Abstract
The relationships of higher order chromatin organization to mammalian gene expression remain incompletely defined. The human Growth Hormone (hGH) multigene cluster contains five gene paralogs. These genes are selectively activated in either the pituitary or the placenta by distinct components of a remote locus control region (LCR). Prior studies have revealed that appropriate activation of the placental genes is dependent not only on the actions of the LCR, but also on the multigene composition of the cluster itself. Here, we demonstrate that the hGH LCR ‘loops’ over a distance of 28 kb in primary placental nuclei to make specific contacts with the promoters of the two GH genes in the cluster. This long-range interaction sequesters the GH genes from the three hCS genes which co-assemble into a tightly packed ‘hCS chromatin hub’. Elimination of the long-range looping, via specific deletion of the placental LCR components, triggers a dramatic disruption of the hCS chromatin hub. These data reveal a higher-order structural pathway by which long-range looping from an LCR impacts on local chromatin architecture that is linked to tissue-specific gene regulation within a multigene cluster.
INTRODUCTION
The human growth hormone (hGH) multigene cluster spans 48 kb within a gene-rich segment of chromosome 17q22–24 (Figure 1A). This gene cluster encompasses five gene paralogs that were generated during primate evolution via reduplications of an ancestral GH gene (1). The most 5′ gene in the cluster, hGH-N, is specifically and robustly expressed in somatotropes of the anterior pituitary. The four remaining genes are transcribed specifically in the multinucleated syncytiotrophoblasts (STB) that constitute the epithelial surface of the placental villi. hCS-A and hCS-B are robustly co-expressed in the STBs during the second and third trimester of pregnancy, while hGH-V is co-expressed in the placenta, but at a 100-fold lower level. hCS-L is a placentally transcribed pseudogene whose function has been ablated by multiple loss-of-function mutations (2–4). The hGH cluster is flanked 5′ by genes expressed specifically in B-cells (CD79b) and skeletal muscle (SCN4A), and flanked 3′ by a testis-specific gene (TCAM1) (Figure 1A). The compact positioning of these eight genes and their regulatory regions suggests a tightly regulated network of transcriptional controls capable of driving robust and mutually exclusive expression in five distinct tissues (5–8).
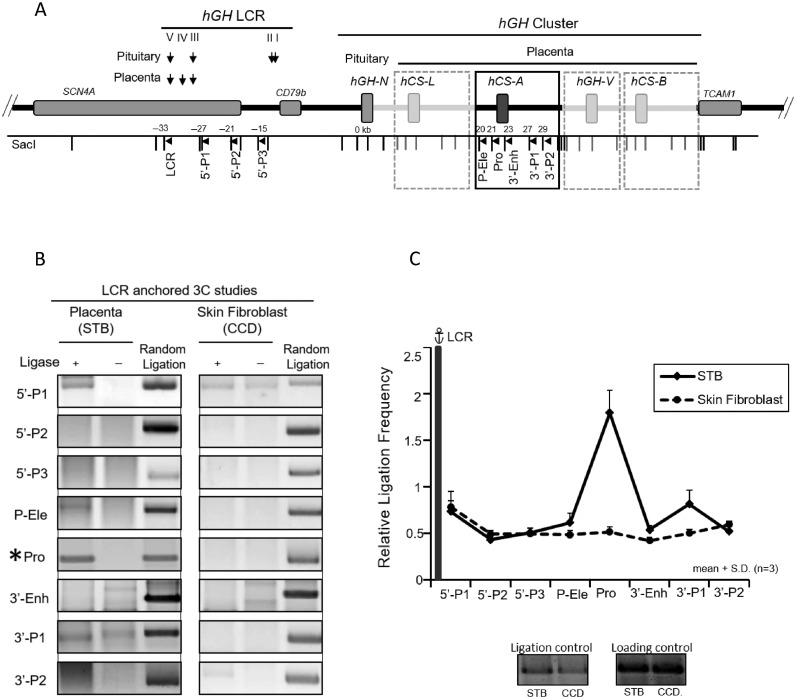
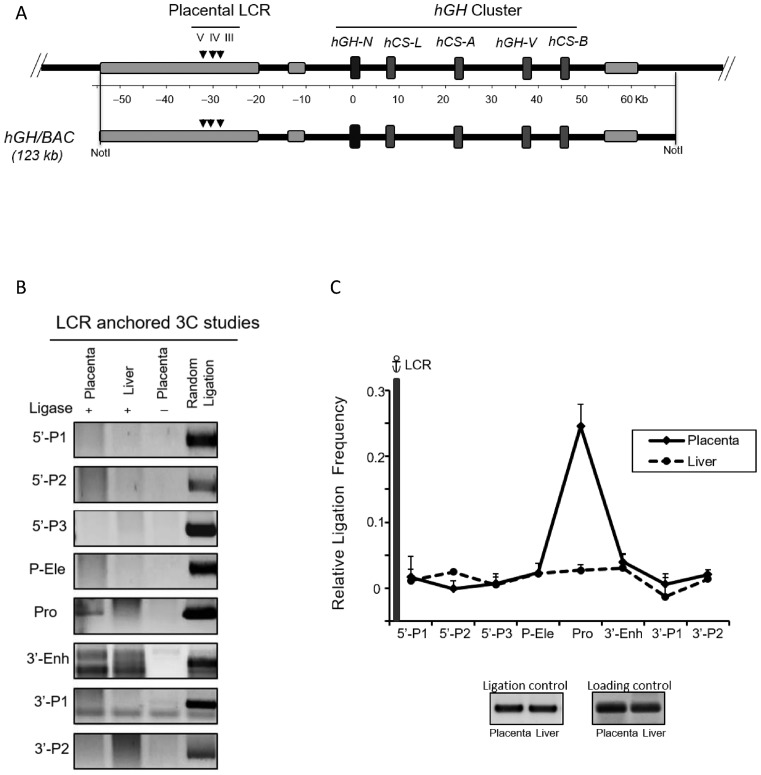
Figure 1.
Looping of the hGH locus control region (HSIII–V) to the hGH gene cluster in primary human placental syncytiotrophoblasts. (A) hGH locus and corresponding 3C mapping strategy. The structures of the hGH gene cluster and flanking regions are displayed in the diagram. The positions of the DNaseI hypersensitive site (HS) that constitute the hGH LCR in the placenta and pituitary are shown. A SacI map is displayed below the diagram; distances (kb) are shown relative to the hGH-N promoter. Primers used in the 3C assays are designed to proceed in the same direction and are indicated by arrowheads under corresponding restriction fragments. A 6 kb SacI fragment encompassing HSIII-V (‘LCR’) was used as an anchor for the assay. The structures of the highly conserved placental gene repeat (PGR) units, corresponding to hCS-L, hCS-A, hGH-V and hCS-B, are labeled in the boxed diagram of the representative hCS-A PGR unit. Of note, the ‘promoter’ primer (Pro) site is conserved among all five genes in the cluster. (B) 3C analysis of the hGH locus using the hGH LCR (HSIII–V) as anchor. Analyses of primary human STB chromatin (left panels) and human skin fibroblasts (CCD; right panels) are displayed. PCR amplicons of the LCR-anchored 3C products were resolved on a 2% argarose gel and stained with SYBR Gold. In each panel, the left lane contains 3C products, the middle lane is generated by a parallel reaction lacking ligase in the 3C assay, and the right lane (control) contains the random ligation products of a SacI-digested hGH/BAC DNA. The only validated 3C product generated in this STB analysis corresponds to the ligation of the LCR anchor and the promoter-containing SacI fragment (‘Pro’) (labeled by *). All other products failed to align with the random ligation control products and failed to be re-cleaved by SacI. (C) The hGH LCR (HSIII-V) loops to the promoter(s) within the hGH cluster. 3C ligation products (as in B) were quantified by PhosphorImager and QuantOne software. The ‘relative ligation frequency’ was determined by the ratio of the 3C ligation product to random ligation product. This ratio was then normalized to the ERCC3 ligation control and loading control (see Materials and Methods). The plot shows the average values from three analyses, each beginning with an independently generated chromatin preparation. The promoter (Pro) primer is perfectly complementary to the promoters of all five of the genes in the cluster. Error bars indicate one standard deviation. These data reveal close proximity of the placental LCR to the promoter region(s) of hGH/hCS gene(s) in STB chromatin (solid line). This proximity was not seen in a parallel analysis of primary skin fibroblasts (dotted line).
The hGH cluster is flanked 5′ by a multicomponent locus control region (LCR) (Figure 1A). HSI and HSII, located at −14.5 kb and −15.5 kb relative to the hGH-N promoter, are formed specifically in somototrope chromatin of the anterior pituitary. In contrast, the formation of HSIV (at −30 kb) is specific to the placental STBs. HSIII and V, at −28 and −32 kb, respectively, appear to be constitutive in the human genome. Thus, the hGH LCR is complex in its structures and tissue specificities and must act over substantial distances to impact on target genes within the hGH cluster.
