Abstract
The tsetse fly Glossina fuscipes fuscipes (Gff) is the insect vector of the two forms of Human African Trypanosomiasis (HAT) that exist in Uganda. Understanding Gff population dynamics, and the underlying genetics of epidemiologically relevant phenotypes is key to reducing disease transmission. Using ddRAD sequence technology, complemented with whole-genome sequencing, we developed a panel of ∼73,000 single-nucleotide polymorphisms (SNPs) distributed across the Gff genome that can be used for population genomics and to perform genome-wide-association studies. We used these markers to estimate genomic patterns of linkage disequilibrium (LD) in Gff, and used the information, in combination with outlier-locus detection tests, to identify candidate regions of the genome under selection. LD in individual populations decays to half of its maximum value (r2max/2) between 1359 and 2429 bp. The overall LD estimated for the species reaches r2max/2 at 708 bp, an order of magnitude slower than in Drosophila. Using 53 infected (Trypanosoma spp.) and uninfected flies from four genetically distinct Ugandan populations adapted to different environmental conditions, we were able to identify SNPs associated with the infection status of the fly and local environmental adaptation. The extent of LD in Gff likely facilitated the detection of loci under selection, despite the small sample size. Furthermore, it is probable that LD in the regions identified is much higher than the average genomic LD due to strong selection. Our results show that even modest sample sizes can reveal significant genetic associations in this species, which has implications for future studies given the difficulties of collecting field specimens with contrasting phenotypes for association analysis.
Keywords: tsetse flies, linkage disequilibrium, association studies, population genomics, ddRAD
African trypanosomiasis negatively impacts both human and animal health in sub-Saharan Africa (Simarro et al. 2012a). In 2008, mortality associated with Human African Trypanosomiasis (HAT or sleeping sickness) ranked ninth out of 25 human infectious and parasitic diseases in Africa (Fevre et al. 2008). The causative agents of HAT are members of the genus Trypanosoma (Kinetoplastida), species T. brucei rhodesiense (Tbr) and T. b. gambiense (Tbg), while Animal African Trypanosomiasis (AAT or Nagana) is caused by T. b. brucei (Tbb), T. congolense, and T. vivax. HAT caused by Tbg is a chronic disease with asymptomatic periods lasting several years, while Tbr infection results in an acute disease with over 80% mortality within the first 6 months if untreated. Over 90% of HAT cases are due to Tbg, which occurs in the northwest regions of Uganda and extends from Central African Republic to Equatorial Guinea. More than 12 million people in eastern and southern Africa, including Uganda, Tanzania, Malawi, Zambia, and Zimbabwe, are at risk for Tbr infection (Simarro et al. 2012a). Intense international interventions have reduced HAT cases to below 10,000 for the first time in 50 yr (Simarro et al. 2011), but many cases likely go undetected in remote regions (Aksoy 2011; Simarro et al. 2011). Since available drugs for treatment are expensive, associated with adverse effects (Simarro et al. 2012b), and exhibit reduced efficacy due to increasing drug resistance in the parasites (Brun et al. 2001), the most effective current control methods involve reduction of the vector populations: tsetse flies that belong to the genus Glossina.
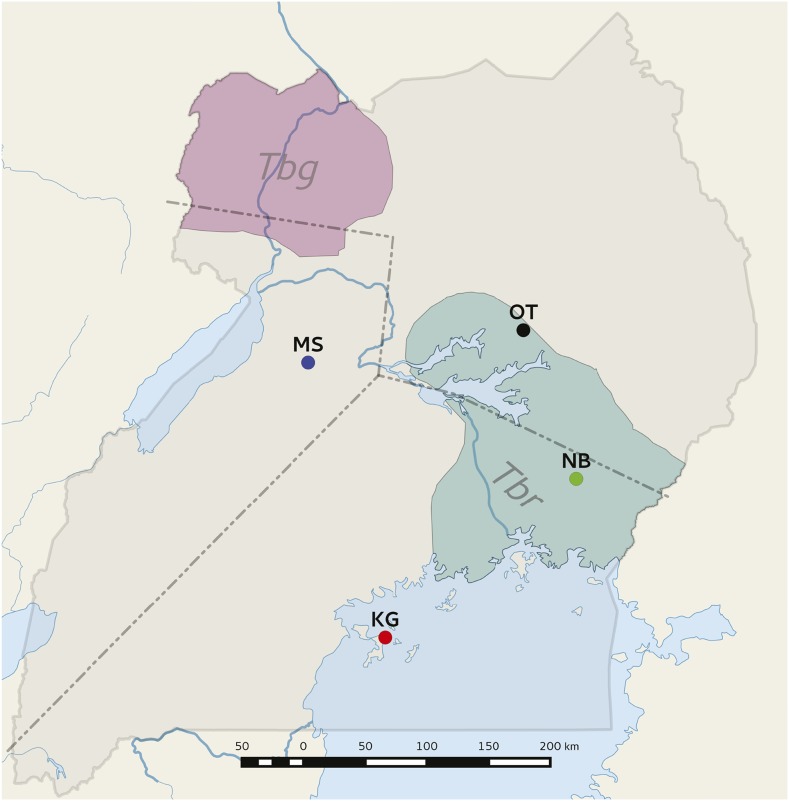
Uganda is the only African country where both forms of HAT exist, with Tbg occurring in the northwest and Tbr in the southeast areas (Figure 1). The two forms of the disease are feared to merge in the near future, as the belt that currently separates them is less than 100 km wide (Picozzi et al. 2005). Given that the pathology, diagnosis, and treatment of HAT for Tbr and Tbg disease vary significantly, the overlap of the two disease belts is expected to complicate HAT control (Welburn et al. 2009). Glossina fuscipes fuscipes (Gff) is the vector for both forms of HAT in Uganda. To understand the biological processes behind disease spread, especially in light of the impending disease merger, we had previously performed population genetics studies on Ugandan populations of Gff and identified three genetically and geographically distinct population clusters (northern, western, and southern Uganda), based on 12–15 microsatellite loci (Hyseni et al. 2012; Echodu et al. 2013; and Figure 1). These genetic clusters display high intercluster Fst values (0.124–0.574), typically associated with insects that differ at the subspecies level, or with the possibility of coexistence of distinct species (Beadell et al. 2010; Hyseni et al. 2012). The northern and southern clusters of Gff meet in a narrow hybrid zone in central Uganda along Lake Kyoga (Beadell et al. 2010). Our results have also shown that Gff populations are genetically stable over time (Beadell et al. 2010; Echodu et al. 2011; Hyseni et al. 2012), that genetic admixing occurs among sites from the same or different clusters within a 100 km radius (Beadell et al. 2010), and that gene flow is not symmetrical, being three times greater from the southeast to northwest than in other directions (Hyseni et al. 2012). Given that Gff is a riverine species that survives in habitats with high humidity (Hargrove 2001a, 2001b; Terblanche et al. 2008; Nash 1933; Rogers 1979), the asymmetric gene flow could be tied to the influence of the Nile River and its tributaries that flow from Lake Victoria in the south through Lake Kyoga to Lake Albert in northern Uganda (Beadell et al. 2010).
Figure 1.
Geographical distribution of Glossina fuscipes fuscipes collections sites in Uganda. Collections from Masindi (MS), Kalangala Island (KG), Otuboi (OT), and Namutumba (NB) were used in this study. Broken lines mark the hypothetical separation of the three main genetic clusters based on microsatellite data (Hyseni et al. 2012; Echodu et al. 2013). Red and green shading indicates the distribution range of Trypanosoma brucei gambiense (Tbg) and T.b. rhodesiense (Tbr).
Although traditional population genetic markers such as mitochondrial and microsatellite loci have provided important insights into the population dynamics of Gff, genome-wide approaches that rely on thousands of polymorphic markers are required to further investigate the patterns of neutral and adaptive genetic variation in wild populations. Genome scans of single copy nucleotide polymorphisms (SNPs) using next-generation DNA sequencing (NGS) can be used to detect signatures of adaptive genetic variation in wild populations without a priori identification of candidate genes (Elmer and Meyer 2011). These methods allow simultaneous screening of thousands of genetic markers, and large numbers of individuals (Storz 2005; Ekblom and Galindo 2011; Seeb et al. 2011; Narum et al. 2013; reviewed by Glenn 2011), and are often coupled with sampling designs and statistical methods that take into account drift and geographic differentiation (Stinchcombe and Hoekstra 2008; Storz 2005; Storz and Wheat 2010; Jones et al. 2012). This approach can be used for both model and nonmodel organisms, regardless of the availability of a reference genome (Bonin et al. 2006; Egan et al. 2008; Hohenlohe et al. 2010; Stapley et al. 2010; Anderson et al. 2012; Nosil and Feder 2012; Orsini et al. 2012).
