Abstract
Cotton seed trichomes are the most important source of natural fibers globally. The major fiber thickness properties influence the price of the raw material, and the quality of the finished product. The recessive immature fiber (im) gene reduces the degree of fiber cell wall thickening by a process that was previously shown to involve mitochondrial function in allotetraploid Gossypium hirsutum. Here, we present the fine genetic mapping of the im locus, gene expression analysis of annotated proteins near the locus, and association analysis of the linked markers. Mapping-by-sequencing identified a 22-bp deletion in a pentatricopeptide repeat (PPR) gene that is completely linked to the immature fiber phenotype in 2837 F2 plants, and is absent from all 163 cultivated varieties tested, although other closely linked marker polymorphisms are prevalent in the diversity panel. This frame-shift mutation results in a transcript with two long open reading frames: one containing the N-terminal transit peptide that targets mitochondria, the other containing only the RNA-binding PPR domains, suggesting that a functional PPR protein cannot be targeted to mitochondria in the im mutant. Taken together, these results suggest that PPR gene Gh_A03G0489 is involved in the cotton fiber wall thickening process, and is a promising candidate gene at the im locus. Our findings expand our understanding of the molecular mechanisms that modulate cotton fiber fineness and maturity, and may facilitate the development of cotton varieties with superior fiber attributes.
Keywords: Cotton fiber cell wall thickness, fiber maturity, frame-shift deletion, im locus, pentatricopeptide repeat (PPR)
Cotton is the world’s most important source of natural fibers for textiles. Cotton breeders have long faced the challenge of simultaneously improving fiber quality and yield (Clement et al. 2014). Among the major fiber properties are thickness-related properties including fineness and maturity, which affect the quality of the produced yarn. Finer fibers allow for more fibers per cross section of yarn, improving yarn tenacity, and delivering a finer yarn for high end garments (Clement et al. 2014). Fiber maturity affects the ability of the yarn to be dyed, and is a measure of the degree of thickening of the cotton fiber cell wall (Bradow et al. 1996). Cotton breeders have found that fiber quality is generally negatively correlated with yield, so a deeper understanding of the genetic mechanisms that control these traits may enable a decoupling of this correlation.
The immature fiber mutant was identified in the early 1970s and is used as a model to understand the development of cotton fiber cells (Kohel et al. 1974). This mutant was identified by thin fibers with reduced cell wall thickening, resulting in nonfluffy bolls of mature cotton. The immature fiber was caused by a single recessive gene, designated im, that was shown to reside on chromosome 3 by aneuploidy tests and genetic marker analysis (Kim et al. 2013a; Kohel et al. 2002; Wang et al. 2013). Analysis of transcription during fiber development in im plants, along with near-isogenic wild-type plants, suggested roles for cell wall, stress response, and respiratory genes in the generation of the mutant phenotype (Kim et al. 2013b; Wang et al. 2014). Interestingly, the identification of altered mitochondrial oxidase pathways successfully predicted differences in reactive oxygen species that were also observed in developing im fibers, supporting a key role for the mitochondria in the development of mature cotton fiber cells (Kim et al. 2013b).
Recently, the release of draft and reference genomes for Gossypium species have accelerated candidate gene discovery for major genes in cotton by mapping-by-sequencing (Thyssen et al. 2014a, 2015). The insertion of a retrotransposon into a homeodomain transcription factor has been proposed to underlie the T1 dominant stem trichome gene (Ding et al. 2015). Another striking mutation affecting the protein sequence of a different homeodomain protein has been linked to the okra leaf phenotype in cotton (Zhu et al. 2015). In this study, we use mapping-by-sequencing, and a newly released draft G. hirsutum genome, to identify a striking 22-bp deletion in a cotton ortholog of a mitochondria targeted pentatricopeptide repeat (PPR) gene (Zhang et al. 2015). This deletion results in a frame shift, which abolishes the ability for the transcript to encode a functional full length protein that contains both the mitochondria-targeting transit peptide and the RNA-binding PPR domains. We found that this deletion is completely linked to the im gene in 2837 F2 plants. Importantly, we also found that it is absent from 163 cultivated wild-type varieties that produce thick and mature fibers, although nearby marker polymorphisms are prevalent in the diversity panel. Therefore, we propose PPR gene Gh_A03G0489 as a candidate gene at the im locus. We expect that alternative alleles of this gene will be useful for developing cotton varieties with superior fiber properties.
Materials and Methods
Plant materials
The plant materials used in this study comprised: 163 cultivated accessions of G. hirsutum in a diversity panel (Supplemental Material, Table S1), and three F2 populations segregating for the immature fiber trait along with their parent lines. The first F2 population (Population 1) was described previously and contained 366 plants (270 wild type: 80 im: 16 no phenotype) that were derived from a cross between G. hirsutum cultivar TM-1 and its near isogenic line (NIL) containing the im gene (Kim et al. 2013a). The second F2 population (Population 2) had the same parents, contained 1880 plants (1299 wild type: 469 im: 112 no phenotype), and was planted in 2013 in a field in Stoneville, MS. The third F2 population (Population 3) derived from a cross between the G. hirsutum cultivar MD 52ne and im mutant, and contained 735 plants (560 wild type: 159 im: 16 no phenotype). This population was grown in a field in Stoneville, MS in 2014. Naturally opened bolls were hand harvested and ginned using a laboratory roller gin. Phenotypes were scored based on lint percentage as before (Kim et al. 2013a). Micronaire (MIC) data were also measured using a high-volume instrument (HVI) for all the plants in Population 3. For Populations 1 and 2, MIC data were measured only for the plants that had marginal lint percentages (in the range 26–29%). Generally, MIC values below 3.5 were considered immature phenotype. Parental im and TM-1 NILs were grown in a field in New Orleans, LA in 2013 for mRNA isolation. Standard conventional field practices were followed at both locations and in all years. DNA was isolated from young leaves as described previously (Fang et al. 2010).
