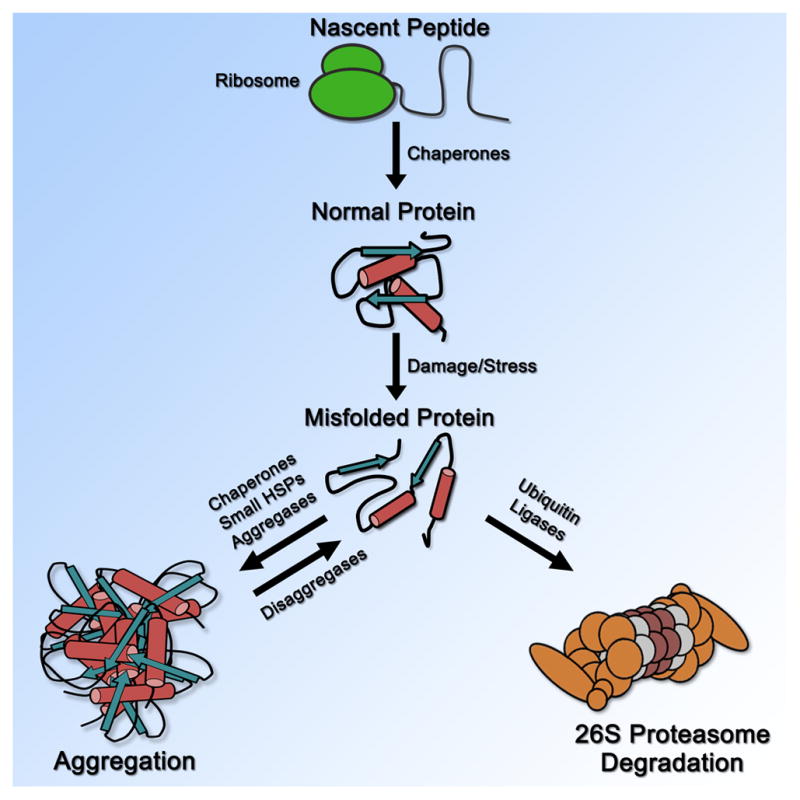
General stages of PQC in the cell from nascent peptide synthesis to degradation.
Overall nuclear protein quality control
Quality control studies in the nucleus have traditionally focused on how the cell maintains the integrity of the nuclear genome and the quality of mRNA prior to export from the nucleus [8,9]. By contrast, our understanding of how the cell maintains the quality of nuclear proteins has lagged behind. The protein aspect of overall nuclear quality control is exceptionally important to consider because an escalating failure to remove or repair misfolded nuclear proteins can lead to a deterioration in the integrity of the nuclear genome and the quality of the mRNA produced. This in turn can impact overall cellular proteostasis due to a reduction in the translational fidelity of the genetic code into its functional complement of proteins. Over the last decade, studies of misfolded nuclear proteins have revealed some of the folding and degradative PQC systems that function in the nucleus and these appear to operate on similar overarching principles as PQC systems in the cytoplasm with chaperones, small heat-shock proteins, and ubiquitin ligases generally managing misfolded nuclear proteins [10–21].
One unique aspect of the nucleus is that it contains a high concentration of DNA, which is organized into chromatin. Due to the negative charge of DNA, many structural chromatin proteins have a considerable positive charge, such as histones. In addition, the nuclear proteome is enriched for proteins that possess low complexity, intrinsically disordered regions [22], suggesting these proteins have a broad capacity for conformational flexibility. Furthermore, chromatin is dynamic and there is ongoing remodeling that involves continuous assembly and disassembly of DNA-RNA-protein complexes [23]. These factors are potentially a considerable source for protein misfolding and aggregation specific to the nucleus.
Unlike the cytoplasm, one key PQC challenge the nucleus does not face to any considerable extent is the need to balance the robustness of PQC in the misfolding of damaged proteins with the PQC in the folding of nascent proteins. That is because nuclear proteins are synthesized in the cytoplasm and are imported into the nucleus through the nuclear pore [24]. Thus, nuclear PQC can be primarily focused on proteins that have become misfolded via damage during or after nuclear import, and nuclear PQC pathways may have evolved to target specific features of damage-induced misfolding that are particularly harmful in the nuclear environment. However, we note there is a possibility that the nucleus engages in limited translation [25], and may have to manage a low level of translational products. Furthermore, failures in cytoplasmic PQC can reduce the amount of correctly folded nuclear proteins, which can subsequently burden nuclear PQC pathways by decreasing the levels of functional nuclear proteins.
PQC degradation in the nucleus
The majority of proteasomes are nuclear localized [26], and the ubiquitin-proteasome system is the main route for misfolded protein degradation in the nucleus [3]. However, a recent study has suggested that nuclear proteins destined for proteasome degradation are first exported from the nucleus and destroyed by cytoplasmic proteasomes [27]. It is not clear if this is specific to a class of proteins that contain nuclear export signals or general for all nuclear proteins. However, it is clear that a number of nuclear-localized ubiquitin ligases have been implicated in the PQC degradation of misfolded nuclear proteins (Table 1). For this review, we will limit our discussion to the major yeast ubiquitin ligases involved in the degradation of misfolded nuclear proteins (Figure 2), as yeast has been the most extensively used organism for studies of nuclear PQC degradation.
Table 1.