Transcriptional activation of hGH-N in pituitary somatotropes relies on coordinate actions of HSI and HSII (9–11). The pituitary-specific POU-homeodomain protein, Pit-1, binds to an array of sites at HSI and generates a localized domain of noncoding transcription coincident with the CD79b locus (12,13). The HSI-linked noncoding transcriptional domain then loops to the hGH-N promoter and activates hGH-N transcription (14). HSI is essential to hGH activation during embryonic pituitary development and to maintenance of hGH-N expression in the adult pituitary (15). Germline deletion of HSI results in loss of long-range looping, a 20-fold decrease in hGH transcription in somatotropes and relaxation of somatotrope specificity. Thus, HSI comprises a powerful pituitary-specific enhancer that works via direct contact (‘looping’) with its target (hGH-N) promoter.
In contrast to the well-defined pathway of hGH-N activation in the pituitary, the basis for the control of hCS expression in the placenta remains unclear. In vitro studies of hCS gene expression have identified promoter-proximal elements that impact on this process. These elements include the promoter TATA box and the promoter-proximal Sp1 binding sites, an initiator element (InrE), as well as a conserved 5′ ‘Placental (P)-element’ and a 3′-enhancer (16–19). Detailed analysis of histone modifications in primary STB chromatin has revealed a zone of chromatin activation (H3/H4 acetylation and H3K4 methylation) encompassing the HSIII–V region (Supplementary Figure S1). In addition, epigenetically defined zones of chromatin activation individually encompass each of the four PGR units within the cluster (Supplementary Figure S1 and boxed segments; Figure 1A) (20). These data suggest a model in which each PGR is activated independently. However, transgenic studies (see below) have revealed that the local determinants defining the individual PGR units are in fact insufficient to accurately drive an isolated hCS gene in vivo and that the combined functions of the LCR in conjunction with the overall structure/organization of the multigene cluster are critical to these controls (8,20,21).
Gene expression from the hGH gene cluster can be accurately modeled in mice. For example, an hGH/BAC transgene, encompassing the entire hGH cluster and the contiguous LCR, expresses hGH-N specifically and robustly in pituitary somatotropes and expresses hCS-A and hCS-B robustly and specifically in the placenta (20). Accurate control of the hCS genes in the placenta is lost when the placental LCR (HSIII–V) is deleted from the hGH/BAC transgene locus. Restriction of hCS gene expression to the placenta is also lost when the multigene cluster is replaced by a single intact hCS-A PGR unit (21). The existence of a placenta-specific LCR component, HSIV and its recruitment of the architectural protein, CCCTC-binding factor (CTCF), further supported the role for the LCR in specifying chromatin architecture at the hGH locus (21). These in vivo functional studies support a model in which accurate control of hCS expression in the placenta relies on the integrated actions of a remote LCR along with the assembly of a functionally-essential chromatin architecture within the cluster itself.
In the present study, we explore chromatin architecture at the native hGH locus in primary human STB chromatin and at the hGH transgene locus in the placentae of mouse embryos. The data from these two sets of analyses are internally consistent in defining a higher-order chromatin organization that juxtaposes (‘loops’) the placental LCR (HSIII–V) with the promoters of the two hGH genes (hGH-N and hGH-V). These long-range contacts exclude the three hCS genes which co-assemble into a tightly packed ‘hCS chromatin hub’. Deletion of the HSIII–V region ablates the long-range looping and triggers a dramatic reconfiguration of gene packing within the multigene locus. This reconfiguration disrupts the hCS chromatin hub in the placenta and is associated with relaxation of hCS transcriptional controls. Thus long-range looping of the placental LCR to the hGH cluster induces a higher-order chromatin architecture within the cluster that is linked to the tight control of hCS transcription.
MATERIALS AND METHODS
Isolation of intact placental nuclei
From transgenic mouse embryos
Two transgenic mouse lines, hGH/BAC and ΔHSIII-V/hGH, were used in this report. These mouse lines were generated as previously described (20,21). Placentas from E18.5 day transgenic embryos were collected and washed in phosphate buffered saline (PBS) (Ca2+ and Mg2+ free). Placental cells were dissociated in cell-free dissociation buffer (GIBCO-BRL) and filtered through a 40 μm cell strainer (Falcon) into 20 ml 10% (v/v) fetal bovine serum (FBS)/Dulbecco's modified Eagle's medium (DMEM). The cell suspensions were fixed in 1% formaldehyde at room temperature (RT) for 10 min followed by the addition of glycine (final concentration 0.125 M) with incubation at room temperature for an additional 5 min. Cells were collected, washed with cold PBS twice and lysed on ice for 10 min in NP-40 buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 0.2% NP-40) and a proteinase inhibitor cocktail (Sigma)) containing 10 mM sodium butyrate. The nuclei were collected by centrifugation, washed twice with RB buffer (0.1 M NaCl, 50 mM Tris-HCl [pH 8.0], 3 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride (PMSF) and 5 mM sodium butyrate [pH 7.0]) and stored in glycerol storage buffer (40% glycerol, 50 mM Tris-HCl [pH 8.3], 5 mM MgCl2, 0.1 mM ethylenediaminetetraacetic acid (EDTA)) containing 20 mM sodium butyrate prior to analysis.
From human placenta
A normal non-identified human term placenta was obtained from the Hospital of the University of Pennsylvania under a protocol approved by the Institutional Review Board (IRB). The STB nuclei were selectively released and isolated as previously described (8,24). Briefly, villous fragments were excised from the placenta and rinsed with cold PBS containing 10 mM sodium butyrate. The fragments were finely minced, suspended in 150 mM NaCl and passed through a 10-gauge screen three times. The resulting sample was collected by centrifugation at 1000 g for 10 min and re-suspended in 150 mM NH4Cl containing 12.5 mg/ml ammonium bicarbonate followed by incubation on ice for 45 min. The released STB nuclei were collected by centrifugation at 1000 g for 10 min and re-suspended in cold RB buffer containing 20 mM sodium butyrate. The sample was passed through a 40 μm nylon filter to remove tissue debris and the presence of intact nuclei was confirmed by microscopic observation. The isolated nuclei were formaldehyde-crosslinked as described above and stored at 80°C in glycerol storage buffer containing 20 mM sodium butyrate prior to analysis.
Chromatin conformation capture (3C) assay
The 3C procedure was performed as previously described with modifications (22). Briefly, 20 μg of formaldehyde-crosslinked nuclei were re-suspended into 0.5 ml restriction enzyme buffer (50 mM potassium acetate, 20 mM tris-acetate [pH 7.9], 10 mM magnesium acetate, 100 μg/ml bovine serum albumin (BSA)) with addition of sodium dodecylsulfate acid (SDS) to the final concentration of 0.3%. After incubation for 1 h at 37°C with shaking at 1200 rpm, Triton X-100 was added to the final concentration of 2%. The sample was incubated for 1 h at 37°C with shaking at 1200 rpm, followed by addition of 800 U of SacI (New England Biolabs). The chromatin was digested overnight at 37°C with continuous shaking at 1200 rpm. After digestion, SDS was added to the final concentration of 2% and the sample was incubated at 65°C for 20 min. Half of the digested chromatin was removed and the crosslinks were reversed by heating at 65°C overnight. DNA was purified from the sample and analyzed to determine digestion efficiency. Only samples digested to greater than 80% were used for 3C analysis. The remaining half of the digested chromatin was diluted into 4 ml of 1X ligation buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 10 mM dithiothreiol (DTT), 0.1 mg/ml BSA and 1 mM ATP). Triton X-100 was added to a final concentration of 1%, followed by incubation for 1 h at 37°C while shaking gently. 4000 U of T4 DNA ligase (New England Biolabs) was added and the ligation reaction was performed for 4 h at 16°C followed by 30 min at room temperature (18–22°C). A control was performed under identical conditions, but without T4 DNA ligase. The ligation reaction was terminated by adding EDTA (10 mM, final concentration). Crosslinks were reversed by incubating the sample at 65°C overnight in the presence of 150 μg proteinase K. A total of 150 μg RNase was added and incubated for 45 min. The DNA samples were extracted with phenol and chloroform, precipitated with ethanol, dissolved in 100 μl of 10 mM Tris [pH 7.5] and analyzed by PCR.