RNA isolation, RNAseq, and RT-qPCR
Three biological replicates of fibers from different developmental time points were collected from the field in New Orleans, LA. Total RNA from 28-d post anthesis (DPA) fiber cells from two replicates were Illumina sequenced by Data2Bio LLC (Ames, IA) with paired-end 101-bp reads. Total RNA from the three biological replicates of 10, 17, and 28-DPA fiber cells were converted to cDNA, and subjected to reverse transcription quantitative polymerase chain reaction (RT-qPCR) as described elsewhere (Naoumkina et al. 2015). Primer sequences for RT-qPCR are provided in Table S2. RNAseq gene expression was quantified using GSNAP and bedtools software, and differential gene expression was evaluated by trimmed mean of M (TMM) normalization and ANOVA analysis as before (Naoumkina et al. 2014). We calculated and present RPKM normalized expression values, although we present P-values derived from the TMM analysis (Mortazavi et al. 2008).
Super bulked segregant analysis sequencing (sBSAseq)
F2 plants from Population 1 were selected for sequencing by a bulked segregant analysis approach (Michelmore et al. 1991; Takagi et al. 2013). Two DNA pools were constructed: a pool of DNA from 80 im fiber and a pool of DNA from 80 wild-type plants. DNA was Illumina sequenced with paired-end 101-bp reads by BGI Americas (Cambridge, MA).
Mapping-by-sequencing, marker design, and fine mapping the im locus
We aligned the sBSAseq total genomic reads, using GSNAP software, to a draft reference genome for G. hirsutum cultivar TM-1 (Wu and Nacu 2010; Zhang et al. 2015). We used vcftools software to call SNPs and indels between the wild type and mutant bulk sequences (Danecek et al. 2011). We generated a histogram by counting the number of SNPs with quality > 50 in 10-Mb intervals. We manually inspected the alignment files to confirm indels in the vicinity of the im locus on chromosome 3, which corresponds to chromosome A03 in the reference genome (Kim et al. 2013a; Zhang et al. 2015). We extracted all reads that mapped to D subgenome chromosomes and realigned them to a reference sequence that contained only the A subgenome chromosomes so that we could identify homeoSNPs near our marker loci. We used the homeoSNPs to design subgenome specific primers essentially as described previously (Thyssen et al. 2014a). Each forward primer ends with the mutant allele SNP, while each reverse primer ends with the A subgenome homeoSNP. Both primers contain an additional mismatch at the third base from the 3′ end, which increases annealing temperature stringency (Drenkard et al. 2000). Primers flanking indels include one subgenome specific primer. The 2981 F2 plants were tested for segregation with the new SNP and indel markers, along with two SSR markers, DPL1071 and SHIN-1511, that were previously linked to the im gene and shown to reside on chromosome 3 (Kim et al. 2013a). We also designed primers to test one SNP from the CottonSNP63k array, i00510GH, on the F2 progeny (Hulse-Kemp et al. 2015). Primer sequences are included in Table S2. The linked indel marker CFBid0001 and three closest markers (CFB5887, CFB5888, and CFBid0002), were tested on a diversity panel of 163 accessions of G. hirsutum. The PCR methods for genotyping SNP markers were described previously (Thyssen et al. 2014a). For genotyping indel markers, the forward primers were fluorescent-labeled at 5′ end with 6-FAM (6-carboxyfluorescein), or HEX (4, 7, 2′, 4′, 5, 7-hexachloro-carboxyfluorescein). Primers were purchased from Sigma Genosys (Woodlands, TX). PCR amplification was according to Fang et al. (2010). Amplified PCR products were separated and measured on an automated capillary electrophoresis system ABI 3730 XL (Applied Biosystems Inc.). GeneScan-400 ROX (Applied Biosystems Inc.) was used as an internal DNA size standard. A genetic linkage map was generated with JoinMap 4.0 software using default parameters (Van Ooijen 2006).
Transcript sequence analysis
Consensus sequences for the im and TM-1 alleles of Gh_A03G0489 were extracted from the RNAseq and sBSAseq alignment files with IGV software (Robinson et al. 2011). Open reading frames were identified using the translate tool software at ExPASy (Gasteiger et al. 2003). The subcellular target and cleavage site of the N-terminal transit peptide were predicted using TargetP software (Emanuelsson et al. 2000; Nielsen et al. 1997). PPR motifs were found using the PROSITE entry PS51375 and the ScanProsite software (De Castro et al. 2006; Sigrist et al. 2013). The RNA-binding site specificity of the PPR sequence was predicted following the method of Yagi et al. (2013). The resulting probability matrix was converted to a sequence logo by Seq2Logo software (Thomsen and Nielsen 2012).