Ubiquitin ligases implicated in nuclear PQC degradation
| Ubiquitin ligase | Species studied | Localization | Key references |
|---|---|---|---|
| San1 | Yeast | Nucleus | [10,12,30] |
| Ubr1 | Yeast/mammals | Cytoplasm/nucleus? | [16,17,37] |
| Doa10/Ssm4 | Yeast | ER membrane/inner nuclear membrane | [19,20,45] |
| Asi complex (Asi1, 2, 3) | Yeast | Inner nuclear membrane | [50,51] |
| Slx5-Slx8 | Yeast | Nucleus | [52,53] |
| PML-IV/RNF4 | Mammals | Nucleus | [54–58] |
| UHRF-1 | Mammals | Nucleus | [59] |
| UHRF-2 | Mammals | Nucleus | [59] |
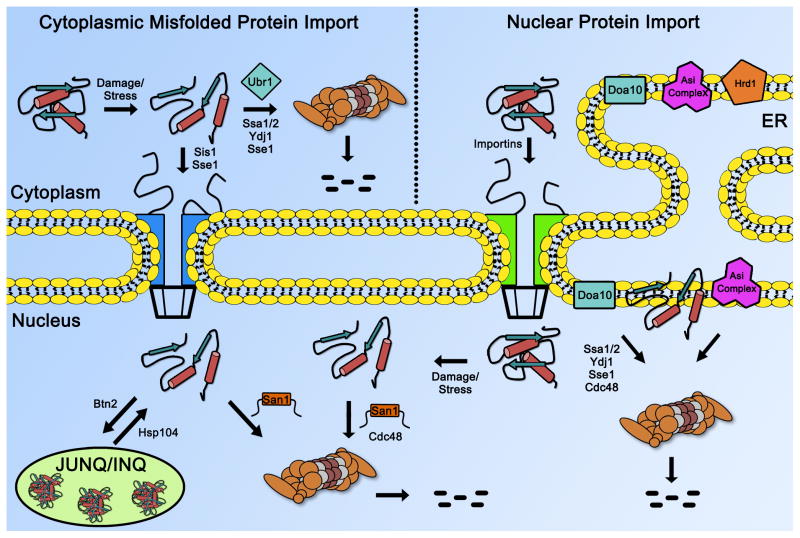
Figure 2. Overall summary of nuclear PQC degradation in yeast.
Major ubiquitin ligases involved in nuclear PQC degradation in yeast are shown in their locations respective to the nuclear membrane. Chaperones involved in different aspects of nuclear PQC are also shown.
The best understood yeast ubiquitin ligase involved in nuclear PQC degradation is San1. Originally discovered as a gene that, when mutated, suppressed the temperature-sensitive mutant phenotypes of sir4-9 and cdc68-1 cells [28,29], San1 was later found to be a ubiquitin ligase [10,30] that is nucleoplasmic localized and specific in targeting misfolded nuclear proteins for ubiquitination and proteasome degradation [10,12]. Since the discovery that San1 is a nuclear PQC ubiquitin ligase [10], additional genetic screens have revealed that loss of San1 leads to the stability of many other temperature-sensitive mutant nuclear proteins [31–34], indicating that degradation of a mutant nuclear protein is a common route to introduce temperature sensitivity. San1’s ability to target misfolded nuclear proteins is attributed to large regions of intrinsic disorder within the N-and C-terminal regions of San1 [12]. These disordered regions possess multiple substrate-interaction regions within a conformationally plastic scaffold, which we propose provides San1 a broad mechanism to target many different kinds of misfolded proteins [12,35]. San1 has been found to recognize exposed hydrophobicity within its misfolded substrates that would have normally been buried within the protein [11]. San1 does not appear to favor a particular type of exposed hydrophobic residue; it only requires that the degree of total exposed hydrophobicity in its substrates is at a threshold that leads to insolubility and aggregation [13], which also correlates in many cases with substrate toxicity [11,13]. San1 does have some substrate overlap with the ubiquitin ligase Ubr1 [15–18], but Ubr1 most likely acts in the cytoplasm rather than the nucleus [16,17,36–39]. San1 has a homolog in fission yeast [40], but no functional equivalent has been identified in metazoans. This is likely due to the extreme intrinsic disorder of San1 [12,35], which allows for a high degree of sequence divergence over evolution [52].
Residing in the ER/inner nuclear membrane, the ubiquitin ligase Doa10 has also been implicated in nuclear PQC degradation [19,41–43]. DOA10 (aka SSM4) was first characterized as an mRNA quality control gene [44]. Mutation to DOA10/SSM4 suppressed the temperature sensitivity of a mutant form of the nuclear Rna14 protein [44]. However, the Doa10 protein is best characterized for its role in ER-associated degradation (ERAD), ubiquitinating both soluble and ER-membrane substrates for degradation by the proteasome [18–20,39,41,45–48]. Similar to San1, Doa10 also recognizes exposed hydrophobicity in its substrates, though the hydrophobicity differs from what San1 recognizes and appears to be presented as an amphipathic helix [19,41,42,45,49]. The exact mechanism by which Doa10 binds its substrates still remains to be elucidated. Doa10 has a mammalian homolog called TEB4/MARCH-VI, but a PQC function has not yet been established for TEB4 [47].
In addition to Doa10, an inner nuclear membrane ubiquitin-ligase complex called Asi (composed of the yeast ubiquitin ligases Asi1 and Asi3, along with Asi2) has recently been implicated in the nuclear PQC degradation of misfolded inner nuclear membrane proteins and soluble transcription factors [50,51]. The Asi complex seems to function in a parallel pathway with the ER-membrane ubiquitin ligase Hrd1 to destroy misfolded nuclear membrane proteins [50,51]. However, more studies are needed to understand the broader role of the Asi complex in nuclear PQC, as well as its substrate specificity at the nuclear membrane.
PQC chaperones in the nucleus
A common characteristic amongst nuclear PQC ubiquitin ligases is that chaperones have been implicated in the degradation of some substrates [15–17,19,41–43,60–62]. A few reports demonstrate that chaperones are required for the ubiquitination of misfolded cytoplasmic proteins destined for nuclear PQC degradation by San1 [15–17]. Another study showed that ubiquitination via San1 does not universally require chaperones [12]. This is a controversial aspect of nuclear PQC degradation because little is known about the precise role chaperones play in nuclear PQC degradation. In Table 2, we outline the chaperones implicated in nuclear PQC.
Table 2.
