Abstract
Eukaryotic DNA replication begins from multiple origins. The origin recognition complex (ORC) binds origin DNA and scaffolds assembly of a prereplicative complex (pre-RC), which is subsequently activated to initiate DNA replication. In multicellular eukaryotes, origins do not share a strict DNA consensus sequence, and their activity changes in concert with chromatin status during development, but mechanisms are ill-defined. Previous genome-wide analyses in Drosophila and other organisms have revealed a correlation between ORC binding sites and the histone variant H3.3. This correlation suggests that H3.3 may designate origin sites, but this idea has remained untested. To address this question, we examined the enrichment and function of H3.3 at the origins responsible for developmental gene amplification in the somatic follicle cells of the Drosophila ovary. We found that H3.3 is abundant at these amplicon origins. H3.3 levels remained high when replication initiation was blocked, indicating that H3.3 is abundant at the origins before activation of the pre-RC. H3.3 was also enriched at the origins during early oogenesis, raising the possibility that H3.3 bookmarks sites for later amplification. However, flies null mutant for both of the H3.3 genes in Drosophila did not have overt defects in developmental gene amplification or genomic replication, suggesting that H3.3 is not essential for the assembly or activation of the pre-RC at origins. Instead, our results imply that the correlation between H3.3 and ORC sites reflects other chromatin attributes that are important for origin function.
Keywords: DNA replication, H3.3, amplicon, chromatin, histone variant
DNA replication in multicellular eukaryotes initiates from multiple origins to ensure timely duplication of the genome (Huberman 1968; Mechali 2010). The right number of origins must be activated exactly once every cell cycle to avoid either under-replication or over-replication of the genome. Defects in origin regulation compromise genome integrity, contribute to cancer, and cause heritable developmental syndromes (Negrini et al. 2010; Bicknell et al. 2011a, 2011b; Guernsey et al. 2011; Kawabata et al. 2011; Hills and Diffley 2014). Despite the importance of origin regulation, much remains unknown about how certain regions of the genome are selected to be active origins. While it is clear that origin activity is strongly influenced by chromatin, molecular mechanisms are not fully defined (Mechali et al. 2013; Hyrien 2015). Here, we investigate how the histone variant H3.3 influences origin function in Drosophila.
The proteins that are required for origin function are conserved from yeast to humans, but, in metazoa, little is known about how specific genomic sites are selected to be origins (Remus and Diffley 2009; Mechali 2010). The origin recognition complex (ORC) binds to origin DNA during G1-phase, and serves as a platform for assembly of a prereplicative complex (pre-RC). A subset of these pre-RCs are then activated by S-phase kinases, resulting in DNA helix unwinding and the recruitment of other proteins to establish active replication forks (Diffley et al. 1995; Fernandez-Cid et al. 2013; Frigola et al. 2013; Weinreich 2015; Yeeles et al. 2015). In recent years, genomic approaches have identified thousands of origins in a variety of organisms including humans (Aladjem 2007; MacAlpine et al. 2010; Cayrou et al. 2011; Eaton et al. 2011; Gilbert 2012). Although some DNA sequence signatures have been reported, a strict origin consensus sequence has not emerged (Vashee et al. 2003; Remus et al. 2004; Cayrou et al. 2011, 2012; Besnard et al. 2012; Comoglio et al. 2015; Foulk et al. 2015). Thus, while it is clear that pre-RC assembly and activation occurs at specific sites in the genome, what specifies these sites remains incompletely defined.
It is now evident that chromatin exerts a strong influence on origin activity. Changes to chromatin during development correlate with different origin activities among cells (Hiratani et al. 2010; Besnard et al. 2012). Origins that are highly active, and that replicate early in S phase, tend to reside in genomic domains with active genes, and are enriched for histone acetylation and other activating chromatin modifications (Aggarwal and Calvi 2004; Danis et al. 2004; Hiratani and Gilbert 2009; MacAlpine et al. 2010; Unnikrishnan et al. 2010; Eaton et al. 2011; Liu et al. 2012). Conversely, origins within heterochromatin are less active and replicate later in S phase, although there are exceptions to this rule (Kim et al. 2003; Hayashi et al. 2009; Schwaiger et al. 2010). In some cases, how specific types of chromatin modifications influence pre-RC assembly and activation have been described, including the dimethylation of lysine 20 on histone H4 (H4K20me2), and acetylation of multiple lysines on histones H3 and H4 (Iizuka and Stillman 1999; Aggarwal and Calvi 2004; Miotto and Struhl 2008, 2010; Tardat et al. 2010; Kuo et al. 2012; Liu et al. 2012; McConnell et al. 2012). The position of nucleosomes also impacts origins, with most ORC binding sites being depleted of nucleosomes, whereas nucleosomes adjacent to some origins may actually assist pre-RC assembly (Lipford and Bell 2001; Eaton et al. 2010, 2011; MacAlpine et al. 2010; Muller et al. 2010; Hoggard et al. 2013; Liu et al. 2015). Despite these advances, the molecular mechanisms by which chromatin impacts origins remain little understood.
Genome-wide studies have shown that another chromatin property that correlates with origins is the histone variant H3.3 (Deal et al. 2010; MacAlpine et al. 2010; Eaton et al. 2011; Stroud et al. 2012). This correlation has led to speculation that nucleosomes containing H3.3 are important for marking sites of pre-RC assembly or activation (Deal et al. 2010; MacAlpine et al. 2010). The H3.3 variant differs from canonical H3 histones (H3.1 and H3.2) by just four to five amino acids depending on organism (Elsaesser et al. 2010; Szenker et al. 2011). Unlike canonical histone genes, which are present in high copy number, there are only two H3.3 genes in most multicellular eukaryotic genomes, including human, mouse, and Drosophila (Elsaesser et al. 2010; Szenker et al. 2011). While canonical H3 expression, and its incorporation into chromatin, is limited to S phase, the H3.3 variant is expressed throughout the cell cycle, and is deposited into chromatin in both a replication-dependent and independent manner (Ahmad and Henikoff 2002; Szenker et al. 2011). Nucleosomes that contain the two-histone variants H3.3 and H2A.z (H2Av in Drosophila) have a dynamic association with DNA, and are enriched at enhancers, promoters, and other highly accessible and active regions of the genome (Jin and Felsenfeld 2007; Henikoff 2009; Jin et al. 2009; Talbert and Henikoff 2010). One possibility is that loci containing these dynamic nucleosomes permit assembly of the pre-RC onto DNA, explaining the correlation of H3.3 with origins. Mutations or altered levels of H3.3, as well as the proteins required for its chromatin deposition, are drivers of pediatric and adult glioblastoma multiforme (GBM) (Schwartzentruber et al. 2012; Wu et al. 2012; Gallo et al. 2015). Given the correlation between H3.3 and origins, the high level of genome instability in these GBM tumors may, in part, be a manifestation of origin dysfunction. Thus, it is crucial to determine whether H3.3 plays a functional role at origins.
In this study, we use developmental gene amplification in somatic follicle cells of the Drosophila ovary as a model to evaluate the function of H3.3 at origins. During amplification, rereplication from origins at six specific loci increases the copy number of genes that are required for egg shell synthesis (Spradling 1981; Calvi 2006; Kim et al. 2011; Liu et al. 2012). These origins are genetically and molecularly well-defined, and provide a number of methodological advantages. Previous evidence indicated that nucleosome acetylation and position are important for ORC binding and amplicon origin function (Aggarwal and Calvi 2004; Hartl et al. 2007; Kim et al. 2011; Liu et al. 2012, 2015; McConnell et al. 2012). Here, we show that H3.3 is highly enriched at amplicon origins, beginning early in oogenesis before amplification, consistent with the idea that dynamic H3.3 nucleosomes may bookmark these sites for later amplicon origin activity. However, analysis of null mutants for histone H3.3 showed no overt defects in amplicon origin activity or genomic DNA replication, suggesting that this histone variant is not essential for the activity of most origins. Although not required for origin function, our results suggest that H3.3 abundance is associated with a chromatin environment that is conducive to origin activity.
Materials and Methods
Fly strains and genetics
All flies were raised at 25° on standard media. The heat shock inducible HS-H3.3A-GFP and HS-H3-GFP (Ahmad and Henikoff 2002), and H3.3 null mutants H3.3B0;H3.3A2*1/CyO-GFP (Sakai et al. 2009) were generous gifts from Kami Ahmad. The two strains with deficiencies encompassing the H3.3A gene were w1118; Df(2L)Exel7022 / CyO and w1118; Df(2L)BSC110 / CyO, and were obtained from the Bloomington Drosophila stock center. The C323:Gal4 was used to drive the expression of P{ w+mC UAS:dacapo} in flies that also had the heat inducible HS-H3.3A-GFP (de Nooij et al. 1996; Lane et al. 1996). Heat induction of HS-H3.3A-GFP was at 37° for 30 min, followed by recovery for 90 min at 25° before fixation for immunostaining or ChIP.
