The yeast Candida albicans causes human infections that have mortality rates approaching 50%. The key to developing improved therapeutics is to understand the host-pathogen interface. A critical interaction is that with macrophages: intracellular Candida triggers the NLRP3/caspase-1 inflammasome for escape through lytic host cell death, but this also activates antifungal responses. To better understand how the inflammasome response to Candida is fine-tuned, we established live-cell imaging of inflammasome activation at single-cell resolution, coupled with analysis of the fungal ERMES complex, a mitochondrial regulator that lacks human homologs. We show that ERMES mediates Candida escape via inflammasome-dependent processes, and our data suggest that inflammasome activation is controlled by the level of hyphal growth and exposure of cell wall components as a proxy for severity of danger. Our study provides the most detailed dynamic analysis of inflammasome responses to a fungal pathogen so far and establishes promising pathogen- and host-derived therapeutic strategies.
KEYWORDS: Candida albicans, macrophage, metabolism, mitochondria
ABSTRACT
The pathogenic yeast Candida albicans escapes macrophages by triggering NLRP3 inflammasome-dependent host cell death (pyroptosis). Pyroptosis is inflammatory and must be tightly regulated by host and microbe, but the mechanism is incompletely defined. We characterized the C. albicans endoplasmic reticulum (ER)-mitochondrion tether ERMES and show that the ERMES mmm1 mutant is severely crippled in killing macrophages despite hyphal formation and normal phagocytosis and survival. To understand dynamic inflammasome responses to Candida with high spatiotemporal resolution, we established live-cell imaging for parallel detection of inflammasome activation and pyroptosis at the single-cell level. This showed that the inflammasome response to mmm1 mutant hyphae is delayed by 10 h, after which an exacerbated activation occurs. The NLRP3 inhibitor MCC950 inhibited inflammasome activation and pyroptosis by C. albicans, including exacerbated inflammasome activation by the mmm1 mutant. At the cell biology level, inactivation of ERMES led to a rapid collapse of mitochondrial tubular morphology, slow growth and hyphal elongation at host temperature, and reduced exposed 1,3-β-glucan in hyphal populations. Our data suggest that inflammasome activation by C. albicans requires a signal threshold dependent on hyphal elongation and cell wall remodeling, which could fine-tune the response relative to the level of danger posed by C. albicans. The phenotypes of the ERMES mutant and the lack of conservation in animals suggest that ERMES is a promising antifungal drug target. Our data further indicate that NLRP3 inhibition by MCC950 could modulate C. albicans-induced inflammation.
IMPORTANCE The yeast Candida albicans causes human infections that have mortality rates approaching 50%. The key to developing improved therapeutics is to understand the host-pathogen interface. A critical interaction is that with macrophages: intracellular Candida triggers the NLRP3/caspase-1 inflammasome for escape through lytic host cell death, but this also activates antifungal responses. To better understand how the inflammasome response to Candida is fine-tuned, we established live-cell imaging of inflammasome activation at single-cell resolution, coupled with analysis of the fungal ERMES complex, a mitochondrial regulator that lacks human homologs. We show that ERMES mediates Candida escape via inflammasome-dependent processes, and our data suggest that inflammasome activation is controlled by the level of hyphal growth and exposure of cell wall components as a proxy for severity of danger. Our study provides the most detailed dynamic analysis of inflammasome responses to a fungal pathogen so far and establishes promising pathogen- and host-derived therapeutic strategies.
INTRODUCTION
Important aspects of microbial pathogenesis involve metabolic adaptation in the host (1–3). The yeast Candida albicans is the most common fungal pathogen in human infections, and it can cause deadly systemic disease (4). For C. albicans, metabolic regulation has wide-ranging consequences for virulence, including interaction with host immunity and resistance to stressors and antifungal therapeutics (reviewed in reference 3). In eukaryotic cells, mitochondria have central functions in energy production and metabolism, and mitochondrial function is necessary for virulence of pathogenic fungi (reviewed in references 5 and 6). Therefore, the development of new antifungal strategies based on metabolic and mitochondrial regulation is promising, but few regulators of these pathways have been characterized in fungal pathogens.
Metabolic adaptation is important for C. albicans during the critical immune interaction with macrophages. C. albicans reprograms its metabolism to suit the nutrient environment in phagocytes (7, 8). Furthermore, metabolism is involved in the transition of C. albicans from yeast to hyphal morphology, which promotes immune evasion by causing host cell lysis (9–12). Recent work from our lab and the Krysan lab has shown that, upon phagocytosis, hyphae rapidly trigger a programmed macrophage cell death mechanism termed pyroptosis (13, 14). Pyroptosis exposes intracellular pathogens to immune attack and rids them of their replication niche (15). Given that C. albicans primarily replicates extracellularly, we have proposed that this fungus “hijacks” pyroptosis to egress and evade intracellular killing (13). Induction of pyroptosis by C. albicans depends on the NLRP3/caspase-1 inflammasome (13, 14). Activation of the NLRP3 inflammasome by Candida needs to be tightly regulated by both pathogen and host. On the pathogen side, filamentous growth has been linked to inflammasome activation and pyroptosis (13, 14, 16). This suggests that C. albicans induces pyroptosis after the expression of virulence traits that may be important for extracellular survival and dissemination under inflammatory conditions, as activation of the NLRP3 inflammasome also triggers antifungal immune responses (reviewed in references 17 and 18). How inflammasome activation by C. albicans is tightly regulated remains to be fully understood, particularly in light of recent studies that showed that factors other than fungal morphology are at play, and yeast cells can also cause inflammasome-dependent macrophage lysis under some conditions (13, 14, 19). On the host side, inflammasome activation by C. albicans needs to be regulated to modulate inflammation in response to commensal or pathogenic fungal growth. Besides pyroptosis, C. albicans triggers other, less-defined forms of macrophage death, most strikingly a second wave of killing that eventually eliminates the entire macrophage population (13, 14). We termed these two stages of killing phase 1 (pyroptotic death) and phase 2 (nonpyroptotic death) (13). Fungal factors coordinating these distinct macrophage death pathways are unknown.
Here, we sought to characterize novel regulators that mediate evasion of macrophages by Candida, focusing on fungal mitochondria, which have so far been largely understudied in this context. For this, we characterized a key mitochondrial regulator, the endoplasmic reticulum (ER)-mitochondrion tethering complex ERMES (20). Complexes such as ERMES, which mediate interactions between organelles by providing “membrane contact sites,” represent hubs that can control cell physiology on a global level (21, 22). The functions of such complexes are poorly understood in eukaryotic pathogens. ERMES is particularly promising in the context of fungal pathogenesis because it is found broadly in fungi but is absent from animals (23), and it could therefore be targeted for antifungal therapy. In support of ERMES being a promising antifungal drug target, a mutant library screen by Merck identified the ERMES subunit MMM1 as being important for C. albicans virulence in the mouse tail vein infection model of candidiasis (24). However, the cellular functions of ERMES in C. albicans and its potential roles in host-pathogen interactions have not been studied so far.
We report here that in C. albicans the activity of ERMES is important for enabling immune evasion via multiple macrophage death mechanisms (phase 1 and phase 2). Key roles of ERMES in C. albicans are the regulation of mitochondrial morphology and enabling optimal growth at host temperature. To further understand the interplay between C. albicans and the inflammasome, we established live-cell imaging to monitor inflammasome activation and macrophage death in parallel, at single-cell resolution and in real time over the entire interaction course of 24 h (i.e., until the entire macrophage culture collapses). We combined this powerful assay with ERMES mutant analysis and a newly described small-molecule inhibitor of NLRP3 that showed promise in treating inflammatory disorders (25). Using these novel tools, we show that the inflammasome not only responds to fungal morphotype but also discriminates hyphae produced by wild-type C. albicans from hyphae produced by the less virulent ERMES mutant. We propose that this discrimination is achieved through a signal threshold response for inflammasome activation that is linked to hyphal growth and cell wall remodeling. Based on our data, we suggest novel host- and pathogen-derived avenues for antifungal drug development and propose that our results and the imaging assay that we established will be broadly applicable to the understanding of dynamic inflammasome responses to fungal pathogens.
RESULTS
C. albicans ERMES is required for macrophage killing and immune evasion.
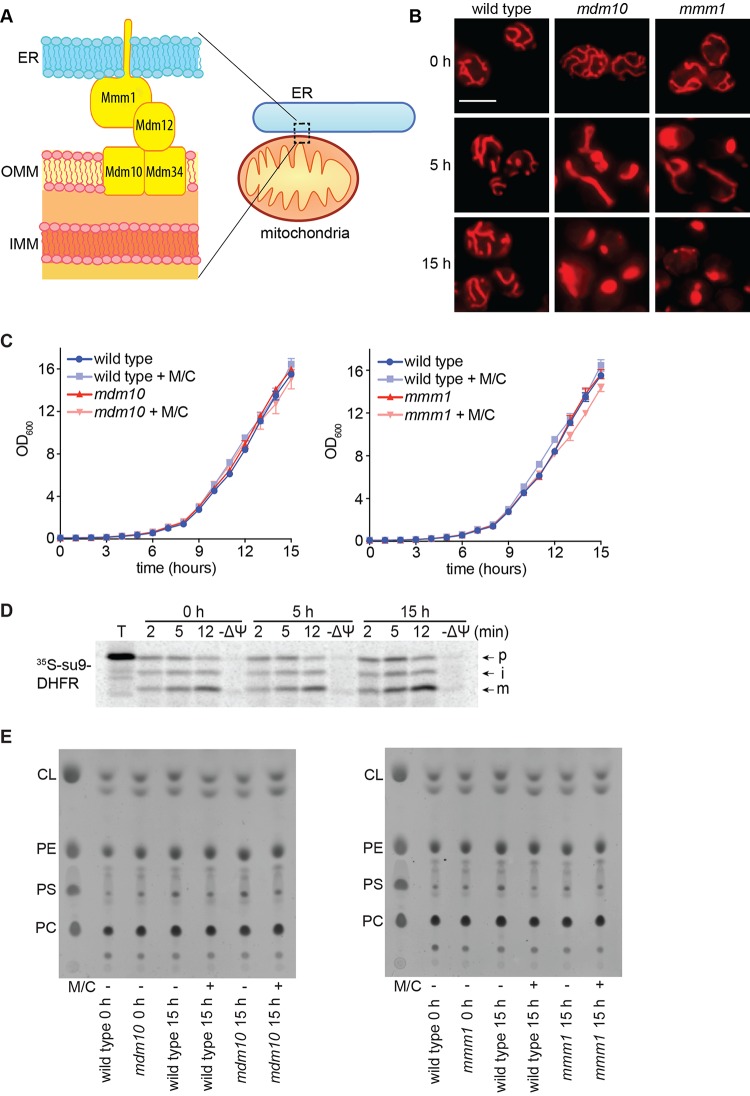
In C. albicans, mitochondrial dysfunction can have large effects on fitness (for example, see reference 26). Therefore, to start delineating the functions of ERMES, we constructed conditional mutants in two ERMES genes: MMM1, which encodes the subunit anchored in the ER, and MDM10, which encodes a subunit located in the mitochondrial outer membrane (Fig. 1A). In these mutants, one allele is deleted and the other one is placed under the MET3 promoter, which is “on” in the absence of methionine and cysteine and “off” in their presence. Gene repression was achieved following addition of methionine and cysteine to the medium (see Fig. S1A in the supplemental material), and microscopy showed that under these conditions ERMES function is inactivated, as both mutants displayed an early and clear defect in mitochondrial morphology (Fig. 1B; see also Fig. S2). Already at 5 h postrepression, loss of mitochondrial tubular network structure was observed, and the defect was even more pronounced after 15 h, with the clear appearance of globular, collapsed mitochondria (Fig. 1B; see also quantification in Fig. S3). This mitochondrial morphology defect is consistent with what is observed in ERMES mutants of Saccharomyces cerevisiae (27–30). Unlike mitochondrial morphology, the growth of the two mutants was not compromised to a considerable degree even after several cell divisions in the 15-h time course (Fig. 1C). Consistent with normal respiration, mitochondria isolated from the mdm10 mutant maintained their membrane potential upon repression, as they imported a substrate normally into the mitochondrial matrix (Fig. 1D). Moreover, steady-state levels of cellular phospholipids in both mutants were the same as those in the wild type at 15 h postrepression, and this included the mitochondrion-specific cardiolipin (Fig. 1E; see quantification in Fig. S4). Studies in S. cerevisiae suggested roles for ERMES not only in mitochondrial morphology but also in mitochondrial lipid homeostasis, fitness, and respiration through maintenance of the mitochondrial genome (20, 27–34). To further address these additional functions of ERMES in C. albicans, we made homozygous deletion mutants in each one of the four ERMES subunits. The four mutants had equivalent phenotypes and displayed large fitness defects with barely viable cells, lack of growth on glycerol, lack of a wild-type mitochondrial network, and altered lipid homeostasis through loss of the mitochondrial phospholipid cardiolipin (see Fig. S5). Collectively, our results show that in C. albicans the earliest defect upon ERMES inactivation is loss of wild-type mitochondrial morphology, while the lipid, respiration, and fitness defects are seen following longer-term inactivation of ERMES. While our data show that, of all the mutant phenotypes tested, mitochondrial morphology is the most sensitive to ERMES gene repression, it is possible that residual ERMES protein levels are present in the conditional mutants, and this could support functions in mitochondrial lipid homeostasis and fitness.
