Abstract
Osteoblasts and bone marrow adipocytes originate from bone marrow mesenchymal stem cells (BMMSCs) and there appears to be a reciprocal relationship between adipogenesis and osteoblastogenesis. Alterations in the balance between adipogenesis and osteoblastogenesis in BMMSCs wherein adipogenesis is increased relative to osteoblastogenesis are associated with decreased bone quality and quantity. Several proteins have been reported to regulate this reciprocal relationship but the exact nature of the signals regulating the balance between osteoblast and adipocyte formation within the bone marrow space remains to be determined. In this review, we focus on the role of Transducin-Like Enhancer of Split 3 (TLE3), which was recently reported to regulate the balance between osteoblast and adipocyte formation from BMMSCs. We also discuss evidence implicating canonical Wnt signalling, which plays important roles in both adipogenesis and osteoblastogenesis, in regulating TLE3 expression. Currently, there is demand for new effective therapies that target the stimulation of osteoblast differentiation to enhance bone formation. We speculate that reducing TLE3 expression or activity in BMMSCs could be a useful approach towards increasing osteoblast numbers and reducing adipogenesis in the bone marrow environment.
1. Introduction
In 2010, more than 10 million Americans over the age of 50 had osteoporosis with another 43 million Americans at risk for the disease [1]. It is estimated that greater than 1.5 million fragility fractures occur each year, with an annual health care cost of at least 14 billion US dollars [2]. By 2025, the health care expenditures for osteoporotic fractures will approach 25.3 billion US dollars [3]. Bone is constantly remodeled through the processes of bone formation by osteoblasts and bone resorption by osteoclasts. Osteoclasts are derived from hematopoietic stem cell precursors of the monocyte/macrophage lineage located in the blood and bone marrow [4]; conversely, osteoblast-lineage cells (osteoblasts and osteocytes) originate from bone marrow mesenchymal stem cells (BMMSCs) [5]. BMMSCs are a multipotent cell type that can give rise not only to osteoblast-lineage cells but also to a range of other cell types, including adipocytes [6] (Figure 1). In some pathological conditions, including senile osteoporosis, the balance between adipocyte and osteoblast differentiation is disrupted in this cell population such that adipocyte differentiation is increased relative to osteoblast differentiation and this is associated with reduced bone mass, increased bone fragility, and increased susceptibility to fracture [7]. Therefore, understanding the molecular mechanism(s) responsible for controlling the balance between osteoblastogenesis and adipogenesis in the adult bone environment is of great significance.
Figure 1.
Bone marrow mesenchymal stem cells differentiate into both adipocytes and osteoblasts. Osteoblast and marrow adipocytes are derived from common progenitors, the bone marrow mesenchymal stem cells. BMMSCs: bone marrow mesenchymal stem cells; OBs: osteoblasts; ADs: adipocytes.
In this review, we will summarize the processes of osteoblast and adipocyte differentiation from BMMSCs, focusing on the role of Transducin-Like Enhancer of Split 3 (TLE3), which was recently reported to regulate osteoblastogenesis and adipogenesis. We also discuss the prospect of bone regenerative therapy by using stem cells.
2. Relationship between Adipogenesis and Osteoblastogenesis
Adipogenesis is driven by a complex and well-orchestrated signalling cascade composed of several key transcription factors, most notably proliferator-activated receptor- (PPAR-) γ and several members of the CCAAT/enhancer-binding family of proteins (C/EBPs) [8]. PPAR-γ is commonly referred to as the master regulator of adipogenesis because no factor has yet been identified that can induce normal adipogenesis in its absence [9].
BMP-SMAD signalling plays an important role in osteoblastogenesis by inducing expression of several critical transcription factors such as RUNX2, Osterix, DLX2, and DLX5 [10–12]. RUNX2 is essential for the commitment of mesenchymal stem cells to the osteoblast lineage and homozygous deletion of Runx2 in mice results in a complete lack of osteoblasts [13, 14]. It appears that adequate RUNX2 is also dosage-dependent since haploinsufficiency of Runx2 in mice or RUNX2 in humans causes hypoplastic clavicles and delayed closure of the fontanelles, defects that are characteristic of cleidocranial dysplasia in humans [15, 16]. RUNX2 controls osteoblast-related genes such as Osterix, collagen I, and osteocalcin [17] and autoregulates the Runx2 gene itself [18].
Several proteins have been reported to regulate both adipogenesis and osteoblastogenesis (Table 1) and, in general, adipogenesis is reciprocally related to osteoblastogenesis in BMMSCs. However, the exact nature of the signals regulating the balance between osteoblast and adipocyte formation within the bone marrow space remains to be determined. In the sections below, we seek to bring attention to TLE3, which is a relatively understudied regulator of osteoblastogenesis and adipogenesis that is a member of the Groucho/TLE family of transcription factors [19].
Table 1.
