SUMMARY
Integrating stress responses across tissues is essential for survival of multicellular organisms. The metazoan nervous system can sense protein misfolding stress arising in different subcellular compartments and initiate cytoprotective transcriptional responses in the periphery. Several subcellular compartments possess a homotypic signal whereby the respective compartment relies on a single signaling mechanism to convey information within the affected cell to the same stress responsive pathway in peripheral tissues. In contrast, we find that the heat shock transcription factor, HSF-1, specifies its mode of transcellular protection via two distinct signaling pathways. Upon thermal stress, neural HSF-1 primes peripheral tissues through the thermosensory neural circuit to mount a heat shock response. Independent of this thermosensory circuit, neural HSF-1 activates the FOXO transcription factor, DAF-16, in the periphery and prolongs lifespan. Thus a single transcription factor can coordinate different stress response pathways to specify its mode of protection against changing environmental conditions.
INTRODUCTION
The long-term health of an organism is linked to its ability to recognize and respond to stresses that arise in its environment. Across evolutionary spectra, organisms have developed complex and highly specialized defense pathways that become transcriptionally activated during times of stress. Often, diverse stress stimuli initiate distinct transcriptional signatures that activate protective and adaptive genes to defend against environmental challenges and restore homeostasis. In metazoa, the upregulation of stress response pathways also requires the coordinated activation of stress response machinery across multiple tissues. Consequently, a hierarchical mode of tissue regulation has evolved in which particular cell types can act as master regulators, initiating protective pathways in peripheral tissues (Wolff et al., 2014).
Organisms are frequently subjected to acute challenges that require a rapid response to potentially lethal conditions. These transient stresses elicit a dramatic cellular reaction with a rewiring of gene expression and a temporary suspension of normal cellular function. Conversely, organisms regularly encounter chronic insults that are not lethal even after long exposures. These prolonged stresses initiate distinct and more sustained responses that allow for the continuance of most normal cellular functions. The cumulative effect of chronic stress over the lifetime of the organism is known to play a causative role in the onset and severity of many age-related diseases (Failla, 1958; Harman, 1956; Orgel, 1963). However, it is unclear how acute stress responses can alleviate the negative effects of the aging process (Lithgow et al., 1995).
Thermal adaptation in metazoans requires the perception, communication, and initiation of a response across the entire organism. The transcription factor HSF-1 is the key regulator of the cellular and organismal response to heat stress and is conserved in all eukaryotes. It is well-established that HSF-1 mediates a protective transcriptional and translational response to acute heat stress through the induced expression of molecular chaperones (Morimoto, 2008). More recently it has been shown in the nematode worm that hsf-1 overexpression in all tissues retards the aging process (Hsu et al., 2003). Thus, mediating stress response pathways by HSF-1 protects against both acute thermal stress and the chronic stresses associated with aging.
In nematodes, thermal adaptation is regulated by a subset of sensory neurons which activate the heat shock response in peripheral tissues (Prahlad et al., 2008). However, the role that HSF-1 plays within the nervous system is not well defined. It is also not clear whether the same sensory neural circuit that controls the heat shock response also controls processes of aging which are tightly associated with heat stress resistance.
RESULTS
Neural overexpression of hsf-1 promotes heat stress resistance and longevity
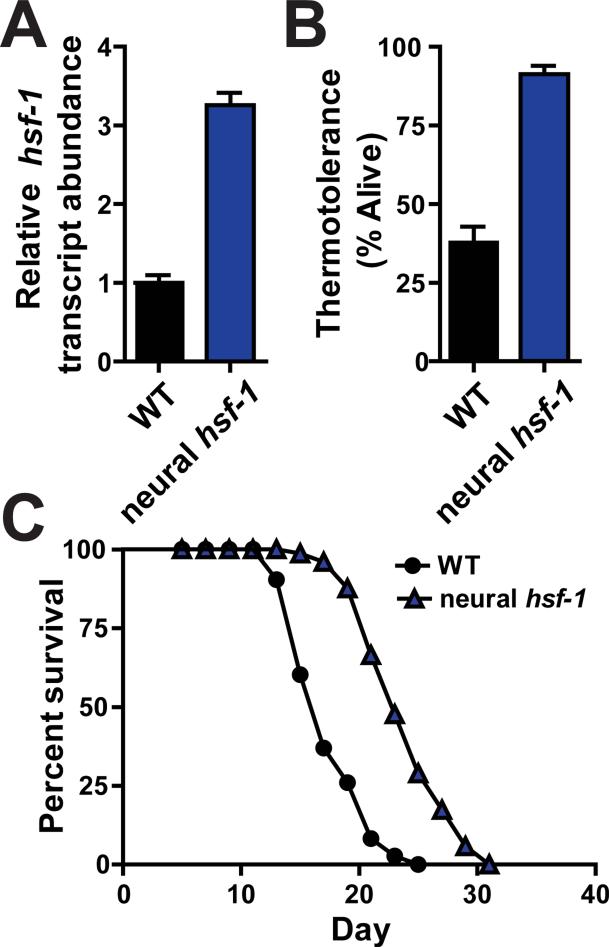
To explore these questions, we examined whether increasing hsf-1 levels exclusively in the worm nervous system was sufficient to mediate protection against acute thermal stress and the aging process. Transgenic worms were generated which ectopically overexpressed hsf-1 throughout the nervous system (Figures 1A, S1A, S1B and S1C). This level of hsf-1 overexpression in neurons was sufficient to extend worm lifespan and protect against heat shock treatments (Figures 1B, 1C, S1D and S1E; Table 1).
Figure 1. Neural overexpression of hsf-1 protects C. elegans against heat stress and aging.
(A) Transcript abundance of hsf-1 determined by quantitative RT-PCR analysis of wild type (N2) and rab-3p::hsf-1 transgenic animals (AGD1289). (B) Thermotolerance of wild type and rab-3p::hsf-1 transgenic worms shifted from permissive (20°) to heat shock (34°) temperatures for 14 hours. (C) Lifespan survival curves of wild type and rab-3p::hsf-1 transgenic strains at permissive temperatures (20°). Lifespan statistics are found in Table 1.
Table 1.
