During vertebrate limb development, Hoxd genes are regulated following a bimodal strategy involving two topologically associating domains (TADs) located on either side of the gene cluster. This work shows that the HOX13 proteins themselves help switch off the telomeric TAD. At the same time, they directly interact with distal enhancers to sustain the activity of the centromeric TAD, thus explaining both the sequential and exclusive operating processes of these two regulatory domains.
Keywords: topologically associating domains (TADs), vertebrate limbs, regulatory landscapes, wrist, ankle, polycomb
Abstract
During vertebrate limb development, Hoxd genes are regulated following a bimodal strategy involving two topologically associating domains (TADs) located on either side of the gene cluster. These regulatory landscapes alternatively control different subsets of Hoxd targets, first into the arm and subsequently into the digits. We studied the transition between these two global regulations, a switch that correlates with the positioning of the wrist, which articulates these two main limb segments. We show that the HOX13 proteins themselves help switch off the telomeric TAD, likely through a global repressive mechanism. At the same time, they directly interact with distal enhancers to sustain the activity of the centromeric TAD, thus explaining both the sequential and exclusive operating processes of these two regulatory domains. We propose a model in which the activation of Hox13 gene expression in distal limb cells both interrupts the proximal Hox gene regulation and re-enforces the distal regulation. In the absence of HOX13 proteins, a proximal limb structure grows without any sign of wrist articulation, likely related to an ancestral fish-like condition.
Vertebrate limbs have been a paradigm in our understanding of the emergence of patterns during evolution and development, in terms of both the molecules involved and the underlying principles (e.g., see Tabin and Wolpert 2007). Developing limb buds indeed implement all major signaling pathways as well as families of transcription factors known for their importance in the building of the embryo. This co-optation of key developmental functions to accompany the evolution of paired limbs was initially observed for the HoxD cluster (Dolle et al. 1989; Lewis and Martin 1989), which, together with the HoxA cluster (Haack and Gruss 1993; Yokouchi et al. 1995b), is critical for the formation of the proximal and distal pieces of our arms and legs (Davis et al. 1995; Fromental-Ramain et al. 1996; Kmita et al. 2005). While the exact function of HOX proteins during limb development remains to be fully assessed, they seem to be involved in the control of bone growth (Zákány and Duboule 1996; Villavicencio-Lorini et al. 2010; Gonzalez-Martin et al. 2014; Kuss et al. 2014) in both complementary and redundant manners (references in Zakany and Duboule 2007).
Hoxa and Hoxd genes are transcribed into precisely delimited domains within the incipient limb buds, which will determine the advent and positioning of future morphologies. For instance, Hoxa13 determines the distal part of the growing limb, the digits, whereas its neighbor, Hoxa11, is a marker of the proximal limb piece, the forearm (Yokouchi et al. 1991; Nelson et al. 1996). The study of the regulations acting over the HoxA cluster to elicit these transcript patterns revealed the presence of long-range global enhancers (Lehoczky and Innis 2008) located within a flanking topologically associating domain (TAD) (Berlivet et al. 2013; Woltering et al. 2014); i.e., a chromatin structure where enhancer–promoter contacts as well as constitutive interactions are privileged (Dixon et al. 2012; Nora et al. 2012; Sexton and Cavalli 2015). In the case of Hoxd genes, genetic and molecular analyses have shown that their complex expression patterns are controlled by the successive implementation of global regulations contained within two flanking TADs, covering the neighboring gene deserts (Andrey et al. 2013).
In early limb buds, enhancers located within the telomeric TAD (T-DOM) control the transcription of a central group of genes, from Hoxd8 to Hoxd11, into the presumptive forearm (Andrey et al. 2013). Soon after, in the most distal aspect of the limb bud, the T-DOM stops operating, and enhancers specific for digit expression, located within the centromeric TAD (C-DOM), become activated (Montavon et al. 2011). Therefore, the complete expression pattern of Hoxd genes involves a switch from a situation in which T-DOM is active and C-DOM is inactive to a state where C-DOM is active and T-DOM is inactive (see Lonfat and Duboule 2015). The existence of two separate and independent regulatory landscapes allows for the appearance of a stripe of nonexpressing cells between the two transcript domains. These cells express low levels of Hox genes and are thought to produce the mesopodial articulation; i.e., the wrist and ankle (Villavicencio-Lorini et al. 2010; Woltering and Duboule 2010). In this context, the switch in TAD regulations is key in the making and positioning of the mesopodium, an essential structure in the evolution of tetrapods, which allowed them to properly articulate their newly acquired digits.
The molecular mechanism controlling the switch from T-DOM to C-DOM at the HoxD locus is hard to study due to the high number of enhancers, their remote locations, and the scarcity of in vivo biological material. However, some evidence suggested that the HOX13 proteins themselves may have a negative impact on the transcription of forearm-specific Hoxa and Hoxd genes. First, the proximal boundary of the Hoxa13 expression domain, which labels the distal end of the forearm, exactly matches the distal boundaries of the cellular domains expressing either Hoxa11 or Hoxd9, Hoxd10, and Hoxd11 (e.g., see Woltering and Duboule 2010). The separation between these expression domains is not observed in fish, where the Hoxa13 and Hoxa11 cellular territories overlap, suggesting an important evolutionary change (van der Hoeven et al. 1996; Metscher et al. 2005; Davis et al. 2007; Tamura et al. 2008). Second, limb buds lacking the function of Hoxa13 showed a slight distalization of Hoxa11 expression, as if this latter gene was derepressed in distal cells (Yokouchi et al. 1995a; Post and Innis 1999). The additional removal of Hoxd13 function clearly enhanced this phenomenon (Sheth et al. 2014; Woltering et al. 2014). Furthermore, the same effect was observed upon Hoxd gene expression, whose transcription was equally gained in more distal domains (Sheth et al. 2014), suggesting that HOX13 proteins may indeed globally repress the T-DOM-dependent transcription of Hoxd genes and thus terminate forearm-specific patterning instructions.
Here, we investigated the possibility that the HOX13 proteins have a direct impact on the transcription of Hoxd genes to repress their transcription in the digital plate, thereby establishing the expression boundary between the proximal and distal limb domain, leading to the positioning of the wrist. We show that HOXA13 binding is enriched at the HoxD locus within both TADs. In the T-DOM, HOXA13 exerts a negative effect by switching off transcriptional activity, an effect genetically enhanced by the presence of HOXD13. In contrast, the binding of HOXA13 to the regulatory islands within the C-DOM is required to sustain transcription in developing digits. This antagonistic effect of HOX13 proteins on the two TADs flanking the HoxD cluster explains why these two regulatory landscapes are never active concomitantly in the same cells during limb development. It also gives a molecular basis to this important switch in regulations, which is necessary for the emergence and positioning of the wrist. We discuss the relevance of these observations in the evolutionary framework of the fin-to-limb transition.
Results
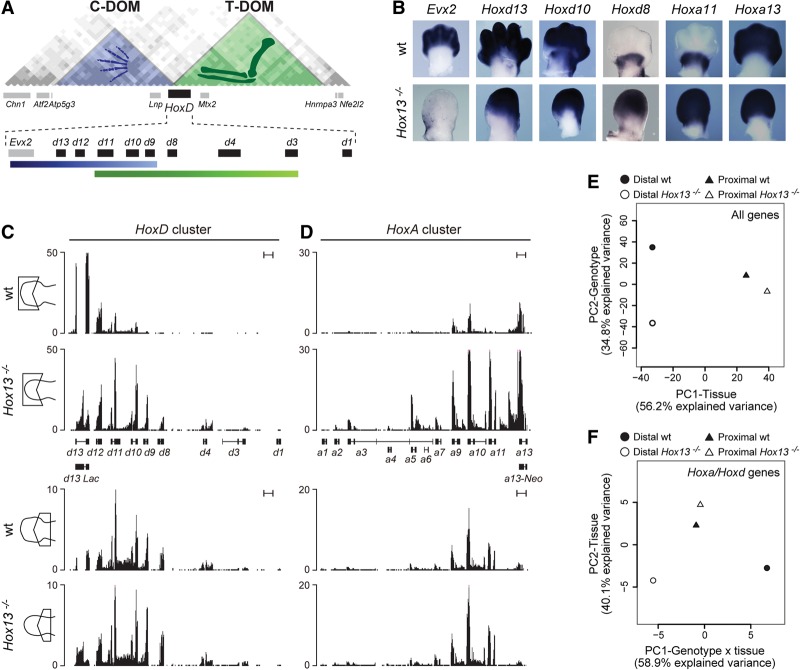
The HoxD cluster is flanked by two large gene deserts (T-DOM and C-DOM), which match the extent of two TADs (Fig. 1A; Dixon et al. 2012). The telomeric T-DOM contains at least two enhancers (CS39 and CS65), which regulate the transcription from Hoxd8 to Hoxd11 into a proximal limb domain (Andrey et al. 2013). The centromeric C-DOM contains at least six enhancers, which participate in the control of Hoxd13 to Hoxd9 transcription into presumptive digits (Montavon et al. 2011). While mice mutant for Hoxd13 display ill-formed digits (Dolle et al. 1993), double mutants for both Hoxd13 and Hoxa13 suffer from severe agenesis of the most distal limb pieces (Fromental-Ramain et al. 1996; Kmita et al. 2005). However, such mutant limbs at embryonic day 12.5 (E12.5) still display a distal cellular field, which clearly resembles a hand plate although smaller in size (Fig. 1B). We asked whether this distal domain was a reduced autopod in molecular terms or instead whether the double loss of function of group 13 genes had lead to an extension of the zeugopodial (forearm) identity due to the absence of distal determinants.
Figure 1.
