Abstract
Studies on melanin-based color variation in a context of natural selection have provided a wealth of information on the link between phenotypic and genetic variation. Here, we evaluated associations between melanic plumage patterns and genetic polymorphism in the Réunion grey white-eye (Zosterops borbonicus), a species in which mutations on MC1R do not seem to play any role in explaining melanic variation. This species exhibits 5 plumage color variants that can be grouped into 3 color forms which occupy discrete geographic regions in the lowlands of Réunion, and a fourth high-elevation form which comprises 2 color morphs (grey and brown) and represents a true color polymorphism. We conducted a comprehensive survey of sequence variation in 96 individuals at a series of 7 candidate genes other than MC1R that have been previously shown to influence melanin-based color patterns in vertebrates, including genes that have rarely been studied in a wild bird species before: POMC, Agouti, TYR, TYRP1, DCT, Corin, and SLC24A5. Of these 7 genes, 2 (Corin and TYRP1) displayed an interesting shift in allele frequencies between lowland and highland forms and a departure from mutation-drift equilibrium consistent with balancing selection in the polymorphic highland form only. Sequence variation at Agouti, a gene frequently involved in melanin-based pigmentation patterning, was not associated with color forms or morphs. Thus, we suggest that functionally important changes in loci other than those classically studied are involved in the color polymorphism exhibited by the Réunion grey white-eye and possibly many other nonmodel species.
Key words: Agouti, Corin, TYRP1, melanin pigments, plumage color, Zosterops
Melanin-based pigmentation is an important component of vertebrate coloration that is often under strong genetic control (Roulin 2004; Hill and McGraw 2006). In laboratory mice (Mus musculus), at least 150 different genes from the melanocortin pathway play some role in shaping patterns of melanin pigment production and deposition (Bennett and Lamoreux 2003). Only a handful of these genes has previously been found to influence body color in other vertebrate species (Hubbard et al. 2010). The melanocortin-1-receptor (MC1R), one of the most studied “color genes,” has been shown to explain color variation in a broad variety of lineages, such as lizards (Rosenblum et al. 2004), Peromyscus mice (Mullen and Hoekstra 2008), humans (Valverde et al. 1995), birds (Theron et al. 2001; Mundy et al. 2004; Haas et al. 2009; Uy et al. 2009; Roulin and Ducrest 2013 for some examples) and even mammoths (Römpler et al. 2006). However, variation in melanin-based color at the intra- or interspecific levels is not always associated with mutations at the MC1R gene, and this seems especially likely in species or populations with complex patterns of eumelanin/phaeomelanin deposition (e.g., MacDougall-Shackleton et al. 2003; Cheviron et al. 2006; Bourgeois et al. 2012). Therefore, understanding the genetic basis of complex melanin-based pigmentation patterns may require a wider exploration of other candidate genes, especially those involved in the melanocortin pathway. This is especially needed in taxa such as birds in which genes involved in melanin-based color variation other than MC1R are particularly understudied. Several genes (e.g., Agouti, TYRP1) known for their influence on mammal pigmentation (Sarangarajan and Boissy 2001; Hoekstra 2006; Nadeau et al. 2007; Steiner et al. 2007; Manceau et al. 2010, Domyan et al. 2014) have a similar effect on plumage color in quail (Coturnix japonica) and chicken (Gallus gallus) (Nadeau et al. 2007, 2008), but it is unknown whether mutations at these genes can be associated with color variation in other, nondomesticated, bird species (but see Lehtonen et al. 2012).
In this study, we evaluated associations between melanistic plumage patterns and genetic polymorphism in the Réunion grey white-eye (Zosterops borbonicus) at a series of 7 candidate genes that have been previously shown to influence melanin-based color patterns in vertebrates (Table 1), including genes that have rarely been studied in wild birds before (but see Poelstra et al. 2013). We chose the Réunion grey white-eye for our study because it is comprised of 5 distinct plumage variants which correspond to 4 geographic forms and differ in the relative extent of melanin-based grey and brown colors in their plumage (Gill 1973). The differences in plumage color traits between these variants appear to have been shaped by natural selection (Cornuault et al. 2015) and are likely controlled by relatively few genes (Gill 1973; Milá et al. 2010). However, amino-acid variation at MC1R plays no role in explaining this pattern of melanic variation (Bourgeois et al. 2012), which may thus reflect a mixture of modifications in the structure and regulation of other genes underlying pigment production. Candidate genes were selected on the basis of 1) their role in pigment synthesis, including melanosome migration (SLC24A5), agonism or antagonism with MC1R (POMC, Agouti), inhibition of other factors (Corin) and required enzymatic reactions for pigment production (TYR, TYRP1, DCT); 2) their likely influence on the switch between eumelanin and phaeomelanin production, either upstream (Corin, Agouti, POMC) or downstream (TYR, TYRP1, DCT, SLC24A5) from MC1R; 3) their known association with pigmentation in vertebrates (Agouti, Corin, enzymatic genes, see description and main references in Table 1). We also tried to obtain sequences from genes responsible for dilution phenotypes in chicken like SLC45A2 (Gunnarsson et al. 2007) or MLPH (Vaez et al. 2008) but their amplification failed in our study species. We screened extensively the Agouti gene, since its large cis-regulatory region (around 100kb) has been shown to be involved in pigmentation patterning in other species. We examined sequence variation at these 7 candidate genes (Agouti, Corin, TYR, TYRP1, DCT, POMC, and SLC24A5) to test the hypothesis that mutations within the focal gene region are associated with variation in melanin-based pigmentation patterns in Z. borbonicus and also tested for evidence of selection.
Table 1.