Here, we used double digestion restriction associated RADSeq (Peterson et al. 2012) in combination with whole genome sequencing (WGS) to develop a SNP toolbox for studying Gff populations in Uganda. We carried out this work using Gff flies from four sampling sites, representative of the three previously described genetic clusters (two from the southern genetic cluster, and one each from the other two clusters; Echodu et al. 2013). The inclusion of multiple genetic populations, which maximizes the representation of the genetic diversity of the Ugandan Gff populations, aims to reduce false positives when looking for adaptive variation by minimizing ascertainment bias (reviewed by Schoville et al. 2012). Since adaptive processes are restricted to specific genes/gene regions, while drift and gene flow would affect all genes equally, the analysis of different genetic backgrounds facilitates the distinction of forces shaping the distribution of neutral and adaptive genetic variation, and thus should help to identify the genetic underpinnings of specific traits (Luikart et al. 2003).
While our main goal was to identify SNPs for future population genomic studies, this dataset provided us with the opportunity to estimate, for the first time, Gff genomic patterns of LD, and to compare the estimates of genetic differentiation from the SNP data with those from previous studies with microsatellite and mtDNA markers. We also used these data to look for associations between SNPs and two epidemiologically relevant traits: susceptibility to Trypanosoma infections, and environmental adaptation. We discuss the epidemiological relevance of this work, and the future research avenues it enables.
Materials and Methods
Glossina fuscipes fuscipes (Gff) samples
Glossina f. fuscipes samples were collected from four sites in Uganda in 2010 and 2011 (Table 1, Figure 1, and Supplemental Material, Table S1): Masindi (MS), Otuboi (OT), Namutumba (NB), and Kalangala Island (KG). Flies from each site included in this study had been previously analyzed using microsatellite and/or mtDNA loci (Beadell et al. 2010; Echodu et al. 2011, 2013; Hyseni et al. 2012). Based on these markers, they belong to three distinct genetic clusters located at the north, south, and north-west of Lake Kyoga (Figure 1). Collections were carried out using biconical traps, and each fly was stored individually in 80% ethanol until DNA extraction. Table 1 lists the number of samples per site, the year of collection, and their Trypanosoma infection status; details for each individual sample are provided in Table S1. All individuals included in this study were adults, but their specific age could not be determined. All samples were collected in 2011, except for the individuals from Kalangala, which were collected in 2010. However, given that the results from previous studies suggest that allelic frequencies are stable over time in Gff populations in Uganda (Echodu et al. 2013), we included this population in the analyses, unless stated otherwise.
Table 1. Sample summary.
| Sequencing Method | No. of Populations | No. of Individuals | Population Name | Year Collected | Symbol | Infected/ Uninfected |
|---|---|---|---|---|---|---|
| Whole-genome Sequencing | 1 | 16 | Namutumba | 2011 | NB | 8/8a |
| ddRAD sequencing | 4 | 48 | Masindi | 2011 | MS | 7/7 |
| Otuboi | 2011 | OT | 7/7 | |||
| Namutumba | 2011 | NB | 8/8 | |||
| Kalangala | 2010 | KG | 2/2 |
Six of these samples were also included in the ddRAD sequencing set (5/1).
DNA extraction and assessment of Trypanosoma infection by PCR
DNA was extracted from carcasses and midguts using the MasterPure Complete DNA Purification Kit (Epicentre Biotechnologies), following the manufacturer’s protocol.
DNA from legs was extracted using the DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer’s instructions. All extractions were stored at –20° until further processing. Each sample was tested for Trypanosoma infection twice via PCR amplification using trypanosome alpha-tubulin primers (trypalphatubF: CTCGACACACTCACTTCTGGAG; trypalphatubR: CGAATTTGTGGTCAATACGAG). This assay was not specific for trypanosomes that are human infective (i.e., Tbr or Tbg), but rather detects the presence of any Trypanosoma species, including T. brucei, T. congolense, and T. vivax. Table 1 reports the number of infected and uninfected flies for each sampling site.
Reduced representation genomic libraries
ddRADseq:
Double-digest restriction site-associated DNA sequencing libraries (ddRAD) were prepared using a modified version of the protocol described in Peterson et al. (2012). Briefly, ∼1 μg of genomic DNA from each Gff individual was digested with restriction enzymes NlaIII and MluCI (NEB). Each individual sample was then labeled with custom Illumina adaptors carrying a unique barcode at the 5′ end of the digested fragments. The fragments were amplified by eight cycles of polymerase chain reaction (PCR) to select for those fragments containing different restriction sites at each end, and increase the total concentration of the desired fragments. After PCR amplification, a library was constructed by pooling the products of 24 individuals and later size-selecting them to a fragment size of 215 bp under the “tight” setting of a Blue Pippin electrophoresis platform (Sage Science). Libraries were sent to the Yale Center for Genome Analysis for 75 bp paired-read sequencing with the Illumina Hi-Seq platform. To achieve the best sequence quality, the complexity of the sequencing lanes was increased by spiking the libraries with a secondary library whose fragments did not begin with the restriction sites used by our ddRAD library. Data are available via BioSample project accession number PRJNA303153, with linked associated short-read sequences and variation data.
Whole-genome-sequencing
Whole genomes of 16 individuals from the NB population were sequenced at the Genome Institute of the Washington University School of Medicine (St Louis, MO) as 100-bp reads using the Illumina HiSeq2000 platform at ∼40 × coverage (Table 1 and Table S1). All sequence data for each of the 16 samples was trimmed using flexbar version 2.4 (Dodt et al. 2012) prior to alignment. Synonymous site identification to perform genetic diversity calculations was performed using the GfusI1.1 gene-build (VectorBase 2014; Giraldo-Calderon et al. 2015) with the variant effect predictor (VEP) tool within Ensembl (Flicek et al. 2014) to determine the genomic context and potential function of all identified variants. The software annotates all SNPs as belonging to one of several functional categories: stop-gained, stop-lost, frameshift-coding, nonsynonymous-coding, splice-site, promoter, 5′ UTR, 3′ UTR, upstream, downstream, intronic; synonymous coding or intergenic. Subsequently, only variants with synonymous effects were parsed for each sample in order to compile a list of synonymous variant sites across the genome assembly. Genetic diversity (π), and Tajima’s D values (5000-bp nonoverlapping windows) were calculated using vcftools v. 0.1.12b (Danecek et al. 2011) and average values obtained within the R software (R Core Team 2013).
Data processing
The ddRAD library raw sequence reads were demultiplexed, quality filtered, and filtered for unambiguous barcodes using “process_radtags” from the Stacks software (Catchen et al. 2011). This dataset was then used to call SNPs using the G. fuscipes reference assembly as described in the next section. To improve the coverage of individuals for which not enough reads were obtained by the original ddRAD run due to the low quality of the sample, we combined the reads with the data from WGS of 16 individuals from the NB population. Combined, our dataset included 58 individuals, as six of them were sequenced with both technologies (Table 1 and Table S1). The software PGDSpider v. 2.0.5.2 (Lischer and Excoffier 2012) was used to convert between file formats for downstream analyses.
Mapping and variant calling and summary statistics
Polymorphic loci were identified from the combined reads of the 58 individuals (ddRAD and WGS) by mapping them against the 2395 supercontigs of the G. fuscipes GfusI1 reference assembly (VectorBase 2014; Giraldo-Calderon et al. 2015) using Bowtie2 v. 2.1.0 (Langmead and Salzberg 2012) in the “very sensitive” option, and Samtools v. 0.1.19 (Li et al. 2009). Variants were called using the bcftools utility from Samtools, and data filtered in vcftools v. 0.1.10 (Danecek et al. 2011) based on genotype depth of coverage (DP > 7) and percentage of missing data allowed (< 30%). Only loci that genotyped in at least 80% of the samples were included in subsequent analysis. Our final variant calling file (vcf format) contained only biallelic SNPs and no indels. Five individuals that did not genotype for at least 80% of SNPs were removed from the analysis, and a second filtering was performed on the remaining 53 individuals, including a minor allele frequency filter (MAF > 0.05). All summary statistics including depth of coverage, SNP density, Hardy-Weinberg equilibrium tests, Tajima’s D, and between populations Fst, were performed using vcftools v. 0.1.12b (Danecek et al. 2011), and processed with the R software (R Core Team 2013). Tajima’s D values were estimated using a nonoverlapping window size of 1000 bp. Tajima’s D values are indicative of the presence and direction of selection in the region. In general, values above 2 are considered significant, with positive values suggesting balancing selection, and negative values the presence of a selective sweep or a bottleneck (Anholt and Mackay 2010).
