Abstract
From embryonic development to cancer metastasis, cell migration plays a central role in health and disease. It is increasingly becoming apparent that cells migrating in three-dimensional (3-D) environments exhibit some striking differences compared with their well-established 2-D counterparts. One key finding is the significant role the nucleus plays during 3-D migration: when cells move in confined spaces, the cell body and nucleus must deform to squeeze through available spaces, and the deformability of the large and relatively rigid nucleus can become rate-limiting. In this review, we highlight recent findings regarding the role of nuclear mechanics in 3-D migration, including factors that govern nuclear deformability, and emerging mechanisms by which cells apply cytoskeletal forces to the nucleus to facilitate nuclear translocation. Intriguingly, the ‘physical barrier’ imposed by the nucleus also impacts cytoplasmic dynamics that affect cell migration and signaling, and changes in nuclear structure resulting from the mechanical forces acting on the nucleus during 3-D migration could further alter cellular function. These findings have broad relevance to the migration of both normal and cancerous cells inside living tissues, and motivate further research into the molecular details by which cells move their nuclei, as well as the consequences of the mechanical stress on the nucleus.
Introduction
In multicellular organisms, cell migration is essential in the development, maintenance and repair of various tissues [1]; it also enables immune cells to survey tissues and to respond to local challenges [2]. At the same time, cell migration drives the tissue invasion and metastasis of cancer cells, which is responsible for the vast majority of cancer deaths [3]. While much of our current knowledge regarding the molecular and biophysical principles of cell migration stems from studying cells moving on 2-D substrates [4], it is now becoming evident that cells migrating in 3-D environments encounter distinct physical challenges. During in vivo migration/invasion, cells must navigate many microstructural obstacles, including extracellular matrix (ECM) networks and neighboring cells. The pore sizes encountered in the interstitial space range from 0.1 to 30 µm in diameter, i.e., comparable to or significantly smaller than the size of the migrating cell [5–7]. Cells have two strategies to penetrate such confining environments: (i) expanding the openings via physical remodeling and/or proteolytic degradation of the ECM [8], or (ii) contorting their shape to accommodate the available spaces [9]. The cell membrane and cytoplasm are able to quickly deform and remodel to penetrate openings less than 1 µm in diameter [10]. In contrast, deformation of the nucleus, the largest and stiffest organelle, presents a more formidable challenge. Here we discuss emerging insights into the intracellular biomechanics and molecular processes involved in translocating the nucleus through tight spaces, including implications on migration efficiency and other biological functions.
The size and rigidity of the nucleus: a physical barrier for cell migration
The nucleus is the largest organelle in the cell, with a diameter between 3–15 µm [11,12], making it substantially larger than many pores encountered during migration in physiological tissues. Furthermore, biophysical measurements of isolated nuclei and intact cells reveal that the nucleus is typically 2- to 10-times stiffer than the surrounding cytoplasm [11]. This combination of large size and relative rigidity of the nucleus led to the hypothesis that the nucleus can impact the cells’ ability to migrate [13]. Early support for this hypothesis came from work on tumor cells migrating through microfabricated channels with precisely defined constrictions [14–16] (see Box 1 for more information on such devices). While moderate confinement results in increased migration speed by allowing cells to employ faster migration modes (e.g., ‘amoeboid migration’ and ‘chimneying’) than during 2-D migration [17], constrictions below approximately 5 µm in diameter require substantial nuclear deformation and result in reduced migration speeds [14–16,18–20]. A seminal study by Friedl, Wolf, and colleagues using a range of cell types demonstrated that nuclear deformability presents a physical limit for the migration through collagen matrices with varying pore sizes [10]. When inhibiting matrix metalloprotease (MMP) activity required to degrade ECM, migration speed declined with decreasing pore size as nuclei had to undergo increasing deformation [10]. At pore sizes smaller than 10% of the non-deformed cross-section of the nucleus, cells reached a ‘nuclear deformation limit’ resulting in complete migration arrest, despite continued protrusion of the cytoplasm [10]. Subsequent studies using a variety of cell lines and experimental assays ranging from microfluidic devices, membranes with defined pores, ECM matrices, and in vivo xenografts have painted a similar picture, in which the deformability of the nucleus limits the cell’s ability to pass through tight spaces, reducing or even stalling migration as the pore size decreases below the cross-section of the nucleus [18–26]. Assessing the role of specific physical factors on cell migration in confined environments, Lautscham and colleagues [20] found that increased nuclear (but not cytoplasmic) volume, increased nuclear stiffness, reduced cell adhesion and lower cell contractility impaired migration through microfluidic constrictions. While the above findings prove common to a large variety of cell lines, including neutrophils, fibroblasts, and tumor cells, the exact degree of confinement necessary to elicit such effects, and the magnitude of the effect, varies with cell type. These differences indicate that variation in nuclear deformability, or the cytoskeletal forces applied to the nucleus, may be important modulators of the ‘nuclear barrier’ effect.
Box 1. Development of tools to study migration in confined environments.
Microfabrication techniques are finding increased application to study cell migration in confined environments, complementing existing approaches such as transwell plates and collagen invasion assays [10,46,77,78]. Besides micropillars, polymer scaffolds, and electrospun matrices [25,26,79,80], microfluidic devices made from polydimethylsiloxane (PDMS) by soft lithography [81,82] have proven particularly powerful in investigating cell migration through tight spaces by providing precisely defined microscale structures and constrictions with cross-sections from 100 µm2 to less than 5 µm2 [15–21,23,24,64,66,83]. These devices, which often include features to apply stable chemotactic gradients [14–16,18,19,24], allow for user defined geometries ranging from simple straight channels [15,16,24,66] to more intricate designs mimicking physiological environments [18–21,23,83]. The migration devices can be functionalized with a variety of ECM proteins to control cell adhesion. Since the devices are made of transparent PDMS and mounted on thin coverslips, they provide superb imaging conditions for live-cell imaging with high spatial and temporal resolution [81,82].
