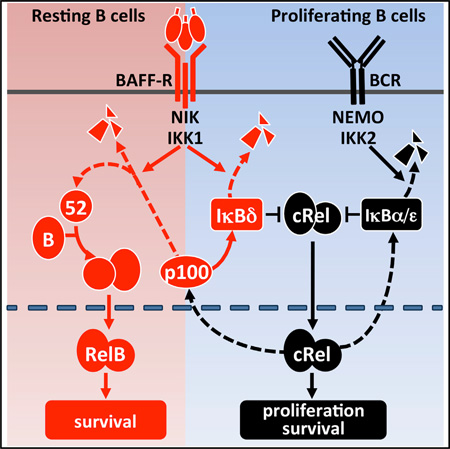
SUMMARY
BAFF, an activator of the noncanonical NFκB pathway, provides critical survival signals during B cell maturation and contributes to B cell proliferation. We found that the NFκB family member RelB is required ex vivo for B cell maturation, but cRel is required for proliferation. Combined molecular network modeling and experimentation revealed Nfkb2 p100 as a pathway switch; at moderate p100 synthesis rates in maturing B cells, BAFF fully utilizes p100 to generate the RelB:p52 dimer, whereas at high synthesis rates, p100 assembles into multimeric IκBsome complexes, which BAFF neutralizes in order to potentiate cRel activity and B cell expansion. Indeed, moderation of p100 expression or disruption of IκBsome assembly circumvented the BAFF requirement for full B cell expansion. Our studies emphasize the importance of p100 in determining distinct NFκB network states during B cell biology, which causes BAFF to have context-dependent functional consequences.
Graphical abstract
INTRODUCTION
Mature follicular B cells are largely responsible for thymus (T)-dependent antigenic responses. Two receptors critical for follicular B cell maintenance and expansion are the B cell antigen receptor (BCR) and the B-cell-activating factor receptor (BAFF-R). BCR is critical for antigen-responsive expansion and maintenance of the mature B cell pool (Lam et al., 1997). BAFF-R (and BAFF) is critical for the survival of maturing transitional B cells (Harless et al., 2001; O’Connor et al., 2004; Schiemann et al., 2001), enhances follicular B cells, enhances antigen-responsive B cell expansion in vitro (Huang et al., 2004; Rickert et al., 2011; Schweighoffer et al., 2013), and strengthens T cell-dependent and independent humoral immune responses (Do et al., 2000; Litinskiy et al., 2002). Indeed, whereas initiation of germinal center formation was found to be independent of BAFF, the B cell responsiveness to antigens (via the BCR) is impaired in BAFF-signaling-deficient mice (Rahman et al., 2003; Vora et al., 2003).
BCR and BAFF-R are known to signal to NFκB via two distinct pathways: the NEMO-dependent “canonical” pathway and the NEMO-independent “noncanonical” pathway, respectively. Activated BCR recruits the Carma1-Bcl10-Malt1-containing complex to the membrane, triggering NEMO ubiquitination and activation of the NEMO-containing IKK complex. This leads to nuclear translocation of preexisting RelA- and cRel-containing NFκB dimers from the latent IκB-inhibited cytoplasmic complexes (Hayden and Ghosh, 2008). BAFF-R stimulation sequesters TRAF3, resulting in the stabilization of NIK and activation of a NEMO-independent IKK1 kinase complex. This stimulates p100 processing to p52 and results in nuclear accumulation of RelB:p52 dimers (Claudio et al., 2002).
Recent studies have begun to address the molecular basis for the functional interactions between BCR and BAFF-R. Tonic BCR signaling and associated canonical pathway activity are critical for the constitutive expression of the Nfkb2 gene generating p100 substrate for NIK/IKK1-dependent processing and production of RelB:p52 dimer in maturing B cells (Cancro, 2009; Stadanlick et al., 2008). Similarly, lymphotoxin-beta receptor-responsive noncanonical pathway activation was found to be dependent on constitutive canonical signaling (Basak et al., 2008). In the context of resting B cells, RelB is a presumed mediator of BAFF’s survival functions dependent on tonic BCR. Extending this model to proliferating B cells suggests that heightened BCR-responsive canonical activity might strengthen BAFF-mediated activation of RelB. In other words, a costimulatory role of BAFF in the expansion of activated B cells might be achieved through RelB-mediated enhanced cell survival. However, there are indications that BAFF may in fact not only enhance cell survival but contribute to cell cycle entry of mature follicular B cells following antigenic stimulation (Allman et al., 2001; Do et al., 2000; Huang et al., 2004; Patke et al., 2006). It is unknown whether this function may also involve NFκB signaling or be entirely mediated by other signaling axes known to be activated by BAFF, such as phosphatidylinositol 3-kinase (PI3K) or mitogen-activated protein kinase/ERK (Jellusova et al., 2013; Mackay and Schneider, 2009; Mackay et al., 2007; Rickert et al., 2011), which are also mediators of BCR signaling (Srinivasan et al., 2009) and potential crosstalk regulators (Schweighoffer et al., 2013).
Here, we addressed the role of the NFκB-signaling system in mediating BAFF’s functions in both maturing as well as proliferating B cells using quantitative cell biology, biochemistry, and mathematical modeling. In particular, we offer genetic evidence that RelB is indeed critical for BAFF-induced survival of maturing B lymphocytes in vitro but the costimulatory effect of BAFF in BCR-triggered population expansion is not based on enhanced B cell survival or elevated RelB activity. Instead, BAFF costimulation augments BCR-triggered cRel activation and the fraction of B cells entering the proliferative program. Quantitative analysis of the NFκB network reveals that cRel hyperactivation is achieved by BAFF neutralizing the inhibitory effect of BCR-induced p100, which was shown to assemble into a multimeric IκBsome (Savinova et al., 2009) with associated IκBδ activity (Basak et al., 2007; Shih et al., 2009).
RESULTS
BAFF-R Enhances BCR-Triggered B Cell Expansion
Prior work has established that BCR and BAFF-R activate distinct NFκB-signaling pathways, the so-called canonical, NEMO-dependent and the noncanonical, NEMO-independent pathways, respectively (Figure 1A). Whereas the primary transcriptional effector of the former is NFκB cRel, which enhances cell survival and may initiate the B-cell-proliferative program, the latter is known to activate RelB. Both receptors also activate other signaling pathways such as PI3K and ERK that contribute to their physiological functions (Mackay et al., 2007; Otipoby et al., 2008; Ramakrishnan et al., 2004).
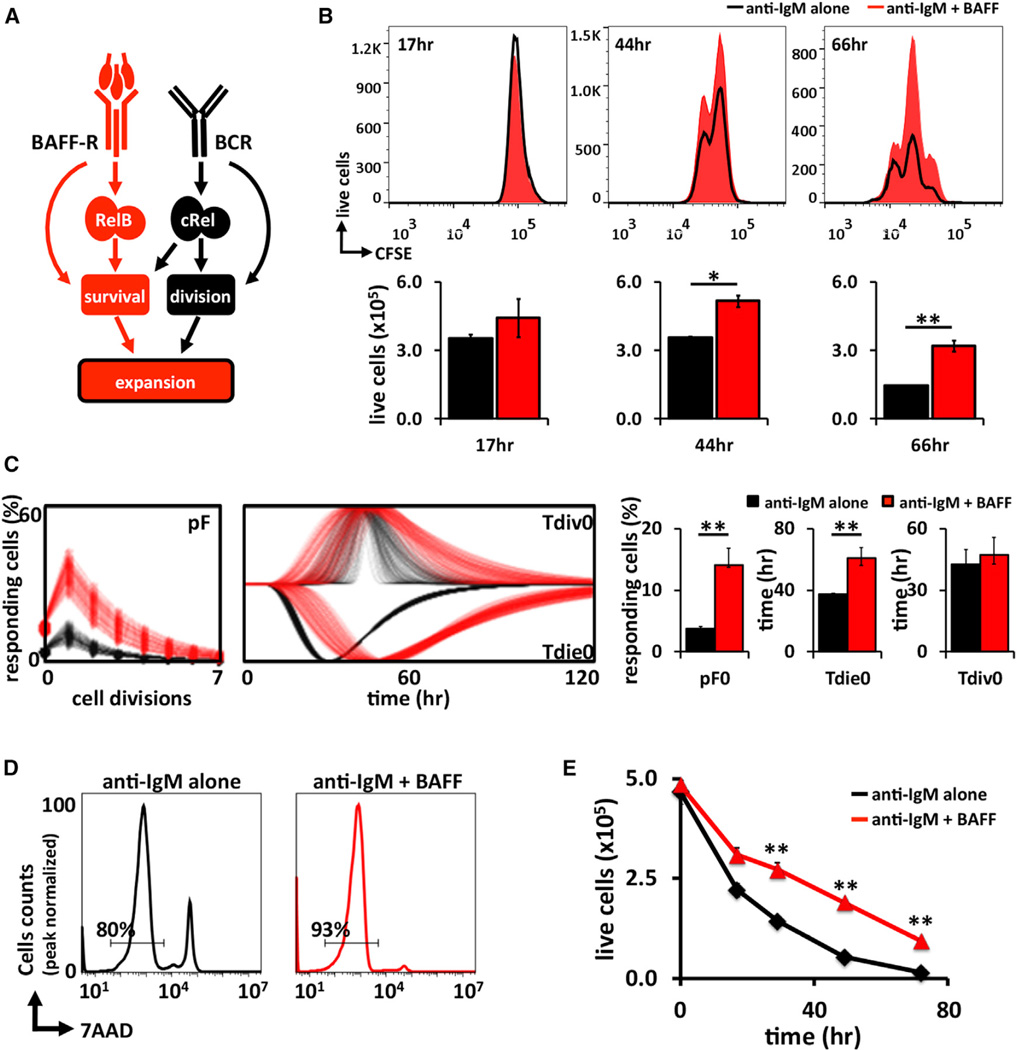
Figure 1. BAFF Enhances BCR-Controlled B Cell Expansion.