Loss of HOX13 functions affects limb patterning and proximal to distal identities. (A) Hi-C data adapted from Dixon et al. (2012) showing the positions and distribution of the two TADs (C-DOM and T-DOM) flanking the HoxD cluster. Black and gray boxes represent Hoxd and unrelated genes, respectively. The blue and green graded lines below indicate the two sets of Hoxd genes, which are expressed in either the distal domain (blue) or the proximal domain (green). C-DOM and T-DOM are depicted with the same colors to indicate the presence of the corresponding enhancer sequences. (B) In situ hybridization analysis showing the expression of different genes (indicated at the top) in the forelimbs of either wild-type (wt) or Hoxa13−/−; Hoxd13−/− (Hox13−/−) double-mutant specimens at E12.5–E13. While some genes maintain their transcription profiles, others are either lost or gained in the distal part. (C,D) Transcription profiles of the HoxA and HoxD clusters in microdissected distal and proximal forelimb domains from either wild-type or Hox13−/− mutant specimens at E12.5. Bar, 5 kb. While the profiles remain the same in control and mutant proximal domains, clear differences are observed in the distal limb. (E,F) Principal component analysis (PCA) of the transcriptomes obtained from proximal and distal forelimb samples dissected out from either wild-type or Hox13−/− mutant specimens. The analysis was carried out by using the log2 transformed FPKM (fragments per kilobase per million mapped fragments) values of either all autosomal genes (E) or the genes present only in the HoxA and HoxD cluster (F). In both cases, the Hox13−/− distal domain (empty circle) does not cluster with either the wild-type counterpart (filled circle) or the wild-type proximal (filled triangle) domain. The separation between wild-type and Hox13−/− distal limb samples is visible on the second PCA axis when considering all autosomal genes (E) and on the first axis when considering only genes located within the HoxA and HoxD clusters (F).
Genetic identity of Hox13 double-mutant limbs
We performed RNA sequencing (RNA-seq) analysis from proximal and distal developing wild-type forelimbs at E12.5 and selected a set of genes showing at least a threefold biased expression level between the proximal and distal domains, referred to here as “proximal” and “distal” genes. We looked at their expression in Hoxa13−/−;Hoxd13−/− double-mutant limbs (termed here Hox13−/−) at E12.5 and obtained somewhat conflicting results. The distal-most aspect of limb buds lacking both HOXA13 and HOXD13 functional proteins due to insertional mutagenesis still contained Hoxa13 and Hoxd13 transcripts, suggesting the persistence of a distal identity (Fig. 1B). Likewise, expression of Hoxd10 and Hoxd11 was maintained distally (Fig. 1B,C), as well as those of other distal genes such as Prrx2 and Lhx2 (Supplemental Fig. S1; Rincon-Limas et al. 1999). On the other hand, some proximal gene expression, including Shox2 and Hoxa11 (Neufeld et al. 2014), was clearly gained within the distal domain, suggestive of an extension of proximal identity into the most distal limb aspect (Fig. 1B,D; Supplemental Fig. S1). Also, a number of genes expressed in the distal limb, including Dbx2, Evx2, and Aldh1a2, showed reduced levels of mRNAs in distal cells of Hox13−/− mutant limbs (Fig. 1B; Supplemental Fig. S1; Shou et al. 2013).
To have a more quantitative assessment, we microdissected proximal and distal pieces from Hox13−/− mutant limbs at E12.5 and produced RNA-seq data sets. Expectedly, the analysis of both HoxA and HoxD clusters revealed no striking difference between control and Hox13−/− mutant proximal limbs; i.e., the domain where neither Hoxa13 nor Hoxd13 is normally expressed (Fig. 1C,D). In the Hox13−/− mutant distal limb, however, the profiles were distinct from their controls and looked more similar to the proximal limb profiles. In the case of HoxD, Hoxd8, Hoxd9, Hoxd10, and Hoxd11 mRNAs were gained, resembling the situation seen in the proximal limb (Fig. 1C). The up-regulation of Hoxd8 in the distal domain of mutant limbs was unexpected, since this gene is normally not strongly regulated by the digit enhancers located within the C-DOM. Likewise, Hoxa11 was gained in the Hox13−/− mutant distal limb, as shown by in situ hybridization (Sheth et al. 2014; Woltering et al. 2014), as well as other more proximal genes like Hoxa5 and Hoxa7 (Fig. 1B,D). Therefore, the expression profiles in the distal limb domain of Hox13−/− mutants were distinct from their control counterparts and generally more related to those of control proximal limbs. Surprisingly, however, the global expression levels of both Hoxd and Hoxa genes in Hox13−/− distal limb domains were higher than in the control proximal domain (Fig. 1B–D; Supplemental Fig. S1), suggesting that HOX13 proteins negatively regulate the mRNA levels of other Hox genes. At the HoxD locus, the Lnp and Evx2 genes, which are localized within the C-DOM on the anti-Hox DNA strand and normally are coexpressed with Hoxd13 in the distal limb, were significantly down-regulated in the Hox13−/− mutant condition (Supplemental Fig. S2).
To assess more generally whether these differences in Hox expression profiles reflected a transformation of the distal domain identity toward a more proximal fate, we compared the transcriptomes using a principal component analysis (PCA). When all autosomal genes were considered (Fig. 1E), the majority of the variability observed among our samples (56.2%) was attributable to the proximal versus distal identity of the analyzed domains. Along this axis, the control and the Hox13−/− mutant proximal limb data sets clustered together and were separated from both the control and Hox13−/− mutant distal data sets, indicating that in the Hox13−/− mutant situation, the distal part of the limb did not simply acquire a proximal identity. The control and Hox13−/− mutant distal domains were nevertheless clearly differentiated along the second axis of the PCA (which accounted for 35% of the total variability), supporting the idea that both functions of Hox13 genes are required for the specification of normal autopods. In contrast, the proximal domains from control and Hox13−/− mutant limbs showed very little difference, as expected from the lack of function of Hox13 genes in this territory.
When the PCA was restricted to the genes, transcripts, and long noncoding RNAs (lncRNAs) present on both strands in the HoxA and HoxD clusters only, most of the variability across samples resulted from the difference between the control and Hox13−/− mutant distal limb domains (Fig. 1F). The first PCA component clearly separated the two distal limb samples, whereas both proximal limb samples clustered together at an intermediate position on the same axis. However, proximal and distal limb identities were still separated along the second PCA component, supporting the observation that the “Hox identity” of the Hox13−/− mutant distal limb territory was not transformed into a proximal identity. These results suggested that both Hoxa and Hoxd genes were more affected by the lack of HOX13 proteins than the bulk of other genes expressed in this distal domain, thus raising the possibility that HOX13 function might be involved in Hox gene regulation within this domain.
Lack of HOX13 proteins has an impact on global HoxD regulation
Hoxd genes up-regulated distally in the absence of HOX13 proteins were precisely those that are strongly coregulated in the wild-type proximal domain (Andrey et al. 2013), suggesting that in the Hox13−/− mutant limbs, the digit enhancers within the C-DOM had changed their realm of action to become able to regulate genes located at a more telomeric position, such as Hoxd8. Alternatively, the telomeric forearm-specific enhancers, normally at work in the proximal domain only, may have lost this restriction in Hox13−/− mutant limbs to continue to exert their regulatory function into the most distal cells. To discriminate between these two scenarios, we used different alleles where the digits enhancers could no longer regulate their Hoxd targets.
The first allele was a large HoxDDel(Atf2-Nsi) deletion, which removes the entire C-DOM [Fig. 2A, Del(Atf2-Nsi)]. In this deletion, expression of Hoxd13 to Hoxd10 was expectedly abolished in the distal domain at E12.5 due to the lack of the appropriate enhancers (Fig. 2A, top), whereas Hoxd10 was still expressed in the proximal part in response to the intact telomeric enhancers (Fig. 2A). However, the combined abrogation of Hoxa13 function induced restoration of both Hoxd13 and Hoxd10 expression in the distal parts of limbs at E12.5 (Fig. 2A, bottom). As anticipated, the distribution of Hoxa13 transcripts did not change in these mutants (data not shown). From this experiment, we concluded that Hoxd13 and Hoxd10 could be transcribed in most distal limb cells in the absence of digit enhancers located within the deleted C-DOM.
Figure 2.
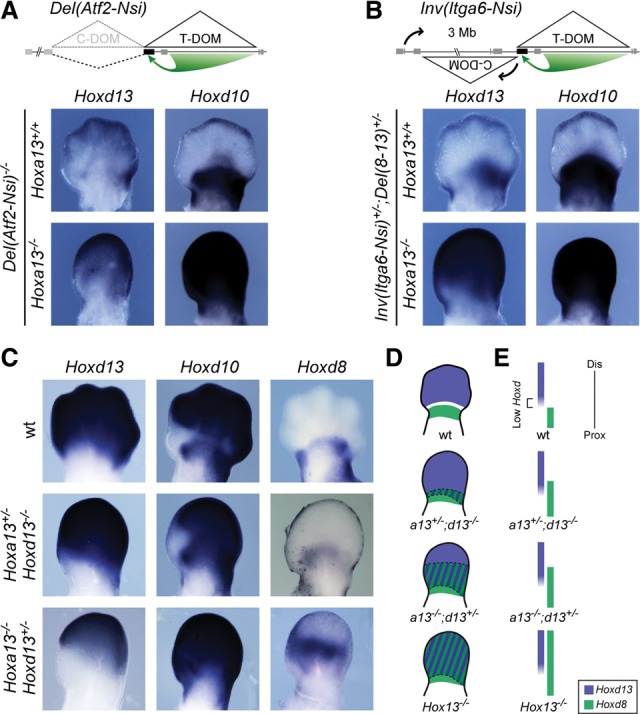
T-DOM drives both proximal and distal expression of Hoxd genes in double Hox13 mutant forelimbs. (A,B) In situ hybridization analysis of E12.5 forelimbs showing the expression of both Hoxd10 and Hoxd13 in two different mutant stocks where the C-DOM is disconnected from the cluster. (A, top) In the HoxDDel(Atf2-Nsi) line the C-DOM is removed through an ∼1-Mb large deletion (dashed line). (B, top) In the HoxDInv(Itga6-Nsi) line, the C-DOM is repositioned several megabases away from the HoxD cluster via a large inversion (black arrows). In both cases, expression of Hoxd10 and Hoxd13 is lost in the distal domain. Expectedly, Hoxd10 remains transcribed proximally, under the control of the T-DOM. (A,B, bottom) However, when the function of Hoxa13 was further removed, expression of both Hoxd10 and Hoxd13 was restored in the distal domain even in the absence of the C-DOM regulation. (C) In situ hybridization analysis of E12.5 forelimbs showing the expression of Hoxd8, Hoxd10, and Hoxd13 in embryos carrying various combinations of mutated alleles (indicated at left). (D,E) A schematic representation shows the C-DOM-driven distal expression domain in blue (e.g., Hoxd13) and the T-DOM-driven proximal expression domain in green (e.g., Hoxd8). In wild type, the domain of low Hoxd expression between Hoxd13- and Hoxd8-expressing regions is represented in white in D and with a bracket in E. In Hox13−/− mutant limbs, the T-DOM controls all Hoxd genes up to the most distal part of the limb (merged domain shown by stripes in D), leading to an ill-defined genetic identity. In such a situation, a presumptive mesopodium (white in D, can no longer appear.