Candidate genes that have been shown to influence melanin-based color patterns in mammals and/or bird model species
| Locus | Chromosome | Expressed in | Physiological roles | Involvement in polymorphism | Pleiotropic effects | References |
|---|---|---|---|---|---|---|
| Agouti | 20 | Dermis, adipose tissues, gonads, heart, liver, muscles | Interacts with MC1R and allows to switch from eumelanin to phaeomelanin production. Interacts with other melanocortin receptors involved in a range of physiological aspects. | Melanic variations in Peromyscus mice. Responsible for the yellow variants in quail and chicken. | + | Minvielle et al. (2007), Hiragaki et al. (2008), Nadeau et al. (2008), Kingsley et al. (2009) |
| Corin | 4 | Dermal papilla, heart | Serine protease expressed in the heart and in dermal papilla. Involved in regulating blood pressure and inhibits Agouti action on MC1R. | Melanic variations in Peromyscus mice. Limits the extent of phaeomelanic band in mice. | + | Yan et al. (1999), Enshell-Seijffers et al. (2008), Manceau et al. (2010) |
| DCT | 1 | Melanocyte | Involved in synthesis of melanin from l-dopaquinone | Responsible for the slaty phenotype in mice | − | Jackson et al. (1992) |
| POMC | 3 | Brain, immune cells, gastro- intestinal tract, dermis, adrenal glands, thyroid, pancreas | Interacts with MC1R and allows to switch to eumelanin production. Interacts with other melanocortin receptors involved in a range of physiological aspects. | KO responsible for obese and yellow mice. Variations in melanocortin levels observed between tawny owls morphs. | ++ | Yaswen et al. (1999), Roulin et al. (2011) |
| SLC24A5 | 10 | Melanocyte | Cation exchanger involved in migration of melanosomes toward keratinocytes | Dilution of pigments in humans, chicken and zebra fish. Down regulated in yellow quails. | − | Lamason et al. (2005), Nadeau et al. (2008) |
| TYR | 1 | Melanocyte | Involved in synthesis of dopaquinone from tyrosine | Responsible for the recessive white phenotype in chicken | − | Sato et al. (2007) |
| TYRP1 | Z | Melanocyte | Involved in synthesis of melanin from l-dopaquinone | Responsible for the roux phenotype in quail. Color variation in flycatchers. | − | Buggiotti (2007), Nadeau et al. (2007) |
Examples of involvement in wild bird species are also given. Chromosome assignation is based on the chicken (Gallus gallus) genome.
Materials and Methods
Bird Materials
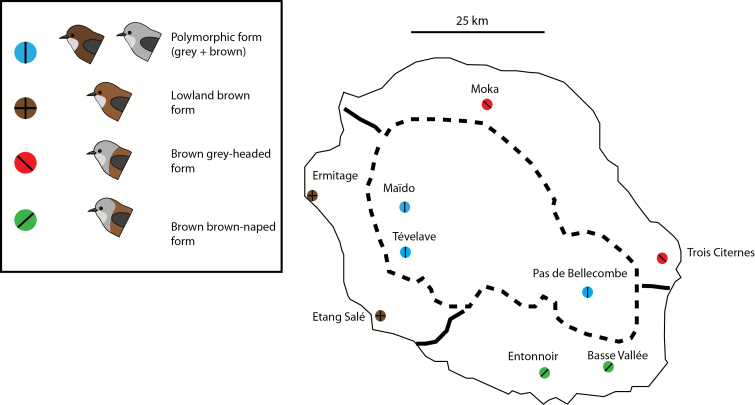
The 5 plumage color variants in the Réunion grey white-eye can be grouped into 4 geographic forms: 3 monomorphic forms occupy discrete geographic regions in the lowlands of Réunion; a fourth form comprises 2 morphs, a grey and a brown, which occur at high elevations in complete sympatry and represents a true color polymorphism (Figure 1; for a description of the plumage color variants, see Gill 1973; Cornuault et al. 2015).
Figure 1.
Map showing localities sampled in this study. For each locality a color code indicates which form is present. Brown circle: lowland brown-headed brown form. Blue circle: highland polymorphic form with grey and brown morphs. Red circle: grey-headed brown form. Green circle: brown-naped brown form. Limits between monomorphic lowland forms are indicated with a hard line; limits between lowland forms and the highland polymorphic form are indicated by a dotted line.
We sampled a total of 96 individuals from 9 different localities (Figure 1, Supplementary Table 1) covering the entire distribution range of Z. borbonicus. For the polymorphic highland form, we sampled 16 individuals (8 grey and 8 brown) at each of 3 localities for a total of 24 individuals per morph across localities. For the 3 lowland forms, we sampled 8 individuals in 2 localities within each form’s range, for a total of 16 individuals per form. For each individual, about 10 µL of blood were collected by gently puncturing the subbrachial vein and conserved in Queen’s lysis buffer (Seutin et al. 1991) before storing at −20 °C.
DNA Isolation, Amplification, and Sequencing
We extracted genomic DNA from blood samples using DNeasy Blood and Tissue kits (Qiagen, Venlo, the Netherlands), following the manufacturer’s instructions for nucleated blood cells. Conserved regions were identified for primer design using 2 avian reference genomes (the chicken: G. gallus and the zebra finch: Taeniopygia guttata). Primers were usually anchored in exons, but we tried whenever possible to obtain intronic sequences since these are known to be more variable. Protocols were optimized until amplification was specific to the targeted loci, with the aim of amplifying 400–1000bp regions for each gene (see Supplementary Table 2 for information on primer sets).
For Agouti, we adopted a different strategy and designed 22 PCR primers to amplify 11 fragments across coding and cis-regulatory regions, spanning around 70kb of the gene. We first obtained sequences from fragments around the largest and most variable marker found in an intronic region of Agouti (Supplementary Figure 1) for all individuals. We then took advantage of the fact that the 2 morphs (grey and brown) of the highland form differ in the overall pattern of melanin deposition throughout the plumage to investigate further a possible role of Agouti. In order to assess a possible role of distant cis-regulatory mutations, we thus obtained sequences from all fragments for all individuals from these 2 morphs.
As TYRP1 is a sex-linked gene, we first conducted molecular sexing of all individuals to correctly determine the number of haploid sequences in our dataset. To do so, we amplified a fragment of CHD genes whose size differs between Z and W copies (Griffiths et al. 1998). After PCR amplification, visualization on a 1% agarose gel allowed sexing by counting the number of bands. One band indicated a ZZ genotype (male), whereas 2 bands indicated a ZW genotype (female).
