Hybrid zones that occur across environmental gradients provide excellent opportunities for studying the maintenance of divergent adaptations in the presence of gene flow. They also provide insight into the biodiversity implications of future species contact and hybridization in a changing world. We studied divergent morphology between two Senecio (ragwort) species that form a natural hybrid zone with respect to elevation on Mount Etna, Italy, using a quantitative trait locus (QTL) mapping approach. We found signals of divergent selection with increased genetic differentiation close to QTLs. Extensive interactions between QTLs and traits suggested a QTL architecture that is resistant to hybridization.
Keywords: Genetic differentiation, hybridization, phenotypic divergence, QTL architecture, QTL interactions, selection, speciation
Abstract
Knowledge of the genetic basis of phenotypic divergence between species and how such divergence is caused and maintained is crucial to an understanding of speciation and the generation of biodiversity. The hybrid zone between Senecio aethnensis and S. chrysanthemifolius on Mount Etna, Sicily, provides a well-studied example of species divergence in response to conditions at different elevations, despite hybridization and gene flow. Here, we investigate the genetic architecture of divergence between these two species using a combination of quantitative trait locus (QTL) mapping and genetic differentiation measures based on genetic marker analysis. A QTL architecture characterized by physical QTL clustering, epistatic interactions between QTLs, and pleiotropy was identified, and is consistent with the presence of divergent QTL complexes resistant to gene flow. A role for divergent selection between species was indicated by significant negative associations between levels of interspecific genetic differentiation at mapped marker gene loci and map distance from QTLs and hybrid incompatibility loci. Within-species selection contributing to interspecific differentiation was evidenced by negative associations between interspecific genetic differentiation and genetic diversity within species. These results show that the two Senecio species, while subject to gene flow, maintain divergent genomic regions consistent with local selection within species and selection against hybrids between species which, in turn, contribute to the maintenance of their distinct phenotypic differences.
Introduction
Speciation commonly proceeds through genetic divergence between populations that ultimately become reproductively isolated from each other due to intrinsic and/or extrinsic breeding barriers (Orr and Turelli 2001; Coyne and Orr 2004; Smadja and Butlin 2011; Nosil and Feder 2012). Phenotypic trait divergence usually accompanies this process, often as a result of adaptation to different environments (Nosil 2012). Understanding how phenotypic trait divergence evolves between populations and is maintained between hybridizing species requires knowledge of the genetic basis of divergent traits and how selection acts on genes controlling these traits (Rieseberg et al. 2003; Lexer et al. 2005; Nosil et al. 2009; Nosil and Feder 2012).
Quantitative trait locus (QTL) analysis is a powerful way of analysing the genetic basis of divergent traits between species (Rieseberg et al. 2003; Lexer et al. 2005; Bouck et al. 2007; Taylor et al. 2012; Rogers et al. 2013). It involves determination of the number and primary effects of QTLs, their genomic locations, the interactions between them (epistasis) and their effects across multiple traits (pleiotropy). The QTL architecture of divergent traits revealed by such analysis is likely to be shaped by divergent selection acting against relatively unfit recombinant hybrid phenotypes (Bierne et al. 2011; Servedio et al. 2011; Abbott et al. 2013; Yeaman 2013), especially where divergence between species occurs in the presence of interspecific gene flow (Via and West 2008; Nosil et al. 2009; Yeaman and Whitlock 2011; Yeaman 2013). This selective scenario could favour the evolution of QTL hotspots, epistasis and pleiotropy as effective means of preserving local adaptation despite gene flow (Whiteley et al. 2008; Gagnaire et al. 2013; Lindtke and Buerkle 2015). Alternatively, recombination and break-up of QTL complexes could be reduced by close physical proximity of QTLs (Yeaman and Whitlock 2011; Jones et al. 2012; Yeaman 2013) or recombination ‘coldspots’ such as near centromeres or chromosomal rearrangements (Turner et al. 2005; Kirkpatrick and Barton 2006; Lowry and Willis 2010; Twyford and Friedman 2015).
Complementary insights into the relationship between QTL architecture and divergent selection can be obtained by investigating genetic diversity and differentiation among mapped molecular marker loci (Rogers and Bernatchez 2007; Stinchcombe and Hoekstra 2008; Gompert et al. 2012; Renaut et al. 2012; Strasburg et al. 2012; Cruikshank and Hahn 2014). Heterogeneous differentiation across the genome is expected to result from divergent selection in the presence of gene flow (Wu 2001; Feder and Nosil 2010) and has been reported in several studies of ecologically divergent pairs of taxa (Turner et al. 2005; Rogers and Bernatchez 2007; Via and West 2008). However, such patterns of differentiation can be highly dependent on the biology and demographic histories of the focal taxa (Jones et al. 2012; Renaut et al. 2012), and their assessment must take account of genetic diversity both within and between focal taxa (Cruikshank and Hahn 2014).
Here, we present a quantitative genetic analysis of divergent traits between two diploid (2n = 20), short-lived perennial, self-incompatible, herbaceous species of Senecio (Asteraceae), S. aethnensis and S. chrysanthemifolius, which grow at elevations above 2000 m and below 1000 m, respectively, on Mount Etna, Sicily. Whereas S. aethnensis produces large flower heads (capitula) and fruits, and entire (spathulate) leaves, S. chrysanthemifolius has smaller flower heads and fruits, and highly dissected (pinnatisect) leaves. The two species hybridize and form a hybrid zone at intermediate elevations on Mount Etna (James and Abbott 2005; Abbott and Brennan 2014). Although connected by hybrid populations, some barriers to interspecific gene flow are apparent in the field. For example, flowering times only partially overlap, with S. chrysanthemifolius flowering 6 weeks earlier (April–June) than S. aethnensis (July–September) (authors’ personal observation). A previous analysis of the hybrid zone showed that leaf shape, flower head structure and fruit structure exhibited steeper clines and/or shifts in cline position relative to a molecular genetic cline (Brennan et al. 2009). This was attributed to both intrinsic and extrinsic environmental selection against hybrids.
An improved understanding of the level of genetic divergence between the two species and the importance of selection in driving genomic divergence recently came from a comparison of their transcriptomes (Chapman et al. 2013). This showed that genome-wide genetic differentiation between the species was low, with only 2.25 % of 8854 loci tested having been subject to divergent selection. Genetic maps for the two Senecio species based on segregation of molecular markers in F2 mapping families (Brennan et al. 2014; Chapman et al. 2016) indicated that large genomic rearrangements were not a cause of reduced fitness in hybrids. However, many markers (∼27 % of 127 maker loci tested, Brennan et al. 2014) exhibited significant transmission ratio distortion (TRD) in the F2 family and clusters of TRD loci (TRDLs) were distributed across multiple linkage groups. This frequency of TRD was similar to that found in other crossing studies involving distinct ‘species’ (e.g. 49 and 33 % in Mimulus and Iris, respectively, Fishman et al. 2001; Taylor et al. 2012). Such extensive genomic incompatibility between the two species would be expected to affect the genetic structure of the hybrid zone on Mount Etna by limiting interspecific gene flow and promoting divergence across the genome. Chapman et al. (2016) further showed that loci exhibiting significant sequence or expression differentiation between the two species had a clustered distribution when placed on the map and several QTLs for species phenotypic differences coincided with these regions.