(A) Schematic of the distinct canonical and noncanonical NFκB pathways identified in B cells. Antigenic stimulation leads to release of cRel-containing dimers into the nucleus, resulting in activation of cell division programs (Grumont et al., 1998, 1999). BAFF activates noncanonical RelB dimers and B cell survival. Thus, this model suggests that BAFF costimulation results in enhanced expansion by augmenting survival of B cells.
(B) In vitro proliferation of CFSE-labeled wild type B cells and stimulated for 3 days with anti-IgM alone (black) or anti-IgM + BAFF ligand (red) and analyzed by FACS. Live cell numbers gated by exclusion of 7AADHi population.
(C) The CFSE proliferation profiles of wild-type B cells stimulated with either anti-IgM or anti-IgM plus BAFF analyzed with FlowMax software tool to calculate the fraction of B cells responding to stimulation (pF), the average time to division of undivided (Tdiv0), and average time to death of undivided (Tdie0) cells.
(D) Representative histograms of cell survival analyzed by 7AAD staining following 24 hr of stimulation by anti-IgM alone (black) or anti-IgM + BAFF ligand (red).
(E) Time course averages of B cells viability stimulated with anti-IgM alone or anti-IgM + BAFF ligand. Live cells were identified by gating out 7AADHi population. FACS plots in (B) and (D) are representative of at least three experiments. Bar graphs in (C) summarize best-fit cellular parameters with the lognormal SD. Error bars in (E) SD; n = 3. *p < 0.05, **p < 0.01 by Student’s t test.
See also Figure S1.
We asked whether BAFF costimulation would enhance BCR-triggered B cell expansion via enhanced cell survival or whether other signaling crosstalk mechanisms might be involved. Using the cell-tracking dye carboxyfluorescein succinimidyl ester (CFSE), we monitored splenic B cell expansion following BCR crosslinking with anti-immunoglobulin M (IgM). Costimulation of BAFF-R increased this B cell expansion (Figures 1B and S1), confirming previous studies (Craxton et al., 2007; Huang et al., 2004; Schweighoffer et al., 2013; Stadanlick et al., 2008). To determine how BAFF affected B cell population dynamics, we employed the software tool FlowMax (Shokhirev and Hoffmann, 2013), which deconvolutes dye dilution data to determine the fraction of cells entering the proliferative program (pF) and the associated cell division (Tdiv) as well as cell survival (Tdie) times. This automated interpretation of CFSE data suggest that BAFF not only increased the lifetime of B cells (from 40 to 61 hr) but also increased the fraction of responding cells entering the proliferative program from 5% to 15% (Figure 1C). However, the time required for first cell division remained relatively constant at 43–48 hr. Indeed, 7-amino-actinomycin D (7AAD) staining revealed fewer dying cells at 24 hr (Figure 1D), but at a longer time course, the number of live cells actually increased with BAFF (Figure 1E), consistent with previous reports that BAFF is able to contribute to cell cycle entry as well (Do et al., 2000; Huang et al., 2004; Patke et al., 2006).
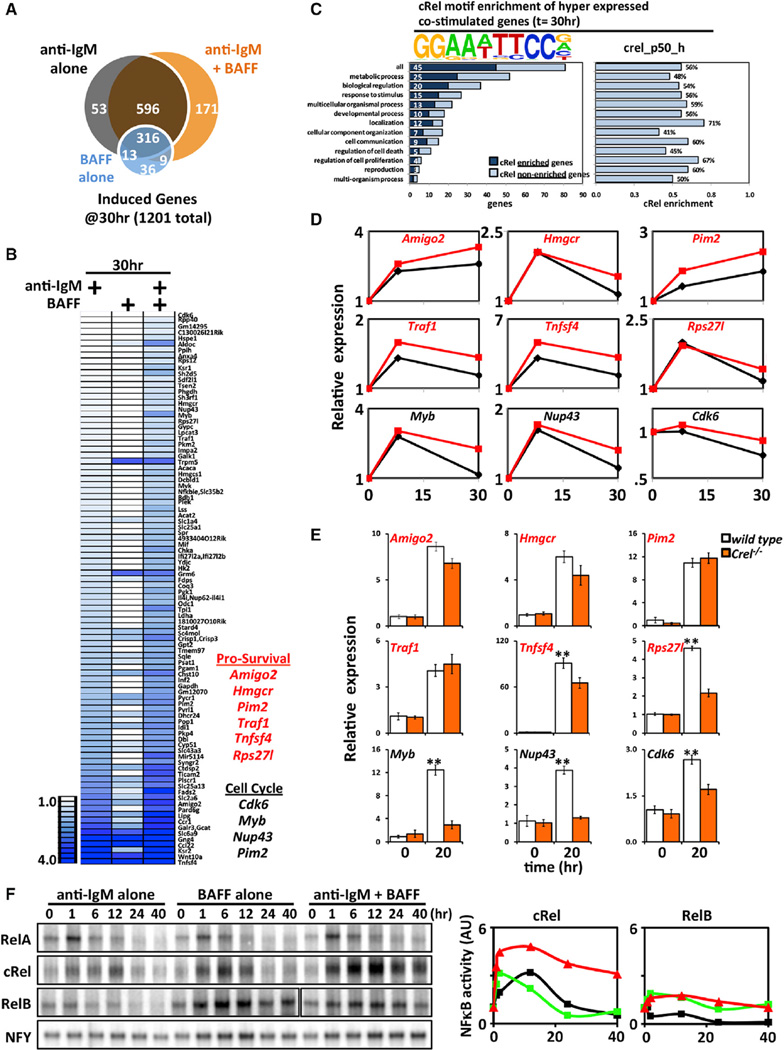
The transcriptomic response to costimulation, as revealed by RNA sequencing (RNA-seq) analysis, included a substantial number of genes (171; Figures 2A and 2B), many of which were dependent on BAFF costimulus (87 were reduced >20.5-fold in single anti-IgM stimulation conditions). Gene ontology analysis revealed terms like “metabolic process” and “biological regulation” scoring highly, whereas regulation of cell death or cell proliferation did not (Figure 2C). Remarkably, motif analysis of the regulatory region (−1,000 to +300 of the transcription start site) of these hyperinduced genes revealed the κB motif as being substantially overrepresented. In particular, using previously characterized NFκB-binding motifs (Siggers et al., 2012), we found that the cRel:p50-binding motif is present in 56% of the proximal regulatory regions of these hyperexpressed genes, regardless of their gene ontology categorization. Of these 81 “late” hyperinduced genes, we identified some candidates involved in prosurvival and cell cycle processes (Figures 2B and 2D). Examination of these hyperexpressed regulators of survival and cell cycle by quantitative PCR revealed that their full expression depended on cRel (Figure 2E).
Figure 2. In the Context of BCR-Stimulated B Cells, BAFF-Induced Hyperexpression Shows a cRel Signature.
(A) Venn diagram of gene sets identified by RNA-seq analysis (NCBI GEO accession number GSE54588) that are induced >2-fold over unstimulated wild-type B cells (t = 0 hr) following 30 hr of stimulation with anti-IgM alone (black), BAFF alone (blue), or anti-IgM and BAFF (orange).
(B) Heatmap of “hyper” expressed genes, defined as induced >log2(0.5) by anti-IgM at 30 hr over unstimulated wild-type B cells (t = 0 hr) and possess an additional >log2(0.5) greater expression in the anti-IgM and BAFF costimulation condition at the same time point. Of these genes, we identified a few genes involved in survival and cell cycle.
(C) Graphic representation of prosurvival and cell cycle genes in RNA-seq analysis in (B). The cRel:p50-binding motif is found overrepresented within the regulatory regions of hyperexpressed genes, regardless of the biological process they are categorized into.
(D) Quantitative PCR of wild-type B cells stimulated with either anti-IgM alone or in costimulation conditions. Relative expression normalized to the basal level is shown.
(E) Quantitative PCR of wild-type and cRel-deficient B cells costimulated with anti-IgM and BAFF for 24 hr. **p < 0.01 denotes significantly higher in wild-type condition than Crel−/− counterpart by Student’s t test.
(F) Time course of NFκB DNA-binding activities in anti-IgM alone, BAFF alone, and anti-IgM plus BAFF costimulated B cells. Nuclear extracts from wild-type B cells activated by indicated stimuli were collected and subjected to RelA, cRel, and RelB EMSA (for cRel EMSA, RelA and RelB antibodies are incubated together with nuclear extracts in order to shift away these NFκB-containing dimers, leaving cRel species untouched; see Experimental Procedures). Signals quantified and graphed relative to their respective resting cells (left). EMSA is representative of five experiments. AU, arbitrary units.
To determine which NFκB activity was hyperinduced, we examined NFκB DNA-binding activities biochemically using electrophoretic mobility shift assays coupled with antibody supershift to ablate all but the NFκB dimer of interest. Anti-IgM stimulation was found to induce a transient cRel activity within the first few hours, followed by a return to basal levels by 24 hr (Figure 2F). BAFF stimulation alone activated cRel similarly and also resulted in long-lasting RelB DNA-binding activity. Upon costimulating BCR and BAFF-R, we observed surprisingly little change in the induced RelB activity but found that cRel DNA-binding activity was enhanced in amplitude at 12 hr as well as prolonged in duration up to 40 hr. Quantitation of the data confirmed that costimulation provides for “superactivated” cRel, but not RelB (Figure 2F, right). Interestingly, RelA-containing dimers are rapidly induced upon solitary anti-IgM or BAFF treatment, but basal levels are restored within roughly 6 hr. And like RelB, these kinetics are nearly identical in costimulated conditions. We examined expression of these NFκB family members at both the mRNA and the protein level (Figures S3A and S3B) but found little to no increase in the costimulation state compared to individual BCR or BAFF-R stimulation, in contrast to the results of the DNA-binding activity analysis.
