Abstract
Dot1/DOT1L (disruptor of telomeric silencing-1) is an evolutionarily conserved histone methyltransferase that methylates lysine 79 located within the globular domain of histone H3. Dot1 was initially identified by a genetic screen as a disruptor of telomeric silencing in Saccharomyces cerevisiae; further, it is the only known non-SET domain containing histone methyltransferase. Methylation of H3K79 is involved in the regulation of telomeric silencing, cellular development, cell-cycle checkpoint, DNA repair, and regulation of transcription. hDot1L-mediated H3K79 methylation appears to have a crucial role in transformation as well as disease progression in leukemias involving several oncogenic fusion proteins. This review summarizes the multiple functions of Dot1/hDOT1L in a range of cellular processes.
Keywords: Chromatin, Histone methylation, DNA repair
1. Introduction
Genetic information within the eukaryotic cell nucleus is organized in a highly conserved DNA–protein complex, the chromatin, which supports and controls the vital functions of the genome. The basic unit of chromatin is the nucleosome, which contains 146 bp of DNA wrapped around a histone octamer containing two copies each of H2A, H2B, H3, and H4 [1]. The basic organization of the eukaryotic genome into chromatin impedes the protein factor accessibility required for gene transcription, DNA replication, recombination, and repair. Therefore, the cell has developed various mechanisms by which the chromatin structure can be regulated to increase access to DNA. These include adenosine triphosphate (ATP)-dependent chromatin remodeling, incorporation of histone variants into nucleosomes, and covalent histone modifications. [2–4] Histone modifications can alter the charge of specific residues, affecting the histone–histone and histone–DNA interactions, and can act as signals to create landing platforms for a diverse array of transcription factors, chromatin remodelers, and DNA-interacting proteins, which drive distinct downstream functions [1,4]. Histone modifications are highly regulated within the cell, and any misregulation is often associated with cancer and other physiological disorders [5]. Among the various known histone modifications, histone methylation has attracted much attention due to its diverse functions, which include transcriptional regulation, heterochromatin formation, X-chromosome inactivation, DNA repair, and cellular differentiation [1]. Histone methylation is carried out by histone methyltransferases (HMTs), which covalently modify specific lysine and arginine residues by transferring methyl groups from S-adenosyl-l-methionine. HMTs fall into two groups based on an evolutionarily conserved catalytic SET domain named after SU(var), Enhancer of Zeste, and Trithorax, the first three proteins identified to harbor this domain in Drosophila [6]. Disruptor of telomeric silencing-1 (hDot1L), which methylates lysine 79 located within the globular domain of histone H3, is unusual as it does not possess a SET domain. Dot1 is involved in a number of key processes ranging from gene expression to DNA-damage response and cell-cycle progression (Fig. 1). This review summarizes the diverse biological functions of Dot1 in various cellular processes [1].
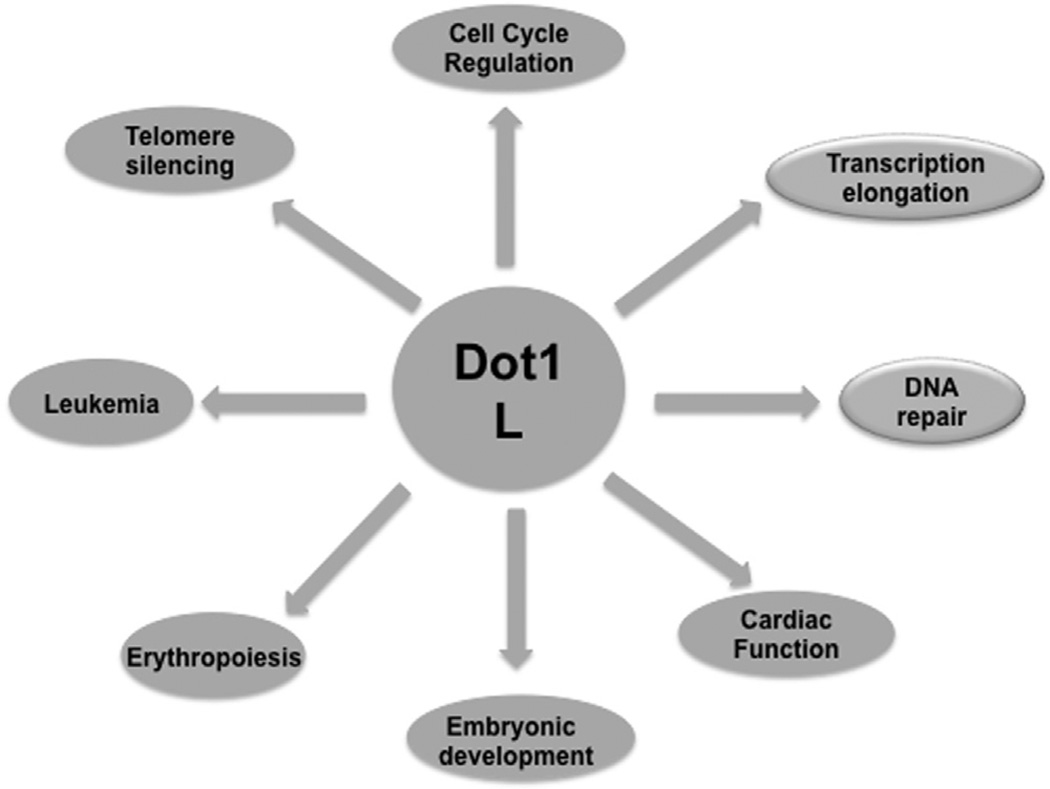
Fig. 1.