PCR reactions (25 µL) were performed using: 5 µL of 5X buffer (Promega®), 0.5 µL of 10 µM dNTPs mix (2.5 µM each), 0.125 µL of Taq (5U/µL, Promega GoTaq® DNA polymerase), 1 µL of each primer (10 µM), 15.4 µL of sterile distilled water, and 2 µL of DNA extract (10ng/µL). The thermocycling profile was as follows: an initial denaturation at 94 °C for 2min, then 40 cycles consisting of a 45s denaturation step at 94 °C, a 45s annealing step at variable temperature, and a 1min extension step at 72 °C. A final elongation step at 72 °C for 5min ended the process. PCR products were visualized on a 1% agarose gel. Both forward and reverse DNA strands were sequenced on an ABI3730XL sequencer (Applied Biosytems) using the same primer pairs as in PCR reactions.
Sequences were edited and aligned unambiguously using Sequencher® and variable sites were checked by eye. Individuals were considered as heterozygous at a given site when we observed double peaks approximately half the height of neighbouring peaks in both strands. Sequences were blasted against zebra finch (T. guttata) sequences to check their identity. Nucleotidic sequences were translated into amino-acid sequences using Mega 5 (Tamura et al. 2011). All sequences were submitted to Genbank (Accession number: KU903288-KU904220).
Data Analyses
The program PHASE (Stephens and Donnelly 2003) implemented in DNAsp (Librado and Rozas 2009) allowed us to reconstruct haplotypes from the population-based genotype data. We tested for deviations from Hardy–Weinberg equilibrium using Arlequin 3.5 (Excoffier and Lischer 2010). We used genotypic data to perform association tests on each of the 6 genes displaying at least 2 segregating sites, following the SCORE test from logistic regression (Clayton et al. 2004) in the package AssotesteR (available at http://gastonsanchez.com/software/; last access: March 2016). This test aims at comparing changes in allele frequencies at several SNP loci from the same genomic region, testing whether they display a significant association with a given phenotype. Statistical significance was assessed with 1000 permutations. As TYRP1 displayed only one polymorphic site, we performed a Fisher’s exact test on the allelic counts at this locus. Tests were run according to 4 distinct configurations, grouping individuals according to 4 phenotypic criteria: head color (brown vs. grey), nape color, back color, and also whether birds were grey or brown when dealing with individuals from the highland form.
To test for population structure, we performed a molecular analysis of variance (AMOVA) on haplotypes inferred by PHASE, with the 5 color variants as groups. As we could observe differences in allele frequencies at some markers between highland and lowland forms, we also performed analyses using data from the polymorphic highland form only, with morph (grey or brown) as a grouping factor. A last analysis was performed to compare the 3 lowland forms. All analyses were carried out using the Arlequin 3.5 package. Significance was assessed based on 10000 permutations.
To test whether selection had a detectable impact on genetic variation, we used the program DnaSP v5.10 (Librado and Rozas 2009) to perform analyses of DNA polymorphism on our data. Nucleotide diversity was estimated by Watterson’s θ w (Watterson 1975) and π, the average number of pairwise nucleotide differences among sequences (Nei and Li 1979).
To identify departures from neutral theory predictions, Tajima’s D (Tajima 1989) was computed for each locus and for each form or morph. This statistic reflects the difference between θ w and π, which are 2 different estimators of θ = 4N e µ that should be correlated positively under neutrality. At mutation-drift equilibrium, the expected value of Tajima’s D is zero, while positive values indicate population reduction or balancing selection, and negative values indicate population expansion or purifying and positive selection. For sequences including exons, we also performed the McDonald–Kreitman (MK) test (McDonald and Kreitman 1991) which compares the patterns of synonymous and nonsynonymous substitutions among and within species. The zebra finch (T. guttata) genome sequence (release 3.2.4) used as an outgroup was obtained from the genomic database Ensembl! (http://www.ensembl.org/index.html; last access: March 2016).
For all tests performed in DNAsp, significance was calculated by 10000 coalescent simulations on the basis of segregating sites and with no recombination to generate the neutral distribution of the statistics. For the MK test, significance was assessed using a 2-tailed Fisher’s exact test.
Results
Sequence Properties
Ninety-six individuals were successfully sequenced at all 7 candidate genes, for a total of approximately 1.3Mb of DNA sequences (ca. 13.5kb per individual). For the whole dataset, we found a total of 66 segregating sites, 24 of which displayed a minor allele frequency greater than 0.05. We did not observe any deviation from Hardy–Weinberg equilibrium across the 9 localities in any of the candidate genes. We obtained the entire coding sequence of the Agouti gene from individuals belonging to the highland form, and partial exonic sequences from all the other genes across all forms (Table 2 and Supplementary Figure 1). Of the 6 cis-regulatory sequences obtained for Agouti, only 3 (consisting of the second, the fourth, and the sixth marker sequenced from 5′, see Supplementary Figure 1) were polymorphic and were included in our analyses. Three indels of 4, 3, and 2bp were found in the intronic region of Agouti, in POMC and SLC24A5 genes, respectively.
Table 2.