Differential Requirements for cRel and RelB in Resting and Proliferating B Cells
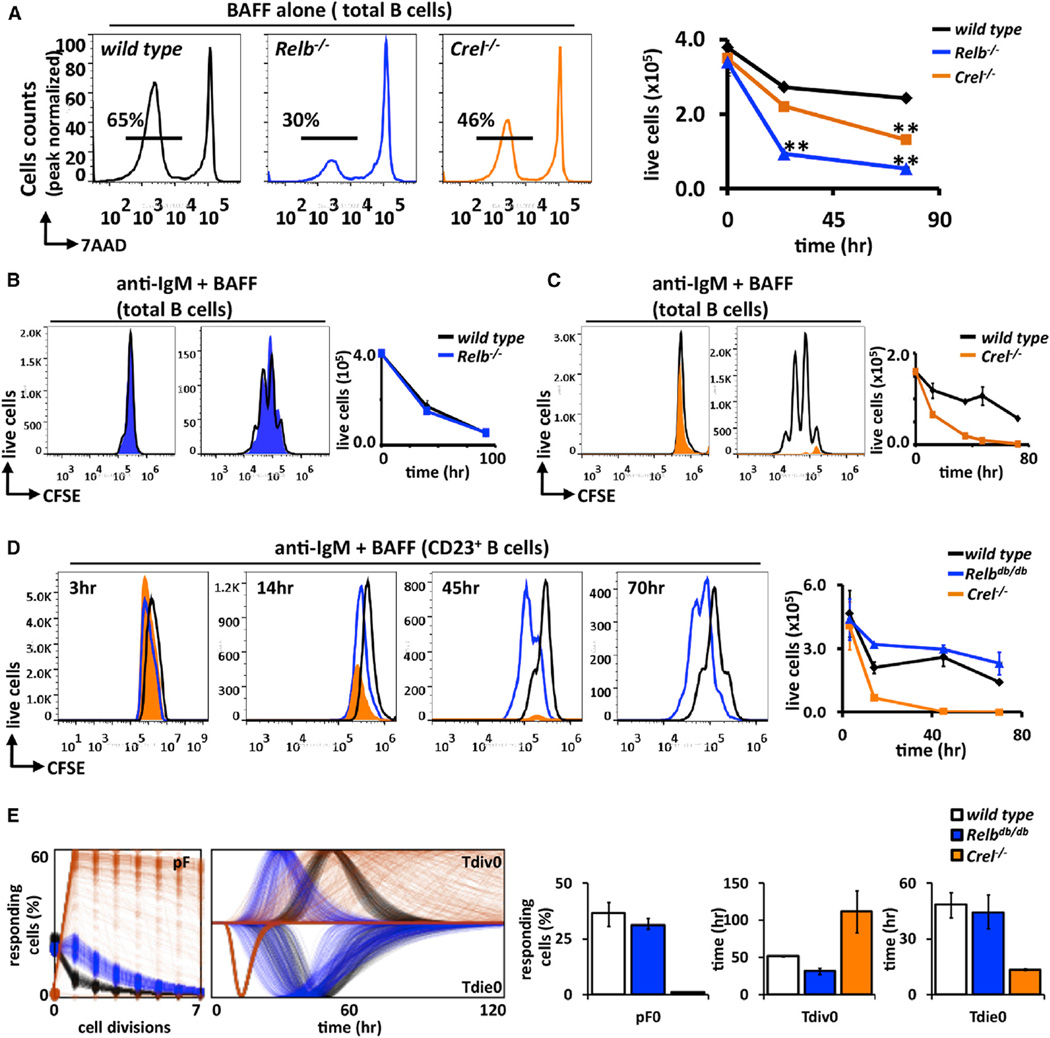
We then investigated the genetic requirement of cRel and RelB NFκB family members in mediating BAFF’s functions in B cells. Mature follicular B cells subsets are the strongest responders to anti-IgM-induced proliferative signals, and their development is unaffected by lack of either cRel or RelB, though RelB−/− mice lack marginal zone B cells (Weih et al., 2001). We first examined in vitro survival of total B cell population cultured in BAFF to find significant impairment in the absence of RelB (Figure 3A) and also a reduction, though less severe, in the absence of cRel. Charting a longer time course confirmed the importance of RelB and, to a lesser extent, cRel in BAFF-R-induced survival of B cells in vitro (Figure 3A, right). We note, however, that in vivo Relb and also Ikk1 appear to be redundant with other pathways, as the respective knockouts do not show deficiencies in B cell numbers (Jellusova et al., 2013). Turning to BAFF’s role as a costimulus during BCR-triggered B cell expansion, fluorescence-activated cell sorting (FACS) analysis of the CFSE-labeled total B cell population revealed that enhanced B cell expansion in response to costimulation did not require RelB (Figure 3B) but was dependent on cRel (Figure 3C), mimicking B cell responses to solitary BCR stimulation (Cheng et al., 2003; Grumont et al., 1998, 1999).
Figure 3. Costimulation of BCR and BAFF-R Result in Activation of cRel and RelB Dimers, but Enhanced Expansion Phenotype Is Dependent Solely on cRel.
(A) Representative FACS plot of B cells derived from cRel (orange) and RelB (blue)-deficient mice cultured with BAFF ligand for 3 days. Percentage of surviving B cells (7AAD−) was assessed by FACS and graphed on left; wild-type (black), Relb−/− (blue) and Crel−/− (orange).
(B and C) Total splenic wild-type, Relb−/−, and Crel−/− B cells are stained with CFSE and then stimulated with anti-IgM plus BAFF. Cell proliferation is examined at the indicated time points by FACS.
(D) Time course for CFSE-labeled magnetically isolated follicular B cells (CD23+) from wild-type, Relbdb/db, and Crel−/− B cells stimulated with anti-IgM + BAFF ligand. Live cells numbers were identified by gating out 7AADHi population and graphed on left.
(E) FlowMax analysis of CFSE proliferation profiles for CD23+ wild-type, Relbdb/db, and Crel−/− B cells in (D). Bar graphs of pF0, Tdiv0, and Tdie0 with the lognormal SD provided.
FACS plots in (A)–(D) are representative of at least two independent experiments. Error bars represent SD; n = 3. *p < 0.05; **p < 0.01 by Student’s t test.
See also Figure S2.
To substantiate these conclusions, we utilized a newly developed mouse model for RelB dysfunction, in which RelB’s ability to bind DNA was inactivated by substitution of three amino acids identified based on protein structure data. These RelBdb/db mice phenocopy RelB−/− in all aspects examined thus far (data not shown). We isolated RelBdb/db follicular B cells to ~85% purity (Figure S2) and compared their proliferative responses with wild-type or Crel−/− CD23+ follicular B cells upon BAFFR+BCR costimulation. Analysis of CFSE-stained follicular B cells revealed substantially enhanced B cell expansion in both wild-type and Relbdb/db, but not in Crel−/−, genotypes (Figure 3D). Total cell number measurements reflect this requirement, as Crel−/− B cells decline much more rapidly than wild-type controls and Relbdb/db shows no obvious deficiency in costimulation conditions (Figure 3D, right). Computational deconvolution of the CFSE data confirmed that cRel is required not only for allowing B cells to enter the proliferative program (pFs) but also for Tdie, whereas RelB deficiency had no discernible effect (Figure 3E).
In sum, these studies indicate a differential requirement for cRel and RelB for BAFF’s physiological function. When B cells are cultured in BAFF, its survival signals are mediated in part by RelB; however, when BAFF functions as a costimulus to BCR, we observed superactivation of cRel, which mediates the enhanced population expansion by increasing the proportion of cells entering the proliferative program.
BAFF Relieves p100/IκBδ Termination of cRel Activity
These observations seemed reminiscent of noncanonical pathway activation of NFκB RelA observed in LTβ-treated fibroblasts (Basak et al., 2007) and led us to hypothesize that p100 not only functions as a precursor for p52 generation via processing but also for oligomerization to form the high-molecular-weight IκBsome complexes that contain IκBδ activity (Figure 4A). When p100 expression is high due to BCR-stimulated RelA and cRel activity, the latter mechanism might lead to post-induction repression of cRel akin to IκBδ-mediated repression of lipopolysaccharide-induced RelA activity in mouse embryonic fibroblasts (MEFs) (Shih et al., 2009). BAFF as a costimulus with IgM might thus neutralize the IκBδ-mediated negative feedback to prolong canonical NFκB responses.