Antibodies, immunostaining, and microscopy
BrdU, antibody and DAPI labeling for Drosophila ovaries were performed as previously described (Calvi and Lilly 2004). α-BrdU (mouse monoclonal, BDB347580; BD Biosciences, San Diego, CA) was used at 1:20. α-GFP (rabbit, Molecular Probes, Life Technologies), and secondary antibodies Alexa 488 anti-rabbit (Invitrogen) were used at 1:500. For EdU labeling, unfixed cells were incubated in 10 µM EdU followed by fixation with formaldehyde. EdU incorporation was detected with Alexa Fluor-488 Azide (Invitrogen) in a copper-catalyzed Click-iT reaction (Buck et al. 2008; Salic and Mitchison 2008). Images were taken with a Leica DMRA2 widefield epifluorescence microscope (Figure S2, and Figure S5, C and D), and a Leica SP5 laser scanning confocal microscope (Figure 2, Figure 6, Figure S1, Figure S4, and Figure S5, A and B).
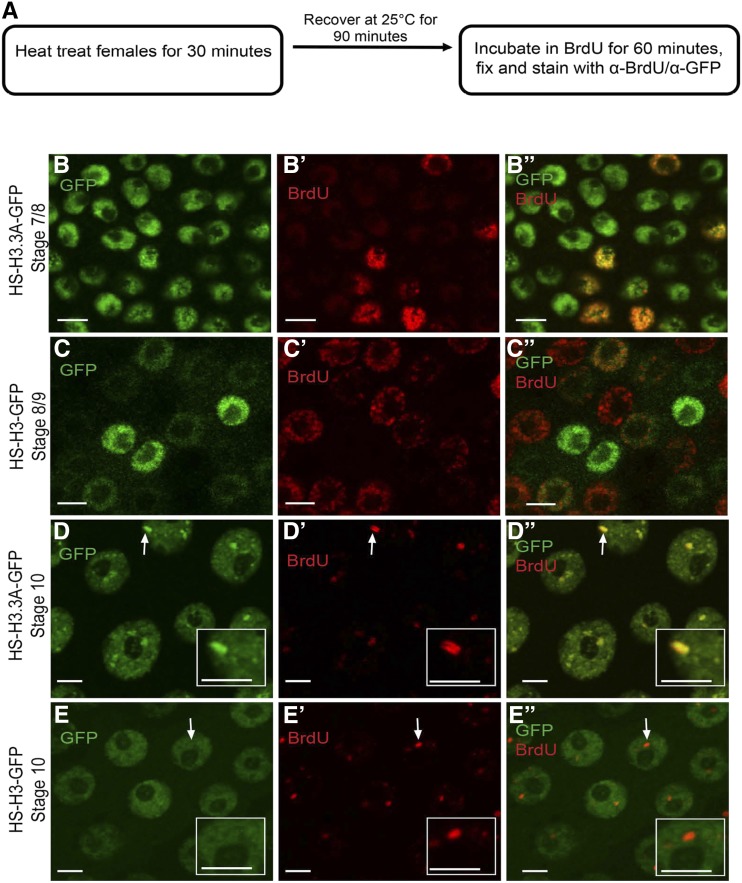
Figure 2.
H3.3 is enriched at the amplicon origins. (A) Experimental scheme for heat induction of H3.3A-GFP and BrdU labeling of follicle cells. (B)–(E”) Anti-GFP (B, C, D, and E), BrdU (B’, C’, D’, and E’), and merged images (B”, C”,D”, and E”) in endocycling (B–C”) and stage 10B amplifying (D–E”) follicle cells that are expressing either the histone variant H3.3A-GFP (B–B” and D–D”) or the canonical histone H3-GFP (C–C” and E–E”), using the protocol described in (A). White arrows in D–D” and E–E” indicate one highly-amplified DAFC-66D locus, which is shown at higher magnification in the insets. The BrdU double bars in D’, D” represent replication forks migrating bidirectionally outward from the origin. See Figure S1 for quantification. Scale bars are 5 µm.
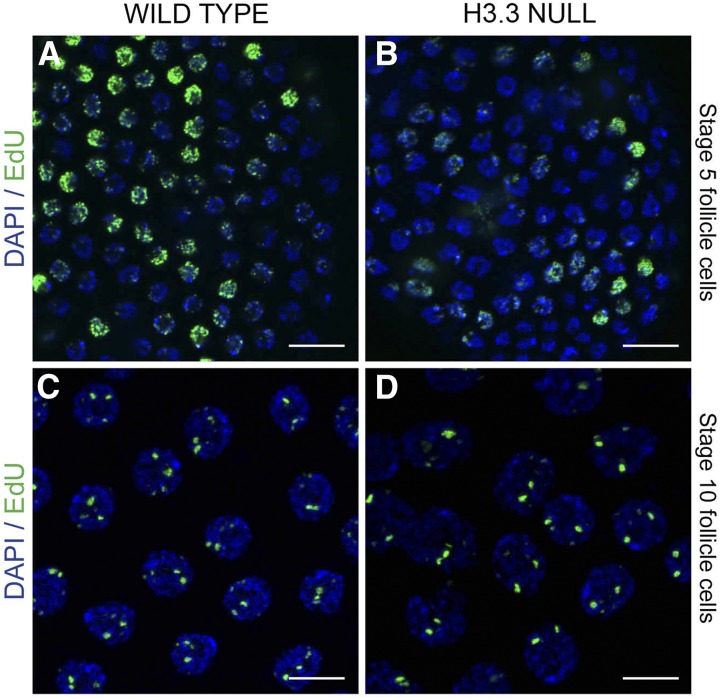
Figure 6.
H3.3 is not essential for genomic DNA replication or developmental gene amplification. (A)–(D) Follicle cell nuclear DNA labeled with DAPI (blue) and for EdU incorporation (green) during genomic replication (A and B) and amplification (C and D) in wild type control (A and C) or H3.3 null females (B and D). The H3.3 null females were homozygous mutant for an H3.3B null allele on the X, and a null H3.3A allele over deficiency on the second, H3.3B0; H3.3A2*1 / Df(2L)BSC110. Scale bars are 10 µm.
Follicle cell nuclear preparation
Follicle cell nuclei were purified from different stage egg chambers as described previously (Liu et al. 2012). Conditioned young adult females were blended by short pulses in cold phosphate-buffered saline buffer in 0.02% Tween-20 in a household blender. Released egg chambers were enriched by serial filtration through 250- to 70-μm meshes. Eggs were then fixed for 15 min at room temperature in 2% formaldehyde solution, followed by quenching with 125 mM glycine, and washing with cold Dulbecco’s phosphate-buffered saline. Stage 1–8 and stage 10 egg chambers were then further manually purified from the crude fractions, and stored at −80° prior to nuclear preparation. Approximately 600–700 stage 10 egg chambers were used for each nuclear preparation. For stage ≤ 8 egg chambers, a tissue volume similar to that of the stage 10 was used. Frozen eggs were thawed on ice, resuspended in mHB buffer (0.34 M sucrose, 15 mM NaCl, 60 mM KCl, 0.2 mM EDTA, 0.2 mM ethylene glycol tetraacetic acid, 0.15 mM spermine, 0.15 mM spermidine in 15 mM Tris-HCl, pH 8.0) supplemented with 0.5% NP-40. The tissue was homogenized in a Kontes 2-ml douncer via 15 strokes with a type A pestle. The lysate was filtrated with a 15-μm Nytex nylon membrane to remove the larger nurse cell nuclei. The filtrate was spun 3 min at 500 × g to pellet follicle nuclei.