Statistical analysis of C. elegans lifespan data.
| Corresponding Figure | Strain, Treatment | Mean Lifespan ± s.e.m. (Median Lifespan in days) | 75th% (days) | Observed/total | % Lifespan increase | p-value log-rank (Mendel-Cox) |
|---|---|---|---|---|---|---|
| 1C | N2 | 17.3+0.4 (17) | 22 | 88/100 | ||
| 1C | AGD1289 (neural hsf-1) | 23.1+0.4 (24) | 27 | 68/101 | 33.5 | <0.0001 |
| 2F | AGD1008 non-transgenic | 18.0+0.4 (18) | 20 | 69/86 | ||
| 2F | AGD1008 (neural hsf-1) | 23.2+0.5 (24) | 26 | 58/88 | 28.8 | <0.0001 |
| 2F | ttx-3(ks5) | 17.5+0.4 (18) | 20 | 88/96 | 2.8 | 0.7231 |
| 2F | AGD1449 (neural hsf-1 ttx-3(ks5)) | 22.0+0.6 (22) | 24 | 59/85 | 22.2 | <0.0001 |
| 3D | N2 | 17.5+0.4 (17) | 21 | 73/100 | ||
| 3D | AGD1289 (neural hsf-1) | 23.9+0.4 (23) | 27 | 70/103 | 36.6 | <0.0001 |
| 3D | daf-16(mu86) | 15.8+0.4 (15) | 19 | 61/100 | −9.7 | 0.0022 |
| 3D | AGD1217 (neural hsf-1 daf-16(mu86)) | 16.6+0.3 (17) | 19 | 72/100 | −5.1 | 0.0578 |
| 4A | AGD1276 (intestinal daf-16) | 16.7+0.5 (17) | 19 | 53/104 | ||
| 4A | AGD1277 (intestinal daf-16, neural hsf-1) | 22.7+0.5 (22) | 26 | 51/110 | 35.9 | <0.0001 |
| 4B | AGD1272 (neural daf-16) | 18.7+0.4 (19) | 22 | 68/108 | ||
| 4B | AGD1273 (neural daf-16, neural hsf-1) | 18.2+0.6 (17) | 22 | 57/105 | −2.6 | 0.6775 |
| 4C | AGD1278 (body-wall muscle daf-16) | 16.8+0.5 (17) | 19 | 71/107 | ||
| 4C | AGD1279 (body-wall muscle daf-16, neural hsf-1) | 18.5+0.6 (19) | 22 | 52/94 | 10.1 | 0.0429 |
| 5A | N2 | 18.5+0.2 (18) | 20 | 90/114 | ||
| 5A | AGD1289 (neural hsf-1) | 27.6+0.4 (28) | 31 | 113/123 | 51.9 | <0.0001 |
| 5A | hsf-1(sy441) | 16.8+0.2 (18) | 18 | 100/114 | ||
| 5A | AGD1471 (hsf-1(sy441), neural hsf-1) | 18.4+0.2 (18) | 20 | 91/120 | 9.5 | <0.0001 |
| S1D | N2 | 17.1+0.4 (16) | 20 | 76/102 | ||
| S1D | AGD1053 (neural hsf-1) | 21.2+0.6 (20) | 25 | 72/100 | 24 | <0.0001 |
| S1D | AGD1054 (neural hsf-1) | 21.4+0.4 (22) | 25 | 76/100 | 25.2 | <0.0001 |
| S1E | N2 | 19.1+0.5 (20) | 22 | 79/102 | ||
| S1E | AGD1441 (neural hsf-1) | 22.2+0.7 (22) | 26 | 62/78 | 16.2 | 0.0002 |
| S3A | N2, vector RNAi | 18.5+0.3 (19) | 21 | 83/109 | ||
| S3A | AGD1289 (neural hsf-1), vector RNAi | 23.0+0.3 (22) | 24 | 94/113 | 24.3 | <0.0001 |
| S3A | N2, hsp16.1 RNAi | 19.6+0.3 (19) | 22 | 92/105 | ||
| S3A | AGD1289 (neural hsf-1), hsp-16.1 RNAi | 23.5+0.3 (24) | 26 | 95/106 | 19.9 | <0.0001 |
| S3B | N2, vector RNAi | 19.3+0.4 (19) | 22 | 68/103 | ||
| S3B | AGD1289 (neural hsf-1), vector RNAi | 23.7+0.4 (25) | 25 | 74/111 | 22.8 | <0.0001 |
| S3B | N2, pha-4 RNAi | 17.6+0.5 (19) | 19 | 43/104 | ||
| S3B | AGD1289 (neural hsf-1), pha-4 RNAi | 22.9+0.4 (22) | 25 | 77/103 | 30.1 | <0.0001 |
| S3C | N2, vector RNAi | 17.0+0.4 (16) | 20 | 90/108 | ||
| S3C,D,E | AGD1289 (neural hsf-1), vector RNAi | 22.4+0.3 (22) | 24 | 57/105 | 31.8 | <0.0001 |
| S3C | N2, xbp-1 RNAi | 18.6+0.4 (18) | 22 | 85/98 | ||
| S3C | AGD1289 (neural hsf-1), xbp-1 RNAi | 23.2+0.4 (24) | 26 | 63/98 | 24.7 | <0.0001 |
| S3D | N2, skn-1 RNAi | 15.3+0.2 (16) | 16 | 89/98 | ||
| S3D | AGD1289 (neural hsf-1), skn-1 RNAi | 22.2+0.3 (22) | 26 | 85/108 | 45.1 | <0.0001 |
| S3E | N2, ubl-5 RNAi | 17.1+0.4 (18) | 20 | 91/100 | ||
| S3E | AGD1289 (neural hsf-1), ubl-5 RNAi | 21.0+0.4 (20) | 24 | 67/91 | 22.8 | <0.0001 |
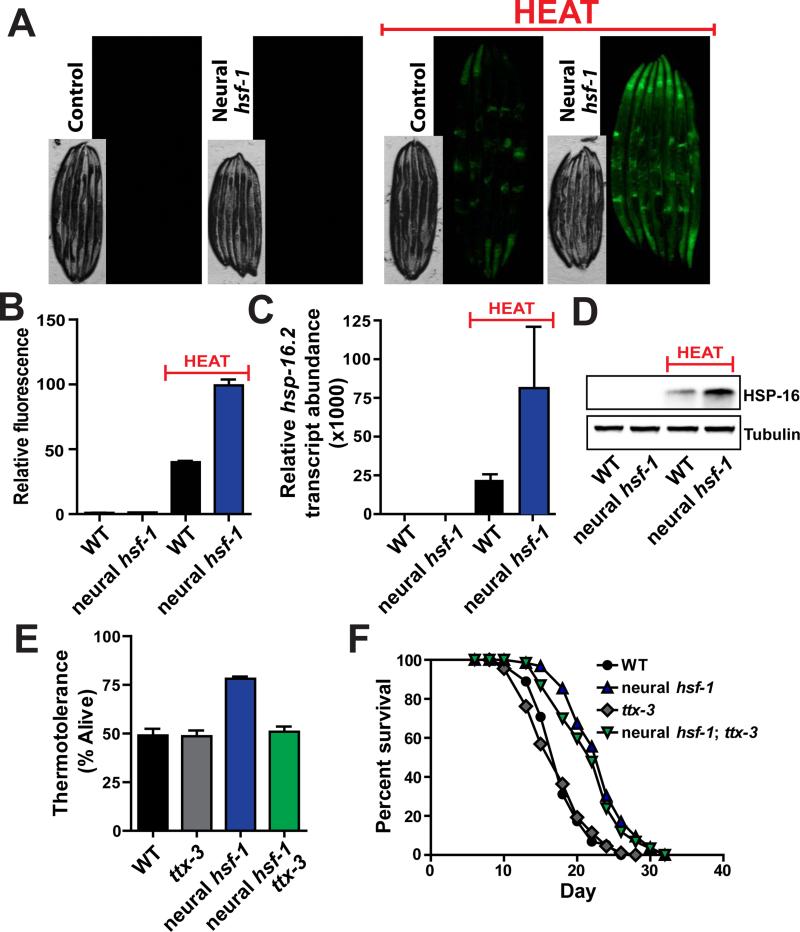
To gain insight into the neural signaling pathways responsible for thermotolerance and longevity assurance, heat shock responsive transcriptional targets were examined under conditions of either acute heat stress or aging. We first utilized a transgenic reporter worm that expresses GFP under the promoter of the HSF-1 target gene, hsp-16.2, a member of the alpha β crystalline family of small chaperones (Link et al., 1999). Upon application of heat stress, animals robustly induced the expression of GFP driven by the hsp-16.2 promoter compared to non-heat treated worms (Figure 2A). Elevating neural hsf-1 expression enhances the heat shock response throughout all worm tissues. These results strongly suggest that HSF-1 activity in neurons communicates to peripheral tissues and enables a more robust transcriptional response to heat stress. Through large-particle flow cytometry, fluorescent quantification of individual worms reveals that heat shock induces expression of hsp-16.2p::GFP over 2-fold higher in worms that overexpressed hsf-1 in the nervous system (Figure 2B and S2A). Similar increases of endogenous hsp-16.2, hsp70a and hsp-70b transcript levels were observed upon heat shock in worms overexpressing neural hsf-1 (Figures 2C, S2B and S2C). Similar expression patterns of HSP-16 protein levels were also observed (Figure 2D). Genome-wide transcriptomics further confirmed that hsf-1 overexpression in the nervous system bolsters transcription of numerous heat responsive genes compared to wild type animals (Figure S2D). The ability of worms overexpressing neural hsf-1 to mount a more robust heat shock response in peripheral tissues is consistent with the hypothesis that thermal protection is conferred by the heat shock response.