Cluster analyses
Two different clustering methods were used to determine the presence of underlying population structure in our samples. First, we performed a Bayesian clustering analysis in fastStructure (Raj et al. 2014), using only loci in Hardy-Weinberg equilibrium (HWE) after Benjamini-Hochberg (BH) correction (P ≤ 0.05) (Benjamini and Hochberg 1995). fastStructure uses variational Bayesian inference, and large numbers of SNPs to assign individuals probabilistically to K numbers of clusters characterized by a set of allele frequencies at each loci, with no a priori information of sample location. This method assumes HWE and no linkage disequilibrium (LD) among loci. In addition to detecting existing population structure, fastStructure provides ancestry estimates of each of the sampled individuals. Ten independent fastStructure runs were conducted with K = 1–5 on all individuals of all populations. The optimal number of K clusters was then determined using the chooseK.py program of fastStructure. Results were plotted with the program DISTRUCT v.1.1 (Rosenberg 2004). Second, we conducted principal component analysis (PCA) of the entire dataset using the Adegenet package v. 1.3.9. (Jombart 2008) available for the R software v. 3.0.1. (R Core Team 2013). PCA provides a simple, low-dimensional, projection of the data by performing single value transformation on multiple possibly correlated variables, allowing for visual identification of existing population structure.
Linkage analysis
Linkage measurements:
Vcftools version 0.1.12 (Danecek et al. 2011) was used to calculate pairwise linkage disequilibrium (LD) as r2 (Hill and Robertson 1968) for all SNP-pairs located on common supercontigs. The “–allow-extra-chr” option was required to handle the number of supercontigs (N = 2395). The KG population was omitted from the analysis due to its low sample size. Unless stated otherwise, all subsequent analysis pertaining to LD used r2.
LD values of all SNP-pairs were compared after binning SNP-pairs by physical distance (bp) to control for unknown rates of recombination in Gff. Bin length was set at 50 bp. To identify SNP-pairs with abnormal LD values, we assigned probabilities to each SNP-pair in the bin. As the distributions of binned SNP-pairs are bounded by 0 and 1 and do not appear to be Normal in shape, and because in many cases the data appear to exhibit peaks at both the lower and upper r2 range, the data were modeled using the probability density function (PDF) of the Beta distribution. A PDF describes the relative likelihood that a random variable will take a particular value, or range of values, given the type of probability distribution. Using the cumulative distribution function (CDF) of the Beta distribution, we can describe the probability that a binned SNP-pair will have an r2 less than or equal to x. It follows that 1 − CDF represents the probability of observing a more extreme value than a particular r2 value. For each set of binned r2 values, the SNP-pairs deemed worthy of further investigation were defined as those where 1 − CDF ≤ 0.01 after BH correction for multiple testing.
The Beta distribution is bounded on the noninclusive interval between 0 and 1, meaning no value is expected to equal exactly 0 or 1. However, there are data in each bin that have been assigned values of 0 or 1. It is likely that these values are not truly 0 or 1 in the discrete binary sense that a coin-flip can produce only a heads or tails result. Therefore, to satisfy the expectations of the Beta distribution, all r2 values for each bin were scaled according to the scheme:
in order to symmetrically shrink the distribution of r2 values to fit between 0 and 1, while not including any values equal to 0 or 1. In the scheme above, let xi stand for the r2 of each SNP-pair in an arbitrary bin, and θ stand for the scaling factor. The results in this paper used θ = 0.999.
Selection analysis
As we hypothesize that SNP variation is associated with phenotypic and environmental variation, we carried out genome scans to identify possible genomic regions under selection associated with the infection status of the fly, assessed via PCR (Trypanosoma infected vs. uninfected), or with local environmental adaptation, determined by the PCA loadings of the environmental parameters corresponding to each geographic location obtained from the WorldClim database (Hijmans et al. 2005; File S1).
We used multiple complementary methods to detect signatures of adaptive change while accounting for drift, geographic structure, and a small sample size. At the individual loci level, we used BayeScan (Fischer et al. 2011), known to perform well with very small sample sizes; and PCAdapt (Duforet-Frebourg et al. 2014), which accounts for population structure and specifically identifies loci associated with environmental adaptation. We also used a haplotype-based approach implemented by hapFLK, (Fariello et al. 2013), which incorporates the hierarchical structure of the populations in the analysis, is robust to the demographic history of the populations, and is capable of detecting incomplete selective sweeps that are difficult to detect with methods that rely on allele frequencies. Details of the selection screens can be found in File S2. Furthermore, we use our estimates of LD to further analyze the BayeScan results to identify loci that might be under selection, but were not recognized as outliers due to insufficient statistical power. This was achieved by applying an LD filter to the dataset of SNPs ranked in BayeScan within the top 10% alpha values. Those SNPs that were part of an SNP-pair with an assigned probability value of P ≤ 0.01, based on a Beta probability distribution, were considered potential candidate loci under selection (see previous section).
Functional annotations and identification of candidate genes
The SNPs identified by each of the above methods were then mapped to the annotated Gff genome and transcriptome to identify potentially relevant genes in the proximity. We searched for genomic regions under selection on individuals from the NB, MS, and OT populations. Population KG was excluded from the analysis due to the low number of individuals sampled.
All putative peptides annotated for Gff in the GfusI1.1 gene-build were obtained from VectorBase (2014); Giraldo-Calderon et al. 2015. The sequences used to compare the Gff peptides against well known/annotated sequences were obtained from UniProt/SwissProt (Boeckmann et al. 2005), used with blastp, and Pfam (Finn et al. 2014), used with hmmscan, as required by ARGOT2 (Falda et al. 2012; Gillis and Pavlidis 2013; Radivojac et al. 2013). The blastp and hmmscan results submitted to ARGOT2 were obtained by performing local searches on the Gff peptides against the UniProt peptide database (obtained on September 8, 2014), and the hidden Markov models (HMM) of the combined protein-domain sets on the Pfam databases (Pfam-A and Pfam-B: obtained on September 8, 2014), respectively. Settings used were as dictated by the ARGOT2 site. The blastp and hmmscan results were uploaded to ARGOT2 servers for analysis after being split into 10 groups (∼2330 peptides per group) to prevent overloading the remote ARGOT2 cluster. The functional annotations were then downloaded and joined back together.
Data availability
The Supplementary Material files contain supplementary Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, and Figure S6, and their legends, as well as Table S4 and the legends of all supplementary tables and supplementary files. File S1 contains the data and analysis of the bioclimatic parameters. File S2 contains details of the selection analyses. File S3 lists the genes located within 1000 bp of SNPs identified by the selection analysis in BayeScan, associated to local environmental adaptation and susceptibility to trypanosome infection. File S4 lists the SNPs identified as outliers in BayeScan during the pairwise population comparisons. The Gff Illumina read sequences produced in this study are available as part of BioSample project accession number PRJNA303153 with linked variation data (ddRAD), and project accession number SAMN02742630 (WGS). Data were deposited at BioSample project under accession numbers PRJNA303153 and SAMN02742630.
Results
Marker discovery
Approximately 15% of the fragments generated after in vitro digestion of Gff genomic DNA using the restriction enzymes NlaIII and MluCI were 172 to 216 bp in length (193 bp on average), the size chosen for constructing the ddRAD libraries. Downstream analysis of the generated ddRAD reads indicates that we achieved an average coverage of 32 × for each individual fly.
We recovered a total of 448,881,370 reads from the 48 individuals subjected to ddRAD sequencing. Quality processing of these reads with the software Stacks (Catchen et al. 2011) yielded a total of 428,673,526 high quality reads (95%) with unambiguous barcodes. Combining the ddRAD reads for the NB population with the WGS reads (see Materials and Methods) allowed us to increase the coverage of individuals of this population that were underrepresented in the ddRAD libraries due to low quality samples. With this strategy we recovered 37 × more SNPs for the NB population than those obtained from the ddRAD seq alone, after the appropriate filtering was performed. The dataset containing the filtered ddRAD reads for all four Ugandan populations, and the filtered WGS reads for the NB population was subsequently used to call variant sites.