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) protocol was as previously described, with slight modifications, and entailed biological replicates from separate isolations of follicle cell nuclei (Liu et al. 2012). Briefly, the prepared follicle cell nuclei were resuspended in nuclear lysis buffer (1% SDS, 1 mM EDTA in 50 mM Tris-Cl, pH 8.0), and subjected to sonication [Diagenode (Bioruptor sonication device), tip probe, high setting, four rounds of 5 min (30-sec on/off) sonication on iced water bath] to a modal size of 450 bp. One-sixth of the sample was saved as input, and the remainder was aliquoted and diluted at least five-fold into IP dilution buffer (0.01% SDS, 1.1% Triton-X 100, 1.1 mM EDTA, 167 mM NaCl in 20 mM Tris-Cl, pH 8.0) to a final volume of 1 ml. H3.3A-GFP ChIP was performed using 2 µl (∼2 µg) of rabbit polyclonal GFP antibody (Molecular Probes, Life Technologies), and Orc2 ChIP used a rabbit polyclonal Orc2 antibody (Austin et al. 1999); normal rabbit serum (1 mg/ml, Jackson ImmunoResearch) was used as a negative control. Incubation with antibody was with nutation at 4° overnight; 30 μl of 50% protein A agarose beads (15918-014; Invitrogen) pretreated with sheared salmon sperm DNA and BSA were added and incubated 1–3 hr at 4° to pull down the chromatin bound to the antibody. The beads were washed two times each with low-salt, high-salt, LiCl, and Tris–EDTA buffers. Elution and reversal of cross-linking was done by heating at 65° overnight with 5 M NaCl in nuclear lysis buffer. Both the input and eluate were treated with 1 μg of RNaseA (11119915001; Roche, Indianapolis, IN), and 50 μg of proteinase K (P8102S; New England BioLabs, Ipswich, MA). DNA was then purified with standard phenol/chloroform procedure and used for qPCR analysis.
qPCR
The enrichment of H3.3A-GFP or ORC at different Drosophila Amplicon in Follicle Cells (DAFCs) was determined by qPCR. Forty cycles of a two-step PCR protocol (denaturation at 95° for 15 sec, and annealing/extension at 62° for 30 sec) were run. The analysis was done on a Stratagene (Santa Clara, CA) Mx3005P machine with SYBR Green Master Mix (600843; Agilent, Santa Clara, CA). For qPCR quantification, the amount of DNA in the pellet as expressed as percentage of input DNA estimated by a standard curve generated from a serial dilution of the input. The values were then normalized to two control, nonorigin, loci at cytogenetic position 64A and 93E/F or to 93E/F alone (Figure 4). Enrichment was also determined at the Drosophila hsp70 locus, as a positive control (Schwartz and Ahmad 2005).
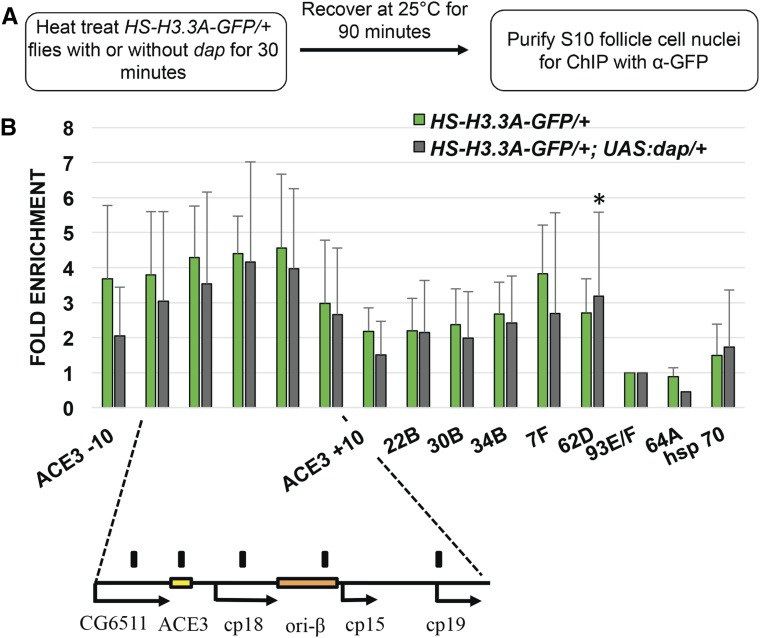
Figure 4.
H3.3 is enriched at amplification origins before the initiation of DNA replication. (A) Experimental scheme for H3.3A-GFP ChIP-qPCR with or without UAS:dacapo (UAS:dap) expression. (B) H3.3 is still enriched at the amplicons when replication initiation is blocked by dap. Anti-GFP ChIP-qPCR analysis of stage 10 follicle cell nuclei after heat induction of H3.3A-GFP from flies with (gray bars) or without (green bars) coexpression of the CDK2 inhibitor dap. Ratio of DNA in the pellet vs. input was normalized to pellet/input ratio at negative control locus 93E/F. The hsp70 locus is a positive control. Note that, because the H3.3A-GFP transgene was hemizygous in these genetic crosses, H3.3A-GFP enrichment at the amplicons is lower than in previous experiments. The DAFC-66D amplicon and position of primers (black bars) are shown below. The other amplicons assayed were DAFC—7F, 22B, 30B, 34B and 62D. The occupancy of H3.3A-GFP was not significantly different from UAS:dap expression, except for DAFC-62D (* P-value of 0.04, as calculated by ratio paired t-test, see Table S2). Values represent the average of three biological replicates, and error bars represent SD.
Statistical analysis
H3.3A-GFP enrichment at DAFC-66D and other amplicons DAFC—7F, 22B, 30B, 34B and 62D was measured as % input in stage 1–8 and stage 10 follicle cell nuclei, and compared to H3.3A-GFP enrichment in those stages at the nonorigin control loci 64A and 93E/F. H3.3A-GFP enrichment at DAFC-66D and other amplicons was also compared in the presence and absence of CDK inhibitor dacapo. The P-values for difference in enrichment over multiple biological replicates was computed by Ratio Paired t-Test using GraphPad Prism version 6.0 for Windows, GraphPad Software (La Jolla, CA; www.graphpad.com).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All data and reagents are available upon request.
Results
H3.3 is enriched at the active amplicon origins
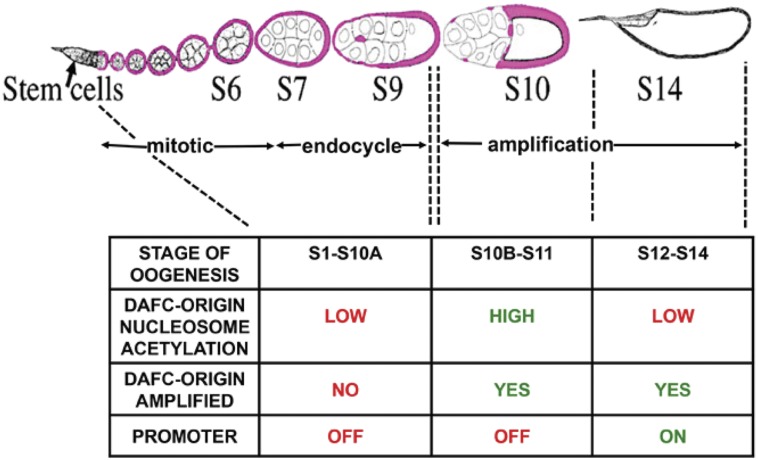
The Drosophila ovary affords a number of advantages for evaluating H3.3 function during DNA replication. The ovarian somatic follicle cells form an epithelial sheet around egg chambers, and undergo three types of DNA replication programs (Figure 1). These DNA replication programs are coordinated with maturation of the egg chambers as they develop through 14 stages as they migrate down the ovariole (Figure 1). Follicle cells proliferate via the canonical mitotic cycle in stages 1–6, periodically duplicate their genome without division during three endocycles (G/S cycle) in stages 7–10A, and then undergo developmental gene amplification in stages 10B–14 (Figure 1) (Calvi and Spradling 1999; Claycomb and Orr-Weaver 2005; Horne-Badovinac and Bilder 2005; Calvi 2006). During gene amplification, genomic replication ceases, and amplicon origins initiate multiple rounds of DNA replication at six loci, resulting in an increase in the DNA copy number of genes required for eggshell synthesis (Spradling 1981; Calvi et al. 1998; Kim et al. 2011).
Figure 1.
Developmental timeline of Drosophila oogenesis, follicle cell cycles, and amplicon origin activity. Egg chambers mature through 14 stages as they migrate posteriorly in the ovariole (from left to right). Somatic follicle cells (pink) surround the germline nurse cells and oocyte in each egg chamber. Follicle cells have three types of DNA replication programs: mitotic cycle (stages 1–6), endocycle (stages 7–10A), and developmental gene amplification (stages 10B–14). The table below shows the status of the amplicon origin, chorion gene promoters, and chromatin at DAFC-66D during different cell cycle programs.
To evaluate the behavior of H3.3 during different types of replication programs, we used a fly strain in which one of the two identical H3.3 genes (H3.3A) is fused to GFP and under the control of the heat-inducible hsp70 promoter (HS-H3.3A-GFP) (Ahmad and Henikoff 2002). We compared it to canonical histone H3 that was also fused to GFP and heat-inducible (HS-H3-GFP). Expression of these transgenes was induced in adult females at 37° for 30 min. Females were allowed to recover for 90 min before ovaries were dissected and incubated in the nucleotide analog BrdU for 60 min, followed by immunofluorescent labeling for BrdU and GFP (Figure 2A).