Figure 2. Neural hsf-1 overexpression enhances heat inducible chaperone expression in all tissues and requires an intact thermosensory circuit for heat protection but not lifespan extension.
(A) Fluorescent microscopy of C. elegans expressing GFP from the hsp-16.2 promoter in control (CL2070) and rab-3p::hsf-1 transgenic animals (AGD1448), at permissive (20°C) and heat shock (34°C) temperatures. (B) Large-particle flow cytometry was used to quantify GFP fluorescence from strains used in (A). (C) Transcript levels of endogenous hsp-16.2 determined by quantitative RT-PCR from day 1 adult wild type and rab-3p::hsf-1 transgenic animals (AGD1289). (D) Western blot analysis of endogenous HSP-16 from strains used in (C). (E) Thermotolerance of WT, ttx-3(ks5), rab-3p::hsf-1 (AGD1289), and rab-3p::hsf-1;ttx-3(ks5) (AGD1449) animals was assessed at 34°C. (F) Lifespan survival was performed at 20°C on strains used in (E). Lifespan statistics are found in Table 1.
Loss of the thermosensory neural circuit disrupts thermotolerance but not longevity
We examined the role that heat-inducible chaperones might play in lifespan determination when hsf-1 is overexpressed in the nervous system. We predicted that induction of heat responsive genes would be correlated with neural hsf-1 overexpression. Perplexingly, only minor differences in heat responsive elements were observed at permissive temperatures between control animals and those overexpressing neural hsf-1 (Figures 2B, 2C, 2D, S2B, S2C and S2D). Thus elevating hsf-1 levels in the nervous system confers longevity without induction of the canonical heat shock responsive chaperones. Furthermore, RNAi knockdown of hsp-16 expression, previously linked to age determination (Walker and Lithgow, 2003), did not alter lifespan extension by neural hsf-1 (Figure S3A; Table 1). These data suggest that hsf-1 overexpression elicits a divergent pathway in response to the aging process. Similar responses have been reported for HSF-1 in the context of reduced insulin/IGF-1 signaling (Hsu et al., 2003).
Because lifespan extension and thermotolerance appear divergent, we speculated that mutations might exist which abolish thermal protection but retain lifespan extension. To test this hypothesis, the thermosensory neural circuit was disrupted and the ability of neural hsf-1 overexpression to enhance heat tolerance and extend lifespan was assessed. Genetic ablation of the AIY interneuron through a ttx-3 mutation severs the thermosensory neural circuit and dampens the activation of heat shock responders in peripheral tissues (Prahlad et al., 2008). As expected, in the ttx-3 mutant background, neural hsf-1 overexpression no longer enhanced heat tolerance (Figure 2E). Surprisingly, neural hsf-1 overexpression was still capable of prolonging lifespan in the ttx-3 mutant (Figure 2F; Table 1). Thus, an intact thermosensory circuit is required for neural hsf-1 overexpressing worms to protect against heat stress but is dispensable for age regulation. More importantly, these data challenge the idea that HSF-1 regulates the aging process through the same heat shock response mechanisms and suggest that HSF-1 initiates an alternative transcriptional pathway to combat the stress of aging.
HSF-1 signals heterotypically to DAF-16 in the periphery for increased longevity
Intrigued by the possible separation of thermotolerance and aging, we sought to identify alternative transcriptional pathways that regulate aging independently of the heat stress response. To gain insight into alternate pathways, we analyzed genes that were significantly upregulated in multiple long-lived, hsf-1 overexpressing strains (Baird et al., 2014). Promoter analysis was performed on these gene data sets to identify possible transcription factor binding elements associated with an alternative cellular process or pathway. As expected, many of the promoters contained HSEs (Heat Shock Elements). We also observed a significant enrichment in DAF-16 Associated Elements (DAEs) (Table S1). The forkhead (FOXO) transcription factor DAF-16 is an essential component in the insulin/IGF-1 signaling cascade. This systemic process enables organisms to maintain glucose and energy homeostasis at optimal levels. HSF-1 has been linked to the insulin signaling pathway and DAF-16 (Chiang et al., 2012; Hsu et al., 2003; Morley and Morimoto, 2004), yet it remains unclear how these essential processes function together.