FIG 1 .
The Candida ERMES has a primary role in mitochondrial morphology. (A) Cartoon of the core ERMES complex as understood in S. cerevisiae. Recent work has shown that Mmm1 and Mdm12 associate as a heterotetramer and that Mdm34 also forms dimers (58), but for simplicity, we do not depict it here. The precise architecture of the entire complex is yet to be understood. ER, endoplasmic reticulum; OMM, outer mitochondrial membrane; IMM, inner mitochondrial membrane. (B) Loss of mitochondrial morphology upon ERMES inactivation monitored at 30°C. Shown are images of representative cells selected from larger microscopy fields depicted in Fig. S2 in the supplemental material. Bar, 5 µm. Quantification is in Fig. S3 in the supplemental material. (C) Growth curves of the indicated strains under permissive or repressive (+ M/C) conditions at 30°C. Shown are averages and the standard errors of the means from 3 biological replicates assayed in the same experiment. OD600, optical density at 600 nm. (D) Mitochondria were prepared from the mdm10 strain before or at 5 h and 15 h after gene repression and incubated with 35S-labeled mitochondrial reporter Su9-dihydrofolate reductase (DHFR) for the indicated times. Mitochondrial membrane potential was dissipated before import in the −ΔΨ lanes. Mitochondria were analyzed by SDS-PAGE and phosphorimaging. p, precursor; i, intermediate; m, mature processed form. (E) Total cellular lipids were extracted following growth under permissive or repressive (+ M/C) conditions for 15 h and separated by thin-layer chromatography. Only phospholipids are shown here: CL, cardiolipin; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PS, phosphatidylserine. Quantification is in Fig. S4 in the supplemental material.
(Fig. S1) Analysis of ERMES gene repression. (A) Cells of the indicated strains were grown under permissive conditions (synthetic medium without methionine and cysteine) followed by addition of 2.5 mM methionine and 0.5 mM cysteine at time zero, and gene repression was monitored by quantitative RT-PCR at the indicated time points. The levels of ERMES genes were normalized to the levels of the RNA polymerase III transcript SCR1 or the rRNA transcript RDN5 or RDN25. Shown are averages and standard errors from 3 biological replicates assayed in the same experiment. (B) MMM1 gene repression in macrophages. The mmm1 mutant and the complemented control strain were grown at 30°C under repressive conditions overnight. Macrophages were infected with C. albicans at the multiplicity of infection of 6 Candida cells to 1 macrophage, with methionine and cysteine included in the tissue culture medium. Gene repression was monitored by quantitative RT-PCR at the indicated time points following coincubation of C. albicans with macrophages and washing of the nonphagocytosed cells. To separate Candida from the macrophages, Trizol treatment was performed to remove lysed macrophages, and subsequently, Candida cell pellets were recovered and RNA was isolated by the hot phenol method. Transcript levels of MMM1 were normalized to the RNA polymerase III transcript SCR1 or the rRNA transcript RDN5. Shown are averages and the standard errors from 4 biological replicates obtained from 2 independent experiments.
(Fig. S2) Mitochondrial morphology defects of ERMES mutant strains. Shown are larger microscopy fields of the MitoTracker Red images displayed in Fig. 1B, without any processing (i.e., color or contrast correction) of the images. Bar, 10 µm.
(Fig. S3) Quantification of the mitochondrial morphology defects after conditional inactivation of ERMES. Relates to Fig. 1B. Percentages of cells with abnormal mitochondria in wild-type and mdm10 (top) and mmm1 (bottom) strains are shown. This was calculated at 0, 5, and 15 h postrepression from 3 independent biological replicates for each of the strains (3 independent colonies) assayed in the same experiment, counting at least 300 cells for each repeat. In parallel, cells were grown under permissive conditions as a control. Impaired mitochondria were quantified based on any mitochondria not resembling the wild-type tubular network as detected by MitoTracker Red staining, and this was subdivided into two distinct categories: a milder phenotype (“shrunken”), where the tubular network was reduced in size and not spanning the entire cell when analyzing multiple planes of focus under the microscope, and a severe phenotype (“collapsed”), consisting of round, enlarged, or severely fragmented mitochondria. The experiment was performed on multiple occasions with equivalent results observed.
(Fig. S4) Quantification of phospholipids after conditional inactivation of ERMES. Relates to Fig. 1E. Lipids were quantified using the Toolbox module of ImageQuant 1D version 7.0, and background signals were subtracted using the local median method performed by the software. Shown are averages and the standard deviations from three biological repeats. The amounts of cardiolipin (CL) and phosphatidylethanolamine (PE) are shown relative to the levels of phosphatidylcholine (PC) in the same sample as an internal control and in comparison to the wild type.
(Fig. S5) Long-term inactivation of ERMES results in loss of fitness, altered mitochondrial network morphology, and phospholipid imbalance. (A) (Left) Wild-type C. albicans and homozygous deletion strains lacking MDM12 (mdm12ΔΔ), MMM1 (mmm1ΔΔ), MDM10 (mdm10ΔΔ), or MDM34 (mdm34ΔΔ) were grown on rich medium containing glucose (YPD) or glycerol (YPG) as a carbon source, at 30°C or 37°C. Tenfold serial dilutions were plated, starting from an optical density at 600 nm (OD600) of 0.5. Images were taken after 3 days. (Right) Complementation of the fitness defects of the homozygous deletion strains for MDM12 (mdm12ΔΔ+12), MMM1 (mmm1ΔΔ+1), and MDM34 (mdm34ΔΔ+34). (B) Representative fluorescence images of the wild-type and ERMES homozygous mutant strains stained with MitoTracker Green dye. Bar, 5 µm. Images grouped together were all taken in parallel in the same experiment. No wild-type mitochondrial network morphology was seen in the homozygous ERMES mutants. Complementation of the mitochondrial network defects is shown for MDM12 (mdm12ΔΔ+12), MMM1 (mmm1ΔΔ+1), and MDM34 (mdm34ΔΔ+34). We noticed that in complemented strains with expression levels of the ERMES genes equivalent to approximately half of wild-type levels (as expected from single-copy gene reintegrants), restoration of mitochondrial morphology defects was partial; clones with expression levels equivalent to 2- to 3-fold over wild-type values displayed significantly better complementation of mitochondrial morphology. Elevated levels of ERMES genes may be necessary for restoring growth to severely sick ERMES mutants. (C) Total cellular lipids were extracted from the indicated strains and separated by thin-layer chromatography (TLC). Standards were used to identify cardiolipin (CL), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylcholine (PC). The right panel displays quantification averages, performed using ImageQuant, from 3 biological repeats. Error bars indicate standard deviations. ***, P < 0.0005; ****, P < 0.0001, compared to wild type using Student’s t test (unpaired, two-tailed). The amounts of CL and PE are shown relative to the levels of PC in the same sample as an internal control and in comparison to the wild type.
(Fig. S6) Effects of methionine and cysteine addition in the macrophage-interaction experiments. Relates to Fig. 2B. Macrophages were infected with the mmm1 mutant and the complemented control strain at an MOI of 6:1 (Candida cells to macrophage). After 1 h of coincubation and washing of nonphagocytosed cells, macrophage medium was included with (+Met/Cys) or without (-Met/Cys) 2.5 mM methionine and 0.5 mM cysteine. Live-cell imaging was then started 3 h later. Shown are averages and standard errors from 2 biological replicates (2 independent colonies of each of the two C. albicans strains) assayed in the same experiment.