Role of hDot1L in various cellular processes: hDot1L is involved in diverse cellular processes such as transcription elongation, DNA repair, cell-cycle regulation, and telomere silencing.
1.1. Dot1 as a Methyltransferase
Dot1 (also known as KMT4) is an evolutionarily conserved enzyme that catalyzes the methylation of histone H3 at lysine 79. It was initially discovered as a gene that disrupts telomeric silencing when overexpressed [7]. Dot1 is the sole enzyme responsible for all forms of H3K79 methylation (monomethylation, dimethylation, and trimethylation) in Saccharomyces cerevisiae, Drosophila melanogaster, and humans, based on the fact that knockout of its gene in these organisms results in complete loss of H3K79 methylation [8]. The protein is conserved in mammals, called Dot1-like protein (DOT1L), and displays enzymatic properties similar to that of its yeast homologue. Dot1 is unique in being the only non-SET domain containing methyltransferase identified to date [8–10]. Two features of this enzyme distinguish it from other known HMTs. First, Dot1 requires a chromatin substrate, and is not active on free histones. Second, the lysine residue modification occurs within the globular region of histone H3, away from the usual histone modification sites that are located in the N- and C-terminal domains. H3K79 is located in the globular domain of histone H3, but it is exposed on the nucleosome surface where it can be methylated by DOT1L. However, this methylation occurs only in a nucleosomal context that is suggestive of a cross talk between H3K79 methylation and other histone modifications [10,11]. Studies regarding activity and regulation of Dot1 in many organisms have revealed roles in different biological processes such as transcription, cell-cycle regulation, and DNA damage response [12].
1.2. Cross talk between histone modifications in regulation of H3K79 methylation
For more than a decade now, chromosomal modifications have been noted to act in concert with each other in a context-dependent manner, which can lead to activation or repression of chromatin-mediated processes. This concept forms the basis of the cross talk between different modifications [13]. The inability of Dot1 to methylate free histone H3 suggests that trans-histone cross talk may regulate Dot1 activity. In yeast, histone H2B is monoubiquitinated at K123 by Rad6 ubiquitin-conjugating E2 enzyme and Bre1 ubiquitin E3 ligase. Both H2B-K123 and H3K79 are located close to each other on the nucleosome surface; further, the interplay between methylation of H3K79 and ubiquitination of H2B-K123 has been suggested because Rad6 (ubiquitin-conjugating enzyme) deletion was found to prevent H3K4 and H3K79 methylation [14–16]. Moreover, mutagenesis of H2B-K123 results in remarkable loss of these two methylations. These results indicate that H2B-K123 ubiquitination is a prerequisite for H3K4 and H3K79 methylation. This effect has been observed to be unidirectional, as Dot1 deletion has no effect on H2B-K123 ubiquitination [16]. Another histone cross-talk pathway that regulates H3K79 methylation levels in S. cerevisiae is the interaction of Dot1 with a short basic patch region of the histone H4 N-terminal tail. Sir3 and Dot1 compete for the same site on histone H4, and the acetylation of H4K16 blocks Sir3 binding and stimulates Dot1 binding and subsequent H3K79 methylation [17].
1.3. H3K79 methylation in transcriptional regulation
Gene expression in higher eukaryotes is intimately linked to DNA accessibility. Normally, DNA remains packaged into a condensed chromatin state inaccessible to the transcriptional machinery. Therefore, for transcription to occur, this state has to be changed by opening up of the chromatin structure. Histone-modifying enzymes are one of the major mediators of this process. Histone-modifying enzymes are targeted to their histone substrates by DNA-bound modulators (activators or repressors). The choice of the particular modulator depends on the given enzyme and the subsequent modification that it performs [3,4]. H3K79 methylation levels show strong correlation with transcriptional activity. In S. cerevisiae, Dot1 and H3K79 trimethylation has been detected in the transcribed regions of active genes [18–20]. In humans, ChIP–chip (chromatin immunoprecipitation coupled with gene expression microarrays) studies demonstrated that H3K79me2/me3 methylation is strongly correlated with gene activity. In Drosophila, ChIP–chip analysis also revealed a correlation between H3K79me2 and active gene transcription [21]. Mutations in the Drosophila Dot1 ortholog grappa lead to polycomb and trithorax group phenotypes, suggesting that H3K79 methylation may influence developmentally regulated gene expression in multicellular eukaryotes [22]. Furthermore, Steger et al. found that H3K79 methylation is linked to gene transcription in mice [23]. In human cells, ChIP–seq (chromatin immunoprecipitation coupled with deep sequencing) experiments indicated that H3K79 methylation is enriched at the transcribed genes [24]. Furthermore, H3K79 methylation is enriched on the histone variant H3.3, which is found at the transcriptionally active loci in Drosophila and mammals [25,26]. Dot1 has also been identified in RNA polymerase II- associated transcription elongation complexes. More recently, DOT1L was found in a complex in Drosophila with members of the Wnt pathway such as AF10, AF17, and AF9. Although P-TEFb was absent from this complex, Dot1L was still required for Wingless target gene expression [27–29]. Furthermore, the Moazed Lab showed that the SIR complex, which mediates the inhibition of transcription from reconstituted chromatin template, was released when H4K16 was acetylated or H3K79 was methylated due to the disruption of the SIR complex association with chromatin [30]. H3K79 methylation could regulate transcription through various mechanisms; H3K79 methylation may be important for the recruitment of transcription machinery or it might act as a landing platform for various transcription activators or it may inhibit the binding of transcription repressors. A proteomic analysis of cell lysate proteins bound to an immobilized unmethylated/methylated H3K79 chromatin template followed my mass spectroscopy analysis would provide important information on the kind of proteins that bind or do not bind to H3K79-methylated chromatin. Methylation of H3K79 has also been implicated in the reactivation of tumor suppressor genes upon DNA demethylation [31]. Furthermore, in human leukemias carrying chromosomal translocations at the mixed lineage leukemia (MLL) genes, mistargeting of hDOT1L leads to transcriptional upregulation [32–35].