Lamins determine nuclear deformability and migration through confined environments
The deformability of the nucleus is largely determined by two components, the nuclear lamin network and chromatin [27,28]. Lamins are type V nuclear intermediate filaments that can be divided into two sub-types, A-type (A, C, C2) and B-type (B1-3) lamins [29–36]. The different lamin subtypes form separate but interdigitating fibrillar networks at the nuclear periphery [37,38]. In addition to regulating nuclear shape and stiffness [27,39–42], they play important roles in chromatin organization, DNA damage repair, and transcriptional regulation [37,43,44]. Cell-stretching and micropipette aspiration experiments indicate that A-type lamins have a larger impact on nuclear stiffness than B-type lamins—nuclear stiffness strongly scales with expression of lamins A/C [39–42], although increased expression of lamin B1 can also increase nuclear rigidity [45]. Consistent with the ‘nuclear barrier’ hypothesis, recent studies found that cells with reduced levels of lamins A/C have more deformable nuclei and migrate faster through tight spaces than control cells with normal lamin A/C levels [18,46]. Conversely, increased expression of lamin A, or expression of a mutant lamin (progerin) that increases nuclear stiffness, impaired transit through narrow constrictions [21,25]. Loss of lamin A/C promotes cell migration through small constrictions by allowing larger nuclear deformation, rather than increased nuclear compression, as the nuclear volume does not decrease during nuclear translocation [19]. These findings have direct physiological and clinical relevance, since downregulation of lamins A/C during granulopoiesis is critical for the ability of neutrophils to pass through micron-sized constrictions [21], and misregulation of lamins is common to many cancers [47] (see Box 2 for more information). Less is known about the role of B-type lamins in 3-D migration. Loss of B-type lamins impairs migration of neurons, which lack A-type lamins, in the developing brain, and this effect is thought to be caused by defects connecting the nuclear interior and cytoplasm [48,49]. Given recent reports that lamin A/C levels and organization can vary in response to substrate stiffness and cytoskeletal tension [41,50,51], it is intriguing to speculate that cells could also dynamically adjust their nuclear stiffness during migration.
Box 2. Lamins, nuclear shape, and disease.
Mutations in lamins cause a large spectrum of human diseases, ranging from muscular dystrophy and dilated cardiomyopathy to premature aging [84,85]. Increasing reports indicate that altered lamin expression (rather than mutations) is found in many cancers, and often correlates with negative clinical outcomes [47,86,87]. For example, reduced expression of lamin A/C has been reported in breast [88,89] and cervical cancer [90], and is correlated with an increased recurrence of stage II and III colon cancer [91] and reduced disease free survival in breast cancer [89]. However, in other cases, increased A-type lamin expression is associated with disease progression, specifically in prostate, colon and ovarian cancers [92,93]. Given the multiple function of lamins, changes in their expression are expected to have pleiotropic effects, affecting not only nuclear stiffness but also proliferation, survival, and gene expression [47,86,87,93,94]. Nonetheless, it is intriguing that more invasive breast cancer cell lines such as Hs578T and MDA-MB-231 are capable of more extensive nuclear deformation than normal and non-malignant controls [95], and move faster through migration devices designed to mimic tight spaces inside the body [15]. Future studies should be directed at characterizing the effects of altered lamin levels on cell migration, as well as other cellular functions, in more detail.
The role of chromatin in nuclear deformability and migration
Chromatin, consisting of DNA wrapped around histone octamers, occupies most of the nuclear interior and contributes to the viscoelastic response of nuclear deformation [28,52]. Chromatin exists in two configurations: (i) open ‘euchromatin’, which is typically transcriptionally active, and (ii) closed, more compact ‘heterochromatin’, which is associated with inactive genes [53]. Promoting euchromatin over heterochromatin organization, for example by treatment with the deacetylase inhibitor trichostatin A (TSA), results in softer and more deformable nuclei [52]. Interestingly, treatment with 5′-deoxy-5′-methylthioadenosine (MTA), a methyltransferase inhibitor that cause de-condensation of chromatin, impairs the migration of the cells through microchannels [15]. It remains unclear whether this counterintuitive effect was due to the increase in nuclear size resulting from chromatin de-condensation, which may counteract reduced nuclear stiffness [20], or due to altered transcriptional regulation, motivating future research on the role of chromatin organization in cell migration through confining constrictions.
Cytoskeletal forces pulling or pushing on the nucleus
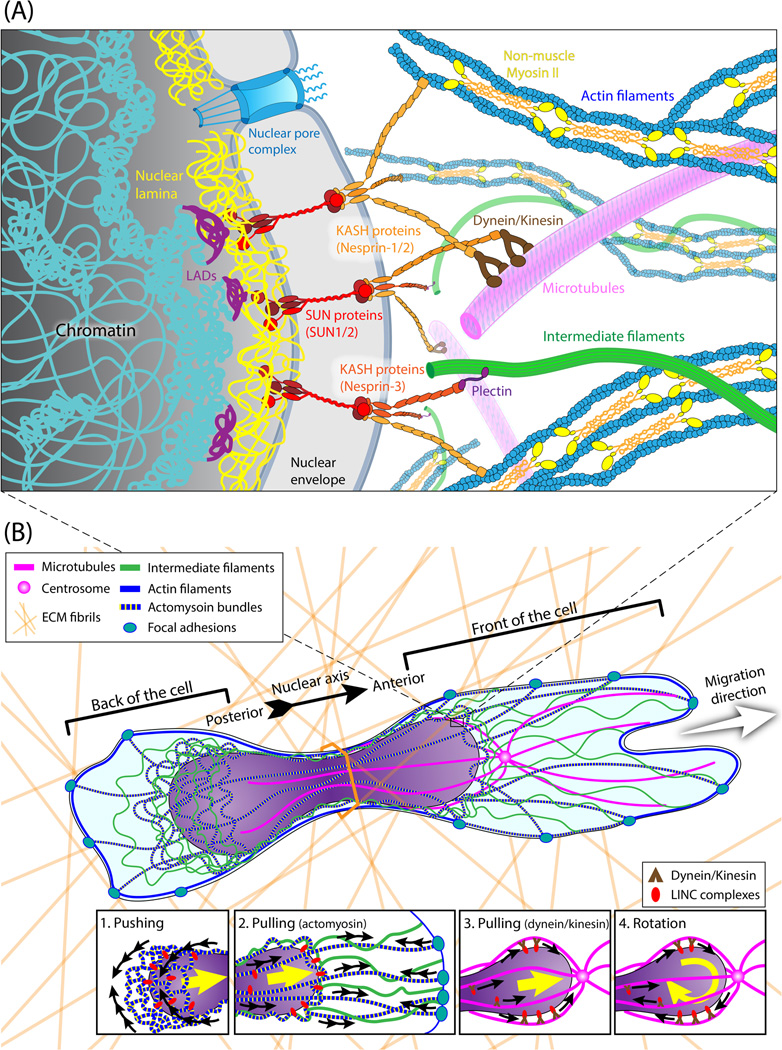
Recent studies have shed light onto the molecular components physically connecting the nucleus and cytoskeleton (Fig. 1A), revealing that the linker of nucleoskeleton and cytoskeleton (LINC) complex is the ‘clutch’ that enables transmission of mechanical force across the nuclear envelope [54]. We refer the reader to excellent recent reviews [55,56] and the article by Burridge et al. in this issue for more details on LINC complex function. However, the mechanism by which cells move the nucleus through confining constrictions, including whether cells predominantly pull or push on the nucleus (Fig. 1B), remains incompletely understood. It is likely that cells apply a varying combination of both mechanisms, depending on the specific conditions.