During mitotic cycles and endocycles (stages 1–10A), both H3.3A-GFP and H3-GFP labeled throughout the nuclei of a subset of follicle cells (Figure 2, B-C”). During amplification stages 10B-13, both H3.3A-GFP and H3-GFP had a low level of pan-nuclear, focal GFP labeling (Figure 2, D and E). In addition, stage 10B-13 follicle cells had several large nuclear foci of H3.3A-GFP, but not H3-GFP (Figure 2, D and E). We wondered whether the larger H3.3A-GFP foci corresponded to amplifying gene loci. During these stages, developmental gene amplification can be seen as foci of BrdU incorporation (Figure 2, D’ and E’) (Calvi et al. 1998; Calvi and Lilly 2004). The H3.3A-GFP foci colocalized with BrdU foci, whereas canonical H3-GFP did not (Figure 2, D-E”, and Figure S1, A-B’). These results suggest H3.3A-GFP is dynamically incorporated into chromatin at active amplicons to levels that exceed that of canonical H3-GFP.
To evaluate H3.3A-GFP incorporation at higher resolution, we analyzed its pattern at the largest BrdU focus. This focus is DAFC-66D, the amplicon that amplifies to highest copy number (∼64-fold), and which is the best characterized (Figure 2D”) (Spradling 1981). At DAFC-66D, ORC is bound during stages 10B-12—a time when origin nucleosomes are hyperacetylated and the origin is active—but the activity is low for nearby promoters of eggshell protein (chorion) genes (Figure 1) (Griffin-Shea et al. 1982; Aggarwal and Calvi 2004; Cavaliere et al. 2008; Kim et al. 2011; Liu et al. 2012). Since the chorion genes at DAFC-66D are not highly expressed during stages 10-11, most of the incorporation of H3.3A-GFP at this time is not mediated by transcription (Griffin-Shea et al. 1982; Cavaliere et al. 2008).
Previous studies have shown that immunolabeling at DAFC-66D has sufficient resolution to distinguish the location of proteins at origins or forks (Loebel et al. 2000; Whittaker et al. 2000; Calvi and Spradling 2001; Claycomb et al. 2002; Nordman et al. 2014; Alexander et al. 2015). During stage 10, the H3.3A-GFP foci had an extended morphology of ∼1.5 µm that was both between, and coincident with, double bars of BrdU incorporation, which represent forks migrating outward from the origin (Figure 2, D-D”, and Figure S1, A and A’). In contrast, fluorescence quantification indicated that the occupancy of canonical H3-GFP at DAFC-66D origins and forks was not different than other nuclear regions (Figure S1B and B’). In stage 12, ORC departs, the origin shuts off, and nearby chorion gene promoters become active (Figure 1) (Austin et al. 1999; Royzman et al. 1999). During this stage, H3.3A-GFP was again distributed both between, and coincident with, the double bars of nucleotide incorporation at the replication forks that continue to migrate outwards from the quiescent origin (Calvi and Spradling 2001) (Figure S1, C and C’, File S1). Comparing the size of the H3.3A-GFP foci to previous multi-color FISH measurements at DAFC-66D suggested that H3.3A-GFP incorporation occurs over a region of at least ∼20 kb, and is not restricted to DAFC-66D DNA replication initiation sites (Calvi and Spradling 2001).
H3.3 is abundant across multiple sites at DAFC-66D
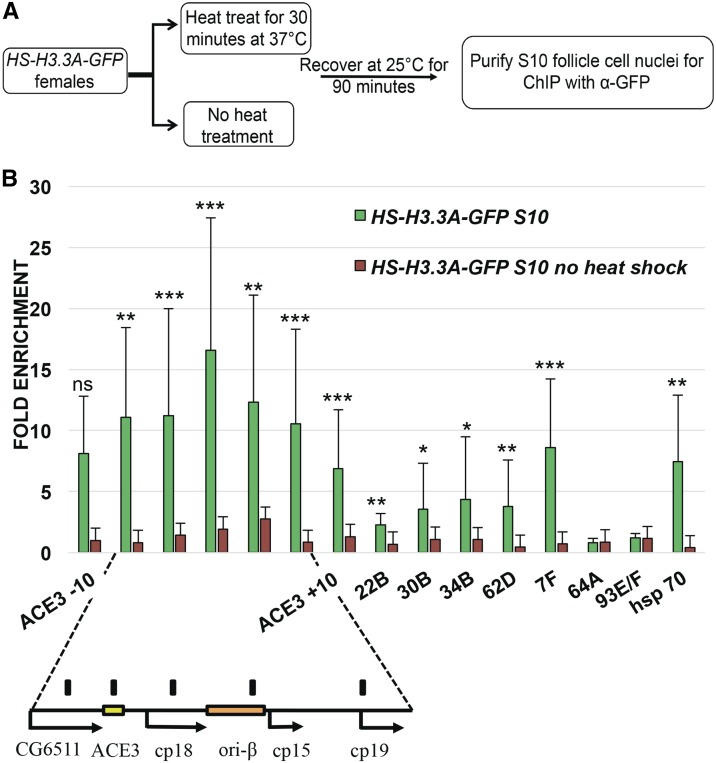
We then used anti-GFP antibodies for ChIP-qPCR to quantify the abundance of H3.3A-GFP at the amplicons to high-resolution. H3.3A-GFP expression was heat induced for 30 min, or not induced in controls, and, 1.5 hr later, stage 10 egg chambers were isolated, formaldehyde fixed, and follicle cell nuclei purified away from larger nurse cell nuclei by filtration, resulting in a high developmental resolution (Figure 3A). Cross-linked follicle cell chromatin was then sheared, and H3.3A-GFP immunoprecipitated with anti-GFP antibodies (Figure 3A). The amount of DAFC-66D DNA in the input and pellet was then assessed by qPCR, which internally controls for developmental amplification of copy number. The input: pellet ratio at the amplicon was then normalized to the input: pellet ratio at two nonorigin control loci at cytogenetic positions 93E/F and 64A. The high-level incorporation of H3.3 during heat-induced transcription of the hsp70Ba locus served as a positive control (Schwartz and Ahmad 2005). The ChIP-qPCR results indicated that H3.3 is significantly enriched at multiple positions across the DAFC-66D locus in stage 10B follicle cells (Figure 3B and Table S1, Table S4). The levels of H3.3 at DAFC-66D (∼6- to 16-fold) were comparable to, or exceeded, the high levels of H3.3 incorporation at the hsp70 transcription unit (∼6-fold) (Figure 3B). H3.3 enrichment was highest in the middle of the DAFC-66D amplicon, coinciding with the two origin regions ACE3 and ori-β, and the 1.5-kb region between them (Figure 3B). ACE3 and ori-β are ORC binding sites, and are required for DAFC-66D origin function, with replication initiation occurring most frequently within ori-β (Delidakis and Kafatos 1988, 1989; Orr-Weaver et al. 1989; Heck and Spradling 1990; Lu et al. 2001). To either side of ACE3 and ori-β, there was a gradient of diminishing H3.3 occupancy spanning a total of ∼20 kb, consistent with the immunofluorescence measurements of H3.3A-GFP foci (Figure 3B and Figure S1, A, A’, C, and C’). In contrast, negative control H3.3A-GFP females that were not heat induced did not have detectable enrichment of DNA in the pellet (Figure 3B). These results suggested that H3.3 is abundant in chromatin across DAFC-66D when this origin is initiating DNA replication in stage 10 follicle cells.
Figure 3.
H3.3 is enriched at amplicon origins. (A) Experimental scheme for H3.3A-GFP ChIP- qPCR for amplification stage 10 follicle cells. (B) Anti-GFP ChIP-qPCR analysis of stage 10 follicle cell nuclei after heat induction of H3.3A-GFP (green bars) or no heat induction control (small orange bars). The y-axis represents fold enrichment of H3.3A-GFP in the pellet relative to input, and then normalized to two nonorigin negative control loci, 93E/F and 64A. H3.3A-GFP deposition into the induced hsp70 transcribed region was a positive control. The DAFC-66D amplicon is shown below, including the essential regions ACE3 and ori-β. Multiple primers were used to quantify H3.3A-GFP chromatin across DAFC-66D, as well as 10 kb to either side of ACE3 (ACE3 –10 and ACE3 +10). The position of these qPCR primers at DAFC-66D are represented by black bars above the map. Also shown is the enrichment of H3.3A-GFP at the other amplicons DAFC—7F, 22B, 30B, 34B, and 62D. H3.3A-GFP was significantly enriched at multiple loci across DAFC 66D and other DAFCs in stage 10 follicle cells compared to the 64A and 93E/F control loci. (* P < 0.05, ** P < 0.01, *** P < 0.001 by ratio paired t-test, see Table S1). The data for heat induced H3.3A-GFP represent an average of at least three biological replicates, while values for no heat induction negative controls represent an average of at least two biological replicates. The error bars represent the range of values.