Here, we investigate further the genetic architecture of phenotypic trait differences and associated divergent selection acting on S. aethnensis and S. chrysanthemifolius by performing a QTL analysis of multiple quantitative traits that distinguish the two species. Our analysis examined additional traits and a larger mapping family relative to the recent study by Chapman et al. (2016), albeit with a reduced number of molecular marker loci. Our study aimed to determine the number and genomic locations of QTLs of relatively large effect controlling phenotypic differences and the extent of epistatic and pleiotropic effects of QTLs that could limit introgression between the two species in the wild. We also conducted genetic differentiation outlier tests on mapped molecular markers in the two species to identify loci under divergent selection and test for associations between outlier loci and QTLs. In addition, we tested whether previously identified hybrid incompatibilities are associated with either QTLs for species differences or highly divergent loci as would be expected under divergent selection.
Methods
Samples
An F2 mapping family (F2AC) of a reciprocal cross between two cross-compatible F1 progeny derived from a reciprocal cross between S. aethnensis (A) and S. chrysanthemifolius (C) was produced as described in Brennan et al. (2014) and used for QTL analysis. This family consisted of 100 individuals of known parental cytotype. For tests of selection based on genetic differentiation, seed was collected from two wild populations of S. aethnensis and three of S. chrysanthemifolius representing the elevational extremes of each species’ range and also the source locations of the mapping family parents (NIC1 and PIC1) [see Supporting Information—Table S1]. Forty-two plants of each species, each representing a separately sampled maternal individual, were raised from this seed in a glasshouse at the same time and under the same conditions as F2AC individuals.
Phenotype measurement
Twenty-five traits were measured on F2AC parents and progeny, and also wild sampled individuals (see Brennan et al. 2009 for a description of traits measured). Extreme outlier values >3 standard deviations from the mean were removed from the datasets for progeny and wild samples of each species prior to analysis. Trait summary statistics were calculated and comparisons between wild sampled S. aethnensis, wild sampled S. chrysanthemifolius and the F2AC mapping family were made using one-way analyses of variance and Mann–Whitney tests. Three traits—capitulum length, ray floret number and selfing rate—were dropped from further analysis after preliminary data exploration found that they showed extreme distributions that could not be satisfactorily resolved with data transformations. Remaining trait measurements were not transformed to become normally distributed before QTL analysis because (i) the expected density distributions of traits with additive effects contributed by multiple loci are not necessarily normally distributed, (ii) the significance of QTL logarithm of odds (LOD) scores can be adequately assessed with data permutation and (iii) estimated sizes of QTL effects are more directly interpretable based on untransformed data (Churchill and Doerge 1994). Cross direction did not significantly influence any trait mean, so this was not required as a cofactor for QTL analysis. Independence between measured traits was examined using paired-trait Spearman correlations, and tests of their significance were performed separately for wild sampled S. aethnensis, wild sampled S. chrysanthemifolius and the F2AC mapping family progeny leading to a subset of 13 highly independent traits being retained for QTL analysis. All tests were performed using R v2.13 software (R Development Core Team 2011).
DNA isolation and genotyping
DNA was extracted from each plant using the method described by Brennan et al. (2009). Plants were genotyped across 127 marker loci comprising 77 amplified fragment length polymorphisms (AFLPs), 8 simple sequence repeats (SSRs) and 42 expressed sequence tag (EST)-SSRs and indel molecular markers as described by Brennan et al. (2014). For ∼10 % of plants (randomly chosen), two independent DNA extracts were made to test for genotyping reliability.
Genetic mapping
A genetic map was constructed from the segregation of genetic markers in the F2AC mapping family as described in Brennan et al. (2014) and Supplementary information. Genotype uncertainty due to scoring of dominant markers was accounted for by using the MapMaker genotype classes C (not a homozygote for the first parental allele) and D (not a homozygote for the second parental allele; Lincoln et al. 1993). The genetic map comprised 10 independent linkage groups with a total length of ∼400 cM. Transmission ratio distortion affected ∼27 % of mapped markers that were clustered into nine TRDLs. Sixty-five mapped loci were included in the QTL analysis after removing 39 loci that did not show F2-like allelic segregation (i.e. each parent had an allele in common) and 23 loci that were located <0.5 cM from the nearest neighbouring marker and which therefore added little extra QTL mapping power.
Quantitative trait locus mapping and analysis
We analysed the data in the form of individual differences from the combined species mean, with the sign altered so that individuals that were more similar to S. aethnensis or S. chrysanthemifolius mean values were positive and negative, respectively. This data transformation preserved effect sizes in original units but had the added advantage of standardizing effect directions according to parental species across all traits. Comparisons with untransformed data showed that LOD scores (base 10 logarithm of odds) were largely unaffected by the transformation. Multiple interval mapping (MIM) was used to identify QTLs because this method has the advantage of simultaneously accounting for multiple QTLs and their interactions (Kao et al. 1999). Multiple interval mapping was performed with QTL cartographer v2.5.10 (Wang et al. 2011) using forward regression with a scanning interval of 3 cM and Bayesian information criterion (BIC-M0) model selection to determine the inclusion of extra QTL or QTL interaction parameters. Initial MIM models were then refined by testing indicated QTLs for significance according to BIC and adding additional QTLs until no further significant model improvement was achieved. Epistatic QTL interactions were also included if BIC was significantly improved. For comparison with MIM, composite interval mapping (CIM; Zeng 1994), a widely used QTL mapping method, was also performed and results obtained from this analysis, which did not differ greatly from those obtained with MIM, are presented in Supplementary information. The potential for TRDLs to influence the QTL results was tested using Spearman rank correlation tests of marker distance to nearest QTL peak against marker χ2 test values for segregation distortion of genotypes, heterozygotes and parental alleles.
Multiple trait CIM (MtCIM) simultaneously analyses multiple trait data and can distinguish between linked QTLs and a single QTL affecting more than one trait through pleiotropy (MtCIM; Jiang and Zeng 1995). Multiple trait CIM analysis was performed using a scanning interval of 3 cM and automatic model selection using forward regression with five cofactor loci outside the test interval window of 10 cM. Significance of QTL LOD scores was tested with 1000 permutations of trait values (Churchill and Doerge 1994). A complementary test of the extent to which QTLs for different traits occupied the same genomic regions applied the ‘sampling without replacement’ method (Paterson 2002). Because the traits examined in this QTL dataset were selected to minimize covariance between them, spurious patterns of QTL coincidence generated by covariance were also assumed to be minimized, avoiding the need for additional statistical correction (Breitling et al. 2008). To perform the ‘sampling without replacement’ test, the genetic map was divided into smaller intervals of equal size corresponding to the mean QTL 2-LOD cM confidence interval of 16.5 cM with intervals chosen to be centred over each linkage group. This level of subdivision of the genetic map generates an optimal proportion of intervals occupied by a QTL for the purposes of this test (Paterson 2002), but the effect of using smaller interval sizes was also tested by repeating the test with 2, 4, 6, 8, 10, 12 and 14 cM interval sizes. A binary matrix describing the presence or absence of QTLs for each trait within intervals was constructed and for each pair of traits, the probability of coincidence (p) was tested according to:
where n is the number of intervals compared, l and s are the number of QTL intervals present in the samples with larger and smaller QTL counts, respectively, and m is the number of paired QTL interval matches present. To test whether QTL coincidence was greater than the null hypothesis of a random distribution of QTLs across the genetic map, the observed mean probability of QTL coincidence across paired-trait comparisons was compared against the distribution obtained from 1000 random permutations of QTL locations. The coincidence between TRDLs and QTLs was also investigated by including TRDL data in this analysis.