Protein chaperones implicated in nuclear PQC
| Chaperone | Species studied | Localization | Key references |
|---|---|---|---|
| Ssa1/Ssa2 (Hsp70) | Yeast | Nucleus | [15–19] |
| Ydj1 (Hsp40) | Yeast | Cytoplasm/nucleus | [15–19] |
| Sse1 (Hsp70) | Yeast | Cytoplasm | [15–18] |
| Sis1 (Hsp40) | Yeast | Cytoplasm/nucleus | [60,65] |
| Sti1 | Yeast/mammals | Cytoplasm/nucleus | [66,67] |
| Btn2 | Yeast | Cytoplasm/nucleus | [68,69] |
| Hsp104 | Yeast | Cytoplasm/nucleus | [70–72] |
| Hsp82/Hsc82 (Hsp90) | Yeast | Cytoplasm/nucleus | [39,67,73] |
| Hsp26 (sHSP) | Yeast/mammals | Cytoplasm/nucleus | [74,75] |
| Hsp42 (sHSP) | Yeast | Cytoplasm | [69] |
| Cdc48/p97 | Yeast/mammals | Cytoplasm/nucleus | [14,41,76,77] |
The linkage between chaperones and PQC degradation comes from a long history of PQC studies in the ER and cytoplasm [6], and has shaped how the PQC field thinks about the compartmental triage of misfolded proteins [63]. Ubr1 targets substrates in what appears to be a chaperone-dependent manner, with the Hsp70 chaperones Ssa1/Ssa2, the Hsp40 chaperone Ydj1, and the Hsp110 chaperone Sse1 required for Ubr1-dependent degradation [15–18]. Doa10 also targets substrates in what appears to be a chaperone-dependent manner, utilizing Ssa1/Ssa2, Ydj1, and the Hsp40 chaperone Sis1 [19,41,43,48,60–62,64]. Although the role of chaperones in San1-mediated degradation remains a subject of open debate [12,15–18], the San1 pathway does utilize the AAA-ATPase chaperone Cdc48/p97 for the degradation of select San1 substrates that are highly insoluble [14]. It may be that chaperone involvement in nuclear PQC degradation is important for partitioning misfolded proteins between insoluble aggregates and available monomers that can be recognized by the nuclear PQC degradation ubiquitin ligases. How chaperones function in nuclear PQC remains an open question. Both Sis1 and Sse1 are implicated in the nuclear transport of certain misfolded cytoplasmic proteins [16,17,65], suggesting that one function for these chaperones may be to facilitate the trafficking of misfolded cytoplasmic proteins into the nucleus where San1 can recognize them.
PQC inclusions in the nucleus
Although each cellular compartment possesses a robust complement of folding and degradative PQC activities, the burden of misfolded proteins can exceed the capacity of the compartment’s PQC systems during stress or with age [78–80]. When the burden of misfolded proteins overwhelms a compartment’s PQC pathways, misfolded proteins can aggregate [78–80]. One way the cell counters the incapacity of PQC pathways is to concentrate misfolded proteins into inclusion bodies that sequester aggregation-prone misfolded proteins. Prominent inclusions include the perinuclear mammalian aggresome [81], the perinuclear/intranuclear yeast JUNQ/INQ [69,82], and the perivacuolar yeast IPOD [82].
The JUNQ/INQ is a primary nuclear inclusion for misfolded protein sequestration in both yeast and mammalian cells [69,82]. Originally identified as perinuclear [82], a recent study found that the deposition is intranuclear and bounded by the nuclear membrane [69]. Furthermore, genotoxic stressors like methyl methanesulfonate (MMS) appear to force important nuclear proteins into this inclusion [83]. Interestingly, san1Δ cells were found to be sensitive to MMS in a large-scale genetic screen [84], suggesting that accumulation of misfolded San1 substrates could contributed to increased DNA damage. Alternatively, MMS can methylate Cys, His, Lys, and Arg residues in proteins [85], which could lead to protein misfolding and the need for San1 to remove MMS-damaged misfolded proteins.
The JUNQ/INQ is dynamic and the misfolded proteins in the JUNQ/INQ are mobile [82]. However, the mobility of misfolded proteins in the JUNQ/INQ can decrease during stress [86,87]. The aggregase Btn2 is important for localizing misfolded proteins to the JUNQ/INQ [69]. Furthermore, Hsp70 chaperones and proteasome subunits have also been shown to localize with the JUNQ [86]. Thus, the JUNQ/INQ is likely to be site where misfolded nuclear proteins are stored until they can be destroyed by ubiquitin-proteasome pathways. An important feature of the JUNQ/INQ is that localization of misfolded proteins to this site prevents their passage from the mother cell to the daughter cell during mitosis [88]. In this way, the quality of the nuclear environment in the daughter can be kept pristine.
In mammalian cells, nuclear inclusions have been studied using proteins with polyglutamine and/or polyalanine-expansions that lead to disorders such as Huntington’s, Kennedy’s, or oculopharyngeal muscular dystrophy (OPMD) [55,89–100]. In most cases, distinct chaperones have also been found to colocalize with the nuclear inclusions [55,89–100], similar to the JUNQ. It currently remains unknown if the nuclear inclusions in these cases are identical to the JUNQ or comprise different structures. Furthermore, different nuclear subcompartments may have their own unique sites of deposition. For example, an inclusion forms within the nucleolus after proteasome inhibition [101], though it is unknown if the nucleolar inclusion is composed of misfolded proteins. Much more work will be needed to understand the breadth and types of inclusions in the nucleus that act as PQC deposition sites.