The proteins regulate adipogenesis and osteoblastogenesis.
| Number | Protein(s) | Function | Assay | Reference(s) |
|---|---|---|---|---|
| 1 | Msx2 | Adipogenesis↓; osteoblastogenesis↑ | In vitro | [66] |
| 2 | Dlk1/Pref-1 | Adipogenesis↓; osteoblastogenesis↑ | In vitro | [67] |
| 3 | TAZ | Adipogenesis↓; osteoblastogenesis↑ | Zebrafish; in vitro | [68] |
| 4 | Wnt10b | Adipogenesis↓; osteoblastogenesis↑ | Knockout mice; transgenic mice | [69] |
| 5 | LIP | Adipogenesis↓; osteoblastogenesis↑ | In vitro | [70] |
| 6 | Dec1 | Adipogenesis↓; osteoblastogenesis↑ | In vitro | [71] |
| 7 | Hemooxygenase-1 | Adipogenesis↓; osteoblastogenesis↑ | In vitro | [72] |
| 8 | ID4 | Adipogenesis↓; osteoblastogenesis↑ | Knockout mice | [73] |
| 9 | Maf | Adipogenesis↓; osteoblastogenesis↑ | Knockout mice | [74] |
| 10 | Pkd1 | Adipogenesis↓; osteoblastogenesis↑ | Knockout mice | [75] |
| 11 | sFRP-1 | Adipogenesis↑; osteoblastogenesis↓ | In vitro | [76] |
| 12 | ZFP467 | Adipogenesis↑; osteoblastogenesis↓ | In vivo injection | [77] |
| 13 | GIT2 | Adipogenesis↓; osteoblastogenesis↑ | Knockout mice | [78] |
| 14 | Wnt6 | Adipogenesis↓; osteoblastogenesis↑ | In vitro | [79] |
| 15 | Wnt10a | Adipogenesis↓; osteoblastogenesis↑ | In vitro | [79] |
| 16 | VEGF | Adipogenesis↓; osteoblastogenesis↑ | Knockout mice | [80] |
| 17 | Semaphorin 3A | Adipogenesis↓; osteoblastogenesis↑ | Knockout mice | [81] |
| 18 | TLE3 | Adipogenesis↑; osteoblastogenesis↓ | In vitro | [19] |
| 19 | S100a16 | Adipogenesis↑; osteoblastogenesis↓ | In vitro | [82] |
| 20 | mTORC2 | Adipogenesis↓; osteoblastogenesis↑ | In vitro | [83] |
| 21 | Adiponectin | Adipogenesis↓; osteoblastogenesis↑ | Knockout mice | [84] |
| 22 | Cysteine dioxygenase type 1 | Adipogenesis↑; osteoblastogenesis↓ | In vitro | [85, 86] |
| 23 | MYSM1 | Adipogenesis↓; osteoblastogenesis↑ | Knockout mice | [87] |
3. Groucho/TLE Family Member
Groucho (Gro)/Transducin-Like Enhancer of Split (TLE) family members are transcriptional cofactors in metazoans that play critical roles during development and cell fate determination, including differentiation into fat and bone cells. The names “Gro” and “TLE” are used interchangeably in the literature and in sequence databases [20] and the Drosophila genome encodes a single Gro while the mouse and human genomes encode four members of each family [21].
Groucho/TLE proteins consist of a five-domain structure [22]: a highly conserved Q domain, which is a glutamine-rich region predicted to form two coiled-coil motifs that facilitates oligomerization of Gro/TLE molecules in vitro [23–25]; a glycine/proline rich (GP) domain, which is essential for interaction of Groucho/TLE proteins with histone deacetylases (HDACs) [23, 24, 26, 27]; a CcN domain, which contains a nuclear localization sequence and putative cdc2 and casein kinase II (protein kinase CK2) phosphorylation sites; a serine/proline rich (SP) domain, which is a region rich in serine/proline residues [22, 28–30]; and a highly conserved WD40 domain, which contains multiple tryptophan and aspartic acid tandem repeats, has been shown by X-ray crystallography to form a β-propeller, and binds many kinds of transcriptional factors [20, 31].
Groucho/TLE proteins do not bind DNA directly but are instead recruited by other transcription factors and are largely considered transcriptional corepressors since they often reduce the activity of a target transcriptional factor. However, the Groucho/TLE family member TLE3 was recently reported to induce the transcriptional activity of PPAR-γ, which is a master transcriptional regulator of adipogenesis [32], suggesting that the Groucho/TLE family may act as corepressors or coactivators in a context-dependent manner.
4. Distribution of TLE3 during Development
During development, TLE3 is expressed in the placenta [33] and homozygous null Tle3 mutant mice are smaller than their heterozygous and wild type littermates. Most homozygous null Tle3 mutant embryos demonstrate severe placental defects and die in utero [34]. TLE3 is also expressed in the developing nervous system where as the neural tube closes, its distribution shifts from the entire width of the neural plate to the dorsal region and ventricular zone; expression in the roof of the mesencephalon and metencephalon remains most pronounced at this stage. TLE3 is also expressed in the dorsal root ganglia and its expression in the newly formed somites becomes restricted to a dorsal, bracket-shaped group of cells corresponding to the dermamyotome [35].