Genetic diversity analysis
Summary population genetic statistics were estimated for all mapped markers genotyped in wild samples of S. aethnensis and S. chrysanthemifolius. The population genetics software used included: Arlequin (Excoffier and Lischer 2010), GenAlEx v6.1 (Peakall and Smouse 2006) and HPrare (Kalinowsky 2005). The estimated statistics for AFLP and other dominantly scored markers were band presence frequency (p; assuming Hardy–Weinberg equilibrium), effective number of alleles (Ne), unbiased heterozygosity (UHe), allelic richness (Ar), private allelic richness (pAr), genetic differentiation among species (FST) and genotypic differentiation (ΦPT). The same statistics, excluding p but including the minor allele frequency (MAF) and inbreeding coefficient (FIS), were calculated for codominantly scored markers.
Patterns of differentiation across loci were investigated to detect both strongly and weakly differentiated outlier loci using BayeScan (Foll and Gaggiotti 2008), which employs Bayesian methods to estimate locus-specific differentiation and to evaluate its probability relative to population-level differentiation. Default starting parameter settings were used, except for a Monte Carlo Markov Chain size of 10 000, thinning interval of 50, ten pilot runs of 10 000 and an additional burn-in of 1 00 000. Outlier loci were identified based on log10 Bayes Factors values greater than one. Outlier analysis was performed with individuals classified according to both species and population. Initial runs suggested that loci with very low MAF were over-represented among outliers. To overcome this problem, only those loci with MAF >0.05 were included in final differentiation analyses, which were conducted separately on datasets comprising 64 codominant loci and 132 dominant loci.
The presence of ‘genomic islands’ of divergence was investigated by testing the genomic clustering of outlier markers with binomial tests that the observed number of neighbouring pairs of significantly selected loci was greater than the expected number of neighbouring paired selected loci given by the square of the observed frequency of selected loci. Genetic differentiation, measured as both FST and ΦPT, was tested for an association with the genetic map distance to the nearest QTL peak and the nearest TRDL peak using Spearman rank correlation tests. Genetic differentiation was further tested for associations with local recombination rate, measured as the genetic map distance to the nearest mapped marker, and with genetic diversity within species, measured as each of UHe, Ar and MAF using Spearman rank correlation tests. Marker loci on linkage groups without QTLs were assigned large QTL distance values of 50 cM in order to include them as part of these association tests.
Results
Quantitative trait locus mapping and analysis
The two parent species, S. aethnensis and S. chrysanthemifolius, differed significantly for 22 of the 25 traits. The exceptions were flowering time, leaf number and selfed seed-set (Traits 1, 3 and 18) [see Supporting Information—Table S2 and Fig. S1]. We surmise that the lack of flowering time difference in the glasshouse compared with field observations reflects the importance of environmental conditions for the expression of this trait. For example, suitable growing conditions at the onset of spring start later in higher elevation S. aethnensis habitat than lower elevation S. chrysanthemifolius habitat. In summary, S. aethnensis differed from S. chrysanthemifolius in being shorter and less branched, possessing smaller, less dissected leaves (i.e. having entire or slightly lobed edges), and fewer but larger capitula that produced larger seed. Significant differences between the mean of the F2AC family and those of one or both parent species were also evident for all traits apart from pollen viability and selfed seed-set (Traits 16 and 18) [see Supporting Information—Table S2]. The means of the F2AC family for all traits were neither significantly higher nor lower than the means of both parents. Paired-trait correlations are summarized in Supporting Information—Table S3. Overall, 4.3, 2.7 and 11 % of pairs of traits were significantly correlated after correction for multiple testing among wild sampled S. aethnensis, wild sampled S. chrysanthemifolius and the F2AC mapping family, respectively. Instances of non-independence between traits were reduced by dropping highly correlated traits and traits used to calculate compound characters, leaving a subset of 13 independent traits for QTL analysis.
Significant QTLs for each trait were detected and characterized by LOD score, map position, two LOD confidence intervals, size of additive, dominance and epistatic effects, and percentage variance explained (PVE). A total of 29 significant QTLs were detected across the 13 traits examined with mean QTL effect size of 15 % (Table 1, Fig. 1). Quantitative trait loci were distributed across all major linkage groups except AC3 and AC6, with one to five QTLs detected for each trait (Fig. 1). The mean PVE of all identified QTLs per trait was 33.5 % (range = 10.0–69.8 %).
Table 1.
Summary QTL results from a MIM analysis of a reciprocal F2 S. aethnensis and S. chrysanthemifolius mapping family. Quantitative trait locus LG and position are the linkage group and maximum likelihood of odds score (LOD) cM position of significant QTLs identified from MIM. Quantitative trait locus cM interval is the 2-LOD cM interval around the maximum LOD value that is indicated in parentheses. The additive and dominance effects are in the same units as trait measures. Positive additive effects support the direction of the species difference and vice versa for negative effects, while positive dominance effects indicate that S. aethnensis alleles are dominant and vice versa for negative effects. The PVE is shown in parentheses. Epistatic interactions show the loci for each trait (numbered in the order they appear in the table) with significant interactions with the additional PVE shown in parentheses.