1.4. Dot1-mediated H3K79 methylation in telomeric silencing
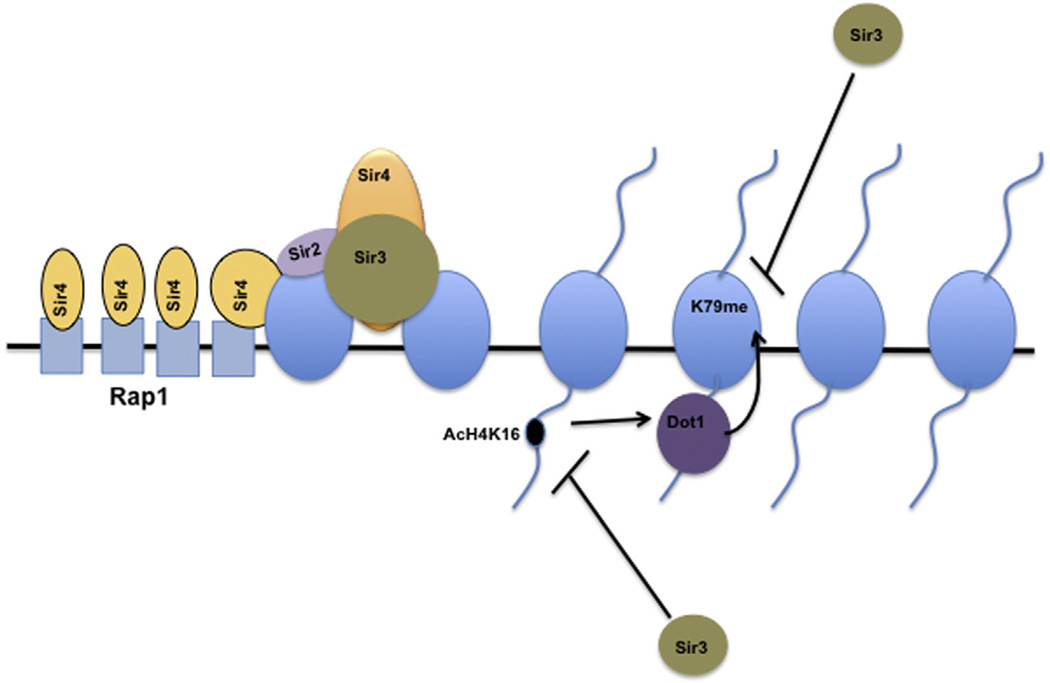
Dot1 was initially discovered in a genetic screen as a gene that disrupts telomeric silencing when overexpressed. In S. cerevisiae, telomeric heterochromatin is initiated at the chromosome ends by Rap1, a DNA-binding protein, binding to a series of 16–20 binding sites. [36,37] Once Rap1 binds, it recruits Sir4 [38], a member of the SIR complex. Sir4 brings the other member of the Sir complex, the Sir2, a nicotinamide adenine dinucleotide (NAD)-dependent H4K16 histone deactylase. Deacetylation of H4K16 facilitates Sir3 binding, which recruits an additional Sir complex and leads to the spreading of heterochromatin [39–41]. A bi-product of the deacetylation reaction O-acetyl-ADP-ribose (AAR) further reinforces the heterochromatin formation and stability [42]. The boundary between heterochromatin and euchromatin is determined by two opposing enzymes Sir2 (H4K16 deacetylase) and Sas2 (H4K16 acetyltransferase) [38,43]. Deletion of Sas2 leads to the spreading of the Sir complex from the heterochromatic regions into the euchromatin, in turn silencing potentially critical genes [38,43]. Other histone modifications act in concert with acetylation to prevent the spreading of the Sir complex into the euchromatin, significant among these is methylation of H3K79. Mutation of H3K79 to alanine or deletion of Dot1 impairs telomeric silencing by disrupting Sir protein localization. ChIP analysis has shown that deletion or overexpression of DOT1 leads to mislocalization of the SIR complex (Sir2, 3,4p) [44,45]. Cells overexpressing Dot1 show detectable H3K79 methylation spreading into silent regions of the chromatin. The presence of H3K79 methylation is considered to displace Sir proteins from silent chromatin, causing silencing defects. Recent studies have shown that a small basic patch encompassing residues R17H18R19 of histone H4 is crucial for Dot1 binding and subsequent H3K79 methylation [17,46]. Dot1-mediated H3K79 methylation is inhibited by the presence of Sir3, indicating that both Dot1 and Sir3 compete for the same site on the H4 tail. The binding of Dot1 to the H4 tail is regulated by the acetylation of H4K16. In vitro studies have shown that Sir3 can bind a H3 peptide that encompasses H3K79, but the presence of mono-, di-, and trimethylation on H3K79 prevents Sir3 binding, suggesting that Dot1 and Sir3 specifically compete for regions on histone H3 and H4 (Fig. 2) [17]. While Dot1p is insensitive to the H3K79 methylation status, Sir3p binds only to unmethylated H3; this competition between Sir3p and Dot1p determines whether chromatin is silenced or expressed. It will be interesting to investigate the role of hDot1L in heterochromatin boundary decisions in higher eukaryotes. Using a URA3 telomere reporter assay, Rossmann et al. [88] showed that the silencing defect in Dot1 deletion cells is due to an imbalance in ribonucleotide reductase (RNR) and a promoter of URA3 at telomere VII-L. In another study, Takahashi et al. [89] showed that the requirement for Dot1 in heterochromatin formation is telomere specific. Nevertheless, substantial data reveal that mutation of H3K79 to alanine causes Sir protein mislocalization and heterochromatin spreading towards the euchromatin. Furthermore, H3K79-methylated peptides or chromatin can block the binding of Sir proteins, indicating a strong interplay between Sir proteins and H3K79 methylation [9,17,18,46,90].