In older mouse embryos expression of TLE3 in the central nervous system (CNS) is observed along the entire length of the brain and spinal cord in the ventricular zone, with the strongest expression in the layer of cells immediately lining the lumen. In the developing eye, TLE3 is located in the lens and the neural layer of the retina. Somatic expression of TLE3 continues in the dermamyotome and in the condensing sclerotome, forming the vertebrae and bones. Faint staining for TLE3 is also observed in the metanephros (embryonic kidney); tissues derived from the pharynx, including Rathke's pouch and the thymic primordial; the lining of the gut and tissues derived from the gut endoderm such as the epithelial walls of the bronchi of the lungs and the liver; and derivatives of the branchial arches such as the dorsum and intrinsic muscles of the tongue and the dental laminae of the tooth primordial [35].
In later stages of mouse development (16.5 days after conception), TLE3 expression is more restricted than at midgestation. For instance, Tle3 mRNA is detected in the ventricular zone and the cortical plate of the cerebral cortex; the colliculus; the cerebellum; the olfactory lobe; nasal epithelia; whisker follicles primordia; epithelial cells of the salivary glands; basal layer of skin and hair follicles; and derivatives of the pharyngeal pouches including the lining of the cochlea, eustachian tube, esophagus, larynx, epiglottis, and the thymus [35]. TLE3 is also expressed by cells of the bone marrow [19] and brown and white adipose tissue [32], with the expression level of TLE3 increasing with adipocyte differentiation [19, 32].
5. TLE3 Enhances Adipocyte Differentiation and Suppresses Osteoblastogenesis
Adipocytes are classically classified into two kinds: white adipocytes and brown adipocytes. White adipocytes are optimized to store energy as triglycerides in large, unilocular lipid droplets. When metabolic needs arise, white adipocytes mobilize energy through hydrolysis of triglycerides and release of free fatty acids into the circulation [36]. White adipocytes express a battery of genes involved in lipid handing, triglyceride biosynthesis, triglyceride mobilization, and endocrine signalling [37–39].
Brown adipocytes derive their color from their high mitochondrial content. Unlike white adipocytes, brown adipocytes store energy primarily to provide an intracellular fuel source for thermogenesis [40]. During cold exposure, brown adipose tissue (BAT) executes a transcriptional program that promotes energy expenditure and thermogenesis. Induction of the gene encoding Mitochondrial Uncoupling Protein-1 (UCP1) is critical for brown fat thermogenesis [41, 42]. It has been thought that Ucp1 expression is restricted to BAT; however, recent studies have demonstrated that Ucp-1-positive cells can be detected even in white adipose tissue under certain circumstances. These cells are called “beige adipocytes” [43] and have characteristics of both white and brown adipose cells: during basal state, beige adipocytes display unilocular morphology similar to white adipocytes, but upon cold stimulation, these cells acquire features of intermediate morphology ultimately resulting in expression of proteins typical for BAT and transformation of stored fat into the small lipid droplets typical for brown adipocytes [44–46].
While the transcriptional determinants of the white and brown adipocyte gene programs are incompletely understood, it is known that PPARγ is the master transcriptional regulator of both white and brown fat differentiation. In support of this, mice deficient in PPARγ lack both types of adipose tissue [9, 47–49]. Villanueva et al. [32] identified TLE3 as a cofactor for PPARγ and it was later confirmed that TLE3 enhances transcriptional activity of PPARγ, thereby inducing adipocyte differentiation of BMMSCs [19, 21]. Additionally, TLE3 disrupts the physical interaction between transcriptional cofactor PRDM16, which was identified as a key factor driving brown adipocyte linage development [43, 50], and PPARγ, thereby suppressing brown-fat-specific genes and inducing white-fat-specific genes; the net result of these effects is impaired fatty acid oxidation and thermogenesis [51]. We predict that TLE3 has some influence on beige adipocyte formation, but further studies are needed to examine this possibility.
Described above, osteoblast-lineage cells and marrow adipocytes are derived from a common progenitor, the BMMSCs. RUNX2 controls osteoblast-related genes and is essential for commitment to the osteoblast lineage [13, 14, 52]. RUNX2 interacts with Groucho/TLE family members, which act as corepressors of RUNX2 activity [53, 54]. For instance, TLE1 and TLE2 repress RUNX2-dependent activation of osteocalcin gene transcription [55]. And TLE3 suppresses BMP2-induced osteoblast differentiation of BMMSCs via recruiting HDAC and repressing RUNX2 transcriptional activity [19].
6. Expression of TLE3 Is Regulated by Canonical Wnt Signalling
The Wnt family of nineteen secreted glycoproteins has a critical role in regulating embryonic development, cell differentiation, and cell fate determination [56]. Wnts transduce two types of intracellular signalling referred to as canonical and noncanonical pathways. Canonical Wnt signalling, that is, signalling mediated by the effector β-catenin, has a key role in adult skeletal homeostasis and bone remodeling [57] by promoting differentiation and maturation of osteoblasts and, thereby, increasing bone formation [58]. In contrast, canonical Wnt signalling suppresses adipocyte differentiation [59].