We validated this observation with a second allele where C-DOM was relocated several megabases away from its target Hoxd genes through an inversion (Fig. 2B). As for the deleted allele, this HoxDInv(Itga6-Nsi) inversion led to the absence of any Hoxd13 to Hoxd10 transcripts in the distal domain at E12.5 [Fig. 2B, Inv(Itga6-Nsi); Tschopp and Duboule 2011] due to the increased enhancer-to-promoter distance. Here again, however, the additional abrogation of Hoxa13 function restored expression in the distal domain at E12.5 (Fig. 2B, bottom), as if digit enhancers were at work. From these results, we concluded that the restored distal expression in these two double mutants was controlled by the T-DOM regulation rather than by the usual digit enhancers located in C-DOM. This implied that T-DOM has the intrinsic capacity to drive Hoxd gene transcription into the digit domain, a capacity normally inhibited by HOX13 proteins. In both the Del(Atf2-Nsi) and Inv(Itga6-Nsi) alleles, the constitutive interactions observed between Hoxd13 and the C-DOM (Montavon et al. 2011) had disappeared and were likely replaced by looser interactions with the new genomic neighborhoods (Andrey et al. 2013), thus allowing the establishment of unusual contacts between Hoxd13 and T-DOM enhancers. These differences in the new genomic neighborhoods due to either the deletion or the inversion of the C-DOM explained the slight variations in the level of ectopic Hoxd13 transcripts now controlled by the T-DOM.
This gain of Hoxd gene expression in the distal domain scored in the absence of HOX13 proteins also occurred in Hoxa13−/−;Hoxd13+/− mutant limbs at E12.5, indicating a dosage effect. However, this gain was much weaker, as seen with Hoxd8 transcripts, which were detected more distally than in control limbs yet not in the most distal aspect of the limbs (Fig. 2C). The restoration of Hoxd10 distal expression in these different mutant configurations produced a single extended domain instead of the two “classical” proximal and distal domains expected from the switch between T-DOM and C-DOM regulation (Figs. 1B, 2). Accordingly, the zone of low Hoxd expression separating the two domains (Fig. 2D,E top, white zone) had disappeared to generate a continuous expression domain (Fig. 2D,E, bottom).
Binding of HOX13 within both the C-DOM and T-DOM
To investigate whether this repressive effect of HOX13 proteins was due to a direct interaction, we performed ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) analysis using an antibody against HOXA13. In E12.5 wild-type distal limb cells, HOXA13 binding was enriched around the HoxD locus at large with a series of peaks telomeric to the HoxD cluster, as identified by using model-based analysis of ChIP-seq (MACS) (Fig. 3A; Zhang et al. 2008). Interestingly, these signals extended approximately over the length of the T-DOM (Fig. 3A), supporting a potential direct negative effect of HOXA13 protein over T-DOM limb enhancers. In particular, a significant peak was scored near the CS39 proximal enhancer (Fig. 3B; Andrey et al. 2013). Some of these signals were also observed at cognate genomic positions when stage HH28 chicken distal limbs (Hamburger and Hamilton 1992) were used in similar ChIP-seq experiments, further validating the significance of peaks (Fig. 3, filled circles below the profiles; Supplemental Fig. S3).
Figure 3.
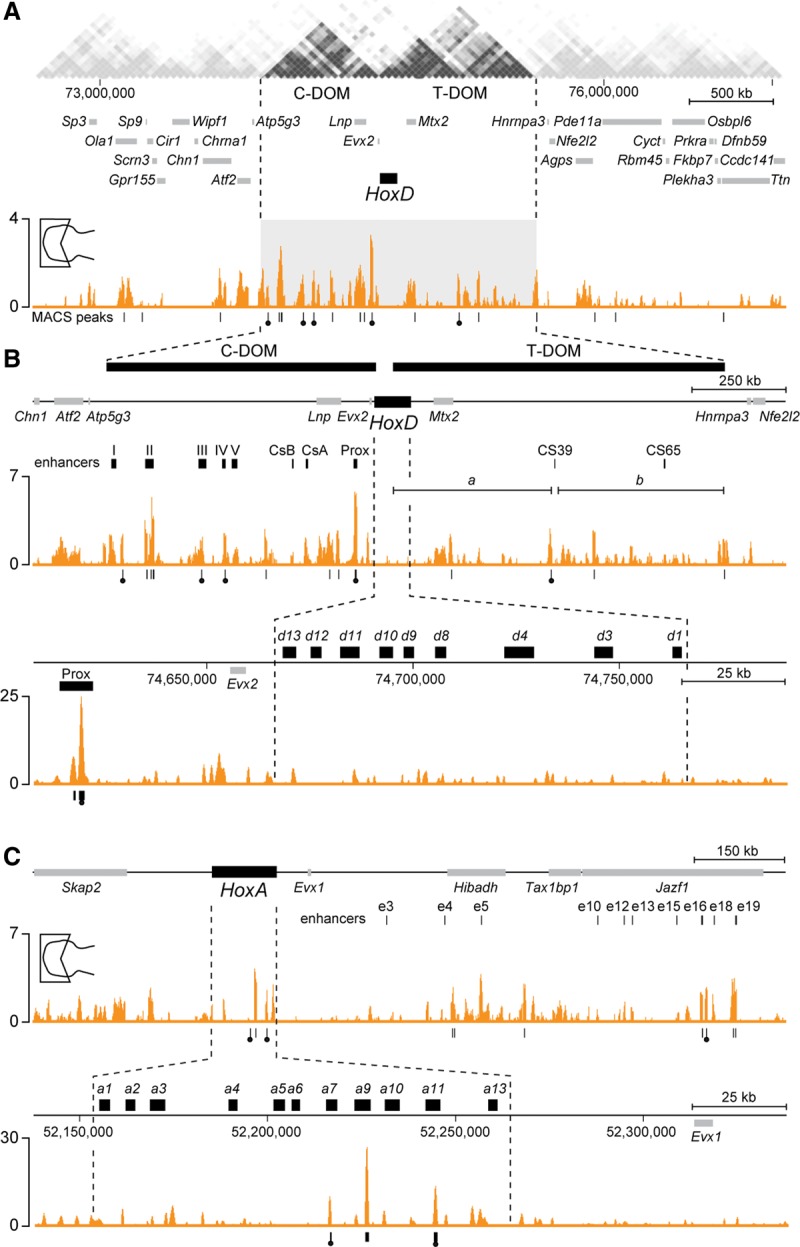
Enrichment of bound HOXA13 protein in both T-DOM and C-DOM regulatory landscapes. (A) HOXA13 ChIP-seq data in the distal domain of the forelimb at E12.5. Enrichment (Y-axis) is shown as the difference of the normalized number of reads between ChIP and input samples. Peaks identified by MACS analysis are represented as vertical traits below the HOXA13-binding profile. Peaks identified by MACS in both the mouse and the chick data sets are marked with filled circles (see also in Supplemental Fig. S3). (Top) A profile over 4 Mb of DNA shows enriched binding sites for HOXA13 in both the C-DOM and the T-DOM (Dixon et al. 2012). (B) Successive close-up views of bound HOXA13 in either both the C-DOM and the T-DOM or the HoxD cluster. Bound HOXA13 was found at previously identified regulatory regions, such as islands II, III, and IV and Prox (top track), whereas no binding was observed in the HoxD cluster (bottom track). (Top track) Regions “a” and “b” represent the two sub-TADs previously described within the T-DOM. (C, top track) The analysis of HOXA13 binding at the HoxA locus also revealed a specific enrichment at previously characterized HoxA digit regulatory elements such as e16 and e19 (Berlivet et al. 2013). (Bottom track) In contrast to HoxD, significant binding was scored within the cluster at the Hoxa7, Hoxa9, and Hoxa11 loci.
Unexpectedly, however, several strong HOXA13 peaks were also found centromeric to HoxD. Again, signals were detected essentially throughout the length of the C-DOM, with peaks observed precisely over most of the previously defined islands necessary for digit regulation (Montavon et al. 2011) in both mouse and chick limbs (Fig. 3B; Supplemental Fig. S3). In contrast to the situation in the T-DOM, this latter observation suggested that in the C-DOM, HOXA13 might be required to maintain or re-enforce the digit regulation. As in the case of the T-DOM, these contacts were not seen in the proximal limb (Supplemental Fig. S4) or much attenuated, likely due to a weak contamination in dissection. Alternatively, some residual HOX13 protein may be present in low amounts due to invading muscle precursor cells (Yamamoto et al. 1998). Interestingly, no HOXA13 binding was scored within the mouse HoxD cluster itself (Fig. 3B).
We also investigated the binding of HOXA13 at the HoxA locus, which contains similar limb regulatory sequences (Lehoczky and Innis 2008; Berlivet et al. 2013). There again, enrichments were found over the regulatory regions that control Hoxa13 during digit development, in particular at the positions of the e16 and e19 digit enhancers (Fig. 3C; Berlivet et al. 2013). In addition and unlike for HoxD, HOXA13 binding was observed within the HoxA cluster itself in both the mouse and chick samples at the Hoxa11, Hoxa9, and Hoxa7 loci (Fig. 3C; Supplemental Fig. S3).