Fig. 2.
H3K79 methylation in telomere silencing: Spreading of SIR complex from heterochromatin to euchromatin is blocked by combined action of H4K16 acetylation and H3K79 methylation. Sas2 acetylates H4K16, which prevents Sir3 from binding to the H4 tail and stimulates Dot1 binding to the H4 N-terminal tail. Methylation of H3K79 by Dot1 further blocks Sir3 binding to the H3K79 region.
1.5. H3K79 methylation in cell-cycle regulation
The cell cycle is a dynamic process that takes place in a highly regulated manner to ensure nuclear information is transmitted to the daughter cells. Apart from the classical players of cell-cycle regulation (cyclins, CDKs, etc.), histone modifications such as H3K79 methylation have recently been proven to be crucial contributors to cell-cycle regulation [47–49]. H3K79 methylation levels undergo dynamic changes during the cell cycle. In S. cerevisiae, the levels of H3K79me3 remain unchanged throughout the cell cycle, whereas the H3K79me2 levels increase gradually from the G1 to S phase and then further in the G2/M phase [50,51]. Similarly in the protozoan parasite Trypanosoma brucei, the levels of H3K76me3 remain unchanged during the cell cycle (in T. brucei, the histone residue H3K76 acts as the target of DOT1), whereas H3K76me1 is detectable in mitosis and H3K76me2 in G2/M phases (T. brucei contains two enzyme homologues of DOT1, that is, DOT1A and DOT1B, the former catalyzing mono- and dimethylation of H3K76 and the latter catalyzing the trimethylation of H3K76) [52,53]. In mice, before the blastocyst stage of preimplantation, H3K79me3 levels are not detectable in either the interphase or the M phase. However, H3K79me2 is detectable in both the interphase and the M phase, with levels being lower in the former and higher in the latter [53]. In contrast to these findings, in human cancer cells such as A549 and HeLa, the level of H3K79me2 decreases from the G1 to G2/M phase, with the levels being highest at the G1/S transition and lowest at G2/M [11,54]. Studies conducted on A549 cells have shown that the levels of both H3K79me2 and H3K79me3 are regulated in a similar manner [54]. Another study in K562 leukemia cells found unchanged levels of H3K79me2 during different phases of the cell cycle [55]. Irrespective of the broad spectrum of H3K79 methylation levels at the G1 phase across different species, H3K79 methylation is suggested to play a role in G1/S transition, as knockout/knockdown mutants of Dot1/hDOT1L in these species cause cell-cycle arrest at the G1 phase [50,54,56–58]. The level of H3K79 methylation, and hence its role in S-phase regulation, also differs among different species. In S. cerevisiae, the CAF-1 (chromatin assembly factor 1) complex is involved in the S-phase deposition of H3K79me2, which might be involved in nucleosome formation. In T. brucei, DOT1A deficiency leads to hypoploidy and replication inhibition, whereas its overexpression leads to hyperploidy [52,53]. Deficiency of hDOT1L in human cancer cells such as HCT116 results in aneuploidy (hypo- and hyperploidy), apoptosis, and nondividing cell deposition in the S phase [53]. In terms of the role of H3K79 methylation in mitosis and meiosis, in S. cerevisiae, DOT1 might be involved in the exit from mitosis [59] and is crucial to the pachytene checkpoint during meiosis, as Dot1 or H3K79 mutants exhibit defective meiotic checkpoint control [60,61]. In mouse fibroblasts, H3K79me3 has been suggested to play a role in mitosis, due to its localization at chromosome boundaries. H3K79 methylation also plays a role in mouse spermatogenesis and oocyte development [62]. However, in the case of mammals, the precise role of H3K79 methylation during mitosis is still unclear. The diverse levels of H3K79 methylation throughout the cell cycle in species as divergent as S. cerevisiae and humans indicates that the mechanism of cell-cycle regulation in relation to H3K79 methylation might differ in these species. Thus, further studies are needed into the mechanisms of cell-cycle regulation in these organisms. In humans, the role of H3K79 methylation in cell-cycle regulation must be extended to normal cell lines and also to in vivo systems, before any roles are assigned to H3K79 methylation in cell-cycle regulation.