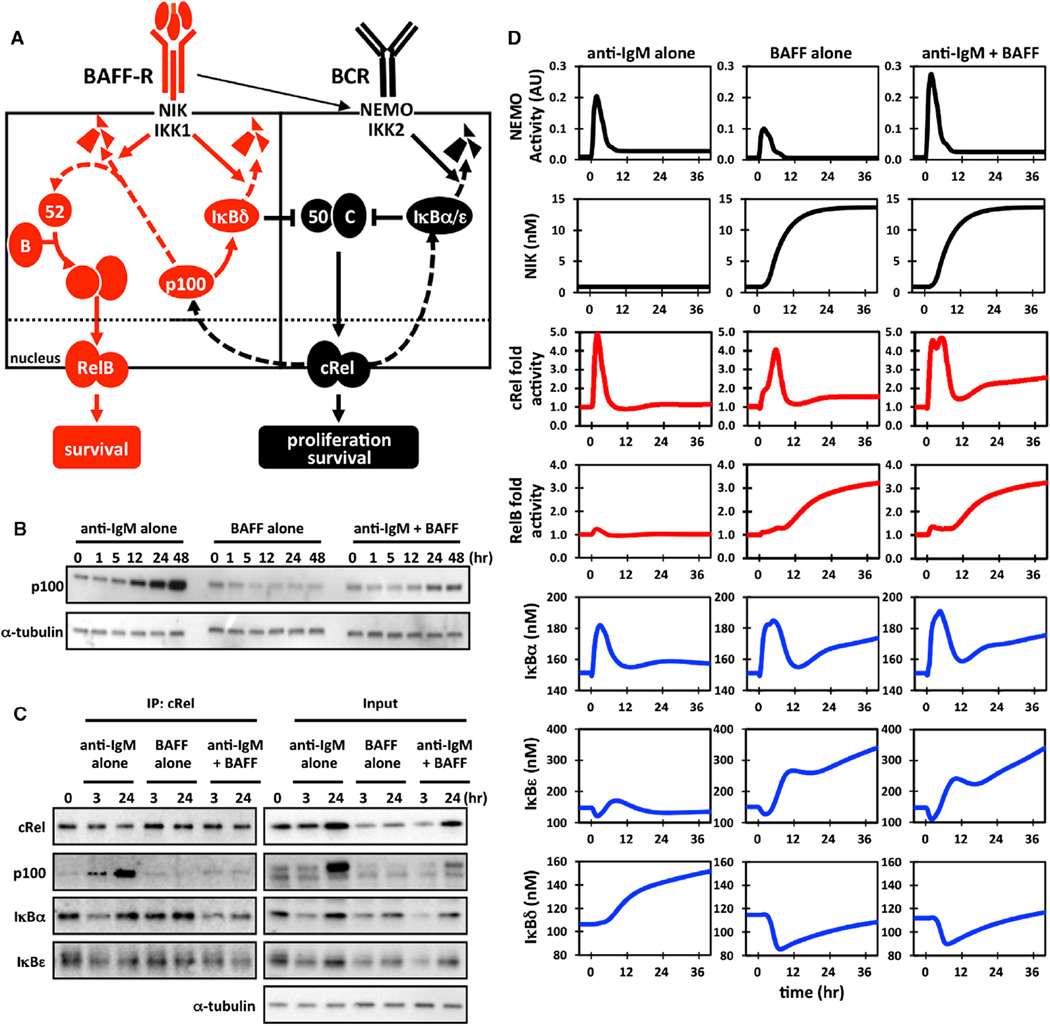
Figure 4. BAFF Releases IκBδ Inhibition of cRel in BCR-Stimulated B Cells.
(A) Proposed mechanism of IκBδ inhibition of cRel in BCR-stimulated B cells. In resting B cells, BAFF-R stimulation activates the NIK/IKK1 kinase complex that results in p100 processing, which allows for RelB:p52 nuclear translocation (Figure 3A). In activated B cells, continuous BCR stimulation activates cRel dimers, resulting in the expression of p100, which functions as a substrate for oligomerization to form the inhibitor IκBδ, which binds cRel, forming a negative feedback loop. During BAFF costimulation, the p100 buildup is prevented, allowing for persistent “superactivated” cRel.
(B) Cytoplasmic protein levels of p100 in stimulated wild-type B cells were measured by immunoblot.
(C) Immunoblots of cRel coimmunoprecipitates prepared from wild-type B cells stimulated with anti-IgM, BAFF ligand, or anti-IgM and BAFF. Samples were normalized to cRel protein levels to allow for comparison of distinct conditions (left). Whole-cell lysates (input) are also presented. IP, immunoprecipitation.
(D) Computational simulations are shown for the NEMO/IKK2-inducing stimulus anti-IgM (left column), for the NIK/IKK1-inducing stimulus BAFF (middle column), and a combination of both (right column), namely NEMO/IKK2 and NIK/IKK1 activity profiles (top, black), NFκB family members cRel and RelB nuclear fold activities (middle, red), and total cellular protein levels of IκBα, IκBε, and ανδ p100 (IκBδ substrate; bottom, blue).
Gel images in (B) and (C) are representative of at least four experiments.
See also Figures S3–S6 and Tables S1–S3.
To examine this hypothesis, we first showed that the three canonical IκBs, IκBα, IκBβ, and IκBε, showed characteristic degradation at early and induction at late time points in response to anti-IgM was unchanged when BAFF was applied as a costimulus (Figure S3C) as was the IKK activity profile (Figure S3D). In contrast, p100 was not subjected to IgM-mediated initial degradation but rather was induced from 12 hr on, and this late induction was almost completely absent in costimulation conditions (Figure 4B). Further, cRel immunoprecipitates revealed substantial p100 interaction following 24 hr of IgM stimulation, but this was largely absent in the costimulation condition (Figure 4C). This time point coincides with the presence of superactivated cRel observed by electrophoretic mobility shift assay (EMSA; Figure 2F). In contrast, IκBα and IκBε interaction with cRel was similar between IgM-stimulated and BAFF-costimulated cells, with similar transient reduction at the early 3 hr time point. In sum, these observations support the hypothesis that p100 forms an inhibitory IκBδ-containing complex, which limits cRel activity during BCR-triggered B cell expansion and which may be neutralized by BAFF-costimulation to prolong cRel activation.
To test this hypothesis more quantitatively (and thus determine whether it provides a sufficient explanation) and develop a research tool that may complement wet lab experimentation, we constructed a mathematical model of the NFκB-signaling system in B cells, capable of responding to both canonical and noncanonical signals induced by BCR and BAFFR. Development of the mathematical model began with delineation of the model’s topology, based on previously established models of lesser scope. We distinguish between reactions that control IκB synthesis and degradation (Figure S4A) and those that control NFκB monomer synthesis, dimerization, and interactions of dimers with IκBs (Figure S4B). The former represents an extension of the previously published four-IκB model (Basak et al., 2007; Shih et al., 2009), and the latter is an extension of a model of RelA and RelB dimers in dendritic cells (Shih et al., 2012) to include cRel as well (see also Tables S1 and S2). Thus, we account for all p50- and p52-containing dimers and the associated 19 NFκB-IκB complexes. The two topologies are connected not only via protein-protein interactions but also via NFκB-dependent synthesis control and the dual roles of the Nfkb2 gene product p100 as a precursor not only for the NFκB dimerization partner p52 but also for the IκBδ activity contained within the multimeric IκBsome. Data in Figures 1, 2, and 3 and quantitation of polypeptides (Figure S5; Table S3) provide an extensive training set to achieve a B-cell-specific parameterization.
To account for cell-to-cell heterogeneity of B cell responses, each condition was simulated with distributed kinase input profiles that may result from heterogeneous BCR-induced signaling (Shinohara et al., 2014; Figure S6); their average conforms to the experimentally measured profiles of IKK activity (Figure S3D). The similarly averaged simulations of IκB and NFκB dynamics show that cRel-containing dimers are transiently induced, peaking at about 5 hr before returning to basal levels in response to BCR or BAFF-R stimulation alone (Figure 4D, third row), whereas RelB-containing dimers are induced late and in a sustained and BAFF-R-dependent manner (Figure 4D, fourth row); these simulation outputs match our biochemical analysis at late times (Figure 3A), though early RelB activity observed experimentally eludes the present version of the model. Finally, simulation of the costimulation scenario (Figure 4D, third column) reveals a RelB profile that is surprisingly unchanged from the BAFF-alone condition and a cRel profile that shows superactivation at both early (1–4 hr) and late times (20–40 hr). Whereas early superactivation coincides with elevated NEMO-IKK activity, late superactivation does not. Instead, late cRel superactivation coincides with a BAFF- and NIK-dependent reduction in p100/IκBδ abundance at late times, which neutralizes its IgM-driven hyperexpression. In sum, mathematical modeling demonstrates that kinetic considerations alone are sufficient to account for context-specific NFκB effectors of BAFF-R signaling without the need to invoke context-specific molecular mechanisms.
p100/IκBδ Limits cRel-Mediated B Cell Expansion
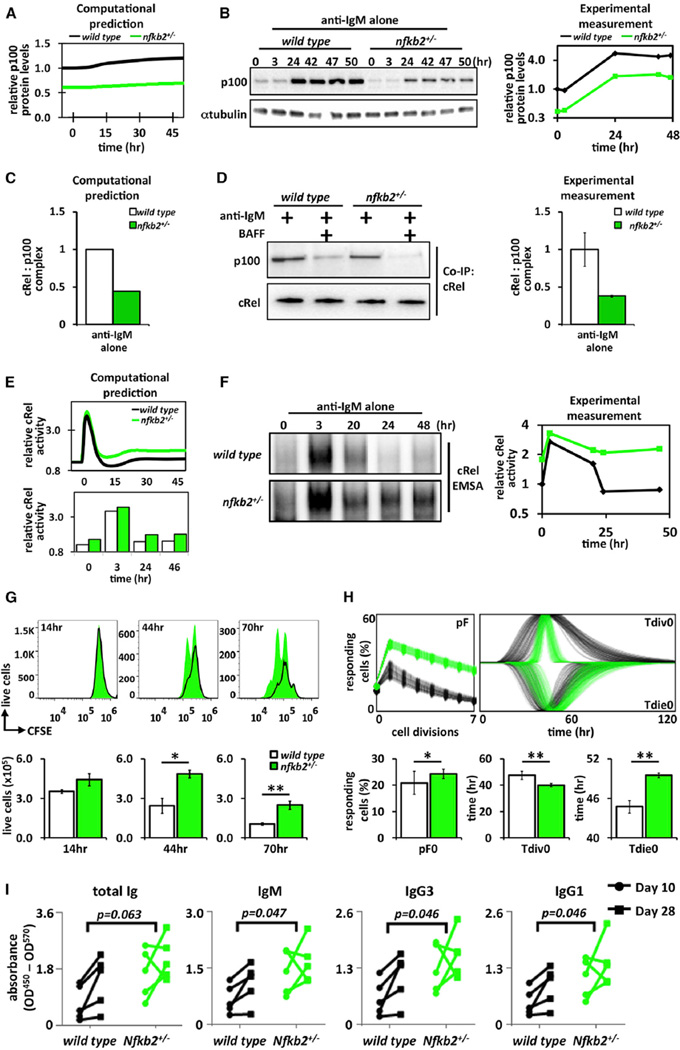
The mathematical model encapsulates the key hypothesis that BAFF functions to relieve IκBδ-mediated termination of cRel activity. Model simulations suggested that a 2-fold reduction in the Nfkb2 mRNA synthesis rate, akin to Nfkb2 heterozygosity, would result in reduced p100 protein in both basal and IgM-stimulated conditions in silico (Figure 5A), which was confirmed in cells (Figure 5B). Unlike Nfkb2−/− mice, we found that Nfkb2+/− mice showed normal follicular B cell populations (Figures S7A and S7B). Further, the model predicted that the amount of inhibited cRel-p100 complex following 24 hr of anti-IgM stimulation would be reduced in Nfkb2+/− B cells compared to the wild-type controls (Figure 5C). Indeed, quantitative cRel coimmunoprecipitation revealed severely diminished p100 interaction in anti-IgM-treated Nfkb2+/− cells akin to those in BAFF-treated wild-type cells (Figure 5D). Next, model simulations predicted that Nfkb2 heterozygosity would enhance cRel activity at late times (24–48 hr; Figure 5E). Biochemical analysis confirmed this prediction, showing increased cRel DNA-binding activity at late times as compared to wild-type cells (Figure 5F). These studies establish the expression level of p100 as a remarkably sensitive parameter in terminating NFκB activity via the presumed multimeric IκBδ.