Disrupting insulin signaling through various mutations in different transduction components enhances longevity (Kenyon, 2011). Moreover, this lifespan extension requires the activation of DAF-16 to drive the pro-longevity transcriptional response. The canonical DAF-16 target gene Superoxide Dismutase-3 (sod-3) exhibits expression levels highly correlative with lifespan extension (Henderson et al., 2006; Sanchez-Blanco and Kim, 2011). Because neural hsf-1 overexpression does not activate a heat shock response at permissive temperatures, we hypothesized that hsf-1 might be capable of activating DAF-16 target genes. Fluorescence was examined in transgenic worms which expressed GFP under the control of the sod-3 promoter. In this manner, temporal and spatial aspects of sod-3 transcriptional activity were analyzed in different worm tissues.
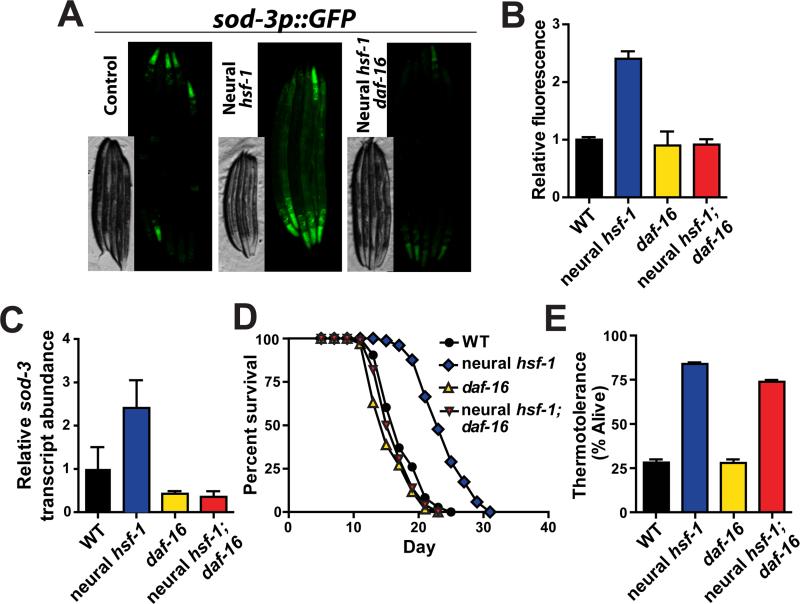
Elevated expression of hsf-1 in the nervous system increased GFP fluorescence in all worm tissues harboring the transcriptional sod-3 reporter (Figure 3A). We observed that overexpression of neural hsf-1 yields twice as much GFP fluorescence (Figure 3B and S4A). GFP expression was not only elevated in the nervous system by neural hsf-1 overexpression but also in peripheral tissues. The daf-16(mu86) null allele prevented neural hsf-1 overexpression from increasing GFP fluorescence of the sod-3 reporter strain (Figure 3A, 3B and S4A). Induction of endogenous sod-3 transcripts by neural hsf-1 overexpression was also daf-16 dependent (Figure 3C). Thus, neural hsf-1 overexpression drives FOXO-dependent activation of sod-3 at permissive temperatures in all worm tissues.
Figure 3. daf-16 is required for neural hsf-1 to induce sod-3 expression in peripheral tissues and extend lifespan, but is dispensable for increased thermotolerance.
(A) Fluorescent microscopy of C. elegans expressing GFP from the sod-3 promoter in control (CF1553), rab-3p::hsf-1 (AGD1198) and rab-3p::hsf-1; daf-16(mu86) (AGD1457) animals at 20°C. (B) Quantification of GFP fluorescence from strains used in (A) as determined by large-particle flow cytometry. (C) Transcript levels of endogenous sod-3 determined by quantitative RT-PCR from day 1 adult WT, daf-16(mu86), rab-3p::hsf-1 (AGD1289) and rab-3p::hsf-1; daf-16(mu86) (AGD1217) animals. (D) Lifespan survival was assessed at 20°C for strains used in (C). Lifespan statistics are found in Table 1. (E) Thermotolerance was determined at 34°C for strains used in (C-D).
Taken together, these results suggest that hsf-1 combats acute heat stress through the activation of a transcellular heat shock network and hsf-1 in the nervous system initiates an independent signal to activate DAF-16 in peripheral tissues to extend lifespan. To disprove this hypothesis, we determined the dependence of daf-16 upon lifespan extension by neural hsf-1. Consistent with separation of thermotolerance and aging, hsf-1 overexpression in the nervous system was incapable of extending worm lifespan in daf-16 mutants (Figure 3D; Table 1). In contrast, stress responsive transcriptions factors such as pha-4, xbp-1, skn-1 and ubl-5 were dispensable for neural hsf-1 mediated lifespan extension (Figure S3B, S3C, S3D and S3E; Table 1). The ability of increased neural hsf-1 to modulate sod-3 levels in a daf-16 dependent manner correlates with its ability to extend animal lifespan. Although daf-16 possesses thermal protective properties (Volovik et al., 2014), daf-16 was not required for neural hsf-1 overexpressing worms to protect against heat stress (Figure 3E). Moreover, expression of the sod-3 stress responsive gene was not heat inducible and likely represents a distinct stress response pathway utilized by neural hsf-1 during the aging process (Figure S4B, S4C and S4D).
The ability of hsf-1 to regulate the heat shock response and aging are separable. Removal of the thermosensory circuit through a ttx-3 mutation blocks heat resistance but has no effect on increased longevity. Conversely, eliminating daf-16 function does not affect thermotolerance but abolishes lifespan extension by neural hsf-1 overexpression. Therefore, distinct signaling events are communicated across an organism from hsf-1 in the nervous system in response to acute heat stress compared to the chronic stress of aging.