Groucho/TLE family members, including TLE3, act as transcriptional corepressors of canonical Wnt signalling via binding to the downstream effectors TCF/LEF and inhibiting Wnt target gene transcription [20, 60–62]. According to Daniels and Weis [62], β-catenin that enters the nucleus upon activation of the Wnt pathway directly competes with Groucho/TLE proteins for TCF/LEF binding to accomplish gene regulation.
Recently, Wnt responsive elements in the TLE3 promoter region were identified through comparative genomic analysis and functional analyses confirmed that expression of TLE3 is increased by Wnt signalling [21]. Given the opposing roles of TLE3 and Wnt signalling in BMMSCs differentiation, this finding suggests that induction of TLE3 by Wnt signalling is part of a negative feedback loop active during osteoblast differentiation and/or a part of a positive feedback loop during adipogenesis, suggesting that TLE3 regulates the cell fate of BMMSCs between osteoblasts and adipocytes (Figure 2).
Figure 2.
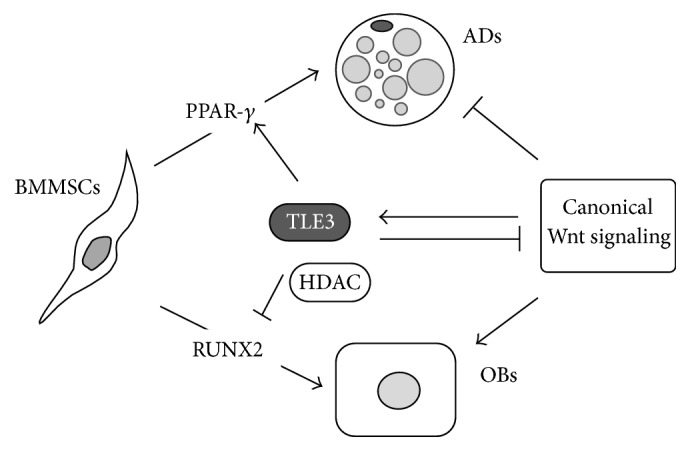
Model for the role of TLE3 in the bone marrow microenvironment. TLE3 directly induces adipogenesis and suppresses osteoblastogenesis of BMMSCs by acting on PPAR-γ and RUNX2, respectively. TLE3 also indirectly induces adipogenesis and suppresses osteoblastogenesis by repressing canonical Wnt signalling, which is capable of inducing osteoblastogenesis and inhibiting adipogenesis. In addition, canonical Wnt signalling induces TLE3 expression, suggesting that the induction of TLE3 by Wnt signalling may be part of a negative feedback loop during osteoblastogenesis and/or a positive feedback loop during adipogenesis in the adult bone marrow microenvironment. BMMSCs: bone marrow mesenchymal stem cells; OBs: osteoblasts; ADs: adipocytes.
7. Prospects for Therapy
Osteoporosis, which is one of the most abundant bone-related diseases, is characterized by low bone mass and microarchitectural deterioration of bone tissue that results in increased bone fragility and susceptibility to fracture [7]. The most commonly prescribed therapeutics are antiresorptives, such as calcitonin, estrogen, and bisphosphonates, that block osteoclast activity as a means to stabilize bone architecture. While efficacious in halting further bone loss, little or no new bone mass is added to the skeleton while on antiresorptive therapy. Recent data on the importance of continuous bone remodeling suggest that overuse of antiresorptives could lead to BRONJ (bisphosphonate-related osteonecrosis of the jaw) [63] and fracture in some patients [64]. Thus, development of new, effective therapies that target enhancing bone formation by stimulating osteoblast differentiation is required.
8. Conclusion
In this review we summarized the cell fate determination and the differentiation of BMMSCs and especially focus on the role of TLE3, which represses osteoblast differentiation and enhances adipocyte formation from BMMSCs. Therefore, we speculate that reducing TLE3 expression or activity in BMMSCs could be a useful approach towards increasing osteoblast numbers and reducing adipogenesis in the bone marrow environment. Recently, a delivery system involving dioleoyl trimethylammonium propane- (DOTAP-) based cationic liposomes attached to six repetitive sequences of aspartate, serine, and serine ((AspSerSer)6) was utilized to deliver siRNAs specifically to bone formation surfaces [65]. Delivery of siRNAs against Tle3 with this delivery system might be useful for reducing mRNA levels of TLE3 in bone without affecting other organs and/or tissues. Thus, developing effective methods of reducing TLE3 expression or activity in bone locally may shed light on novel bone formation therapies.