PCR analysis of 3C ligation products
The PCR was performed using 2% of the purified DNA from each 3C ligation reaction (see above). One nanogram of a ligation reaction containing SacI-digested hGH/BAC plasmid was used for the random ligation control. The primers for the PCR are listed in Supplementary Table S1. The annealing temperatures were specifically adjusted for each primer set to optimize PCR efficiency. The PCR products were separated on 2% agarose gels and stained with SYBR Gold (Invitrogen). In a set of control studies, each of the PCR products was purified, and re-cleaved or sequenced to confirm the predicted ligation product. The intensities of the bands were analyzed by PhosphorImager and quantified by QuantOne software. Two primer pairs of a ubiquitously expressed ERCC3 gene, one located within two adjacent SacI-fragments and another located within a single SacI-fragment, were used as the ligation control and loading control respectively. The ligation frequency was determined as a ratio of the 3C ligation product to the corresponding product in the random ligation control. This ratio was then normalized to the ERCC3 ligation control and loading control.
Sequencing assay
Specific sets of purified DNA after 3C procedure (above) were analyzed by sequencing assay. The desired sets of ligation products were amplified by PCR. The generated amplicons were purified by gel extraction (Qiagen), subcloned into pGEM 7Z vectors (Promega) and the structure of each cloned insert was determined by DNA sequencing.
CTCF-chromatin immunoprecipitation (ChIP)
Crosslinked nuclei (prepared as above) were pelleted and suspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1% SDS, 10 mM sodium butyrate and 0.1 mM PMSF) and incubated on ice for 10 min. The lysates were then sheared by sonication (Sonic Dismembrator, Fisher Scientific) to an average size of 200 to 1000 bp and the soluble chromatin was concentrated using Centricon-10 (Amicon Inc.) and taken up in IP buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 mM EDTA, 0.01% SDS, 1% Triton X-100, 10 mM sodium butyrate and 0.1 mM PMSF). An aliquot of each sample was removed as ‘input’ and used in PCR analysis. The remainder of the soluble chromatin was incubated at 4°C overnight with 20 μl of CTCF antibody (Upstate Biotech) or preimmune IgG (Upstate Biotech). Immune complexes were isolated by incubation with 60 μl of Protein A/G-agarose (Millipore) for 2 h at 4°C. The complexes were serially washed in 1 ml low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [8.0] and 150 mM NaCl), 1 ml of the same buffer but with high salt (500 mM NaCl), 1 ml of LiCl buffer (250 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA and 10 mM Tris-HCl [pH 8.0]) and twice with TE (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA). The complexes were eluted with two 250 μl aliquots of elution buffer (1% SDS and 0.1 M NaHCO3) at RT for 15 min. DNAs were isolated by reversing the crosslinks (65°C for 6 h in the presence of 0.2 M NaCl and 2 μl of 10 mg/ml RNase A) and subsequently digested with 1 μl of 20 mg/ml proteinase K at 45°C for 12 h. The purified DNAs were purified by QIAquick spin column (Qiagen) and analyzed by PCR with specified primer pairs [21]. The amplified products were resolved by electrophoresis on 2% agarose gels. DNAs were visualized by fluorescence staining with SYBR Gold (Invitrogen) and quantified by PhosphorImager analysis. Band intensities were expressed relative to the signal obtained from 0.01% of input, and normalized with positive control (defined as 100) using a primer set that amplified CTCF binding site in the Igf2/H19 imprinting control region. All signals were demonstrated to be proportional to the amount of DNA input.
RESULTS
The hGH LCR (HSIII–V) loops to the hGH gene cluster in human placental STB chromatin
To explore the relationship of higher-level chromatin organization to placental expression of the hCS genes, we determined the configuration of the hGH locus in primary human placental STB. This was accomplished by standard chromatin conformation capture (3C) (22). The large multinucleate STB cells were selectively released from human term placental villi by osmotic lysis (23). The STB chromatin was formaldehyde crosslinked and digested with SacI DNA endonuclease. As a control for tissue specificity, a parallel 3C study was carried out on chromatin from primary human skin fibroblasts. SacI digestion isolates the LCR (HSIII–V) region on a 6 kb fragment and generates an array of fragments that represent repeated sites within the four PGR units (Figure 1A). These fragments include three determinants that are relevant to transcriptional activity of PGRs based on in vivo and in vitro studies (16,18,19); the promoters (‘Pro’), the structurally conserved P-elements (‘P-Ele’) located 2 kb 5′ to each PGR promoter, and a structurally conserved enhancer (‘3-Enh’) located 2 kb 3′ to each of the three hCS genes (A, B and L).
The SacI fragment encompassing HSIII–V (‘LCR’) was used as the anchor for the initial 3C assay (Figures 1B and C). Analysis of 3C ligation products generated from the STB chromatin revealed robust interaction of the LCR anchor with SacI fragments encompassing the hGH/hCS gene promoters. The absence of interactions between the LCR anchor and sites in the region between the LCR and the gene cluster (5’P-1, 5’P-2 or 5’P-3 primer) suggested that the interaction(s) of the LCR (HSIII–V) with the gene promoter(s) effectively ‘loops out’ the intervening 28 kb region. The parallel analysis of fibroblast chromatin lacked evidence of higher-order interactions of the cluster with the LCR. These data led us to conclude that specific long-range interaction(s) were established between the hGH LCR (HSIII–V) and the hGH/CS gene promoter(s) in STB chromatin (Figures 1B and C).
The placental LCR (HSIII–V) specifically loops to the hGH-N and hGH-V promoters within the multigene cluster
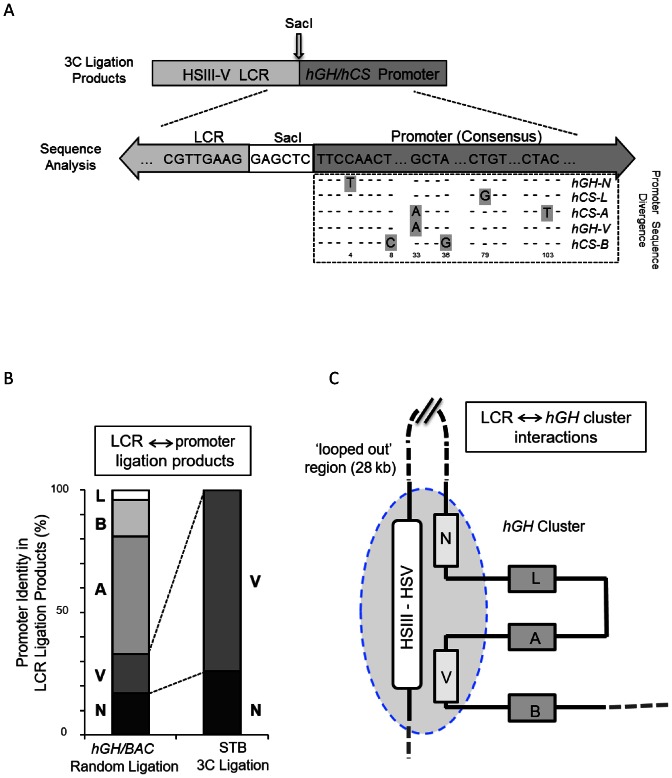
To map the interactions of the hGH LCR and gene promoter(s) in greater detail, the 3C ligation products corresponding to the LCR-promoter looping interactions with the Pro fragments were cloned and sequenced. These analyses confirmed the presence of LCR and Pro fragments linked by a SacI recognition site in each product (Figure 2A). The sequence analysis of the ligation products generated from the SacI-digested hGH/BAC plasmid (random ligation control) revealed ligations of the LCR to each of the five promoters (Figure 2B, left) with 70% of these random ligations occurring between the LCR and the hCS promoter fragments (hCS-A, -B and -L). In marked contrast, LCR contacts in the STB chromatin were entirely restricted to the promoters of the two hGH genes (N and V) (Figure 2B, right). Thus the 3C analysis of primary STB chromatin demonstrates looping of the hGH LCR (HSIII–V) into close proximity with the hGH-N and hGH-V promoters. Most remarkably, we observed that in this configuration the two robustly-expressed placental hCS genes, hCS-A and hCS-B, are excluded from these placenta-specific LCR contacts (Figure 2C).
Figure 2.