Intestinal daf-16 is sufficient to extend lifespan by neural hsf-1 overexpression
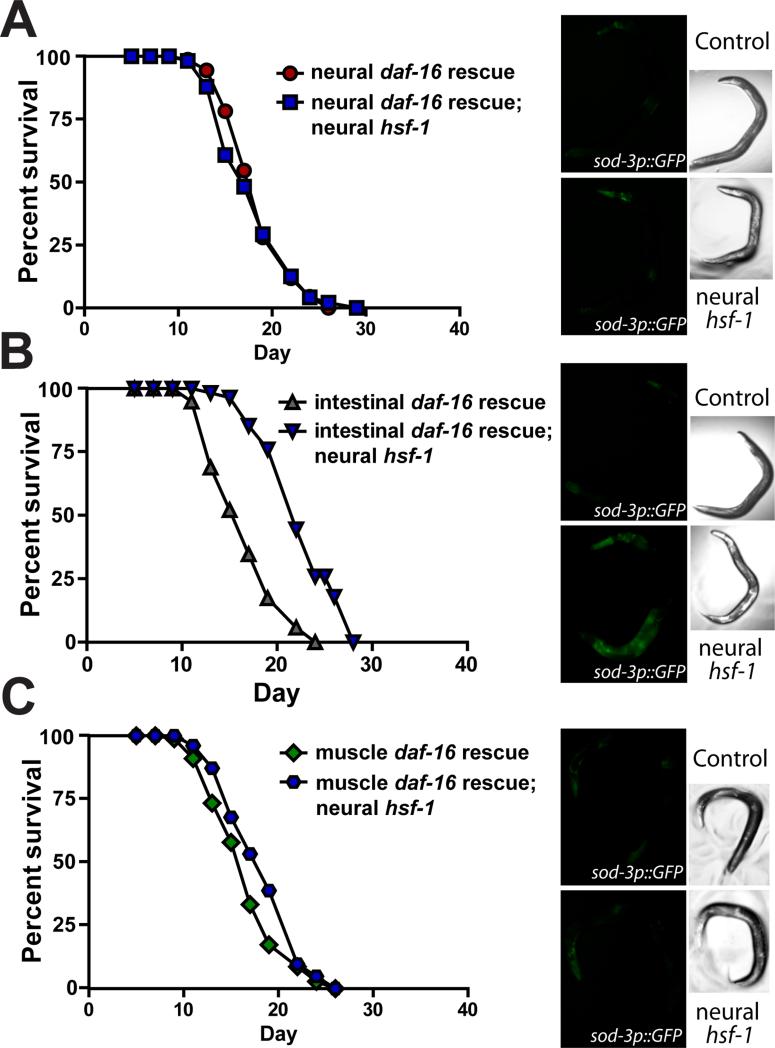
Neural hsf-1 regulates daf-16 in peripheral tissues; however, it is not clear in what tissues daf-16 is required for lifespan extension. By rescuing daf-16 expression in individual worm tissues in an otherwise null daf-16(mu86) mutant, we tested whether restoring daf-16 activity in a particular tissue was sufficient to drive the lifespan extension by neural hsf-1 overexpression. Expression of daf-16 was rescued in the nervous system, intestine and body-wall muscle (Libina et al., 2003). Rescuing expression of daf-16 in the nervous system was not sufficient for neural hsf-1 overexpression to extend lifespan or enhance GFP fluorescence of the sod-3 transcriptional reporter (Figure 4A, S4E and Table 1). This demonstrates that daf-16 in the nervous system is not required for neural hsf-1 to communicate to peripheral tissues. In contrast, daf-16 expression exclusively in the intestine enabled neural hsf-1 overexpression to both extend lifespan and modestly enhance GFP fluorescence in the sod-3 transcriptional reporter (Figure 4B, S4E and Table 1). Additionally, neural hsf-1 acted specifically through the intestine, as daf-16 expression in body-wall muscles did not extend lifespan or induce the sod-3 reporter (Figure 4C, S4E and Table 1). Therefore, neural hsf-1 functions cell non-autonomously in lifespan regulation via a signaling mechanism that requires the activity of daf-16 in intestinal cells. DAF-16 has previously been shown to play an important role in the worm intestine to influence lifespan (Libina et al., 2003). The communication between neural hsf-1 and daf-16 in the periphery does not require daf-16 in the nervous system, indicating that hsf-1 regulates daf-16 in a transcellular manner (Figure 5B).
Figure 4. Neural overexpression of hsf-1 requires daf-16 in the intestine to activate sod-3 and extend lifespan.
(A-C) Lifespan survival curves and representative sod-3p::GFP fluorescent micrographs of rab-3p::hsf-1 transgenic animals with different tissue-specific daf-16 rescues in an otherwise daf-16(mu86) mutant background. Expression of daf-16 is ectopically restored in individual tissues of daf-16(mu86) null animals including the (A) nervous system (AGD1273), (B) intestine (AGD1277), and (C) body-wall muscle (AGD1279). Lifespan statistics are found in Table 1.
Figure 5. hsf-1 is required in peripheral tissues for neural hsf-1 overexpressing worms to extend lifespan.
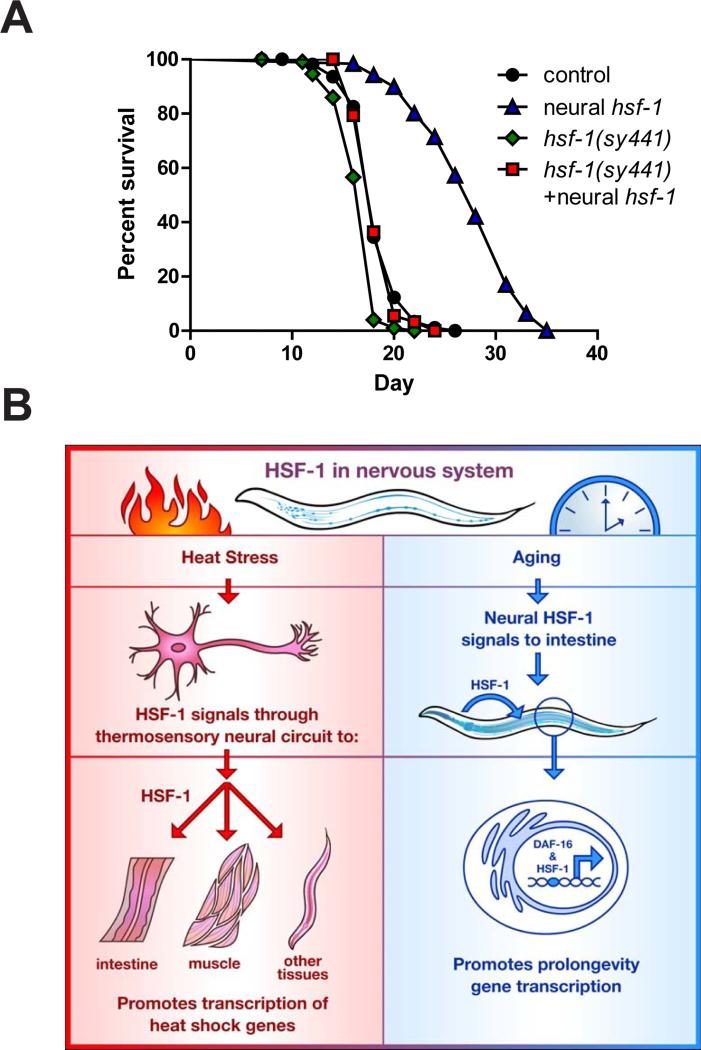
(A) Lifespan survival curves of WT and rab-3p::hsf-1 overexpressing nematodes (AGD1289) in the presence or absence of the hypomorphic hsf-1(sy441) mutation (AGD1471). Lifespan statistics are found in Table 1. (B) Model depicting how heterotypic signals by neural hsf-1 separate thermal protection from age regulation. Under thermal stress, HSF-1 in the nervous system signals to peripheral tissues through the thermosensory neural circuit and enhances the heat shock response to protect worms (left model). Conversely, neural hsf-1 functions independently of the thermosensory circuit to generate a transcellular signal which activates DAF-16 and HSF-1 in the intestine and drives pro-longevity gene expression (right model).