| Trait ID number: trait (units) | QTL LG and position (cM) | QTL cM interval (max LOD) | Additive effect (PVE) | Dominant effect (PVE) | Total PVE | Epistatic interactions (PVE) |
|---|---|---|---|---|---|---|
| 1: Time from first true leaf to flowering (days) | AC1; 41.3 | 36.6–44.5 (6.19) | −3.41 (5.7) | −2.31 (1.3) | 7 | 2 × 3 (5.4) |
| AC2; 9.3 | 6.3–11.3 (9.22) | −5.27 (14.3) | 0.13 (0) | 14.3 | ||
| AC10A; 0 | 0–0.5 (15.35) | 9.02 (46.2) | −3.56 (2.3) | 48.5 | ||
| 7: Primary stem midleaf auricle width (mm) | AC10B; 3 | 0–4.2 (2.21) | 0.52 (7.3) | 0.41 (2.7) | 10 | |
| 8: Primary inflorescence capitulum number (count) | AC2; 11.3 | 0–14.2 (2.5) | 1.29 (8.5) | 0.96 (3.6) | 12.1 | |
| AC5A; 0 | 0–6.9 (2.57) | −1.34 (9) | 0.36 (0.4) | 9.4 | ||
| AC10A; 0 | 0–10 (4.89) | 2.14 (20.7) | −0.43 (−0.1) | 20.6 | ||
| 9: Primary capitulum pedicel length (cm) | AC8A; 11.4 | 0–27.5 (2.5) | 0.26 (11.7) | −0.07 (0.4) | 12.1 | |
| 11: Primary capitulum disc diameter (mm) | AC4; 23.6 | 12.5–36.3 (3.52) | 0.05 (6.4) | 0.08 (9.8) | 16.2 | |
| AC9; 0 | 0–15 (2.13) | −0.05 (6.2) | 0.03 (1.2) | 7.4 | ||
| 15: Mean pollen number (per 3/40 florets) | AC10B; 3 | 0–4.2 (2.03) | 7.79 (6.2) | −8.94 (3.8) | 10 | |
| 16: Mean pollen viability (proportion) | AC1; 0.9 | 0–5.5 (5.4) | 0.01 (−1.2) | −0.19 (25.9) | 24.7 | |
| 19: Mean fruit length (mm) | AC1; 18 | 2.4–39.6 (2.89) | −0.21 (−5.8) | 0.36 (26.1) | 20.3 | |
| 20: Mean pappus length (mm) | AC1; 23 | 8.5–36.6 (4.6) | 0.15 (5.9) | 0.32 (12) | 17.9 | |
| AC5B; 0 | 0–9.5 (2.09) | 0.2 (5.6) | 0.01 (0) | 5.6 | ||
| AC8A; 13.6 | 0–27.5 (3.43) | 0.27 (10.6) | 0.12 (1.3) | 11.9 | ||
| 21: Primary stem node length, height to leaf number ratio (cm) | AC4; 6 | 0–41.3 (2.8) | 0.09 (8.6) | 0.1 (2.9) | 11.5 | |
| AC5A; 0 | 0–6.9 (6.37) | −0.12 (18) | −0.01 (0.1) | 18.1 | ||
| AC10A; 4.1 | 0–10 (11.57) | 0.16 (30.1) | −0.11 (2) | 32.1 | ||
| 22: Branch number to leaf number (proportion) | AC4; 26.6 | 13.5–41.3 (4.3) | 0.1 (9.9) | −0.17 (12.1) | 22 | |
| 24: Primary capitulum ray display area (mm2) | AC1; 15 | 6.5–25 (6.75) | 4.69 (3.3) | 3.37 (1.9) | 5.2 | 1 × 3 (5.1) |
| AC4; 30.3 | 4–41.3 (2.07) | 6.9 (6.2) | 0.25 (0.1) | 6.3 | 1 × 4 (7.8) | |
| AC7A; 4.6 | 0–14.2 (6.45) | 5.79 (2.1) | 16.83 (7.1) | 9.2 | ||
| AC8A; 15.1 | 2.5–24.5 (5.5) | 8.13 (8) | 5.67 (−0.2) | 7.8 | ||
| AC10A; 2.8 | 0–8.6 (8.01) | 15.43 (20.9) | 0.54 (0.2) | 21.1 | ||
| 25: Primary stem midleaf dissection, perimeter to area ratio (per mm) | AC4; 3 | 0–41.3 (2.02) | 0.04 (6.9) | 0.04 (2.9) | 9.8 | 2 × 3 (10.1) |
| AC8A; 6 | 0–10 (7.29) | 0.04 (7.7) | 0.05 (6.3) | 14 | ||
| AC10A; 0 | 0–10 (9.01) | 0.06 (21.1) | 0.01 (0.5) | 21.6 | ||
| AC10B; 3 | 0–4.2 (2.6) | 0.02 (3.9) | 0.04 (4.4) | 8.3 |
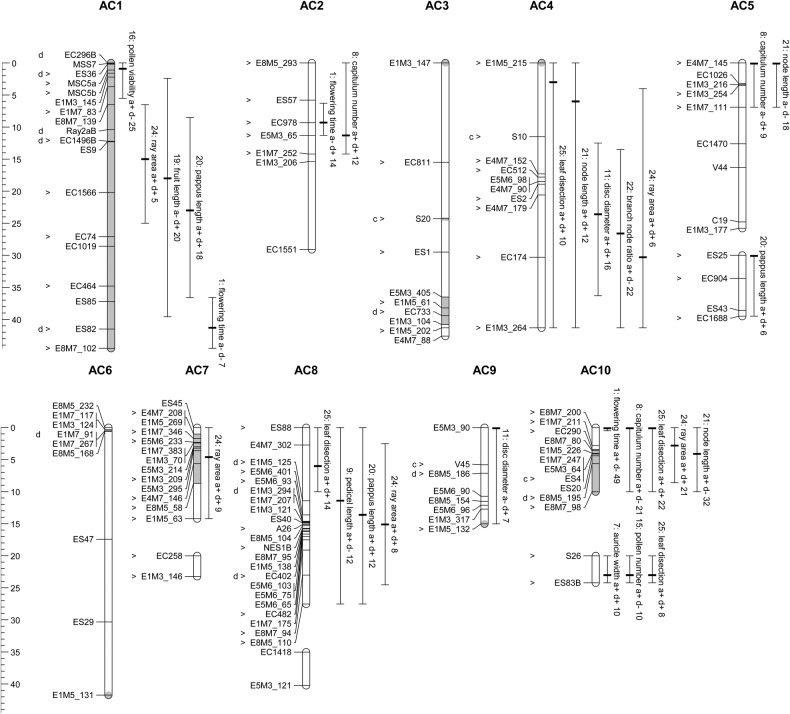
Figure 1.
Genetic map of a reciprocal F2 S. aethnensis and S. chrysanthemifolius mapping family showing quantitative trait loci identified by MIM and marker loci that were significantly divergent or convergent between species. Map distances in Kosambi centiMorgans are shown in the scale bar to the left of linkage groups. Linkage groups are represented by vertical bars with mapped locus positions indicated with horizontal lines. Weakly linked linkage groups (<4 LOD or >20 cM) that probably belong to the same chromosome are aligned vertically. Grey shading on linkage groups indicates regions exhibiting significant TRDLs. Locus names are listed to the left of linkage groups and mapped QTLs are listed to the right. ‘c’ or ‘d’ listed to the left of locus names indicates if locus was identified to be significantly convergent or divergent based on genetic differentiation analysis across sample populations, while the greater than symbol to the left of locus names indicates if the locus was included in QTL analysis. Quantitative trait loci were identified by MIM with significance testing by BIC model comparisons. Quantitative trait loci 2-LOD ranges are indicated with vertical lines with a bold horizontal line indicating the highest LOD score position. Quantitative trait locus summary information includes trait names, ‘a’ or ‘d’ each followed by ‘+’ or ‘−’ indicating additive or dominance effects and their direction of effect supporting or opposing the observed species difference, respectively, and the PVE.
Four pairs of QTLs exhibited significant epistatic interaction effects with a mean PVE of 7.1 % (range = 5.1–10.1; Table 1). The MtCIM analysis of all traits identified three significant and three almost significant (within 1 LOD of the permutation threshold of 14.82 LOD) pleiotropic loci with multiple trait effects (Table 2) [see Supporting Information—Table S5]. These potential pleiotropic loci overlapped with the 2-LOD intervals of 14/29 of the individual trait QTLs, with up to four traits affected at each site (Table 1). Thus, 14 QTLs for eight traits exhibited pleiotropic effects. The ‘sampling without replacement’ method using the 16.5 cM interval size found four trait pairs, auricle width and pollen number, capitulum number and node length, capitulum number and flowering time, and node length and leaf dissection, that showed significantly coincident QTL locations [see Supporting Information—Table S6]. Sampling without replacement analyses using a range of shorter interval sizes found similar evidence for coincident QTL locations, but failed to find any previously identified TRDLs that were significantly coincident with trait QTLs [see Supporting Information—Table S6].
Table 2.