Data availability
All relevant data are within the paper and the associated Supplemental Material files. Table S1 contains detailed descriptions of all studied cotton accessions. Table S2 contains all of the primers designed for this research. Figure S2 contains the consensus coding sequences obtained for the im and TM-1 alleles of PPR Gh_A03G0489.
Results
The immature fiber mutant phenotype
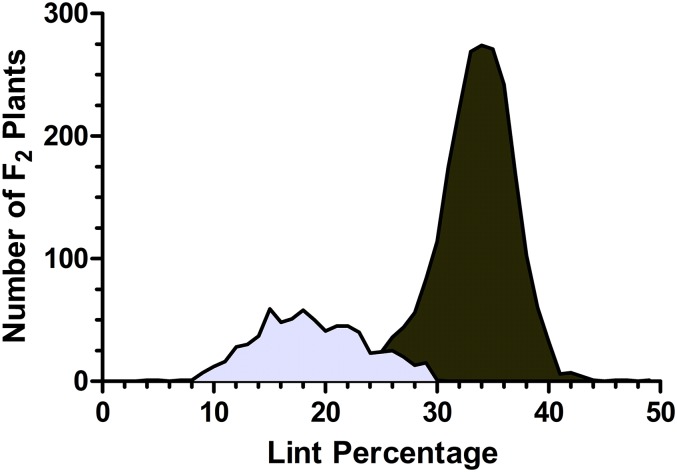
As before, we were able to treat the im phenotype as a qualitative character based on lint percentage and MIC (Kim et al. 2013a). Plants from segregating F2 plants were scored as wild type or im mutant using a threshold of 25% lint, and marginal plants were determined based on MIC value. The populations segregated 3:1, confirming that im phenotype is controlled by a single recessive locus (Figure 1). Comprehensive analysis of im fiber properties has been reported elsewhere (Kim et al. 2013a, 2013b).
Figure 1.
Lint percentage in 2837 F2 plants segregating for the immature fiber trait. Wild-type plants are represented by the dark gray histogram, while im plants are light gray. Plants producing less than 25% lint were scored as immature fiber, and plants with 26–29% lint were further evaluated by micronaire (MIC).
Mapping-by-sequencing
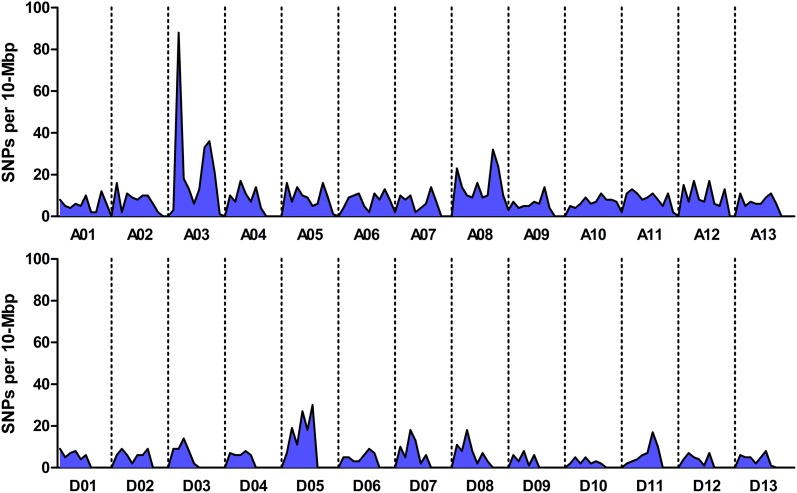
Comparison of the sequences obtained from sBSAseq clearly identified a region of chromosome 3 (Chr. A03) with high nucleotide polymorphism between the im mutant and wild-type bulks of F2 plants (Figure 2). The striking peak on Chr. A03 identified 88 high quality SNPs between the 10th and 20th Mb of the reference Chr. A03 (Zhang et al. 2015). This region is consistent with earlier reports, and contains sequences that match SSRs that were previously linked to the im gene (Figure 3) (Kim et al. 2013a; Wang et al. 2013).
Figure 2.
Distribution of SNPs in sBSAseq data from bulked immature and wild-type F2 plants. The chromosomes are named according to the draft reference Gossypium hirsutum cv. TM-1 genome.
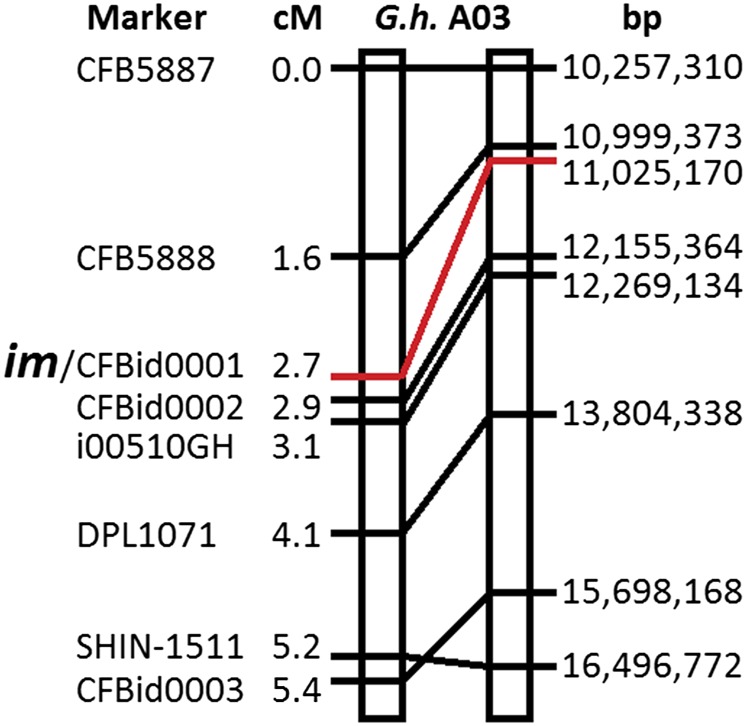
Figure 3.