Although daf-16 activity is required in the intestine for neural hsf-1 overexpressing worms to extend lifespan, it is unclear whether hsf-1 also functions in the peripheral, non-neural tissues to mediate longevity assurance. Thus hsf-1 activity in peripheral tissues was reduced through a hypomorphic hsf-1(sy441) allele in which the carboxy-terminal transactivation domain has been removed through a premature stop codon (Hajdu-Cronin et al., 2004). Overexpressing full length hsf-1 in the nervous system was not able to extend the lifespan of the hypomorphic hsf-1(sy441) animals (Figure 5A and Table 1). These results indicate that hsf-1 activity in the nervous system is not sufficient to increase lifespan on its own, but rather communication of neural hsf-1 to peripheral cells expressing hsf-1 is essential for longevity assurance.
DISCUSSION
Stress encompasses a wide spectrum of insults for which cells have evolved highly specialized responses to both combat the immediate stress and initiate recovery mechanisms. With the evolution of multicellularity, organisms developed a hierarchical mode of stress response regulation across tissues. This normally includes a master tissue, which can both sense the particular stress and transmit the appropriate response to the pertinent peripheral tissues. Cell non-autonomous signaling of stress responses includes numerous mechanisms, such as the insulin signaling pathway (Libina et al., 2003), germ-line ablation (Hsin and Kenyon, 1999), mitochondrial unfolded protein response (UPR) (Durieux et al., 2011; Owusu-Ansah et al., 2013), endoplasmic reticulum UPR (Deng et al., 2013; Taylor and Dillin, 2013) and the heat shock response (Prahlad et al., 2008). To date, each signaling mechanism arises in a particular tissue and functions in a homotypic manner whereby a single signaling cascade conveys stress responsive cues to all pertinent tissues. Consistent with these responses, neural hsf-1 signaling directly regulates hsf-1 targets in peripheral tissues to mount the heat shock response upon acute stress. In contrast, neural hsf-1 initiates a distinct response under aging to coordinate daf-16 activity in peripheral tissues in addition to hsf-1. Thus, hsf-1 communicates in a heterotypic manner to regulate different stress response pathways. Given the vast array of stresses that hsf-1 has been reported to protect against (Hsu et al., 2003; Morimoto, 2008), it is reasonable that hsf-1 can optimally tailor stress response machinery to combat either acute or chronic insults.
Links between hsf-1 and components of the insulin/IGF-1 signaling pathway exists in multiple experimental paradigms including aging, proteotoxicity and thermotolerance (Hsu et al., 2003; McColl et al., 2010; Morley and Morimoto, 2004). From these reports, hsf-1 function has been modeled to reside downstream of, or in parallel with, the insulin signaling pathway. Herein, we provide evidence suggesting that hsf-1 functions upstream of daf-16 signaling. Furthermore, mutations abolishing distinct types of neural vesicular release, unc-13 and unc-31, extend worm lifespan in a daf-16 dependent manner (Ailion et al., 1999; Gems and Riddle, 2000). We observe that animals harboring the unc-13 or unc-31 mutation induce sod-3 expression (Figure S4F), suggesting that reduced neural secretion of insulin-like peptides activates daf-16 in distal tissues to promote longevity. Because of this functional redundancy, the signaling mechanism by which neural hsf-1 communicates to the periphery remains unclear, but could include any of the 39 insulin-like peptides found in C. elegans.
Within the nervous system, hsf-1 could function at multiples steps to modulate insulin signaling; either by regulating the production, processing, trafficking or secretion of insulin peptides during conditions of chronic stress. Future studies are needed to elucidate the molecular mechanisms by which hsf-1 overexpression in neurons could possibly regulate insulin biogenesis. Expanding these concepts to mammalian systems, it will be interesting to understand how bolstering hsf-1 activity in the brain influences more localized insulin secretion within the nervous system versus insulin biogenesis in pancreatic β-cells. Either course of action could profoundly affect energy homeostasis and provide a novel, long-term mode of diabetes intervention.
The process of aging is due, in part, to protein misfolding events and a general deterioration in the quality of the proteome. In support of this hypothesis, metastable proteins that can fold and function in youthful cells begin to misfold upon aging, losing functionality (Ben-Zvi et al., 2009). A similar phenomenon appears in numerous age-onset neurodegenerative disorders, in which the aging brain can no longer maintain disease-linked proteins in properly folded, functional states and misfolding leads to multimerization of the disease proteins and neural death (Douglas and Dillin, 2010; Morimoto, 2008). In these studies, the levels of hsf-1 and its chaperone target genes directly correlated with the age-onset of different neurodegenerative models. Thus, by extension, it was hypothesized that hsf-1 regulates the aging process by modulating chaperone network components to directly influence the folding state of the proteome (Morimoto, 2008). Our data suggests an alternate method by which hsf-1 can regulate the aging process. In this model, hsf-1 activates the FOXO transcription factor, which initiates a pro-longevity stress response that is distinct from the heat shock response. Although we cannot entirely exclude the possible involvement of the chaperone network in lifespan extension by neural hsf-1 overexpression, we did not observe an enhancement in heat responsive elements. Further investigation is needed to uncover stress responsive genes within the DAF-16 activation cascade that are responsible for longevity assurance.