Figure 1. Schematic overview of the physical connections between the nucleus and cytoskeleton, and their roles in moving the nucleus through confined spaces.
(A) At the nuclear periphery, chromatin interacts with lamins at lamina-associated domains (LADs). SUN domain proteins (SUN1 and SUN2) are anchored to the nuclear lamina and other components of the nuclear interior by their C-terminus. The N-terminal luminal long stalks and SUN domains of SUN1/2 form trimers that interact with KASH domain proteins located in the outer nuclear membrane (nesprin-1/-2/-3, along with the cell-type specific nesprin-4 and KASH5), forming the LINC complex [101]. The strong interaction between SUN domain trimers and the KASH domains provide the basis to mechanically couple the nuclear interior with the cytoskeleton—nesprins interact directly with actomyosin bundles, or indirectly with microtubules and intermediate filaments via intermediary proteins (such as kinesin, dynein, plectin) [55]. Mechanical force transmission via nucleo-cytoskeletal coupling may also trigger mechanotransduction events, ranging from the recruitment of lamins to the LINC complex to changes in chromatin organization and gene expression, which may further impact cell migration processes. (B) Cytoskeletal organization and dynamics during migration in confined 3-D environments. As the cell passes through narrow pores, the nucleus separates the cell into front and back ends. The insets depict proposed mechanisms by which the cytoskeleton translocates the nucleus through confined spaces, including pushing via actomyosin contraction at the posterior of the nucleus (1), pulling via actomyosin contraction facilitated by intermediate filaments (2), pulling via microtubule-associated motors (3), and rotation via microtubule-associated motors (4).
Pulling the nucleus forward
In most migrating cells, the nucleus is positioned rearward, i.e., in the back of the cell [55]. Recent studies indicate that actomyosin contractility, possibly in combination with intermediate filaments [57,58], physically pull the nucleus forward during 2-D and 3-D migration [10,59]. In 3-D lamellipodial-based migration, actomyosin contractility and integrin-mediated traction at the leading edge are required to translocate the nucleus through narrow constrictions [10]. During 3-D lobopodial-based migration, non-muscle myosin IIA (NMIIA)-containing actomyosin bundles work with vimentin filaments to pull the nucleus forward by binding to nesprin-3α of the LINC complex via plectin [58,60]. Whereas microtubule associated motors are dispensable for nuclear translocation in 2-D migration [59], the microtubule-associated motors dynein/kinesin, which can directly bind to LINC complex proteins [56], are essential for interkinetic nuclear migration in neurons [61]. Dynein and kinesis are also required for nuclear positioning in multinucleated myotubes, where nuclei are squeezed, rotated and translocated to form proper myotube structure [62]. Thus, it is possible that microtubule-associated motors also participate in nuclear translocation during 3D cell migration.
Pushing the nucleus forward
Actomyosin contraction at the back of the cell is required for squeezing the leukocyte nucleus through narrow pores [63]. In breast and brain cancer cells, non-muscle myosin IIB (NMIIB) is recruited to the perinuclear cytoskeleton and posterior of the cell, possibly via nesprin-2, where it applies pushing forces to the nucleus to facilitate nuclear translocation through confining 3-D environments [64,65]. Depletion of NMIIB, but not NMIIA, impairs migration of breast cancer cells through microfluidic constrictions and dense collagen matrices [64], suggesting a specific role of perinuclear NMIIB actin networks in squeezing the nucleus through constrictions. Nonetheless, further studies are required to determine whether NMIIB-dependent actomyosin contraction constitutes a general mechanism for nuclear translocation, whether it also involves nesprin-3α and vimentin filaments, and whether increased pulling forces can compensate for impaired pushing forces and vice versa.
Physical compartmentalization by the nucleus
During migration through confined spaces, the nucleus takes up most of the space within the constriction, thus leaving little or no room for cytoplasmic transport around the nucleus and effectively dividing the cytoplasm into “front” and “back” compartments (Fig. 1B). This intracellular compartmentalization plays an important role in two mechanisms: osmotic pressure difference caused by water permeation, and intracellular pressure generated by the nucleus as an active ‘piston’ [60,66]. Polarized distribution of Na+/H+ pumps and aquaporins on the plasma membrane at the front and back of the cell alters water permeation and can drive migration and nuclear translocation, even when actin polymerization is inhibited [66]. However, since this osmotic pressure-based mechanism has only been described in tube-like microchannels that restrict fluid flow to the front and back of the cells, its relevance in more physiological environments still needs to be examined. Work by the Yamada group [60] reveals that during lobopodial-based migration, actomyosin-mediated pulling of the nucleus compresses the front of the cell, similar to a piston in a cylinder, building up higher intracellular pressure within this compartment and driving the formation of new lobopodial protrusions at the leading edge. The nucleus may similarly serve as a counterbalance for the directional protrusion of invadopodia, causing visible indentation of the basal nuclear surface [67]. Taken together, these studies illustrate how cells can take advantage of the large and rigid nucleus to promote cell migration in 3-D environments.