Genetic and physical map of the immature genetic locus based on 2981 F2 plants. Genetic distances are shown in centiMorgans (cM), and markers are also located on chromosome A03 of the draft reference G. hirsutum cv. TM-1 genome.
Fine linkage mapping
To explore linkage between sequence polymorphisms in the sequencing data on Chr. A03 and the im gene, we scored two new SNP markers (CFB5887 and CFB5888), three new indel markers (CFBid0001, CFBid0002, and CFBid0003), a SNP from the CottonSNP63k array (i00510GH), and two SSRs (DPL1071 and SHIN-1511) on 2981 segregating F2 plants. Although they lacked phenotypes, 144 of these plants were included to determine accurate genetic distances between the markers. Generally, our genetic markers were spaced each megabase of physical distance, and were found to be about a centiMorgan apart, giving a 1 cM/Mb ratio in this region (Figure 3). All 708 plants that displayed the im phenotype were homozygous for the allele of the CFBid0001 marker that is present in the im NIL that was a parent in each F2 population. Furthermore, no wild-type plant was homozygous for the im allele of CFB0001, indicating complete linkage of this marker with the immature fiber phenotype in 2837 F2 plants (Figure 3). Of the closely linked markers, only CFBid0001 is located in the coding sequence of a gene, and is a 22-bp deletion in a PPR gene, Gh_A03G0489.
Gene expression analysis
We examined the expression of all annotated proteins between markers CFB5887 and CFBid0002, which corresponds to approximately 1-Mb of sequence on either side of the completely linked marker, CFBid0001 (Figure 3). Of the 38 annotated proteins in this interval, only 15 were detected at > 1 RPKM in either mutant or wild type fibers at 28 DPA. Of these, only one, Gh_A03G0468, which has similarity to protoheme IX farnesyltransferase, was significantly differentially expressed (P < 0.05) (Table 1). We tested all 38 genes for relative transcript abundance by RT-qPCR in 10, 17, and 28-DPA fiber cells, but were able to detect expression of only 28 genes. None of these genes showed significant, twofold differential expression between immature and wild-type fibers at any of the time points, including Gh_A03G0468 and the PPR gene Gh_ A03G0489 (Figure S1).
Table 1. Gene expression near the im locus in 28-DPA fiber cells by RNAseq.
| Gene/Marker | Position Chr. A03 | TM-1 RPKM | im RPKM | log2 (im/TM-1) | P-value | TAIR Ortholog | Description |
|---|---|---|---|---|---|---|---|
| CFB5887 | 10,257,310 | Flanking SNP marker | |||||
| Gh_A03G0468 | 10,292,732 | 6.4 | 1.5 | −2.1 | 0.024 | None | Protoheme IX farnesyltransferase |
| Gh_A03G0469 | 10,294,222 | 1.2 | 1.4 | 0.2 | 0.052 | AT5G50580 | SUMO-activating enzyme 1B |
| Gh_A03G0473 | 10,352,804 | 1.5 | 1.3 | −0.2 | 0.752 | AT3G26890 | None |