The Gff GfusI1 reference assembly (VectorBase 2014; Giraldo-Calderon et al. 2015) used for variant calling extends ∼375 Mbp, broken down into 2395 supercontigs that range in size from 886 bp to 3,329,503 bp (mean = 156,482 bp, and median = 19,838 bp). A total of 5,246,046 SNPs were obtained after mapping the combined dataset to this reference (see Materials and Methods). Further filtering for biallelic SNPs, with a minimum depth of coverage of 7 ×, and present in at least 80% of the samples, resulted in a set of 153,663 SNPs. Individuals KG10_030, MS11_0017, MS11_0050, NB11_056, and NB11_062 were removed from subsequent analyses because they genotyped for < 80% of the SNPs called. This dataset, containing 53 individual flies, was filtered once more to remove any sites that were polymorphic only in the individuals removed. The final set included 73,297 SNPs, with an average coverage of 41.89 × ± 17.46 × per site, ranging from 18.76 × to 102.78 × (median = 40.31 ×). The final dataset was used for the downstream analyses described below.
We identified SNPs in 1389 of the assembled Gff supercontigs (58% of the total), with an average of two SNPs for every 10,000 bp. The number of SNPs per supercontig ranged from zero to 25 at the 10,000 bp window size (median = 1 SNP / 10,000 bp). Analysis using a window size of 1000 bp yielded an average of 0.2 SNPs / 1000 bp, with some supercontigs having no SNPs, and 26 contigs having 10 or more.
Genetic diversity and differentiation
Observed individual heterozygosities (Ho) ranged from 0.1111 to 0.3435 (mean Ho = 0.2156 ± 0.0477). Average heterozygosities per population were HoKG = 0.4631, HoOT = 0.3404, HoNB = 0.3203, and HoMS = 0.2968. Average Tajima’s D across all populations and considering 1000 bp nonoverlapping windows (regardless of whether they contain SNPs or not) was 0.0526. Mean Tajima’s D values considering only those 1000 bp windows that contain at least one SNP was 0.4055, with absolute values larger than |2| detected in 2684 of the windows, and 77 of them displaying values larger than |3| (Table S2 and Figure S1).
Estimates of genome-wide genetic diversity calculated from the WGS data available for the NB population were π = 0.00056, with an average Tajima’s D of –0.0782. When only synonymous sites were considered, these values were π = 0. 0.0001, with an average Tajima’s D of 0.0828.
Values of between-population genetic differentiation (Fst) are shown in Table 2. Genetic differentiation was higher between KG and OT (Fst = 0.3214); and between NB and OT (Fst = 0.3015). The lowest degree of differentiation was found between KG and NB (Fst = 0.0853). The average pairwise Fst = 0.2877 (weighted Fst = 0.3409).
Table 2. Pairwise Fst values between populations of Glossina fuscipes fuscipes from Uganda.
| NB | OT | MS | KG | |
|---|---|---|---|---|
| NB | 0 | 0.42626 | 0.21029 | 0.18811 |
| OT | 0.30145 | 0 | 0.34275 | 0.47684 |
| MS | 0.16772 | 0.24805 | 0 | 0.26597 |
| KG | 0.08532 | 0.32144 | 0.14583 | 0 |
Fst values are above the diagonal; weighted Fst values are below the diagonal. Weighted Fst is calculated in vcftools v. 0.1.12b (Danecek et al. 2011), using the Weir and Cockerham (1984) estimator to correct for samples size.
We identified 6180 loci that deviated from HWE after Bonferroni (Bf) correction (PBf ≤ 0.05), and 25,300 loci that deviated from HWE when BH correction was applied instead (PBH ≤ 0.05). These latter loci were not considered for the Bayesian clustering analysis in fastStructure, which assumes HWE (see next section). When populations were analyzed individually, no loci deviated from HWE in KG, MS, and OT after either Bf or BH correction was applied. However, in the NB population, 278 (Bf) or 936 (BH) loci were out of HWE.
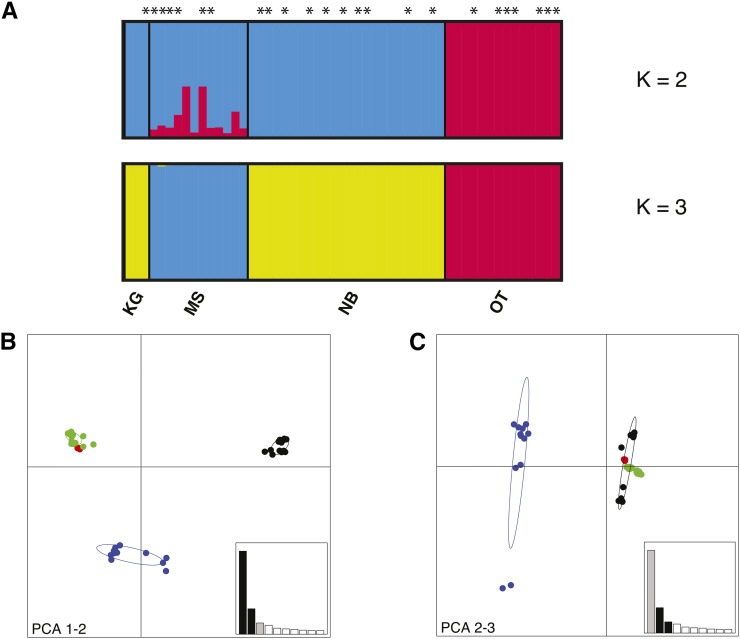
Bayesian clustering analysis in fastStructure (Raj et al. 2014) suggests K = 3 as the most likely number of genetic groups, with KG clustering together with NB (Figure 2A). Clustering is based on geography rather than infection status at both K = 2 and K = 3 (Figure 2A). Although this analysis was run using only loci in HWE to meet the assumptions of the test, the pattern remained unchanged when all 73,297 loci of the dataset were included (Figure S2). PCA of the data shows the same pattern as the Bayesian clustering (Figure 2B), with the KG population grouping with NB. PC1 separates these two populations from OT and MS. OT is placed with KG and NB by PC3, leaving MS as an independent cluster (Figure 2B).
Figure 2.
Population structure in Glossina fuscipes fuscipes (Gff) from Uganda. (A) Genetic membership bar plot based on the 47,997 SNPs that met HWE expectations after BH correction obtained using fastStructure (Raj et al. 2014). Each vertical bar represents a single individual. The height of each color represents the probability of assignment to each of K clusters. Individuals infected with Trypanosoma are indicated with and asterisk (*). Plots of K = 2 and K = 3 are shown for all sampled flies in all four populations (N = 53). (B)–(C) Principal component analysis (PCA) plots of Gff populations based on all 73,297 SNPs. Each dot represents an individual. The 95% inertia ellipses are shown for each population. (B) PC1 vs. PC2. (C) PC2 vs. PC3. Locations are identified by different colors. Variance explained by the different components = PCA1: 0.3%, PCA 2: 0.09%, PCA3: 0.04%. KG, Kalangala Island (Red); MS, Masindi (Blue); OT, Otuboi (Black); and NB, Namutumba (Green).
Linkage disequilibrium
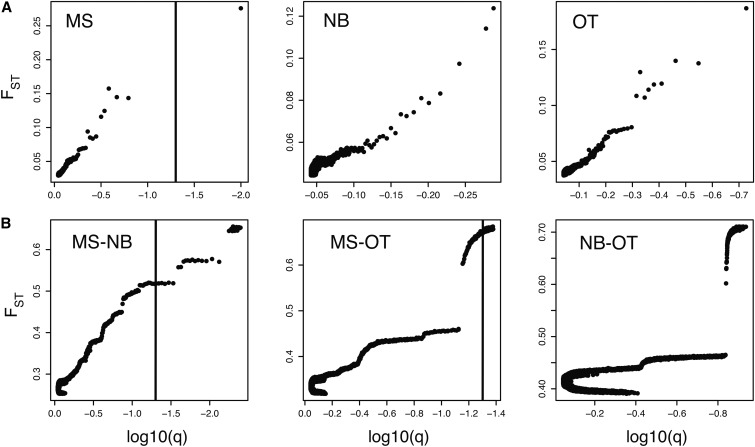
The mean bin-wise LD in Gff has a maximum value at r2max = 0.4018, and decays with physical distance to reach half of this maximum value (r2max/2 = 0.2009) near 708 bp (Figure S3). When considering individual populations with smaller sample size, r2max increases (Figure 3), and r2 reaches values between 0.5 and 0.6. The analyses of individual populations also show variation in the number of base pairs at which LD decayed by half (r2max/2): NB = 1359 bp, OT = 2334 bp, and MS = 2429 bp; Figure 3). KG was excluded from the analysis due to low sample size.
Figure 3.