Results from selection tests in highland (N = 48) and lowland (N = 48) forms
| Locus | Length | S | Number of sequenced exons | Cumulative length of exons | Nonsynonymous mutations | Synonymous mutations | Number of alleles | π | ϴw | Tajima’s D | MK test |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Highland polymorphic form only | |||||||||||
| Agouti (complete) | 3522 | 12 | 3 | 417 | 1 | 0 | 16 | 0.000218* | 0.000664 | −1.79** | — |
| Agouti cis 2 | 346 | 1 | 0 | 0 | 0 | 0 | 2 | 0.000061 | 0.000565 | −1.04 | — |
| Agouti cis 4 | 228 | 1 | 0 | 0 | 0 | 0 | 2 | 0.000874 | 0.000676 | 0.34 | — |
| Agouti cis 6 | 267 | 1 | 0 | 0 | 0 | 0 | 2 | 0.000682 | 0.000739 | −0.09 | — |
| Corin | 702 | 2 | 2 | 239 | 0 | 0 | 2 | 0.00146** | 0.00056 | 2.46** | — |
| DCT | 512 | 3 | 1 | 301 | 1 | 1 | 4 | 0.000904 | 0.00115 | −0.39 | NS |
| POMC | 379 | 7 | 1 | 379 | 5 | 2 | 9 | 0.002123 | 0.00364 | −0.99 | NS |
| SLC | 652 | 5 | 3 | 256 | 0 | 2 | 8 | 0.000828 | 0.001797 | −1.22 | NS |
| TYR | 767 | 2 | 1 | 767 | 2 | 0 | 3 | 0.000233 | 0.00051 | −0.85 | NS |
| TYRP1 | 589 | 1 | 1 | 382 | 1 | 0 | 2 | 0.00086* | 0.00035 | 1.78*** | NS |
| Lowland forms | |||||||||||
| Agouti (partial) | 960 | 12 | 0 | 0 | 0 | 0 | 12 | 0.00079 | 0.00243 | −1.80* | — |
| Agouti cis 2 | 346 | 0 | 0 | 0 | 0 | 0 | 1 | — | — | — | — |
| Agouti cis 4 | 228 | 1 | 0 | 0 | 0 | 0 | 2 | 0.00144 | 0.00068 | 1.3 | — |
| Agouti cis 6 | 267 | 1 | 0 | 0 | 0 | 0 | 2 | 0.00053 | 0.00074 | −0.32 | — |
| Corin | 702 | 3 | 2 | 239 | 0 | 0 | 4 | 0.0006 | 0.00083 | −0.5 | — |
| DCT | 512 | 10 | 1 | 301 | 2 | 4 | 10 | 0.00144* | 0.0038 | −1.60* | NS |
| POMC | 379 | 10 | 1 | 379 | 8 | 2 | 17 | 0.00307 | 0.00518 | −1.05 | NS |
| SLC | 652 | 6 | 3 | 256 | 1 | 2 | 10 | 0.00107 | 0.00183 | −0.95 | NS |
| TYR | 767 | 4 | 1 | 767 | 2 | 2 | 5 | 0.00014** | 0.00102 | −1.73** | NS |
| TYRP1 | 589 | 1 | 1 | 382 | 1 | 0 | 2 | 0.0001 | 0.00036 | −0.9 | NS |
S, Number of segregating sites; π: Nei & Li’s nucleotide diversity; ϴw: Watterson’s theta; NS: non-significant. Selection was assessed with Tajima’s D and positive selection was assessed in coding regions with a McDonald–Kreitman (MK) test. Significance levels: *P < 0.05, **; P < 0.01, ***; P < 0.001. Significant values after Bonferroni correction for multiple testing are indicated in bold.
Association between Genes and Plumage Color
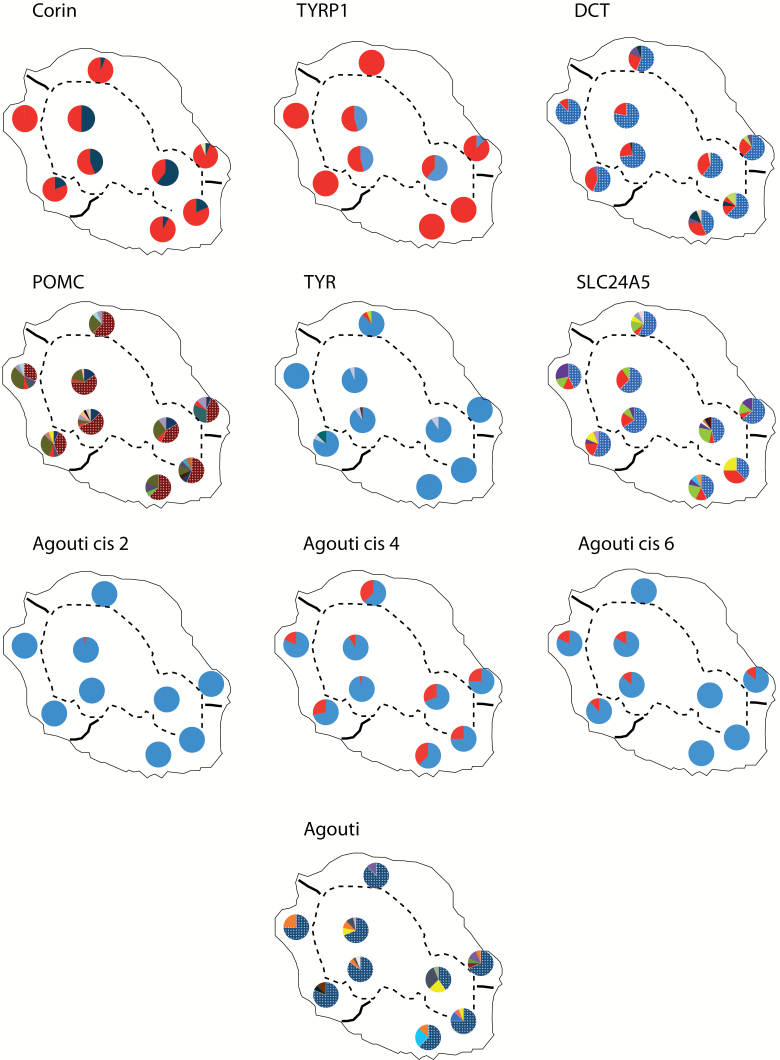
We did not find any significant association between genes and plumage color differences, except for TYRP1, where a significant association was detected with back color (P = 0.001). This association disappears when comparing brown and grey birds from the altitudinal form only (Table 3), meaning that these alleles could be linked to color variation with altitude. However, it might also be due to a population structure effect. The spatial distribution of haplotypes among forms or between morphs in the polymorphic highland form did not reveal any obvious shift in frequencies, except for Corin and TYRP1, for which there was a clear difference in frequency between lowland and highland forms. Again, this might reflect a population structure effect rather than a phenotype–genotype association, as shown by significant patterns of differentiation for both Corin and TYRP1 (Figure 2; Table 4)
Table 3.