| Gh_A03G0475 | 10,381,659 | 3.1 | 2.9 | −0.1 | 0.685 | AT4G00650 | FRIGIDA-like protein |
| Gh_A03G0480 | 10,491,490 | 1.4 | 1.2 | −0.2 | 0.840 | AT5G51050 | Mitochondrial substrate carrier family protein |
| Gh_A03G0483 | 10,599,666 | 3.0 | 2.3 | −0.4 | 0.193 | AT5G51020 | CRUMPLED LEAF |
| Gh_A03G0484 | 10,616,903 | 16.8 | 15.9 | −0.1 | 0.660 | AT1G01630 | Sec14p-like phosphatidylinositol transfer protein |
| CFB5888 | 10,999,373 | Flanking SNP marker | |||||
| Gh_A03G0489 | 11,022,670 | 2.4 | 1.0 | −1.3 | 0.164 | AT1G64580 | Pentatricopeptide repeat (PPR) protein, mitochondrial |
| CFBid0001 | 11,025,170 | im-Linked 22-bp deletion in exon of PPR | |||||
| Gh_A03G0491 | 11,449,872 | 12.7 | 9.6 | −0.4 | 0.192 | AT5G50920 | Chaperone protein ClpC, chloroplastic |
| Gh_A03G0492 | 11,458,109 | 4.0 | 4.3 | 0.1 | 0.139 | AT2G42490 | Copper methylamine oxidase |
| Gh_A03G0493 | 11,495,867 | 29.6 | 26.1 | −0.2 | 0.933 | AT4G24690 | NBR1, a selective autophagy substrate |
| Gh_A03G0495 | 11,523,884 | 2.9 | 1.8 | −0.7 | 0.676 | None | None |
| Gh_A03G0496 | 11,525,162 | 1.1 | 0.3 | −1.9 | 0.126 | AT4G24700 | None |
| Gh_A03G0498 | 11,626,203 | 14.3 | 13.5 | −0.1 | 0.457 | AT4G20360 | RAB GTPase homolog E1B |
| Gh_A03G0500 | 11,796,370 | 1.7 | 1.9 | 0.2 | 0.529 | AT5G62350 | Pectin methylesterase inhibitor superfamily protein |
| CFBid0002 | 12,155,364 | Flanking deletion marker | |||||
| Gh_A03G0506 | 12,155,624 | 0.0 | 0.0 | 1.6 | 0.559 | AT1G12260 | NAC 007 |
Only annotated proteins with >1 RPKM and a select cell wall gene are presented. Nearby marker locations are indicated
Association of markers in diversity panel
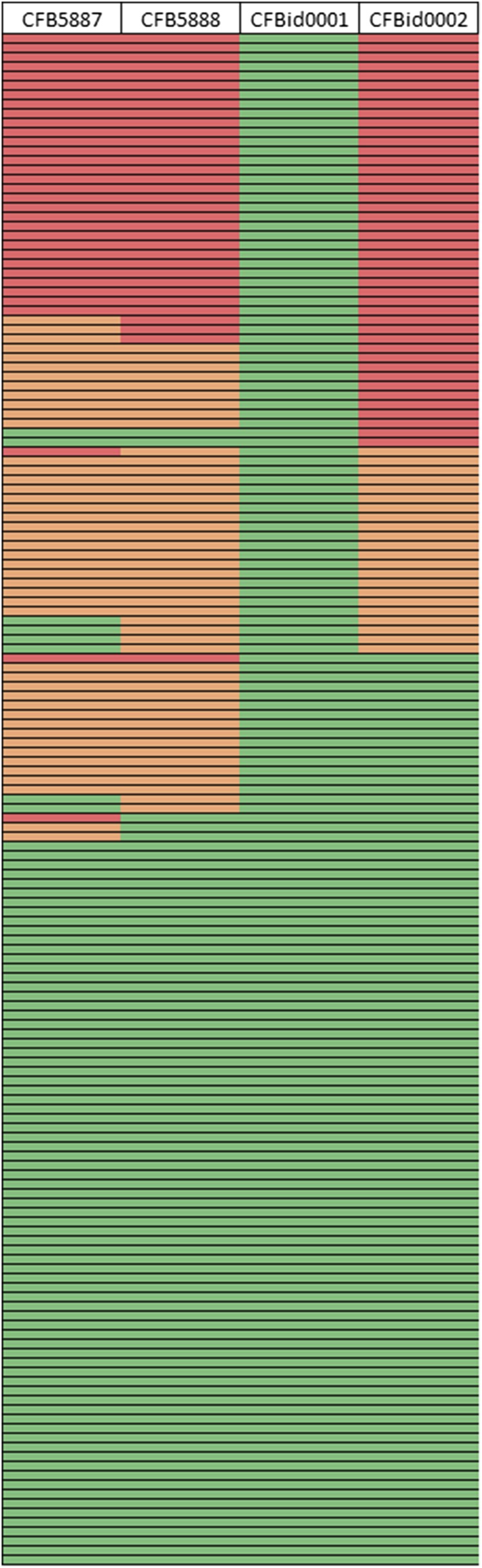
To ascertain if any of our closely linked markers could be the causative mutation that controls the immature phenotypic trait, we tested the completely linked marker CFBid0001, and three closely linked markers (CFB5887, CFB5888, and CFBid0002) on a diversity panel of 163 cultivated wild-type varieties (Figure 4 and Table S1). Of these varieties, not one contained an im-type allele of CFBid0001/Gh_A03G0489, including 30 lines that were homozygous for the im-type allele of all three nearby markers. These 30 varieties originate from diverse countries, including the USA, China, Australia, Pakistan, India, and Uzbekistan (Table S1). An additional 56 varieties contained im-type alleles in at least one of the three nearby markers (Figure 4 and Table S1).
Figure 4.
Association of closely linked genetic markers in a diversity panel of 163 accessions of G. hirsutum. Each row is an independent variety described in Table S1, and the columns are labeled with the respective genetic markers. Homozygocity for the im-type allele is shown in red, homozygocity for the TM-1-type allele is shown in green, and heterozygocity for a marker is indicated with orange.