We also evaluated H3.3A-GFP enrichment at five other amplicon loci (DAFC-7F, DAFC-22B, DAFC-30B, DAFC-34B, and DAFC-62D), which amplify to lower final DNA copy number (Kim et al. 2011) (Table S4). H3.3A-GFP was significantly enriched at these amplicon loci, but to a lesser extent than at DAFC-66D (Figure 3B). Among these other amplicons, the DAFC-7F locus amplifies most highly (∼16-fold DNA copy number), and was the most enriched for H3.3A-GFP (∼9-fold), while the amplicons that amplify 4- to 8-fold in DNA copy number had lower, but significant, enrichment of H3.3A-GFP (∼2- to 4-fold) relative to nonorigin control loci (Figure 3B and Table S1) (Claycomb and Orr-Weaver 2005; Kim et al. 2011). Thus, including DAFC-66D (∼64-fold amplified), there was a correlation between the abundance of H3.3 in origin chromatin and the number of times the origin initiates DNA replication. These data indicate that H3.3 is highly abundant in the chromatin of all the amplicons when these origins are initiating replication in stage 10.
H3.3 deposition occurs before DNA replication initiates
In addition to its replication-independent deposition, it is known that H3.3 is deposited during chromatin assembly behind the replication fork (Ahmad and Henikoff 2002; Xu et al. 2010; Dunleavy et al. 2011; Frey et al. 2014). It remained unclear, therefore, whether the abundance of H3.3 at the amplicons represented replication-dependent incorporation, or whether H3.3 is also present at the origin before the initiation of DNA replication. To address this, we expressed the Cyclin E/CDK2 inhibitor dacapo (dap) beginning in stage 9 follicle cells, which blocks CDK activation of the pre-RC, but not pre-RC assembly, and strongly inhibits amplification (Figure 4A) (de Nooij et al. 1996; Lane et al. 1996; Calvi et al. 1998; Liu et al. 2012). We then compared the H3.3A-GFP ChIP-qPCR signal from cells hemizygous for H3.3A-GFP and that were, or were not, expressing dap (Figure 4B). The results showed that dap expression blocked replication but did not significantly reduce the H3.3A-GFP signal at the amplicons (Figure 4B and Figure S2). The distribution of H3.3A-GFP also appeared similar between dap-expressing and nonexpressing follicle cells, with highest abundance at the center of DAFC-66D (Figure 4B). These results suggest that much of the H3.3A-GFP incorporation at the amplicons occurs before the initiation of replication, and is not dependent on active replication forks.
H3.3 is enriched at DAFC-66D origin early in oogenesis before amplification
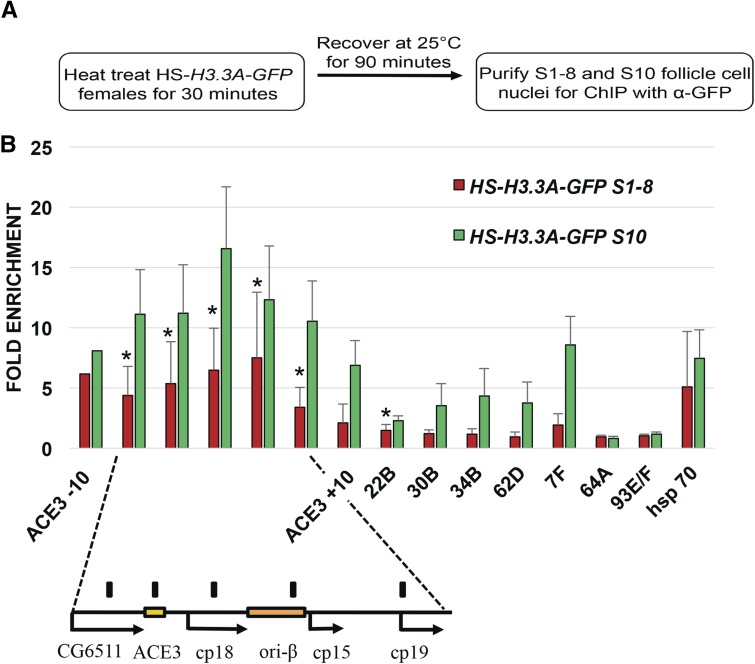
The dap results indicated that H3.3A-GFP is present at the amplicons before activation of the pre-RC in stage 10 follicle cells. This raised the question as to whether H3.3 is enriched at the amplicons in follicle cells during early oogenesis before the onset of amplification. To address this, we repeated the H3.3A-GFP ChIP on stage 1–8 follicle cells, a time in oogenesis that is at least 24 hr before the onset of amplification (Calvi et al. 1998; Horne-Badovinac and Bilder 2005) (Figure 5A). The results indicated that H3.3A-GFP was significantly enriched at DAFC-66D relative to control loci in preamplification stage 1–8 follicle cells (∼4- to 6-fold), although to a lesser extent than in stage 10 (∼6- to 16-fold) (Figure 5B). Enrichment of H3.3A-GFP at the other amplicons was not significantly different from control loci during stages 1–8, except for DAFC-22B (Figure 5B). These data indicate that the deposition of H3.3 at the DAFC-66D origin begins during early oogenesis before the onset of amplification.
Figure 5.
H3.3 is enriched at amplicon origins in early oogenesis before developmental amplification. (A) Experimental scheme for H3.3A-GFP ChIP-qPCR for stage 1–8 follicle cell nuclei. (B) Anti-GFP ChIP-qPCR analysis of stage 1–8 follicle cell nuclei after heat induction of HS-H3.3A-GFP (red bars). The relative abundance of H3.3A-GFP in stage 10 (green bars), when amplicon origins are active, was assessed in parallel from the same ovary preparations and is replotted from Figure 3B for comparison. During stages 1–8, H3.3A-GFP was significantly enriched at multiple loci across DAFC 66D and at DAFC 22B relative to the two nonorigin control loci 64A and 93E (* P < 0.05, ** P < 0.01 by ratio paired t-test). H3.3A-GFP occupancy was significantly lower during stages 1–8 than stage 10 (see Table S3 for P-values). Values represent average of at least three biological replicates, and error bars represent SD, except for ACE3 –10, which is based on two replicates.
The combined evidence from follicle cells during stages 1–8, and those expressing dap, suggested that pre-RC activation is not required for H3.3 deposition at DAFC-66D. We wondered whether H3.3 is deposited at the origin before pre-RC assembly, the first step of which is binding of ORC to DNA (Mechali 2010). Several lines of evidence suggest that the activity of the DAFC-66D origin is specific to late stage follicle cells. Data from modENCODE indicated that DAFC-66D does not initiate DNA replication during the S phase of several Drosophila cell lines, and is not significantly occupied by ORC in cell lines, embryos, or larval salivary glands (MacAlpine et al. 2010; modENCODE Consortiumet al. 2010; Eaton et al. 2011; Sher et al. 2012; Boley et al. 2014). Moreover, during the stage 9–10 transition at the onset of amplification, there is a significant increase in ORC occupancy at DAFC-66D, which is dependent on the acquisition of new histone acetylation at the origin (Austin et al. 1999; Aggarwal and Calvi 2004; Hartl et al. 2007; Kim et al. 2011; Liu et al. 2012; McConnell et al. 2012). However, the binding of ORC to DAFC-66D in stage 1–8 follicle cells has not been directly examined. To address this question, we used antibodies against the Orc-2 subunit for ChIP-qPCR on stages 1–8 and stage 10 follicle cells (Austin et al. 1999). While the mean ORC abundance was lower in stages 1–8 vs. stage 10, the variance in the data do not permit us to conclude that this difference is significant (Figure S3). Therefore, while all published evidence suggests that pre-RC binding and origin activity at DAFC-66D is specific to follicle cells in late oogenesis, our data do not permit us to conclude that H3.3 is bound before ORC in stages 1–8.
H3.3 is not essential for genomic replication or developmental gene amplification
The enrichment of H3.3 at DAFC-66D early in oogenesis raised the possibility that H3.3 may mark the locus for amplification during later oogenesis. This idea is consistent with the correlation between H3.3 and ORC binding sites from several genome-wide studies, which led to suggestions that H3.3 may be important for origin function (Deal et al. 2010; MacAlpine et al. 2010). To test this idea, we determined whether mutations in H3.3 impaired follicle cell genomic replication or developmental amplification. We used a strain that is null mutant for the two H3.3 genes in the Drosophila genome on the X and second chromosome (H3.3B0 ; H3.3A2*1), which was previously reported to be homozygous semilethal (Sakai et al. 2009). To control for cryptic second site mutations, we crossed this strain to two different strains that have second chromosome deletions spanning the H3.3A gene [Df(2L)Exel7022 and Df(2L) BSC110]. For both crosses, most H3.3B0; H3.3A2*1/ Df(2L) null flies were lethal before adulthood, with only a few rare escaper adults that were male and female sterile. This lethality was not a result of cryptic mutations on the X chromosome in these crosses because the single-mutant H3.3B0 / H3.3B0 homozygous females and H3.3B0 / Y hemizygous males were viable (data not shown). To determine if the adult female sterility is caused by impaired DNA replication or amplification, we labeled ovaries from the surviving double-mutant H3.3 null females with the nucleotide analog EdU. H3.3 mutant ovaries had frequent follicle cell death and degeneration of midstage egg chambers, a stress response known as the vitellogenic checkpoint (Figure S4, B and C) (McCall 2004; Pritchett et al. 2009). This stress response resulted in fewer EdU-labeled H3.3 mutant than wild type follicle cells. However, the quality of EdU incorporation into follicle cell nuclei during mitotic cycle and endocycle S phases appeared similar between wild type and H3.3 mutant ovaries (Figure 6, A and B, and Figure S5, A and B).