Results from the SCORE association tests comparing birds displaying changes in head, nape, and back color (N = 96 individuals)
| Comparison | Agouti | Corin | DCT | POMC | SLC24A5 | TYR | TYRP1a |
|---|---|---|---|---|---|---|---|
| Head: brown vs. grey | 0.52 | 0.08 | 0.25 | 0.97 | 0.56 | 0.81 | 0.213 |
| Nape: brown vs. grey | 0.78 | 0.45 | 0.74 | 0.82 | 0.98 | 0.27 | 0.059 |
| Back: brown vs. grey | 0.12 | 0.18 | 0.88 | 0.38 | 0.90 | 0.26 | 0.001 |
| Highland form: brown vs. grey | 0.54 | 0.13 | 0.18 | 0.38 | 0.46 | 0.43 | 0.309 |
The fourth test compared highland grey and brown morphs (N = 48 individuals). Significant P-value is highlighted in bold.
aFor TYRP1, a Fisher’s exact test was performed as there was a single polymorphic site in the sequence.
Figure 2.
Maps showing spatial patterns of allele frequencies for the 7 candidate genes and the 3 Agouti cis-regulatory regions (2nd, 4th, and 6th marker) into which polymorphism was detected.
Table 4.
Hierarchical AMOVA at all markers used in this study
| Marker | A priori grouping | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| All 5 plumage color variants | Highland grey and brown morphs | Lowland forms | |||||||
| F CT | F SC | F ST | F CT | F SC | F ST | F CT | F SC | F ST | |
| Agouti cis 2 | −0.010 | 0.009 | −0.001 | 0 | 0 | 0 | 0 | 0 | 0 |
| Agouti cis 4 | 0.017 | 0.034 | 0.051 | 0.002 | 0.123* | 0.124* | −0.007 | −0.033 | −0.040 |
| Agouti cis 6 | 0.023 | 0.026 | 0.049 | 0.037 | 0.043 | 0.079 | 0.059 | −0.007 | 0.052 |
| Agouti partial | 0.009 | 0.119*** | 0.127*** | −0.023 | 0.115** | 0.095** | −0.033 | 0.105*** | 0.076*** |
| Agouti complete | — | — | — | −0.030 | 0.099** | 0.072** | — | — | — |
| Corin | 0.224** | 0.001 | 0.225*** | 0.008 | −0.021 | −0.013 | −0.036 | 0.048 | 0.014 |
| DCT | −0.003 | 0.008 | 0.006 | 0.016 | 0.021 | 0.037 | −0.013 | 0.004 | −0.009 |
| POMC | 0.009 | 0.010 | 0.019 | −0.016 | 0.018 | 0.002 | 0.010 | 0.001 | 0.011 |
| SLC24A5 | 0.001 | 0.036* | 0.037* | −0.005 | 0.046 | 0.041 | 0.016 | 0.026 | 0.042 |
| TYR | 0.012 | 0.020 | 0.031 | −0.016 | 0.017 | 0.001 | 0.006 | 0.018 | 0.024 |
| TYRP1 | 0.292* | 0.019 | 0.306*** | −0.029 | 0.009 | −0.020 | 0.010 | 0.038 | 0.048 |
Analyses were conducted using plumage color variant, highland color morph, or lowland color form as grouping factors. F CT, F SC, F ST stand for genetic differences among a priori defined groups, among localities within groups, and among localities, respectively. Significance levels: *P < 0.05, **P < 0.01, ***P < 0.001. Significant values are highlighted in bold.
Tests of Selective Neutrality
Coalescence simulations showed that Agouti coding regions displayed significantly lower nucleotide diversity (π) than expected under a neutral model before Bonferroni correction (Table 2). On the other hand, Corin and TYRP1 displayed significantly higher nucleotide diversity than neutral expectations, but only for samples belonging to the polymorphic highland form. As noted previously, this change in diversity was not associated to a particular plumage color, both grey and brown morphs displaying the same allele frequencies (Table 4). The number of segregating sites was generally higher in the 3 lowland forms than in the polymorphic highland form.
A few genes displayed significant deviation from neutrality (Table 2). Among these markers, the region of Agouti including the 3 coding exons and 2 introns showed a tendency toward negative values for Tajima’s D, but none of the tests was significant after Bonferroni correction. In contrast, Corin and TYRP1 displayed a strong and significant trend toward a positive Tajima’s D indicating population reduction or balancing selection. However, this pattern was found only in polymorphic highland populations. The MK test could not be performed on Agouti and Corin as no polymorphic synonymous mutation on coding sequences could be observed, but was always nonsignificant for other markers.
Discussion
We report here a comprehensive screening of the variation in a series of candidate genes involved in melanin-based pigmentation in a wild passerine bird displaying a complex melanic polymorphism not explained by MC1R variation. Although we did not sequence the whole coding sequence of each gene, except for Agouti, we would expect that any gene involved in color variation should be a target of selection, since divergent selection seems to have shaped plumage color patterns (Cornuault et al. 2015). Moreover, it has been suggested that the genetic determinism of color variation is quite simple. At high altitude, crosses between grey individuals produce both grey and brown offsprings, whereas crosses between brown parents produce only brown offsprings (Gill 1973). This observation suggests a role for a few loci of major effect, at least for the 2 sympatric morphs at high elevation. This would result in detecting more easily association between plumage patterns and genetic variants even at markers that do not include the selected sites, though genetic hitchhiking (Barton 2000). However, we found no evidence across all genes that sequence variation is associated with plumage color differences in Z. borbonicus. This result suggests that variation at other genes besides Agouti, POMC, SLC24A5, Corin, DCT, TYRP1, and TYR must be involved in melanic plumage color variation. In particular, we found no evidence that Agouti, a promising candidate gene for melanin-based pigmentation patterns (e.g., Nadeau et al. 2008; Manceau et al. 2011), experienced disruptive selection that could have driven plumage color divergence between forms or between morphs within the highland form. However, the large cis-regulatory region in Agouti and the low variability in our cis-regulatory markers leave the possibility of undetected selection far from the sites we studied, especially if selection is relatively weak.