The hGH LCR (HSIII–V) selectively loops to the hGH-N and hGH-V promoters in primary human placental STBs. (A) Approach to specifying interactions of the hGH LCR with individual hGH/hCS promoters. 3C-ligation products generated between the SacI fragments containing the hGH LCR (anchor) and the gene promoter (Pro) fragments in the random ligation control (SacI-digested hGH/BAC plasmid) and in STB chromatin (as in Figure 1) were cloned and individually sequenced. The identities of the LCR anchor fragments and SacI recognition sequence (GAGCTC) were confirmed in each ligation product and the specific identity of each linked promoter fragment was determined via unique distinguishing sequences (as highlighted in the promoter sequence alignment). (B) The LCR interacts specifically with the hGH-N and hGH-V promoters. The relative distributions of each of the five gene promoters ligated to the placental LCR (as determined in A, above) from the random ligation of SacI-digested hGH/BAC DNA and from the 3C analysis of human placental STB chromatin are displayed as indicated. These data represent the sequence analysis of 96 and 104 subcloned ligation products, respectively. (C) Schematic of interactions between the hGH LCR and the hGH gene cluster in primary human STB cells. This model is based on the 3C analysis using the LCR anchor primer. The specificity of promoter identities (as in A) and their relative frequencies (as in B) are represented with HSIII–V LCR being situated in proximity to the hGH-N and hGH-V promoters. The 28 kb region between the hGH LCR and the hGH-N promoter lacks interactions with the LCR and is therefore represented as a ‘looped-out’ segment. The hCS-L, hCS-A and hCS-B genes are all excluded from LCR interactions and are similarly shown as ‘looped-out’ from the hub of LCR interactions with the two GH genes.
The hCS genes within the hGH cluster are organized into a tightly packed ‘chromatin hub’ in human placental chromatin
Our prior studies have revealed that conversion of the hGH multigene cluster into a single gene (hCS-A PGR) unit, results in a dramatic loss of hCS transcriptional control (21). The hCS expression in this single-gene transgenic locus is sensitive to site-of-integration effects as reflected in a loss of strict copy-number-dependent expression in the placenta and substantial ectopic expression in multiple tissues. These data suggested that the internal structure of the hGH multigene cluster is critical to placenta-specific controls over hCS expression. To define the internal organization of the intact cluster we re-analyzed the STB 3C library using the 3′-P2 anchor fragment located at the 3′ terminus of each PGR unit and 8 kb 3′ to the corresponding promoters (Figure 1A). This site was used as anchor as it is structurally conserved in all three hCS genes and is enriched for CTCF binding (ENCODE database (24)). The analysis revealed isolated interaction(s) between the 3′-P2 fragment and the P-element(s) located 2 kb 5′ of each of the four PGR promoters. These interactions were not detected in a parallel analysis of skin fibroblast chromatin (Figures 3A and B). These data reveal the presence of short-range interactions within the hGH multigene cluster in STB chromatin.
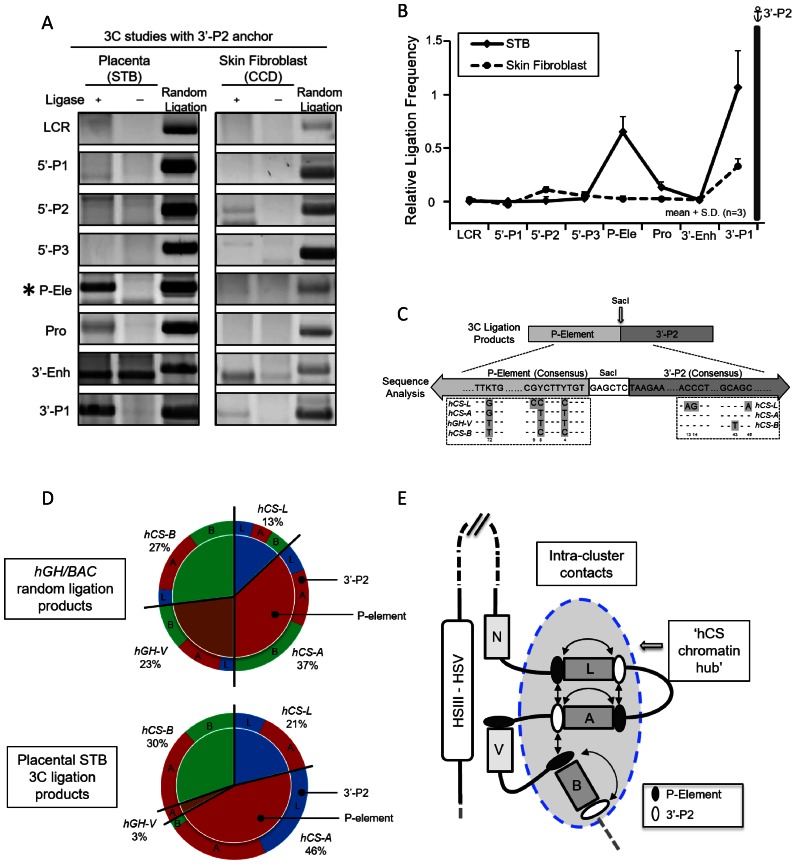
Figure 3.
Structural analysis of interactions within the hGH cluster in primary human placental syncytiotrophoblasts. (A) 3C analysis of hGH locus using the 3′-P2 anchor fragment located at the 3′ end of the PGR unit. The 3′-P2 anchor fragment (see Figure 1A) is conserved in all three hCS genes (hCS-A, -B and -L). The analysis of the PCR assay of ligation products is organized as in Figure 1B. (B) 3C analysis reveals proximity of the 3′-P2 anchor with the conserved P-elements. Ligation frequencies (calculated as in Figure 1C) of each indicated sites are displayed in the graph. The interactions between the P-element and the 3′-P2 site in the primary human placental STB cells (solid line) were not observed in primary human skin fibroblasts (dotted line). (C) Specification of interactions amongst 3′-P2 and P-element fragments. The 3′-P2 anchor/P-element 3C ligation products were cloned and sequenced and the specific identity of each interaction was determined. Diagnostic sequence divergences in the P-elements and the 3′-P2 regions were used to identify specific interactions within the PGR units as highlighted inside the dashed boxes. The sequencing analysis confirmed the accurate ligation of the P-element fragment and 5′-end of 3’P-2 fragment across the SacI restriction site in all products tabulated. (D) Quantitation of ligation frequencies between specified 3′-P2 and P-element sites. The relative ligation frequencies are displayed in the compound pie charts. The top graph represents products from a random ligation of SacI-digested hGH/BAC plasmids and the bottom graph represents the ligation frequencies within the placental STB chromatin locus. The numbers of clones analyzed were 96 and 102, respectively. The random ligation control data reveals the P-elements from each PGR unit were evenly distributed in their interaction frequencies (top, inner circle) and each P-element interacted with different 3′-P2 fragments (top, outer circle) at approximately the same frequency. In contrast, the analysis of the STB sample revealed that the P-elements from three hCS genes were involved in 97% of the ligations, while only 3% involved the hGH-V (bottom, inner circle). P-element interactions with 3′-P2 fragments were also highly selective; there was a complete absence of interactions between hCS-L and hCS-B, and only trace evidence for interactions of the P-element of hCS-A with 3′-P2 fragment of hCS-B (bottom, outer circle). (E) Model of higher-order interactions within the hGH cluster in primary human placental syncytiotrophoblast chromatin. This diagram integrates the results of the 3C analysis internal to the hGH gene cluster (as shown in D). The interactions between P-element and 3′-P2 regions within the hGH cluster are represented by double-headed arrows. The selective long-range contacts of the placental LCR (HSIII–V) with the promoters of the hGH-N and hGH-V genes (Figure 2) appear to sequester these two genes away from the ‘hub’ of interactions internal to the cluster.
Due to the conserved structures of the PGR units, and to the conserved sizes of the SacI fragments encompassing the three 3’P2 sites and the four P-Ele sites (Figure 1A), the 3C analysis could not specify the exact architecture of the intra-cluster contacts. To increase the level of resolution of the analysis, we therefore sequenced the individual 3′-P2/P-Ele ligation products. The nucleotide divergences in each of the corresponding fragments were sufficient to yield unambiguous assignment of each component within the ligation product (Figure 3C). The random ligation of SacI-digested hGH/BAC plasmid DNA revealed all possible pairing combinations of four P-elements and three 3′-P2 anchors (Figure 3D, top). In contrast, the analysis of the STB chromatin revealed that the P-elements of the three hCS genes contributed to 97% of the ligation products (Figure 3D, bottom). Of note, the contribution of the P-element 5′ to the hGH-V gene decreased from 23% in the random ligation control to 3% in the STB chromatin analysis. In addition, each of the three hCS P-elements interacted with distinct subsets of 3′-P2 sites (Figure 3D, bottom). A schematic summary of these interactions in STB cells reveals close packing of the three hCS genes mediated by 3′-P2 interactions with the P-element. These interactions generate an ‘hCS chromatin hub’. The exclusion of the hGH-N andhGH-V genes from this hub is fully consistent with the selective sequestration of their promoters by the placental LCR looping interactions (Figures 2 and 3E). Furthermore, the absence of interactions between hCS-L and hCS-B and the minimal interactions of hCS-A (P-element) with hCS-B (3′-P2) suggests that the hCS-B gene occupies a relatively unconstrained position at the periphery of the hCS chromatin hub structure (Figures 3D and E).