(Fig. S7) Characterization of hyphal growth of the mmm1 mutant. (A) Relates to Fig. 2E. Mitochondrial network structure of hyphae was visualized by staining with MitoTracker Red dye. Representative images of several hyphae made by the mmm1 mutant and the complemented control strain are shown in the left panel. The right panel contains a higher magnification of a single hyphal cell with bright-field overlay. In both images, the scale bar represents 10 µm. (B) Relates to Fig. 5C. Hyphal length distribution in RPMI medium after 3 h of growth. Germ tubes or hyphal filaments were over 5 µm. Each cell measured is depicted by a line, with length shown on the y axis and the measurements ranked in order from smallest to largest on the x axis. Three biological replicates were performed. Two repeats are shown here, and the third repeat is shown in Fig. 5C. (C) Relates to Fig. 5E. Hyphal length distribution in macrophages following 1 h of coincubation and washes was measured in 2 independent experiments with equivalent results (n = 200 fungal cells per strain). One experiment is shown here, and the other one is shown in Fig. 5E. Download Figures S1 to S7, PDF file, 8.5 MB (8.7MB, pdf) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
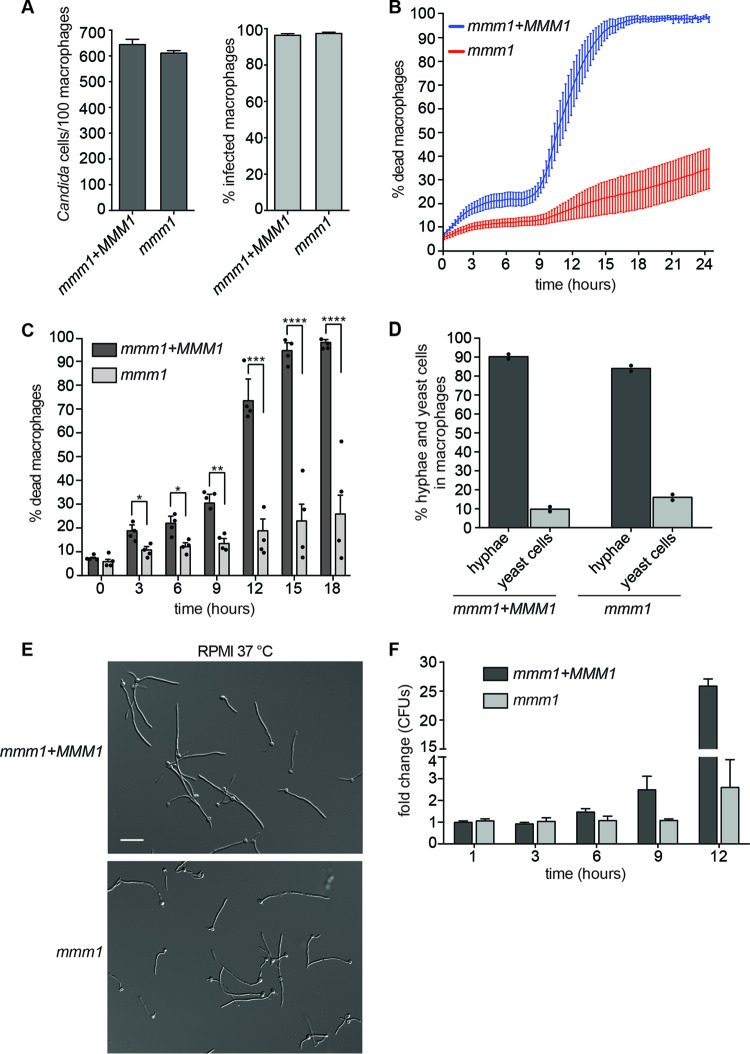
Next, we tested how ERMES might be important for Candida-macrophage interactions. The experiments were done under repressive conditions (in the presence of methionine and cysteine), using the conditional mmm1 mutant and the complemented mmm1+MMM1 strain as the control. Quantitative reverse transcription-PCR (RT-PCR) analysis showed that repression of the MMM1 gene was maintained in macrophages for at least 15 h (see Fig. S1B in the supplemental material). Addition of methionine and cysteine to the medium did not change the progression of macrophage killing by the control C. albicans strain (see Fig. S6). Moreover, the macrophage killing curve obtained under these conditions was comparable to our previous results in medium without additional supplementation with methionine and cysteine (13). The mmm1 mutant was phagocytosed normally by bone marrow-derived macrophages (BMDMs) (Fig. 2A), but live-cell imaging showed that its ability to kill macrophages was severely compromised, with macrophage death reaching only ≈30% by 24 h (Fig. 2B and C; see also Movies S1 and S2). As a control, we show that derepression of the MMM1 gene by omission of methionine and cysteine from the medium during the C. albicans-macrophage interaction experiment resulted in higher macrophage killing by the ERMES mutant than that obtained under repressive conditions (see Fig. S6). Despite highly compromised host cell killing, the mmm1 mutant was able to undergo hyphal morphogenesis in macrophages (Fig. 2D). Hyphal formation by the mutant was also evident in macrophage growth medium in vitro (Fig. 2E; see also Fig. S7A), and the mutant hyphae continued to grow over the course of the macrophage experiment (see Movie S2). The mmm1 mutant maintained viability in macrophages for at least 12 h postinfection (Fig. 2F). In the first 6 h postinfection, Candida CFU derived from infected macrophages were similar between the mmm1 mutant and the control strain (Fig. 2F). At 9 h postinfection, some increase in CFU was seen for the control strain, and at 12 h postinfection, control strain CFU clearly increased, while the increase in mutant CFU was diminished (Fig. 2F). This is consistent with substantial escape of the control Candida strain from macrophages, as most fungal growth under these conditions is seen after hyphae lyse macrophages and egress into the surrounding medium (see Movie S1). In this study and previously, we observed that after escape from macrophages into extracellular medium, growth of yeast-form cells coincides with phase 2, pyroptosis-independent macrophage death (see Movie S1) (13). Unlike in control samples, in infections with the mmm1 mutant no substantial yeast growth was observed in the medium at later time points (compare Movies S1 and S2 in the supplemental material).
FIG 2 .
ERMES is required for macrophage killing and fungal escape. (A) The phagocytic index (number of Candida cells per 100 macrophages) and the percentage of infected macrophages were determined at 1 h postphagocytosis. The MOI was 6 Candida cells to 1 macrophage. Shown are averages and the standard errors of the means from 3 independent experiments. (B) Macrophage cell death over time. Time zero is the start of live-cell imaging, after coincubation of C. albicans with macrophages for 1 h and washing of nonphagocytosed cells. Shown are averages from 4 independent experiments and the standard errors. See also Movies S1 and S2 in the supplemental material. (C) Bar graphs of selected time points from panel B, with each experimental data point shown in the scatter plot overlay. Statistical significance was determined by unpaired t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Quantification of hyphal formation in macrophages after 1 h of coincubation and washes. Data are from 2 independent experiments (shown separately as dot points and the mean), and 200 cells were measured for each strain per experiment. “Hyphae” represent germ tubes plus hyphal filaments. (E) Hyphal formation in repressive RPMI medium, 3 h at 37°C. Bar, 20 µm. (F) Fungal CFU were determined at the indicated time points following phagocytosis. After macrophage lysis, fungal cells were plated onto medium permissive for growth of the mmm1 mutant. Shown are the averages and standard errors of the means from 3 independent experiments. CFU fold change was calculated by normalizing to the control strain at 1 h.
Murine bone marrow-derived macrophages (BMDMs) infected with the control C. albicans strain (mmm1+MMM1). To create the video files, using Fiji software, the TIFF hyperstack was exported as an AVI file with JPEG compression at 3.0 frames per s. For all data analysis, the images were kept in TIFF format. Download Movie S1, AVI file, 14.2 MB (14.5MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Murine bone marrow-derived macrophages (BMDMs) infected with the mmm1 mutant strain. The file was created as described for Movie S1. Download Movie S2, AVI file, 11 MB (11.2MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Monitoring C. albicans-induced inflammasome activation and pyroptosis at single-cell level and in real time.
The uncoupling of hyphal morphogenesis and the ability to cause macrophage death seen in the mmm1 mutant was striking. Previous studies have reported on mutants that had much milder phenotypes (13), or macrophage death was assessed only at one, early time point (4 or 5 h postinfection) (14, 19, 35). NLRP3 inflammasome-dependent pyroptosis is a dominant mechanism of C. albicans-induced macrophage death early in infection (13, 14). The severe and prolonged defect in macrophage killing by the mmm1 mutant hyphae, coupled with the results from the work of Becker et al. showing that the mmm1 mutant is avirulent in the murine systemic candidiasis model (24), suggested that the NLRP3 inflammasome response is not only regulated on the basis of fungal morphotype but more sensitively tailored to the pathogenicity of the C. albicans strain. We reasoned that the mmm1 mutant might be a useful tool to understand how this could be achieved.
So far, inflammasome activation by fungal pathogens has been studied only in bulk macrophage populations. These lack dynamic spatiotemporal resolution and do not allow for a direct correlation between hyphal morphogenesis, inflammasome activation, and macrophage cell death in response to infection. To understand these processes in greater detail, we established live-cell imaging of Candida-induced NLRP3 inflammasome activation and pyroptosis at the single-cell level, in real time, and over the entire interaction time of approximately 24 h (i.e., until essentially all macrophages are killed by C. albicans) (Fig. 3A). This allowed us to study the process at unprecedented resolution. For this, we utilized macrophages expressing fluorescently labeled inflammasome subunit ASC (ASC-Cerulean) (36, 37). Under default conditions, ASC is uniformly dispersed in the cytoplasm, but it becomes concentrated in a single speck upon inflammasome activation. The involvement of the NLRP3 inflammasome was addressed by using MCC950, the novel small-molecule inhibitor of NLRP3 (25). Control samples were treated with an inactive compound, MCC6642 (25). Macrophages treated with heat-killed Candida and the compounds MCC950 and MCC6642 survived normally in the assay, demonstrating that these molecules do not have adverse effects on macrophage cell survival. ASC speck formation was monitored dynamically over time by live-cell microscopy, and macrophage cell death was monitored simultaneously using the membrane-impermeant DNA-staining dye DRAQ7. Initial experiments revealed that imaging in one plane of focus was not sufficient to capture all ASC speck formation events. Therefore, to ensure that all forming ASC specks were captured in multiple planes of focus, z-stacks spaced 8.5 µm part, totaling 42.5 µm, were taken in time-lapse images.
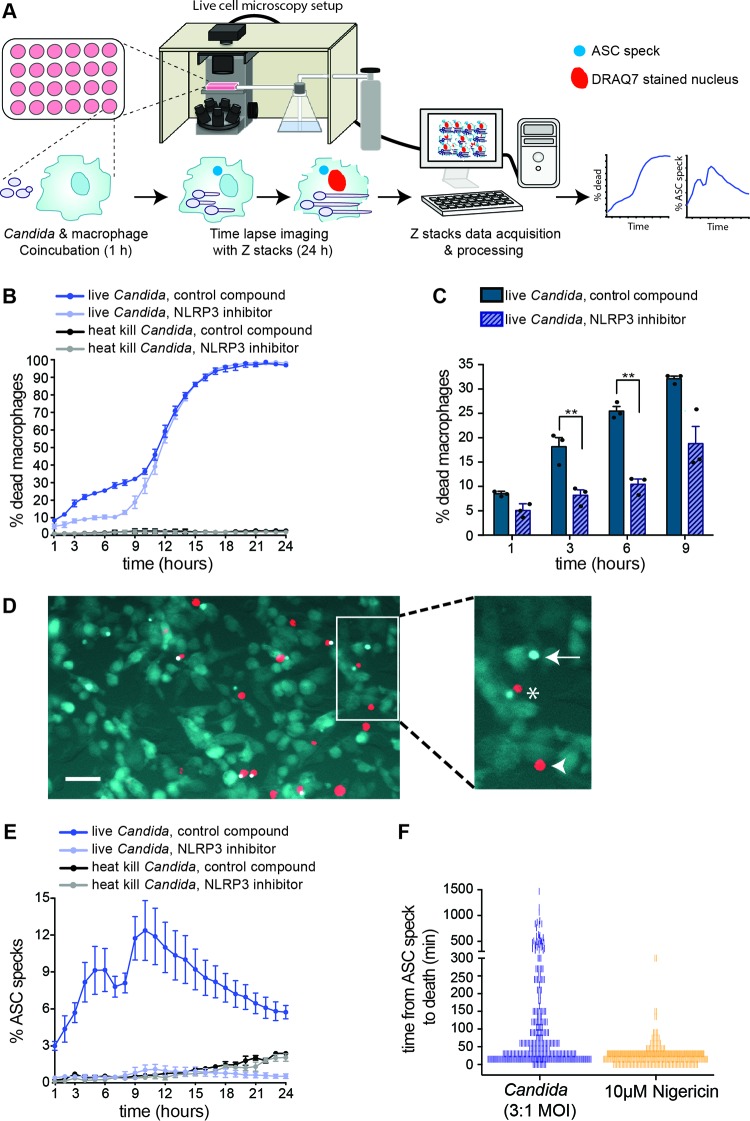
FIG 3 .