Characterization of PPR Gh_ A03G0489 transcripts
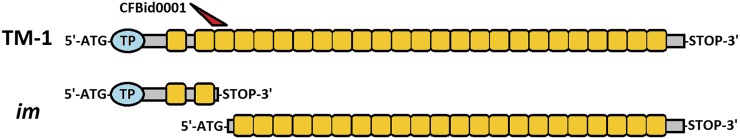
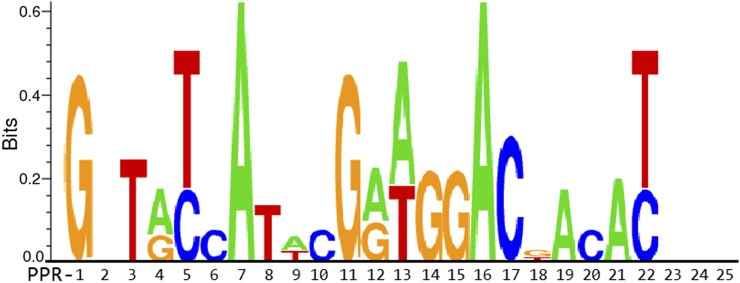
To predict the consequences of the 22-bp deletion in PPR Gh_A03G0489, we first obtained the full-length consensus coding sequences for the im and TM-1 alleles from our short read RNAseq and sBSAseq alignments (Figure S2). Next, we identified open reading frames (ORFs) in each transcript. The wild-type transcript contained a single ORF that corresponded to the full-length protein sequence [1023 amino acids (aa)], as expected, while the im transcript contained two ORFs longer than 100 aa (Figure S3). One of these ORFs begins at the canonical ATG but, because of the frame-shift deletion, terminates after 184 aa. The second ORF initiates from a downstream ATG and contains the C-terminal 813 aa of the reference protein (Figure S3). Since the annotation for the protein identifies it as an ortholog of a mitochondria-targeted PPR protein, we used the ORFs, a reference PPR motif, and a transit peptide database to further characterize the proteins. Like its orthologs, Gh_A03G0489, has an N-terminal transit peptide that is predicted to confer mitochondrial targeting (Table S3). The full-length protein sequence contains 25 PPR-like repeats, with 24 in immediate succession (Figure 5 and Table S3). Of the two ORFs in the im transcript, one contains the transit peptide sequence and the first two PPR repeats, while the other long ORF contains the C-terminal 22 PPRs. Since the crystal structure of PPR proteins bound to RNA has been solved and PPR binding preferences identified, we followed the method of Yagi et al. (2013) to identify the critical nucleotide specifying residues in each repeat and predict the nucleic acid binding motif of wild-type Gh_A03G0489, which we present as a sequence logo and position-specific weight matrix (Figure 6 and Table S3).
Figure 5.
Open reading frames in immature (im) and wild-type (TM-1) transcripts of Gh_ A03G0489. The N-terminal mitochondrial transit peptide is labeled with TP. The location of the im-linked 22-bp deletion, CFBid0001, is indicated. The RNA-binding pentatricopeptide repeats (PPR) are indicated with squares with rounded corners.
Figure 6.
Predicted mitochondrial RNA-binding motif for PPR gene Gh_A03G0489. The nucleotide specified by each of the 25 PPR domains is shown. The height of each letter is proportional to the expected frequency of binding. For simplicity, only positive bit scores are shown. For a complete position-specific weight matrix, see Table S3.
Discussion
Mapping-by-sequencing of the immature fiber (im) gene
We took advantage of the recently released draft reference genome for G. hirsutum cultivar TM-1 to identify a region of diversity between two bulked pools of F2 plants that were segregating for the im gene (Figure 2). Within the 10-Mb interval that contained the highest density of SNPs, we also identified indels, which, along with the SNPs, we used to design genetic markers to test on individual F2 plants from a total population of 2981 (Figure 3). Of these, 2837 plants produced enough fiber to be phenotyped. One deletion marker, CFBid0001, was completely linked to the phenotype in the segregating progeny (Figure 3). This 22-bp deletion is a striking frame-shift mutation in the coding sequence of a PPR gene, Gh_A03G0489 (Figure S2 and Figure S3).
Diversity panel
A nearby, but not completely linked, marker, CFBid0002, is a deletion less than 300 bp upstream of Gh_A03G0506, which is an ortholog of NAC 007, a transcription factor that orchestrates secondary cell wall development in Arabidopsis (Table 1) (Ooka et al. 2003; Zhou et al. 2014). Because of the altered secondary cell wall thickness in fiber cells of plants with the immature fiber phenotype, we also considered this mutation as an appealing candidate near the im locus. Since the causative mutation for the immature fiber trait results in an obvious phenotype, it should be absent from most, if not all, cultivated G. hirsutum varieties. When we tested the four closest markers on a panel of 163 cultivars, we found that the deletion in the NAC promoter was prevalent, but the CFBid0001 deletion in the PPR coding sequence was completely absent (Figure 4 and Table S2). Additionally, the two nearby SNP markers, CFB5887 and CFB5888, were present in many varieties. The presence of im-type alleles of three of the markers in the same varieties suggests that these are mutations of little agronomic consequence that reside together in a haplotype that has long been part of the global cotton germplasm, and are examples of identity by descent. Conversely, the lack of the im allele of the CFBid0001 marker in the diversity panel suggests that this mutation is relatively recent and is absent from cultivated germplasm. Indeed, this evidence is consistent with the history of the immature fiber mutation (Kohel et al. 1974).