1.6. H3K79 methylation in DNA repair
DNA damage and double-strand breaks (DSBs) can lead to serious cellular consequences, if not repaired rapidly and accurately, including tumor suppressor gene inactivation, oncogene activation, promotion of carcinogenesis, as well as mutation and loss of genetic material [1,64]. A number of studies performed in yeast and mammalian cells have established a link between H3K79 methylation and DNA damage repair. The first evidence was binding of the tandem tudor domain (a chromodomain) of human 53BP1 (ortholog of Rad9 in yeast) protein to methylated H3K79 and its recruitment to DNA DSBs [63]. In line with these findings, the mutation of histone H3 lysine 79 or depletion of hDOT1L inhibited the recruitment of 53BP1 to DSBs as H3K79 methylation is impaired in both cases. Mutation of the tudor domain of 53BP1 also has the same effect. The interaction between 53BP1 and methylated H3K79 is evolutionarily conserved, as the same interaction has been detected between Rad9 and methylated H3K79 in S. cerevisiae [65]. Interaction between methylated H3K79 and Rad9 inhibits the production of single-stranded DNA (ssDNA) at DSBs and at uncapped telomeres, indicating a role in controlling DNA resection at damaged sites as part of the repair process by homologous recombination [66]. As H3K79 methylation levels remain unchanged upon DNA damage, it has been proposed that the DSB passively relaxes higher-order chromatin structures around the break site, allowing 53BP1 to access methylated H3K79 [63]. Docked 53BP1 at the damage site could, in turn, recruit additional proteins to activate the checkpoint response [63]. In S. cerevisiae, recruitment of Rad9 to methylated H3K79 and initiation of a 53BP1-like cascade have also been proposed [65,67]. Dot1-mediated H3K79 methylation and Rad9 recruitment also play an integral role in the repair of DNA damage in the late G2 phase by regulating resection to limit the amount of ssDNA produced during non-homologous end joining (NHEJ) [66]. Similar to H3K79 methylation, a novel regulatory mechanism was recently uncovered wherein ATM-mediated MOF phosphorylation was found to mediate the DNA DSB repair pathway choice according to the cell-cycle position [68]. In fact, a wide range of histone modifications have been shown to play a role in the repair of DNA DSBs, and they have been reviewed in detail elsewhere [69]. In addition to DSBs, Dot1-mediated H3K79 methylation also plays a critical role in repairing ultraviolet (UV)-induced DNA damage [67], as loss of H3K79 methylation results in UV hypersensitivity [66,71]. Dot1 also helps maintain genome integrity after DNA damage by negative regulation of translesion synthesis (TLS) [72– 74]. One important histone modification that may also coordinate with H3K79 methylation in regulating heterochromatin in higher eukaryotes is acetylation of H4K16, catalyzed by MOF histone acetyltransferase. Mice deficient in Purkinje cell MOF display impaired motor coordination and a reduced life span, symptoms that are alleviated by treatment with deacetylase inhibitors [70]. As H4K16 acetylation is associated with chromatin relaxation, it is worth investigating the presence of any cross talk between H3K79 methylation and H4K16 acetylation in regulating neuronal function.
1.7. H3K79 methylation and leukemia
MLL (also known as ALL or HRX) is a gene present in mammals that plays a crucial role in regulating segment identity specifying homeobox (Hox) gene cluster [75,76]. This gene is the homolog of Drosophila Trithorax (Trx), and both are members of an evolutionarily conserved family of proteins termed as the trithorax group (trxG). Trithorax-group proteins are positive regulators of developmental gene expression. Their action is opposed by the repressive activity of Polycomb-group (PcG) genes. MLL contains a number of functional motifs, but the two most important motifs are the N-terminal motif, which facilitates DNA-binding function, and the C-terminal SET domain, which methylates H3K4 [77,78]. Methylation of H3K4 is believed to play an important role in the positive regulatory function of MLL, as this modification is associated with transcriptional activation [77–80]. MLL also has an important role in hematopoietic development [81,82]. Chromosomal translocations are one of the major causes of acute leukemias, and MLL has emerged as the most commonly translocated gene found in leukemia. Due to chromosomal translocation, the N-terminal of MLL becomes fused in frame to one of a large number (>50) of fusion partners [83]. In most cases (approximately 80%), this fusion occurs between the N-terminal MLL and either AF4, AF6, AF9, AF10, or ENL [83]. Due to the DNA-binding properties of the N- terminal MLL motif, these fusion proteins are always nuclear and bind to target genes controlled by MLL irrespective of the normal location of the C-terminal partner [84]. Two of the fusion chimeras of MLL, that is, MLL-AF10 and MLL-ENL, have been shown to recruit hDOT1L. This phenomenon has been demonstrated to promote DOT1L-mediated methylation of H3K79 on the HOXA9 promoter, which contributes to increased expression of Hoxa9 in AML (acute myeloid leukemia) [30,85,86]. This aberrant recruitment of hDOT1L to the promoters of MLL target genes seems to be a common feature of many oncogenic MLL fusion chimeras, as a number of MLL fusion chimeras such as MLL-AF10, MLL-AF4, and MLL-AF9 have been found to be associated with hDot1L in nuclear complexes [27,30,85–87]. Although hDOT1L is not genetically altered in the disease per se, its mislocated enzymatic activity is a direct consequence of the chromosomal translocation that affects MLL patients. Thus, hDOT1L has been proposed to be a catalytic driver of leukemogenesis in this disease. The enzymatic activity of hDOT1L is critical to the pathogenesis of MLL, because methyltransferase-deficient hDot1L is capable of suppressing growth of MLL_fusion-transformed cells. hDot1L-mediated H3-K79 methylation of MLL target genes in leukemogenesis implies that the HMTase activity of hDot1L can be targeted for therapeutic intervention. Thus, identifying small molecules capable of disrupting the interaction between hDOT1L and AF10 or inhibiting the HMTase activity of hDOT1L may lead to novel treatments for leukemia (Fig. 3). Recently, EPZ004777 was identified as a small-molecule inhibitor of hDOT1L. This compound inhibits cellular H3K79 methylation, blocks leukemogenic gene expression, and selectively kills cultured cells bearing MLL translocations. However, EPZ004777 was found to have poor pharmacological properties. Thus, further studies are needed to discover potent inhibitors of hDOT1L for both biological and therapeutic applications.