Real-time, single-cell-resolution analysis of C. albicans triggering the NLRP3 inflammasome and macrophage pyroptosis. (A) Live-cell imaging assay. ASC-Cerulean macrophages were infected with 3 Candida cells per macrophage. The assays were performed in the presence of the NLRP3 inhibitor MCC950 or the inactive small molecule MCC6642 (25), used at 10 µM concentrations. For all panels, the data shown here are the same as the data for the complemented mutant strain (mmm1+MMM1) and heat-killed Candida in Fig. 4. We show them here separately for clarity of the assay. These experiments were performed in the presence of methionine and cysteine in the medium for direct comparison with the mmm1 mutant, as shown in Fig. 4. (B) Macrophage cell death quantified from live-cell imaging in the same experiments as ASC speck formation in panel E. Shown are averages from 3 biological repeats and the standard error. See also Movies S3 and S4 in the supplemental material. (C) Bar graph of selected time points from the curves in panel B, with each experimental data point shown in the scatter plot overlay. Statistical significance was calculated by paired t test. **, P < 0.01. (D) Representative image of ASC speck formation upon recognition of C. albicans. Bar, 40 µm. Asterisk, macrophage pyroptosis detected by DRAQ7 as red nuclear fluorescence, in proximity to an ASC-Cerulean speck. Arrow, macrophage displaying inflammasome activation but not yet dead. Arrowhead, pyroptosis-independent death. (E) Quantification of ASC speck formation over time. Shown are averages from 3 biological repeats and the standard error. See also Movies S3 and S4. (F) Time to macrophage death following inflammasome activation by Candida or treatment with 10 µM nigericin. Individual macrophages (n = 300) were monitored from the moment of ASC speck formation until the appearance of DRAQ7 fluorescence. Each tally represents a single macrophage count and is reported in 15-min increments at the lower end of the scale.
ASC-Cerulean macrophages infected with the control C. albicans strain (mmm1+MMM1), under control conditions (treatment with the inactive compound MCC6642). The file was created as described for Movie S1. Download Movie S3, AVI file, 2.7 MB (2.7MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ASC-Cerulean macrophages infected with the control C. albicans strain (mmm1+MMM1) and treated with the NLRP3 inhibitor MCC950. The file was created as described for Movie S1. Download Movie S4, AVI file, 2.6 MB (2.7MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Given that ASC-Cerulean is an immortalized cell line in which NLRP3 and ASC are overexpressed (37), it was important to optimize the infection conditions to closely mimic the response observed in primary BMDMs. Optimization identified that at a multiplicity of 3 Candida cells to 1 macrophage, ASC-Cerulean-expressing macrophages behaved similarly to primary BMDMs (13), as 20 to 30% of macrophages were killed in the first 9 h of infection, and all host cells were dead by 24 h (Fig. 3B and C). Infection with C. albicans readily caused ASC speck formation (labeled with an arrow), followed by death as determined by the appearance of red, DRAQ7-stained nuclei in the same cell (labeled with an asterisk) (Fig. 3D; see also Movie S3 in the supplemental material). Some macrophages died without ASC speck formation within 9 h postinfection (Fig. 3D, arrowhead), in line with our observation that inactivation of pyroptosis does not block all C. albicans-induced macrophage death in the early stage of infection (13). The number of ASC specks increased over time and peaked at 10 h postinfection, after which inflammasome activation stopped (Fig. 3E).
Some ASC specks are lost shortly after macrophage death, meaning that the total number of ASC speck-positive macrophages can be underestimated if considering only those cells that are positive at any given time point. Therefore, to have a clearer estimate of the total number of ASC speck-positive macrophages (i.e., the total number of macrophages that activated the inflammasome), we counted the percentage of dead macrophages that previously displayed an ASC speck during the first 10 h postinfection (n = 200). Based on this, we estimated that ≈22% of macrophages contained an ASC speck during the first 10 h of infection; in other words, inflammasome activation by C. albicans is heterogeneous and occurs in only a portion of infected macrophages even in this relatively long time course of 10 h. Approximately ≈30 to 35% of macrophages are killed within the first 10 h (Fig. 3B), showing that the majority of DRAQ7-positive macrophages at this time had induced pyroptosis via the NLRP3/ASC/caspase-1 inflammasome. In addition to heterogeneous inflammasome activation, large differences were observed between individual macrophages in the timing of macrophage death post-ASC speck formation, ranging from 15 min to 24 h (Fig. 3F). This means that a small number of macrophages did not readily die after the inflammasome had been activated, but rather that death was observed only after the transition into phase 2 (pyroptosis-independent) death. As a control, we treated macrophages with the bacterial toxin nigericin, a known activator of the NLRP3 inflammasome, and observed immediate and more uniform ASC speck formation in the macrophage population, and in the majority of macrophages death occurred within 45 min (Fig. 3F). The NLRP3 inhibitor MCC950 blocked C. albicans-induced inflammasome activation and pyroptosis (Fig. 3E; see also Movie S4 in the supplemental material), and, accordingly, macrophage killing by C. albicans in the first 9 h of interaction was significantly reduced (Fig. 3B and C). Consistent with MCC950 blocking C. albicans-induced pyroptosis, the macrophage killing curve obtained in the presence of MCC950 closely mimics what we previously observed with C. albicans infecting macrophages that are inactivated for the pyroptotic caspases 1 and 11 (13).
Inflammasome activation in response to the mmm1 mutant.
To gain insight into how the inflammasome responds to hyphae produced by the avirulent mmm1 mutant, we used the assay with ASC-Cerulean-expressing macrophages described above (Fig. 4; of note, the data for the complemented mmm1+MMM1 strain and heat-killed Candida shown in Fig. 4 are the same as the data for these control strains shown in Fig. 3). Upon infection of macrophages with the mmm1 mutant, inflammasome activation was severely delayed for up to 10 h (Fig. 4A and B; see also Movie S5 in the supplemental material). As in BMDMs, this was accompanied by lower rates of cell death of mutant-infected macrophages (Fig. 4C and D). Treatment with the NLRP3 inhibitor abrogated the difference between the mmm1 mutant and the control strain in macrophage killing in the first 9 h (Fig. 4C and D; see also Movie S6). This shows that reduced macrophage killing is largely due to the inability of the mutant to trigger NLRP3-dependent pyroptosis for a prolonged time.
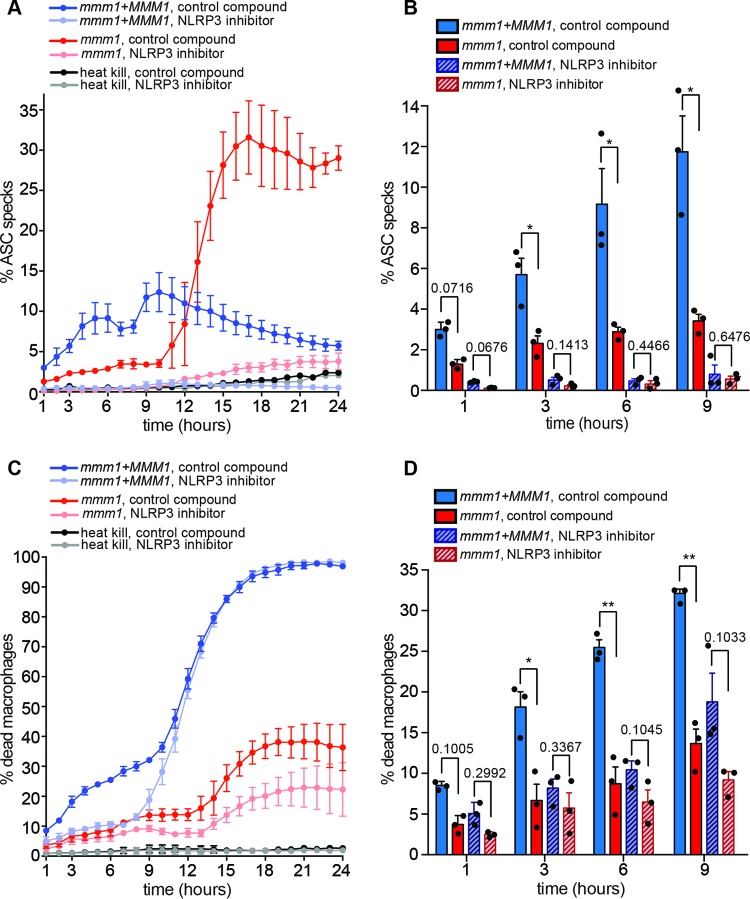
FIG 4 .
Inflammasome activation in response to the mmm1 mutant is indicative of a signal threshold response. The movies are uploaded as Movies S3 and S4 (control strain without or with the NLRP3 inhibitor) and Movies S5 and S6 (mmm1 mutant strain without or with the NLRP3 inhibitor) in the supplemental material. The data with the control and heat-killed Candida presented here are the same as those in Fig. 3. In all experiments, the control and the mmm1 mutant strain were analyzed in parallel for direct comparison. (A) Quantification of ASC speck formation over time, in the presence of control (inactive) compound or the NLRP3 inhibitor MCC950. Macrophages infected with heat-killed C. albicans are negative controls. Shown are averages and the standard errors from 3 independent experiments. (B) Bar graph of selected time points from the curves in panel A, with each experimental data point shown in the scatter plot overlay. *, P < 0.05 (unpaired t test with Welch’s correction). (C) Quantification of macrophage death by DRAQ7 fluorescence. (D) Bar graph of selected time points from the curves in panel C, with each experimental data point shown in the scatter plot overlay. *, P < 0.05; **, P < 0.01 (unpaired t test with Welch’s correction).
ASC-Cerulean macrophages infected with the mmm1 mutant strain under control conditions (treatment with the inactive compound MCC6642). The file was created as described for Movie S1. Download Movie S5, AVI file, 1.7 MB (1.7MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ASC-Cerulean macrophages infected with the mmm1 mutant and treated with the NLRP3 inhibitor MCC950. The file was created as described for Movie S1. Download Movie S6, AVI file, 1.9 MB (1.9MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Unexpectedly, after 10 h of infection, macrophages infected with the mmm1 mutant showed a sharp increase in ASC speck formation, surpassing what is seen with the control strain (Fig. 4A). More than 30% of macrophages contained ASC specks by 16 h (the highest ASC speck value observed was 31.58%). This exacerbated ASC speck formation was almost entirely blocked by the NLRP3 inhibitor MCC950 (Fig. 4A), demonstrating that it is due to NLRP3 activation. The increase in ASC specks was mirrored by an increase in macrophage cell death starting at 13 h and reaching ≈35% by 24 h (the highest observed death value was 38.29% [Fig. 4C]). The NLRP3 inhibitor partially rescued mmm1-induced macrophage death (Fig. 4C and D).