Summary QTL results from a MtCIM analysis of a reciprocal F2 S. aethnensis and S. chrysanthemifolius mapping family. Locus LG and peak cM are the linkage group and maximum likelihood odds score (LOD) cM position of a locus that affects the expression of multiple traits. Locus 2-LOD interval is the 2-LOD interval around the peak for the locus with the maximum LOD value indicated in parentheses. Overlapping quantitative trait loci from the MIM analysis are shown for comparison [see Table 1 and Supporting Information—Table S4 for more details about QTLs].
| Locus LG, peak cM position | Locus 2-LOD interval (peak LOD) | Overlapping QTL LOD intervals |
|---|---|---|
| AC1, 0.9 | 0–6.5 (14.52) | 16: Mean poor pollen |
| AC1, 29.9 | 26.9–32.9 (14.35) | 19: Mean fruit length 20: Mean pappus length |
| AC2, 5.8 | 1.5–11.3 (14.1) | 1: Time from first true leaf to flowering 8: Primary inflorescence capitulum number |
| AC4, 3 | 1.8–4.2 (17.69) | 21: Primary stem node length 24: Primary capitulum ray display area 25: Primary stem midleaf dissection |
| AC5A, 3 | 0–6.9 (17.24) | 8: Primary inflorescence capitulum number 21: Primary stem node length |
| AC10A, 8.6 | 3.4–10 (28.6) | 8: Primary inflorescence capitulum number 21: Primary stem node length 24: Primary capitulum ray display area 25: Primary stem midleaf dissection |
Genetic diversity analysis
Both species exhibited similar levels of genetic diversity, with the highest diversity recorded for anonymous SSRs, followed in turn by EST-SSRs, EST-indels and AFLPs, and other dominant markers [see Supporting Information—Tables S7 and S8]. Overall, inbreeding coefficients were not significantly different from zero in either species indicative of random mating (FIS = 0.02 and 0.06 in S. aethnensis and S. chrysanthemifolius, respectively) [see Supporting Information—Table S7]. The two species were significantly genetically differentiated across all marker types with overall FST of 0.28 and 0.31 observed for dominant markers and codominant markers, respectively [see Supporting Information—Tables S7 and S8].
Bayesian analyses of species differentiation showed that 4.7 % of codominant markers, but 0 % of dominant markers, were divergent outliers and that the same percentages of each marker type were significantly convergent outliers (Table 3). When population information was included in these analyses, the tests were more sensitive and identified 7.8 and 5.3 % of significantly divergent codominant and dominant markers, respectively, and 4.7 and 0.8 % of significantly convergent codominant and dominant markers, respectively (Fig. 1, Table 3) [see Supporting Information—Table S9]. Significant outlier loci were distributed across most linkage groups of the genetic map (Fig. 1) and showed no evidence of clustering according to a one-way binomial test of an excess of neighbouring pairs of outlier markers (P = 0.1754). However, significant negative associations between measures of species differentiation for marker loci and the genetic map distance from the nearest QTL peak were present (Fig. 2). Similarly, there was evidence for negative associations between marker gene differentiation and genetic map distance from the nearest TRDL (Fig. 2). A significant negative association between genetic differentiation and low recombination in the form of genetic map distance to closest neighbouring mapped locus was also found (Fig. 2). Also, significant negative associations were present between genetic differentiation between species and the various intraspecific genetic diversity measures (Fig. 3). In general, all of these associations were stronger for codominant than for dominant markers.
Table 3.
Summary of numbers of marker loci identified as significantly divergent or convergent outliers between S. aethnensis and S. chrysanthemifolius. Samples tested included all samples scored according to species (Species), populations (Populations) or data subsets of only S. aethnensis populations or only S. chrysanthemifolius populations. Only polymorphic loci with minor allele frequencies >0.05 were included in analyses. In the case of dominant loci, allele frequency was calculated assuming within-population Hardy–Weinberg equilibrium. Loci were considered significantly divergent or convergent with log10 Bayes Factor statistics >1.
| Samples tested | No. codominant loci tested | No. dominant loci tested | No. codominant loci divergent (%) | No. codominant loci convergent (%) | No. dominant loci divergent (%) | No. dominant loci convergent (%) |
|---|---|---|---|---|---|---|
| Species | 64 | 132 | 3 (4.7) | 3 (4.7) | 0 (0.0) | 0 (0.0) |
| Populations | 64 | 132 | 5 (7.8) | 3 (4.7) | 7 (5.3) | 1 (0.8) |
| S. aethnensis | 53 | 115 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| S. chrysanth emifolius | 61 | 110 | 1 (1.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
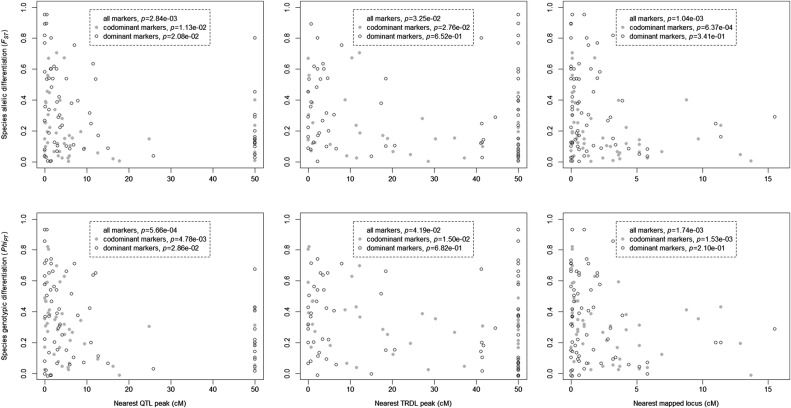
Figure 2.
Relationships between genetic differentiation and genetic map distance to the nearest QTL peak, the nearest TRDL or the nearest mapped marker. Presented P values summarize Spearman rank correlation tests. All significant associations were negative. Sample sizes were 48 codominant loci and 63 dominant loci. Loci on linkage groups without a QTL or TRDL peak were assigned an unlinked genetic map distance of 50 cM.
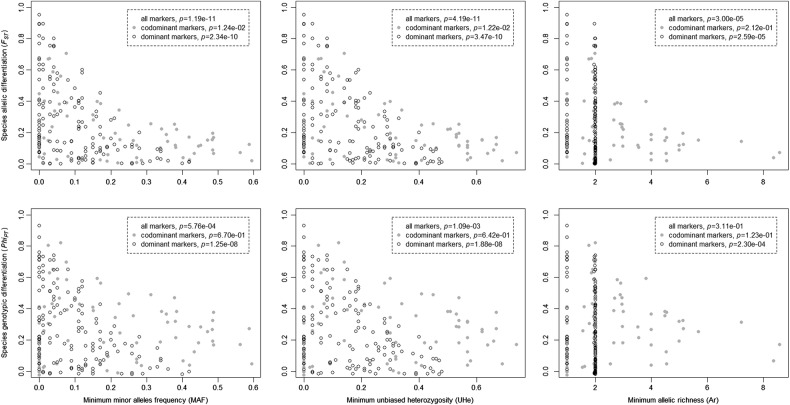
Figure 3.
Relationships between genetic differentiation and genetic diversity of wild sampled S. aethnensis and S. chrysanthemifolius. Presented P values summarize Spearman rank correlation tests. All significant associations were negative. Sample sizes were 65 codominant loci and 145 dominant loci.