The absence of a regulatory switch in Hox13 mutant limbs
We assessed the functional states of both the T-DOM and the C-DOM in the absence of HOX13 proteins by comparing the distribution of chromatin modifications between wild-type control and double Hox13−/− mutant proximal and distal limbs at E12.5 (Fig. 4). We looked at both the acetylation of H3K27 (H3K27ac; a mark associated with enhancer and transcriptional activity) and the trimethylation of the same residue (H3K27me3; a modification associated with polycomb-dependent silencing). In the wild-type distal sample, H3K27ac was enriched over the C-DOM, illustrating the intense activity of this regulatory landscape during digit development, with robust peaks over the previously defined islands (Fig. 4A, track 1). In contrast, the low level of this mark over the T-DOM reflected its silent state in such distal cells (Andrey et al. 2013). In the proximal limb, the opposite profile was scored, with only traces of H3K27ac over the C-DOM and a concentration of these modified histones at the telomeric end of the T-DOM, corresponding to one of the two subinteraction domains contained within the T-DOM (Fig. 4, bracket b). At E12.5, indeed, the level of H3K27ac detected in these proximal cells started to decrease while remaining high in the latter part of the telomeric landscape (Andrey et al. 2013).
Figure 4.
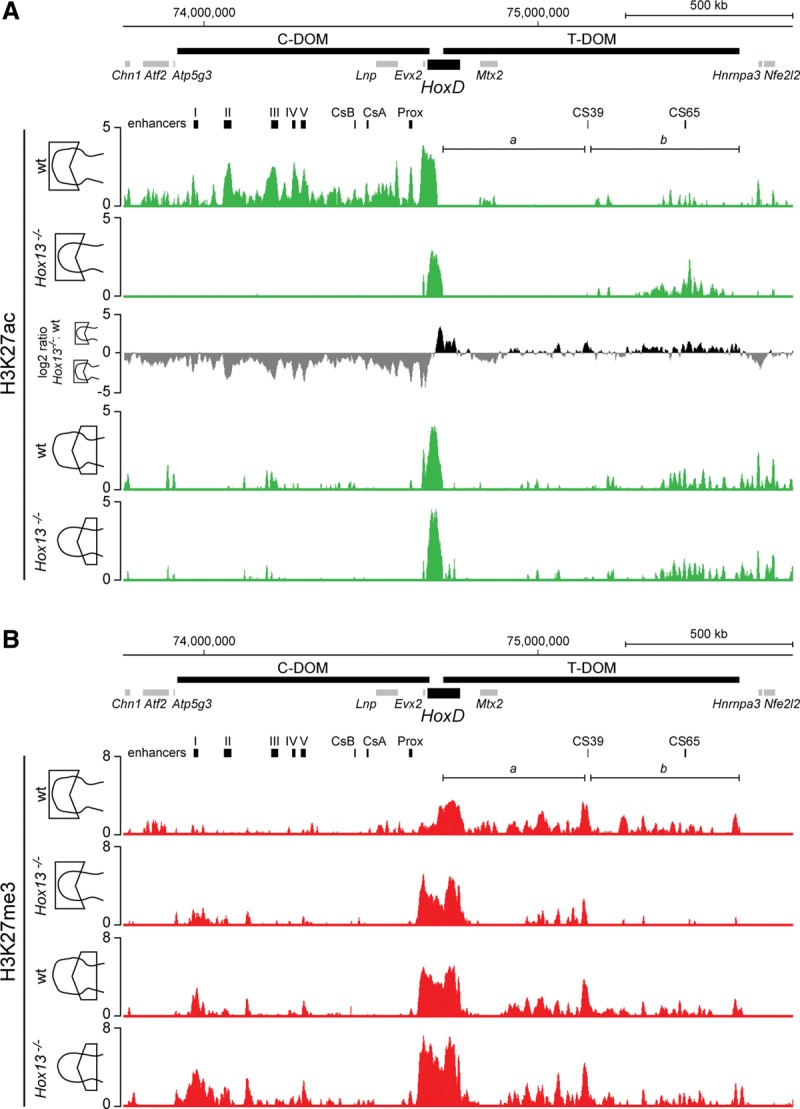
Lack of HOX13 function prevents the regulatory switch from the T-DOM to the C-DOM. (A) Comparison of H3K27ac profiles in the distal and proximal parts of forelimbs obtained from either wild-type or Hox13−/− double-mutant specimens at E12.5. While H3K27ac peaks are distributed throughout the C-DOM in the control track (top), distal Hox13−/− mutant limbs displayed an almost complete absence of H3K27ac signals (second track). In the latter, however, signals appeared within a region of the T-DOM, similar to the profiles observed in the proximal sample (tracks below). The log2 ratio of the normalized number of reads between H3K27ac of Hox13−/− mutant and wild-type distal limbs is shown by positive (black) and negative (gray) values, respectively. (B) Distribution of H3K27me3 marks in distal and proximal forelimbs of either wild-type or Hox13−/− mutant specimens at E12.5. In the distal part of the Hox13−/− mutant, H3K27me3 enrichment of the T-DOM was dramatically reduced, in particular within subdomain “b,” corresponding to the increased enrichment of H3K27ac (shown in A). In contrast, H3K27me3 enrichment around the C-DOM-located islands I and II was slightly increased, in agreement with the loss of H3K27ac (shown in A). Enrichment (Y-axis) is shown as the log2 ratio of the normalized number of reads between ChIP and input samples, except for the third track in A. The localizations of genes and regulatory islands are shown at the top of each panel.
In Hox13−/− mutant distal limbs, no enrichment of H3K27ac was detected over the entire centromeric region (Fig. 4A, track 2), indicating that this regulatory landscape had not been activated. However, H3K27ac marks were clearly enhanced in the T-DOM precisely at the place where these marks were scored in proximal cells; i.e., in cells implementing the T-DOM regulation (Fig. 4A, bracket b). As expected, the H3K27ac profiles were comparable between mutant and control proximal limbs, which do not express any Hox13 gene (Fig. 4A, tracks 3, 4). These observations suggested that in distal limb cells lacking HOX13 function, the T-DOM regulation was no longer interrupted and continued to exert its regulatory activity, whereas the C-DOM regulation was either not implemented at all or at least not maintained after an initial burst.
We verified this conclusion by producing the profile of H3K27me3, which antagonizes the acetylation of the same residue (Tie et al. 2009). Here again, the profiles of control and Hox13−/− proximal limb domains did not significantly differ from one another (Fig. 4B, bottom lines). Signals were detected in the T-DOM, in particular in the region where few H3K27ac marks were scored (Fig. 4A,B; bracket a). In contrast, signals were weaker in the region where H3K27ac was detected (Fig. 4A,B; bracket b), indicating the progressive termination of T-DOM function as normally observed in E12.5 proximal limbs (Andrey et al. 2013). In control distal cells, these repressive marks equally accumulated over the T-DOM after it had switched off. Strikingly, this accumulation was not scored in the Hox13−/− mutant distal domain (Fig. 4B, brackets b), further indicating that the telomeric regulation was kept abnormally active in distal cells.
The distribution of both H3K27ac and H3K27me3 modifications over the HoxD cluster itself supported this interpretation. The H3K27ac profile in the control proximal domain mostly encompassed Hoxd8 to Hoxd11, with significant signals also detected over Hoxd12, Hoxd13, and Evx2 (Supplemental Fig. S5). In the Hox13−/− specimen, this profile was globally conserved, with the exception of Hoxd13 and Evx2 displaying somewhat weaker signals. In the distal limb domain, however, the H3K27ac profile was virtually identical to that of the control proximal domain (Supplemental Fig. S5), with a strong reduction over the Evx2–Hoxd13 region and a gain for the Hoxd8 and Hoxd9 regions, which are normally strong targets of T-DOM regulation.
In wild-type proximal limbs, H3K27me3 marks showed a bimodal distribution with strong coverage of both the 3′ and 5′ extremities of the cluster; i.e., those subgroups of genes not transcribed at this stage. In between, the active Hoxd8 to Hoxd11 region contained fewer of these marks. On the other hand, the control distal domain was heavily decorated with H3K27me3 from Hoxd1 to Hoxd8, whereas the part of the cluster responding to the C-DOM regulation in digits was mostly devoid of it (from Hoxd9 to Hoxd13) (Supplemental Fig. S5). In the mutant condition, this latter profile drastically changed to appear like the control proximal profile, suggesting again that the C-DOM regulation was inactive, in particular considering the robust H3K27me3 coverage of Hoxd13, Hoxd12, and Evx2 (Supplemental Fig. S5B, track 2). In mutant distal cells, however, the H3K27me3 coverage over the Hoxd12–Hoxd13 region was less compact than in mutant proximal cells (Supplemental Fig. S5), coinciding with the weak distal expression of Hoxd13 unexpectedly controlled by T-DOM regulation (see Fig. 2A,B). In contrast, H3K27me3 coverage was comparable in the Hoxd1 to Hoxd4 region of distal versus proximal mutant cells.
The distribution of H3K27ac and H3K27me3 marks over the HoxA cluster in control and mutant cells also suggested that a “proximal” type of regulation was maintained in distal cells lacking HOX13 function. This was best illustrated by both an increased acetylation of the Hoxa1 to Hoxa7 region and a decreased acetylation of the Hoxa11 to Hoxa13 region in mutant distal cells when compared with control, thus resembling the H3K27ac profiles of both control and mutant proximal cells (Supplemental Fig. S6).