Acknowledgments
The authors thank the members of the Division of Molecular Signaling and Biochemistry Department of Health Promotion, Kyushu Dental University, the Department of Oral and Maxillofacial Surgery, Faculty of Medicine, Saitama Medical University, the Division of Pathophysiology, Research Center for Genomic Medicine, Saitama Medical University, and the Harvard School of Dental Medicine for their valuable comments and discussion.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Wright N. C., Looker A. C., Saag K. G., et al. The recent prevalence of osteoporosis and low bone mass in the united states based on bone mineral density at the femoral neck or lumbar spine. Journal of Bone and Mineral Research. 2014;29(1):2520–2526. doi: 10.1002/jbmr.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blume S. W., Curtis J. R. Medical costs of osteoporosis in the elderly Medicare population. Osteoporosis International. 2011;22(6):1835–1844. doi: 10.1007/s00198-010-1419-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raisz L. G., Elderkin A. L., Schargorodski L., et al. A call to action: developing and implementing a national action plan to improve bone health. Osteoporosis International. 2009;20(11):1805–1806. doi: 10.1007/s00198-009-1041-8. [DOI] [PubMed] [Google Scholar]
- 4.Teitelbaum S. L. Bone resorption by osteoclasts. Science. 2000;289(5484):1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 5.Owen M. Marrow stromal stem cells. Journal of Cell Science. 1988;(10):63–76. doi: 10.1242/jcs.1988.supplement_10.5. [DOI] [PubMed] [Google Scholar]
- 6.Chamberlain G., Fox J., Ashton B., Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. STEM CELLS. 2007;25(11):2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- 7.Harvey N., Dennison E., Cooper C. Osteoporosis: impact on health and economics. Nature Reviews Rheumatology. 2010;6(2):99–105. doi: 10.1038/nrrheum.2009.260. [DOI] [PubMed] [Google Scholar]
- 8.Rosen E. D., Walkey C. J., Puigserver P., Spiegelman B. M. Transcriptional regulation of adipogenesis. Genes and Development. 2000;14(11):1293–1307. [PubMed] [Google Scholar]
- 9.Tontonoz P., Hu E., Spiegelman B. M. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 10.Xu S. C., Harris M. A., Rubenstein J. L. R., Mundy G. R., Harris S. E. Bone morphogenetic protein-2 (BMP-2) signaling to the Col2α1 gene in chondroblasts requires the homeobox gene Dlx-2. DNA and Cell Biology. 2001;20(6):359–365. doi: 10.1089/10445490152122479. [DOI] [PubMed] [Google Scholar]
- 11.Nakashima K., Zhou X., Kunkel G., et al. The novel zinc finger-containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108(1):17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 12.Miyama K., Yamada G., Yamamoto T. S., et al. A BMP-inducible gene, Dlx5, regulates osteoblast differentiation and mesoderm induction. Developmental Biology. 1999;208(1):123–133. doi: 10.1006/dbio.1998.9197. [DOI] [PubMed] [Google Scholar]
- 13.Komori T., Yagi H., Nomura S., et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 14.Otto F., Thornell A. P., Crompton T., et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–771. doi: 10.1016/S0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 15.Mundlos S., Otto F., Mundlos C., et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89(5):773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 16.Lee B., Thirunavukkarasu K., Zhou L., et al. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nature Genetics. 1997;16(3):307–310. doi: 10.1038/ng0797-307. [DOI] [PubMed] [Google Scholar]
- 17.Stein G. S., Lian J. B., Van Wijnen A. J., et al. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene. 2004;23(24):4315–4329. doi: 10.1038/sj.onc.1207676. [DOI] [PubMed] [Google Scholar]
- 18.Drissi H., Luc Q., Shakoori R., et al. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. Journal of Cellular Physiology. 2000;184(3):341–350. doi: 10.1002/1097-4652(200009)184:3<341::aid-jcp8>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 19.Kokabu S., Nguyen T., Ohte S., et al. TLE3, transducing-like enhancer of split 3, suppresses osteoblast differentiation of bone marrow stromal cells. Biochemical and Biophysical Research Communications. 2013;438(1):205–210. doi: 10.1016/j.bbrc.2013.07.054. [DOI] [PubMed] [Google Scholar]
- 20.Jennings B. H., Ish-Horowicz D. The Groucho/TLE/Grg family of transcriptional co-repressors. Genome Biology. 2008;9(1):p. 205. doi: 10.1186/gb-2008-9-1-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kokabu S., Sato T., Ohte S., et al. Expression of TLE3 by bone marrow stromal cells is regulated by canonical Wnt signaling. FEBS Letters. 2014;588(4):614–619. doi: 10.1016/j.febslet.2013.12.031. [DOI] [PubMed] [Google Scholar]
- 22.Stifani S., Blaumueller C. M., Redhead N. J., Hill R. E., Artavanis-Tsakonas S. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nature Genetics. 1992;2(2):119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- 23.Chen G., Nguyen P. H., Courey A. J. A role for Groucho tetramerization in transcriptional repression. Molecular and Cellular Biology. 1998;18(12):7259–7268. doi: 10.1128/mcb.18.12.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinto M., Lobe C. G. Products of the grg (Groucho-related gene) family can dimerize through the amino-terminal Q domain. The Journal of Biological Chemistry. 1996;271(51):33026–33031. doi: 10.1074/jbc.271.51.33026. [DOI] [PubMed] [Google Scholar]