Biological consequences of nuclear deformation during 3-D cell migration
The nucleus is not only a passive mechanical element, but also houses the cell’s genomic information and is the site of DNA replication, transcription, and RNA processing. Thus, the substantial mechanical forces and deformation incurred during migration through tight spaces may have severe biological consequences that impact cellular function and viability.
Influences on cell survival and genomic stability
Recent studies have produced conflicting results on the effect of cell migration through tight spaces on cell viability. Whereas one study found increased apoptosis after migration through small (3 µm) pores, particularly in lamin A/C-deficient cells [46], others reported no noticeable increase in cell death during confinement [17–19,21]. These apparent differences may reflect differences in cell type or mechanical confinement (e.g., porous membranes vs. microfluidic devices and collagen matrices). Interestingly, pharmacological inhibition of the heat shock protein 90 (HSP90), which facilitates DNA damage repair [68], results in increased cell death after passage through 3-µm diameter pores, suggesting that cells may have suffered DNA damage during migration that causes apoptosis when not adequately repaired [46]. Mechanical stress alone can induce DNA damage [69], but it remains to be seen whether the mechanical stress incurred during 3-D cell migration is sufficient to cause DNA damage.
Influences on mechanotransduction signaling and gene expression
Mechanical stress on the nucleus during migration may also trigger non-lethal changes, which could further affect cell migration and cellular function. Recent studies in which isolated nuclei were subjected to mechanical stress suggest that the nucleus itself can act as a mechanosensitive element. Force application via the LINC complex induces rapid phosphorylation of the inner nuclear membrane protein emerin, leading to recruitment of lamin A/C to the nuclear envelope and nuclear stiffening [70]. Shearing isolated nuclei causes partial unfolding of lamin A/C, exposing cryptic binding sites that could initiate mechanotransduction events [41]. Furthermore, changes in the mechanical microenvironment and force application to intact cells can induce chromatin remodeling [71,72] and dissociation of nuclear protein complexes [73], which could affect both nuclear deformability and gene expression (see [74] and [75] for a detailed discussion of nuclear mechanotransduction). While these reports suggest that nuclear deformation during 3-D migration could impact nuclear organization, chromatin remodeling, and gene expression, direct experimental evidence for this hypothesis is still missing.
Conclusions and Outlook
The nucleus has long been recognized as a central hub for genomic information and its processing. Work published over the last few years makes it clear that one has to also consider the physical impact of the nucleus on cellular function, particularly during migration in 3-D environments. The nucleus, with its large volume and relative rigidity, acts as physical barrier when cells encounter pore sizes smaller than the nuclear diameter, resulting in reduced migration efficiency or even complete migration arrest. The extent of this nuclear barrier effect is largely driven by the nuclear size and stiffness, which is governed by the levels of the nuclear envelope proteins lamin A/C and chromatin organization. These findings are particularly relevant to immune cells and to invading cancer cells, which move through tissues with pore sizes smaller than the size of the nucleus and often have altered expression of lamins and other nuclear envelope proteins. Despite recent advances, many open questions remain. For example, can cells dynamically adjust their nuclear stiffness to facilitate cell migration through tight spaces, possibly by phosphorylation and/or degradation of nuclear lamins? Are some cells particularly well-suited for 3-D migration, either by having more deformable nuclei or by pulling/pushing harder on the nucleus? What are the precise molecular mechanisms by which cells translocate the nucleus through tight constrictions? And what are the biological consequences resulting from the large nuclear deformations, which could include changes in chromatin organization, DNA damage, and altered gene expression? Such mechanically induced events could not only affect migration itself, e.g., by altering nuclear stiffness and cytoskeletal dynamics, but also impact various other cellular functions and even viability. To drive new discoveries, it will be crucial to combine new imaging tools, such as fluorescence resonance energy transfer (FRET)-based intracellular force probes [76], with microfabricated environments that mimic physiological environments while providing defined geometries and enhanced live-cell imaging conditions, as well as single cell based assays to measure cell viability, gene expression, and epigenetic modification. Further insights into the role of the nucleus in 3-D migration will not only improve our understanding of the physical constraints during migration in physiological environments, but may ultimately lead to new strategies to better target invasive cancer cells and to reduce or eliminate metastatic spreading.
Table 1.
Overview of relevant molecular components involved in nuclear mechanics and migration in confining environments.
| Components | Functions | References |
|---|---|---|
| Lamins |
|
[25] [37] [39] [40] [54] |
| Chromatin |
|
[28] [52] [96] [97] |
| LINC complexes |
|
[54] [55] [56] |
| Actomyosin bundles and non-muscle myosin II (NMII) |
|
[10] [57] [60] [63] [64] |
| Intermediate filaments |
|
[60] [98] |
| Integrins |
|
[10] [99] |
| Microtubules and dynein/kinesin motor proteins |
|
[55] [62] [100] |
Acknowledgments
The authors apologize to all authors whose work could not be cited due to space constraints. The authors thank Emily Bell and Peter DelNero for discussion and critical reading of the manuscript. This work was supported by awards from the National Institutes of Health [R01 HL082792], the National Science Foundation [CBET-1254846], the National Cancer Institute [U54CA143876], and the Department of Defense [Breast Cancer Idea Award BC102152], as well as a NSF Graduate Research Fellowship to ALM [DGE-1144153]. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended readings
- 1.Munjal A, Lecuit T. Actomyosin networks and tissue morphogenesis. Development. 2014;141:1789–1793. doi: 10.1242/dev.091645. [DOI] [PubMed] [Google Scholar]
- 2.Weninger W, Biro M, Jain R. Leukocyte migration in the interstitial space of nonlymphoid organs. Nat. Rev. Immunol. 2014;14:232–246. doi: 10.1038/nri3641. [DOI] [PubMed] [Google Scholar]
- 3.Chaffer CL, Weinberg Ra. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 4.Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doerschuk CM, Beyers N, Coxson HO, Wiggs B, Hogg JC. Comparison of neutrophil and capillary diameters and their relation to neutrophil sequestration in the lung. J. Appl. Physiol. 1993;74:3040–3045. doi: 10.1152/jappl.1993.74.6.3040. [DOI] [PubMed] [Google Scholar]
- 6.Stoitzner P, Pfaller K, Stössel H, Romani N. A close-up view of migrating Langerhans cells in the skin. J. Invest. Dermatol. 2002;118:117–125. doi: 10.1046/j.0022-202x.2001.01631.x. [DOI] [PubMed] [Google Scholar]
- 7.Weigelin B, Bakker G-J, Friedl P. Intravital third harmonic generation microscopy of collective melanoma cell invasion. IntraVital. 2012;1:32–43. doi: 10.4161/intv.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stetler-Stevenson WG, Aznavoorian S, Liotta LA. Tumor cell interactions with the extracellular matrix during invasion and metastasis. Annu. Rev. Cell Biol. 1993;9:541–573. doi: 10.1146/annurev.cb.09.110193.002545. [DOI] [PubMed] [Google Scholar]
- 9.Wolf K, Mazo I, Leung H, Engelke K, Von Andrian UH, Deryugina EI, Strongin AY, Bröcker EB, Friedl P. Compensation mechanism in tumor cell migration: Mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolf K, te Lindert M, Krause M, Alexander S, te Riet J, Willis aL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. This is the first study to directly demonstrate the concept of the nucleus as a physical barrier during 3-D cell migration, including in physiological collagen networks. Using a variety of cell lines, the authors show that when proteolytic ECM degradation is inhibited, cell migration speed through collagen matrices decreases with decreasing pore size, and cell migration is completely arrested when cell reach a ‘nuclear deformability limit’.