Discussion
Quantitative trait locus architecture
Quantitative trait locus analysis identified 1–5 QTLs per trait and 29 QTLs in total for the 13 independent traits examined that distinguish the two Senecio species. In addition to resolving the primary effects of individual QTLs, MIM and MtCIM analyses provided evidence for epistatic interactions between four pairs of QTLs and possible pleiotropic effects at six loci affecting eight traits (Tables 1 and 2). Sampling without replacement tests indicated that QTL map locations were significantly clustered across the genetic map, with significant physical associations evident for four trait pairs [see Supporting Information—Table S5]. Chapman et al. (2016) reported similar clustering of QTLs for species differences in an independent mapping study of S. aethnensis and S. chrysanthemifolius. However, their study did not investigate patterns of epistasis and pleiotropy. Regardless of whether the observed interactions between QTLs are due to epistasis or pleiotropy or physical linkage, they indicate that different traits are not genetically independent and that divergent selection acting on one trait would therefore also affect other traits.
A QTL architecture involving extensive physical and epistatic interactions between QTLs, together with pleiotropic effects of individual QTLs, should limit introgression between the two Senecio species on Mount Etna since hybridization would tend to break up gene complexes that control the expression of adaptive phenotypes in each species (Fenster et al. 1997; Turelli et al. 2001). The complex genomic architecture of interspecific divergence revealed in Senecio might reflect the evolutionary outcome of selection for non-independence of different QTLs controlling traits under divergent selection (Kirkpatrick and Barton 2006; Nosil et al. 2009; Nosil and Feder 2012; Yeaman 2013). Limiting recombination seems to be the crucial factor permitting interacting QTLs to evolve into divergent co-adapted QTL complexes in the presence of gene flow. This can be achieved either through chromosomal rearrangement that causes recombination between rearranged regions to become deleterious (Feder et al. 2003; Lowry and Willis 2010; Twyford and Friedman 2015) or by evolution towards increased physical proximity (coincidence) through locally biased persistence, establishment or translocation of QTLs (Via and West 2008; Nosil et al. 2009; Yeaman 2013). Genetic mapping indicates that S. aethnensis and S. chrysanthemifolius are not distinguished by major genome rearrangements (Brennan et al. 2014), which instead emphasizes the importance of QTL coincidence for this system (our results and those of Chapman et al. 2016).
While TRDLs were not significantly coincident with QTLs for any trait according to the ‘sampling without replacement’ method, a QTL affecting pollen viability co-located with a TRDL of large effect in linkage group AC1 (Fig. 1). This finding is of interest as it adds to the result previously reported by Chapman et al. (2016) of co-localization of TRDLs with QTLs affecting F2 hybrid necrosis. Hybrid incompatibilities, such as decreased F2 pollen viability and hybrid necrosis and their associated TRDLs, are expected to limit introgression across large genomic regions allowing further divergence of these regions during speciation (Barton and Bengtsson 1986; Barton and de Cara 2009).
Non-random patterns of divergence across the genome
Levels of molecular genetic diversity were similar in wild samples of both Senecio species, while genetic differentiation between species was moderate. Genetic diversity decreased from estimates based on anonymous SSRs to EST-SSRs, to EST-indels to AFLPs, corresponding to the expected ability of each marker type to resolve allelic variation [see Supporting Information—Tables S6 and S7]. Low levels of genetic differentiation between the two species were also reported by Muir et al. (2013), Osborne et al. (2013) and Chapman et al. (2013), based on surveys of microsatellite and sequence variation. We identified a small percentage of loci that were either significantly divergent or convergent (up to 7.8 %) between species, dependent on the marker set analysed (Table 3) [see Supporting Information—Table S9]. This value is slightly greater than the 2.25 % of outliers from a study of 8854 loci recorded by Chapman et al. (2013) based on a comparison of the two species’ transcriptomes, but the two findings are probably within the bounds of error given the different numbers of loci examined. More discussion about the functions of significantly divergent or convergent loci is provided in the Supplementary information. Inevitably, the 196 marker loci for which patterns of differentiation were compared to detect significant divergence between species in the present study provide only a very coarse-grained perspective across the whole genome, and many of the true genetic targets of selection will not have been surveyed.
Reduced effective gene flow in the vicinity of selected loci is often used to explain significantly differentiated loci and ‘islands of divergence’ (Wu 2001; Feder and Nosil 2010). In support of this hypothesis, significant associations were found between interspecific genetic differentiation and genetic map distance to QTLs and TRDLs (Fig. 2). These associations were negative with more highly differentiated loci positioned closer to QTLs or TRDLs. These results support previous findings that selection against hybridization is important for maintaining species distinctiveness across the Senecio hybrid zone on Mount Etna (Brennan et al. 2009; Chapman et al. 2013, 2016). However, independently of gene flow, within-species directional selection can also generate the same pattern of divergence via species-specific reductions in diversity (Cruikshank and Hahn 2014). The latter is amplified when it occurs in regions of low recombination as it causes longer genomic regions to be affected by selection at linked markers. In accordance with these hypotheses and in agreement with the findings of Chapman et al. (2016), we also found evidence for intraspecific selection in the form of significant negative associations between interspecific differentiation and local recombination, and between interspecific differentiation and intraspecific genetic diversity (Figs 2 and 3). It is plausible that S. aethnensis and S. chrysanthemifolius experience distinct localized selection pressures related to the very different environments they occupy at different elevations on Mount Etna. Such within-species selection would be expected to reduce within-species genetic diversity in the genomic regions experiencing selection. These findings, therefore, suggest a role for environment-specific extrinsic selection in maintaining the cline with elevation on Mount Etna. While this pattern of diversity might also signal past periods of isolation facilitating divergence, other genetic studies suggest that gene flow between the two species has probably been continuous throughout their history (Chapman et al. 2013; Osborne et al. 2013; Filatov et al. 2016).
Conclusions
Our study shows that phenotypic divergence across the elevational gradient on Mount Etna involves divergence of multiple quantitative traits controlled by numerous interacting genes (QTLs). A breakdown in the complex genetic architecture of these traits following hybridization would be expected to reduce the fitness of most hybrid offspring and therefore contribute to introgression barriers between the two Senecio species. Our combined analyses of genetic differentiation, QTLs and TRDLs emphasize that divergence is non-randomly distributed across the genomes of these species and that both selection against hybrids between species and locally maladapted individuals within species will act to maintain phenotypic divergence between the two species in the face of gene flow.
Sources of Funding
The research was funded by a Natural Environment Research Council (NERC) Grant NE/D014166/1 to R.J.A. as principal investigator. A.C.B. was supported during part of the writing of this paper by funding from European Commission 7th Framework Programme Capacities Work Programme (FP7-REGPOT 2010-1), Grant No. 264125 EcoGenes.
Contributions by the Authors
R.J.A., A.C.B. and S.J.H. designed the research. A.C.B. performed the experiments and analysis. A.C.B. wrote the first draft and A.C.B., R.J.A. and S.J.H. contributed to revisions.
Conflict of Interest Statement
None declared.
Supporting Information
The following additional information is available in the online version of this article —
Table S1. Information on wild sampled populations of S. aethnensis and S. chrysanthemifolius.
Table S2. Summary quantitative trait results for S. aethnensis, S. chrysanthemifolius and a reciprocal F2 S. aethnensis and S. chrysanthemifolius mapping family.
Table S3. Paired-trait correlations in (a) F2AC progeny, (b) Senecio aethnensis, (c) S. chrysanthemifolius and (d) all three samples.
Table S4. Comparison of summary QTL results for a CIM and MIM analysis of a reciprocal F2 S. aethnensis and S. chrysanthemifolius mapping family.
Table S5. Summary QTLs results from a MtCIM analysis compared with single trait QTL analyses of a reciprocal F2 S. aethnensis and S. chrysanthemifolius mapping family.