- 25.Song H., Hasson P., Paroush Z., Courey A. J. Groucho oligomerization is required for repression in vivo. Molecular and Cellular Biology. 2004;24(10):4341–4350. doi: 10.1128/MCB.24.10.4341-4350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grbavec D., Lo R., Liu Y., Stifani S. Transducin-like enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with hairy/enhancer of split proteins, and is expressed during neuronal development. European Journal of Biochemistry. 1998;258(2):339–349. doi: 10.1046/j.1432-1327.1998.2580339.x. [DOI] [PubMed] [Google Scholar]
- 27.Brantjes H., Roose J., van de Wetering M., Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Research. 2001;29(7):1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyasaka H., Okabe S., Ishiguro K., Uchida T., Hirokawa N. Interaction of the tail domain of high molecular weight subunits of neurofilaments with the COOH-terminal region of tubulin and its regulation by tau protein kinase II. The Journal of Biological Chemistry. 1993;268(30):22695–22702. [PubMed] [Google Scholar]
- 29.Fisher A. L., Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes and Development. 1998;12(13):1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 30.Parkhurst S. M. Groucho: making its Marx as a transcriptional co-repressor. Trends in Genetics. 1998;14(4):130–132. doi: 10.1016/s0168-9525(98)01407-3. [DOI] [PubMed] [Google Scholar]
- 31.Pickles L. M., Roe S. M., Hemingway E. J., Stifani S., Pearl L. H. Crystal structure of the C-terminal WD40 repeat domain of the human Groucho/TLE1 transcriptional corepressor. Structure. 2002;10(6):751–761. doi: 10.1016/S0969-2126(02)00768-2. [DOI] [PubMed] [Google Scholar]
- 32.Villanueva C. J., Waki H., Godio C., et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metabolism. 2011;13(4):413–427. doi: 10.1016/j.cmet.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama H., Liu Y., Stifani S., Cross J. C. Developmental restriction of Mash-2 expression in trophoblast correlates with potential activation of the Notch-2 pathway. Developmental Genetics. 1997;21(1):21–30. doi: 10.1002/(SICI)1520-6408(1997)21:1<21::AID-DVG3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 34.Metzger D. E., Gasperowicz M., Otto F., Cross J. C., Gradwohl G., Zaret K. S. The transcriptional co-repressor Grg3/Tle3 promotes pancreatic endocrine progenitor delamination and β-cell differentiation. Development. 2012;139(8):1447–1456. doi: 10.1242/dev.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leon C., Lobe C. G. Grg3, a murine groucho-related gene, is expressed in the developing nervous system and in mesenchyme-induced epithelial structures. Developmental Dynamics. 1997;208(1):11–24. doi: 10.1002/(SICI)1097-0177(199701)208:1<11::AID-AJA2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 36.Zechner R., Kienesberger P. C., Haemmerle G., Zimmermann R., Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. Journal of Lipid Research. 2009;50(1):3–21. doi: 10.1194/jlr.r800031-jlr200. [DOI] [PubMed] [Google Scholar]
- 37.Coleman R. A., Bell R. M. Selective changes in enzymes of the sn-glycerol 3-phosphate and dihydroxyacetone-phosphate pathways of triacylglycerol biosynthesis during differentiation of 3T3-L1 preadipocytes. Journal of Biological Chemistry. 1980;255(16):7681–7687. [PubMed] [Google Scholar]
- 38.Cook K. S., Min H. Y., Johnson D., et al. Adipsin: a circulating serine protease homolog secreted by adipose tissue and sciatic nerve. Science. 1987;237(4813):402–405. doi: 10.1126/science.3299705. [DOI] [PubMed] [Google Scholar]
- 39.Halaas J. L., Gajiwala K. S., Maffei M., et al. Weight-reducing effects of the plasma protein encoded by the obese gene. Science. 1995;269(5223):543–546. doi: 10.1126/science.7624777. [DOI] [PubMed] [Google Scholar]
- 40.Smith R. E., Roberts J. C. Thermogenesis of brown adipose tissue in cold-acclimated rats. American Journal of Physiology. 1964;206:143–148. doi: 10.1152/ajplegacy.1964.206.1.143. [DOI] [PubMed] [Google Scholar]
- 41.Bouillaud F., Ricquier D., Thibault J., Weissenbach J. Molecular approach to thermogenesis in brown adipose tissue: cDNA cloning of the mitochondrial uncoupling protein. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(2):445–448. doi: 10.1073/pnas.82.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobsson A., Stadler U., Glotzer M. A., Kozak L. P. Mitochondrial uncoupling protein from mouse brown fat. Molecular cloning, genetic mapping, and mRNA expression. The Journal of Biological Chemistry. 1985;260(30):16250–16254. [PubMed] [Google Scholar]