We found an interesting pattern of increased polymorphism at TYRP1 and Corin in the polymorphic highland form as well as important shifts in allele frequencies between this form and the 3 lowland forms, although no relation between alleles and plumage color was observed. The shift in allele frequencies might indicate a change in selective regime between lowland and highland forms for TYRP1 and Corin. Apart for these 2 loci, no significant differentiation between highland and lowland forms could be detected in this work. Previous studies nonetheless showed genetic differentiation between these forms (Milá et al. 2010), as well as small-scale differentiation between populations (as highlighted in Bertrand et al. 2014). Only a more detailed study at the genomic scale would bring insights about how common such outliers are under a scenario of pure drift. Changes in allele frequencies may be linked to the tendency for highland forms to display darker plumage (as first documented by Gloger in 1833), with melanic plumage color patterns conveying information on physiological adaptations to altitude.
Melanin-based plumage color phenotypes can be superficially similar, yet do not necessarily rely on the same physiological mechanisms and thus might not share the same genetic basis (e.g., Poelstra et al. 2013, 2014). The popularity of loci such as Agouti or MC1R, which are often considered as “optimally pleiotropic” genes (Kopp 2009) due to their limited physiological impact outside color variation (Ducrest et al. 2008; Hubbard et al. 2010), may have led to an assessment bias inflating their importance. Focusing on such genes does not reflect the whole range of processes underlying and maintaining melanic color variation. First, pleiotropic effects may be under positive selection as was shown in the barn owl (Tyto alba) where high concentrations in melanocortin enhance immune response and influence coloration (Roulin et al. 2011). Second, it is likely that mutations in cis-regulatory regions control and fine-tune gene expression in space and time during development, thus limiting any negative impact on fitness, as was shown for Agouti in mice (Steiner et al. 2007; Manceau et al. 2011). Such mutations also seem to explain changes in spot position on Drosophila wings (Gompel et al. 2005), or the plasticity of mimetic patterns in Heliconius butterflies (Joron et al. 2006). Third, selective sweeps or chromosomal rearrangements might increase the frequency of mutations having an impact on color if those mutations are found close to loci involved in local adaptation. If negative effects due to mutations on genes involved in color patterning are counterbalanced by advantageous mutations linked to local adaptation or alternative mating strategies, then color polymorphism might maintain even if color genes with broad pleiotropic effects are involved. Such association between genes with different physiological function may occur in white-throated sparrows, where an inversion on chromosome 2 was found to be responsible for both color and behavioral variation (Thomas et al. 2008).
Bird studies having shown the involvement of MC1R mostly focused on species displaying discrete color polymorphism with mutations associated with color variation across the whole body and rarely on situations where variants differ in the patterns of melanin deposition across the body (see Bourgeois et al. 2012 for a summary). Color pattern evolution in species with complex patterns of melanin deposition may be due to local changes in the expression of pigmentation genes rather than amino acid mutations (e.g., Manceau et al. 2011). This is strongly suggested by recent results in crows, where local variation in gene expression has been found to be correlated with the presence or absence of grey and black color patches (Poelstra et al. 2015). Since color variation in Z. borbonicus consists of local changes in pigment deposition, it seems likely that the mutations responsible for color pattern differences between variants and morphs lie in the 5′ cis-regulatory region of one or a few genes that remain to be identified.
The lack of obvious association between plumage color and major genes in the melanocortin pathway in Z. borbonicus shows the existence of untapped genetic complexity in the genetic basis of melanin-based coloration in birds and perhaps other vertebrates. It also confirms that mechanisms of plumage color evolution are more diverse than implied by studies of discrete melanic/nonmelanic polymorphisms and their study will greatly benefit from the application of next-generation sequencing to candidate gene finding in nonmodel species, for example, through genome-wide association mapping (Ellegren et al. 2012; Bourgeois et al. 2013; Poelstra et al. 2014).
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
Y.B., J.B., B.D., and J.C. were supported by MESR (Ministère de l’Enseignement Supérieur et de la Recherche) PhD scholarships. The research was supported by Fondation pour la Recherche sur la Biodiversité (FRB) and PEPS-CNRS grants to CT, a National Geographic Society grant to BM, and the “Laboratoire d’Excellence” TULIP (ANR-10-LABX-41).
Data Availability
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.47dq3
Supplementary Material
Acknowledgments
Ben Warren, Guillaume Gélinaud, Dominique Strasberg, Juli Broggi, Magali Thierry, René-Claude Billot, Jean-Michel Probst, Isabelle Henry, Vincent Leconte, Marc Salamolard, and Benoît Lequette provided valuable help with fieldwork and logistics. We thank Philipp Heeb for comments on an earlier version of the manuscript and the Réunion National Park for granting permission to conduct fieldwork. We also thank Patricia Jargeat and Emeline Lhuillier for much advice in the lab, Hopi Hoekstra and her lab members for kindly sharing their expertise in pigmentation genes. We thank 3 anonymous reviewers for their comments which improved an earlier version of the manuscript.