PQC interplay between the nucleus and cytoplasm
Finally, it is important to consider that there will likely be significant interplay between nuclear and cytoplasmic PQC pathways as there is an intimate and dynamic communication between the two compartments through the nuclear pore. One interesting type of PQC interplay that has emerged in the last few years is that misfolded proteins are not always excluded from the nucleus if they are first generated in the cytoplasm [15–18]. This seems counterintuitive because cytoplasmic PQC pathways should prevent misfolded proteins from entering the nucleus where they can do harm. Why are these ‘cytoplasmic’ misfolded proteins imported into the nucleus when they should be managed in the cytoplasm? There are two possibilities. First, it could be that some misfolded cytoplasmic proteins are below the ~40kDa passive diffusion limit of the nuclear pore [24]. This is the case for a number of misfolded cytoplasmic proteins studied [15–18]. Second, it could be there are active mechanisms to import misfolded cytoplasmic proteins to the nucleus if nuclear PQC degradation is more robust than cytoplasmic PQC degradation. Consistent with active import, it has been shown that the Hsp40 chaperone Sis1 is required for the nuclear localization and degradation of a misfolded cytoplasmic protein in yeast [65]. In addition, deletion of the small heat shock protein Hsp42 leads to the nuclear localization of misfolded cytoplasmic proteins [69], indicating that there are also retention mechanisms in the cytoplasm to prevent nuclear import. How transport and retention pathways coordinate to allow or disallow nuclear import of misfolded proteins is a new frontier for nuclear PQC studies.
Another possibility for interplay between cytoplasmic and nuclear PQC systems is that there may be conditions where cytoplasmic PQC pathways become overloaded and unable to buffer the cytoplasm against misfolded protein accumulation. For example, it has been shown that expression of toxic polyQ proteins in worms causes loss of function for chaperone-dependent, metastable proteins [102]. In addition, protein aggregation in the cytoplasm can interfere with nucleocytoplasmic transport [103]. It is possible that the nucleus serves as a back-up PQC compartment for cytoplasmic PQC when the cytoplasm can’t handle a high burden of misfolded proteins.
Lastly, some nuclear pore proteins are exceptionally long-lived in non-dividing cells [104,105], and nuclear pores been found to deteriorate with age potentially allowing larger proteins to passively enter the nucleus [106]. Thus, nuclear PQC pathways might become increasingly important with age as nuclear pore selectivity breaks down. This has significant ramifications for human health because at least 15 diseases have the common characteristic that they show greater level of misfolded protein aggregation in the nucleus that increases with age, and many of these proteins are not found in the nucleus at youthful ages [7].
Concluding remarks
We have gained foundational knowledge about nuclear PQC during the last decade. However, important pieces are still missing from the nuclear PQC puzzle. For example, it is not yet known if there is a mammalian equivalent to yeast San1. In addition, nuclear PQC functions for the mammalian homologs of yeast Doa10 and Ubr1 have yet to be shown. It will also be crucial to determine the particular features of misfolded proteins that are recognized by individual nuclear PQC degradation pathways, and how the pathways bind these features. Understanding the features of misfolding will also help delineate why chaperones are required for the nuclear PQC degradation of some substrates but not others. From a more holistic view, it will be important to explore nuclear PQC as a network and discover how the nucleus regulates the levels of its PQC systems in response to the burden of misfolded proteins. In the ER, the unfolded protein response (UPR) is crucial for protecting the ER environment during stress [107]. Evidence that a nuclear UPR might exist comes from the increased expression of nuclear chaperone and stress response genes when the San1 function is compromised [10]. Particular chaperone genes include those that encode for Ssa4, Hsp26, and Hsp104, all of which become enriched in the nucleus upon exposure of cells to protein misfolding stressors [70–72,74,108–110]. Revealing how nuclear PQC systems are coordinately regulated to protect the nuclear environment will be essential to understanding how these systems might fail during aging and lead to nuclear aggregation diseases.
Acknowledgments
We tried our best to cite all primary literature pertaining specifically to nuclear PQC. We apologize to our colleagues if we unintentionally missed their relevant studies due to space constrictions. This work was supported by a NIH/NIGMS training grant 5T32 GM007750 to R.D.J. and a NIH/NIA grant R01AG031136 to R.G.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
* of special interest
** of outstanding interest
- 1.Aguzzi A, O’Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat Rev Drug Discov. 2010;9:237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- 2.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Ulrich Hartl F. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 3.Fredrickson EK, Gardner RG. Selective destruction of abnormal proteins by ubiquitin-mediated protein quality control degradation. Semin Cell Dev Biol. 2012;23:530–537. doi: 10.1016/j.semcdb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hyttinen JM, Amadio M, Viiri J, Pascale A, Salminen A, Kaarniranta K. Clearance of misfolded and aggregated proteins by aggrephagy and implications for aggregation diseases. Ageing Res Rev. 2014;18C:16–28. doi: 10.1016/j.arr.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Park C, Cuervo AM. Selective Autophagy: Talking with the UPS. Cell Biochem Biophys. 2013 doi: 10.1007/s12013-013-9623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–252. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Woulfe J. Nuclear bodies in neurodegenerative disease. Biochim Biophys Acta. 2008;1783:2195–2206. doi: 10.1016/j.bbamcr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Eberle AB, Visa N. Quality control of mRNP biogenesis: networking at the transcription site. Semin Cell Dev Biol. 2014;32:37–46. doi: 10.1016/j.semcdb.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 9.Heyer WD. Regulation of recombination and genomic maintenance. Cold Spring Harb Perspect Biol. 2015;7:a016501. doi: 10.1101/cshperspect.a016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10**.Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. This study identified San1 as the first ubiquitin ligase that is known to function in PQC degradation in the nucleus. [DOI] [PubMed] [Google Scholar]