To directly assess whether H3.3 is required for origin activity, we used EdU labeling to evaluate developmental gene amplification. Among the stage 10B-13 egg chambers that were produced in the H3.3 null females, incorporation of EdU into amplicon foci was similar between H3.3 null and wild type (∼15 egg chambers total over five biological replicates) (Figure 6, C and D, Figure S5, C and D, and Figure S6). The H3.3 null females also laid eggs with normal shells, further indicating that amplification of eggshell gene copy number is not greatly impaired (≥ five females tested for each of four biological replicates, data not shown). These results suggest that H3.3 is not essential for origin function during genomic DNA replication or developmental gene amplification.
Discussion
We have investigated the function of H3.3 at origins using the unique methods afforded by the gene amplification system in Drosophila. Our results indicate that H3.3 is abundant at amplicon origins, consistent with the results of previous genome-wide studies that found a correlation between origins and H3.3 (Deal et al. 2010; MacAlpine et al. 2010; Eaton et al. 2011; Stroud et al. 2012). H3.3 was abundant at the DAFC-66D amplicon when replication initiation was blocked, and also early in oogenesis before the onset of amplification, indicating that the high level of H3.3 at this amplicon is not simply a result of deposition behind the replication forks during amplification. Importantly, however, H3.3 was not essential for origin function during amplification or genomic replication. These results resolve a long-standing question raised by genome-wide studies about the possible function of H3.3 at origins. Although not required, the high levels of H3.3 at origins suggest that it may reflect open chromatin attributes that are important for origin function.
Our data indicate that pre-RC binding sites at amplicon origins are enriched for H3.3—a property that is shared with origins that initiate genomic replication. Although H3.3 was not essential for amplicon origin function, it remains formally possible that it is required at a subset of other origins during genomic replication. It is known that cells license more origins than are used, and that dormant origins can initiate DNA replication during times of replication stress (Ge et al. 2007; Kawabata et al. 2011). Indeed, when we began this study, we considered that replication stress may contribute to the slow development and high degree of lethality of H3.3 null animals. The H3.3 null females had somewhat fewer EdU labeled follicle cells during genomic replication stages. This is likely an indirect effect of the high levels of cell death and egg chamber degeneration caused by the vitellogenic checkpoint response in midoogenesis because the quality of nuclear EdU labeling during genomic replication of H3.3 null cells appeared similar to wild type. The incorporation of EdU into amplicon foci also appeared similar between mutant and wild type, and the H3.3 null females produced eggs with normal shells. This observation is significant because it has been shown that amplification is a sensitized genetic system for detection of defects in proteins that are required for the function of all origins (Calvi 2006). For example, females homozygous for hypomorphic, missense alleles of the MCM6 subunit of the replicative helicase have normal genomic replication, but reduce amplification from 64-fold to 16-fold, which is manifest as undetectable amplicon foci in most cells, a severe thin eggshell, and female sterility (Schwed et al. 2002). It remains possible that H3.3 null females have a small decrease in final DNA copy number at the amplified loci, something we were unable to measure by qPCR because of the paucity of late stage egg chambers. The normal size and intensity of amplicon foci, however, suggest that there is not a major quantitative effect on origin efficiency. We favor the interpretation, therefore, that the oogenesis stress response in H3.3 null females is likely the result of the known effects of H3.3 deficits on transcription or chromatin maintenance, and that H3.3 is not required for the function of most origins (Sakai et al. 2009; Ray-Gallet et al. 2011; Szenker et al. 2011; Schneiderman et al. 2012).
In contrast to our results, other studies have shown that H3.3 is required for DNA replication at specific developmental times and during stress. In chicken DT40 cells, H3.3 is required for normal fork progression after UV irradiation, suggesting it may have a function at the fork during DNA repair or lesion bypass (Frey et al. 2014). Moreover, deficits in maternal H3.3, or its chaperone HIRA, result in DNA replication defects during the first zygotic S phase of mouse embryogenesis (Lin et al. 2014). Although we did not detect a difference in genomic replication or developmental amplification in H3.3 null animals, it appears that H3.3 may have a distinct requirement for DNA replication after DNA damage and during early development (Sakai et al. 2009; Lin et al. 2014; Wen et al. 2014).
One caveat to our results for H3.3 at amplicons is that H3.3A-GFP was overexpressed. This H3.3A-GFP transgene, however, is a validated system that has been used to show H3.3 chromatin deposition at other loci (Ahmad and Henikoff 2002; Schwartz and Ahmad 2005; Sakai et al. 2009). Our data support, therefore, that the amplicon origins have dynamic H3.3 exchange that is significantly higher than nonorigin loci.
Why is there a genome-wide correlation between H3.3 and origins? We favor the interpretation that this correlation reflects an open chromatin state that is conducive to pre-RC assembly. It is known that H3.3 nucleosomes have a dynamic association with DNA, and that their locations correspond to “nucleosome-depleted regions” (NDRs) because they are extracted under standard conditions for micrococcal nuclease (MNase) mapping (Jin and Felsenfeld 2007; Henikoff 2009; Jin et al. 2009). In fact, our recent MNase-Seq mapping in follicle cells indicated that the ORC binding sites ACE3 and ori-β at DAFC-66D are NDRs (Liu et al. 2015). Thus, the correlation between H3.3 and origins may reflect the ability of ORC to compete with dynamic H3.3-containing nucleosomes for binding to DNA. Our recent analysis also suggested, however, that ORC binds NDRs at DAFC-66D in part because it favors DNA sequences and structures that are disfavored by canonical nucleosomes, a result that may be generalizable genome-wide (Segal et al. 2006; Comoglio et al. 2015; Liu et al. 2015). Therefore, the correlation between ORC and H3.3 occupancy may reflect the fact that both are enriched at genomic regions with DNA sequences and structures that are disfavored by canonical nucleosomes (Ray-Gallet et al. 2011; Schneiderman et al. 2012; Comoglio et al. 2015; Liu et al. 2015).
It is important to stress that, while most ORC binding sites are NDRs and enriched for H3.3, many H3.3-containing NDRs are not ORC binding sites (Deal et al. 2010; Liu et al. 2015). This is further evidence that H3.3 is not instructive for specifying ORC binding sites and origins. It is clear that other chromatin attributes do have a major impact on origin function. For example, histone acetylation at DAFC-66D promotes ORC binding in stage 10 follicle cells and correlates with early-replicating and efficient origins during genomic replication (Aggarwal and Calvi 2004; Eaton et al. 2011). Both histone acetylation and ORC binding at DAFC-66D occurs over a ∼20 kb domain, with ORC binding likely occurring within the NDRs across the locus (Kim et al. 2011; Liu et al. 2012). Our ChIP-qPCR and immunofluorescent quantification indicated that H3.3 nucleosomes are also deposited over this same ∼20 kb domain, also likely within MNase defined “NDRs.” These data suggest, therefore, that this extended H3.3 distribution may be characteristic of a larger, open epigenome domain that is permissive for pre-RC assembly or activation during amplification. It will be interesting to determine whether this epigenome domain of nucleosome acetylation, ORC binding, and H3.3 reflects a change in higher order chromatin architecture that is conducive for origin function.
Our findings may also have broader relevance to understanding the pediatric gliomas that are caused by mutations in H3.3 and its chaperones (Schwartzentruber et al. 2012; Wu et al. 2012). If H3.3 also plays a minor role at mammalian origins, other functions of H3.3 should be emphasized to define the molecular mechanisms of those childhood cancers (Goldberg et al. 2010; Ray-Gallet et al. 2011; Schneiderman et al. 2012; Liu et al. 2014; Elsasser et al. 2015; Jang et al. 2015).
Supplementary Material
Acknowledgments
We thank K. Ahmad and I. Hariharan for fly strains and T. Orr-Weaver for Orc2 antibodies. We also thank K. Cook and K. Matthews of the Bloomington Drosophila Stock Center, J. Powers of the Indiana Light Microscopy Imaging Center, M. Dixon for technical support, and J. Liu and all members of Calvi lab for helpful advice and comments. This research was supported by the National Institutes of Health grants R01 GM612290 and R01 GM061290-S1 to B.R.C.