PPR genes affect mitochondrial gene expression
The PPR gene family consists of 450 genes in Arabidopsis and has been shown to play critical roles in all stages of organellar gene expression (Barkan and Small 2014). PPR proteins are specifically targeted to chloroplasts and mitochondria, where they bind single-stranded RNA (O’Toole et al. 2008; Barkan and Small 2014; Yin et al. 2013). Different PPR proteins associate with transcription and translation machineries and are involved in various aspects of organellar mRNA processing, including splicing, cleavage, and editing (Barkan and Small 2014). Since the crystal structure of the PPR domain bound to RNA has been solved, a “PPR-code” has been proposed based on the nucleotide specifying residues in each repeat (Yagi et al. 2013; Yin et al. 2013; Gully et al. 2015). Our analysis of transcripts in im mutant and wild-type parental NILs indicates that the 22-bp deletion should abolish the function of the PPR gene Gh_A03G0489. Of the reading frames that are present in the mutant transcript, one contains essentially only the transit peptide, while another contains most of the RNA-binding domain, but importantly lacks the transit peptide (Figure 5 and Figure S3). N-terminal transit peptides are required for import to chloroplasts and mitochondria, and can be specific to either or both (Emanuelsson et al. 2000). Analysis of Gh_A03G0489 indicates that it contains a mitochondria-specific transit peptide and 25 PPR repeats with a specific predicted RNA binding motif (Figure 6 and Table S3).
Role of mitochondria in the development of cotton fiber properties
Our earlier work strongly implicated mitochondrial function in the development of the immature fiber mutant phenotype. We previously observed alteration of transcript levels of 12 genes involved in the cytochrome c oxidase respiration pathway, and activation of the alternative oxidase respiration machinery in mitochondria of the im mutant (Kim et al. 2013b). As the results of biochemical analysis in addition to the cotton microarray data, we suggested that deregulations of mitochondrial respiration in the im mutant fibers could cause energy deprivation that might reduce the degree of wall thickness of the im mutant fibers (Kim et al. 2013b). The verification of the significant upregulation (321-fold) of alternative oxidase (Gh_A12G2493/ Gh_D12G2621) by RNA-seq in the actively developing im mutant fibers at 28 DPA consistently indicated mitochondrial dysfunction in the im mutant fibers. Similarly, in Arabidopsis, disruption of the mitochondria-targeted PPR40 gene by T-DNA insertional mutagenesis interrupted the mitochondrial cytochrome pathway and activated the alternative pathway (Zsigmond et al. 2008). Our group has also identified altered mitochondrial gene expression in a different cotton fiber mutant, underscoring the importance of mitochondria in cotton fiber development and inviting future study (Thyssen et al. 2014b). Here, we have presented evidence of complete linkage of a frame-shifted PPR gene Gh_A03G0489 to the im allele that directly supports the association between mitochondrial dysfunction and mutant fiber phenotypes (Kim et al. 2013b). Further work is required to identify the specific RNA targets of the im candidate PPR gene Gh_A03G0489 in cotton mitochondria, and the direct functional consequence. Although mitochondrial dysfunction can clearly compromise fiber quality, it is not a foregone conclusion that wild-type mitochondrial function can be altered to improve fiber quality. However, researchers can already begin mining cotton genetic diversity for alternative alleles of PPR genes in the hope of fine-tuning maturity and fineness by affecting mitochondrial gene expression, to the benefit of cotton breeders, growers, and consumers.
Supplementary Material
Acknowledgments
We appreciate Mrs. Sheron Simpson and Dr. Brian Scheffler at the Genomics and Bioinformatics Research Unit, USDA-ARS at Stoneville, MS, for their support in SSR and indel marker analysis. We appreciate Dr. Marina Naoumkina of the Cotton Fiber Bioscience Unit, USDA-ARS-SRRC in New Orleans, LA, for assistance with statistical analysis. This project was financially supported by the USDA-ARS CRIS project no. 6054-21000-017-00D and Cotton Incorporated project no. 12-199. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture, which is an equal opportunity provider and employer.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.027649/-/DC1
Communicating editor: A. H. Paterson
Literature Cited
- Barkan A., Small I., 2014. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Bradow, J., G. Davidonis, O. Hinojosa, L. Wartelle, K. Pratt et al., 1996 Environmentally induced variation in cotton fiber maturity and related yarn and dyed knit defects. Proceedings Beltwide Cotton Conferences, Nashville, TN, January 9–12, 1996: Vol 2, pp 1279–1284. [Google Scholar]
- Clement J., Constable G., Liu S., 2014. Increasing cotton seed fibre density as a breeding strategy to improve fibre fineness. Field Crops Res. 160: 81–89. [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27(15): 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro E., Sigrist C. J., Gattiker A., Bulliard V., Langendijk-Genevaux P. S., et al. , 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34(suppl 2): W362–W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M., Ye W., Lin L., He S., Du X., et al. , 2015. The hairless stem phenotype of cotton (Gossypium barbadense) is linked to a copia-like retrotransposon insertion in a homeodomain-leucine zipper gene (HD1). Genetics 201(1): 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard E., Richter B. G., Rozen S., Stutius L. M., Angell N. A., et al. , 2000. A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiol. 124(4): 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G., 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300(4): 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fang D. D., Xiao J., Canci P. C., Cantrell R. G., 2010. A new SNP haplotype associated with blue disease resistance gene in cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 120(5): 943–953. [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R. D., et al. , 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31(13): 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gully B. S., Cowieson N., Stanley W. A., Shearston K., Small I. D., et al. , 2015. The solution structure of the pentatricopeptide repeat protein PPR10 upon binding atpH RNA. Nucleic Acids Res. 43: 1918–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulse-Kemp A.M., Lemm J., Plieske J., Ashrafi H., Buyyarapu R., et al. , 2015. Development of a 63K SNP array for cotton and high-density mapping of intra- and inter-specific populations of Gossypium spp. G3 (Bethesda) 5(6):1187–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Moon H. S., Delhom C. D., Zeng L., Fang D. D., 2013a Molecular markers associated with the immature fiber (im) gene affecting the degree of fiber cell wall thickening in cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 126(1): 23–31. [DOI] [PubMed] [Google Scholar]