- 43.Kajimura S., Seale P., Kubota K., et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-β transcriptional complex. Nature. 2009;460(7259):1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park A., Kim W. K., Bae K. H. Distinction of white, beige and brown adipocytes derived from mesenchymal stem cells. World Journal of Stem Cells. 2014;6(1):33–42. doi: 10.4252/wjsc.v6.i1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waldén T. B., Hansen I. R., Timmons J. A., Cannon B., Nedergaard J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. American Journal of Physiology-Endocrinology and Metabolism. 2012;302(1):E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- 46.Wu J., Boström P., Sparks L. M., et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barak Y., Nelson M. C., Ong E. S., et al. PPARγ is required for placental, cardiac, and adipose tissue development. Molecular Cell. 1999;4(4):585–595. doi: 10.1016/S1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 48.Rosen E. D., Sarraf P., Troy A. E., et al. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Molecular Cell. 1999;4(4):611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 49.Tontonoz P., Hu E., Graves R. A., Budavari A. I., Spiegelman B. M. mPPARγ2: tissue-specific regulator of an adipocyte enhancer. Genes & Development. 1994;8(10):1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 50.Seale P., Bjork B., Yang W., et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villanueva C. J., Vergnes L., Wang J., et al. Adipose subtype-selective recruitment of TLE3 or prdm16 by PPARγ specifies lipid storage versus thermogenic gene programs. Cell Metabolism. 2013;17(3):423–435. doi: 10.1016/j.cmet.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 53.Westendorf J. J. Transcriptional co-repressors of Runx2. Journal of Cellular Biochemistry. 2006;98(1):54–64. doi: 10.1002/jcb.20805. [DOI] [PubMed] [Google Scholar]
- 54.McLarren K. W., Lo R., Grbavec D., Thirunavukkarasu K., Karsenty G., Stifani S. The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. Journal of Biological Chemistry. 2000;275(1):530–538. doi: 10.1074/jbc.275.1.530. [DOI] [PubMed] [Google Scholar]
- 55.Javed A., Guo B., Hiebert S., et al. Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(alpha)/AML/PEBP2(alpha)) dependent activation of tissue-specific gene transcription. Journal of Cell Science. 2000;113(12):2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- 56.Moon R. T., Bowerman B., Boutros M., Perrimon N. The promise and perils of Wnt signaling through β-catenin. Science. 2002;296(5573):1644–1646. doi: 10.1126/science.1071549. [DOI] [PubMed] [Google Scholar]
- 57.Monroe D. G., McGee-Lawrence M. E., Oursler M. J., Westendorf J. J. Update on Wnt signaling in bone cell biology and bone disease. Gene. 2012;492(1):1–18. doi: 10.1016/j.gene.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosen V. Harnessing the parathyroid hormone, Wnt, and bone morphogenetic protein signaling cascades for successful bone tissue engineering. Tissue Engineering—Part B: Reviews. 2011;17(6):475–479. doi: 10.1089/ten.teb.2011.0265. [DOI] [PubMed] [Google Scholar]
- 59.Ross S. E., Hemati N., Longo K. A., et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289(5481):950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- 60.Cavallo R. A., Cox R. T., Moline M. M., et al. Drosophila Tcf and Groucho interact to repress wingless signalling activity. Nature. 1998;395(6702):604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 61.Roose J., Molenaar M., Peterson J., et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395(6702):608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 62.Daniels D. L., Weis W. I. β-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nature Structural and Molecular Biology. 2005;12(4):364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 63.Marx R. E. Pamidronate (Aredia) and zoledronate (Zometa) induced avascular necrosis of the jaws: a growing epidemic. Journal of Oral and Maxillofacial Surgery. 2003;61(9):1115–1117. doi: 10.1016/s0278-2391(03)00720-1. [DOI] [PubMed] [Google Scholar]
- 64.Lenart B. A., Lorich D. G., Lane J. M. Atypical fractures of the femoral diaphysis in postmenopausal women taking alendronate. The New England Journal of Medicine. 2008;358(12):1304–1306. doi: 10.1056/nejmc0707493. [DOI] [PubMed] [Google Scholar]
- 65.Zhang G., Guo B., Wu H., et al. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nature Medicine. 2012;18(2):307–314. doi: 10.1038/nm.2617. [DOI] [PubMed] [Google Scholar]
- 66.Cheng S.-L., Shao J.-S., Charlton-Kachigian N., Loewy A. P., Towler D. A. Msx2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. Journal of Biological Chemistry. 2003;278(46):45969–45977. doi: 10.1074/jbc.m306972200. [DOI] [PubMed] [Google Scholar]
- 67.Abdallah B. M., Jensen C. H., Gutierrez G., Leslie R. G. Q., Jensen T. G., Kassem M. Regulation of human skeletal stem cells differentiation by Dlk1/Pref-1. Journal of Bone and Mineral Research. 2004;19(5):841–852. doi: 10.1359/jbmr.040118. [DOI] [PubMed] [Google Scholar]