CTCF is selectively recruited to HSIV in primary STB chromatin
The selective looping of the hGH LCR (HSIII–V) to the promoters of the two hGH genes in placental chromatin was unexpected. This configuration juxtaposes the placental LCR with a GH gene that is tightly repressed in the placenta (hGH-N) and its paralog (hGH-V) that is expressed in the placenta, but at very low levels. This chromatin organization contrasts markedly with that in the pituitary where the sommatotrope-specific LCR determinants (HSI,II), containing the powerful HSI enhancer element, loop directly to its target hGH-N gene and robustly activates its transcription. These observations suggest that the placental LCR (HSIII–V) impacts on a regulatory function linked to cluster architecture rather than to direct transcriptional activation.
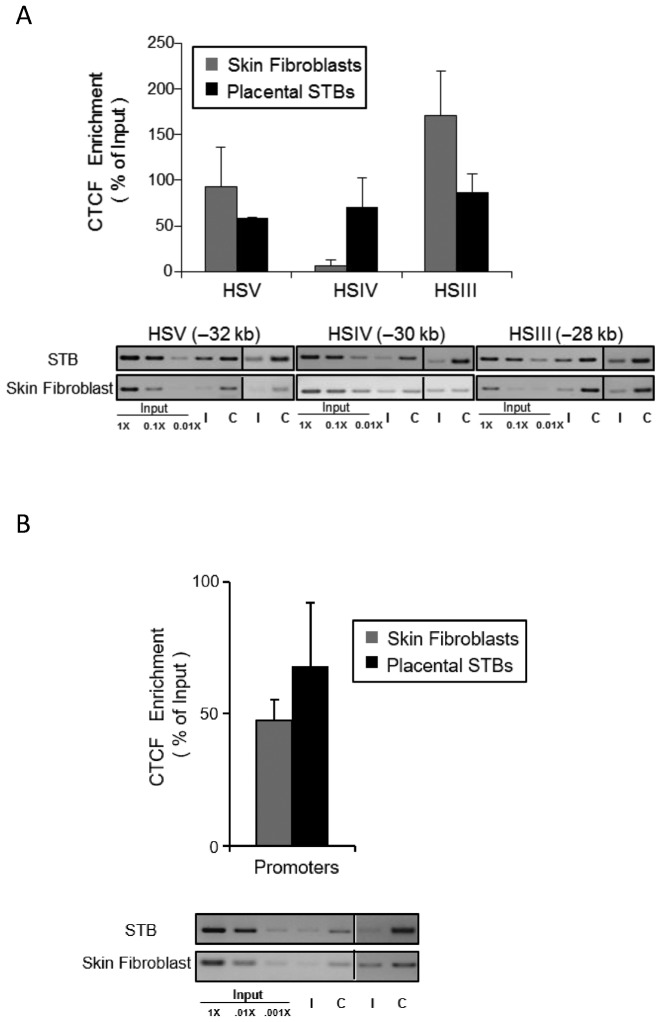
Genome-wide studies indicate that multiple aspects of chromatin architecture, and a subset of long-range chromatin interactions, are established and/or stabilized by one or more ‘architectural proteins’. Prominent among these proteins is the multi-zinc finger protein CTCF (25,26). Of note, we have demonstrated tissue-specific occupancy of CTCF at HSIV in the E18.5 placenta of hGH/BAC transgenic mice (21). To extend these observations to the native locus, we assessed CTCF occupancy in human STB chromatin. These CTCF ChIP data confirmed CTCF occupancy at HSIV in the primary human placenta and its absence in human fibroblast chromatin (Figure 4A). In contrast, CTCF is constitutively recruited to HSIII, HSV and all five hGH/CS promoters in the cluster both in STBs and in skin fibroblasts (Figures 4A and B, and Supplementary Figure S2). The placenta-specific occupancy of CTCF at HSIV is consistent with a model in which HSIV contributes to the formation of a specific chromatin architecture in the placenta that supports robust expression of hCS from the hGH locus.
Figure 4.
CTCF occupancy at HSIV is specific to primary human placental syncytiotrophoblasts. (A) CTCF is selectively recruited to HSIV in primary human STBs. CTCF recruitment within the HSIII–V LCR was assessed in the human STB chromatin by ChIP analysis in parallel with analysis of primary human skin fibroblasts. A serial dilution of input DNA was used to confirm that the semi-quantitative PCR was in a linear range in each assay. A pre-immune IgG antibody established nonspecific background levels generated by the IP; IgG antibody (I); CTCF antibody (C). DNAs isolated from each IP pellet was assessed with primer sets corresponding to the predicted CTCF binding sites at HSIII, HSIV and HSV. The PCR analysis showed that CTCF proteins were recruited to the HSIII and HSV in both tissues while CTCF enrichment at HSIV was STB-specific. (B) Constitutive recruitment of CTCF to the hGH/hCS promoters. The ChIP assay demonstrates CTCF occupancy at hGH/hCS promoter regions. This occupancy, in combination with similar results in multiple other tissues (ENCODE), confirms that promoter occupancy by CTCF in the cluster is a constitutive property of this region. Additional sequence analysis of PCR amplicons demonstrated CTCFs are enriched at all five promoters within the cluster (Supplementary Figure S2).
The chromatin organization of the hGH/BAC transgene in the mouse placenta accurately models the architecture of the native hGH locus in the human placenta
The higher-order structure of the hGH locus in human placental STB chromatin is organized on the basis of both long range (Figures 1 and 2) and local (Figure 3) interactions. We next wished to assess the interdependence of these interactions by determining whether the architecture internal to the cluster is dependent on the long-range looping. To validate the use of transgenic mouse models for this investigation, we first determined whether the chromatin contacts identified between the LCR and hGH cluster in primary human STB cells were accurately recapitulated at the hGH/BAC transgenic locus in the mouse placenta. The 123 kb hGH/BAC transgene contains the entire hGH cluster along with extended 5′- and 3′-flanking regions (Figure 5A). We have previously demonstrated that HSIII, IV and V are formed within the hGH/BAC transgene locus in the E18.5 mouse placenta and that hCS-A and hCS-B are specifically and robustly expressed at this time (20,21). 3C analysis of crosslinked chromatin isolated from E18.5 day hGH/BAC placenta demonstrated interactions between the LCR HSIII–V region (anchor fragment) and gene promoter(s) (Figures 5B and C). An independent reciprocal 3C analysis using the ‘Pro’ anchor fragment confirmed this observation (Supplementary Figure S3). In addition, the promoter fragment failed to make contact with the region between the LCR and hGH cluster. These findings were fully consistent with the 3C mapping in STB chromatin (Figure 1C). Furthermore, sequence analysis of the 3C ligation products generated from the hGH/BAC transgene locus in the mouse placenta demonstrated the same set of interactions within the hGH/BAC transgene locus and the formation of the same ‘hCS chromatin hub’ as observed in the STB chromatin (Supplementary Figure S4). Thus, the hGH/BAC transgene locus establishes the same long distance 3D chromatin organization and the same architecture internal to the hGH gene cluster as detected at the native locus in primary human STB cells.
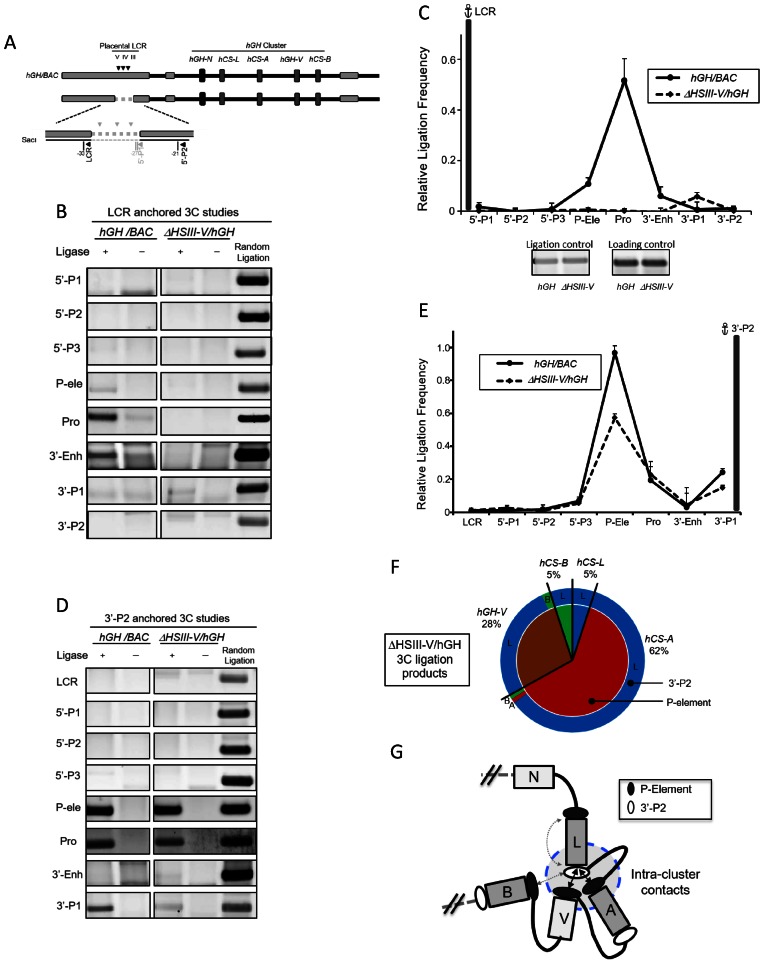
Figure 5.