Figure 5. IκBδ Limits the Proliferative Capacity of BCR-Stimulated B Cells.
(A) Model simulation of fold p100 protein levels (log2) in wild-type (black) and Nfkb2 heterozygous (green) B cells stimulated with anti-IgM alone.
(B) Immunoblot of p100 protein expression in wild-type and Nfkb2+/− B cells stimulated with anti-IgM alone. Blots were quantified and plotted (log2, right).
(C) Computational prediction of p100:cRel complex following 24 hr of anti-IgM stimulation in wild-type (black) and Nfkb2+/− (green) B cells.
(D) Representative immunoblot blot for coimmunoprecipitation of p100:cRel complex from wild-type and Nfkb2+/− B cells following 24 hr of stimulation with either anti-IgM alone or anti-IgM plus BAFF ligand. Samples normalized to cRel protein levels (bottom). Immunoblots were quantified and graphed (right).
(E) Model simulation of nuclear cRel protein levels in wild-type and Nfkb2 heterozygous B cells stimulated with anti-IgM alone (log2).
(F) cRel DNA-binding activities in wild-type and Nfkb2 heterozygous B cells induced by anti-IgM alone were monitored by EMSA. EMSA was quantified and graphed (log2, right).
(G) Time course of cell proliferation for CD23+ wild-type (black) and Nfkb2+/− (green) B cells labeled with CFSE and 7AAD.
(H) FlowMax analysis of CFSE proliferation assay in (G).
(I) T-dependent Ag responses in Nfkb2+/− mice immunized with NP-KLH, and serum levels of anti-NP total Ig and specific Ig isotypes at 10 and 28 days postimmunization measured by ELISA (n = 5). Indicated p values are calculated by two-way ANOVA.
Unless stated, all experiments in Figure 5 used total splenic B cells. Immunoblots and FACS plot for (B), (D), (F), and (G) are representative of at least three experiments. Error bars in (D) SD; n = 3. Bar graphs in (H) summarize best-fit cellular parameters with the lognormal SD; *p < 0.05; **p < 0.01.
See also Figure S7.
To examine the cell biological consequence of enhanced cRel activation in Nfkb2+/− B cells, we undertook CFSE dye dilution studies, which revealed that anti-IgM-stimulated Nfkb2+/− B cells showed enhanced B cell expansion over wild-type controls (Figure 5G). Computational deconvolution (Figure 5H) showed that this increased expansion was primarily due to increases in the fraction of cell entering the proliferative program (pF), moderately for generation zero (pF0) but more so for subsequent generations (pF2, pF3, pF4, etc.). Further, survival times (Tdie0) increased, mimicking costimulation conditions. Our biochemical analysis, combined with enhanced proliferative capacity, document that Nfkb2 heterozygous B cells possess a heightened BCR responsiveness in vitro. To test whether this phenotype holds true in vivo, we immunized Nfkb2+/− mice with 4-hydroxy-3-nitrophenyl keyhole limpet hemocyanin (NP-KLH) to induce a T-dependent antigen response and then examined antibody levels in sera (Figure 5I). We found a significant increase of NP-specific IgM, IgG3, and IgG1 levels in Nfkb2+/− mice compared with wild-type 10 and 28 days postimmunization. We also note an increase in total Ig Nfkb2+/− mice. These data are noteworthy because Nfkb2-knockouts, which lack not only IκBδ (deficient in Nfkb2+/−) but also survival-associated RelB:p52 (present in Nfkb2+/−), show reduced mature B cells and diminished antibody responses (Caamaño et al., 1998, Franzoso et al., 1998). Together, our data support the notion that the IκBδ-containing IκBsome plays a key role in terminating antigen-triggered B-proliferative responses and humoral immunity by limiting the duration of cRel transcription factor activity.
Nfkb1/p105 Stabilizes IκBδ to Control B Cell Expansion
To further test the model that the IκBδ-containing IκBsome is critical for terminating cRel-driven B-cell-proliferative responses, we sought ways to perturb IκBsome formation. Previous biochemical studies described an assembly pathway for high-molecular-weight IκBsome involving mutually stabilizing interactions between the Nfkb1 and Nfkb2 gene products p105 and p100 (Savinova et al., 2009). In addition, p50 and p52 have been described to compete for a limited pool of activation-domain-containing NFκB proteins, RelA, cRel, and RelB (Basak et al., 2008). Both mechanisms determine the balance between IκBsome formation by p100 and its processing to p50/p52. These interdependencies in p100 and p105 (Figure 6A) allow for compensation for the absence of p50 by p52 (Hoffmann et al., 2003) but with concomitant loss of p100 and lymphotoxin-beta receptor responsiveness in Nfkb1−/− MEFs (Basak et al., 2008) and lymph node formation in Nfkb1−/− mice (Lo et al., 2006).
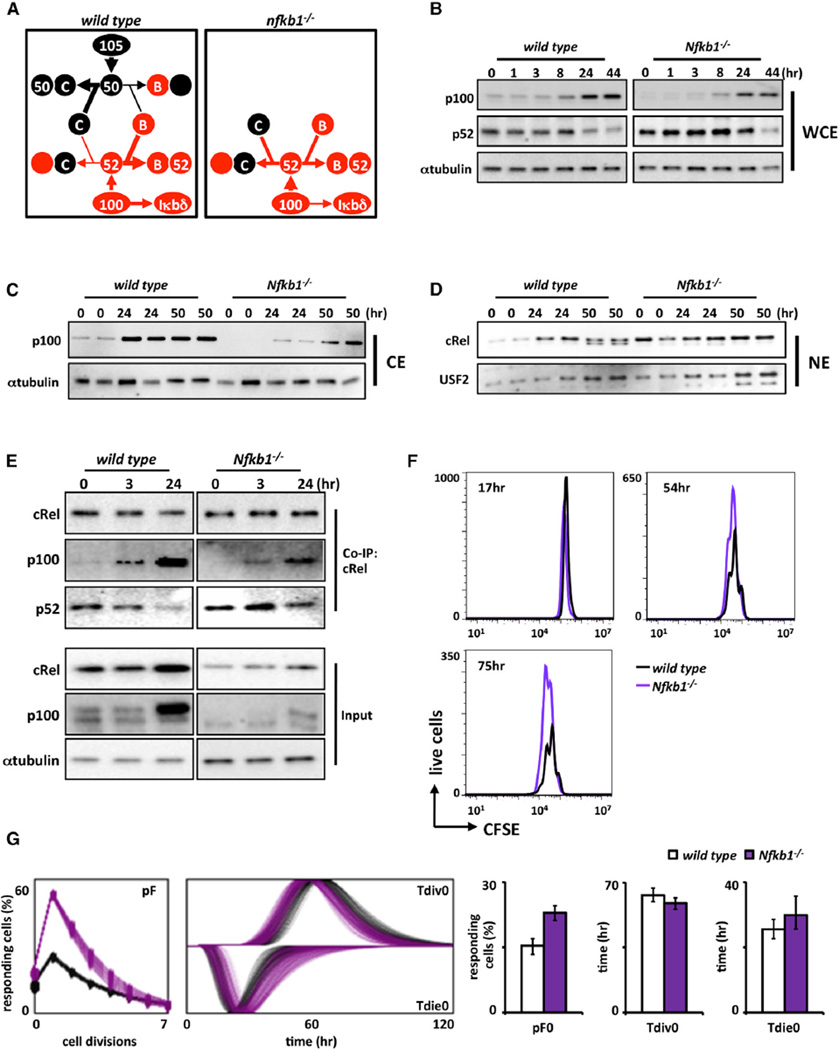
Figure 6. Nfkb1/p105 Limits the Proliferative Capacity of B Cells by Stabilizing p100/IκBδ.