The rate of linkage disequilibrium (LD) varies with the Glossina fuscipes fuscipes population. Pairwise LD between SNPs located in the same supercontig was estimated from all individuals of all populations using Vcftools v. 0.1.12 (Danecek et al. 2011) as r2. Each “dot” represents the mean LD for that set of binned SNP-pairs. The color of the dot illustrates the number of SNP-pairs contributing to the mean; the color scale is shown in the right vertical bar. The blue line is a loess regression line of best fit, and the purple line corresponds to r2max/2. (A) Masindi, MS; (B) Namutumba, NB; and (C) Otuboi, OT.
After applying an LD filter based on a Beta probability distribution to identify potential LD outliers, we obtained 24,372 out of 6,454,294 SNP-pairs (0.38%) with corrected P values ≤ 0.01 (see Materials and Methods). Rather than looking at each of these individual genomic locations, we then used this information to sort potential SNPs of interest from the genome screen conducted in BayeScan, as relevant SNPs might have not been identified as outliers in this analysis due to the lack of statistical power consequence of the small sample size (see following section).
Selection analyses
Susceptibility to infection:
Table S1 shows the Trypanosoma spp. infection status of the flies from the four Gff collection sites included in this study. BayeScan analysis (Fischer et al. 2011) of infected and uninfected individuals did not detect any Fst outlier loci when all locations were grouped together (Figure S4). Additional LD filtering of the top 10% BayeScan alpha values from this comparison, followed by a genome scan for genes located within 1000 bp of any SNP in the pair, identified a total of 340 genes (136 with functional annotations; File S3). Among the most prevalent biological functions of these genes were: trans-membrane transport, transcription regulation, rRNA processing, DNA-replication, and metabolic processing. In the molecular function (MF) domain, some of the frequently occurring motifs were related to zinc ion binding, DNA binding, transferase activity, and oxidoreductase activity.
When each location was analyzed individually (Figure 4A), we found one outlier locus in the MS dataset, located at JFJR01006593.1: 85294. No annotated genes were identified within a 1000-bp window of this SNP. We explored these results further by analyzing SNPs ranked within the top 5% and 10% alpha cutoff and that were common among each individual population analysis (Table S3). We identified 10 and 43 SNPs at the 5% and 10% alpha cutoff, respectively, and carried out a screen for putative genes under selection based on proximity, using a window of 1000 bp. This screen identified 20 genes, including transcription factors, and genes associated with circadian rhythms and membrane integrity. The genes and their distance to each SNP are found in File S3.
Figure 4.
Bayescan Fst posterior probability plots. A screen for Fst outlier loci was performed on (A) each of three Glossina fuscipes fuscipes (Gff) populations from Uganda included in this study (both infected and uninfected flies), to identify SNPs associated with susceptibility to infection by Trypanosome; and (B) pairwise population comparisons, to identify SNPs associated with environmental local adaptation. The vertical line corresponds with the false discovery rate tradeoff value (FDR = 0.05) used to call outliers in BayeScan (Fischer et al. 2011). The population or pair of populations analyzed in each plot is indicated by a legend at the top left corner. MS, Masindi; OT, Otuboi; NB, Namutumba. Note that the KG (Kalangala Island) population was not included in this analysis due to an insufficient number of individuals.
Local environmental adaptation:
We searched for SNPs associated with local environmental adaptation by searching for Fst outlier loci in pairwise comparisons contrasting samples from the three distinct genetic clusters from northern, southern, and western Uganda, as these populations experience different environmental regimes that differ mainly in precipitation and seasonality (File S1). Using BayeScan we did not identify any SNPs between NB and OT, but found 40 outlier loci between MS and NB, and 119 outlier loci between MS and OT (Figure 4B; File S4). Two of these outliers were common to both pairwise comparisons (Scaffold150:13771 and Scaffold150:13772; File S3) and are adjacent to each other, falling within a 100 bp window that includes three SNPs, and has a Tajima’s D value of 1.25 (Table S2). A genomic scan around these SNPs identified genes GFUI009284 and GFUI009279 within a region 1000 bp; unfortunately, no functional annotations were available for any of these genes (File S3). Eight of the 40 outlier loci between MS and NB also passed the LD filter, with two of these SNPs located within genes, and one located 405 bp apart, none of them with functional annotations (File S3). Of the 119 outlier SNPs from the MS vs. OT comparison, 60 had LD levels above the threshold, with 35 genes located within 1000 bp of them (File S3). We could not carry out this analysis for the NB vs. OT comparison because no outliers were detected by the BayeScan method.
PCadapt (Duforet-Frebourg et al. 2014) was used as an independent method to identify SNPs specifically associated to local adaptation. This analysis identified six SNPs as contributors to the two main factors discriminating among three genetic clusters (Table S4 and Figure S5), clusters that are consistent with the ones detected by the fastStructure and the PCA analyses (Figure 2), and match the geographical location of the sampling sites (Figure 1). Interestingly, two of these SNPs (Scaffold150: 13772 and 13773) had nearby annotated genes that were also identified using BayeScan: GFUI009284 and GFUI009279 (File S3). Only one of the six SNPs identified by PCadapt was part of an SNP-pair that passed the LD filter (Scaffold368: 310507), but no genes were located within 1000 bp of either SNP of the LD pair.
The haplotype-based analysis conducted in Hapflk (Fariello et al. 2013) on all populations, with K = 3 a priori number of clusters to reflect the population structure of the dataset (Figure 2), did not find any region with significant signatures of selection (Figure S6).
Discussion
Marker development, genetic diversity, and LD
Using a combination of ddRAD sequencing and WGS, we identified 73,297 variable sites across the genome of Glossina fuscipes fuscipes flies from Uganda. Advantages of using ddRADs include that they are randomly distributed across the genome, and that they are affordable to obtain enough read coverage to detect polymorphic loci even when at lower frequencies because only a fraction of the genome is being sequenced. Due to the low quality of six of the original NB samples, we combined the ddRAD dataset with WGS data, achieving higher sequence coverage for our original (low quality) samples while adding data from 10 additional individuals of the NB population to the dataset. The depth of coverage achieved by our method (∼40 ×) provides us with a high probability of observing both alleles of an individual, if in a heterozygote state, assuming random detection of each allele. The use of two restriction enzymes during the ddRAD library preparation, in contrast to similar techniques (Miller et al. 2007; Baird et al. 2008) that randomly sheared the genome, provides consistency in marker recovery increasing the chances that the same loci will be sequenced across all individuals, thus reducing the amount of missing data.
We identified an average of two SNPs for every 10,000 bp of the Gff genome covered, and used them to estimate LD in this species. This information is needed because associations between genes and phenotypic traits depend upon the existence of nonrandom associations between linked loci to identify causative genetic variants. This is the first study to estimate LD across the Gff genome. In fact, it is the first study to provide LD estimates from any Glossina species. LD decay needs to be considered when designing GWAS, as it affects the number of markers and samples required to achieve the power needed to identify the genetic basis of phenotypic variants. Organisms with rapid LD decay will require many more markers to detect associations, while those with slow LD decay will require less markers but provide less resolution for mapping the associations. The theoretical maximum value that LD measured as r2 can reach is r2max = 0.43051 due to its dependence on allele frequencies (VanLiere and Rosenberg 2008); the LD estimated across three Gff populations is consistent with this expectation (Figure S3). Average LD in Gff decays in half (r2max / 2 = 0.2009) at a distance of 708 bp. In comparison, Drosophila melanogaster achieves this r2 value (0.2) at 10 bp in autosomes, and 30 bp in the X chromosome (Mackay et al. 2012), Anopheles arabiensis at 200 bp (Marsden et al. 2014), and honeybee at 500 bp (Wallberg et al. 2014). The average size of LD blocks in Anopheles gambiae are estimated to be ∼40 bp (Wang et al. 2015).
The higher LD observed in Gff might be a consequence of a smaller effective population size (Ne), and higher level of genetic structuring compared to either Anopheles or Drosophila populations. The average genome-wide genetic diversity (π) of the NB population, calculated from WGS data, is consistent with a small population size. The value of π for this population is one or two orders of magnitude lower (π = 0.00056) than reported values for Drosophila (0.0047–0.0114: Ometto et al. 2005; Fabian et al. 2012; Mackay et al. 2012), Anopheles gambiae (0.0043–0.0208: Stump et al. 2005; Chang et al. 2012), or An. Arabiensis (0.0020–0.0064: Stump et al. 2005; Cohuet et al. 2008; Marsden et al. 2014). Likewise, if one considers only synonymous sites, the value of π = 0.0001, compared to the corresponding value estimated in Drosophila of π = 0.0112 (Mackay et al. 2012).