Looping between the hGH LCR (HSIII–V) and the hGH/CS promoters is accurately modeled at the hGH/BAC transgene locus in the mouse placenta. (A) Diagram of the native hGH locus and the corresponding hGH/BAC transgene. A NotI released 123 kb fragment of human genomic DNA, encompassing hGH gene cluster along with its 5′ LCR and 3′-flanking region, represents the extent of the hGH/BAC transgene. Landmarks are as in Figure 1A. hGH/BAC transgenic mouse lines were generated from the released 123 kb NotI fragment (20). (B) 3C analyses of the hGH/BAC locus in transgenic mouse placenta and liver. Chromatin from placenta (E18.5) and adult liver of hGH/BAC transgenic mice were analyzed by 3C using the LCR anchor fragment (as in Figure 1). PCR-amplified ligation products are displayed as in Figure 1B. (C) hGH LCR loops to promoters within the hGH/BAC transgene locus in mouse placenta. 3C ligation products (as in B) were quantified by PhosphorImager and by QuantOne software (see Materials and Methods). The data were quantified and normalized as in Figure 1C. The looping of the hGH LCR (HSIII–V) to promoters within the hGH/BAC transgene cluster is identical to that observed in primary human STB cells (Figure 1C).
The tissue specificity of the interactions at the hGH/BAC transgene locus was assessed by promoter-anchored 3C analyses of transgenic mouse kidney, testis, liver and spleen (Supplementary Figure S3). These studies confirmed that the long-range interactions between the LCR and the promoter were specific to the placenta. Of note, novel sets of interactions were identified in each of the four non-placental tissues from the viewpoint of the ‘Pro’ anchor, suggesting that the interactions at this locus are highly sensitive to tissue-specific influences and further supporting the specificity of the placental configuration.
Formation of the ‘hCS chromatin hub’ within the hGH cluster is dependent on long-range looping from the hGH LCR
The relationship of the LCR looping to the configuration within the hGH cluster was investigated by deletion of the placental LCR from the otherwise intact hGH/BAC transgene (Figures 1A and 6A) (see Materials and Methods). E18.5 day placental chromatin was isolated from an ΔHSIII-V/hGH transgenic mouse and subjected to 3C analysis. The 5′ terminus of the ‘LCR’ SacI fragment that was used as the 3C anchor in analysis of the native cluster was retained in the ΔHSIII-V/hGH transgene so as to maintain a mapping procedure that was fully consistent with the analysis of the native cluster (see Materials and Methods). As expected, deletion of the LCR determinants ablated long-range looping interactions in the ΔHSIII-V/hGH transgene locus (Figures 6B and C). Remarkably, the 3C analysis of the chromatin architecture internal to the cluster using the 3′-P2 fragment as the anchor probe (Figures 6D and E) revealed a dramatic alteration in the higher order interactions (Figure 6F versus Figure 3D). In the absence of placental LCR (HSIII-V), the intra-cluster contacts were predominantly formed between the 3′-P2 region of hCS-L and the P-element regions of hCS-A or hGH-V (Figure 6G). In addition, the extensive 5′-to-3′ self-interactions within individual hCS-A or hCS-B gene units that contribute to the compact hCS chromatin hub conformation in the native hGH locus are fully ablated within the ΔHSIII-V/hGH locus. The shift in the architecture of the cluster subsequent to LCR deletion is summarized in Figure 7. These results lead us to conclude that the three-dimensional chromatin organization within the activated hGH cluster that forms the hCS chromatin hub in STB chromatin is driven by the actions of the looped placental LCR.
Figure 6.
The configuration within hGH multigene cluster is dependent on the placental LCR (HSIII–V). (A) Diagram of ΔHSIII-V/hGH transgene. A 6 kb DNA segment containing CTCF-binding sites in HSIII, IV and V was deleted from the hGH/BAC and the resultant ΔHSIII-V/hGH BAC was used to generate a set of transgenic mouse lines (see Materials and Methods). The ‘LCR primer’ site located at the 5′ terminus of the LCR SacI-fragment was retained intact to facilitate mapping. (B) 3C analysis of ΔHSIII-V/hGH transgene conformaton. The chromatin interactions with LCR anchor fragment in the wild-type (left panels) and ΔHSIII-V/hGH transgenic loci (right panels) were assessed by 3C (displayed as in Figure 1B). (C) Deletion of the HSIII–V region results in loss of long-range looping of the remote 5′ region to the hGH cluster. Quantification of the PCR products is as described in Figure 1C. The LCR-promoter interactions established in the hGH transgenic locus (solid line) are eliminated in the ΔHSIII-V/hGH transgene (dashed line). (D) 3C analyses of the placental hGH chromatin locus using the 3′-P2 fragment anchor site. The analyses of the native hGH/BAC transgenic locus (left panels) and ΔHSIII-V/hGH transgenic locus (displayed as in Figure 1B). (E) Chromatin contacts are established between 3′-P2 and P-element in the hGH/BAC transgene lacking HSIII–V region. The plotted 3C ligation frequencies of each site to the 3′-P2 anchor fragment were calculated based on PCR products (from D). Interactions between P-element and 3′-P2 regions observed in the wild-type hGH/BAC transgene (solid line) are formed in the ΔHSIII-V/hGH transgene locus despite the loss of LCR elements and long-range looping (dashed line). (F) Gene-specific interactions of the 3′-P2 and P-element within the hGH cluster undergo a dramatic shift secondary to the loss of LCR looping. The ligation products between the P-element and the 3′-P2 anchor were cloned and sequenced (as in Figures 3C and D). In contrast to the gene specific interactions in the intact hGH/BAC transgene, the analysis of the ΔHSIII-V/hGH transgenic locus demonstrated that 96% of interactions are linked to the 3′-P2 of hCS-L (outer circle). In additional, most of these 3′-P2 interactions are with the P-elements (inner circle) of hCS-A (62%) and hGH-V(28%). Only 5% of the interactions are with P-element of hCS-B or hCS-L. (G) Model of interactions within the hGH cluster in the ΔHSIII-V/hGH transgenic placenta. The diagram integrates the 3C (D and E) and sequencing analyses (F) internal to the hGH gene cluster in the ΔHSIII-V/hGH transgenic locus in the mouse placenta. Two double-headed arrows represent the major intra-cluster contacts between P-element and 3′-P2 regions. The minor frequency interactions between hCS-L and hCS-B and within hCS-L are indicated by the dashed lines. Ligation frequencies less than 1% are not shown.
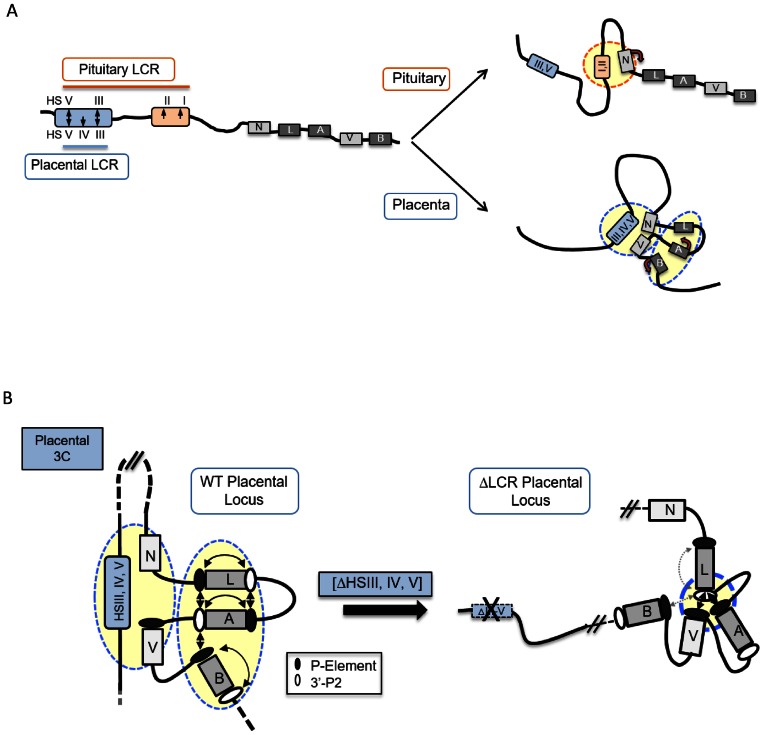
Figure 7.