- 11**.Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol Biol Cell. 2011;22:2384–2395. doi: 10.1091/mbc.E11-03-0256. This study revealed that San1 specifically recognizes exposed hydrophobicity in its misfolded protein substrates, and the exposed hydrophobicity can often correlate with the toxicity of the misfolded protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. This study found that San1 uses regions of high disorder to recognize its substrates. In addition, it demonstrated through in vitro and E. coli experiments that chaperones aren’t a necessity for San1-mediated ubiquitination of misfolded proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Fredrickson EK, Gallagher PS, Clowes Candadai SV, Gardner RG. Substrate recognition in nuclear protein quality control degradation is governed by exposed hydrophobicity that correlates with aggregation and insolubility. J Biol Chem. 2013;288:6130–6139. doi: 10.1074/jbc.M112.406710. This study demonstrated that San1 recognizes the exposed hydrophobicity with misfolded nuclear proteins that is at or above the threshold that induces protein insolubility and aggregation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Gallagher PS, Clowes Candadai SV, Gardner RG. The requirement for Cdc48/p97 in nuclear protein quality control degradation depends on the substrate and correlates with substrate insolubility. J Cell Sci. 2014;127:1980–1991. doi: 10.1242/jcs.141838. This study found that the AAA-ATPase Cdc48 was involved in the degradation of ubiuitinated San1 substrates. However, the involvement was not universal and was only required for select San1 substrates that were highly insoluble and aggregation-prone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Guerriero CJ, Weiberth KF, Brodsky JL. Hsp70 Targets a Cytoplasmic Quality Control Substrate to the San1p Ubiquitin Ligase. Journal of Biological Chemistry. 2013;288:18506–18520. doi: 10.1074/jbc.M113.475905. This study demonstrated that a misfolded cytoplasmic protein was targted by San1. It also found that chaperones required for the degradation of this San1 substrate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. This is one of the first studies revealing that misfolded cytoplasmic proteins could be targeted by San1 by virtue of their unexpected nuclear localization. Chaperones were involved in the degradation of these substrates. It was also discovered that Ubr1 is another ubiquitin ligase that is involved in the degradation of these substrates. Ubr1 was previously found to be involved primarily in N-end rule degradation. It is likely that Ubr1 functions in the cytoplasm, upstream of San1-dependent nuclear PQC degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Prasad R, Kawaguchi S, Ng DT. A nucleus-based quality control mechanism for cytosolic proteins. Mol Biol Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. This is also one of the first studies revealing that misfolded cytoplasmic proteins could be targeted by San1 by virtue of their unexpected nuclear localization. Chaperones were also implicated in the degradation of these substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Prasad R, Kawaguchi S, Ng DT. Biosynthetic mode can determine the mechanism of protein quality control. Biochem Biophys Res Commun. 2012;425:689–695. doi: 10.1016/j.bbrc.2012.07.080. This study demonstrated that additional misfolded cytoplasmic substrates are targeted by San1. [DOI] [PubMed] [Google Scholar]
- 19**.Furth N, Gertman O, Shiber A, Alfassy OS, Cohen I, Rosenberg MM, Doron NK, Friedler A, Ravid T. Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol Biol Cell. 2011;22:4726–4739. doi: 10.1091/mbc.E11-05-0463. This study revealed the features of misfolding recognized by the ER/nuclear membrane ubiquitin ligase Doa10. This study is interesting because the determinants for Doa10 recognition appear to be different than for San1 recognition, even though both recognize hydrophobicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. This study found that localization of Doa10 to the inner nuclear membrane was required for its degradation of the MATalpha2 transcription factor, which also displays an amphipathic helix to Doa10 for recognition. [DOI] [PubMed] [Google Scholar]
- 21.Mijaljica D, Prescott M, Devenish RJ. Nibbling within the nucleus: turnover of nuclear contents. Cell Mol Life Sci. 2007;64:581–588. doi: 10.1007/s00018-007-6395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng F, Na I, Kurgan L, Uversky VN. Compartmentalization and Functionality of Nuclear Disorder: Intrinsic Disorder and Protein-Protein Interactions in Intra-Nuclear Compartments. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langst G, Manelyte L. Chromatin Remodelers: From Function to Dysfunction. Genes (Basel) 2015;6:299–324. doi: 10.3390/genes6020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossman E, Medalia O, Zwerger M. Functional architecture of the nuclear pore complex. Annu Rev Biophys. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- 25.Pederson T. The persistent plausibility of protein synthesis in the nucleus: process, palimpsest or pitfall? Curr Opin Cell Biol. 2013;25:520–521. doi: 10.1016/j.ceb.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Wojcik C, DeMartino GN. Intracellular localization of proteasomes. Int J Biochem Cell Biol. 2003;35:579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Madura K. Degradation of specific nuclear proteins occurs in the cytoplasm in Saccharomyces cerevisiae. Genetics. 2014;197:193–197. doi: 10.1534/genetics.114.163824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schnell R, D’Ari L, Foss M, Goodman D, Rine J. Genetic and molecular characterization of suppressors of SIR4 mutations in Saccharomyces cerevisiae. Genetics. 1989;122:29–46. doi: 10.1093/genetics/122.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Q, Johnston GC, Singer RA. The Saccharomyces cerevisiae Cdc68 transcription activator is antagonized by San1, a protein implicated in transcriptional silencing. Mol Cell Biol. 1993;13:7553–7565. doi: 10.1128/mcb.13.12.7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dasgupta A, Ramsey KL, Smith JS, Auble DT. Sir antagonist 1 (San1) is a ubiquitin ligase. J Biol Chem. 2004;279:26830–26838. doi: 10.1074/jbc.M400894200. [DOI] [PubMed] [Google Scholar]