References
- Barton NH. 2000. Genetic hitchhiking. Philos Trans R Soc Lond B Biol Sci. 355:1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DC, Lamoreux ML. 2003. Review—pigment gene focus the color loci of mice—a genetic century. Pigment Cell Res. 16:333–344. [DOI] [PubMed] [Google Scholar]
- Bertrand JAM, Bourgeois YXC, Delahaie B, Duval T, García-Jiménez R, Cornuault J, Heeb P, Milá B, Pujol B, Thébaud C. 2014. Extremely reduced dispersal and gene flow in an island bird. Heredity. 112:190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois YX, Bertrand JA, Thébaud C, Milá B. 2012. Investigating the role of the melanocortin-1 receptor gene in an extreme case of microgeographical variation in the pattern of melanin-based plumage pigmentation. PLoS One. 7:e50906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois YX, Lhuillier E, Cézard T, Bertrand JA, Delahaie B, Cornuault J, Duval T, Bouchez O, Milá B, Thébaud C. 2013. Mass production of SNP markers in a nonmodel passerine bird through RAD sequencing and contig mapping to the zebra finch genome. Mol Ecol Resour. 13:899–907. [DOI] [PubMed] [Google Scholar]
- Buggiotti L. 2007. Avian evolutionary genomics : studies of Ficedula flycatchers [PhD thesis]. [Turku (Finland)]: University of Turku. [Google Scholar]
- Cornuault J, Delahaie B, Bertrand JAM, Bourgeois YXC, Milá B, Heeb P, Thébaud C. 2015. Morphological and plumage color variation in the Réunion grey white-eye. Aves: Zosterops borbonicus): assessing the role of selection. Biol Jour Linn Soc. 114:459–473. [Google Scholar]
- Cheviron ZA, Hackett SJ, Brumfield RT. 2006. Sequence variation in the coding region of the melanocortin-1 receptor gene (MC1R) is not associated with plumage variation in the blue-crowned manakin (Lepidothrix coronata). Proc Biol Sci. 273:1613–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D, Chapman J, Cooper J. 2004. Use of unphased multilocus genotype data in indirect association studies. Genet Epidemiol. 27:415–428. [DOI] [PubMed] [Google Scholar]
- Domyan ET, Guernsey MW, Kronenberg Z, Krishnan S, Boissy RE, Vickrey AI, Rodgers C, Cassidy P, Leachman SA, Fondon JW, et al. 2014. Epistatic and combinatorial effects of pigmentary gene mutations in the domestic pigeon. Curr Biol. 24:459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducrest A-L, Keller L, Roulin A. 2008. Pleiotropy in the melanocortin system, coloration and behavioural syndromes. Trends Ecol Evol. 23:502–510. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Smeds L, Burri R, Olason PI, Backström N, Kawakami T, Künstner A, Mäkinen H, Nadachowska-Brzyska K, Qvarnström A, et al. 2012. The genomic landscape of species divergence in Ficedula flycatchers . Nature. 491:756–760. [DOI] [PubMed] [Google Scholar]
- Enshell-Seijffers D, Lindon C, Morgan BA. 2008. The serine protease Corin is a novel modifier of the Agouti pathway. Development. 135:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10:564–567. [DOI] [PubMed] [Google Scholar]
- Gill FB. 1973. Intra-island variation in the mascarene white-eye Zosterops borbonica . Ornithol Monogr. 12:1–66. [Google Scholar]
- Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila . Nature. 433:481–487. [DOI] [PubMed] [Google Scholar]
- Griffiths R, Double MC, Orr K, Dawson RJ. 1998. A DNA test to sex most birds. Mol Ecol. 7:1071–1075. [DOI] [PubMed] [Google Scholar]
- Gunnarsson U, Hellström AR, Tixier-Boichard M, Minvielle F, Bed’hom B, Ito S, et al. 2007. Mutations in SLC45A2 cause plumage color variation in chicken and Japanese quail. Genetics. 175:867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas F, Pointer MA, Saino N, Brodin A, Mundy NI, Hansson B. 2009. An analysis of population genetic differentiation and genotype-phenotype association across the hybrid zone of carrion and hooded crows using microsatellites and MC1R. Mol Ecol. 18:294–305. [DOI] [PubMed] [Google Scholar]
- Hill GE and McGraw KJ. 2006. Bird coloration. Volume I. Mechanisms and Measurements. Cambridge (MA): Harvard University Press. [Google Scholar]
- Hiragaki T, Inoue-Murayama M, Miwa M, Fujiwara A, Mizutani M, Minvielle F, Ito S. 2008. Recessive black is allelic to the yellow plumage locus in Japanese quail and associated with a frameshift deletion in the ASIP gene. Genetics. 178:771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE. 2006. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity. 97: 222–234. [DOI] [PubMed] [Google Scholar]
- Hubbard JK, Uy JA, Hauber ME, Hoekstra HE, Safran RJ. 2010. Vertebrate pigmentation: from underlying genes to adaptive function. Trends Genet. 26:231–239. [DOI] [PubMed] [Google Scholar]
- Jackson IJ, Chambers DM, Tsukamoto K, Copeland NG, Gilbert DJ, Jenkins NA, et al. 1992. A second tyrosinase-related protein, TRP-2, maps to and is mutated at the mouse slaty locus. EMBO J. 11:527–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joron M, Jiggins CD, Papanicolaou A, McMillan WO. 2006. Heliconius wing patterns: an evo-devo model for understanding phenotypic diversity. Heredity (Edinb). 97:157–167. [DOI] [PubMed] [Google Scholar]
- Kingsley EP, Manceau M, Wiley CD, Hoekstra HE. 2009. Melanism in peromyscus is caused by independent mutations in Agouti. PLoS One. 4:e6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A. 2009. Metamodels and phylogenetic replication: a systematic approach to the evolution of developmental pathways. Evolution. 63:2771–2789. [DOI] [PubMed] [Google Scholar]
- Lamason RL, Mohideen M-APK, Mest JR, Wong AC, Norton HL, Aros MC, et al. 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science. 310:1782–1786. [DOI] [PubMed] [Google Scholar]
- Lehtonen PK, Laaksonen T, Artemyev V, Belskii E, Berg PR, Both C, et al. 2012. Candidate genes for color and vision exhibit signals of selection across the pied flycatcher (Ficedula hypoleuca) breeding range. Heredity. 108:431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- MacDougall-Shackleton EA, Blanchard L, Igdoura SA, Gibbs HL. 2003. Unmelanized plumage patterns in Old World leaf warblers do not correspond to sequence variation at the melanocortin-1 receptor locus (MC1R). Mol Biol Evol. 20:1675–1681. [DOI] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Linnen CR, Rosenblum EB, Hoekstra HE. 2010. Convergence in pigmentation at multiple levels: mutations, genes and function. Philos Trans R Soc Lond B Biol Sci. 365:2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Mallarino R, Hoekstra HE. 2011. The developmental role of Agouti in color pattern evolution. Science. 331:1062–1065. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. 1991. Adaptive protein evolution at the Adh locus in Drosophila . Nature. 351:652–654. [DOI] [PubMed] [Google Scholar]