Table S6. (a) ‘Sampling without replacement’ test results for paired-trait QTL coincidence, (b) permutation tests of overall paired-trait QTL coincidence using different QTL and TRDL datasets and genetic map interval sizes.
Table S7. Summary population genetic statistics for AFLPs and other dominantly scored molecular genetic markers from S. aethnensis and S. chrysanthemifolius samples.
Table S8. Summary population genetic statistics for codominantly scored molecular genetic markers from S. aethnensis and S. chrysanthemifolius samples.
Table S9. Expressed sequence tag loci showing evidence for divergent or convergent selection between S. aethnensis and S. chrysanthemifolius.
Figure S1. Boxplots summarizing quantitative trait results for S. aethnensis, S. chrysanthemifolius and a reciprocal F2 mapping family that were included in the QTL analysis. Trait numbers in titles correspond to the trait numbering system of Table 1. Bold horizontal lines indicate median values. Boxes indicate 25–75 percentile range. Lines indicate the range of values within 1.5 times the upper and lower quartiles, respectively. Points indicate values more extreme than 1.5 times the upper and lower quartiles. Asterisks indicate the trait values of the mapping family parents. No mapping family parental values were available for flowering time as these individuals were vegetatively propagated for comparison with their progeny.
Figure S2. Genetic map of a reciprocal F2 S. aethnensis and S. chrysanthemifolius mapping family showing quantitative trait loci identified by CIM and marker loci that were significantly divergent or convergent between species. Map distances in Kosambi centiMorgans are shown in the scale bar to the left of linkage groups. Linkage groups are represented by vertical bars with mapped locus positions indicated with horizontal lines. Weakly linked linkage groups (<4 LOD or >20 cM) that probably belong to the same chromosome are aligned vertically. Grey shading on linkage groups indicates regions exhibiting significant TRDLs. Locus names are listed to the left of linkage groups and mapped QTLs are listed to the right. ‘c’ or ‘d’ listed to the left of locus names indicates if that locus was identified as significantly convergent or divergent based on genetic differentiation analysis across sample populations, while greater than symbol to the left of locus names indicates if the locus was included in QTL analysis. Quantitative trait loci were identified by CIM with significance determined if the LOD score exceeded the 0.95 quantile of 1000 data permutations. Quantitative trait loci 2-LOD interval ranges are indicated with vertical lines with a bold horizontal line indicating the highest LOD score position. Quantitative trait locus summary information includes trait names, ‘a’ or ‘d’ each followed by ‘+’ or ‘−’ indicating additive or dominance effects and their direction of effect supporting or opposing the observed species difference, respectively, and the PVE.
Supplementary information. Additional text describing the genetic map, TRD analysis, composite interval mapping, QTL sign tests, genetic diversity analyses and discussion of the functions of significantly divergent and convergent loci.
Acknowledgements
We thank David Forbes for technical assistance and Ai-Lan Wang for help with measurement of quantitative traits. We thank the managing editor, Diana Wolf and anonymous reviewers for their suggestions to improve earlier versions of this paper.
Literature Cited
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierrne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez-Rodriguez P, Möst M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Väinölä R, Wolf JBW, Zinner D. 2013. Hybridization and speciation. Journal of Evolutionary Biology 26:229–246. 10.1111/j.1420-9101.2012.02599.x [DOI] [PubMed] [Google Scholar]
- Abbott RJ, Brennan AC. 2014. Altitudinal gradients, plant hybrid zones and evolutionary novelty. Philosophical Transactions of the Royal Society Series B 369:20130346 10.1098/rstb.2013.0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N, Bengtsson BO. 1986. The barrier to genetic exchange between hybridising populations. Heredity 57:357–376. 10.1038/hdy.1986.135 [DOI] [PubMed] [Google Scholar]
- Barton NH, De Cara MAR. 2009. The evolution of strong reproductive isolation. Evolution 63:1171–1190. 10.1111/j.1558-5646.2009.00622.x [DOI] [PubMed] [Google Scholar]
- Bierne N, Welch J, Loire E, Bonhomme F, David P. 2011. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Molecular Ecology 20:2044–2072. 10.1111/j.1365-294X.2011.05080.x [DOI] [PubMed] [Google Scholar]
- Bouck A, Wessler SR, Arnold ML. 2007. QTL analysis of floral traits in Louisiana Iris hybrids. Evolution 61:2308–2319. 10.1111/j.1558-5646.2007.00214.x [DOI] [PubMed] [Google Scholar]
- Breitling R, Li Y, Tesson BM, Fu J, Wu C, Wiltshire T, Gerrits A, Bystrykh LV, De Haan G, Su AI, Jansen RC. 2008. Genetical genomics: spotlight on QTL hotspots. PLoS Genetics 4:e1000232 10.1371/journal.pgen.1000232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan AC, Bridle JR, Wang A-L, Hiscock SJ, Abbott RJ. 2009. Adaptation and selection in the Senecio (Asteraceae) hybrid zone on Mount Etna, Sicily. New Phytologist 183:702–717. 10.1111/j.1469-8137.2009.02944.x [DOI] [PubMed] [Google Scholar]
- Brennan AC, Hiscock SJ, Abbott RJ. 2014. Interspecific crossing and genetic mapping reveal intrinsic genomic incompatibility between two Senecio species that form a hybrid zone on Mount Etna, Sicily. Heredity 113:195–204. 10.1038/hdy.2014.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Hiscock SJ, Filatov DA. 2013. Genomic divergence during speciation driven by adaptation to altitude. Molecular Biology and Evolution 30:2553–2567. 10.1093/molbev/mst168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Hiscock SJ, Filatov DA. 2016. The genomic bases of morphological divergence and reproductive isolation driven by ecological speciation in Senecio (Asteraceae). Journal Evolutionary Biology 29:98–113. 10.1111/jeb.12765 [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138:963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 2004. Speciation. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Cruikshank TE, Hahn MW. 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Molecular Ecology 23:3133–3157. 10.1111/mec.12796 [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10:564–567. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- Feder JL, Nosil P. 2010. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution 64:1729–1747. 10.1111/j.1558-5646.2009.00943.x [DOI] [PubMed] [Google Scholar]
- Feder JL, Roethele JB, Filchak K, Niedbalski J, Romero-Severson J. 2003. Evidence for inversion polymorphism related to sympatric host race formation in the apple maggot fly, Rhagoletis pomonella . Genetics 163:939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CB, Galloway LF, Chao L. 1997. Epistasis and its consequences for the evolution of natural populations. Trends in Ecology and Evolution 12:282–286. 10.1016/S0169-5347(97)81027-0 [DOI] [PubMed] [Google Scholar]
- Filatov DA, Osborne OG, Papadopulos AS. 2016. Demographic history of speciation in a Senecio altitudinal hybrid zone on Mt. Etna. Molecular Ecology, in press 10.1111/mec.13618 [DOI] [PubMed] [Google Scholar]
- Fishman L, Kelly AJ, Morgan E, Willis JH. 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159:1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993. 10.1534/genetics.108.092221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire P-A, Normandeau E, Pavey SA, Bernatchez L. 2013. Mapping phenotypic, expression and transmission ratio distortion QTL using RAD markers in the Lake Whitefish (Coregonus clupeaformis). Molecular Ecology 22:3036–3048. 10.1111/mec.12127 [DOI] [PubMed] [Google Scholar]