A relatively small population size of Gff populations has been also previously reported in the literature. Estimated values of Ne for Ugandan Gff populations range from 33 to 310 when estimated from microsatellites (Hyseni et al. 2012), and fall below 500 individuals when estimated from mitochondrial data (Beadell et al. 2010). In contrast, Ne values estimated for An. arabiensis and An. gambiae range among thousands of individuals (Taylor et al. 1993; Hodges et al. 2013), and Drosophila populations in Africa have been estimated to have a Ne of over a million (Charlesworth 2009). Likewise, Gff genetic clusters are geographically structured at the local scale (Figure 1 and Figure 2; Beadell et al. 2010; Echodu et al. 2011; Hyseni et al. 2012; Aksoy et al. 2013), while fine geographic structure is not evident in African populations of Anopheles (Lanzaro et al. 1998; Moreno et al. 2007), and Drosophila (Hamlin et al.and Veuille 1999; Dieringer et al. 2005; Pool and Aquadro 2006; Pool et al. 2012).
Given the slow LD decay found in this study for wild Gff populations, the number of markers required to perform GWAS may need to be modified depending on the strength of selection acting on the trait of interest. Traits that are under strong selection, like those related to local environmental adaptation, may generate larger LD regions and thus might require less markers than those under a weaker selection regime, or those involving complex genotypic interactions, like susceptibility to trypanosome infection. For example, LD measured within a region of 57 kb around the Drosophila delta gene, which affects bristle variation and is under selection, found values of r2 of 0.3 to extend up to 5 kb (Long et al. 1998). LD blocks around the para gene, which provides insecticide resistance in An. arabiensis, extended for 2.4 Mb (Wang et al. 2015).
Another factor to take into account while selecting markers for GWAS is the fact that different populations of the same species may differ in their degree of genomic LD depending on their age and demographic histories. Populations established from a small number of founders after a bottleneck would have longer tracks of LD relative to those established from a large number of individuals (Reich et al. 2001). Conversely, older populations are likely to have shorter LD tracks because the genome has experienced recombination for a longer period of time. When LD was measured for the individual Gff populations, we found an r2max value larger than that obtained from all the individuals combined (r2max = 0.5–0.6 vs. 0.4). This may be explained if the smaller sample size of the individual populations results in the estimation of more homogeneous allele frequencies. The data also shows that LD decayed faster in NB than in OT and MS (Figure 3), with MS having the slowest rate of genome LD decay overall reaching half of r2max at 2429 bp (compared to r2max / 2 = 1359 bp in NB and 2334 bp in OT). A plausible explanation for this pattern could be that MS has undergone a recent bottleneck. This is consistent with the bottleneck evidence found by Beadell et al. (2010) and Echodu et al. (2011) using microsatellite data. Microsatellite data also points to some evidence of a bottleneck in OT, but the data are not conclusive (Echodu et al. 2011). The shorter LD tracks observed in NB could also be explained by the age of the population. Using genetic assignment tests on microsatellites it has been inferred that Gff in Uganda migrates from the southeast to the northwest (Aksoy et al. 2013), which would be consistent with NB being the oldest population from the group. Interpopulation differences in genomic LD were obscured when we estimated LD from all populations combined, and could explain why genetic associations were more likely to be identified when the analyzed datasets included the MS population, which had the strongest LD from the populations analyzed.
Population structure
Genetic clustering of the four Ugandan Gff populations is based on geography, rather than on infection state (Figure 2A). Clustering analysis on the SNP dataset groups the four Ugandan populations in three distinct genetic clusters (Figure 2, A–C), and suggests that MS, NB, and KG are less differentiated from each other than to OT, consistent with results from mitochondrial and microsatellite data (Beadell et al. 2010; Hyseni et al. 2012; Echodu et al. 2013). Pairwise Fst values calculated from microsatellites (Echodu et al. 2013) suggest a similar level of genetic differentiation between OT, NB, and MS, while values estimated from SNPs suggest that OT is more genetically differentiated from NB than it is from MS. The SNP data also suggest that MS is closer to NB than it is to OT.
Subsequent work expanding the sample size and spatial breath of Ugandan Gff collections to accurately capture the SNP diversity across the species range will allow the identification of unique SNPs signatures that can subsequently be used to develop low cost genotyping tools to efficiently identify population of origin. This will provide insights into the difference between patterns of genetic differentiation found between the SNP and microsatellite/mtDNA datasets—differences that are currently based on the comparison with the SNP dataset of only four populations.
Regions under selection
The Gff genome is estimated to have ∼20,749 genes (VectorBase 2014: Glossina fuscipes Gfusl1 assembly; Giraldo-Calderon et al. 2015). Using our SNP dataset, we performed a genomic screen to search for genes that may impact the vector competence of flies to trypanosome infection and to local environmental adaptation.
With our current individual sample size we identified one locus in the MS population with signatures of selection in relation to the infection status of the flies (Figure 4A). The complexity of the parameters that determined the infection state of a fly might have complicated detection of loci associated with this trait, especially at this low sample size. However, the power of detection is expected to significantly increase with the number of individuals genotyped. In humans, who have a genome 6 × larger than that of Gff, the slope of the power curve plateaus when the number of individuals reaches the thousands when dealing with genes with large magnitude effects and using ∼300–500K markers (Klein 2007). The percentage of trypanosome-infected flies varies widely across populations in Uganda (0–40%; Alam et al. 2012). In order to have significant power to detect additional loci under selection for this phenotype we need to screen many more samples.
Another caveat is the effect of age of the fly at the moment of capture, as younger flies are less likely to have ingested an infected blood meal compared to older flies and thus might not have been exposed to the parasite yet. Although all the flies included in this study were adults, we did not control for age. Future studies should account for these parameters in order to increase the probability of identifying genetic variants related to susceptibility of infection by trypanosomes. Furthermore, establishing an accurate diagnostic assay for the specific trypanosome species infecting the flies (i.e., infections with HAT vs. AAT causing parasites), would both increase the power of the analysis and provide specific information on human disease risk.
The second genomic screen seeking loci involved in local environmental adaptation identified over 150 SNPs associated with geographic location. Using two independent methods, pairwise-population comparison in BayeScan (Fischer et al. 2011; Figure 4B) and PCadapt (Duforet-Frebourg et al. 2014; \ure S5), we identified regions under selection between populations in the west (MS), and east (NB and OT) of Uganda. Of particular interest is the genomic region around Scaffold150, positions 13771–13773, where three SNPs were identified as potential targets of selection. These SNPs are proximate to genes GFUI009284 and GFUI009279, which unfortunately lack homologs or a functional annotation (File S3, File S4, and Table S4).
Analysis of the bioclimatic parameters for each of the populations (Hijmans et al. 2005; File S1) indicates that the seasonality of precipitation as well as temperature and precipitation during the wettest month of the year are different in MS compared to the eastern populations (NB and OT). Tsetse flies are associated with riverine habitats and vegetation thickets along rivers, which they use to get relief from heat and desiccation and to seek hosts upon which to feed (Dyer et al. 2011). Given their life history, and that they are extremely sensitive to temperature and precipitation (Nash 1933; Hargrove 2001a, 2001b; Terblanche et al. 2008), we can hypothesize that the genomic region identified as relevant to local environmental adaptation might, for example, regulate the ability to resist desiccation.
Epidemiological relevance and future research
Gff flies cause major public health concern and economic losses in Uganda due to the pathogenic parasites they transmit. We have developed a set of 73,297 genotyping markers across the genome of Gff, provided information on LD patterns, conducted a preliminary study of the pattern of genetic differentiation revealed by these markers, and performed a pilot study looking for gene regions under selection using a variety of methods that account for drift and population structure, and incorporate the information from the LD analyses.
Specifically, we searched for genomic regions responsible for tsetse’s resistance/susceptibility phenotype to trypanosome infections and to different environmental adaptations. Identifying genetic regions associated with Trypanosoma infections could inform about (a) the genetic basis for resistance to trypanosomes in natural Gff populations, and (b) if different Gff genotypes vary in their transmission ability of Tbg and Tbr parasite species. This in turn could provide an immediate mechanism of action against the spread of the disease via the introduction of refractory genes into wild populations. Knowledge of the environmental parameters involved in generating and maintaining the genetic differences among Gff populations will contribute to develop more realistic suitability maps for this vector than currently available, as the integration of the ecological and evolutionary axes of divergence will likely increase their predictive power, and thus our ability to forecast changes in the vector distribution in response to impeding change in climatic conditions. The results of this study demonstrate the efficacy of our approach, and provide baseline data for future work to look at the genetic underpinning of these epidemiologically important traits.