(A) Proposed schematic of Nfkb1 and Nfkb2 protein fates in wild-type and Nfkb1−/− B cells. In normal wild-type B cells, p105 and p100 mainly function in the NFκB-signaling system as precursors of p50 and p52. p105 is processed into p50 and dimerizes predominantly with cRel and RelA and, to a lesser extent, RelB. p100 processing yields p52, which in turn dimerizes with RelB primarily. A second function of p100 is to form the inhibitory molecule IκBδ (left). In Nfkb1−/− B cells, p105/p50 absence leaves cRel and RelA molecules without their primary binding partner. This in turn shifts the available pool of p100 to be processed into p52 in order to overcome the loss of p50; as a result, less p100 is accessible to form IκBδ (right).
(B) Immunoblots of p52 whole-cell expression in wild-type and Nfkb1−/− B cells stimulated with anti-IgM. WCE, whole-cell extract.
(C) Immunoblots of cytoplasmic p100 expression in wild-type and Nfkb1−/− B cells stimulated with anti-IgM. CE, cytoplasmic extract.
(D) Immunoblot of nuclear cRel for wild-type and Nfkb1−/− B cells stimulated with anti-IgM. Nuclear marker USF2 is used as a loading control. NE, nuclear extract.
(E) Immunoblots of immunoprecipitates to monitor p100/IκBδ (and p52) associated with cRel during BCR stimulation time course. cRel was immunoprecipitated from B cell whole-cell extracts and prepared at indicated time points. Whole-cell lysates (input) are also shown; α-tubulin serves as loading control.
(F) FACS analysis for in vitro proliferation of wild type (black) and Nfkb1−/− (purple) B cells labeled with CFSE and stimulated for 3 days with anti-IgM alone. Live-cell numbers gated by exclusion of 7AADHi population.
(G) FlowMax analysis of CFSE proliferation profiles for wild-type (black) versus Nfkb1−/− (purple) B cells in (F).
Immunoblots and FACS plots for (B)–(F) are representative of at least three experiments. Bar graphs in (G) summarize best-fit cellular parameters with the lognormal SD.
See also Figure S7.
We examined whether Nfkb1 might be required for efficient IκBδ function during IgM-triggered B cell expansion. Examination of B cell populations in Nfkb1-deficient mice was similar to wild-type controls (Figure S7C), and our quantitative gene-expression analysis confirmed that the steady-state and inducible levels of Nfkb2 mRNA are similar in Nfkb1−/− and wild-type B cells (Figure S7D). However, immunoblot analysis of Nfkb1−/− B cell extracts indicated lower levels of p100 protein and elevated levels of p52 (Figure 6B). This decrease in the available p100 pool was also evident in the cytoplasmic fraction (Figure 6C). Immunoblotting nuclear fractions, we detected elevated cRel levels in BCR-stimulated Nfkb1−/− B cells over wild-type counterparts (Figure 6D). These data suggest that Nfkb1 deficiency destabilizes p100-containing IκBδ to neutralize its inhibition of cRel activity. Coimmunoprecipitation analysis confirmed that p100 was bound to a lesser extent to cRel in anti-IgM-treated Nfkb1−/− B cells than wild-type controls (Figure 6E).
Next, we examined BCR-triggered B cell expansion with CFSE dye dilution analysis. FACS analysis revealed enhanced proliferative responses in Nfkb1−/− B cells compared to wild-type controls (Figure 6F). Computational deconvolution identified that the phenotype was primarily due to a greater fraction of cells entering the proliferative program with only minor changes in cell division or survival times (Figure 6G). Although Nfkb1/p50 is a known dimerization partner of cRel, our data suggest that Nfkb1’s primary role in antigen-responsive B cell expansion is to limit the proliferative program by Nfkb1/p105-stabilizing IκBδ.
DISCUSSION
The tumor necrosis factor (TNF) superfamily member BAFF has diverse roles during B lymphocyte development, homeostasis, and pathogen-responsive expansion. Using an iterative systems biology approach, we identified two distinct molecular mechanisms and transcriptional effectors that mediate them. Whereas BAFF’s survival function in maturing B cells is mediated by the noncanonical NFκB pathway effector RelB, its function as a costimulus in antigen-responsive B cell expansion is mediated by superactivation of cRel that enhances the fraction of cells entering the proliferative program. Our studies reveal Nfkb2 /p100 as the critical pathway switch, as it functions as the precursor not only for protein processing to p52 but also for IκBδ and IκBsome assembly. Our data support a model in which the multimeric IκBδ activity limits the cRel-driven-proliferative program, and BAFF activation of the noncanonical pathway neutralizes the IκBδ negative feedback loop.
Previous studies established that BAFF engages the noncanonical NFκB-signaling pathway, which activates NFκB activity via NEMO-independent kinases NIK and IKK1. A hallmark of noncanonical NFκB activity is RelB:p52 activation. Although NIK, cRel, and RelA have been shown to be required for normal B cell homeostasis, no genetic evidence pointed to the necessity of RelB. Here, we showed that BAFF-induced survival of resting splenocytes in vitro is indeed RelB dependent (Figure 3B).
However, when employing BAFF as a costimulus during antigen-receptor-triggered B cell expansion, we found that RelB was dispensable (Figure 3D), prompting us to search for other relevant effectors. One clue was provided by the unbiased, software-aided analysis of the CFSE dye dilution data of BCR and BAFF/BCR-costimulated B cells: enhanced expansion was largely due to an increased fraction of cells entering the cell division program, particularly at later generations. B cell proliferation is known to be mediated by cRel (Grumont et al., 1998), and we found dramatic superactivation of cRel in costimulation conditions but little enhancement in RelB activity. Further, our transcriptomic studies identified enrichment of cRel-binding regulatory motif (Figures 2A and 2C) and genetic cRel dependency (Figure 2E) in genes with enhanced expression during costimulation.
The TNF superfamily receptors, BAFF-R and transmembrane activator and CAML interactor (TNFRSF13B) (TACI), are expressed on all B cell subsets and mediate BAFF function (Shulga-Morskaya et al., 2004). Whereas TACI is known to activate the canonical NFκB pathway (Marsters et al., 2000), recent studies have also implicated BAFF-R (Enzler et al., 2006) and shown that BAFF stimulation of Taci−/− B cells also leads to IκBα degradation and canonical pathway activation (Shinners et al., 2007). Although early cRel activation (3–8 hr) may be attributed to such canonical activation by TACI and BAFF-R, late cRel activity (24–48 hr) only appears in costimulation conditions (Figure 3A).
How does BAFF-R allow for late superactivation of cRel in proliferating B cells? One mechanism might be the generation of a cRel:p52 dimer as a result of enhanced p100 processing; we indeed saw increased amounts of p52 bound to cRel in coimmunoprecipitation experiments (data not shown), though this remained a minor component of the DNA-protein complex found in EMSAs. The primary mechanism appeared to be the derepression of cRel activity via the previously described IκBδ activity, which results from oligomerized Nfkb2 p100 (Basak et al., 2007) within the high-molecular-weight IκBsome (Savinova et al., 2009). Indeed, progressive maturation of the IκBsome to a high-molecular-weight complex with potent NFκB inhibitor function begins with RelA-responsive expression of both p100 and p105 and requires their mutual interactions (Basak et al., 2007; Savinova et al., 2009; Shih et al., 2009).
By describing the signaling network in mathematical terms, we could show that known molecular mechanisms and kinetic rate constants (experimentally determined or inferred within a reasonable range) are sufficient to account for the observed molecular and genetic phenomena in both maturing and proliferating B cells. Further, using computational simulations, we developed hypotheses that distinguished the IκBsome model from the alternative RelB:p52 model. Testing these hypotheses experimentally, we showed (1) that reduced p100 expression levels (with Nfkb2 heterozygous B cells) result in elevated levels of nuclear cRel at late times and increased B cell proliferation (rather than decreased proliferation that would be expected if RelB:p52 was the relevant effector) and (2) that disruption of the IκBsome assembly pathway (due to p105 deficiency) would also relieve cRel inhibition, resulting in enhanced expansion and mimicking the costimulated state. Our results do not contradict previous work that tonic canonical NFκB signaling in resting B cells is required for noncanonical RelB:p52 activation (Shinners et al., 2007; Stadanlick et al., 2008). The quantitative description of the network clarifies that low-RelA/cRel activity is required to generate p100 as a substrate for processing to p52 (as well as RelB; Basak et al., 2008) for RelB:p52 generation (Shinners et al., 2007; Stadanlick et al., 2008) but that elevated p100 production, as it occurs in proliferating conditions, results in an excess of p100 molecules that then assemble into the IκBsome, which BAFF in turn neutralizes.
Our work here on the IκBsome’s role in lymphocyte expansion may be a description of its physiological significance. Future studies may implicate it in other physiological functions. Indeed, phenotypes associated with C-terminal deletions of the Nfkb1 and Nfkb2 genes reported many years ago (Caamaño et al., 1998; Migliazza et al., 1994) may best be understood in terms of disrupted IκBsome assembly. Further, cancer-associated mutations in B cell lymphoma or multiple myeloma affecting the noncanonical signaling pathway likely function by disrupting the normal buffering functions of the IκBsome (Busino et al., 2012; Hailfinger et al., 2009; Lim et al., 2012).
EXPERIMENTAL PROCEDURES
B Cell Isolation, Proliferation, and Survival Analysis
Spleens were harvested from C57Bl6 wild-type or littermate Relb−/−, RelBdb/db, Crel−/−, Nfkb1−/−, and Nfkb2+/− mice. For B cell isolation, homogenized splenocytes were incubated with anti-CD43 microbeads for 15 min at room temperature, washed with FACS buffer, and separated on LS column (Miltenyi Biotec). Purity was confirmed to be ~92%–95%. Marginal Zone and Follicular B Cell Isolation Kit (Miltenyi Biotec) was used for separation of follicular B cells from whole splenocytes, with a purity of 83%–87% CD23+ follicular B cells (Figure S2). Complete media consisted of RPMI-1640, 10 mM HEPES, 0.055 mM β-mercaptoethanol, penicillin and streptomycin, and l-glutamine. B cells were stimulated with 5 µg/ml of Fab anti-mouse IgM (Jackson Immunology Research) and/or 50 ng/ml recombinant mouse BAFF (R&D Systems). In vitro proliferation assay was performed using CFSE tracking dye as described (Hawkins et al., 2007). Data were collected on a C6 Accuri (BD Biosciences) or FACSCalibur and analyzed with FlowJo software and FlowMax (Shokhirev and Hoffmann, 2013). B cell survival was determined by 7ADD staining using FlowJo for data analysis.