- Gompert Z, Parchman TL, Buerkle CA. 2012. Genomics of isolation in hybrids. Philosophical Transactions of the Royal Society Series B 367:439–450. 10.1098/rstb.2011.0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JK, Abbott RJ. 2005. Recent, allopatric, homoploid hybrid speciation: the origin of Senecio squalidus (Asteraceae) in the British Isles from a hybrid zone on Mount Etna, Sicily. Evolution 59:2533–2547. 10.1111/j.0014-3820.2005.tb00967.x [DOI] [PubMed] [Google Scholar]
- Jiang C, Zeng ZB. 1995. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140:1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, Birney E, Searle S, Schmutz J, Grimwood J, Dickson MC, Myers RM, Miller CT, Summers BR, Knecht AK, Brady SD, Zhang H, Pollen AA, Howes T, Amemiya C, Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team, Lander ES, Di Palma F, Lindblad-Toh K, Kingsley DM. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484:55–61. 10.1038/nature10944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowsky ST. 2005. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes 5:187–189. 10.1111/j.1471-8286.2004.00845.x [DOI] [Google Scholar]
- Kao C-H, Zeng ZB, Teasdale RD. 1999. Multiple interval mapping for quantitative trait loci. Genetics 152:1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. 2006. Chromosome inversions, local adaptation and speciation. Genetics 173:419–434. 10.1534/genetics.105.047985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexer C, Rosenthal DM, Raymond O, Donovan LA, Rieseberg LH. 2005. Genetics of species differences in the wild annual sunflowers, Helianthus annuus and H. petiolaris. Genetics 169:2225–2239. 10.1534/genetics.104.031195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln SE, Daly MJ, Lander ES. 1993. Constructing genetic linkage maps with MAPMAKER/EXP version 3.0: A tutorial and reference manual, 3rd edn, Technical Report Cambridge, MA: Whitehead Institute for Biomedical Research. [Google Scholar]
- Lindtke D, Buerkle CA. 2015. The genetic architecture of hybrid incompatibilities and their effect on barriers to introgression in secondary contact. Evolution 69:1987–2004. 10.1111/evo.12725 [DOI] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biology 8:e1000500 10.1371/journal.pbio.1000500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir G, Osborne OG, Sarasa J, Hiscock SJ, Filatov DA. 2013. Recent ecological selection on regulatory divergence is shaping clinal variation in Senecio on Mount Etna. Evolution 67:3032–3042. 10.1111/evo.12062 [DOI] [PubMed] [Google Scholar]
- Nosil P. 2012. Ecological speciation. Oxford, UK: Oxford University Press. [Google Scholar]
- Nosil P, Feder JL. 2012. Genomic divergence during speciation: causes and consequences. Philosophical Transactions of the Royal Society Series B 367:332–342. 10.1098/rstb.2011.0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortíz-Barrientos D. 2009. Divergent selection and heterogeneous genomic divergence. Molecular Ecology 18:375–402. 10.1111/j.1365-294X.2008.03946.x [DOI] [PubMed] [Google Scholar]
- Orr HA, Turelli M. 2001. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55:1085–1094. 10.1111/j.0014-3820.2001.tb00628.x [DOI] [PubMed] [Google Scholar]
- Osborne OG, Batstone TE, Hiscock SJ, Filatov DA. 2013. Rapid speciation with gene flow following the formation of Mt. Etna. Genome Biology and Evolution 5:1704–1715. 10.1093/gbe/evt127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH. 2002. What has QTL mapping taught us about plant domestication? New Phytologist 154:591–608. 10.1046/j.1469-8137.2002.00420.x [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6:288–295. 10.1111/j.1471-8286.2005.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical; Computing. [Google Scholar]
- Renaut S, Maillet N, Normandeau E, Sauvage C, Derome N, Rogers SM, Bernatchez L. 2012. Genome-wide patterns of divergence during speciation: the lake whitefish case study. Philosophical Transactions of the Royal Society Series B 367:354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C. 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216. 10.1126/science.1086949 [DOI] [PubMed] [Google Scholar]
- Rogers SM, Bernatchez L. 2007. The genetic architecture of ecological speciation and the association with signatures of selection in natural lake whitefish (Coregonus sp. Salmonidae) species pairs. Molecular Biology and Evolution 24:1423–1438. 10.1093/molbev/msm066 [DOI] [PubMed] [Google Scholar]
- Rogers SM, Mee JA, Bowles E. 2013. The consequences of genomic architecture on ecological speciation in postglacial fishes. Current Zoology 59:53–71. 10.1093/czoolo/59.1.53 [DOI] [Google Scholar]
- Servedio MR, Van Doorn GS, Kopp M, Frame AM, Nosil P. 2011. Magic traits in speciation: ‘magic’ but not rare? Trends in Ecology and Evolution 26:389–397. 10.1016/j.tree.2011.04.005 [DOI] [PubMed] [Google Scholar]
- Smadja CM, Butlin RK. 2011. A framework for comparing processes of speciation in the presence of gene flow. Molecular Ecology 20:5123–5140. 10.1111/j.1365-294X.2011.05350.x [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. 2008. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity 100:158–170. 10.1038/sj.hdy.6800937 [DOI] [PubMed] [Google Scholar]
- Strasburg JL, Sherman NA, Wright KM, Moyle LC, Willis JH, Rieseberg LH. 2012. What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Philosophical Transactions of the Royal Society Series B 367:364–373. 10.1098/rstb.2011.0199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SJ, Rojas LD, Ho SW, Martin NH. 2012. Genomic collinearity and the genetic architecture of floral differences between the homoploid hybrid species Iris nelsonii and one of its progenitors, Iris hexagona. Heredity 110:63–70. 10.1038/hdy.2012.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Barton NH, Coyne JA. 2001. Theory and speciation. Trends in Ecology and Evolution 16:330–343. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biology 3:e285 10.1371/journal.pbio.0030285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyford AD, Friedman J. 2015. Adaptive divergence in the monkey flower Mimulus guttatus is maintained by a chromosomal inversion. Evolution 69:1476–1486. 10.1111/evo.12663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S, West J. 2008. The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Molecular Ecology 17:4334–4345. 10.1111/j.1365-294X.2008.03921.x [DOI] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng Z-B. 2011. Windows QTL Cartographer 2.5. Raleigh, NC: Department of Statistics, North Carolina State University. [Google Scholar]
- Whiteley AR, Derome N, Rogers SM, St-Cyr J, Laroche J, Labbe A, Nolte A, Renaut S, Jeukens J, Bernatchez L. 2008. The phenomics and expression quantitative trait locus mapping of brain transcriptomes regulating adaptive divergence in Lake Whitefish species pairs (Coregonus sp.). Genetics 180:147–164. 10.1534/genetics.108.089938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I. 2001. The genic view of the process of speciation. Journal of Evolutionary Biology 14:851–865. 10.1046/j.1420-9101.2001.00335.x [DOI] [Google Scholar]
- Yeaman S. 2013. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proceedings of the National Academy of Sciences of the USA 110:E1743–E1751. 10.1073/pnas.1219381110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman S, Whitlock MC. 2011. The genetic architecture of adaptation under migration–selection balance. Evolution 65:1897–1911. 10.1111/j.1558-5646.2011.01269.x [DOI] [PubMed] [Google Scholar]
- Zeng Z-B. 1994. Precision mapping of quantitative trait loci. Genetics 136:1457–1468. 10.1111/j.1558-5646.2011.01269.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.