HOX13-dependent modifications in three-dimensional (3D) interactions
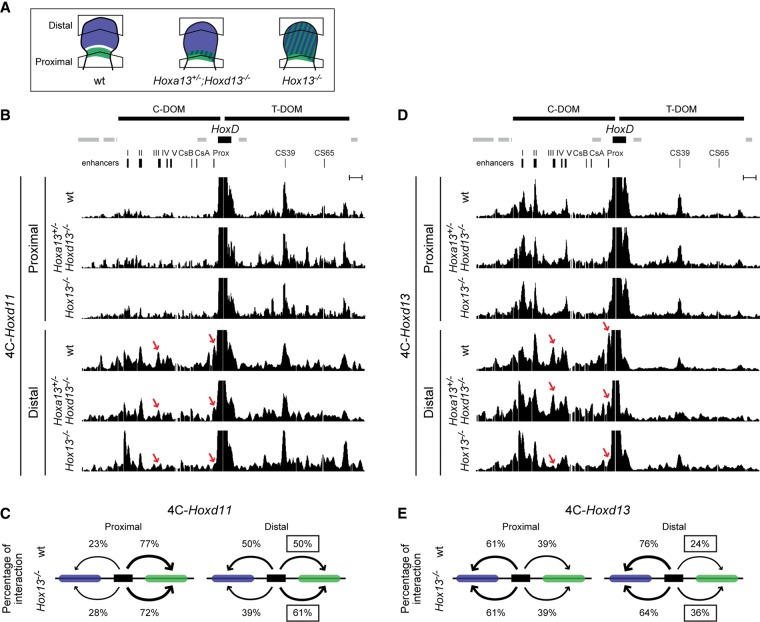
The regulation of Hoxd13-to-Hoxd9 transcription by C-DOM enhancers in developing digits involves strong physical interactions. Among these contacts, those with the island III and Prox sequences are hallmarks of Hoxd13 transcriptional activation (Montavon et al. 2011). By using circularized chromosome conformation capture (4C) coupled with next-generation sequencing (4C-seq), interactions between Hoxd13 or Hoxd11 and these two sequences were indeed scored in all cases where the material was derived from developing digits but absent from control negative tissues, which display only constitutive nonproductive contacts with Hoxd13 (Andrey et al. 2013; Lonfat et al. 2014). We thus looked for these contacts in double-mutant distal limbs at E12.5 as an ultimate assessment of the nonfunctionality of the C-DOM in these cells. Because of the reduced size of the distal domain in double Hox13 mutant limbs (see Figs. 1, 2), we restricted our microdissections to the most proximal and most distal pieces of the developing limbs to prevent cross-contamination between cells (Fig. 5A).
Figure 5.
Reallocation of interactions from the C-DOM to the T-DOM in limbs lacking HOX13 function. (A) Schemes showing the dissection strategies used for sampling. The color code is as in Figure 2D. (B,D) 4C-seq tracks representing contacts established by Hoxd11 (B) and Hoxd13 (D) in proximal and distal limb samples (indicated at the top) of either wild-type or various Hox13 mutant combinations at E12.5 (indicated at the left). The extents of both the C-DOM and the T-DOM are shown as thick black lines at the top of B, together with the positions of genes. The previously described regulatory elements are indicated below (in black). The red arrows in the distal samples point to specific contacts detected between genes normally expressed in the distal part under the control of the C-DOM (Andrey et al. 2013). These interactions with island III and Prox, which are hallmarks of the C-DOM operating in limbs, are no longer detected in distal samples lacking all HOX13 function. (C,E) Schemes representing the percentages of 4C contacts for either Hoxd11 (C) or Hoxd13 (E) with the C-DOM (blue rectangle) and the T-DOM (green rectangle) in the various samples. The HoxD cluster is shown as a black rectangle. While virtually no difference was observed in the proximal samples, both Hoxd11 and Hoxd13 in the Hox13−/− double-mutant distal sample lost contacts with the C-DOM and gained contacts with the T-DOM, in agreement with their transcription now controlled by this latter regulatory landscape.
In control proximal cells, Hoxd11 mostly contacted the T-DOM, with a particularly strong interaction with region CS39. In Hox13−/− double mutants, the interaction profiles were globally comparable (Fig. 5B, top). In distal cells, however, the Hoxd11 interaction profile changed drastically in mutant versus control samples (Fig. 5B, bottom), as the reported shift in contacts from the T-DOM toward the C-DOM (Andrey et al. 2013) was no longer observed. A quantification of contacts indicated 77% of telomeric contacts for Hoxd11 in control proximal cells and 50% in distal cells, showing that Hoxd11 had reallocated almost 30% of its contacts toward the C-DOM in digit cells (Fig. 5C). Within the C-DOM, interactions were scored with the islands III and Prox sequences (Fig. 5B, red arrows; enlargement in Supplemental Fig. S7), indicative of an active transcriptional state.
In contrast, mutant distal cells did not show these interactions (Supplemental Fig. S7). Instead, contacts were re-enforced within the T-DOM, further indicating that Hoxd11 had remained regulated by this landscape rather than shifting toward the C-DOM. The transcriptional inactivity of C-DOM regulatory sequences was corroborated by the interaction profile of Hoxd13 (Fig. 5D). In control distal cells, Hoxd13 showed the expected robust contacts with the C-DOM, including strong interactions with both region III and Prox (Fig. 5D, red arrows). In the double Hox13 mutant limb cells, however, while the bulk of interactions remained over the C-DOM, as in control negative cells like brain tissue (Montavon et al. 2011), the specific interactions with region III and Prox had virtually disappeared along with the transcriptional off state of the C-DOM. Interestingly, contacts established by Hoxd13 with the T-DOM were slightly increased (from 24% to 36%) (Fig. 5E), in agreement with the weak expression of this gene in distal mutant cells (Fig. 1B,C). As expected, the interaction profiles of Hoxd13 in control and mutant proximal cells were comparable.
We also assessed the impact of HOX13 dosage on the control of this regulatory switch by looking at the interaction profiles of intermediate allelic combinations. We analyzed both Hoxa13−/−;Hoxd13+/− (data not shown) and Hoxa13+/−;Hoxd13−/− (Fig. 5) mutant limbs. In these mutants, autopods were also severely impaired, yet a distal domain devoid of the Hoxd8 transcript was still identified (Fig. 2C,D), unlike in full Hox13−/− embryos (Figs. 1B, 2D), indicating that the switch had been achieved at least partially. Accordingly, we observed only slight and mostly quantitative modifications in the interaction profiles. For example, weaker yet present contacts between both Hoxd11 and Hoxd13 and region III and Prox were scored in mutant distal cells, showing that the C-DOM was still at work, although with a lower transcriptional capacity. Although a single dose of HOX13 protein was sufficient to sustain the C-DOM regulatory program in both cases, it failed to fully implement this complex regulation. This quantitative effect was similar when the last Hox13 copy left was either Hoxa13 or Hoxd13 (Fig. 5; data not shown), suggesting a close to equal function of both proteins in this context. From these experiments, we concluded that both proteins cooperate in a dose-dependent manner to repress the activity of the T-DOM and/or maintain transcription driven by the C-DOM.
Distal repression of the T-DOM is not targeted to enhancer sequences
While the pattern of HOXA13 occupancy within the C-DOM was rather specific to known regulatory islands, peaks in the T-DOM were mostly concentrated in subdomain “b” (Fig. 3B), where most of the Hoxd11 contacts were gained when 4C-seq was carried out with double-mutant distal limbs (Fig. 5B). However, HOXA13 binding was not observed in either of the two known CS39 and CS65 proximal limb enhancers. Instead, the most significant HOXA13 signal was located near CS39, matching the CS38 sequence, which contains the transcription start sites of the two opposite lncRNAs: Hotdog and Twin of Hotdog (Delpretti et al. 2013). This indicated that HOX13 repression of T-DOM regulation may not involve direct binding to forearm-specific enhancers.
To test this hypothesis, we produced lines of LacZ reporter transgenic mice carrying random integrations of either the CS39 or the CS65 forearm-specific enhancers, reasoning that such sequences may escape repression in distal limbs when placed outside of the T-DOM context due to the absence of direct HOXA13 binding. In both cases, after an initial phase of expression in the proximal limb bud, LacZ staining extended up to the most distal limb aspects at E12.5 (Fig. 6), suggesting that the transgenes had escaped the HOXA13-dependent repression normally associated with T-DOM. These experiments showed that both CS39 and CS65 enhancers have the capacity to drive expression of a reporter gene throughout the developing limb buds. However, this capacity is normally inhibited in distal limb cells due to the action of HOX13 proteins over the T-DOM.
Figure 6.
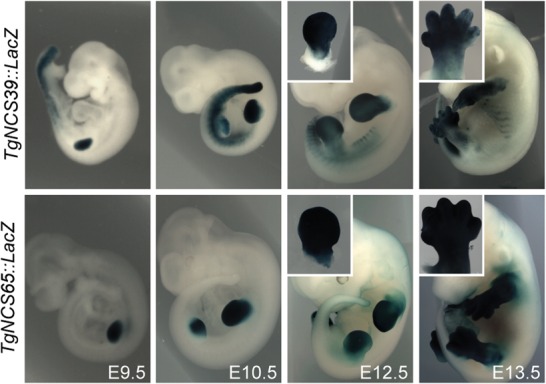
The T-DOM CS39 and CS65 sequences are global rather than proximal limb enhancers. Mouse stable transgenic lines carrying a LacZ reporter construct under the control of either the CS39 (top) or the CS65 (bottom) enhancers. In both cases, expression was scored throughout the developing limbs from the budding stage at E9.5 (left) to at least E13.5 (right).
Discussion
TAD switching and the wrist boundary
The presumptive wrist region corresponds to an area of the developing limb bud where Hoxd gene expression is minimal. As a result, the ossification of cartilage condensations does not process along the proximal to distal axis (Gonzalez-Martin et al. 2014), leading to the nonelongated mesopodial bone morphologies (Villavicencio-Lorini et al. 2010). This cellular zone devoid of Hoxd gene transcripts results from the bimodal regulatory strategy investigated in this study. Here, we show that the boundary between the wrist and the future forearm is fixed by the activation of Hox13 genes, which repress the transcription of Hoxd genes, likely through direct binding within the T-DOM. The HOXA13 protein switches off the T-DOM regulation and hence prevents proximal Hoxd genes from being transcribed more distally, including in the wrist region. A complete repression also requires HOXD13 combination, as shown by the fact that either Hoxa13−/− or Hoxd13−/− single-mutant animals have genuine yet slightly affected wrists (Dolle et al. 1993; Fromental-Ramain et al. 1996), whereas double-mutant limbs do not display any structure reminiscent of this articulation (Fromental-Ramain et al. 1996; Kmita et al. 2005).
HOX13 proteins as C-DOM activators?
Our results suggest that HOX13 proteins are essential to fully activate and/or sustain the regulatory activity of the C-DOM. Their combined abrogation leads to the absence of TAD switching, with the C-DOM never becoming detectably active. While the underlying mechanism of action is elusive, our ChIP-seq results indicate that the HOXA13 protein occupies most of the previously determined digit regulatory islands (Montavon et al. 2011). Nevertheless, the Hoxd13 transcript domain does not perfectly match that of Hoxa13, with Hoxd13 being slightly less proximal (see Woltering and Duboule 2010), indicating that HOXA13 alone is not sufficient to activate Hoxd13 and that other factors are required to initiate the transcription of Hoxd genes in distal cells. Also, Hoxa13−/− mutant mice display normal Hoxd13 transcription. In addition, some of the C-DOM islands bound by HOXA13 establish only constitutive contacts with the target HoxD cluster and the HOXA13 signals extended over the size of these islands, usually several kilobases in size. This generally protein-dense aspect is quite distinct from a classical DNA-binding protein profile under the same experimental conditions.