Supplementary Material
Acknowledgments
We thank C. Heffelfinger for his help with the sequence analysis and N.P. Havill and J.R. Powell for providing helpful comments. This work was supported by awards from the National Institutes of Health (R01 AI068932) and Fogarty International Center (D43 TW007391 and R03 TW008755).
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.027235/-/DC1
Communicating editor: S. I. Wright
Literature Cited
- Alam U., Hyseni C., Symula R. E., Brelsfoard C., Wu Y., et al. , 2012. Implications of microfauna-host interactions for trypanosome transmission dynamics in Glossina fuscipes fuscipes in Uganda. Appl. Environ. Microbiol. 78(13): 4627–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy S., 2011. Sleeping sickness elimination in sight: time to celebrate and reflect, but not relax. PLoS Negl. Trop. Dis. 5(2): e1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy S., Caccone A., Galvani A. P., Okedi L. M., 2013. Glossina fuscipes populations provide insights for human African trypanosomiasis transmission in Uganda. Trends. Parasitol. 29(8): 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. L., Rodriguez M. A., Braasch I., Amores A., Hohenlohe P., et al. , 2012. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS One 7(7): e40701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt R. R. H., Mackay T. F. C., 2010. Principles of Behavioral Genetics, Academic Press, Elsevier Inc., New York. [Google Scholar]
- Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadell J. S., Hyseni C., Abila P. P., Azabo R., Enyaru J. C. K., et al. , 2010. Phylogeography and population structure of Glossina fuscipes fuscipes in Uganda: Implications for control of tsetse. PLoS Negl. Trop. Dis. 4(3): e636 . 10.1371/journal.pntd.0000636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumond M. A., Balding D. J., 2004. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13: 969–980. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300. [Google Scholar]
- Boeckmann B., Blatter M.-C., Famiglietti L., Hinz U., Lane L., et al. , 2005. Protein variety and functional diversity: Swiss-Prot annotation in its biological context. C. R. Biol. 328: 882–899. [DOI] [PubMed] [Google Scholar]
- Bonin A., Taberlet P., Miaud C., Pompanon F., 2006. Explorative genome scan to detect candidate loci for adaptation along a gradient of altitude in the common frog (Rana temporaria). Mol. Biol. Evol. 23(4): 773–783. [DOI] [PubMed] [Google Scholar]
- Brun R., Schumacher R., Schmid C., Kunz C., Burri C., 2001. The phenomenon of treatment failures in Human African Trypanosomiasis. Trop. Med. Int. Health 6(11): 906–914. [DOI] [PubMed] [Google Scholar]
- Catchen J. M., Amores A., Hohenlohe P., Cresko W., Postlethwait J. H., 2011. Stacks: building and genotyping loci de novo from short-read sequences. G3 (Bethesda) 1(3): 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 2009. Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10(3): 195–205. [DOI] [PubMed] [Google Scholar]
- Cohuet A., Krishnakumar S., Simard F., Morlais I., Koutsos A., et al. , 2008. SNP discovery and molecular evolution in Anopheles gambiae, with special emphasis on innate immune system. BMC Genomics 9(227): 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The Variant Call Format and VCFtools. J. Bioinform. 27(15): 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt M., Roehr J. T., Ahmed R., Dieterich C., 2012. Flexbar□flexible barcode and adapter processing for next-generation sequencing platforms. MDPI Biology. 1(3): 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieringer D., Nolte V., Schlötterer C., 2005. Population structure in African Drosophila melanogaster revealed by microsatellite analysis. Mol. Ecol. 14(2): 563–573. [DOI] [PubMed] [Google Scholar]
- Duforet-Frebourg N., Bazin E., Blum M. G. B., 2014. Genome scans for detecting footprints of local adaptation using a Bayesian factor model. Mol. Biol. Evol. 31: 2483–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer N. A., Ravel S., Choi K.-S., Darby A. C., Causse S., et al. , 2011. Cryptic diversity within the major trypanosomiasis vector Glossina fuscipes revealed by molecular markers. PLoS Negl. Trop. Dis. 5(8): e1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echodu R., Beadell J. S., Okedi L. M., Hyseni C., Aksoy S., et al. , 2011. Temporal stability of Glossina fuscipes fuscipes populations in Uganda. Parasit. Vectors 4: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echodu R., Sistrom M., Hyseni C., Enyaru J., Okedi L., et al. , 2013. Genetically distinct Glossina fuscipes fuscipes populations in the Lake Kyoga Region of Uganda and its relevance for Human African Trypanosomiasis. BioMed Res. Int. 2013: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan S. P., Nosil P., Funk D. J., 2008. Selection and genomic differentiation during ecological speciation: isolating the contributions of host association via a comparative genome scan of Neochlamisus bebbianae leaf beetles. Evolution. 62(5): 1162–1181. [DOI] [PubMed] [Google Scholar]
- Ekblom R., Galindo J., 2011. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity (Edinb) 107(1): 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer K. R., Meyer A., 2011. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol. Evol. 26(6): 298–306. [DOI] [PubMed] [Google Scholar]
- Fabian D. K., Kapun M., Nolte V., Kofler R., Schmidt P. S., et al. , 2012. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol. Ecol. 21(19): 4748–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falda M., Toppo S., Pescarolo A., Lavezzo E., Di Camillo B., et al. , 2012. Argot2: a large scale function prediction tool relying on semantic similarity of weighted Gene Ontology terms. BMC Bioinformatics 13(Suppl 4): S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fariello M. I., Boitard S., Naya H., SanCristobal M., Servin B., 2013. Detecting signatures of selection through haplotype differentiation among hierarchically structured populations. Genetics 193(3): 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fevre E. M., Odiit M., Coleman P. G., Woolhouse M. E., Welburn S. C., 2008. Estimating the burden of rhodesiense sleeping sickness during an outbreak in Serere, eastern Uganda. BMC Public Health 8: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., et al. , 2014. Pfam: the protein families database. Nucleic Acids Res. 42: D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. C., Foll M., Excoffier L., Heckel G., 2011. Enhanced AFLP genome scans detect local adaptation in high-altitude populations of a small rodent (Microtus arvalis). Mol. Ecol. 20: 1450–1462. [DOI] [PubMed] [Google Scholar]
- Flicek P., Amode M. R., Barrell D., Beal K., Billis K., et al. , 2014. Ensembl 2014. Nucleic Acids Res. 42(Database issue): D749–D55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J., Pavlidis P., 2013. Characterizing the state of the art in the computational assignment of gene function: lessons from the first critical assessment of functional annotation (CAFA). BMC Bioinformatics. BioMed Central Ltd. 14: S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo-Calderon G. I., Emrich S. J., MacCallum R. M., Maslen G., Dialynas E., et al. , 2015. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43: D707–D713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn T. C., 2011. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 11(5): 759–769. [DOI] [PubMed] [Google Scholar]
- Hamlin M., Veuille M., 1999. Population structure among African and derived populations of Drosophila simulans: evidence for ancient subdivision and recent admixture. Genetics 153(1): 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove J. W., 2001a Factors affecting density-independent survival of an island population of tsetse flies in Zimbabwe. Entomol. Exp. Appl. 100(2): 151–164. [Google Scholar]
- Hargrove J. W., 2001b The effect of temperature and saturation deficit on mortality in populations of male Glossina m. morsitans (Diptera: Glossinidae) in Zimbabwe and Tanzania. Bull. Entomol. Res. 91(2): 79–86. [PubMed] [Google Scholar]
- Hijmans R. J., Cameron S. E., Parra J. L., Jones P. G., Jarvis A., 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25: 1965–1978. [Google Scholar]
- Hill W. G., Robertson A., 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 226–231. [DOI] [PubMed] [Google Scholar]
- Hodges T. K., Athrey G., Deitz K. C., Overgaard H. J., Matias A., et al. , 2013. Large fluctuations in the effective population size of the malaria mosquito Anopheles gambiae s.s. during vector control cycle. Evol Appl. 6(8): 1171–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe P. A., Phillips P. C., Cresko W. A., 2010. Using population genomics to detect selection in natural populations: key concepts and methodological considerations. Int. J. Plant Sci. 171(9): 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyseni C., Kato A. B., Okedi L. M., Masembe C., Ouma J. O., et al. , 2012. The population structure of Glossina fuscipes fuscipes in the Lake Victoria basin in Uganda: implications for vector control. Parasit. Vectors 5: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T., 2008. Adegenet: an R package for the multivariate analysis of genetic markers. J. Bioinform. 24: 1403–1405. [DOI] [PubMed] [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E., et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484(7392): 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ., 2007. Power analysis for genome-wide association studies. BMC Genet. 8: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzaro G., Touré Y. T., Carnahan J., Zheng L., Dolo G., et al. , 1998. Complexities in the genetic structure of Anopheles gambiae populations in west Africa as revealed by microsatellite DNA analysis. Proc. Natl. Acad. Sci. USA 95(24): 14260–14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence alignment/map (SAM) format and SAMtools. J. Bioinform. 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischer H. E. L., Excoffier L., 2012. PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. J. Bioinform. 28: 298–299. [DOI] [PubMed] [Google Scholar]