- Kim H. J., Tang Y., Moon H. S., Delhom C. D., Fang D. D., 2013b Functional analyses of cotton (Gossypium hirsutum L.) immature fiber (im) mutant infer that fiber cell wall development is associated with stress responses. BMC Genomics 14(1): 889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohel R., Quisenberry J., Benedict C., 1974. Fiber elongation and dry weight changes in mutant lines of cotton. Crop Sci. 14(3): 471–474. [Google Scholar]
- Kohel R., Stelly D., Yu J., 2002. Tests of six cotton (Gossypium hirsutum L.) mutants for association with aneuploids. J. Hered. 93(2): 130–132. [DOI] [PubMed] [Google Scholar]
- Michelmore R. W., Paran I., Kesseli R., 1991. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88(21): 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5(7): 621–628. [DOI] [PubMed] [Google Scholar]
- Naoumkina M., Thyssen G., Fang D. D., Hinchliffe D. J., Florane C., et al. , 2014. The Li2 mutation results in reduced subgenome expression bias in elongating fibers of allotetraploid cotton (Gossypium hirsutum L.). PLoS One 9(3): e90830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina M., Thyssen G. N., Fang D. D., 2015. RNA-seq analysis of short fiber mutants Ligon-lintless-1 (Li1) and -2 (Li2) revealed important role of aquaporins in cotton (Gossypium hirsutum L.) fiber elongation. BMC Plant Biol. 15: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht J., Brunak S., von Heijne G., 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10(1): 1–6. [DOI] [PubMed] [Google Scholar]
- Ooka H., Satoh K., Doi K., Nagata T., Otomo Y., et al. , 2003. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10(6): 239–247. [DOI] [PubMed] [Google Scholar]
- O’Toole N., Hattori M., Andres C., Iida K., Lurin C., et al. , 2008. On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25(6): 1120–1128. [DOI] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29(1): 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C. J., De Castro E., Cerutti L., Cuche B. A., Hulo N., et al. , 2013. New and continuing developments at PROSITE. Nucleic Acids Res. 41: D344–D347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H., Abe A., Yoshida K., Kosugi S., Natsume S., et al. , 2013. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74(1): 174–183. [DOI] [PubMed] [Google Scholar]
- Thomsen M. C. F., Nielsen M., 2012. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 40(W1): W281–W287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyssen G. N., Fang D. D., Turley R. B., Florane C., Li P., et al. , 2014a Next generation genetic mapping of the Ligon-lintless-2 (Li 2) locus in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 127(10): 2183–2192. [DOI] [PubMed] [Google Scholar]
- Thyssen G. N., Song X., Naoumkina M., Kim H.-J., Fang D. D., 2014b Independent replication of mitochondrial genes supports the transcriptional program in developing fiber cells of cotton (Gossypium hirsutum L.). Gene 544(1): 41–48. [DOI] [PubMed] [Google Scholar]
- Thyssen G. N., Fang D. D., Turley R. B., Florane C., Li P., et al. , 2015. Mapping-by-sequencing of Ligon-lintless-1 (Li 1) reveals a cluster of neighboring genes with correlated expression in developing fibers of Upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 128: 1703–1712. [DOI] [PubMed] [Google Scholar]
- Van Ooijen J., 2006. JoinMap 4 Software for the calculation of genetic linkage maps in experimental populations, Kyazma BV, Wageningen, Netherlands. [Google Scholar]
- Wang C., Zhang T., Guo W., 2013. The mutant gene negatively affects many aspects of fiber quality traits and lint percentage in cotton. Crop Sci. 53(1): 27–37. [Google Scholar]
- Wang C., Lv Y., Xu W., Zhang T., Guo W., 2014. Aberrant phenotype and transcriptome expression during fiber cell wall thickening caused by the mutation of the Im gene in immature fiber (im) mutant in Gossypium hirsutum L. BMC Genomics 15(1): 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. D., Nacu S., 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26(7): 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T., 2013. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One 8(3): e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P., Li Q., Yan C., Liu Y., Liu J., et al. , 2013. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504(7478): 168–171. [DOI] [PubMed] [Google Scholar]
- Zhang T., Hu Y., Jiang W., Fang L., Guan X., et al. , 2015. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33: 531–537. [DOI] [PubMed] [Google Scholar]
- Zhou J., Zhong R., Ye Z.-H., 2014. Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS One 9(8): e105726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.-H., Zhang J., Liu D., Stiller W., Liu D., et al. , 2015. Integrated mapping and characterization of the gene underlying the okra leaf trait in Gossypium hirsutum L. J. Exp. Bot. 67: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsigmond L., Rigó G., Szarka A., Székely G., Ötvös K., et al. , 2008. Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol. 146(4): 1721–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the paper and the associated Supplemental Material files. Table S1 contains detailed descriptions of all studied cotton accessions. Table S2 contains all of the primers designed for this research. Figure S2 contains the consensus coding sequences obtained for the im and TM-1 alleles of PPR Gh_A03G0489.