- 11.Lammerding J. Mechanics of the nucleus. Compr. Physiol. 2011;1:783–807. doi: 10.1002/cphy.c100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martins RP, Finan JD, Guilak F, Lee Da. Mechanical regulation of nuclear structure and function. Annu. Rev. Biomed. Eng. 2012;14:431–455. doi: 10.1146/annurev-bioeng-071910-124638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr. Opin. Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tong Z, Balzer EM, Dallas MR, Hung WC, Stebe KJ, Konstantopoulos K. Chemotaxis of cell populations through confined spaces at Single-Cell resolution. PLoS One. 2012;7:1–10. doi: 10.1371/journal.pone.0029211. This was one of the first studies to use a microfluidic device, including a self-sustaining chemotactic gradient, to study cell migration on a single-cell level. The device enabled the authors to test the effect of various extracellular matrix coatings applied to the microfluidic channels, as well as to deliver different phramcological drugs to the cells and to monitor the consequences on migration.
- 15. Fu Y, Chin LK, Bourouina T, Liu AQ, VanDongen AMJ. Nuclear deformation during breast cancer cell transmigration. Lab Chip. 2012;12:3774–3778. doi: 10.1039/c2lc40477j. This was one of the first studies to use a microfluidic device to study cell migration through confined spaces. The authors found that nuclear deformation was a rate-limiting step in breast cancer cell migration. When the authors treated the cells with an methyltransferase inhibitor, causing de-condensation of chromatin, they observed a reduction in cell migration, thereby linking changes in chromatin organization to cell migration efficiency.
- 16.Balzer EM, Tong Z, Paul CD, Hung WC, Stroka KM, Boggs AE, Martin SS, Konstantopoulos K. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–4056. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y-J, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuzé M, Takaki T, Voituriez R, Piel M. Confinement and Low Adhesion Induce Fast Amoeboid Migration of Slow Mesenchymal Cells. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 18. Davidson PM, Denais C, Bakshi MC, Lammerding J. Nuclear Deformability Constitutes a Rate-Limiting Step During Cell Migration in 3-D Environments. Cell. Mol. Bioeng. 2014;7:293–306. doi: 10.1007/s12195-014-0342-y. This study provides a detailed characterization of the effects of different levels of lamin A/C on migration through confined spaces. The authors examine lamin A/C-deficient and wild-type mouse embryo fibroblasts migrating through a novel microfluidic devices with precisely defined constrictions, finding that while in wild-type cells migration speed decreases when the cells encounter constrictions smaller than 25 µm2, cells completley lacking lamins A/C show no reduction in normalized migration speed, and heterozygous cells with 50% of normal lamin A/C levels show an intermediate phenotype.
- 19.Davidson PM, Sliz J, Isermann P, Denais CM, Lammerding J. Design of a microfluidic device to quantify dynamic intra- nuclear deformation during cell migration through confining environments. Integr. Biol. 2015;7:1534–1546. doi: 10.1039/c5ib00200a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lautscham LA, Kämmerer C, Lange JR, Kolb T, Mark C, Schilling A, Strissel PL, Strick R, Gluth C, Rowat AC, et al. Migration in Confined 3D Environments Is Determined by a Combination of Adhesiveness, Nuclear Volume, Contractility, and Cell Stiffness. Biophys. J. 2015;109:900–913. doi: 10.1016/j.bpj.2015.07.025. This study clearly provides a detailed characterization of multiple physical factors contributing to efficient 3-D cell migration in confined environments. Using microfluidic constriction channels and collagen matrix assays with multiple cell lines, the authors show that the velocity of cell migration through confining constrictions depends not only on cell stiffness, but also on nuclear volume, cell adhesiveness and contractility.
- 21. Rowat AC, Jaalouk DE, Zwerger M, Ung WL, Eydelnant IA, Olins DE, Olins AL, Herrmann H, Weitz DA, Lammerding J. Nuclear Envelope Composition Determines the Ability of Neutrophil-type Cells to Passage through Micron-scale Constrictions. J. Biol. Chem. 2013;288:8610–8618. doi: 10.1074/jbc.M112.441535. This study provided the first functional evidence that the downregulation of lamin A/C during granulopoiesis facilitates the transit of neutrophils in micron-sized constrictions during perfusion and migration, highlighting the physiological relevance of nuclear deformability and changes in lamin levels.