- Long A. D., Lyman R. F., Langley C. H., Mackay T. F., 1998. Two sites in the Delta gene region contribute to naturally occurring variation in bristle number in Drosophila melanogaster. Genetics 149(2): 999–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G., England P. R., Tallmon D., Jordan S., Taberlet P., 2003. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4: 981–994. [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482(7384): 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden C. D., Lee Y., Kreppel K., Weakley A., Cornel A., et al. , 2014. Diversity, differentiation, and linkage disequilibrium: prospects for association mapping in the malaria vector Anopheles arabiensis. G3 (Bethesda) 4(1): 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. R., Dunham J. P., Amores A., Cresko W. A., Johnson E. A., 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 17: 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno M., Salgueiro P., Vicente J. L., Cano J., Berzosa P., et al. , 2007. Genetic population structure of Anopheles gambiae in Equatorial Guinea. Malar. J. 6(1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narum S. R., Buerkle C. A., Davey J. W., Miller M. R., Hohenlohe P. A., 2013. Genotyping-by-sequencing in ecological and conservation genomics. Mol. Ecol. 22(11): 2841–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash T. A. M., 1933. A statistical analysis of the climatic factors influencing the density of Tsetse flies, Glossina morsitans Westw. J. Anim. Ecol. 2(2): 197–203. [Google Scholar]
- Nosil P., Feder J. L., 2012. Genomic divergence during speciation: causes and consequences. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367(1587): 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ometto L., Glinka S., De Lorenzo D., Stephan W., 2005. Inferring the effects of demography and selection on Drosophila melanogaster populations from a chromosome-wide scan of DNA variation. Mol. Biol. Evol. 22(10): 2119–2130. [DOI] [PubMed] [Google Scholar]
- Orsini L., K.ISpanier, and L. De Meester, 2012. Genomic signature of natural and anthropogenic stress in wild populations of the waterflea Daphnia magna: validation in space, time and experimental evolution. Mol. Ecol. 21(9): 2160–2175. [DOI] [PubMed] [Google Scholar]
- Peterson B.K., Weber J.N, Kay E.H, Fisher H.S, Hoekstra H.E, 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 7(5):e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picozzi K., Fèvre E. M., Odiit M., Carrington M., Eisler M. C., et al. , 2005. Sleeping sickness in Uganda: a thin line between two fatal diseases. BMJ 331(7527): 1238–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Aquadro C. F., 2006. History and structure of sub-Saharan populations of Drosophila melanogaster. Genetics 174: 915–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Surgino R. P., Stevens K., Cardeno C. M., et al. , 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8(12): e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2013. ISBN 3–900051–07–0. http://www.R-project.org/
- Radivojac P., Clark W. T., Oron T. R., Schnoes A. M., Wittkop T., et al. , 2013. A large-scale evaluation of computational protein function prediction. Nat. Methods 10: 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Stephens M., Pritchard J. K., 2014. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197: 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D. E., Cargill M., Bolk S., Ireland J., Sabeti P. C., et al. , 2001. Linkage disequilibrium in the human genome. Nature 411(6834): 199–204. [DOI] [PubMed] [Google Scholar]
- Rogers D., 1979. Tsetse population dynamics and distribution: a new analytical approach. J. Anim. Ecol. 48: 825–849. [Google Scholar]
- Rosenberg N. A., 2004. DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes 4: 137–138. [Google Scholar]
- Schoville S. D., Bonin A., François O., Lobreaux S., Melodelima C., et al. , 2012. Genetic variation on the landscape: methods and cases. Annu. Rev. Ecol. Evol. Syst. 43(1): 23–43. [Google Scholar]
- Seeb J. E., Carvalho G., Hauser L., Naish K., Roberts S., et al. , 2011. Single-nucleotide polymorphism (SNP) discovery and applications of SNP genotyping in nonmodel organisms. Mol. Ecol. Resour. 11(Suppl 1): 1–8. [DOI] [PubMed] [Google Scholar]
- Simarro P. P., Diarra A., Ruiz Postigo J. A., Franco J. R., Jannin J. G., 2011. The human African trypanosomiasis control and surveillance programme of the World Health Organization 2000–2009: the way forward. PLoS Negl. Trop. Dis. 5(2): e1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro P. P., Cecchi G., Franco J. R., Paone M., Diarra A., et al. , 2012a Estimating and mapping the population at risk of sleeping sickness. PLoS Negl. Trop. Dis. 6(10): e1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro P. P., Franco J., Diarra A., Postigo J. A., Jannin J., 2012b Update on field use of the available drugs for the chemotherapy of human African trypanosomiasis. Parasitology 139(7): 842–846. [DOI] [PubMed] [Google Scholar]
- Stapley J., Reger J., Feulner P. G., Smadja C., Galindo J., et al. , 2010. Adaptation genomics: the next generation. Trends Ecol. Evol. 25(12): 705–712. [DOI] [PubMed] [Google Scholar]
- Stinchcombe J. R., Hoekstra H. E., 2008. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity (Edinb) 100(2): 158–170. [DOI] [PubMed] [Google Scholar]
- Storz J. F., 2005. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol. Ecol. 14(3): 671–688. [DOI] [PubMed] [Google Scholar]
- Storz J. F., Wheat C. W., 2010. Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution. 64(9): 2489–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump A. D., Fitzpatrick M. C., Lobo N. F., Traore S., Sagnon N., et al. , 2005. Centromere-proximal differentiation and speciation in Anopheles gambiae. Proc. Natl. Acad. Sci. USA 102(44): 15930–15935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C. E., Toure Y. T., Coluzzi M., Petrarcca V., 1993. Effective population size and persistence of Anopheles arabiensis during the dry season in West Africa. Med. Vet. Entomol. 7(4): 351–357. [DOI] [PubMed] [Google Scholar]
- Terblanche J. S., Clusella-Trullas S., Deere J. A., Chown S. L., 2008. Thermal tolerance in a south-east African population of the tsetse fly Glossina pallidipes (Diptera, Glossinidae): implications for forecasting climate change impacts. J. Insect Physiol. 54(1): 114–127. [DOI] [PubMed] [Google Scholar]
- VanLiere J. M., Rosengerg N. A., 2008. Mathematical properties of the r2 measure of linkage disequilibrium. Theor. Popul. Biol. 74(1): 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VectorBase 2014 http://www.vectorbase.org, Glossina fuscipes fuscipes, IAEA, GfusI1.
- Wallberg A., Han F., Wellhagen G., Dahle B., Kawata M., et al. , 2014. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat. Genet. 46(10): 1081–1088. [DOI] [PubMed] [Google Scholar]
- Wang X., Afrane Y. A., Yan G., Li J., 2015. Constructing a genome-wide LD map of wild A. gambiae using next-generation sequencing. BioMed Research International. 2015: 238139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S., Cockerham C. C., 1984. Estimating F-statistics for the analysis of population structure. Evolution 38(6): 1358–1370. [DOI] [PubMed] [Google Scholar]
- Welburn S. C., Maudlin I., Simarro P. P., 2009. Controlling sleeping sickness—a review. Parasitology 136(14): 1943–1949. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Supplementary Material files contain supplementary Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, and Figure S6, and their legends, as well as Table S4 and the legends of all supplementary tables and supplementary files. File S1 contains the data and analysis of the bioclimatic parameters. File S2 contains details of the selection analyses. File S3 lists the genes located within 1000 bp of SNPs identified by the selection analysis in BayeScan, associated to local environmental adaptation and susceptibility to trypanosome infection. File S4 lists the SNPs identified as outliers in BayeScan during the pairwise population comparisons. The Gff Illumina read sequences produced in this study are available as part of BioSample project accession number PRJNA303153 with linked variation data (ddRAD), and project accession number SAMN02742630 (WGS). Data were deposited at BioSample project under accession numbers PRJNA303153 and SAMN02742630.