The positive effect of HOX13 proteins in implementing the C-DOM regulation may not be very selective. Instead, the fact that the signals tend to colocalize with all DNA segments in contact with the target Hoxd13 gene may reflect an impact on the general architecture of the C-DOM itself. Alternatively, the architecture of the C-DOM may favor the recognition of low-affinity binding sites by HOX13 proteins. In any case, it is likely that the same effect will be observed wherever the C-DOM is active. For instance, Hoxa13/Hoxd13 mutant mice lack external genitals (Kondo et al. 1997; Warot et al. 1997), and Hoxd13 transcription in the genital bud indeed depends on enhancers localized within the same C-DOM (Lonfat et al. 2014).
The HOXA13-binding specificity was controlled by using stage-comparable chicken proximal and distal wing buds. While both the enrichments at the HoxD and HoxA loci and some conspicuous peaks were conserved between species, some differences were observed. For example, MACS analysis revealed that, while the C-DOM regulatory islands III and IV and Prox were bound in both species, region II was bound only in the mouse sample. Likewise, binding to the e16 enhancer at the HoxA locus (Berlivet et al. 2013) was detected only in mice. These differences might reflect either intrinsic properties of the antibody or technical issues. They may also underlie some of the genuine differences observed in the expression of these genes during limb bud development in both mammals and birds (Dolle et al. 1989; Nelson et al. 1996).
HOX13 proteins as T-DOM repressors?
Within the T-DOM, the pattern of HOXA13 binding was somewhat more diffuse and mostly concentrated on the sub-TAD “b.” This subdomain preferentially interacts with Hoxd genes expressed in the forearm region, whereas subdomain “a” contacts the most 3′-located genes on the cluster such as Hoxd1 (Andrey et al. 2013), which do not have any reported function in limb development. Accordingly, in double Hoxa13−/−;Hoxd13−/− mutant distal limbs, the gain of Hoxd11 contacts reallocated from the C-DOM to the T-DOM were scored within subdomain “b,” illustrating the correlation between the presence of HOXA13 protein and the transcriptional inactivity of this region in wild-type distal cells. This region was also rich in H3K27ac up to E12.5 in control proximal limbs (Andrey et al. 2013) and abnormally maintained H3K27ac in Hoxa13−/−;Hoxa13−/− mutant distal cells. Therefore, in control distal cells, T-DOM regulation is terminated and repressed in conjunction with the presence of HOX13 proteins bound over the DNA interval, which is normally the most active in contacting the main Hoxd9, Hoxd10, and Hoxd11 target genes in proximal cells.
While the negative impact of HOX13 proteins on T-DOM regulation was documented by our genetic and molecular approaches, neither the CS39 nor the CS65 enhancers were bound by HOXA13. However, a robust binding was scored over CS38, a conserved DNA sequence next to CS39. Both CS39 and CS65 enhancers contribute to the transcription of Hoxd target genes in the proximal limb, whereas they are normally unable to control the same genes in distal cells (Andrey et al. 2013). When introduced at ectopic genomic positions, these enhancers were nevertheless capable of driving expression distally, showing their intrinsic capacity to control transcription throughout the developing limbs even in presence of physiological amounts of HOXA13 and HOXD13 proteins. We conclude that the restrictive action of these proteins in distal cells is not mediated by direct binding to the enhancers themselves. On the other hand, this distal repression cannot come from the binding of HOX13 directly to the target Hoxd genes because these genes are fully transcribed by the C-DOM enhancers in the same distal cells. Therefore, we conclude that HOX13 proteins terminate T-DOM regulation by binding to this chromatin structure yet not necessarily to the silenced enhancer sequences, raising the possibility that the T-DOM is a global unit of regulation that could be switched on or off by factors not specific to any of the existing enhancer sequences.
The presence of HOXA13 bound at some of the activating enhancers at the HoxA locus (Berlivet et al. 2013) suggests that a similar positive effect may be at work. However, unlike HoxD, the HoxA cluster itself displayed at least three significant sites bound by HOXA13, in particular one covering the Hoxa11 transcription unit, which is silent in distal cells, suggesting that HOX13 proteins may also have a direct effect on some Hoxa target genes. HOXA13 binding was also observed near Hoxa7 and Hoxa9, two genes up-regulated in the distal limbs of Hox13−/− mutants, suggesting that HOX13 proteins may help restrict the expression of these genes proximally by acting on local regulatory elements.
The mechanism of TAD switching
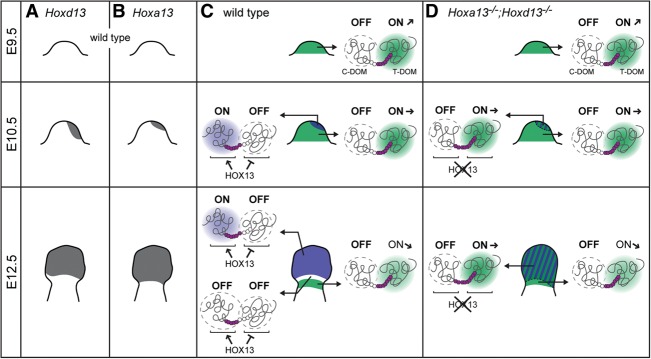
From this set of experiments, we propose a hypothesis for the bimodal Hoxd gene regulation during limb development. In the incipient bud, the T-DOM regulates the progressive transcriptional activity of Hoxd8 to Hoxd11, which help organize and grow the proximal structures (Tarchini and Duboule 2006), whereas the C-DOM is inactive (Fig. 7C, top). At E10.5, along with distal growth and the release of secreted factors by the distal apical ectodermal ridge (AER), the Shh system and its feedback loop are fully implemented, partly under the control of HOX proteins present in the early bud (Knezevic et al. 1997; Tarchini et al. 2006; Zeller et al. 2009; Sheth et al. 2013). In turn, this signaling system triggers the activation of the C-DOM, leading to the appearance of Hoxd13 transcripts in the most posterior and distal aspect of the bud (Fig. 7C, blue in the middle panel). The HOXD13 protein—along with HOXA13, also activated in the most distal cells—represses the activity of the T-DOM in distal cells, while, at the same time, likely in conjunction with AER factors, it sustains the positive activity of the C-DOM over Hoxd13 to Hoxd10 (Montavon et al. 2008). At E12.5, the domain of C-DOM activity has extended distally, where the concentrations of AER secreted factors are high, and some proximal cells where the C-DOM was initially active stop operating this regulation. In these presumptive cells of the mesopodium, both the T-DOM and the C-DOM are inactive, leading to only residual amounts of HOX protein. Cartilage models will fail to elongate and produce the wrist (Fig. 7B, bottom; Villavicencio-Lorini et al. 2010; Woltering and Duboule 2010; Gonzalez-Martin et al. 2014).
Figure 7.
HOX13 function in the control of the regulatory switch at the HoxD locus. (A,B) Schematic Hoxd13 (A) and Hoxa13 (B) expression at E9.5, E10.5, and E12.5. Expression domains are in gray. (C) In the wild-type situation, the T-DOM operates in the early E9.5 limb bud. In E10.5 proximal limb cells, the T-DOM continues to drive Hoxd gene expression yet with a weaker activity. At this stage, distal cells (blue) individualize where Hox13 genes start be transcribed. In these cells, HOX13 proteins both switch off the T-DOM and sustain strong transcription of Hoxd13 by the C-DOM. In E12.5 limbs, proximal cells still weakly implement T-DOM regulation, while cells operating C-DOM regulation have expanded distally, likely in response to distal factors released by the AER. This expansion generates a zone of low Hox gene activity (white), which gives rise to the mesopodium (wrist and ankle). (D) In Hoxa13−/−;Hoxd13−/− double-mutant limbs, the T-DOM normally operates in proximal cells at E9.5. Transcription of Hoxd13 is initiated in distal cells at E10.5 (shaded blue cells), but, in the absence of HOX13 proteins, the C-DOM remains switched off, and the T-DOM continues to drive Hoxd genes in the most distal cells. The active C-DOM and T-DOM are shown by blue and green clouds, respectively. Active and inactive Hoxd genes are depicted with purple or white hexagons, respectively. The font size and arrows next to “ON” illustrate the various levels of T-DOM activity.
In the double Hox13−/− mutant condition, the system initiates normally in the early bud, with the T-DOM in an “on” state (Fig. 7D, top), up to the start of Hoxd13 transcription, which either does not occur or occurs at a very low level. In the absence of both HOX13 proteins, the T-DOM continues to operate in most distal cells, whereas the C-DOM remains inactive. In the absence of C-DOM function, T-DOM enhancers are even capable of activating Hoxd13 transcription, although at a rather low level. The continuous action of T-DOM regulation produces a distal aspect of the limb bud lacking a clear “proximal” or “distal” identity (Fig. 7D, green and blue shaded areas), at least when considering our principal component analyses and the phenotype of such mutant fetuses. While they clearly show the presence of a smaller “digital plate” at an early stage, digits and more proximal structures are not subsequently formed (Fromental-Ramain et al. 1996). From this, we conclude that, while Hox genes are critical for the development of distal structures, they are not the sole genetic determinants of a “distal” limb identity.
The absence of a switch between TADs makes the appearance of HOX-negative cells impossible due to the nonrepression of T-DOM activity and concurrent nonsegregation of proximal and distal expression domains (Sheth et al. 2014). Consequently, such mutant limbs do not develop any clear mesopodial-like structures (Fromental-Ramain et al. 1996). In this view, the opposite functions of HOX13 proteins on the T-DOM and C-DOM appears as a parsimonious solution to make these regulations exclusive from one another, an essential prerequisite for the offset between proximal and distal expression domains and hence the development of the mesopodial articulation. While the molecular mechanisms underlying these antagonistic activities are unclear, they may involve various cofactors, as reported in other cases of transcription factors capable of such a dual activity (e.g., Tetreault et al. 2009).