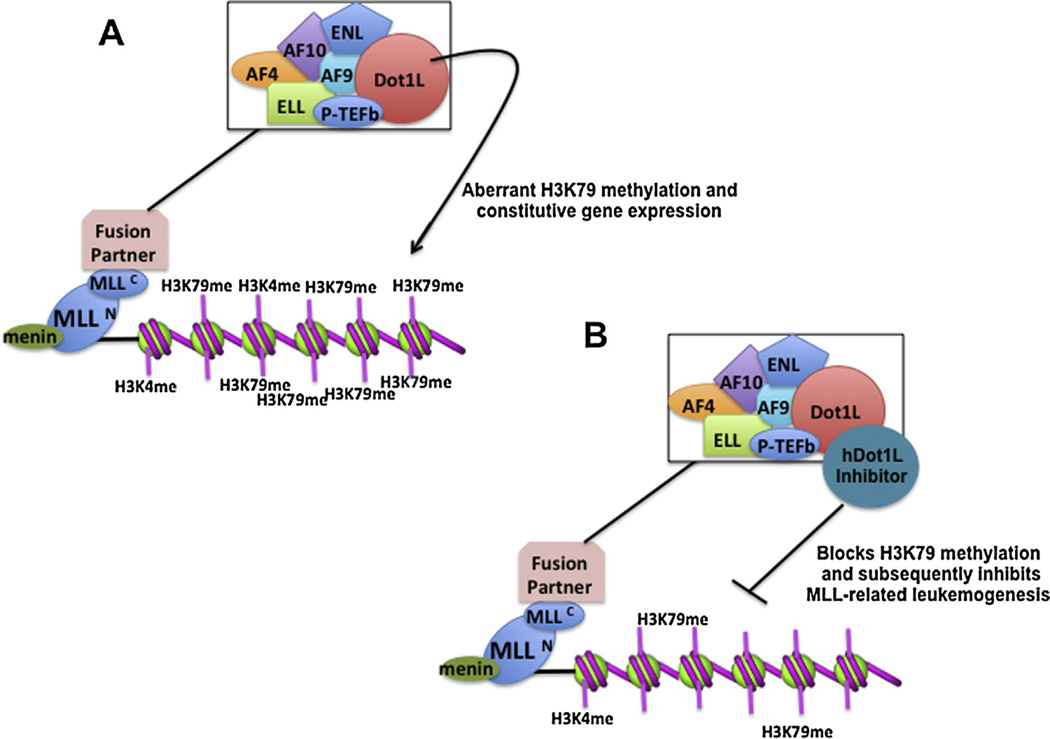
Fig. 3.
Model for leukemogenesis mediated by aberrant DOT1L H3K79 methylation. (A) MLL gene undergoes chromosomal translocations and fusion with a number of fusion proteins such as AF9, AF10, and hDot1L. This leads to mistargeting of hDot1L to MLL-regulated genes such as Hox genes. Aberrant H3K79 methylation and the subsequent activation of the target genes result in leukemic transformation. (B) Inhibition of hDot1L HMTase activity with small molecules would prevent aberrant H3K79 methylation and inhibition of MLL-related leukemogenesis.
2. Conclusions and future perspectives
hDot1L is emerging as a crucial non-SET domain-containing HMT involved in a diverse range of biological processes including telomeric silencing, cellular development, cell-cycle checkpoint, DNA repair, and transcription regulation. hDot1L has also been linked to MLL-related leukemogenesis, with a crucial role in transformation as well as disease progression in leukemias involving several oncogenic fusion proteins. Fusion of hDOT1L and MLL protein domains leads to mistargeting of hDOT1L to the transcriptional targets of MLL fusion proteins, such as the Hoxa cluster. The resultant aberrant hypermethylation of H3K79 at the gene site leads to constitutive transcriptional activation followed by leukemic transformation. Therefore, inhibition of hDOT1L enzymatic activity may be a promising strategy for the treatment of MLL fusion-related leukemias. Further insight into hDot1L can be obtained through biochemical characterization of hDot1L complexes and their components within the cell, discovery of non-histone substrates of hDOT1L, identification of H3K79 methylation-regulated transcriptional activators or repressors, and studies into the role of Dot1L–Sir3 competition in higher eukaryotes. Furthermore, H3K79 is demethylated in both mice and flies during early development, suggesting the presence of a demethylase. The discovery of H3K79 demethylase would be a major step towards a more complete understanding of the role of hDot1L in both normal and tumor cell biological processes.
Acknowledgments
The authors thank the members of Bhat Laboratory and Dr. Ajazul Hamid Wani for their fruitful discussions. The work is supported by the Ramanujan Fellowship grant from DST, India, and a UGC start-up grant from the University Grants Commission, New Delhi, India, and RO1 CA129537 and RO1 CA154320, USA.
Footnotes
The authors declare that there are no conflicts of interest.