Footnotes
Supplemental material is available online at http://www.g3journal.org/cgi/data/g3.116.028068/DC1/1
Communicating editor: R. A. Sclafani
Literature Cited
- Aggarwal B. D., Calvi B. R., 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376. [DOI] [PubMed] [Google Scholar]
- Ahmad K., Henikoff S., 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9: 1191–1200. [DOI] [PubMed] [Google Scholar]
- Aladjem M. I., 2007. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet. 8: 588–600. [DOI] [PubMed] [Google Scholar]
- Alexander J. L., Barrasa M. I., Orr-Weaver T. L., 2015. Replication fork progression during re-replication requires the DNA damage checkpoint and double-strand break repair. Curr. Biol. 25: 1654–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin R. J., Orr-Weaver T. L., Bell S. P., 1999. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 13: 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard E., Babled A., Lapasset L., Milhavet O., Parrinello H., et al. , 2012. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 19: 837–844. [DOI] [PubMed] [Google Scholar]
- Bicknell L. S., Bongers E. M., Leitch A., Brown S., Schoots J., et al. , 2011a Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 43: 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell L. S., Walker S., Klingseisen A., Stiff T., Leitch A., et al. , 2011b Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat. Genet. 43: 350–355. [DOI] [PubMed] [Google Scholar]
- Boley N., Wan K. H., Bickel P. J., Celniker S. E., 2014. Navigating and mining modENCODE data. Methods 68: 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck S. B., Bradford J., Gee K. R., Agnew B. J., Clarke S. T., et al. , 2008. Detection of S-phase cell cycle progression using 5-ethynyl-2′-deoxyuridine incorporation with click chemistry, an alternative to using 5-bromo-2′-deoxyuridine antibodies. Biotechniques 44: 927–929. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., 2006. Developmental gene amplification, pp. 233–255 in DNA Replication and Human Disease, edited by M. L. DePamphilisCold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Calvi B. R., Spradling A. C., 1999. Chorion gene amplification in Drosophila: a model for metazoan origins of DNA replication and S-phase control. Methods 18: 407–417. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., Spradling A. C., 2001. The nuclear location and chromatin organization of active chorion amplification origins. Chromosoma 110: 159–172. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., Lilly M. A., 2004. Fluorescent BrdU labeling and nuclear flow sorting of the Drosophila ovary, pp. 203–213 in Drosophila Cytogenetics Protocols, edited by D. Henderson, Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Calvi B. R., Lilly M. A., Spradling A. C., 1998. Cell cycle control of chorion gene amplification. Genes Dev. 12: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaliere V., Bernardi F., Romani P., Duchi S., Gargiulo G., 2008. Building up the Drosophila eggshell: first of all the eggshell genes must be transcribed. Dev. Dyn. 237: 2061–2072. [DOI] [PubMed] [Google Scholar]
- Cayrou C., Coulombe P., Vigneron A., Stanojcic S., Ganier O., et al. , 2011. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 21: 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C., Coulombe P., Puy A., Rialle S., Kaplan N., et al. , 2012. New insights into replication origin characteristics in metazoans. Cell Cycle 11: 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claycomb J. M., Orr-Weaver T. L., 2005. Developmental gene amplification: insights into DNA replication and gene expression. Trends Genet. 21: 149–162. [DOI] [PubMed] [Google Scholar]
- Claycomb J. M., MacAlpine D. M., Evans J. G., Bell S. P., Orr-Weaver T. L., 2002. Visualization of replication initiation and elongation in Drosophila. J. Cell Biol. 159: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoglio F., Schlumpf T., Schmid V., Rohs R., Beisel C., et al. , 2015. High-resolution profiling of Drosophila replication start sites reveals a DNA shape and chromatin signature of metazoan origins. Cell Reports 11: 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danis E., Brodolin K., Menut S., Maiorano D., Girard-Reydet C., et al. , 2004. Specification of a DNA replication origin by a transcription complex. Nat. Cell Biol. 6: 721–730. [DOI] [PubMed] [Google Scholar]
- Deal R. B., Henikoff J. G., Henikoff S., 2010. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328: 1161–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delidakis C., Kafatos F. C., 1988. Deletion analysis of cis-acting elements for chorion gene amplification in Drosophila melanogaster. Cancer Cells 6: 311–315. [Google Scholar]
- Delidakis C., Kafatos F. C., 1989. Amplification enhancers and replication origins in the autosomal chorion gene cluster of Drosophila. EMBO J. 8: 891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nooij J. C., Letendre M. A., Hariharan I. K., 1996. A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell 87: 1237–1247. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H., Dowell S. J., Harwood J., Rowley A., 1995. Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle. J. Cell Sci. Suppl. 19: 67–72. [DOI] [PubMed] [Google Scholar]
- Dunleavy E. M., Almouzni G., Karpen G. H., 2011. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus 2: 146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton M. L., Galani K., Kang S., Bell S. P., MacAlpine D. M., 2010. Conserved nucleosome positioning defines replication origins. Genes Dev. 24: 748–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton M. L., Prinz J. A., MacAlpine H. K., Tretyakov G., Kharchenko P. V., et al. , 2011. Chromatin signatures of the Drosophila replication program. Genome Res. 21: 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsaesser S. J., Goldberg A. D., Allis C. D., 2010. New functions for an old variant: no substitute for histone H3.3. Curr. Opin. Genet. Dev. 20: 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S. J., Noh K. M., Diaz N., Allis C. D., Banaszynski L. A., 2015. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cid A., Riera A., Tognetti S., Herrera M. C., Samel S., et al. , 2013. An ORC/Cdc6/MCM2–7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol. Cell 50: 577–588. [DOI] [PubMed] [Google Scholar]
- Foulk M. S., Urban J. M., Casella C., Gerbi S. A., 2015. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 25: 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A., Listovsky T., Guilbaud G., Sarkies P., Sale J. E., 2014. Histone H3.3 is required to maintain replication fork progression after UV damage. Curr. Biol. 24: 2195–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J., Remus D., Mehanna A., Diffley J. F., 2013. ATPase-dependent quality control of DNA replication origin licensing. Nature 495: 339–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo M., Coutinho F. J., Vanner R. J., Gayden T., Mack S. C., et al. , 2015. MLL5 orchestrates a cancer self-renewal state by repressing the histone variant H3.3 and globally reorganizing chromatin. Cancer Cell 28: 715–729. [DOI] [PubMed] [Google Scholar]
- Ge X. Q., Jackson D. A., Blow J. J., 2007. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 21: 3331–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D. M., 2012. Replication origins run (ultra) deep. Nat. Struct. Mol. Biol. 19: 740–742. [DOI] [PubMed] [Google Scholar]
- Goldberg A. D., Banaszynski L. A., Noh K. M., Lewis P. W., Elsaesser S. J., et al. , 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin-Shea R., Thireos G., Kafatos F. C., 1982. Organization of a cluster of four chorion genes in Drosophila and its relationship to developmental expression and amplification. Dev. Biol. 91: 325–336. [DOI] [PubMed] [Google Scholar]
- Guernsey D. L., Matsuoka M., Jiang H., Evans S., Macgillivray C., et al. , 2011. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat. Genet. 43: 360–364. [DOI] [PubMed] [Google Scholar]
- Hartl T., Boswell C., Orr-Weaver T. L., Bosco G., 2007. Developmentally regulated histone modifications in Drosophila follicle cells: initiation of gene amplification is associated with histone H3 and H4 hyperacetylation and H1 phosphorylation. Chromosoma 116: 197–214. [DOI] [PubMed] [Google Scholar]
- Hayashi M. T., Takahashi T. S., Nakagawa T., Nakayama J., Masukata H., 2009. The heterochromatin protein Swi6/HP1 activates replication origins at the pericentromeric region and silent mating-type locus. Nat. Cell Biol. 11: 357–362. [DOI] [PubMed] [Google Scholar]
- Heck M. M., Spradling A. C., 1990. Multiple replication origins are used during Drosophila chorion gene amplification. J. Cell Biol. 110: 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., 2009. Labile H3.3+H2A.Z nucleosomes mark ‘nucleosome-free regions.’ Nat. Genet. 41: 865–866. [DOI] [PubMed] [Google Scholar]
- Hills S. A., Diffley J. F., 2014. DNA replication and oncogene-induced replicative stress. Curr. Biol. 24: R435–R444. [DOI] [PubMed] [Google Scholar]
- Hiratani I., Gilbert D. M., 2009. Replication timing as an epigenetic mark. Epigenetics 4: 93–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I., Ryba T., Itoh M., Rathjen J., Kulik M., et al. , 2010. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 20: 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard T., Shor E., Muller C. A., Nieduszynski C. A., Fox C. A., 2013. A link between ORC-origin binding mechanisms and origin activation time revealed in budding yeast. PLoS Genet. 9: e1003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne-Badovinac S., Bilder D., 2005. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Dev. Dyn. 232: 559–574. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., 1968. Visualization of replicating mammalian and T4 bacteriophage DNA. Cold Spring Harb. Symp. Quant. Biol. 33: 509–524. [DOI] [PubMed] [Google Scholar]