Evolution of Hox regulation in the transformation of paired appendages
During fin bud development, the transcript domains of Hoxa13 and Hoxd13 overlap with those of Hoxa11 or Hoxd11 (Sordino et al. 1995; van der Hoeven et al. 1996; Metscher et al. 2005; Ahn and Ho 2008), suggesting that the overall regulatory strategy is different and that fish lack structures homologous to tetrapod digits (see Woltering and Duboule 2010). However, a late distal phase of Hox13 expression was detected in several fish species (Davis et al. 2007; Freitas et al. 2007; Johanson et al. 2007), leading to the proposal that the mammalian biphasic Hox regulation described above is an ancestral gnathostome character (Shubin et al. 2009). Our present work shows that distal expression for Hox13 genes can be achieved without the proper digit regulatory controls, provided that the negative impact of HOX13 proteins on the system is abrogated, which is naturally the case in fish, as shown by their early coexpression with Hox11 genes. In fact, the strict correlation proposed in tetrapods between the expression of Hoxd13 and digit formation (Dolle et al. 1993) applies to only the protein and not the mRNAs because, in double Hox13 mutant buds, Hoxd13 transcripts can be located distally, resembling the wild-type situation yet in a domain that does not elicit digit formation. In this case, Hoxd13 distal transcription is controlled by the “proximal system.”
In this context, one may hypothesize that either one or both ancestral fish TADs, which were observed at the HoxD locus (Woltering et al. 2014; Acemel et al. 2016), may have triggered rather general, proximal, and distal transcription of Hoxd genes in the budding fins (e.g., Gehrke et al. 2015) without any negative “cross-talk” between them. At some point during the evolution of tetrapods, the negative effect of HOX13 proteins over the T-DOM and their positive re-enforcement through the C-DOM appeared either via protein sequence modifications or through variations in the properties of these TADs. This step, along with the repression of Hoxa11 by Hoxa13, leads to the two-phase, bimodal regulation described at the HoxD locus (Sheth et al. 2014; Woltering et al. 2014). Such a bimodal regulation allowed the self-enhanced transcription of Hoxd13, which, like Hoxa13, has the ability to stimulate growth and produce cartilage models and long bones of reduced size (Yokouchi et al. 1995a; Freitas et al. 2012), leading to digits. This also provided the mechanistic opportunity to offset the two domains of transcription and, as a consequence, produce the mesopodial articulation (Villavicencio-Lorini et al. 2010; Woltering and Duboule 2010; Andrey et al. 2013).
Materials and methods
Animal experimentation
All experiments were performed in agreement with the Swiss law on animal protection (LPA) under license number GE 81/14 (to D. Duboule). Chick embryos from a White Leghorn strain were incubated at 37.5°C and staged according to Hamburger and Hamilton (1992).
In situ hybridization
Whole-mount in situ hybridizations were performed as described in Woltering et al. (2014).
RNA extraction
Total RNA was extracted from individual pairs of wild-type or mutant proximal and distal forelimbs using the RNeasy microkit (Qiagen) following the manufacturer's instructions. A total of 100 ng of pure total RNA was amplified following standard Illumina procedure for polyA-selected RNA and sequenced on a HiSeq sequencer with a read length of 100 base pairs (bp). An in silico mutant version of the genome, including both the Hoxd13/LacZ and the Hoxa13/Neomycin alleles, was assembled, annotated, and used as the reference genome to map the Hoxa13−/−;Hoxd13−/− transcriptomes with the mouse GCRm38 assembly. The mapping and FPKM (fragments per kilobase per million mapped fragments) calculations of expressed transcripts were performed using TopHat and Cufflinks implemented in a local Galaxy platform.
4C-seq
4C-seq was performed as described in Noordermeer et al. (2014). Pairs of proximal or distal forelimbs and hindlimbs were individually fixed with 2% formaldehyde, lysed, and stored at −80°C. After genotyping, pools of eight proximal or distal limbs were digested with NlaIII and DpnII as primary and secondary restriction enzymes, respectively, and ligation steps were performed using highly concentrated T4 DNA ligase (Promega, M1794). Inverse PCRs for amplification were carried out using primers for the Hoxd11 and Hoxd13 viewpoints (Noordermeer et al. 2011). PCR products were multiplexed and sequenced using a HiSeq sequencer from Illumina, and post-processing (demultiplexing, mapping, and 4C analysis) was conducted on the Bioinformatics and Biostatistics Core Facility HTSstation (http://htsstation.epfl.ch) (Noordermeer et al. 2011; David et al. 2014). Data were plotted on University of California at Santa Cruz (UCSC) genome bioinformatics site and smoothed with a window size of 11 fragments. A tentative relative quantification of the signal spanning both the HoxD telomeric and centromeric deserts was performed as described in Andrey et al. (2013). This quantification was not absolute and only reflected the balance of contacts between the two domains for each sample.
Mutant and transgenic mice
Mice mutant for either the Hoxa13 or the Hoxd13 gene were those described in Fromental-Ramain et al. (1996) and Kondo et al. (1998). The mice carrying a deletion [HoxDDel(Nsi-Atf2)] or inversion [HoxDInv(Nsi-Itga6)] of the C-DOM were described in Montavon et al. (2011) and Tschopp and Duboule (2011), respectively. To establish stable transgenic lines carrying either the CS39 or the CS65 enhancers, we generated a scaffold vector carrying the β-globin minimal promoter and the LacZ coding sequence (pSK-LacZ). The genomic region containing the regulatory elements CS39 (mm10, chromosome 2: 75,147,318–75,148,561) and CS65 (mm10, chromosome 2: 75,439,366–75,440,449) were PCR-amplified using specific primers and cloned into the pSK-LacZ vector. In both cases, the insert carrying the enhancer, β-globin minimal promoter, and LacZ coding sequence was excised from the vector backbone by digestion with KpnI–NotI. The fragment was gel-purified using the QIAquick gel extraction kit (Qiagen) and injected into fertilized oocytes. Three independent founders showing robust and reproducible LacZ expression in their offspring were selected for each construct. The presence of the transgene was assessed by PCR.
ChIP-seq
Microdissected limb tissues from mouse and chick embryos were cross-linked with 1% formaldehyde/PBS for 18 min. Chromatin was sheared and used for each immunoprecipitation with antibodies against HOXA13 (Yokouchi et al. 1995a), H3K27ac (Abcam, ab4729), and H3K27me3 (Merck Millipore, 07-449), respectively. For ChIP-seq, at least 5 ng of purified DNA was used to make libraries according to the manufacturer's protocol (Illumina). The material was sequenced with 100-bp single-end reads on an Illumina HiSeq according to the manufacturer's specifications.
Accession numbers
RNA-seq, ChIP-seq, and 4C-seq data sets are available from the NCBI Gene Expression Omnibus repository under accession number GSE79261. The control ChIP-seq data for H3K27ac in proximal limb can be found under GSM1104588. Any other information for HOXA13 ChIP-seq data and ChIP-seq data from Hoxa13−/−;Hoxd13−/− embryos is available on request.
Supplementary Material
Acknowledgments
We thank F. Darbellay and R. Schep for their help in some experiments, and J.-M. Matter as well as members of the Duboule laboratories and M. Ros for sharing material and discussions. We also thank S. Gitto and T.H. Nguyen Huynh for help with mutant stocks. We thank the Geneva Genomics Platform (University of Geneva) and the Bioinformatics and Biostatistics Core Facility of the Ecole Polytechnique Fédérale in Lausanne for their assistance. This work was supported by funds from the Ecole Polytechnique Fédérale, the University of Geneva, the Swiss National Research Fund, European Research Council grants RegulHox, and the Claraz Foundation (to D.D). N.Y.-K. was supported by the Naito Foundation. S.Y. and A.K were supported by the “Integrative Graduate Education and Research in Green Natural Sciences” program (Ministry of Education, Culture, Sports, Science and Technology, Japan) and a Grant-in-Aid for Scientific Research on Innovative Areas (25124703; Ministry of Education, Culture, Sports, Science and Technology, Japan), respectively.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.281055.116.
Freely available online through the Genes & Development Open Access option.