References
- 1.Altaf M, Saksouk Nehme, Cote Jacques. Histone modifications in response to DNA damage. Mutat. Res. 2007;618:81–90. doi: 10.1016/j.mrfmmm.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Altaf M, Auger A, Covic M, Cote J. Connection between histone H2Avariants and Chromatin remodeling complexes. Biochem. Cell. Biol. 2009;87(1):35–50. doi: 10.1139/O08-140. [DOI] [PubMed] [Google Scholar]
- 3.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007 Feb;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Egger Gerda, Liang Gangning, Aparicio Aparicio, Jones Peter A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2016;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 6.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005 Nov;6(11):838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 7.Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.VanLeeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 9.Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir Protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacoste N, Utley RT, Hunter JM, Poirier GG, Côte J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 2002 Aug;277(34):30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- 11.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 14.Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt CD, Allis DF, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;41:8–498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 15.Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 16.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 17.Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Côté J. interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell. 2007;28(6):1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pokholok DK, Harbison CT, Levine S, Cole M, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 20.Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Schubeler D. The histone modification pattern of active genes revealed through genome—wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, Schedl P. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics. 2005;169:173–184. doi: 10.1534/genetics.104.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Blobel GA, Vakoc CR. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 2008;28(8):2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and posttranslational modifications associated with mammalian histone H3 variants. J. Biol. Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 26.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3:3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 28.Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signalling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A, Zhou R, Nesvizhskii A, Chinnaiyan A, Hess JL, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson A, Li G, Sikorski TW, Buratowski S, Woodcock D. Reconstitution of heterochromatin-dependent transcriptional gene silencing. Mol. Cell. 2009 Sep;35(25):769–781. doi: 10.1016/j.molcel.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacinto FV, Ballestar E, Esteller M. Impaired recruitment of the histone methyltransferase DOT1L contributes to the incomplete reactivation of tumor suppressor genes upon DNA demethylation. Oncogene. 2009;28:4212–4224. doi: 10.1038/onc.2009.267. [DOI] [PubMed] [Google Scholar]
- 32.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 33.Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guenther M, Lawton L, Rozovskaia T, Frampton G, Levine S, Volkert T, Croce C, Nakamura T, Canaani E, Young R. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y. Leukaemic transformation by CALM-AF10 involves up regulation of Hoxa5 by hDOT1L. Nat. Cell. Biol. 2006;8:1017–1024. doi: 10.1038/ncb1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buchman AR, Lue NF, Kornberg RD. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol. Cell. Biol. 1988;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8(19):2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 38.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32(3):378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 39.Imai S, Johnson FB, Marciniak RA, McVey M, Park PU, Guarente L. Sir2: an NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb. Symp. Quant. Biol. 2000;65:297–302. doi: 10.1101/sqb.2000.65.297. [DOI] [PubMed] [Google Scholar]
- 40.Johnson LM, Adams G. Fisher, Grunstein M. Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J. 1992;11(6):2201–2209. doi: 10.1002/j.1460-2075.1992.tb05279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson JS, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369(6477):245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 42.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121(4):515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 43.Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32(3):370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- 44.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 45.Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 46.Fingerman IM, Li HC, Briggs SD. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 2007;21(16):2018–2029. doi: 10.1101/gad.1560607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu D, Bai J, Duan Q, Costa M, Dai W. Covalent modifications of histones during mitosis and meiosis. Cell Cycle. 2009;8:3688–3694. doi: 10.4161/cc.8.22.9908. [DOI] [PubMed] [Google Scholar]
- 49.Sawicka A, Seiser C. Histone H3 phosphorylation- a versatile chromatin modification for different occasions. Biochimie. 2012;94:2193–2201. doi: 10.1016/j.biochi.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell cycle regulation of H3K79 dimethylation. Mol. Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou H, Madden BJ, Muddiman DC, Zhang Z. Chromatin assembly factor 1 interacts with histone H3 methylated at lysine 79 in the processes of epigenetic silencing and DNA repair. Biochemistry. 2006;45:2852–2861. doi: 10.1021/bi0521083. [DOI] [PubMed] [Google Scholar]