3D chromatin architectures at the hGH multigene locus in the pituitary and placenta. (A) Distinct patterns of long-range LCR looping in the pituitary and placenta parallel tissue-specific gene expression from the hGH gene cluster in the two tissues. During the activation of pituitary hGH locus, the pituitary-specific HSI enhancer directly interacts with its target hGH-N promoter via chromatin looping (14). In contrast, in the placenta the HSIII–V LCR region loops to two hGH promoters, displacing the activated hCS genes into a tightly packed ‘hCS chromatin hub’. (B) The organization of hGH gene cluster in placental chromatin is driven by long-range LCR interactions. HSIII–V LCR interactions segregate the hGH-N and hGH-V genes from the hCS gene hub within the intact hGH locus in the placenta. Deletion of the placental LCR (HSIII–V) triggers a dramatic reconfiguration within the cluster that disrupts the ‘hCS chromatin hub’ with a corresponding loss of hCS transcriptional control.
DISCUSSION
Transcriptional controls are major drivers of cellular differentiation. Mechanisms of transcriptional control in eukaryotes are heavily dependent upon defined modifications of histone structures and on the assembly of chromatin interaction networks (25,26,27–29). 3C-based studies have linked temporal and spatial controls of transcription with 3D architectures in the mammalian genome (29–33). In the present report, we map the long-range interactions and local chromatin conformations at the hGH multigene cluster in the human placenta and in transgenic mice. These studies, in conjunction with our prior studies in the pituitary, highlight the distinct pathways by which a locus control region can interact with target gene(s) in a multigene cluster to establish tissue-specific transcriptional controls (Figure 7A).
The expression of hGH-N in pituitary somatotropes is tightly linked to looping of the pituitary-specific HSI,II enhancer determinant from the hGH LCR to the target hGH-N promoter (Figure 7A, top). This looping plays a direct and essential role in robust and specific activation of hGH-N gene expression within the hGH gene cluster (14,15). We now describe an entirely distinct scenario played by the hGH LCR in the placental expression of the hCS genes (Figure 7A, bottom). In this case the placental LCR unit, comprising HSIII–V, loops to the promoters of two genes that are either not expressed at all (hGH-N) or minimally expressed (hGH-V) (Figures 1C and 2B). Thus the looping in this case excludes the most robustly expressed genes, hCS-A and hCS-B. The chromatin architecture established by this long-range looping effectively segregates the two GH genes from the three hCS genes, clustering the three hCS genes in a tightly packed ‘hCS chromatin hub’. Thus, in the pituitary, transcription is enhanced by direct juxtaposition of a potent enhancer element of the LCR to the target hGH-N promoter, while in the placenta transcription is supported by LCR-mediated organization of the cluster into a functional chromatin configuration.
We have previously demonstrated the critical role played by the HSIII–V LCR in insulating the hGH transgene loci from site-of-integration effects and its apparent linkage to the placenta-specific binding of CTCF to HSIV (21). The CTCF protein participates in a wide variety of transcriptional regulatory pathways, including gene activation, repression and chromatin insulator/boundary functions (34,35). Genome-wide analyses have also linked CTCF, along with other ‘architectural’ proteins such as cohesin and mediator, to the assembly of long-range chromatin interactions within the mammalian genome. These proteins are thought to stabilize 3D chromatin architectures and are assumed to be of intrinsic importance to coordinating the expression of dispersed loci throughout the genome (25,26,29,36). Genome-wide surveys suggest that ∼30–60% of CTCF-binding sites are cell-type-specific and may play critical role in cell-linage programming via their impact on these chromatin networks (27,36,37). Our current analysis reveals that CTCF is recruited to the placenta-specific HSIV in the native setting of primary human STB nuclei (Figure 4A)(8). These data are consistent with our prior demonstration of placenta-specific recruitment of CTCF to HSIV in the hGH/BAC transgene locus (21). This specific recruitment of CTCF to the placenta-specific HSIV may play a central role in the apparent boundary function mediated by the looping of the placental LCR (HSIII–V) to the hGH cluster in the placenta and the formation of a fully effective and controlled organization of hCS genes (21).
The detailed analysis of chromatin contacts internal to the activated hGH cluster in the placenta, both in human STB cells and in the transgenic mouse placenta, demonstrates high frequency of interactions that bridge the 5′ and 3′ flanking regions of each of the three individual hCS units via P-element looping to the 3′-P2. High-frequency interactions were also identified among the three hCS genes via interactions of a P-element with the 3′-P2 segment of neighboring hCS genes. Of note, these interactions among the hCS genes which form the ‘hCS chromatin hub’ exclude the poorly expressed PGR unit, hGH-V, as well as the pituitary-restricted hGH-N gene (Figures 3D and E). These two hGH genes are selectively sequestered by specific interactions with the looped placental LCR.
The 3′-P2 regions are 8 kb downstream of each of the three hCS genes and P-elements are 2 kb upstream of the three hCS genes as well as hGH-V. We investigated the CTCF enrichment at these regions and observed CTCF occupancy at the 3′-P2 regions and at the P-elements in both placental and non-placental (skin fibroblasts) tissues (Supplementary Figure S5). These data are consistent with the more broad based surveys of CTCF occupancy in the ENCODE database (see Supplementary Figure S5). Thus, while CTCF at P-element and 3′-P2 may be involved in the organization of hCS chromatin hub, the tissue-specificity of this organization clearly relies on one or more placenta-specific determinants as well. The dependence upon HSIII–V region of the LCR for the placenta-specific packing of the cluster and placental specificity of CTCF occupancy at HSIV supports a model in which HSIV CTCF occupancy plays a critical role in this organization. Further defining the basis for the unique chromatin packaging within the STB cells and the relationship(s) of this packing to the evolutionary origins human hGH multigene cluster will be the subject of our future studies.
The current data describe the assembly of a higher-order chromatin network in which the expressed hCS genes are assembled into a tightly packed chromatin hub while the long-range LCR interactions segregate the hGH-N and hGH-V genes away from these intra-cluster hCS gene interactions (Figure 3E). Importantly, the interdependence of these two sets of interactions is confirmed by the dramatic shift in the cluster configuration in the transgene locus lacking the LCR (summarized in Figure 7B).
The P-element is conserved in all PGR units and has been linked in a number of studies to placenta-specific expression (16,19,38). Remarkably, the mechanisms that underlie its function(s) have remained unclear. The results of the present study suggest the P-elements may be exerting their activity at the level of chromatin organization by generating contacts among the hCS genes that pack them into a functional unit. Although the hGH-V gene is also flanked by a P-element, its interaction with the LCR appears to be sufficient to sequester it from the tight-packing hCS gene interactions. This sequestration may contribute to the comparatively weak expression of the hGH-V gene in the placenta (<1% of hCS (20)). The deletion of the LCR HSIII–V region appears to release hGH-V from its native LCR contacts, allowing it to ‘invade’ and disrupt the hCS chromatin hub. This hGH-V shift displaces hCS-B from the hub and generates a new architecture that is now heavily reliant on robust interactions with the hGH-V P-element (Figure 7B). This shift in local structure is accompanied by a significant relaxation in the stringent control of hCS transgene expression in the placenta (21).
The specificity of the 3D chromatin organization within placental hGH locus was confirmed by 3C studies of the intact hGH/BAC transgene locus in four non-placental tissues (Supplementary Figure S3). These studies revealed a variety of promoter-linked interactions within the transgenic locus that were all distinct from those in the placenta. These divergent chromatin configurations may reflect the influences of the elements that drive tissue-specific expression in adjacent genes (e.g. skeletal muscle SCN4A and B-cell CD79b upstream, or testis TCAM1 downstream). Alternatively, they may reflect the formation of a variety of actively repressing or nonfunctional default chromatin structures.
The structural mapping in this report, when combined with our prior functional mapping studies (21) strongly support a model in which long-range and local chromatin conformations interact in establishing tight control of hCS gene expression in the placenta. However, it is apparent that the basis for repression of hCS expression in non-placental tissues remains undefined. We have previously demonstrated that deletion of the HSIII–V region sensitizes hCS expression to site-of-integration effects in the placenta and triggers its ectopic expression in a number of non-placental tissues. The same relaxation of controls over hCS expression was observed when we converted the multigene cluster to a single hCS-A PGR unit. These observations suggest that the LCR determinants and the structure of the cluster play a role in the effective insulation of the hCS genes ectopic expression. Thus, HSV and HSIII, which are formed in a constitutive manner, may serve independent and critical roles in these additional constraints on hCS expression.