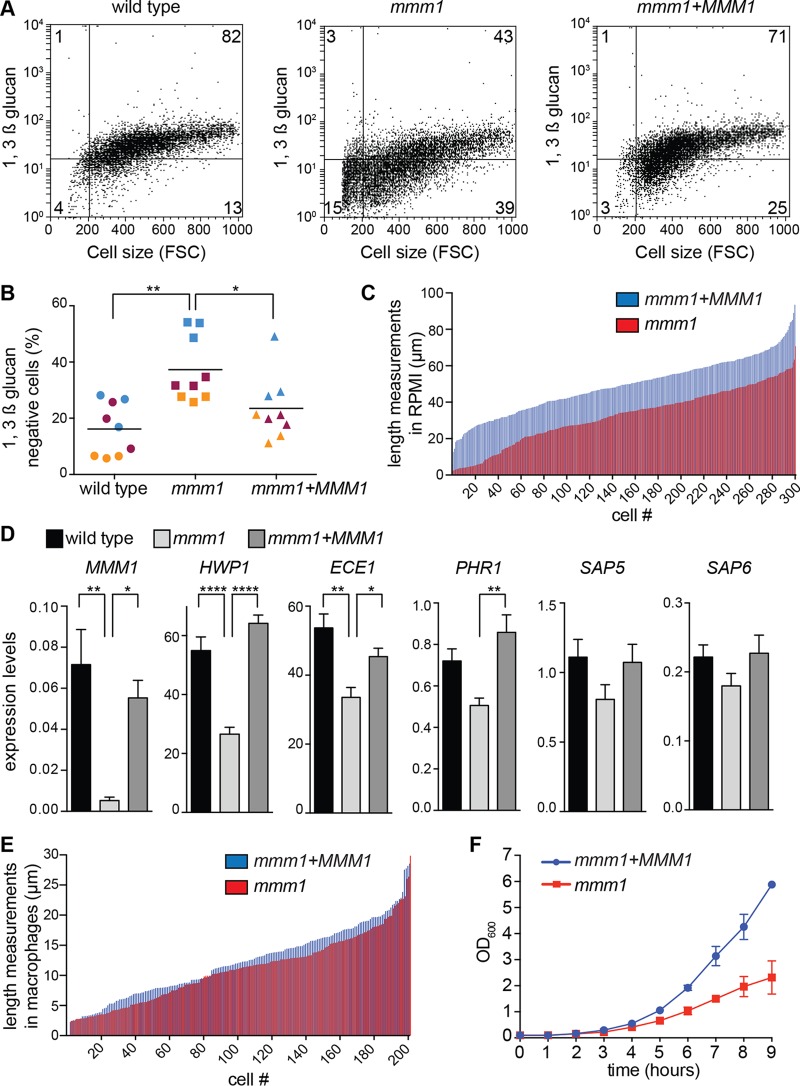
The kinetics of ASC speck formation in infections with the mmm1 mutant shows that, in response to a strain of reduced pathogenicity, inflammasome activation can be delayed by several hours. Importantly, inflammasome activation in response to the mmm1 mutant was not blocked and eventually occurred sharply. The kinetics suggested that signals derived from hyphae activate the inflammasome by a threshold mechanism, reminiscent of the recently described “digital” mode of activation for caspase-1 in response to various signals (38). In this scenario, in infections with the mmm1 mutant, the threshold is reached much later than in controls. Mitochondrial activity is needed for cell wall integrity in C. albicans (39–41; reviewed in references 5 and 6), and cell wall components, including 1,3-β-glucan and cell wall mannosylation, correlate with inflammasome activation and pyroptosis (13, 19, 42–44). We therefore hypothesized that the mmm1 mutant shows delayed cell wall restructuring during hyphal morphogenesis, leading to reduced numbers of exposed cell surface molecules that could be providing the signal for inflammasome activation. Consistent with this proposition, the percentage of cells that were negative for exposed 1,3-β-glucan was significantly higher in hyphal cultures of the mmm1 mutant than in controls (Fig. 5A and B). We further noticed that the population of shorter 1,3-β-glucan-negative cells was larger in the mmm1 mutant (Fig. 5A, lower left and right quadrants of dot plots). Since the formation of hyphae is linked to NLRP3 inflammasome activation, presumably due to changes to the fungal cell surface or cell physiology that provide the required signal, we addressed the expression of some of the genes induced in hyphal cells compared to yeast. Figure 5D shows that the expression levels of hypha-specific genes HWP1 and ECE1 and the cell wall glycosidase PHR1 were reduced in a statistically significant manner in mmm1 mutant hyphae (Fig. 5D). The mmm1 mutant hyphae were of normal morphology (Fig. 2E), but our data in Fig. 5A suggested that the mutant displayed reduced hyphal elongation. Measurements showed that the distribution of hyphal lengths was similar to that in controls, but there was a shift toward shorter lengths in macrophages and a very clear difference in the length of hyphal filaments in in vitro cultures (Fig. 5C and E; see also Fig. S7B and C in the supplemental material). The more pronounced defect in hyphal lengths in vitro than in macrophages is due to a longer time of hyphal growth in vitro (in macrophages, cell lengths were determined after 1 h of coincubation, as it is difficult to determine cell lengths at later time points when extensive hyphal growth occurs; in vitro, the length of the filaments was determined at 3 h post-induction of hyphal growth). At host temperature (37°C), growth retardation for the mmm1 mutant was observed after 4 h (Fig. 5F). Collectively, these results show that MMM1 is not required for hyphal morphogenesis per se. However, MMM1 is needed to establish wild-type levels of hyphal growth, elongation, and cell wall remodeling that are required to rapidly reach the signal threshold for inflammasome activation by C. albicans and enable fungal escape through pyroptosis.
FIG 5 .
ERMES impacts on the exposure of pathogen-associated molecular patterns during hyphal growth. (A) Hyphae were grown for 3 h in RPMI medium under repressive conditions, and surface-exposed 1,3-β-glucan was analyzed by flow cytometry following staining with the 1,3-β-glucan antibody. The experiment was repeated 3 times with equivalent results (see also panel B). Shown are representative dot plots from one biological replicate for each strain, plotting 1,3-β-glucan staining versus cell size (forward scatter [FSC]), with the percentage of cells shown for each quadrant. The upper left and right quadrants are glucan-positive cells, while the lower left and right quadrants are glucan-negative cells. (B) Percentage of cells that are negative for exposed 1,3-β-glucan. n is 9 from 3 independent experiments with 3 biological replicates each. The biological replicates analyzed together are shown by the same color. The line represents the mean. *, P < 0.05; **, P < 0.01 (one-way analysis of variance, followed by Tukey’s multiple-comparison test). (C) Hyphal length after 3 h in RPMI medium. Each measured cell is depicted by a line, with length shown on the y axis and the measurements ranked in order from smallest to largest on the x axis. Three biological replicates were performed. One repeat is shown here, and the other two are in Fig. S7B in the supplemental material. (D) Hyphal gene expression after 3 h in repressive RPMI medium. Shown is the ratio of gene expression normalized to SCR1. Error bars represent standard errors of the averages from 6 biological replicates assayed in 2 independent experiments. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (one-way analysis of variance followed by Tukey’s multiple-comparison test). (E) Hyphal length distribution in macrophages following 1 h of coincubation and washes. Data were analyzed in 2 independent experiments with equivalent results (n = 200 fungal cells per strain). One experiment is shown here, and the other is shown in Fig. S7C in the supplemental material. (F) Growth at 37°C in repressive medium. Shown are the averages and the standard errors of the means from 3 biological replicates assayed in the same experiment. OD600, optical density at 600 nm.
DISCUSSION
Roles of ERMES and mitochondrial morphology in fungal virulence.
Here, we report the first detailed characterization of the ERMES complex in a pathogenic fungal species, showing how this mitochondrial hub is important for the ability of C. albicans to kill macrophages for immune escape. Following conditional inactivation of ERMES, changes to mitochondrial morphology were seen first. Moreover, they could be uncoupled from fitness, respiratory, and lipid perturbations that were seen only after longer-term inactivation of the complex. Therefore, our data suggest that the primary role of C. albicans ERMES is in maintaining mitochondrial shape. Our characterization suggests that wild-type mitochondrial network shape is important for differentiating hyphae that can trigger pyroptotic macrophage death. While the mmm1 mutant was able to form hyphae both in liquid tissue culture medium and in macrophages, these mutant hyphae were unable to trigger NLRP3-dependent pyroptosis for a very long time of approximately 10 h. Hyphal filaments made by the mmm1 mutant were of normal morphology, and overall length distribution was similar to that of the control, but with a shift toward shorter lengths, indicating reduced hyphal growth and elongation. Compared to the control, fitness differences were not obvious for several hours in macrophages. In wild-type filaments, mitochondrial tubules extend throughout the hyphal cell, while in the ERMES mutants, the “giant” spherical mitochondrial morphology means that large parts of the hyphal filaments are devoid of mitochondria (see Fig. S7A in the supplemental material). Proper mitochondrial distribution in the filaments could serve to power hyphal elongation, as hyphal growth depends more prominently on mitochondrial metabolism than does yeast growth (45). The mitochondrial defects in the mmm1 mutant further had more important contributions to growth at 37°C than at 30°C with glucose as the carbon source (Fig. 1 and 5), and the mmm1 mutant was unable to grow on glycerol plates at 37°C, consistent with mitochondrial dysfunction (data not shown). Hyphal populations from the mmm1 mutant displayed overall surface changes, with smaller amounts of exposed 1,3-β-glucan and moderately reduced expression of HWP1 and PHR1 encoding a hypha-specific cell wall adhesin and a cell wall-remodeling enzyme, respectively, as well as ECE1, a hypha-specific gene that was recently shown to encode a toxin that can cause damage to epithelia (46). These changes in cell wall structure and hyphal gene expression are likely to lead to severely delayed inflammasome activation, delayed pyroptosis, and lack of fungal escape, as both glucan and cell wall protein mannosylation have been implicated in inflammasome activation and pyroptosis by C. albicans (13, 19, 42–44). Importantly, while severely delayed, inflammasome activation by the mmm1 mutant eventually occurred and it paradoxically surpassed what is seen in macrophages infected with the control strain. Exacerbated inflammasome activation was dependent on NLRP3, and all host killing by the mmm1 mutant at later time points in infection was by pyroptosis. To our knowledge, the mmm1 mutant phenotype of delayed inflammasome activation and pyroptosis followed by an exacerbated response has not been reported for any other C. albicans mutants that showed inflammasome activation defects. This illustrates the power of our live-cell microscopy assay to dynamically monitor inflammasome activation and pyroptosis in parallel. The implications of this result for the mechanism of inflammasome activation by C. albicans are further discussed below. In addition to being unable to rapidly activate macrophage pyroptosis, the mmm1 mutant was also unable to trigger the pyroptosis-independent phase 2 of macrophage killing, as almost all macrophage death in mutant infections was by pyroptosis, after which death plateaued. While the mmm1 mutant survived intracellularly in macrophages for extended periods of time and formed hyphae, growth was reduced later in infection. Therefore, the presence of even substantial intracellular hyphae is not sufficient to trigger the phase 2, caspase-1-independent form of cell death; this requires rapidly growing, persistent filaments. Collectively, our data show that ERMES coordinates short-term survival strategies of C. albicans by triggering rapid macrophage pyroptosis, with longer-term effects by ensuring optimal growth at host temperature and the ability to trigger multiple mechanisms of macrophage cell death. Our characterization further explains the role of MMM1 in systemic virulence in mutant library screens (24).
Insight into the NLRP3 inflammasome response to C. albicans.
Characterization of the ERMES mutant, coupled with real-time single-cell imaging of inflammasome activation, showed that (i) NLRP3 inflammasome activation by C. albicans is heterogeneous in the macrophage population; (ii) the inflammasome response is sensitively tailored to hyphal growth, and we propose that this is achieved via threshold activation; (iii) macrophages can be sensitized to NLRP3 inflammasome activation; and (iv) the new NLRP3 inhibitor MCC950 blocks inflammasome activation and pyroptosis following Candida infection. MCC950 is promising for treating diseases associated with pathogenic inflammation (25), and based on our data, we suggest that MCC950 could be explored for modulating Candida-induced inflammation that might be contributing to disease.