References
- Acemel RD, Tena JJ, Irastorza-Azcarate I, Marletaz F, Gomez-Marin C, de la Calle-Mustienes E, Bertrand S, Diaz SG, Aldea D, Aury JM, et al. 2016. A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat Genet 48: 336–341. [DOI] [PubMed] [Google Scholar]
- Ahn D, Ho RK. 2008. Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: implications for the evolution of vertebrate paired appendages. Dev Biol 322: 220–233. [DOI] [PubMed] [Google Scholar]
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340: 1234167. [DOI] [PubMed] [Google Scholar]
- Berlivet S, Paquette D, Dumouchel A, Langlais D, Dostie J, Kmita M. 2013. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet 9: e1004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David FP, Delafontaine J, Carat S, Ross FJ, Lefebvre G, Jarosz Y, Sinclair L, Noordermeer D, Rougemont J, Leleu M. 2014. HTSstation: a Web application and open-access libraries for high-throughput sequencing data analysis. PLoS One 9: e85879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. 1995. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375: 791–795. [DOI] [PubMed] [Google Scholar]
- Davis MC, Dahn RD, Shubin NH. 2007. An autopodial-like pattern of Hox expression in the fins of a basal actinopterygian fish. Nature 447: 473–476. [DOI] [PubMed] [Google Scholar]
- Delpretti S, Montavon T, Leleu M, Joye E, Tzika A, Milinkovitch M, Duboule D. 2013. Multiple enhancers regulate Hoxd genes and the Hotdog lncRNA during cecum budding. Cell Rep 5: 137–150. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolle P, Izpisua-Belmonte JC, Falkenstein H, Renucci A, Duboule D. 1989. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature 342: 767–772. [DOI] [PubMed] [Google Scholar]
- Dolle P, Dierich A, LeMeur M, Schimmang T, Schuhbaur B, Chambon P, Duboule D. 1993. Disruption of the Hoxd-13 gene induces localized heterochrony leading to mice with neotenic limbs. Cell 75: 431–441. [DOI] [PubMed] [Google Scholar]
- Freitas R, Zhang G, Cohn MJ. 2007. Biphasic Hoxd gene expression in shark paired fins reveals an ancient origin of the distal limb domain. PLoS One 2: e754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas R, Gomez-Marin C, Wilson JM, Casares F, Gomez-Skarmeta JL. 2012. Hoxd13 contribution to the evolution of vertebrate appendages. Dev Cell 23: 1219–1229. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, Chambon P. 1996. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122: 2997–3011. [DOI] [PubMed] [Google Scholar]
- Gehrke AR, Schneider I, de la Calle-Mustienes E, Tena JJ, Gomez-Marin C, Chandran M, Nakamura T, Braasch I, Postlethwait JH, Gomez-Skarmeta JL, et al. 2015. Deep conservation of wrist and digit enhancers in fish. Proc Natl Acad Sci 112: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Martin MC, Mallo M, Ros MA. 2014. Long bone development requires a threshold of Hox function. Dev Biol 392: 454–465. [DOI] [PubMed] [Google Scholar]
- Haack H, Gruss P. 1993. The establishment of murine Hox-1 expression domains during patterning of the limb. Dev Biol 157: 410–422. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. 1992. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn 195: 231–272. [DOI] [PubMed] [Google Scholar]
- Johanson Z, Joss J, Boisvert CA, Ericsson R, Sutija M, Ahlberg PE. 2007. Fish fingers: digit homologues in sarcopterygian fish fins. J Exp Zool Mol Dev Evol 308: 757–768. [DOI] [PubMed] [Google Scholar]
- Kmita M, Tarchini B, Zakany J, Logan M, Tabin CJ, Duboule D. 2005. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 435: 1113–1116. [DOI] [PubMed] [Google Scholar]
- Knezevic V, De Santo R, Schughart K, Huffstadt U, Chiang C, Mahon KA, Mackem S. 1997. Hoxd-12 differentially affects preaxial and postaxial chondrogenic branches in the limb and regulates Sonic hedgehog in a positive feedback loop. Development 124: 4523–4536. [DOI] [PubMed] [Google Scholar]
- Kondo T, Zakany J, Innis JW, Duboule D. 1997. Of fingers, toes and penises. Nature 390: 29. [DOI] [PubMed] [Google Scholar]
- Kondo T, Zákány J, Duboule D. 1998. Control of colinearity in AbdB genes of the mouse HoxD complex. Mol Cell 1: 289–300. [DOI] [PubMed] [Google Scholar]
- Kuss P, Kraft K, Stumm J, Ibrahim D, Vallecillo-Garcia P, Mundlos S, Stricker S. 2014. Regulation of cell polarity in the cartilage growth plate and perichondrium of metacarpal elements by HOXD13 and WNT5A. Dev Biol 385: 83–93. [DOI] [PubMed] [Google Scholar]
- Lehoczky JA, Innis JW. 2008. BAC transgenic analysis reveals enhancers sufficient for Hoxa13 and neighborhood gene expression in mouse embryonic distal limbs and genital bud. Evol Dev 10: 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J, Martin P. 1989. Vertebrate development. Limbs: a pattern emerges. Nature 342: 734–735. [DOI] [PubMed] [Google Scholar]
- Lonfat N, Duboule D. 2015. Structure, function and evolution of topologically associating domains (TADs) at HOX loci. FEBS Lett 589: 2869–2876. [DOI] [PubMed] [Google Scholar]
- Lonfat N, Montavon T, Darbellay F, Gitto S, Duboule D. 2014. Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science 346: 1004–1006. [DOI] [PubMed] [Google Scholar]
- Metscher BD, Takahashi K, Crow K, Amemiya C, Nonaka DF, Wagner GP. 2005. Expression of Hoxa-11 and Hoxa-13 in the pectoral fin of a basal ray-finned fish, Polyodon spathula: implications for the origin of tetrapod limbs. Evol Dev 7: 186–195. [DOI] [PubMed] [Google Scholar]
- Montavon T, Le Garrec J-F, Kerszberg M, Duboule D. 2008. Modeling Hox gene regulation in digits: reverse collinearity and the molecular origin of thumbness. Genes Dev 22: 346–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. 2011. A regulatory archipelago controls Hox genes transcription in digits. Cell 147: 1132–1145. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Morgan BA, Burke AC, Laufer E, DiMambro E, Murtaugh LC, Gonzales E, Tessarollo L, Parada LF, Tabin C. 1996. Analysis of Hox gene expression in the chick limb bud. Development 122: 1449–1466. [DOI] [PubMed] [Google Scholar]
- Neufeld SJ, Wang F, Cobb J. 2014. Genetic interactions between Shox2 and Hox genes during the regional growth and development of the mouse limb. Genetics 198: 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. 2011. The dynamic architecture of Hox gene clusters. Science 334: 222–225. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Schorderet P, Joye E, Chabaud F, Duboule D. 2014. Temporal dynamics and developmental memory of 3D chromatin architecture at Hox gene loci. Elife 3: e02557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post LC, Innis JW. 1999. Altered Hox expression and increased cell death distinguish Hypodactyly from Hoxa13 null mice. Int J Dev Biol 43: 287–294. [PubMed] [Google Scholar]
- Rincon-Limas DE, Lu CH, Canal I, Calleja M, Rodriguez-Esteban C, Izpisua-Belmonte JC, Botas J. 1999. Conservation of the expression and function of apterous orthologs in Drosophila and mammals. Proc Natl Acad Sci 96: 2165–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Cavalli G. 2015. The role of chromosome domains in shaping the functional genome. Cell 160: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Sheth R, Gregoire D, Dumouchel A, Scotti M, Pham JM, Nemec S, Bastida MF, Ros MA, Kmita M. 2013. Decoupling the function of Hox and Shh in developing limb reveals multiple inputs of Hox genes on limb growth. Development 140: 2130–2138. [DOI] [PubMed] [Google Scholar]
- Sheth R, Bastida MF, Kmita M, Ros M. 2014. ‘Self-regulation,’ a new facet of Hox genes’ function. Dev Dyn 243: 182–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou S, Carlson HL, Perez WD, Stadler HS. 2013. HOXA13 regulates Aldh1a2 expression in the autopod to facilitate interdigital programmed cell death. Dev Dyn 242: 687–698. [DOI] [PubMed] [Google Scholar]
- Shubin N, Tabin C, Carroll S. 2009. Deep homology and the origins of evolutionary novelty. Nature 457: 818–823. [DOI] [PubMed] [Google Scholar]
- Sordino P, van der Hoeven F, Duboule D. 1995. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature 375: 678–681. [DOI] [PubMed] [Google Scholar]
- Tabin C, Wolpert L. 2007. Rethinking the proximodistal axis of the vertebrate limb in the molecular era. Genes Dev 21: 1433–1442. [DOI] [PubMed] [Google Scholar]
- Tamura K, Yonei-Tamura S, Yano T, Yokoyama H, Ide H. 2008. The autopod: its formation during limb development. Dev Growth Differ 50: S177–S187. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Duboule D. 2006. Control of Hoxd genes’ collinearity during early limb development. Dev Cell 10: 93–103. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Duboule D, Kmita M. 2006. Regulatory constraints in the evolution of the tetrapod limb anterior-posterior polarity. Nature 443: 985–988. [DOI] [PubMed] [Google Scholar]
- Tetreault N, Champagne MP, Bernier G. 2009. The LIM homeobox transcription factor Lhx2 is required to specify the retina field and synergistically cooperates with Pax6 for Six6 trans-activation. Dev Biol 327: 541–550. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp P, Duboule D. 2011. A regulatory ‘landscape effect’ over the HoxD cluster. Dev Biol 351: 288–296. [DOI] [PubMed] [Google Scholar]
- van der Hoeven F, Sordino P, Fraudeau N, Izpisua-Belmonte JC, Duboule D. 1996. Teleost HoxD and HoxA genes: comparison with tetrapods and functional evolution of the HoxD complex. Mech Dev 54: 9–21. [DOI] [PubMed] [Google Scholar]
- Villavicencio-Lorini P, Kuss P, Friedrich J, Haupt J, Farooq M, Turkmen S, Duboule D, Hecht J, Mundlos S. 2010. Homeobox genes d11–d13 and a13 control mouse autopod cortical bone and joint formation. J Clin Invest 120: 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warot X, Fromental-Ramain C, Fraulob V, Chambon P, Dolle P. 1997. Gene dosage-dependent effects of the Hoxa-13 and Hoxd-13 mutations on morphogenesis of the terminal parts of the digestive and urogenital tracts. Development 124: 4781–4791. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Duboule D. 2010. The origin of digits: expression patterns versus regulatory mechanisms. Dev Cell 18: 526–532. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Noordermeer D, Leleu M, Duboule D. 2014. Conservation and divergence of regulatory strategies at Hox loci and the origin of tetrapod digits. PLoS Biol 12: e1001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Gotoh Y, Tamura K, Tanaka M, Kawakami A, Ide H, Kuroiwa A. 1998. Coordinated expression of Hoxa-11 and Hoxa-13 during limb muscle patterning. Development 125: 1325–1335. [DOI] [PubMed] [Google Scholar]
- Yokouchi Y, Sasaki H, Kuroiwa A. 1991. Homeobox gene expression correlated with the bifurcation process of limb cartilage development. Nature 353: 443–445. [DOI] [PubMed] [Google Scholar]
- Yokouchi Y, Nakazato S, Yamamoto M, Goto Y, Kameda T, Iba H, Kuroiwa A. 1995a. Misexpression of Hoxa-13 induces cartilage homeotic transformation and changes cell adhesiveness in chick limb buds. Genes Dev 9: 2509–2522. [DOI] [PubMed] [Google Scholar]
- Yokouchi Y, Sakiyama J, Kuroiwa A. 1995b. Coordinated expression of Abd-B subfamily genes of the HoxA cluster in the developing digestive tract of chick embryo. Dev Biol 169: 76–89. [DOI] [PubMed] [Google Scholar]
- Zákány J, Duboule D. 1996. Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature 384: 69–71. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. 2007. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev 17: 359–366. [DOI] [PubMed] [Google Scholar]
- Zeller R, Lopez-Rios J, Zuniga A. 2009. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat Rev Genet 10: 845–858. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.