EXPERIMENTAL PROCEDURES
C. elegans Strains and Maintenance
All strains were maintained at 15°C on the E. coli strain, OP50. The following strains were used in this work: wild-type (N2), ttx-3(ks5), daf-16(mu86), hsf-1(sy441), daf-2(e1370), AM101 (rmIS110[gref-1p::Q40::YFP]; AGD440 (N2; uthEx457[rab-3p::tdTomato); rol-6(su1006)]); AGD1008 (uthEx663[rab-3p::hsf-1; myo-2p::tdTomato]), AGD1441 (uthEx741[rgef-1p::hsf-1; myo-2p::tdTomato]), AGD1289 (uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]), AGD1053 (uthIS365[rab-3p::hsf-1; myo-2p::tdTomato]), AGD1054 (uthIS366[rab-3p::hsf-1; myo-2p::tdTomato]), AGD1449 (ttx-3(ks5); uthEx663[rab-3p::hsf-1; myo-2p::tdTomato]), AGD1471 (hsf-1(sy441); uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]), CL2070 (dvln70[pCL25 (hsp-16.2p::GFP); pRF4(rol-6)]), AGD1448 (dvln70[pCL25 (hsp-16.2p::GFP); pRF4(rol-6)]; uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]), CF1553 (muls84[pAD76(sod-3p::GFP)]), AGD709 (daf-16(mu86); muls84[pAD76(sod-3p::GFP)]), AGD1198 (muls84[pAD76(sod-3p::GFP)]; uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]), AGD1457 (daf-16(mu86); muls84[pAD76(sod-3p::GFP)]; uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]; AGD1217 (daf-16(mu86); uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]; AGD1272 (daf-16(mu86); muEx169[unc-119p::GFP::daf-16, rol-6(su1006)]); AGD1273 (daf-16(mu86); muEx169[unc-119p::GFP::daf-16, rol-6(su1006)]; uthIS368[rab-3p::hsf-1; myo-2p::tdTomato] ); AGD1276 (daf-16(mu86); muEx211[pNL213(ges-1p::GFP::daf-16), rol-6(su1006)]); AGD1277 (daf-16(mu86); muEx211[pNL213(ges-1p::GFP::daf-16), rol-6(su1006)]; uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]), AGD1278 (daf-16(mu86); muEx212[pNL212(myo-3p::GFP::daf-16), rol-6(su1006)]), AGD1279 (daf-16(mu86); muEx212[pNL212(myo-3p::GFP::daf-16), rol-6(su1006)]; uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]), AGD1309 (daf-16(mu86); muEx169[unc-119p::GFP::daf-16, rol-6(su1006)]; muls84[pAD76(sod-3p::GFP)]), AGD1313 (daf-16(mu86); muEx169[unc-119p::GFP::daf-16, rol-6(su1006)]; uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]; muls84[pAD76(sod-3p::GFP)]), AGD1311 (daf-16(mu86); muEx211[pNL213(ges-1p::GFP::daf-16), rol-6(su1006)]; muls84[pAD76(sod-3p::GFP)]), AGD1315 (daf-16(mu86); muEx211[pNL213(ges-1p::GFP::daf-16), rol-6(su1006)]; uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]; muls84[pAD76(sod-3p::GFP)]), AGD1312 (daf-16(mu86); muEx212[pNL212(myo-3p::GFP::daf-16), rol-6(su1006)]; muls84[pAD76(sod-3p::GFP)]), AGD1316 (daf-16(mu86); muEx212[pNL212(myo-3p::GFP::daf-16), rol-6(su1006)]; uthIS368[rab-3p::hsf-1; myo-2p::tdTomato]; muls84[pAD76(sod-3p::GFP)]), AGD1475 (unc-13(e450); muIs84[pAD76(sod-3p::GFP)]), AGD1476 (unc-31(e928); muIs84[pAD76(sod-3p::GFP)])
Wild-type (N2), ttx-3(ks5) and daf-16(mu86), CF1553 and CL2070 strains were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). For generation of transgenic overexpression strains, hsf-1 cDNA was inserted downstream of the neural promoters rab-3 or gref-1 and upstream of the unc-54 3’ untranslated region (UTR). Neural hsf-1 DNA plasmid constructs were injected at 2 ng/μl along with a co-injection marker (myo-2p::tdTomato) at 10 ng/μl to make transgenic overexpression worms.
Western Blot Analysis
Age synchronized worms were cultivated on nematode growth (NG) plates containing the E. coli strain, OP50, at 20°C until day 1 adulthood. Worm were washed off the plate with M9 buffer pre-heated to 34°C, collected and incubated in a 34°C water bath for 15 minutes. Worms were centrifuged at 1000 × g for 30 sec and move back to NG plates seeded with OP50 bacteria at 20°C. Worms were allowed 1.5 hrs of recovery at 20°C before worms were collected and frozen in liquid nitrogen for further processing.
Worm extracts were generated by glass bead disruption in non-denaturing lysis buffer [150mM NaCl, 50mM Hepes at pH 7.4, 1mM EDTA, 1% Triton X100, protease inhibitor cocktail without EDTA (Roche)]. Crude lysates were subject to centrifugation at 10,000 × g at 4°C for 5 mins. The supernatant was supplemented with 2x SDS sample buffer containing [50mM Tris-Cl at pH 6.8, 2 mM EDTA, 4% glycerol, 2% SDS, Coomassie Blue, protease inhibitor cocktail without EDTA (Roche)]. Samples were boiled for 10 minutes and resolved by SDS-PAGE. Proteins levels were monitored by standard immuno-blotting procedures with α-Hsp-16.2 (kind gift from Lithgow Lab) and α-tubulin (Sigma T6074) antibodies.
Transcript Analysis
Total RNA was isolated from synchronized populations at day 1 of adulthood using Qiazol (Qiagen) and then further purified with the RNeasy mini kit (Qiagen). cDNA was synthesized using the QuantiTect kit (Qiagen). SybrGreen was used for quantitative PCR as described in the SsoAdvanced SYBR Green Supermix protocol (BioRad). Experiments were repeated with three biological repeats and analyzed using the comparative Ct method. Internal controls utilized a geometric mean of cdc-42, pmp-3 and Y45F10D.4. The Roche Universal ProbeFinder online tool was used to design primers. Primer sequences as follows:
cdc-42 forward 5’- AGGAACGTCTTCCTTGTCTCC -3’
cdc-42 reverse 5’- GGACATAGAAAGAAAAACACAGTCAC -3’
pmp-3 forward 5’- CGGTGTTAAAACTCACTGGAGA -3’
pmp-3 reverse 5’- TCGTGAAGTTCCATAACACGA -3’
Y45F10D.4 forward 5’- AAGCGTCGGAACAGGAATC -3’
Y45F10D.4 reverse 5’- TTTTTCCGTTATCGTCGACTC -3’
hsf-1 forward 5’- TTTGCATTTTCTCGTCTCTGTC -3’
hsf-1 reverse 5’- TCTATTTCCAGCACACCTCGT -3’
hsp-16.2 forward 5’- TCCATCTGAGTCTTCTGAGATTGTTA -3’
hsp-16.2 reverse 5’- TGGTTTAAACTGTGAGACGTTGA -3’
hsp-70a (C12C8.1) forward 5’- CGGTATTTATCAAAATGGAAAGGTT -3’
hsp-70a (C12C8.1) reverse 5’- TACGAGCGGCTTGATCTTTT -3’
hsp-70b (F44E5.4) forward 5’- TGCACCAATCTGGACAATCT -3’
hsp-70b (F44E5.4) reverse 5’- TCCAGCAGTTCCAGGATTTC -3’
pat-10 forward 5’- TCGAGGAGTTCTGGGAGTTG -3’
pat-10 reverse 5’- TTGTAGATCAGCGATTTTAAAGGA -3’
sod-3 forward 5’- CACTGCTTCAAAGCTTGTTCA -3’
sod-3 reverse 5’- ATGGGAGATCTGGGAGAGTG -3’
RNA for global sequencing analysis was prepared using Illumina TruSeq RNA Sample Prep Kit (Illumina). Paired-end sequencing was performed on an Illumina HiSeq 2000 and data was analyzed with CLC Genomics Workbench 7.0.4 software.