Although the majority of macrophages were infected at the multiplicity of infection (MOI) used (3 Candida cells per macrophage), only a proportion of up to 22% of them activated the NLRP3 inflammasome. We noticed that the number of C. albicans cells phagocytosed by a single macrophage varied in the population. This could be related to distinct inflammasome activation in individual macrophages, and future experiments will address this. Our observation is in line with a recent report showing that another NLRP3-inflammasome activator, silica crystals, caused caspase-1 activation in only a fraction of host cells (38). In contrast, treatment with the potassium ionophore nigericin triggers a more uniform NLRP3 inflammasome activation (37). The molecular mechanism leading to NLRP3 inflammasome activation by any stimuli remains ill defined, but it is thought that lysosome rupture contributes in the case of silica crystals and C. albicans hyphae. Activation by nigericin could be more direct due to rapid potassium efflux. Therefore, signals derived from lysosomes might require a threshold to activate the NLRP3 inflammasome that is reached distinctly in individual macrophages and could depend on signals derived from the pathogen and/or on factors present in only a subpopulation of macrophages. Activation of the NLRP3 inflammasome by C. albicans is a double-edged sword, as it triggers lytic pyroptosis that enables escape but also activates antifungal responses. Heterogeneous responses in infected macrophages could modulate these contrasting processes, with potential benefits to pathogen or host.
The dynamics of inflammasome activation by the mmm1 mutant showed that infection with a less virulent strain leads to a long delay in inflammasome activation despite the formation of hyphae. Importantly, the mmm1 mutant hyphae eventually triggered the inflammasome response (Fig. 4 and model in Fig. 6). The kinetics of the inflammasome response to the mmm1 mutant is consistent with a signal threshold response. Another possible explanation is that, at later time points postphagocytosis, the mmm1 mutant hyphae express alternative signals that activate the NLRP3 inflammasome by a different mechanism than what is observed with the control strain. We favor the signal threshold model, as the mmm1 mutant hyphae displayed quantitative changes in hyphal growth/filament length, in surface-exposed 1,3-β-glucan, and in the expression of hyphal genes. Consistent with our proposition for a signal threshold model are recent data of caspase-1 activation in response to Salmonella, or signals such as the NLRP3 activator silica, which showed “digital” or threshold signaling (38). Host responses to C. albicans in epithelial cells have been shown to depend on fungal cell numbers, also suggesting threshold signaling (47). Our data suggest that, to reach the threshold for NLRP3 inflammasome activation, hyphae need to elongate persistently, accompanied by cell wall remodeling and exposure of pathogen-associated molecular patterns (PAMPs), such as 1,3-β-glucan or mannosylated cell wall proteins (Fig. 6). Relevant to this is a recent report of a distinct structure of glucan derived from C. albicans hyphae compared to yeast and more potent stimulation by hyphal glucan of interleukin-1β (IL-1β) secretion as a proxy for caspase-1 activation (48). As discussed in a recent review, threshold mechanisms of signaling allow for sensitive responses tailored to the level of threat, thereby minimizing noisy inflammatory responses that could be detrimental to the host (49). In the case of C. albicans, this means that not only does the inflammasome discriminate yeast from hyphal morphology (16) but the response is more sensitively fine-tuned to hyphal growth levels, which could allow for tight regulation.
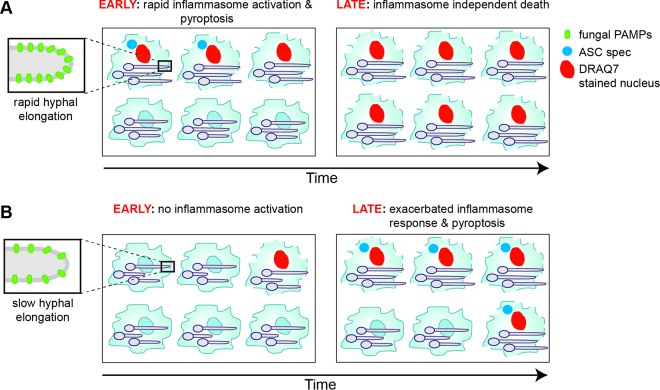
FIG 6 .
Model of inflammasome activation by C. albicans strains of distinct hyphal growth. In strains with robust filamentation and high virulence, inflammasome activation occurs rapidly upon phagocytosis by macrophages, leading to host cell pyroptosis. In infection by a strain with reduced virulence (mmm1 mutant), a longer time is needed to trigger the inflammasome, but eventual sensitization of host cells occurs, and inflammasome activation is seen in a larger number of macrophages in the population than that with the control strain with more robust filamentation. These differences could be explained by a signal threshold mode of inflammasome activation that depends on hyphal growth, cell wall remodeling, and exposure of fungal pathogen-associated molecular patterns (PAMPs).
The kinetics of inflammasome activation by the mmm1 mutant showed sensitization of the response and exacerbated activation after a prolonged delay (Fig. 4). This result should be considered in the light of differences in filamentation robustness, growth rates, and virulence potential within a collection of C. albicans clinical isolates (50). In this set, SC5314, the parent of most laboratory strains, including our own, is on the extreme end, being a highly filamentous strain (50). It could be that the dynamics of inflammasome activation that we see with the mmm1 mutant, with the prolonged delay, sensitization, and hyperactivation, is representative of some clinical strains of C. albicans as well as of the response at a low multiplicity of infection, as is likely to be the case in clinical situations.
Conclusions and outlook.
Our study lays the foundation for exploring the roles of ERMES, and mitochondrial dynamics processes more generally, in fungal immune evasion and virulence across diverse human fungal pathogens. Evidence suggests that disrupting mitochondrial morphology might be a global antifungal strategy. In addition to C. albicans (this study and reference 51), mitochondrial morphology has been implicated in virulence-related processes in Cryptococcus gattii (52) and Aspergillus fumigatus (53). Due to its absence in humans (23) and a key role in maintaining the tubular mitochondrial network structure, ERMES is particularly promising for antifungal drug discovery. Our data further develop a new understanding of how the dynamic inflammasome response to C. albicans is fine-tuned to reflect the pathogenic state of the fungus. The single-cell-resolution, parallel live-cell imaging of the inflammasome response and pyroptotic death that we established can be extended in the future to multiple fungal strains, as well as studying the impact of genetic and pharmacological manipulation of host and pathogen pathways on immune interactions.
MATERIALS AND METHODS
C. albicans strains and growth conditions.
The C. albicans strains used in this study are derivatives of BWP17 and are listed in Table S1 in the supplemental material. Primers for strain construction are listed in Table S2. Methods for strain construction and growth conditions are detailed in Text S1 in the supplemental material.
Supplemental materials and methods. Download Text S1, PDF file, 0.1 MB (155.6KB, pdf) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microscopy.
Detailed microscopy methods are given in Text S1 in the supplemental material. Images were taken with a 100× objective using an Olympus BX60 fluorescence microscope equipped with Spot Advanced Software (Spot Imaging, Sterling Heights, MI). Mitochondrial network morphology was imaged following staining with MitoTracker dyes. Hyphal formation was assessed in liquid RPMI medium under repressive conditions (with 2.5 mM methionine and 0.5 mM cysteine), after 3 h at 37°C. The lengths of filaments were measured using Fiji (http://www.fiji.sc/Fiji).
Macrophage interaction assays.
Experiments involving animals were approved by the Monash University Animal Ethics Committee, in accordance with the guidelines and policies in the Australian code for the care and use of animals for scientific purposes provided by the Australian National Health and Medical Research Council (approval numbers SOBS-2010-M-49 and MARP-2011-086). Murine bone marrow-derived macrophages (BMDMs) were obtained essentially as described in reference 13; please see Text S1 in the supplemental material for a detailed description. For these experiments, ERMES gene repression was initiated by patching C. albicans colonies overnight on repressive medium plates at 30°C, after which C. albicans cells were resuspended in phosphate-buffered saline (PBS) and counted and macrophages were infected at a multiplicity of infection (MOI) of 6:1 (Candida cells to macrophage). Four biological replicates were performed, and the data were analyzed in GraphPad Prism. The immortalized mCerulean-tagged ASC inflammasome reporter macrophages were a gift from Eicke Latz (36). Live-cell imaging was set up as described above, and the MOI was 3:1 (Candida cells to macrophage). The acquisition of time-lapse images, methods for processing, and counting of ASC speck formation are described in Text S1 in the supplemental material. All macrophage experiments (with BMDMs and ASC-Cerulean macrophages) were done in the presence of 2.5 mM methionine and 0.5 mM cysteine in the medium to allow for MMM1 gene repression. Heat-killed Candida cells were incubated at 80°C for 1 h prior to addition to macrophages. For experiments including drug treatments, 10 µM MCC950 or 10 µM MCC6642 (both made as 10 mM stocks in dimethyl sulfoxide [DMSO]) or 10 µM nigericin (Invitrogen) was included at the same time as addition of Candida.
Mitochondrial isolation and protein import assays.
Isolation of mitochondria and mitochondrial protein import assays were performed as previously described (54). In order to improve detection, ImageJ was used to apply a contrast alteration to the entire phosphorimage scan, in a manner that maintains the linear relationship of the gray tones in the image.
Quantitative RT-PCR analysis.
All primers used for quantitative RT-PCR are listed in Table S3 in the supplemental material, and some are further described in references 55 and 56. Growth conditions and experimental setup are described in the figure legends, and the methods are further detailed in the supplemental material. Data analysis was done using LinReg software (57).
Phospholipid analysis.
For lipid extractions, conditional ERMES mutants or homozygous deletion mutants were grown as described in Text S1 in the supplemental material and the figure legends. The procedure for lipid extraction is detailed in Text S1. Lipids were normalized according to protein levels and separated by thin-layer chromatography (TLC). Standards were from Avanti Polar Lipids. Lipids were quantified using the Toolbox module of ImageQuant 1D version 7.0, and background signals were subtracted using the local median method performed by the software.
Analysis of 1,3-β-glucan by flow cytometry.
1,3-β-Glucan on hyphal cells was quantified as previously described (13).
Statistical analysis.
Statistical analysis was performed using GraphPad Prism software, and the relevant tests used are indicated in the figure legends. Biological repeats were from cultures obtained from independent colonies of the indicated C. albicans strains. For the experiments with bone marrow-derived macrophages, different mice were used for the independent experiments.
(Table S1) C. albicans strains used in the study.
(Table S2) Strain construction primers.
(Table S3) Quantitative RT-PCR primers. Download Tables S1 to S3, PDF file, 0.1 MB (107.9KB, pdf) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Paul Harrison, David Powell, and Luka Traven for advice on statistics and data analysis and Trevor Lithgow for help with the cartoon in Fig. 1. We thank Eicke Latz for the ASC-Cerulean-labeled macrophages. We acknowledge the invaluable support of the Monash MicroImaging facility and, in particular, Keith Schulze for help with the analysis of ASC speck formation data.
Funding Statement
In addition to the grants listed above, this research was supported by a Pfizer Australian Research Fellowship (to T.B.), the Monash Researcher Accelerator Grant (MRA) to A.T., and an Australian Postgraduate Award (APA) to V.L.H.