- Milá B, Warren BH, Heeb P, Thébaud C. 2010. The geographic scale of diversification on islands: genetic and morphological divergence at a very small spatial scale in the Mascarene grey white-eye (Aves: Zosterops borbonicus). BMC Evol Biol. 10:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle F, Gourichon D, Ito S, Inoue-Murayama M, Rivière S. 2007. Effects of the dominant lethal yellow mutation on reproduction, growth, feed consumption, body temperature, and body composition of the Japanese quail. Poult Sci. 86:1646–1650. [DOI] [PubMed] [Google Scholar]
- Mullen LM, Hoekstra HE. 2008. Natural selection along an environmental gradient: a classic cline in mouse pigmentation. Evolution. 62:1555–1570. [DOI] [PubMed] [Google Scholar]
- Mundy NI, Badcock NS, Hart T, Scribner K, Janssen K, Nadeau NJ. 2004. Conserved genetic basis of a quantitative plumage trait involved in mate choice. Science. 303:1870–1873. [DOI] [PubMed] [Google Scholar]
- Nadeau NJ, Minvielle F, Ito S, Inoue-Murayama M, Gourichon D, Follett SA, et al. 2008. Characterization of Japanese quail yellow as a genomic deletion upstream of the avian homolog of the mammalian ASIP (agouti) gene. Genetics. 178:777–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau NJ, Mundy NI, Gourichon D, Minvielle F. 2007. Association of a single-nucleotide substitution in TYRP1 with roux in Japanese quail (Coturnix japonica). Anim Genet. 38:609–613. [DOI] [PubMed] [Google Scholar]
- Nei M, Li WH. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 76:5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelstra JW, Ellegren H, Wolf JB. 2013. An extensive candidate gene approach to speciation: diversity, divergence and linkage disequilibrium in candidate pigmentation genes across the European crow hybrid zone. Heredity (Edinb). 111:467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelstra JW, Vijay N, Bossu CM, Lantz H, Ryll B, Müller I, Baglione V, Unneberg P, Wikelski M, Grabherr MG, Wolf JBW. 2014. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science. 344:1410–1414. [DOI] [PubMed] [Google Scholar]
- Poelstra JW, Vijay N, Hoeppner MP, Wolf JBW. 2015. Transcriptomics of color patterning and coloration shifts in crows. Mol Ecol. 24:4617–4628. doi: 10.1111/mec.13353. [DOI] [PubMed] [Google Scholar]
- Römpler H, Rohland N, Lalueza-Fox C, Willerslev E, Kuznetsova T, Rabeder G, et al. 2006. Nuclear gene indicates coat-color polymorphism in mammoths. Science. 313:62. [DOI] [PubMed] [Google Scholar]
- Rosenblum EB, Hoekstra HE, Nachman MW. 2004. Adaptive reptile color variation and the evolution of the Mc1r gene. Evolution. 58:1794–1808. [DOI] [PubMed] [Google Scholar]
- Roulin A. 2004. The evolution, maintenance and adaptive function of genetic colour polymorphism in birds. Biol Rev Camb Philos Soc. 79:815–848. [DOI] [PubMed] [Google Scholar]
- Roulin A, Emaresi G, Bize P, Gasparini J, Piault R, Ducrest AL. 2011. Pale and dark reddish melanic tawny owls differentially regulate the level of blood circulating POMC prohormone in relation to environmental conditions. Oecologia. 166:913–921. [DOI] [PubMed] [Google Scholar]
- Roulin A, Ducrest AL. 2013. Genetics of colouration in birds. Semin Cell Dev Biol. 24:594–608. [DOI] [PubMed] [Google Scholar]
- Sarangarajan R, Boissy RE. 2001. Tyrp1 and oculocutaneous albinism type 3. Pigment Cell Res. 6:437–444. [DOI] [PubMed] [Google Scholar]
- Sato S, Otake T, Suzuki C, Saburi J, Kobayashi E. 2007. Mapping of the recessive white locus and analysis of the tyrosinase gene in chickens. Poult Sci. 86:2126–2133. [DOI] [PubMed] [Google Scholar]
- Seutin G, White BN, Boag PT. 1991. Preservation of avian blood and tissue samples for DNA analyses. Can J Zool. l69:82–90. [Google Scholar]
- Steiner CC, Weber JN, Hoekstra HE. 2007. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 5:e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. 2003. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 73:1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theron E, Hawkins K, Bermingham E, Ricklefs RE, Mundy NI. 2001. The molecular basis of an avian plumage polymorphism in the wild: a melanocortin-1-receptor point mutation is perfectly associated with the melanic plumage morph of the bananaquit, Coereba flaveola . Curr Biol. 11:550–557. [DOI] [PubMed] [Google Scholar]
- Thomas JW, Cáceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, et al. 2008. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics. 179:1455–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uy JA, Moyle RG, Filardi CE, Cheviron ZA. 2009. Difference in plumage color used in species recognition between incipient species is linked to a single amino acid substitution in the melanocortin-1 receptor. Am Nat. 174:244–254. [DOI] [PubMed] [Google Scholar]
- Vaez M, Follett SA, Bed’hom B, Gourichon D, Tixier-Boichard M, Burke T. 2008. A single point-mutation within the melanophilin gene causes the lavender plumage colour dilution phenotype in the chicken. BMC Genet. 9:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. 1995. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 11:328–330. [DOI] [PubMed] [Google Scholar]
- Watterson GA. 1975. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 7:256–276. [DOI] [PubMed] [Google Scholar]
- Yan W, Sheng N, Seto M, Morser J, Wu Q. 1999. Corin, a mosaic transmembrane serine protease encoded by a novel cDNA from human heart. J Biol Chem. 274:14926–14935. [DOI] [PubMed] [Google Scholar]
- Yaswen L, Diehl N, Brennan MB, Hochgeschwender U. 1999. Obesity in the mouse model of pro-opiomelanocortin deficiency responds to peripheral melanocortin. Nat Med. 5:1066–1070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.47dq3