Supplementary Material
Acknowledgments
The authors thank the University of Pennsylvania Transgenic and Chimeric Mouse Facility (supported by National Institutes of Health [P30's DK019525, DK050306, CA016520]) for generation of the transgenic mice for our studies.
Footnotes
Present address: Yu-Cheng Tsai, Research Center for Tumor Medical Science, China Medical University, Taichung, Taiwan.
FUNDING
National Institutes of Health (NIH) [R01 HD/DK25147 and R01 HD/DK046737 to S.A.L and N.E.C.]. Funding for open access charge: NIH [R01 HD/DK25147 and R01 HD/DK046737 to S.A.L and N.E.C.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Barsh G.S., Seeburg P.H., Gelinas R.E. The human growth hormone gene family: structure and evolution of the chromosomal locus. Nucleic Acids Res. 1983;11:3939–3958. doi: 10.1093/nar/11.12.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McWilliams D., Boime I. Cytological localization of placental lactogen messenger ribonucleic acid in syncytiotrophoblast layers of human placenta. Endocrinology. 1980;107:761–765. doi: 10.1210/endo-107-3-761. [DOI] [PubMed] [Google Scholar]
- 3.Liebhaber S.A., Urbanek M., Ray J., Tuan R.S., Cooke N.E. Characterization and histological localization of human growth hormone-variant gene expression in the placenta. J. Clin. Invest. 1989;83:1985–1991. doi: 10.1172/JCI114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacLeod J.N., Lee A.K., Liebhaber S.A., Cooke N.E. Developmental control and alternative splicing of the placentally expressed transcripts from the human growth hormone gene cluster. J. Biol. Chem. 1992;267:14219–14316. [PubMed] [Google Scholar]
- 5.Yoo E.J., Cajiao I., Kim J.S., Kimura A.P., Zhang A., Cooke N.E., Liebhaber S. A. Tissue-specific chromatin modifications at a multigene locus generate asymmetric transcriptional interactions. Mol. Cell. Biol. 2006;26:5569–5579. doi: 10.1128/MCB.00405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennani-Baiti I.M., Jones B.K., Liebhaber S.A., Cooke N.E. Physical linkage of the human growth hormone gene cluster and the skeletal muscle sodium channel alpha-subunit gene (SCN4A) on chromosome 17. Genomics. 1995;29:647–652. doi: 10.1006/geno.1995.9954. [DOI] [PubMed] [Google Scholar]
- 7.Kurihara M., Shiraishi A., Satake H., Kimura A.P. A conserved noncoding sequence can function as a spermatocyte-specific enhancer and a bidirectional promoter for a ubiquitously expressed gene and a testis-specific long noncoding RNA. J. Mol. Biol. 2014;426:3069–3093. doi: 10.1016/j.jmb.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 8.Jones B.M., Monks B.R., Liebhaber S.A., Cooke N.E. The human growth hormone gene is regulated by a multicomponent locus control region. Mol. Cell. Biol. 1995;15:7010–7021. doi: 10.1128/mcb.15.12.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elefant F., Cooke N.E., Liebhaber S.A. Targeted recruitment of histone acetyltransferase activity to a locus control region. J. Biol. Chem. 2000;275:13827–13834. doi: 10.1074/jbc.275.18.13827. [DOI] [PubMed] [Google Scholar]
- 10.Ho Y., Elefant F., Cooke N.E., Liebhaber S.A. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell. 2002;9:291–302. doi: 10.1016/s1097-2765(02)00447-1. [DOI] [PubMed] [Google Scholar]
- 11.Fleetwood M.R., Ho Y., Cooke N.E., Liebhaber S.A. DNase I hypersensitive site II of the human growth hormone locus control region mediates an essential and distinct long-range enhancer function. J. Biol. Chem. 2012;287:25454–25465. doi: 10.1074/jbc.M112.365825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shewchuk B.M., Asa S.L., Cooke N.E., Liebhaber S.A. Pit-1 binding sites at the somatotrope-specific DNase I hypersensitive sites I, II of the human growth hormone locus control region are essential for in vivo hGH-N gene activation. J. Biol. Chem. 1999;274:35725–35733. doi: 10.1074/jbc.274.50.35725. [DOI] [PubMed] [Google Scholar]
- 13.Ho Y., Elefant F., Liebhaber S. A., Cooke N. E. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell. 2006;23:365–375. doi: 10.1016/j.molcel.2006.05.041. [DOI] [PubMed] [Google Scholar]
- 14.Ho Y., Tadevosyan A., Liebhaber S.A., Cooke N.E. The juxtaposition of a promoter with a locus control region transcriptional domain activates gene expression. EMBO Rep. 2008;9:891–898. doi: 10.1038/embor.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho Y., Shewchuk B.M., Liebhaber S.A., Cooke N.E. Distinct chromatin configurations regulate the initiation and the maintenance of hGH gene expression. Mol. Cell Biol. 2013;33:1723–1734. doi: 10.1128/MCB.01166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nachtigal M.W., Nickel B.E., Cattini P.A. Pituitary-specific repression of placental members of the human growth hormone gene family. J. Biol. Chem. 1993;286:8473–8479. [PubMed] [Google Scholar]
- 17.Jiang S.W., Shepard A.R., Eberhardt N.L. An initiator element is required for maximal human chorionic somatomammotropin gene promoter and enhancer function. J. Biol. Chem. 1995;270:3683–3692. doi: 10.1074/jbc.270.8.3683. [DOI] [PubMed] [Google Scholar]
- 18.Jacquemin P., Martial J.A., Davidson I. Human TEF-5 is preferentially expressed in placenta and binds to multiple functional elements of the human chorionic somatomammotropin-B gene enhancer. J. Biol. Chem. 1997;272:12928–12937. doi: 10.1074/jbc.272.20.12928. [DOI] [PubMed] [Google Scholar]
- 19.Elefant F., Su Y., Liebhaber S.A., Cooke N.E. Patterns of histone acetylation suggest dual pathways for gene activation by a bifunctional locus control region. EMBO. 2000;19:6814–6822. doi: 10.1093/emboj/19.24.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura A.P., Sizova D., Handwerger S., Cooke N.E., Liebhaber S.A. Epigenetic activation of the human growth hormone gene cluster during placental cytotrophoblast differentiation. Mol. Cell Biol. 2007;27:6555–6568. doi: 10.1128/MCB.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai Y-C., Cooke N.E., Liebhaber S.A. Tissue specific CTCF occupancy and boundary function at the human growth hormone locus. Nucleic Acids Res. 2014;42:4906–4921. doi: 10.1093/nar/gku139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 23.Kimura A., Liebhaber S.A., Cooke N.E. Epigenetic modifications at the human growth hormone locus predict distinct roles for histone acetylation and methylation in placental gene activation. Mol. Endocrinol. 2004;18:1018–1032. doi: 10.1210/me.2003-0468. [DOI] [PubMed] [Google Scholar]
- 24.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merkenschlager M., Odom D.T. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 26.Ong C.T., Corces V.G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 28.Harmston N., Lenhard B. Chromatin and epigenetic features of long-range gene regulation. Nucleic Acids Res. 2013;41:7185–7199. doi: 10.1093/nar/gkt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gómez-Díaz E., Corces V.G. Architectural proteins: regulators of 3D genome organization in cell fate. Trends Cell Biol. 2014;24:703–711. doi: 10.1016/j.tcb.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J., Sankaran V.G., Ni M., Menne T.F., Puram R.V., Kim W., Orkin S.H. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010;24:783–798. doi: 10.1101/gad.1897310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marsman J., Horsfield J.A. Long distance relationships: enhancer-promoter communication and dynamic gene transcription. Biochim. Biophys. Acta. 2012;1819:1217–1227. doi: 10.1016/j.bbagrm.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Kim A., Dean A. Chromatin loop formation in the β-globin locus and its role in globin gene transcription. Mol. Cells. 2012;34:1–5. doi: 10.1007/s10059-012-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng W., Rupon J.W., Krivega I., Breda L., Motta I., Jahn K.S., Reik A., Gregory P.D., Rivella S., Dean A., et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace J.A., Felsenfeld G. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Junier I., Dale R.K., Hou C., Képès F., Dean A. CTCF-mediated transcriptional regulation through cell type-specific chromosome organization in the β-globin locus. Nucleic Acids Res. 2012;40:7718–7727. doi: 10.1093/nar/gks536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuddapah S., Jothi R., Schones D.E., Roh T.Y., Cui K., Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen E.Y., Liao Y.C., Smith D.H., Barrera-Saldaña H.A., Gelinas R.E., Seeburg P.H. The human growth hormone locus: Nucleotide sequence, biology, and evolution. Genomics. 1989;4:479–497. doi: 10.1016/0888-7543(89)90271-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.