REFERENCES
- 1.Rohmer L, Hocquet D, Miller SI. 2011. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol 19:341–348. doi: 10.1016/j.tim.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baruch M, Belotserkovsky I, Hertzog BB, Ravins M, Dov E, McIver KS, Le Breton YS, Zhou Y, Cheng CY, Hanski E. 2014. An extracellular bacterial pathogen modulates host metabolism to regulate its own sensing and proliferation. Cell 156:97–108. doi: 10.1016/j.cell.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown AJ, Brown GD, Netea MG, Gow NA. 2014. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol 22:614–622. doi: 10.1016/j.tim.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 5.Calderone R, Li D, Traven A. 2015. System-level impact of mitochondria on fungal virulence: to metabolism and beyond. FEMS Yeast Res 15:fov027. doi: 10.1093/femsyr/fov027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shingu-Vazquez M, Traven A. 2011. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryot Cell 10:1376–1383. doi: 10.1128/EC.05184-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiménez-López C, Lorenz MC. 2013. Fungal immune evasion in a model host-pathogen interaction: Candida albicans versus macrophages. PLoS Pathog 9:e1003741. doi: 10.1371/journal.ppat.1003741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenz MC, Bender JA, Fink GR. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell 3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949. doi: 10.1016/S0092-8674(00)80358-X. [DOI] [PubMed] [Google Scholar]
- 10.Jiménez-López C, Collette JR, Brothers KM, Shepardson KM, Cramer RA, Wheeler RT, Lorenz MC. 2013. Candida albicans induces arginine biosynthetic genes in response to host-derived reactive oxygen species. Eukaryot Cell 12:91–100. doi: 10.1128/EC.00290-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vylkova S, Lorenz MC. 2014. Modulation of phagosomal pH by Candida albicans promotes hyphal morphogenesis and requires Stp2p, a regulator of amino acid transport. PLoS Pathog 10:e1003995. doi: 10.1371/journal.ppat.1003995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh S, Navarathna DH, Roberts DD, Cooper JT, Atkin AL, Petro TM, Nickerson KW. 2009. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect Immun 77:1596–1605. doi: 10.1128/IAI.01452-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uwamahoro N, Verma-Gaur J, Shen HH, Qu Y, Lewis R, Lu J, Bambery K, Masters SL, Vince JE, Naderer T, Traven A. 2014. The pathogen Candida albicans hijacks pyroptosis for escape from macrophages. mBio 5:e00003-00014. doi: 10.1128/mBio.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wellington M, Koselny K, Sutterwala FS, Krysan DJ. 2014. Candida albicans triggers NLRP3-mediated pyroptosis in macrophages. Eukaryot Cell 13:329–340. doi: 10.1128/EC.00336-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joly S, Ma N, Sadler JJ, Soll DR, Cassel SL, Sutterwala FS. 2009. Cutting edge: Candida albicans hyphae formation triggers activation of the Nlrp3 inflammasome. J Immunol 183:3578–3581. doi: 10.4049/jimmunol.0901323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. 2012. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 10:112–122. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Netea MG, Joosten LA, van der Meer JW, Kullberg BJ, van de Veerdonk FL. 2015. Immune defence against Candida fungal infections. Nat Rev Immunol 15:630–642. doi: 10.1038/nri3897. [DOI] [PubMed] [Google Scholar]
- 19.O’Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, Cowen LE. 2015. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun 6:6741. doi: 10.1038/ncomms7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornmann B, Walter P. 2010. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J Cell Sci 123:1389–1393. doi: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murley A, Nunnari J. 2016. The emerging network of mitochondria-organelle contacts. Mol Cell 61:648–653. doi: 10.1016/j.molcel.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wideman JG, Gawryluk RM, Gray MW, Dacks JB. 2013. The ancient and widespread nature of the ER-mitochondria encounter structure. Mol Biol Evol 30:2044–2049. doi: 10.1093/molbev/mst120. [DOI] [PubMed] [Google Scholar]
- 24.Becker JM, Kauffman SJ, Hauser M, Huang L, Lin M, Sillaots S, Jiang B, Xu D, Roemer T. 2010. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc Natl Acad Sci U S A 107:22044–22049. doi: 10.1073/pnas.1009845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coll RC, Robertson AA, Chae JJ, Higgins SC, Muñoz-Planillo R, Inserra MC, Vetter I, Dungan LS, Monks BG, Stutz A, Croker DE, Butler MS, Haneklaus M, Sutton CE, Núñez G, Latz E, Kastner DL, Mills KH, Masters SL, Schroder K, Cooper MA, O’Neill LA. 2015. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med 21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu Y, Jelicic B, Pettolino F, Perry A, Lo TL, Hewitt VL, Bantun F, Beilharz TH, Peleg AY, Lithgow T, Djordjevic JT, Traven A. 2012. Mitochondrial sorting and assembly machinery subunit Sam37 in Candida albicans: insight into the roles of mitochondria in fitness, cell wall integrity, and virulence. Eukaryot Cell 11:532–544. doi: 10.1128/EC.05292-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sogo LF, Yaffe MP. 1994. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol 126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger KH, Sogo LF, Yaffe MP. 1997. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol 136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess SM, Delannoy M, Jensen RE. 1994. MMM1 encodes a mitochondrial outer membrane protein essential for establishing and maintaining the structure of yeast mitochondria. J Cell Biol 126:1375–1391. doi: 10.1083/jcb.126.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Youngman MJ, Hobbs AE, Burgess SM, Srinivasan M, Jensen RE. 2004. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J Cell Biol 164:677–688. doi: 10.1083/jcb.200308012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murley A, Lackner LL, Osman C, West M, Voeltz GK, Walter P, Nunnari J. 2013. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife 2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. 2001. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol 152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boldogh IR, Nowakowski DW, Yang HC, Chung H, Karmon S, Royes P, Pon LA. 2003. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol Biol Cell 14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osman C, Haag M, Potting C, Rodenfels J, Dip PV, Wieland FT, Brügger B, Westermann B, Langer T. 2009. The genetic interactome of prohibitions: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J Cell Biol 184:583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wellington M, Koselny K, Krysan DJ. 2012. Candida albicans morphogenesis is not required for macrophage interleukin 1beta production. mBio 4:e00433-12. doi: 10.1128/mBio.00433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, Engels G, Brenker C, Nordhoff M, Mirandola SR, Al-Amoudi A, Mangan MS, Zimmer S, Monks BG, Fricke M, Schmidt RE, Espevik T, Jones B, Jarnicki AG, Hansbro PM, Busto P, Marshak-Rothstein A, Hornemann S, Aguzzi A, Kastenmuller W, Latz E. 2014. The adaptor ASC has extracellular and “prionoid” activities that propagate inflammation. Nat Immunol 15:727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stutz A, Horvath GL, Monks BG, Latz E. 2013. ASC speck formation as a readout for inflammasome activation. Methods Mol Biol 1040:91–101. doi: 10.1007/978-1-62703-523-1_8. [DOI] [PubMed] [Google Scholar]
- 38.Liu T, Yamaguchi Y, Shirasaki Y, Shikada K, Yamagishi M, Hoshino K, Kaisho T, Takemoto K, Suzuki T, Kuranaga E, Ohara O, Miura M. 2014. Single-cell imaging of caspase-1 dynamics reveals an all-or-none inflammasome signaling response. Cell Rep 8:974–982. doi: 10.1016/j.celrep.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 39.Dagley MJ, Gentle IE, Beilharz TH, Pettolino FA, Djordjevic JT, Lo TL, Uwamahoro N, Rupasinghe T, Tull DL, McConville M, Beaurepaire C, Nantel A, Lithgow T, Mitchell AP, Traven A. 2011. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol Microbiol 79:968–989. doi: 10.1111/j.1365-2958.2010.07503.x. [DOI] [PubMed] [Google Scholar]
- 40.She X, Khamooshi K, Gao Y, Shen Y, Lv Y, Calderone R, Fonzi W, Liu W, Li D. 2015. Fungal-specific subunits of the Candida albicans mitochondrial complex I drive diverse cell functions including cell wall synthesis. Cell Microbiol 17:1350–1364. doi: 10.1111/cmi.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.She X, Zhang L, Chen H, Calderone R, Li D. 2013. Cell surface changes in the Candida albicans mitochondrial mutant goa1Δ are associated with reduced recognition by innate immune cells. Cell Microbiol 15:1572–1584. doi: 10.1111/cmi.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar H, Kumagai Y, Tsuchida T, Koenig PA, Satoh T, Guo Z, Jang MH, Saitoh T, Akira S, Kawai T. 2009. Involvement of the NLRP3 inflammasome in innate and humoral adaptive immune responses to fungal beta-glucan. J Immunol 183:8061–8067. doi: 10.4049/jimmunol.0902477. [DOI] [PubMed] [Google Scholar]
- 43.Kankkunen P, Teirilä L, Rintahaka J, Alenius H, Wolff H, Matikainen S. 2010. (1,3)-Beta-glucans activate both dectin-1 and NLRP3 inflammasome in human macrophages. J Immunol 184:6335–6342. doi: 10.4049/jimmunol.0903019. [DOI] [PubMed] [Google Scholar]
- 44.Cheng SC, van de Veerdonk FL, Lenardon M, Stoffels M, Plantinga T, Smeekens S, Rizzetto L, Mukaremera L, Preechasuth K, Cavalieri D, Kanneganti TD, van der Meer JW, Kullberg BJ, Joosten LA, Gow NA, Netea MG. 2011. The dectin-1/inflammasome pathway is responsible for the induction of protective T-helper 17 responses that discriminate between yeasts and hyphae of Candida albicans. J Leukoc Biol 90:357–366. doi: 10.1189/jlb.1210702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guedouari H, Gergondey R, Bourdais A, Vanparis O, Bulteau AL, Camadro JM, Auchère F. 2014. Changes in glutathione-dependent redox status and mitochondrial energetic strategies are part of the adaptive response during the filamentation process in Candida albicans. Biochim Biophys Acta 1842:1855–1869. doi: 10.1016/j.bbadis.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 46.Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, Murciano C, Blagojevic M, Thavaraj S, Förster TM, Hebecker B, Kasper L, Vizcay G, Iancu SI, Kichik N, Häder A, Kurzai O, Luo T, Kruger T, Kniemeyer O, Cota E, Bader O, Wheeler RT, Gutsmann T, Hube B, Naglik JR. 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532:64–68. doi: 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moyes DL, Runglall M, Murciano C, Shen C, Nayar D, Thavaraj S, Kohli A, Islam A, Mora-Montes H, Challacombe SJ, Naglik JR. 2010. A biphasic innate immune MAPK response discriminates between the yeast and hyphal forms of Candida albicans in epithelial cells. Cell Host Microbe 8:225–235. doi: 10.1016/j.chom.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowman DW, Greene RR, Bearden DW, Kruppa MD, Pottier M, Monteiro MA, Soldatov DV, Ensley HE, Cheng SC, Netea MG, Williams DL. 2014. Novel structural features in Candida albicans hyphal glucan provide a basis for differential innate immune recognition of hyphae versus yeast. J Biol Chem 289:3432–3443. doi: 10.1074/jbc.M113.529131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu H. 2013. Higher-order assemblies in a new paradigm of signal transduction. Cell 153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirakawa MP, Martinez DA, Sakthikumar S, Anderson MZ, Berlin A, Gujja S, Zeng Q, Zisson E, Wang JM, Greenberg JM, Berman J, Bennett RJ, Cuomo CA. 2015. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res 25:413–425. doi: 10.1101/gr.174623.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas E, Roman E, Claypool S, Manzoor N, Pla J, Panwar SL. 2013. Mitochondria influence CDR1 efflux pump activity, Hog1-mediated oxidative stress pathway, iron homeostasis, and ergosterol levels in Candida albicans. Antimicrob Agents Chemother 57:5580–5599. doi: 10.1128/AAC.00889-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voelz K, Johnston SA, Smith LM, Hall RA, Idnurm A, May RC. 2014. 'Division of labour' in response to host oxidative burst drives a fatal Cryptococcus gattii outbreak. Nat Commun 5:5194. doi: 10.1038/ncomms6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neubauer M, Zhu Z, Penka M, Helmschrott C, Wagener N, Wagener J. 2015. Mitochondrial dynamics in the pathogenic mold Aspergillus fumigatus: therapeutic and evolutionary implications. Mol Microbiol 98:930–945. doi: 10.1111/mmi.13167. [DOI] [PubMed] [Google Scholar]