- 22.Guzman A, Ziperstein MJ, Kaufman LJ. The effect of fibrillar matrix architecture on tumor cell invasion of physically challenging environments. Biomaterials. 2014;35:6954–6963. doi: 10.1016/j.biomaterials.2014.04.086. [DOI] [PubMed] [Google Scholar]
- 23.Mak M, Reinhart-King Ca, Erickson D. Elucidating mechanical transition effects of invading cancer cells with a subnucleus-scaled microfluidic serial dimensional modulation device. Lab Chip. 2013;13:340–348. doi: 10.1039/c2lc41117b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malboubi M, Jayo A, Parsons M, Charras G. An open access microfluidic device for the study of the physical limits of cancer cell deformation during migration in confined environments. Microelectron. Eng. 2015;144:42–45. doi: 10.1016/j.mee.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Booth-Gauthier Ea, Du V, Ghibaudo M, Rape AD, Dahl KN, Ladoux B. Hutchinson- Gilford progeria syndrome alters nuclear shape and reduces cell motility in three dimensional model substrates. Integr. Biol. 2013;5:569–577. doi: 10.1039/c3ib20231c. This was the first study to assess the effect of a disease-causing lamin A mutation on cell migration through confining environments. The authors studied cells expressing progerin, the mutant form of lamin A responsible for Hutchinson-Gilford progeria syndrome, moving through an array of PDMS micropillars with decreasing spacing. Progerin expressing cells, which have stiffer nuclei, were were less motile, and eventually incapable of migrating through the decreasing spaces between the pillars, compared to normal control cells.
- 26.Greiner AM, Jäckel M, Scheiwe AC, Stamow DR, Autenrieth TJ, Lahann J, Franz CM, Bastmeyer M. Multifunctional polymer scaffolds with adjustable pore size andchemoattractant gradients for studying cell matrix invasion. Biomaterials. 2014;35:611–619. doi: 10.1016/j.biomaterials.2013.09.095. [DOI] [PubMed] [Google Scholar]
- 27.Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 2004;117:4779–4786. doi: 10.1242/jcs.01357. [DOI] [PubMed] [Google Scholar]
- 28.Dahl KN, Engler AJ, Pajerowski JD, Discher DE. Power-Law Rheology of Isolated Nuclei with Deformation Mapping of Nuclear Substructures. Biophys. J. 2005;89:2855–2864. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher DZ, Chaudhary N, Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. U. S. A. 1986;83:6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKeon FD, Kirschner MW, Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- 31.Furukawa K, Inagaki H, Hotta Y. Identification and cloning of an mRNA coding for a germ cell-specific A-type lamin in mice. Exp. Cell Res. 1994;212:426–430. doi: 10.1006/excr.1994.1164. [DOI] [PubMed] [Google Scholar]
- 32.Peter M, Kitten GT, Lehner CF, Vorburger K, Bailer SM, Maridor G, Nigg Ea. Cloning and sequencing of cDNA clones encoding chicken lamins A and B1 and comparison of the primary structures of vertebrate A- and B-type lamins. J. Mol. Biol. 1989;208:393–404. doi: 10.1016/0022-2836(89)90504-4. [DOI] [PubMed] [Google Scholar]
- 33.Vorburger K, Lehner CF, Kitten GT, Eppenberger HM, Nigg EA. A second higher vertebrate B-type lamin. cDNA sequence determination and in vitro processing of chicken lamin B2. J. Mol. Biol. 1989;208:405–415. doi: 10.1016/0022-2836(89)90505-6. [DOI] [PubMed] [Google Scholar]
- 34.Lin F, Worman HJ. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics. 1995;27:230–236. doi: 10.1006/geno.1995.1036. [DOI] [PubMed] [Google Scholar]
- 35.Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- 36.Machiels BM, Zorenc AHG, Endert JM, Kuijpers HJH, Van Eys GJJM, Ramaekers FCS, Broers JLV. An alternative splicing product of the lamin A/C gene lacks exon 10. J. Biol. Chem. 1996;271:9249–9253. doi: 10.1074/jbc.271.16.9249. [DOI] [PubMed] [Google Scholar]
- 37.Shimi T, Pfleghaar K, Kojima S, Pack C-G, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, et al. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA, Zheng Y, Jaqaman K, Goldman RD. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell. 2015;26:4075–4086. doi: 10.1091/mbc.E15-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins a and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 41.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PCDP, Pinter J, Pajerowski JD, Spinler KR, Shin J-W, Tewari M, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schäpe J, Prausse S, Radmacher M, Stick R. Influence of lamin A on the mechanical properties of amphibian oocyte nuclei measured by atomic force microscopy. Biophys. J. 2009;96:4319–4325. doi: 10.1016/j.bpj.2009.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho CY, Lammerding J. Lamins at a glance. J. Cell Sci. 2012;125:2087–2093. doi: 10.1242/jcs.087288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 45.Ferrera D, Canale C, Marotta R, Mazzaro N, Gritti M, Mazzanti M, Capellari S, Cortelli P, Gasparini L. Lamin B1 overexpression increases nuclear rigidity in autosomal dominant leukodystrophy fibroblasts. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2014;28:3906–3918. doi: 10.1096/fj.13-247635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PCDP, Ivanovska IL, Discher DE. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 2014;204:669–682. doi: 10.1083/jcb.201308029. This is the first study to demonstrate that while reduced levels of lamin A/C can promote migration through narrow constrictions, the increased nuclear deformability/fragility makes cells more sensitive to migration-induced mechanical stress. Using fibronectin-coated transwell assays and mouse xenografts, the authors show that intermediate A:B-type lamin ratio promotes cell migration both in vitro and in vivo, while deep knockdown of A-type lamin results in increased apoptosis in cells migrating through 3-µm diameter pores, and cell survival can be further diminished by treatment with an HSP90 inhibitor.
- 47. Hutchison CJ. Do lamins influence disease progression in cancer? Adv. Exp. Med. Biol. 2014;773:593–604. doi: 10.1007/978-1-4899-8032-8_27. The author provides a comprehensive review that summarizes and compares recent findings of altered lamin expression in a wide variety of cancers, while also discussing potential mechanisms by which lamins may impact cancer progression and metastasis.