- 31.Addinall SG, Downey M, Yu M, Zubko MK, Dewar J, Leake A, Hallinan J, Shaw O, James K, Wilkinson DJ, et al. A genomewide suppressor and enhancer analysis of cdc13–1 reveals varied cellular processes influencing telomere capping in Saccharomyces cerevisiae. Genetics. 2008;180:2251–2266. doi: 10.1534/genetics.108.092577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arlow T, Scott K, Wagenseller A, Gammie A. Proteasome inhibition rescues clinically significant unstable variants of the mismatch repair protein Msh2. Proc Natl Acad Sci U S A. 2013;110:246–251. doi: 10.1073/pnas.1215510110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Estruch F, Peiro-Chova L, Gomez-Navarro N, Durban J, Hodge C, Del Olmo M, Cole CN. A genetic screen in Saccharomyces cerevisiae identifies new genes that interact with mex67-5, a temperature-sensitive allele of the gene encoding the mRNA export receptor. Mol Genet Genomics. 2009;281:125–134. doi: 10.1007/s00438-008-0402-x. [DOI] [PubMed] [Google Scholar]
- 34.Lewis MJ, Pelham HR. Inefficient quality control of thermosensitive proteins on the plasma membrane. PLoS One. 2009;4:e5038. doi: 10.1371/journal.pone.0005038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fredrickson EK, Clowes Candadai SV, Tam CH, Gardner RG. Means of self-preservation: how an intrinsically disordered ubiquitin-protein ligase averts self-destruction. Mol Biol Cell. 2013;24:1041–1052. doi: 10.1091/mbc.E12-11-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Eisele F, Wolf DH. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. This study was also one of the first to demonstrate a PQC degradation role for Ubr1, which was previously found to be involved in N-end rule degradation. [DOI] [PubMed] [Google Scholar]
- 37**.Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, Sultana R, Wu K, Johnson J, Cyr DM, Caplan AJ. Ubr1 and Ubr2 Function in a Quality Control Pathway for Degradation of Unfolded Cytosolic Proteins. Mol Biol Cell. 2010;21:2102–2116. doi: 10.1091/mbc.E10-02-0098. This study was also one of the first to demonstrate a PQC degradation role for Ubr1, which was previously found to be involved in N-end rule degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Summers DW, Wolfe KJ, Ren HY, Cyr DM. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One. 2013;8:e52099. doi: 10.1371/journal.pone.0052099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theodoraki MA, Nillegoda NB, Saini J, Caplan AJ. A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. J Biol Chem. 2012;287:23911–23922. doi: 10.1074/jbc.M112.341164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuo Y, Kishimoto H, Tanae K, Kitamura K, Katayama S, Kawamukai M. Nuclear protein quality is regulated by the ubiquitin-proteasome system through the activity of Ubc4 and San1 in fission yeast. J Biol Chem. 2011;286:13775–13790. doi: 10.1074/jbc.M110.169953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. This study demonstrated that different Doa10 substrates had distinct requirements for Cdc48/p97 for their degradation depending upon their relationship to the ER membrane. Membrane-bound substrates required Cdc48, whereas soluble substrates did not. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alfassy OS, Cohen I, Reiss Y, Tirosh B, Ravid T. Placing a disrupted degradation motif at the C terminus of proteasome substrates attenuates degradation without impairing ubiquitylation. J Biol Chem. 2013;288:12645–12653. doi: 10.1074/jbc.M113.453027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandart E, Dufour ME, Lacroute F. Inactivation of SSM4, a new Saccharomyces cerevisiae gene, suppresses mRNA instability due to rna14 mutations. Mol Gen Genet. 1994;245:323–333. doi: 10.1007/BF00290112. [DOI] [PubMed] [Google Scholar]
- 45**.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. This study was the first to identify Doa10 as an ER/nuclear membrane PQC ubiquitin ligase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 47.Kreft SG, Wang L, Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J Biol Chem. 2006;281:4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- 48.Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilon T, Chomsky O, Kulka RG. Degradation signals recognized by the Ubc6p-Ubc7p ubiquitin-conjugating enzyme pair. Mol Cell Biol. 2000;20:7214–7219. doi: 10.1128/mcb.20.19.7214-7219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Foresti O, Rodriguez-Vaello V, Funaya C, Carvalho P. Quality control of inner nuclear membrane proteins by the Asi complex. Science. 2014;346:751–755. doi: 10.1126/science.1255638. This study was one of the first to discover the Asi ubiquitin ligase complex that functions in the PQC degradation of misfolded inner nuclear membrane proteins. The Asi complex along with Doa10 are now two PQC ubiquitin ligases that function at the inner nuclear membrane. [DOI] [PubMed] [Google Scholar]
- 51**.Khmelinskii A, Blaszczak E, Pantazopoulou M, Fischer B, Omnus DJ, Le Dez G, Brossard A, Gunnarsson A, Barry JD, Meurer M, et al. Protein quality control at the inner nuclear membrane. Nature. 2014;516:410–413. doi: 10.1038/nature14096. This study was one of the first to discover the Asi ubiquitin ligase complex that functions in the PQC degradation of misfolded inner nuclear membrane proteins. The Asi complex along with Doa10 are now two PQC ubiquitin ligases that function at the inner nuclear membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z, Prelich G. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol. 2009;29:1694–1706. doi: 10.1128/MCB.01470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie Y, Rubenstein EM, Matt T, Hochstrasser M. SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor. Genes Dev. 2010;24:893–903. doi: 10.1101/gad.1906510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu L, Gao YS, Tousson A, Shah A, Chen TL, Vertel BM, Sztul E. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol Biol Cell. 2005;16:4905–4917. doi: 10.1091/mbc.E05-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Janer A, Martin E, Muriel MP, Latouche M, Fujigasaki H, Ruberg M, Brice A, Trottier Y, Sittler A. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J Cell Biol. 2006;174:65–76. doi: 10.1083/jcb.200511045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gartner A, Muller S. PML, SUMO, and RNF4: guardians of nuclear protein quality. Mol Cell. 2014;55:1–3. doi: 10.1016/j.molcel.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 57.Sahin U, Ferhi O, Jeanne M, Benhenda S, Berthier C, Jollivet F, Niwa-Kawakita M, Faklaris O, Setterblad N, de The H, et al. Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J Cell Biol. 2014;204:931–945. doi: 10.1083/jcb.201305148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo L, Giasson BI, Glavis-Bloom A, Brewer MD, Shorter J, Gitler AD, Yang X. A cellular system that degrades misfolded proteins and protects against neurodegeneration. Mol Cell. 2014;55:15–30. doi: 10.1016/j.molcel.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iwata A, Nagashima Y, Matsumoto L, Suzuki T, Yamanaka T, Date H, Deoka K, Nukina N, Tsuji S. Intra-nuclear degradation of polyglutamine aggregates by the ubiquitin proteasome system. J Biol Chem. 2009;284:9796–9803. doi: 10.1074/jbc.M809739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shiber A, Breuer W, Brandeis M, Ravid T. Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol Biol Cell. 2013;24:2076–2087. doi: 10.1091/mbc.E13-01-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 62.Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 64.Shiber A, Ravid T. Chaperoning proteins for destruction: diverse roles of Hsp70 chaperones and their co-chaperones in targeting misfolded proteins to the proteasome. Biomolecules. 2014;4:704–724. doi: 10.3390/biom4030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65**.Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, Hipp MS, Hayer-Hartl M, Hartl FU. PolyQ Proteins Interfere with Nuclear Degradation of Cytosolic Proteins by Sequestering the Sis1p Chaperone. Cell. 2013;154:134–145. doi: 10.1016/j.cell.2013.06.003. This study demonstrated that the Hsp40 chaperone Sis1 is important for the nuclear localization of a misfolded cytoplasmic protein. To this point, it was unclear how misfolded cytoplasmic proteins entered the nucleus. Involvement of Sis1 indicates it is a chaperone-dependent process. [DOI] [PubMed] [Google Scholar]
- 66.Longshaw VM, Chapple JP, Balda MS, Cheetham ME, Blatch GL. Nuclear translocation of the Hsp70/Hsp90 organizing protein mSTI1 is regulated by cell cycle kinases. J Cell Sci. 2004;117:701–710. doi: 10.1242/jcs.00905. [DOI] [PubMed] [Google Scholar]
- 67*.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. This study uncovered chaperone pathways that each distinctly behave in the refolding or degradation of the tumor suppressor VHL. It demonstrated that different classes of chaperones can iperate in the different aspects of PQC. [DOI] [PubMed] [Google Scholar]
- 68*.Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell. 2012;23:3041–3056. doi: 10.1091/mbc.E12-03-0194. This study revealed that Btn2 can interact with Sis1 to assist in localization of misfolded proteins to the JUNQ/INQ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69**.Miller SB, Ho CT, Winkler J, Khokhrina M, Neuner A, Mohamed MY, Guilbride DL, Richter K, Lisby M, Schiebel E, et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015;34:778–797. doi: 10.15252/embj.201489524. This study found that misfolded proteins that were previously found to localize to the juxtanuclear quality control inclusion (or JUNQ) are not in a juxtanuclear inclusion, but in an intranuclear inclusion (or INQ) that is bound by the nuclear membrane. This would indicate that partioning in cellular inclusions happens between the cytoplasm and nucleus. This would place further importance on understanding the mechanisms that allow misfolded cytoplasmic proteins to enter the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawai R, Fujita K, Iwahashi H, Komatsu Y. Direct evidence for the intracellular localization of Hsp104 in Saccharomyces cerevisiae by immunoelectron microscopy. Cell Stress Chaperones. 1999;4:46–53. doi: 10.1054/csac.1999.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 72.Tkach JM, Glover JR. Nucleocytoplasmic trafficking of the molecular chaperone Hsp104 in unstressed and heat-shocked cells. Traffic. 2008;9:39–56. doi: 10.1111/j.1600-0854.2007.00666.x. [DOI] [PubMed] [Google Scholar]
- 73.Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rossi JM, Lindquist S. The intracellular location of yeast heat-shock protein 26 varies with metabolism. J Cell Biol. 1989;108:425–439. doi: 10.1083/jcb.108.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willsie JK, Clegg JS. Small heat shock protein p26 associates with nuclear lamins and HSP70 in nuclei and nuclear matrix fractions from stressed cells. J Cell Biochem. 2002;84:601–614. [PubMed] [Google Scholar]
- 76.Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell. 2011;41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Richardson A, Gardner RG, Prelich G. Physical and genetic associations of the Irc20 ubiquitin ligase with Cdc48 and SUMO. PLoS One. 2013;8:e76424. doi: 10.1371/journal.pone.0076424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 79.Kikis EA, Gidalevitz T, Morimoto RI. Protein homeostasis in models of aging and age-related conformational disease. Adv Exp Med Biol. 2010;694:138–159. doi: 10.1007/978-1-4419-7002-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taylor RC, Dillin A. Aging as an event of proteostasis collapse. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82**.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. This study demonstrated that misfolded proteins in yeast and mammalian cells can partition between two major inclusions: the perivacuolar insoluble protein deposit (or IPOD) and the juxtanuclear quality control compartment (or JUNQ). Partitioning of a misfolded protein to each distinct inclusion varied depending on the ubiquitination state of the protein and its ability to form amyloid-like aggregates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83**.Gallina I, Colding C, Henriksen P, Beli P, Nakamura K, Offman J, Mathiasen DP, Silva S, Hoffmann E, Groth A, et al. Cmr1/WDR76 defines a nuclear genotoxic stress body linking genome integrity and protein quality control. Nat Commun. 2015;6:6533. doi: 10.1038/ncomms7533. This study discovered that many nuclear proteins can localize to the JUNQ/INQ after exposure of cells to a genotoxic stress (MMS) or a proteomic stress (inhibition of proteasome activity). It has uncovered native nuclear proteins that may be susceptible to misfolding and are targted for deposition to the JUNQ/INQ when their degradation is compromised. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- 85.Boffa LC, Bolognesi C. Nuclear proteins damage by alkylating agents with different degrees of carcinogenicity. Chem Biol Interact. 1985;55:235–245. doi: 10.1016/s0009-2797(85)80131-9. [DOI] [PubMed] [Google Scholar]