- Hyrien O., 2015. Peaks cloaked in the mist: the landscape of mammalian replication origins. J. Cell Biol. 208: 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M., Stillman B., 1999. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274: 23027–23034. [DOI] [PubMed] [Google Scholar]
- Jang C. W., Shibata Y., Starmer J., Yee D., Magnuson T., 2015. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 29: 1377–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Felsenfeld G., 2007. Nucleosome stability mediated by histone variants H3.3 and H2A. Z. Genes Dev 21: 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Zang C., Wei G., Cui K., Peng W., et al. , 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 41: 941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T., Luebben S. W., Yamaguchi S., Ilves I., Matise I., et al. , 2011. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell 41: 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. C., Nordman J., Xie F., Kashevsky H., Eng T., et al. , 2011. Integrative analysis of gene amplification in Drosophila follicle cells: parameters of origin activation and repression. Genes Dev. 25: 1384–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. M., Dubey D. D., Huberman J. A., 2003. Early-replicating heterochromatin. Genes Dev. 17: 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo A. J., Song J., Cheung P., Ishibe-Murakami S., Yamazoe S., et al. , 2012. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484: 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M. E., Sauer K., Wallace K., Jan Y. N., Lehner C. F., et al. , 1996. Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell 87: 1225–1235. [DOI] [PubMed] [Google Scholar]
- Lin C. J., Koh F. M., Wong P., Conti M., Ramalho-Santos M., 2014. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev. Cell 30: 268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford J. R., Bell S. P., 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7: 21–30. [DOI] [PubMed] [Google Scholar]
- Liu J., McConnell K., Dixon M., Calvi B. R., 2012. Analysis of model replication origins in Drosophila reveals new aspects of the chromatin landscape and its relationship to origin activity and the prereplicative complex. Mol. Biol. Cell 23: 200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Zimmer K., Rusch D. B., Paranjape N., Podicheti R., et al. , 2015. DNA sequence templates adjacent nucleosome and ORC sites at gene amplification origins in Drosophila. Nucleic Acids Res. 43(18): 8746–8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., McEachron T. A., Schwartzentruber J., Wu G., 2014. Histone H3 mutations in pediatric brain tumors. Cold Spring Harb. Perspect. Biol. 6: a018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebel D., Huikeshoven H., Cotterill S., 2000. Localisation of the DmCdc45 DNA replication factor in the mitotic cycle and during chorion gene amplification. Nucleic Acids Res. 28: 3897–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Zhang H., Tower J., 2001. Functionally distinct, sequence-specific replicator and origin elements are required for Drosophila chorion gene amplification. Genes Dev. 15: 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine H. K., Gordan R., Powell S. K., Hartemink A. J., MacAlpine D. M., 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 20: 201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall K., 2004. Eggs over easy: cell death in the Drosophila ovary. Dev. Biol. 274: 3–14. [DOI] [PubMed] [Google Scholar]
- McConnell K. H., Dixon M., Calvi B. R., 2012. The histone acetyltransferases CBP and Chameau integrate developmental and DNA replication programs in Drosophila ovarian follicle cells. Development 139: 3880–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M., 2010. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat. Rev. Mol. Cell Biol. 11: 728–738. [DOI] [PubMed] [Google Scholar]
- Mechali M., Yoshida K., Coulombe P., Pasero P., 2013. Genetic and epigenetic determinants of DNA replication origins, position and activation. Curr. Opin. Genet. Dev. 23: 124–131. [DOI] [PubMed] [Google Scholar]
- Miotto B., Struhl K., 2008. HBO1 histone acetylase is a coactivator of the replication licensing factor Cdt1. Genes Dev. 22: 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B., Struhl K., 2010. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol. Cell 37: 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modENCODE Consortium. Roy S., Ernst J., Kharchenko P. V., Kheradpour P., et al. , 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P., Park S., Shor E., Huebert D. J., Warren C. L., et al. , 2010. The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev. 24: 1418–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Gorgoulis V. G., Halazonetis T. D., 2010. Genomic instability—an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 11: 220–228. [DOI] [PubMed] [Google Scholar]
- Nordman J. T., Kozhevnikova E. N., Verrijzer C. P., Pindyurin A. V., Andreyeva E. N., et al. , 2014. DNA copy-number control through inhibition of replication fork progression. Cell Reports 9: 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Johnston C. G., Spradling A. C., 1989. The role of ACE3 in Drosophila chorion gene amplification. EMBO J. 8: 4153–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchett T. L., Tanner E. A., McCall K., 2009. Cracking open cell death in the Drosophila ovary. Apoptosis 14: 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Gallet D., Woolfe A., Vassias I., Pellentz C., Lacoste N., et al. , 2011. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol. Cell 44: 928–941. [DOI] [PubMed] [Google Scholar]
- Remus D., Diffley J. F., 2009. Eukaryotic DNA replication control: lock and load, then fire. Curr. Opin. Cell Biol. 21: 771–777. [DOI] [PubMed] [Google Scholar]
- Remus D., Beall E. L., Botchan M. R., 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. EMBO J. 23: 897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I., Austin R. J., Bosco G., Bell S. P., Orr-Weaver T. L., 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 13: 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A., Schwartz B. E., Goldstein S., Ahmad K., 2009. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol. 19: 1816–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A., Mitchison T. J., 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 105: 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiderman J. I., Orsi G. A., Hughes K. T., Loppin B., Ahmad K., 2012. Nucleosome-depleted chromatin gaps recruit assembly factors for the H3.3 histone variant. Proc. Natl. Acad. Sci. USA 109: 19721–19726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger M., Kohler H., Oakeley E. J., Stadler M. B., Schubeler D., 2010. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res. 20: 771–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B. E., Ahmad K., 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J., Korshunov A., Liu X. Y., Jones D. T., Pfaff E., et al. , 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231. [DOI] [PubMed] [Google Scholar]
- Schwed G., May N., Pechersky Y., Calvi B. R., 2002. Drosophila minichromosome maintenance 6 is required for chorion gene amplification and genomic replication. Mol. Biol. Cell 13: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Fondufe-Mittendorf Y., Chen L., Thastrom A., Field Y., et al. , 2006. A genomic code for nucleosome positioning. Nature 442: 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher N., Bell G. W., Li S., Nordman J., Eng T., et al. , 2012. Developmental control of gene copy number by repression of replication initiation and fork progression. Genome Res. 22: 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., 1981. The organization and amplification of two chromosomal domains containing Drosophila chorion genes. Cell 27: 193–201. [DOI] [PubMed] [Google Scholar]
- Stroud H., Otero S., Desvoyes B., Ramirez-Parra E., Jacobsen S. E., et al. , 2012. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109: 5370–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szenker E., Ray-Gallet D., Almouzni G., 2011. The double face of the histone variant H3.3. Cell Res. 21: 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P. B., Henikoff S., 2010. Histone variants–ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11: 264–275. [DOI] [PubMed] [Google Scholar]
- Tardat M., Brustel J., Kirsh O., Lefevbre C., Callanan M., et al. , 2010. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat. Cell Biol. 12: 1086–1093. [DOI] [PubMed] [Google Scholar]
- Unnikrishnan A., Gafken P. R., Tsukiyama T., 2010. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat. Struct. Mol. Biol. 17: 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S., Cvetic C., Lu W., Simancek P., Kelly T. J., et al. , 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17: 1894–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M., 2015. Molecular biology: DNA replication reconstructed. Nature 519: 418–419. [DOI] [PubMed] [Google Scholar]
- Wen D., Banaszynski L. A., Liu Y., Geng F., Noh K. M., et al. , 2014. Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc. Natl. Acad. Sci. USA 111: 7325–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker A. J., Royzman I., Orr-Weaver T. L., 2000. Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes Dev. 14: 1765–1776. [PMC free article] [PubMed] [Google Scholar]
- Wu G., Broniscer A., McEachron T. A., Lu C., Paugh B. S., et al. , 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 44: 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Long C., Chen X., Huang C., Chen S., et al. , 2010. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328: 94–98. [DOI] [PubMed] [Google Scholar]
- Yeeles J. T., Deegan T. D., Janska A., Early A., Diffley J. F., 2015. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519: 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. All data and reagents are available upon request.