- 48.Coffinier C, Chang SY, Nobumori C, Tu Y, Farber Ea, Toth JI, Fong LG, Young SG. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5076–5081. doi: 10.1073/pnas.0908790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young SG, Jung H-J, Lee JM, Fong LG. Nuclear Lamins and Neurobiology. Mol. Cell. Biol. 2014;34:2776–2785. doi: 10.1128/MCB.00486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ihalainen TO, Aires L, Herzog FA, Schwartlander R, Moeller J, Vogel V. Differential basal-to-apical accessibility of lamin A/C epitopes in the nuclear lamina regulated by changes in cytoskeletal tension. Nat. Mater. 2015;14:1252–1261. doi: 10.1038/nmat4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buxboim A, Swift J, Irianto J, Spinler KR, Dingal PCDP, Athirasala A, Kao Y-RC, Cho S, Harada T, Shin J-W, et al. Matrix Elasticity Regulates Lamin-A,C Phosphorylation and Turnover with Feedback to Actomyosin. Curr. Biol. 2014;24:1909–1917. doi: 10.1016/j.cub.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eissenberg JC, Elgin SCR. eLS. John Wiley & Sons, Ltd; 2001. Heterochromatin and Euchromatin. [Google Scholar]
- 54.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152:1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 2015;208:11–22. doi: 10.1083/jcb.201409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dupin I, Sakamoto Y, Etienne-Manneville S. Cytoplasmic intermediate filaments mediate actin-driven positioning of the nucleus. J. Cell Sci. 2011;124:865–872. doi: 10.1242/jcs.076356. [DOI] [PubMed] [Google Scholar]
- 58.Ketema M, Kreft M, Secades P, Janssen H, Sonnenberg A. Nesprin-3 connects plectin and vimentin to the nuclear envelope of Sertoli cells but is not required for Sertoli cell function in spermatogenesis. Mol. Biol. Cell. 2013;24:2454–2466. doi: 10.1091/mbc.E13-02-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu J, Kent Ia, Shekhar N, Chancellor TJ, Mendonca A, Dickinson RB, Lele TP. Actomyosin pulls to advance the nucleus in a migrating tissue cell. Biophys. J. 2014;106:7–15. doi: 10.1016/j.bpj.2013.11.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345:1062–1065. doi: 10.1126/science.1256965. This is the first study to demonstrate that the nucleus can act as a “piston” to drive 3-D cell migration and cytoplasmic protrusion at the leading edge. By measuring the intracellular pressure at the front and back of the migrating cell, the authors show that the nucleus is pulled forward by actomyosin bundles and vimentin filaments, which pressurizes the front compartment to drive lobopodia formation at the leading edge.
- 61.Tsai J-W, Lian W-N, Kemal S, Kriegstein AR, Vallee RB. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat. Neurosci. 2010;13:1463–1471. doi: 10.1038/nn.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson MH, Holzbaur ELF. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J. Cell Sci. 2012;125:4158–4169. doi: 10.1242/jcs.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lämmermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Söldner R, Hirsch K, Keller M, Förster R, Critchley DR, Fässler R, et al. Rapid leukocyte migration by integrinindependent flowing and squeezing. Nature. 2008;453:51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 64. Thomas DG, Yenepalli A, Denais CM, Rape A, Beach JR, Wang Y-L, Schiemann WP, Baskaran H, Lammerding J, Egelhoff TT. Non-muscle myosin IIB is critical for nuclear translocation during 3D invasion. J. Cell Biol. 2015;210:583–594. doi: 10.1083/jcb.201502039. This is the first study that demonstrates a specific role of NMIIB’s in nuclear transloation during 3-D cell migration. Using a microfluidic migration device, traction force microscopy, and collagen invasion assays, the authors show that during 3-D migration, NMIIA is critical for traction force generation at front end of cells, while NMIIB at the back of the cell is crucial for pushing the nucleus through constrictions.
- 65.Ivkovic S, Beadle C, Noticewala S, Massey SC, Swanson KR, Toro LN, Bresnick AR, Canoll P, Rosenfeld SS. Direct inhibition of myosin II effectively blocks glioma invasion in the presence of multiple motogens. Mol. Biol. Cell. 2012;23:533–542. doi: 10.1091/mbc.E11-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stroka KM, Jiang H, Chen S-H, Tong Z, Wirtz D, Sun SX, Konstantopoulos K. Water permeation drives tumor cell migration in confined microenvironments. Cell. 2014;157:611–623. doi: 10.1016/j.cell.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Revach O-Y, Weiner A, Rechav K, Sabanay I, Livne A, Geiger B. Mechanical interplay between invadopodia and the nucleus in cultured cancer cells. Sci. Rep. 2015;5:9466. doi: 10.1038/srep09466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dote H, Burgan WE, Camphausen K, Tofilon PJ. Inhibition of hsp90 compromises the DNA damage response to radiation. Cancer Res. 2006;66:9211–9220. doi: 10.1158/0008-5472.CAN-06-2181. [DOI] [PubMed] [Google Scholar]
- 69.Mayr M, Hu YH, Hainaut P, Xu QB. Mechanical stress-induced DNA damage and racp38MAPK signal pathways mediate p53-dependent apoptosis in vascular smooth muscle cells. Faseb J. 2002;16:1423–1425. doi: 10.1096/fj.02-0042fje. [DOI] [PubMed] [Google Scholar]
- 70. Guilluy C, Osborne LD, Van LL, Sharek L, Superfine R, Garcia-Mata R, Burridge K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat.Cell Biol. 2014;16:376–381. doi: 10.1038/ncb2927. This is the first study to provide direct evidence that the nucleus itself can act as a mechanosensor. The authors use magnetic beads coated with an anti-nesprin antibody to apply mechanical force to isolated nuclei, while measuring the induced bead displacement and assessing changes in nuclear envelope proteins. The experiments reveal that force application to the nucleus via the LINC complex causes nuclear stiffening, phosphorylation of emerin by Src, recruitment of lamin A/C to the nuclear envelope. These nuclear mechanotransduction processes require lamin A/C and emerin, but are independent of chromatin and nuclear actin rearrangments.