- 52.Janzen CJ, Hake SB, Lowell JE, Cross GA. Selective di- or trimethylation of histone H3 lysine 76 by two DOT1 homologs is important for cell cycle regulation in Trypanosoma brucei. Mol. Cell. 2006;23:497–507. doi: 10.1016/j.molcel.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 53.Gassen A, Brechtefeld D, Schandry N, Artega-Salas JM, Israel L, Imhof A, Janzen CJ. DOT1A-dependent H3K76 methylation is required for replication regulation in Trypanosomabrucei. Nucleic Acids Res. 2012;40:10302–10311. doi: 10.1093/nar/gks801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim W, Kim R, Park G, Park JW, Kim JE. Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J. Biol. Chem. 2012;287:5588–5599. doi: 10.1074/jbc.M111.328138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu H, Maunakea AK, Martin MM, Huang L, Zhang Y, Ryan M, Kim R, Lin CM, Zhao K, Aladjem MI. Methylation of histone on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle. PLoS Genet. 2013;9(6):e1003542. doi: 10.1371/journal.pgen.1003542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117:6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feng Y, Yang Y, Ortega MM, Copeland JN, Zhang M, Jacob JB, Fields TA, Vivian JL, Fields PE. Early mammalian erythropoiesis requires the Dot1L methyltransferase. Blood. 2010;116:4483–4491. doi: 10.1182/blood-2010-03-276501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hwang WW, Madhani HD. Nonredundant requirement for multiple histone modifications for the early anaphase release of the mitotic exit regulator Cdc14 from nucleolar chromatin. PLoS Genet. 2009;5(8):e1000588. doi: 10.1371/journal.pgen.1000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.San-Segundo PA, Roeder GS. Role for the silencing protein Dot1 in meiotic checkpoint control. Mol. Biol. Cell. 2000;11:3601–3615. doi: 10.1091/mbc.11.10.3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ontoso D, Acosta I, van Leeuwen F, Freire R, San-Segundo PA. Dot1-dependent histone H3K79 methylation promotes activation of the Mek1 meiotic checkpoint effector kinase by regulating the Hop1 adaptor. PLoS Genet. 2013;9(1):e1003262. doi: 10.1371/journal.pgen.1003262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ooga M, Inoue A, Kageyama S, Akiyama T, Nagata M, Aoki F. Changes in H3K79 methylation during preimplantation development in mice. Biol. Reprod. 2008;78:413–424. doi: 10.1095/biolreprod.107.063453. [DOI] [PubMed] [Google Scholar]
- 63.Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 64.Nnakwe Chinyone C, Altaf Mohammad, Cote Jacques, Kron Stephen J. Dissection of Rad9 BRCT domain function in the mitotic checkpoint response to telomere uncapping. DNA Repair. 2009;8(12):1452–1461. doi: 10.1016/j.dnarep.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6–Bre1 and H3 methylation by Dot1. J. Biol. Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 68.Gupta Arun, Hunt Clayton R, Hegde Muralidhar L, Chakraborty Sharmistha, Udayakumar Durga, Horikoshi Nobuo, Singh Mayank, Ramnarain Deepti B, Hittelman Walter N, Namjoshi Sarita, Asaithamby Aroumougame, Hazra Tapas K, Ludwig Thomas, Pandita Raj K, Tyler Jessica K, Pandita Tej K. MOF phosphorylation by ATM regulates 53BP1-mediated double-strand break repair pathway choice. Cell Rep. 2014 Jul;8(10):177–189. doi: 10.1016/j.celrep.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hunt Clayton R, Ramnarain Deepti, Horikoshi Nobuo, Iyengar Puneeth, Pandita Raj K, Shay Jerry W, Pandita Tej K. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat. Res. 2013;179:383–392. doi: 10.1667/RR3308.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar R, Hunt CR, Gupta A, Nannepaga S, Pandita RK, Shay JW, Bachoo R, Ludwig T, Burns DK, Pandita TK. Purkinje cell specific males absent on the first (mMof) gene deletion results in an ataxia telangiectasia like neurological phenotype and backward walking in mice. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3636–3641. doi: 10.1073/pnas.1016524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bostelman LJ, Keller AM, Albrecht AM, Arat A, Thompson JS. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2007;6:383–395. doi: 10.1016/j.dnarep.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 72.Conde F, San-Segundo PA. Role of Dot1 in the response to alkylating DNA damage in Saccharomyces cerevisiae: regulation of DNA damage tolerance by the error-prone polymerases Polzeta/Rev1. Genetics. 2008;179:1197–1210. doi: 10.1534/genetics.108.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conde F, Ontoso D, Acosta I, Gallego-Sanchez A, Bueno A, San- Segundo PA. Regulation of tolerance to DNA alkylating damage by Dot1 and Rad53 in Saccharomyces cerevisiae. DNA Repair (Amst) 2010;9:1038–1049. doi: 10.1016/j.dnarep.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 74.Levesque N, Leung GP, Fok AK, Schmidt TI, Kobor MS. Loss of H3 K79 trimethylation leads to suppression of Rtt107-dependent DNA damage sensitivity through the translesion synthesis pathway. J. Biol. Chem. 2010;285:35113–35122. doi: 10.1074/jbc.M110.116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 76.Daser A, Rabbitts TH. Extending the repertoire of the mixed lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18:965–974. doi: 10.1101/gad.1195504. [DOI] [PubMed] [Google Scholar]
- 77.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 78.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 79.Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 80.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 81.Hess JL, Yu BD, Li B, Hanson R, Korsmeyer SJ. Defects in yolk sac hematopoiesis in Mll-null embryos. Blood. 1997;90:1799–1806. [PubMed] [Google Scholar]
- 82.Jude CD, Climer L, Xu D, Artinger E, Fisher JK, Ernst P. Unique and independent roles for MLL in adult hematopoietic stem cells and progenitors. Cell Stem Cell. 2007;1:324–337. doi: 10.1016/j.stem.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 84.Dimartino JF, Cleary ML. Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br. J. Haematol. 1999;106:614–626. doi: 10.1046/j.1365-2141.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 85.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, Su L, Xu G, Zhang Y. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 86.Zeisig DT, Bittner CB, Zeisig BB, Garcia-Cuellar MP, Hess JL, Slany RK. The eleven-nineteen-leukemia protein ENL connects nuclear MLL fusion partners with chromatin. Oncogene. 2005;24:5525–5532. doi: 10.1038/sj.onc.1208699. [DOI] [PubMed] [Google Scholar]
- 87.Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. J. Biol. Chem. 2006;281:1–18059. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rossmann MP, Luo W, Tsaponina O, Chabes A, Stillman B. A common telomeric gene silencing assay is affected by nucleotide metabolism. Mol. Cell. 2011 Apr;42(8):127–136. doi: 10.1016/j.molcel.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takahashi Yoh-Hei, Schulze Julia M, Jackson Jessica, Hentrich Thomas, Seidel Chris, Jaspersen Sue L, Kobor Michael S, Shilatifard A. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol. Cell. 2011;42:118–126. doi: 10.1016/j.molcel.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]