Biochemical Assays
Immunoblotting analysis, EMSAs, and kinase assays were conducted with standard methods as described previously (Shih et al., 2012; Werner et al., 2005) and antibodies specific for RelA (Santa Cruz Biotechnology; sc372), RelB (sc226), cRel (sc71), IκBα (sc371), IκBε (sc7155), α-tubulin (sc226), p100, and p105 (Dr. Nancy Rice). For EMSAs focusing on cRel-DNA-binding activity, nuclear extracts are preincubated with RelA and cRel antibodies for 15 min on ice prior to addition of radiolabeled probe to ablate their respective DNA-binding activities. Similarly, nuclear extracts were preincubated with RelA and cRel antibodies or RelB and cRel antibodies when RelB- or RelA-DNA-binding activity was the focus, respectively. RNAeasy Mini Kit (QIAGEN) was used to purify RNA. cDNA synthesis performed with iScript cDNA Synthesis Kit (Bio-Rad). Quantitative RT-PCR was performed with SYBR Green PCR Master Mix reagent using the Δ(ΔCt) method with β-actin as normalization control, relative to unstimulated signals in wild-type B cells to derive fold induction.
RNA-Seq Analysis
Total RNA was isolated from 50 ng/ml BAFF-stimulated wild-type B cells following 8 and 30 hr of culture. mRNA was extracted from 2 µg total RNA using oligo (dT) magnetic beads and fragmented at high temperature using divalent cations. cDNA libraries were generated using the IlluminaTruSeq kits, and quantitation was performed using the Roche Light Cycler 480. Sequencing was performed on Illumina’s HiSeq 2000, according to manufacturer’s recommendations, and prepared for RNA sequencing analysis by BIOGEM core facility at University of California, San Diego. Reads were aligned to the mouse mm10 genome and RefSeq genes (Kent et al., 2002; Waterston et al., 2002) with Tophat (Trapnell et al., 2009). Cufflinks CummRbund (Trapnell et al., 2012) was used to ascertain differential expression of genes. Gene differential fragments per kilobase of transcript per million mapped reads were obtained from the cuffdiff program in Tuxedo RNA-seq analysis suite.
Immunization
The primary immune response to T-dependent Ag was assessed after intraperitoneal immunization with 50 µg of NP-KLH precipitated in alum. Sera were collected 10 and 28 days after challenge and tested by ELISA. Briefly, plates were coated with 10 µg/ml NP-BSA (Biosearch Technologies) and sera were assessed at 1/100,000 dilution. Bound antibodies (Abs) were detected with HRP-conjugated Abs to total mouse Ig or specific mouse isotypes (eBioscience and Santa Cruz Biotechnology).
Computational Modeling
The ordinary differential equations model of multi-NFκB control was adapted from previous iteration of the B cell IκB-NFκB-signaling model (Alves et al., 2014) and a previous version of dendritic cell IκB-NFκB-signaling model (Shih et al., 2012) to account for activities of RelA:p50, RelA:p52, RelB:p50, RelB:p52, cRel:p50, and cRel:p52 dimers (Table S1). Parameterization of the reaction of the model (Table S2) included quantitative measurements of B cell protein abundances (Table S3). The model describes the single-cell response within a population of B cells; variability of NFκB response among cells is assumed to be due to variability (log normally distributed) in the activation of NEMO-IKK2. Based on FACS experiments, the fraction of 10% BCR-responsive cells was derived. In response BAFF stimulation, we assume all cells accumulate NIK and have low log-normally distributed NEMO-IKK2 activity. Hence, costimulation results in 20% cells with NEMO-IKK2 activity, and 80% have BAFF-induced NEMO-IKK2 activity. To allow comparison with experimental measurements of the bulk population, we graphed the average of 1,000 simulations of single B cells. The ordinary differential equations were solved numerically using MATLAB version R2013a (The MathWorks) with subroutine ode15s. Following system equilibration, anti-IgM and BAFF stimulation was simulated with numerical NEMO-IKK input curves (adapted from Werner et al., 2005) and by decreasing the NIK degradation rate constant by 30-fold at 5 hr. MATLAB model codes are available upon request.
Animal Use
The animal protocols for this study were approved by the University of California, San Diego Animal Care and Use Committee. Generation of the RelBdb/db mouse strain containing mutations of residues required for DNA-binding R117A/Y120A/E123A will be described elsewhere.
Supplementary Material
In Brief.
Almaden et al. show that BAFF, an activator of the noncanonical NFκB pathway, provides critical B cell survival signals via RelB or contributes to B cell proliferation via cRel. An iterative systems biology approach revealed Nfkb2 p100 as a pathway switch that directs noncanonical NFkB signaling to either RelB:p52 activation or potentiation of cRel:p50.
Highlights.
BAFF enhances in vitro survival of maturing B cells and aids B cell proliferation
BAFF’s activation of RelB is required for enhanced cell survival, but not division
In proliferating B cells, BAFF neutralizes p100-mediated termination of cRel
Disrupting IκBδ expression/assembly mimics BAFF’s costimulation of B cell expansion
Acknowledgments
We thank V. Shih, B. Alves, and M. Asagiri for experimental advice and S. Hedrick, C. Murre, and A. Goldrath for critical discussions. This study was funded by NIAID R01 AI083453, NCI R01 CA141722, NIGMS R01 GM071573, P01 GM071862, P50 GM085764 (to A.H.), NSF GTRF (to J.C.D.-T. and M.N.S.), and a Cell and Molecular Genetics Training Grant (to J.V.A.).
Footnotes
ACCESSION NUMBERS
The RNA-seq data presented in this publication have been deposited to the NCBI Gene Expression Omnibus and are accessible under accession number GSE54588.
Supplemental Information includes seven figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.11.024.
AUTHOR CONTRIBUTIONS
J.V.A., R.T., R.C.R., and A.H. designed the study. J.V.A. carried out all B cell experimental work; R.T. the computational modeling work; Y.C.L., J.V.A., and D.O. the immunization study; and S.B. quantitative immunoblotting. K.A.N. produced the mouse strain harboring the RelB DNA-binding mutation. H.B. and J.V.A. carried out the gene expression analysis. M.N.S. and J.V.A. carried out the CFSE data analysis with FlowMax. J.C.D.-T. contributed to the computational work. J.V.A., R.T., and A.H. wrote the manuscript.