- 86.Weisberg SJ, Lyakhovetsky R, Werdiger AC, Gitler AD, Soen Y, Kaganovich D. Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proc Natl Acad Sci U S A. 2012;109:15811–15816. doi: 10.1073/pnas.1205829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, Qian SB. Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes. Mol Biol Cell. 2011;22:3277–3288. doi: 10.1091/mbc.E11-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012;2:738–747. doi: 10.1016/j.celrep.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 89.Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Abel A, Walcott J, Woods J, Duda J, Merry DE. Expression of expanded repeat androgen receptor produces neurologic disease in transgenic mice. Hum Mol Genet. 2001;10:107–116. doi: 10.1093/hmg/10.2.107. [DOI] [PubMed] [Google Scholar]
- 91.Jana NR, Tanaka M, Wang G, Nukina N. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet. 2000;9:2009–2018. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 92.Bailey CK, Andriola IF, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2002;11:515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- 93.Adachi H, Katsuno M, Minamiyama M, Sang C, Pagoulatos G, Angelidis C, Kusakabe M, Yoshiki A, Kobayashi Y, Doyu M, et al. Heat shock protein 70 chaperone overexpression ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model by reducing nuclear-localized mutant androgen receptor protein. J Neurosci. 2003;23:2203–2211. doi: 10.1523/JNEUROSCI.23-06-02203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ishihara K, Yamagishi N, Saito Y, Adachi H, Kobayashi Y, Sobue G, Ohtsuka K, Hatayama T. Hsp105alpha suppresses the aggregation of truncated androgen receptor with expanded CAG repeats and cell toxicity. J Biol Chem. 2003;278:25143–25150. doi: 10.1074/jbc.M302975200. [DOI] [PubMed] [Google Scholar]
- 95.Latouche M, Fragner P, Martin E, El Hachimi KH, Zander C, Sittler A, Ruberg M, Brice A, Stevanin G. Polyglutamine and polyalanine expansions in ataxin7 result in different types of aggregation and levels of toxicity. Mol Cell Neurosci. 2006;31:438–445. doi: 10.1016/j.mcn.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 96.Seidel K, Meister M, Dugbartey GJ, Zijlstra MP, Vinet J, Brunt ER, van Leeuwen FW, Rub U, Kampinga HH, den Dunnen WF. Cellular protein quality control and the evolution of aggregates in spinocerebellar ataxia type 3 (SCA3) Neuropathol Appl Neurobiol. 2012;38:548–558. doi: 10.1111/j.1365-2990.2011.01220.x. [DOI] [PubMed] [Google Scholar]
- 97.Bao YP, Cook LJ, O’Donovan D, Uyama E, Rubinsztein DC. Mammalian, yeast, bacterial, and chemical chaperones reduce aggregate formation and death in a cell model of oculopharyngeal muscular dystrophy. J Biol Chem. 2002;277:12263–12269. doi: 10.1074/jbc.M109633200. [DOI] [PubMed] [Google Scholar]
- 98.Corbeil-Girard LP, Klein AF, Sasseville AM, Lavoie H, Dicaire MJ, Saint-Denis A, Page M, Duranceau A, Codere F, Bouchard JP, et al. PABPN1 overexpression leads to upregulation of genes encoding nuclear proteins that are sequestered in oculopharyngeal muscular dystrophy nuclear inclusions. Neurobiol Dis. 2005;18:551–567. doi: 10.1016/j.nbd.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 99.Chartier A, Benoit B, Simonelig M. A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. EMBO J. 2006;25:2253–2262. doi: 10.1038/sj.emboj.7601117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tavanez JP, Bengoechea R, Berciano MT, Lafarga M, Carmo-Fonseca M, Enguita FJ. Hsp70 chaperones and type I PRMTs are sequestered at intranuclear inclusions caused by polyalanine expansions in PABPN1. PLoS One. 2009;4:e6418. doi: 10.1371/journal.pone.0006418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Latonen L, Moore HM, Bai B, Jaamaa S, Laiho M. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene. 2011;30:790–805. doi: 10.1038/onc.2010.469. [DOI] [PubMed] [Google Scholar]
- 102.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- 103.Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, Patra M, Tatzelt J, Mann M, Winklhofer KF, Hartl FU, et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–176. doi: 10.1126/science.aad2033. [DOI] [PubMed] [Google Scholar]
- 104.Savas JN, Toyama BH, Xu T, Yates JR, 3rd, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, 3rd, Hetzer MW. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154:971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106*.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. This study found that the selectivity of the nuclear pore decreases with age, allowing previously excluded cytoplasmic proteins to enter into the nucleus. This has important ramifications for protein misfolding in the nucleus and how loss of nuclear prore selectivity could contribute to age-dependent nuclear aggregation diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5:a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quan X, Rassadi R, Rabie B, Matusiewicz N, Stochaj U. Regulated nuclear accumulation of the yeast hsp70 Ssa4p in ethanol-stressed cells is mediated by the N-terminal domain, requires the nuclear carrier Nmd5p and protein kinase C. FASEB J. 2004;18:899–901. doi: 10.1096/fj.03-0947fje. [DOI] [PubMed] [Google Scholar]
- 109.Quan X, Tsoulos P, Kuritzky A, Zhang R, Stochaj U. The carrier Msn5p/Kap142p promotes nuclear export of the hsp70 Ssa4p and relocates in response to stress. Mol Microbiol. 2006;62:592–609. doi: 10.1111/j.1365-2958.2006.05395.x. [DOI] [PubMed] [Google Scholar]
- 110.Chughtai ZS, Rassadi R, Matusiewicz N, Stochaj U. Starvation promotes nuclear accumulation of the hsp70 Ssa4p in yeast cells. J Biol Chem. 2001;276:20261–20266. doi: 10.1074/jbc.M100364200. [DOI] [PubMed] [Google Scholar]