Promoter analysis
We used RSAT (Thomas-Chollier et al., 2011) to ask for overrepresented sequences of length 6, 7, or 8 upstream of our ORF start sites, within 1.5kb or until the preceding ORF, whichever was closer. e-value is a multiple testing corrected estimate of the probability of this degree of overrepresentation.
Lifespan analysis
Lifespan experiments were conducted at 20°C as previously described (Wilkinson et al., 2012) and a minimum of 3 independent experiments was performed under every condition. Worms were fed different E. coli, OP50 or HT115 for experiments involving RNAi knockdown of gene expression. Tissue-specific daf-16 rescue lifespans were performed on OP50. The pre-fertile period of adulthood was considered day 0. Worms were transferred to fresh plates every second day until day 12. To prevent excessive worm censorship, 5-fluoro-2′-deoxyuridine, FUDR, was supplemented into growth media of lifespan experiments involving the hypomorphic hsf-1(sy441). Lifespan analysis on Prism 6 and JMP software was used for statistical analysis to determine significance calculated using the log-rank (Mantel-Cox) method.
Thermotolerance assay
Synchronized day 1 adult worms were placed at 34°C for 12-14 hrs on plates spotted with OP50 E. coli or HT115 for RNAi. Worms were then scored for viability. At least 80 worms were used per genotype and experiments were repeated at least three times. Prism 6 software was used for statistical analysis.
RNAi feeding
Worms were fed from hatch HT115 E. coli containing an empty vector control or expressing double-stranded RNA. RNAi strains were taken from the Vidal library if present, or the Ahringer library if absent from the Vidal library. All RNAi clones were sequence verified prior to use and knockdown verified previously (Carrano et al., 2009; Durieux et al., 2011; Panowski et al., 2007; Taylor and Dillin, 2013).
Microscopy and Fluorescence analysis
For fluorescence microscopy, worms were anesthetized with 10mM levaminsole and images were captured using a Leica DM6000 B microscope and Hamamatsu ORCA-ER camera. We also used a COPAS Biosort (Union Biometrica) to measure individual day 1 worm length, width and GFP fluorescence. At least 500 worms were measured per genotype and pooled in three biological replicates. We normalized fluorescence by worm size to compare between genotypes.
Supplementary Material
ACKNOWLEDGMENTS
We thank the laboratory of Dr. G. Lithgow for generously sharing the HSP-16 antibody and Kenyon lab for providing tissue specific daf-16 rescue strains. We also thank the Illustrator, Lindsay Daniele, for the graphical abstract. Some of the nematode strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota), which is supported by the NIH – Office of Research Infrastructure Programs (P40 OD010440). This work was supported by NIH grants (R37AG024365, R01AG042679 and R01ES021667). P.M.D. was funded by the NIH/NIA (1K99AG042495) and the George E. Hewitt Foundation for Medical Research. N.A.B. was funded by a postdoctoral fellowship from the Salk Center for Nutritional Genomics from the Leona M. & Harry B. Helmsley Charitable Trust. S.C.W. is grateful for the generous support of the Glenn Foundation for Medical Research. M.A.M. has been supported by NIH training grant (T32AG000266). B.K.K. is supported by NIH (R01AG025549) and (R01AG043080).
Footnotes
AUTHOR CONTRIBUTIONS
P.M.D, N.A.B, M.S.S. and S.U. performed the experiments and analyzed the data. M.A.M and B.K.K. assisted P.M.D. on bioinformatics analysis. P.M.D, N.A.B., S.C.W and A.D. wrote the manuscript. P.M.D. and N.A.B contributed equally.
REFERENCES
- Ailion M, Inoue T, Weaver CI, Holdcraft RW, Thomas JH. Neurosecretory control of aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1999;96:7394–7397. doi: 10.1073/pnas.96.13.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Douglas PM, Simic MS, Grant AR, Moresco JJ, Wolff SC, Yates JR, 3rd, Manning G, Dillin A. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science. 2014;346:360–363. doi: 10.1126/science.1253168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Liu Z, Dillin A, Hunter T. A conserved ubiquitination pathway determines longevity in response to diet restriction. Nature. 2009;460:396–399. doi: 10.1038/nature08130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Ching TT, Lee HC, Mousigian C, Hsu AL. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell. 2012;148:322–334. doi: 10.1016/j.cell.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wang ZV, Tao C, Gao N, Holland WL, Ferdous A, Repa JJ, Liang G, Ye J, Lehrman MA, et al. The Xbp1s/GalE axis links ER stress to postprandial hepatic metabolism. J Clin Invest. 2013;123:455–468. doi: 10.1172/JCI62819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Dillin A. Protein homeostasis and aging in neurodegeneration. J Cell Biol. 2010;190:719–729. doi: 10.1083/jcb.201005144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failla G. The aging process and cancerogenesis. Ann N Y Acad Sci. 1958;71:1124–1140. doi: 10.1111/j.1749-6632.1958.tb46828.x. [DOI] [PubMed] [Google Scholar]
- Gems D, Riddle DL. Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics. 2000;154:1597–1610. doi: 10.1093/genetics/154.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu-Cronin YM, Chen WJ, Sternberg PW. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics. 2004;168:1937–1949. doi: 10.1534/genetics.104.028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Philos Trans R Soc Lond B Biol Sci. 2011;366:9–16. doi: 10.1098/rstb.2010.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Link CD, Cypser JR, Johnson CJ, Johnson TE. Direct observation of stress response in Caenorhabditis elegans using a reporter transgene. Cell Stress Chaperones. 1999;4:235–242. doi: 10.1379/1466-1268(1999)004<0235:doosri>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, Bush AI, Kapahi P, Lithgow GJ. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 2010;12:260–272. doi: 10.1016/j.cmet.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc Natl Acad Sci U S A. 1963;49:517–521. doi: 10.1073/pnas.49.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Blanco A, Kim SK. Variable pathogenicity determines individual lifespan in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002047. doi: 10.1371/journal.pgen.1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39:W86–91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volovik Y, Moll L, Marques FC, Maman M, Bejerano-Sagie M, Cohen E. Differential regulation of the heat shock factor 1 and DAF-16 by neuronal nhl-1 in the nematode C. elegans. Cell Rep. 2014;9:2192–2205. doi: 10.1016/j.celrep.2014.11.028. [DOI] [PubMed] [Google Scholar]
- Walker GA, Lithgow GJ. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. 2003;2:131–139. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson DS, Taylor RC, Dillin A. Analysis of aging in Caenorhabditis elegans. Methods Cell Biol. 2012;107:353–381. doi: 10.1016/B978-0-12-394620-1.00012-6. [DOI] [PubMed] [Google Scholar]
- Wolff S, Weissman JS, Dillin A. Differential scales of protein quality control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.