- 68.Hong J.-H., Hwang E. S., McManus M. T., et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309(5737):1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 69.Bennett C. N., Longo K. A., Wright W. S., et al. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(9):3324–3329. doi: 10.1073/pnas.0408742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hata K., Nishimura R., Ueda M., et al. A CCAAT/enhancer binding protein β isoform, liver-enriched inhibitory protein, regulates commitment of osteoblasts and adipocytes. Molecular and Cellular Biology. 2005;25(5):1971–1979. doi: 10.1128/mcb.25.5.1971-1979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iwata T., Kawamoto T., Sasabe E., et al. Effects of overexpression of basic helix-loop-helix transcription factor Dec1 on osteogenic and adipogenic differentiation of mesenchymal stem cells. European Journal of Cell Biology. 2006;85(5):423–431. doi: 10.1016/j.ejcb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 72.Barbagallo I., Vanella A., Peterson S. J., et al. Overexpression of heme oxygenase-1 increases human osteoblast stem cell differentiation. Journal of Bone and Mineral Metabolism. 2010;28(3):276–288. doi: 10.1007/s00774-009-0134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tokuzawa Y., Yagi K., Yamashita Y., et al. Id4, a new candidate gene for senile osteoporosis, acts as a molecular switch promoting osteoblast differentiation. PLoS Genetics. 2010;6(7) doi: 10.1371/journal.pgen.1001019.e1001019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishikawa K., Nakashima T., Takeda S., et al. Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. The Journal of Clinical Investigation. 2010;120(10):3455–3465. doi: 10.1172/jci42528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qiu N., Cao L., David V., Darryl Quarles L., Xiao Z. Kif3a deficiency reverses the skeletal abnormalities in Pkd1 deficient mice by restoring the balance between osteogenesis and adipogenesis. PLoS ONE. 2010;5(12, article e15240) doi: 10.1371/journal.pone.0015240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taipaleenmäki H., Abdallah B. M., AlDahmash A., Säämänen A.-M., Kassem M. Wnt signalling mediates the cross-talk between bone marrow derived pre-adipocytic and pre-osteoblastic cell populations. Experimental Cell Research. 2011;317(6):745–756. doi: 10.1016/j.yexcr.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 77.Quach J. M., Walker E. C., Allan E., et al. Zinc finger protein 467 is a novel regulator of osteoblast and adipocyte commitment. The Journal of Biological Chemistry. 2011;286(6):4186–4198. doi: 10.1074/jbc.m110.178251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X., Liao S., Nelson E. R., et al. The cytoskeletal regulatory scaffold protein GIT2 modulates mesenchymal stem cell differentiation and osteoblastogenesis. Biochemical and Biophysical Research Communications. 2012;425(2):407–412. doi: 10.1016/j.bbrc.2012.07.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cawthorn W. P., Bree A. J., Yao Y., et al. Wnt6, Wnt10a and Wnt10b inhibit adipogenesis and stimulate osteoblastogenesis through a β-catenin-dependent mechanism. Bone. 2012;50(2):477–489. doi: 10.1016/j.bone.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y., Berendsen A. D., Jia S., et al. Intracellular VEGF regulates the balance between osteoblast and adipocyte differentiation. The Journal of Clinical Investigation. 2012;122(9):3101–3113. doi: 10.1172/jci61209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hayashi M., Nakashima T., Taniguchi M., Kodama T., Kumanogoh A., Takayanagi H. Osteoprotection by semaphorin 3A. Nature. 2012;485(7396):69–74. doi: 10.1038/nature11000. [DOI] [PubMed] [Google Scholar]
- 82.Li D., Zhang R., Zhu W., et al. S100A16 inhibits osteogenesis but stimulates adipogenesis. Molecular Biology Reports. 2013;40(5):3465–3473. doi: 10.1007/s11033-012-2413-2. [DOI] [PubMed] [Google Scholar]
- 83.Sen B., Xie Z., Case N., et al. MTORC2 regulates mechanically induced cytoskeletal reorganization and lineage selection in marrow-derived mesenchymal stem cells. Journal of Bone and Mineral Research. 2014;29(1):78–89. doi: 10.1002/jbmr.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu Y., Tu Q., Valverde P., et al. Central adiponectin administration reveals new regulatory mechanisms of bone metabolism in mice. American Journal of Physiology—Endocrinology and Metabolism. 2014;306(12):E1418–E1430. doi: 10.1152/ajpendo.00048.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deng P., Chen Y., Ji N., et al. Cysteine dioxygenase type 1 promotes adipogenesis via interaction with peroxisome proliferator-activated receptor gamma. Biochemical and Biophysical Research Communications. 2015;458(1):123–127. doi: 10.1016/j.bbrc.2015.01.080. [DOI] [PubMed] [Google Scholar]
- 86.Zhao X., Deng P., Feng J., et al. Cysteine dioxygenase type 1 inhibits osteogenesis by regulating Wnt signaling in primary mouse bone marrow stromal cells. Scientific Reports. 2016;6 doi: 10.1038/srep19296.19296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li P., Yang Y. M., Sanchez S., et al. Deubiquitinase MYSM1 is essential for normal bone formation and mesenchymal stem cell differentiation. Scientific Reports. 2016;6 doi: 10.1038/srep22211.22211 [DOI] [PMC free article] [PubMed] [Google Scholar]