- 71.Li Y, Chu JS, Kurpinski K, Li X, Bautista DM, Yang L, Paul Sung KL, Li S. Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophys. J. 2011;100:1902–1909. doi: 10.1016/j.bpj.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iyer KV, Pulford S, Mogilner a, Shivashankar GV. Mechanical activation of cells induces chromatin remodeling preceding MKL nuclear transport. Biophys. J. 2012;103:1416–1428. doi: 10.1016/j.bpj.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poh Y-C, Shevtsov SP, Chowdhury F, Wu DC, Na S, Dundr M, Wang N. Dynamic forceinduced direct dissociation of protein complexes in a nuclear body in living cells. Nat. Commun. 2012;3:866. doi: 10.1038/ncomms1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaminski A, Fedorchak GR, Lammerding J. The cellular mastermind(?)- mechanotransduction and the nucleus. Prog. Mol. Biol. Transl. Sci. 2014;126:157–203. doi: 10.1016/B978-0-12-394624-9.00007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guilluy C, Burridge K. Nuclear mechanotransduction: forcing the nucleus to respond. Nucleus. 2015;6:19–22. doi: 10.1080/19491034.2014.1001705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cost A-L, Ringer P, Chrostek-Grashoff A, Grashoff C. How to Measure Molecular Forces in Cells: A Guide to Evaluating Genetically-Encoded FRET-Based Tension Sensors. Cell. Mol. Bioeng. 2015;8:96–105. doi: 10.1007/s12195-014-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Haeger A, Krause M, Wolf K, Friedl P. Cell jamming: Collective invasion of mesenchymal tumor cells imposed by tissue confinement. Biochim. Biophys. Acta - Gen. Subj. 2014;1840:2386–2395. doi: 10.1016/j.bbagen.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 79.Meehan S, Nain AS. Role of Suspended Fiber Structural Stiffness and Curvature on Single-Cell Migration, Nucleus Shape, and Focal-Adhesion-Cluster Length. Biophys. J. 2014;107:2604–2611. doi: 10.1016/j.bpj.2014.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qin S, Ricotta V, Simon M, Clark RaF, Rafailovich MH. Continual Cell Deformation Induced via Attachment to Oriented Fibers Enhances Fibroblast Cell Migration. PLoS One. 2015;10:e0119094. doi: 10.1371/journal.pone.0119094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duncombe TA, Tentori AM, Herr AE. Microfluidics: reframing biological enquiry. Nat. Rev. Mol. Cell Biol. 2015;16:554–567. doi: 10.1038/nrm4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lautenschläger F, Piel M. Microfabricated devices for cell biology: all for one and one for all. Curr. Opin. Cell Biol. 2013;25:116–124. doi: 10.1016/j.ceb.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 83.Lange JR, Steinwachs J, Kolb T, Lautscham LA, Harder I, Whyte G, Fabry B. Microconstriction Arrays for High-Throughput Quantitative Measurements of Cell Mechanical Properties. Biophys. J. 2015;109:26–34. doi: 10.1016/j.bpj.2015.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schreiber KH, Kennedy BK. When Lamins Go Bad: Nuclear Structure and Disease. Cell. 2013;152:1365–1375. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Davidson PM, Lammerding J. Broken nuclei – lamins, nuclear mechanics, and disease. Trends Cell Biol. 2014;24:247–256. doi: 10.1016/j.tcb.2013.11.004. This review provides a concise overview of the importance of lamins in health and disease, integrating lamin’s various roles on nuclear structure, mechanotransduction signaling, and gene regulation.
- 86.Krause M, Wolf K. Cancer cell migration in 3D tissue: Negotiating space by proteolysis and nuclear deformability. Cell Adh. Migr. 2015;9:357–366. doi: 10.1080/19336918.2015.1061173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Denais C, Lammerding J. Nuclear mechanics in cancer. Adv. Exp. Med. Biol. 2014;773:435–470. doi: 10.1007/978-1-4899-8032-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Capo-chichi CD, Cai KQ, Smedberg J, Azar PG-A, Godwin AK, Xu X-X. Original Article Materials and Methods Tumor specimens, human primary Small interfering RNA transfection (shRNA and siRNA) Flow cytometry analysis Probes for Northern blot and antibodies for. Chin. J. Cancer. 2011;30:415–425. [Google Scholar]
- 89.Wazir U, Ahmed MH, Bridger JM, Harvey A, Jiang WG, Sharma AK, Mokbel K. The clinicopathological significance of lamin A/C, lamin B1 and lamin B receptor mRNA expression in human breast cancer. Cell. Mol. Biol. Lett. 2013;18:595–611. doi: 10.2478/s11658-013-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Capo-chichi CD, Aguida B, Chabi NW, Cai QK, Offrin G, Agossou VK, Sanni A, Xu X-X. Lamin A/C deficiency is an independent risk factor for cervical cancer. Cell. Oncol. 2015 doi: 10.1007/s13402-015-0252-6. [DOI] [PubMed] [Google Scholar]
- 91.Belt EJT, Fijneman RJA, van den Berg EG, Bril H, Delis-van Diemen PM, Tijssen M, van Essen HF, de Lange-de Klerk ESM, Beliën JAM, Stockmann HBAC, et al. Loss of lamin A/C expression in stage II and III colon cancer is associated with disease recurrence. Eur. J. Cancer. 2011;47:1837–1845. doi: 10.1016/j.ejca.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 92.Willis ND, Cox TR, Rahman-Casañs SF, Smits K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M, Wilson RG, de Bruïne A, et al. Lamin A/C is a risk biomarker in colorectal cancer. PLoS One. 2008;3:e2988. doi: 10.1371/journal.pone.0002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chow K-H, Factor RE, Ullman KS. The nuclear envelope environment and its cancer connections. Nat. Rev. Cancer. 2012;12:196–209. doi: 10.1038/nrc3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mitchell MJ, Denais C, Chan MF, Wang Z, Lammerding J, King MR. Lamin A/C deficiency reduces circulating tumor cell resistance to fluid shear stress. Am. J. Physiol. - Cell Physiol. 2015;309:C736–C746. doi: 10.1152/ajpcell.00050.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chiotaki R, Polioudaki H, Theodoropoulos P a. Differential nuclear shape dynamics of invasive and non-invasive breast cancer cells are associated with actin cytoskeleton organization and stability. 2014;295:287–295. doi: 10.1139/bcb-2013-0120. [DOI] [PubMed] [Google Scholar]
- 96.Jain N, Iyer KV, Kumar A, Shivashankar GV. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc. Natl. Acad. Sci. U. S. A. 2013;110:11349–11354. doi: 10.1073/pnas.1300801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, Van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petrie RJ, Yamada KM. Fibroblasts Lead the Way: A Unified View of 3D Cell Motility. Trends Cell Biol. 2015;25:666–674. doi: 10.1016/j.tcb.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 2012;197:439–455. doi: 10.1083/jcb.201201124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Levy JR, Holzbaur ELF. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 2008;121:3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sosa Ba, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]