- 54.Hewitt VL, Heinz E, Shingu-Vazquez M, Qu Y, Jelicic B, Lo TL, Beilharz TH, Dumsday G, Gabriel K, Traven A, Lithgow T. 2012. A model system for mitochondrial biogenesis reveals evolutionary rewiring of protein import and membrane assembly pathways. Proc Natl Acad Sci U S A 109:E3358–E3366. doi: 10.1073/pnas.1206345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verma-Gaur J, Qu Y, Harrison PF, Lo TL, Quenault T, Dagley MJ, Bellousoff M, Powell DR, Beilharz TH, Traven A. 2015. Integration of posttranscriptional gene networks into metabolic adaptation and biofilm maturation in Candida albicans. PLoS Genet 11:e1005590. doi: 10.1371/journal.pgen.1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naglik JR, Rodgers CA, Shirlaw PJ, Dobbie JL, Fernandes-Naglik LL, Greenspan D, Agabian N, Challacombe SJ. 2003. Differential expression of Candida albicans secreted aspartyl proteinase and phospholipase B genes in humans correlates with active oral and vaginal infections. J Infect Dis 188:469–479. doi: 10.1086/376536. [DOI] [PubMed] [Google Scholar]
- 57.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.AhYoung AP, Jiang J, Zhang J, Khoi Dang X, Loo JA, Zhou ZH, Egea PF. 2015. Conserved SMP domains of the ERMES complex bind phospholipids and mediate tether assembly. Proc Natl Acad Sci U S A 112:E3179–E3188. doi: 10.1073/pnas.1422363112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Fig. S1) Analysis of ERMES gene repression. (A) Cells of the indicated strains were grown under permissive conditions (synthetic medium without methionine and cysteine) followed by addition of 2.5 mM methionine and 0.5 mM cysteine at time zero, and gene repression was monitored by quantitative RT-PCR at the indicated time points. The levels of ERMES genes were normalized to the levels of the RNA polymerase III transcript SCR1 or the rRNA transcript RDN5 or RDN25. Shown are averages and standard errors from 3 biological replicates assayed in the same experiment. (B) MMM1 gene repression in macrophages. The mmm1 mutant and the complemented control strain were grown at 30°C under repressive conditions overnight. Macrophages were infected with C. albicans at the multiplicity of infection of 6 Candida cells to 1 macrophage, with methionine and cysteine included in the tissue culture medium. Gene repression was monitored by quantitative RT-PCR at the indicated time points following coincubation of C. albicans with macrophages and washing of the nonphagocytosed cells. To separate Candida from the macrophages, Trizol treatment was performed to remove lysed macrophages, and subsequently, Candida cell pellets were recovered and RNA was isolated by the hot phenol method. Transcript levels of MMM1 were normalized to the RNA polymerase III transcript SCR1 or the rRNA transcript RDN5. Shown are averages and the standard errors from 4 biological replicates obtained from 2 independent experiments.
(Fig. S2) Mitochondrial morphology defects of ERMES mutant strains. Shown are larger microscopy fields of the MitoTracker Red images displayed in Fig. 1B, without any processing (i.e., color or contrast correction) of the images. Bar, 10 µm.
(Fig. S3) Quantification of the mitochondrial morphology defects after conditional inactivation of ERMES. Relates to Fig. 1B. Percentages of cells with abnormal mitochondria in wild-type and mdm10 (top) and mmm1 (bottom) strains are shown. This was calculated at 0, 5, and 15 h postrepression from 3 independent biological replicates for each of the strains (3 independent colonies) assayed in the same experiment, counting at least 300 cells for each repeat. In parallel, cells were grown under permissive conditions as a control. Impaired mitochondria were quantified based on any mitochondria not resembling the wild-type tubular network as detected by MitoTracker Red staining, and this was subdivided into two distinct categories: a milder phenotype (“shrunken”), where the tubular network was reduced in size and not spanning the entire cell when analyzing multiple planes of focus under the microscope, and a severe phenotype (“collapsed”), consisting of round, enlarged, or severely fragmented mitochondria. The experiment was performed on multiple occasions with equivalent results observed.
(Fig. S4) Quantification of phospholipids after conditional inactivation of ERMES. Relates to Fig. 1E. Lipids were quantified using the Toolbox module of ImageQuant 1D version 7.0, and background signals were subtracted using the local median method performed by the software. Shown are averages and the standard deviations from three biological repeats. The amounts of cardiolipin (CL) and phosphatidylethanolamine (PE) are shown relative to the levels of phosphatidylcholine (PC) in the same sample as an internal control and in comparison to the wild type.
(Fig. S5) Long-term inactivation of ERMES results in loss of fitness, altered mitochondrial network morphology, and phospholipid imbalance. (A) (Left) Wild-type C. albicans and homozygous deletion strains lacking MDM12 (mdm12ΔΔ), MMM1 (mmm1ΔΔ), MDM10 (mdm10ΔΔ), or MDM34 (mdm34ΔΔ) were grown on rich medium containing glucose (YPD) or glycerol (YPG) as a carbon source, at 30°C or 37°C. Tenfold serial dilutions were plated, starting from an optical density at 600 nm (OD600) of 0.5. Images were taken after 3 days. (Right) Complementation of the fitness defects of the homozygous deletion strains for MDM12 (mdm12ΔΔ+12), MMM1 (mmm1ΔΔ+1), and MDM34 (mdm34ΔΔ+34). (B) Representative fluorescence images of the wild-type and ERMES homozygous mutant strains stained with MitoTracker Green dye. Bar, 5 µm. Images grouped together were all taken in parallel in the same experiment. No wild-type mitochondrial network morphology was seen in the homozygous ERMES mutants. Complementation of the mitochondrial network defects is shown for MDM12 (mdm12ΔΔ+12), MMM1 (mmm1ΔΔ+1), and MDM34 (mdm34ΔΔ+34). We noticed that in complemented strains with expression levels of the ERMES genes equivalent to approximately half of wild-type levels (as expected from single-copy gene reintegrants), restoration of mitochondrial morphology defects was partial; clones with expression levels equivalent to 2- to 3-fold over wild-type values displayed significantly better complementation of mitochondrial morphology. Elevated levels of ERMES genes may be necessary for restoring growth to severely sick ERMES mutants. (C) Total cellular lipids were extracted from the indicated strains and separated by thin-layer chromatography (TLC). Standards were used to identify cardiolipin (CL), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylcholine (PC). The right panel displays quantification averages, performed using ImageQuant, from 3 biological repeats. Error bars indicate standard deviations. ***, P < 0.0005; ****, P < 0.0001, compared to wild type using Student’s t test (unpaired, two-tailed). The amounts of CL and PE are shown relative to the levels of PC in the same sample as an internal control and in comparison to the wild type.
(Fig. S6) Effects of methionine and cysteine addition in the macrophage-interaction experiments. Relates to Fig. 2B. Macrophages were infected with the mmm1 mutant and the complemented control strain at an MOI of 6:1 (Candida cells to macrophage). After 1 h of coincubation and washing of nonphagocytosed cells, macrophage medium was included with (+Met/Cys) or without (-Met/Cys) 2.5 mM methionine and 0.5 mM cysteine. Live-cell imaging was then started 3 h later. Shown are averages and standard errors from 2 biological replicates (2 independent colonies of each of the two C. albicans strains) assayed in the same experiment.
(Fig. S7) Characterization of hyphal growth of the mmm1 mutant. (A) Relates to Fig. 2E. Mitochondrial network structure of hyphae was visualized by staining with MitoTracker Red dye. Representative images of several hyphae made by the mmm1 mutant and the complemented control strain are shown in the left panel. The right panel contains a higher magnification of a single hyphal cell with bright-field overlay. In both images, the scale bar represents 10 µm. (B) Relates to Fig. 5C. Hyphal length distribution in RPMI medium after 3 h of growth. Germ tubes or hyphal filaments were over 5 µm. Each cell measured is depicted by a line, with length shown on the y axis and the measurements ranked in order from smallest to largest on the x axis. Three biological replicates were performed. Two repeats are shown here, and the third repeat is shown in Fig. 5C. (C) Relates to Fig. 5E. Hyphal length distribution in macrophages following 1 h of coincubation and washes was measured in 2 independent experiments with equivalent results (n = 200 fungal cells per strain). One experiment is shown here, and the other one is shown in Fig. 5E. Download Figures S1 to S7, PDF file, 8.5 MB (8.7MB, pdf) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Murine bone marrow-derived macrophages (BMDMs) infected with the control C. albicans strain (mmm1+MMM1). To create the video files, using Fiji software, the TIFF hyperstack was exported as an AVI file with JPEG compression at 3.0 frames per s. For all data analysis, the images were kept in TIFF format. Download Movie S1, AVI file, 14.2 MB (14.5MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Murine bone marrow-derived macrophages (BMDMs) infected with the mmm1 mutant strain. The file was created as described for Movie S1. Download Movie S2, AVI file, 11 MB (11.2MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ASC-Cerulean macrophages infected with the control C. albicans strain (mmm1+MMM1), under control conditions (treatment with the inactive compound MCC6642). The file was created as described for Movie S1. Download Movie S3, AVI file, 2.7 MB (2.7MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ASC-Cerulean macrophages infected with the control C. albicans strain (mmm1+MMM1) and treated with the NLRP3 inhibitor MCC950. The file was created as described for Movie S1. Download Movie S4, AVI file, 2.6 MB (2.7MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ASC-Cerulean macrophages infected with the mmm1 mutant strain under control conditions (treatment with the inactive compound MCC6642). The file was created as described for Movie S1. Download Movie S5, AVI file, 1.7 MB (1.7MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ASC-Cerulean macrophages infected with the mmm1 mutant and treated with the NLRP3 inhibitor MCC950. The file was created as described for Movie S1. Download Movie S6, AVI file, 1.9 MB (1.9MB, avi) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods. Download Text S1, PDF file, 0.1 MB (155.6KB, pdf) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(Table S1) C. albicans strains used in the study.
(Table S2) Strain construction primers.
(Table S3) Quantitative RT-PCR primers. Download Tables S1 to S3, PDF file, 0.1 MB (107.9KB, pdf) .
Copyright © 2016 Tucey et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.