REFERENCES
- Allman D, Lindsley RC, DeMuth W, Rudd K, Shinton SA, Hardy RR. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J. Immunol. 2001;167:6834–6840. doi: 10.4049/jimmunol.167.12.6834. [DOI] [PubMed] [Google Scholar]
- Alves BN, Tsui R, Almaden J, Shokhirev MN, Davis-Turak J, Fujimoto J, Birnbaum H, Ponomarenko J, Hoffmann A. IκBε is a key regulator of B cell expansion by providing negative feedback on cRel and RelA in a stimulus-specific manner. J. Immunol. 2014;192:3121–3132. doi: 10.4049/jimmunol.1302351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Kim H, Kearns JD, Tergaonkar V, O’Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Shih VF, Hoffmann A. Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system. Mol. Cell. Biol. 2008;28:3139–3150. doi: 10.1128/MCB.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Millman SE, Scotto L, Kyratsous CA, Basrur V, O’Connor O, Hoffmann A, Elenitoba-Johnson KS, Pagano M. Fbxw7α- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat. Cell Biol. 2012;14:375–385. doi: 10.1038/ncb2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caamaño JH, Rizzo CA, Durham SK, Barton DS, Raventós-Suárez C, Snapper CM, Bravo R. Nuclear factor (NF)-kappa B2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J. Exp. Med. 1998;187:185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancro MP. Signalling crosstalk in B cells: managing worth and need. Nat. Rev. Immunol. 2009;9:657–661. doi: 10.1038/nri2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Hsia CY, Leone G, Liou HC. Cyclin E and Bcl-xL cooperatively induce cell cycle progression in c-Rel−/− B cells. Oncogene. 2003;22:8472–8486. doi: 10.1038/sj.onc.1206917. [DOI] [PubMed] [Google Scholar]
- Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat. Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- Craxton A, Draves KE, Clark EA. Bim regulates BCR-induced entry of B cells into the cell cycle. Eur. J. Immunol. 2007;37:2715–2722. doi: 10.1002/eji.200737327. [DOI] [PubMed] [Google Scholar]
- Do RK, Hatada E, Lee H, Tourigny MR, Hilbert D, Chen-Kiang S. Attenuation of apoptosis underlies B lymphocyte stimulator enhancement of humoral immune response. J. Exp. Med. 2000;192:953–964. doi: 10.1084/jem.192.7.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzler T, Bonizzi G, Silverman GJ, Otero DC, Widhopf GF, Anzelon-Mills A, Rickert RC, Karin M. Alternative and classical NF-kappa B signaling retain autoreactive B cells in the splenic marginal zone and result in lupus-like disease. Immunity. 2006;25:403–415. doi: 10.1016/j.immuni.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Franzoso G, Carlson L, Poljak L, Shores EW, Epstein S, Leonardi A, Grinberg A, Tran T, Scharton-Kersten T, Anver M, et al. Mice deficient in nuclear factor (NF)-kappa B/p52 present with defects in humoral responses, germinal center reactions, and splenic microarchitecture. J. Exp. Med. 1998;187:147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumont RJ, Rourke IJ, O’Reilly LA, Strasser A, Miyake K, Sha W, Gerondakis S. B lymphocytes differentially use the Rel and nuclear factor kappaB1 (NF-kappaB1) transcription factors to regulate cell cycle progression and apoptosis in quiescent and mitogen-activated cells. J. Exp. Med. 1998;187:663–674. doi: 10.1084/jem.187.5.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumont RJ, Rourke IJ, Gerondakis S. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 1999;13:400–411. doi: 10.1101/gad.13.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, Penas EM, Dierlamm J, Chan WC, Staudt LM, Thome M. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harless SM, Lentz VM, Sah AP, Hsu BL, Clise-Dwyer K, Hilbert DM, Hayes CE, Cancro MP. Competition for BLyS-mediated signaling through Bcmd/BR3 regulates peripheral B lymphocyte numbers. Curr. Biol. 2001;11:1986–1989. doi: 10.1016/s0960-9822(01)00598-x. [DOI] [PubMed] [Google Scholar]
- Hawkins ED, Hommel M, Turner ML, Battye FL, Markham JF, Hodgkin PD. Measuring lymphocyte proliferation, survival and differentiation using CFSE time-series data. Nat. Protoc. 2007;2:2057–2067. doi: 10.1038/nprot.2007.297. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Di Liberto M, Cunningham AF, Kang L, Cheng S, Ely S, Liou HC, Maclennan IC, Chen-Kiang S. Homeostatic cell-cycle control by BLyS: Induction of cell-cycle entry but not G1/S transition in opposition to p18INK4c and p27Kip1. Proc. Natl. Acad. Sci. USA. 2004;101:17789–17794. doi: 10.1073/pnas.0406111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellusova J, Miletic AV, Cato MH, Lin WW, Hu Y, Bishop GA, Shlomchik MJ, Rickert RC. Context-specific BAFF-R signaling by the NF-κB and PI3K pathways. Cell Rep. 2013;5:1022–1035. doi: 10.1016/j.celrep.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KP, Kühn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- Lim KH, Yang Y, Staudt LM. Pathogenetic importance and therapeutic implications of NF-κB in lymphoid malignancies. Immunol. Rev. 2012;246:359–378. doi: 10.1111/j.1600-065X.2012.01105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litinskiy MB, Nardelli B, Hilbert DM, He B, Schaffer A, Casali P, Cerutti A. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat. Immunol. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo JC, Basak S, James ES, Quiambo RS, Kinsella MC, Alegre ML, Weih F, Franzoso G, Hoffmann A, Fu YX. Coordination between NF-kappaB family members p50 and p52 is essential for mediating LTbetaR signals in the development and organization of secondary lymphoid tissues. Blood. 2006;107:1048–1055. doi: 10.1182/blood-2005-06-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay F, Schneider P. Cracking the BAFF code. Nat. Rev. Immunol. 2009;9:491–502. doi: 10.1038/nri2572. [DOI] [PubMed] [Google Scholar]
- Mackay F, Silveira PA, Brink R. B cells and the BAFF/APRIL axis: fast-forward on autoimmunity and signaling. Curr. Opin. Immunol. 2007;19:327–336. doi: 10.1016/j.coi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Marsters SA, Yan M, Pitti RM, Haas PE, Dixit VM, Ashkenazi A. Interaction of the TNF homologues BLyS and APRIL with the TNF receptor homologues BCMA and TACI. Curr. Biol. 2000;10:785–788. doi: 10.1016/s0960-9822(00)00566-2. [DOI] [PubMed] [Google Scholar]
- Migliazza A, Lombardi L, Rocchi M, Trecca D, Chang CC, Antonacci R, Fracchiolla NS, Ciana P, Maiolo AT, Neri A. Heterogeneous chromosomal aberrations generate 3′ truncations of the NFKB2/lyt-10 gene in lymphoid malignancies. Blood. 1994;84:3850–3860. [PubMed] [Google Scholar]
- O’Connor BP, Raman VS, Erickson LD, Cook WJ, Weaver LK, Ahonen C, Lin LL, Mantchev GT, Bram RJ, Noelle RJ. BCMA is essential for the survival of long-lived bone marrow plasma cells. J. Exp. Med. 2004;199:91–98. doi: 10.1084/jem.20031330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otipoby KL, Sasaki Y, Schmidt-Supprian M, Patke A, Gareus R, Pasparakis M, Tarakhovsky A, Rajewsky K. BAFF activates Akt and Erk through BAFF-R in an IKK1-dependent manner in primary mouse B cells. Proc. Natl. Acad. Sci. USA. 2008;105:12435–12438. doi: 10.1073/pnas.0805460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patke A, Mecklenbräuker I, Erdjument-Bromage H, Tempst P, Tarakhovsky A. BAFF controls B cell metabolic fitness through a PKC beta-and Akt-dependent mechanism. J. Exp. Med. 2006;203:2551–2562. doi: 10.1084/jem.20060990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman ZS, Rao SP, Kalled SL, Manser T. Normal induction but attenuated progression of germinal center responses in BAFF and BAFF-R signaling-deficient mice. J. Exp. Med. 2003;198:1157–1169. doi: 10.1084/jem.20030495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan P, Wang W, Wallach D. Receptor-specific signaling for both the alternative and the canonical NF-kappaB activation pathways by NF-kappaB-inducing kinase. Immunity. 2004;21:477–489. doi: 10.1016/j.immuni.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Rickert RC, Jellusova J, Miletic AV. Signaling by the tumor necrosis factor receptor superfamily in B-cell biology and disease. Immunol. Rev. 2011;244:115–133. doi: 10.1111/j.1600-065X.2011.01067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savinova OV, Hoffmann A, Ghosh G. The Nfkb1 and Nfkb2 proteins p105 and p100 function as the core of high-molecular-weight heterogeneous complexes. Mol. Cell. 2009;34:591–602. doi: 10.1016/j.molcel.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemann B, Gommerman JL, Vora K, Cachero TG, Shulga-Morskaya S, Dobles M, Frew E, Scott ML. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 2001;293:2111–2114. doi: 10.1126/science.1061964. [DOI] [PubMed] [Google Scholar]
- Schweighoffer E, Vanes L, Nys J, Cantrell D, McCleary S, Smithers N, Tybulewicz VL. The BAFF receptor transduces survival signals by co-opting the B cell receptor signaling pathway. Immunity. 2013;38:475–488. doi: 10.1016/j.immuni.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih VF, Kearns JD, Basak S, Savinova OV, Ghosh G, Hoffmann A. Kinetic control of negative feedback regulators of NF-kappaB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proc. Natl. Acad. Sci. USA. 2009;106:9619–9624. doi: 10.1073/pnas.0812367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih VF, Davis-Turak J, Macal M, Huang JQ, Ponomarenko J, Kearns JD, Yu T, Fagerlund R, Asagiri M, Zuniga EI, Hoffmann A. Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-κB pathways. Nat. Immunol. 2012;13:1162–1170. doi: 10.1038/ni.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinners NP, Carlesso G, Castro I, Hoek KL, Corn RA, Woodland RT, Scott ML, Wang D, Khan WN. Bruton’s tyrosine kinase mediates NF-kappa B activation and B cell survival by B cell-activating factor receptor of the TNF-R family. J. Immunol. 2007;179:3872–3880. doi: 10.4049/jimmunol.179.6.3872. [DOI] [PubMed] [Google Scholar]
- Shinohara H, Behar M, Inoue K, Hiroshima M, Yasuda T, Nagashima T, Kimura S, Sanjo H, Maeda S, Yumoto N, et al. Positive feedback within a kinase signaling complex functions as a switch mechanism for NF-κB activation. Science. 2014;344:760–764. doi: 10.1126/science.1250020. [DOI] [PubMed] [Google Scholar]
- Shokhirev MN, Hoffmann A. FlowMax: A Computational Tool for Maximum Likelihood Deconvolution of CFSE Time Courses. PLoS ONE. 2013;8:e67620. doi: 10.1371/journal.pone.0067620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga-Morskaya S, Dobles M, Walsh ME, Ng LG, MacKay F, Rao SP, Kalled SL, Scott ML. B cell-activating factor belonging to the TNF family acts through separate receptors to support B cell survival and T cell-independent antibody formation. J. Immunol. 2004;173:2331–2341. doi: 10.4049/jimmunol.173.4.2331. [DOI] [PubMed] [Google Scholar]
- Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, Bulyk ML. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol. 2012;13:95–102. doi: 10.1038/ni.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, Rajewsky K. PI3 kinase signals BCR-dependent mature B cell survival. Cell. 2009;139:573–586. doi: 10.1016/j.cell.2009.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadanlick JE, Kaileh M, Karnell FG, Scholz JL, Miller JP, Quinn WJ, 3rd, Brezski RJ, Treml LS, Jordan KA, Monroe JG, et al. Tonic B cell antigen receptor signals supply an NF-kappaB substrate for prosurvival BLyS signaling. Nat. Immunol. 2008;9:1379–1387. doi: 10.1038/ni.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vora KA, Wang LC, Rao SP, Liu ZY, Majeau GR, Cutler AH, Hochman PS, Scott ML, Kalled SL. Cutting edge: germinal centers formed in the absence of B cell-activating factor belonging to the TNF family exhibit impaired maturation and function. J. Immunol. 2003;171:547–551. doi: 10.4049/jimmunol.171.2.547. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weih DS, Yilmaz ZB, Weih F. Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J. Immunol. 2001;167:1909–1919. doi: 10.4049/jimmunol.167.4.1909. [DOI] [PubMed] [Google Scholar]
- Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.