Abstract
The parasegment-specific expression of the three Drosophila Bithorax complex homeotic genes is orchestrated by nine functionally autonomous regulatory domains. Functional autonomy depends upon special elements called boundaries or insulators that are located between each domain. The boundaries ensure the independent activity of each domain by blocking adventitious interactions with initiators, enhancers and silencers in the neighboring domains. However, this blocking activity poses a regulatory paradox--the Bithorax boundaries are also able to insulate promoters from regulatory interactions with enhancers and silencers and six of the nine Bithorax regulatory domains are separated from their target genes by at least one boundary element. Here we consider several mechanisms that have been suggested for how the Bithorax regulatory domains are able to bypass intervening boundary elements and direct the appropriate parasegment-specific temporal and spatial expression of their target gene.
Keywords: Bithorax complex, insulator, boundary elements, boundary bypass, enhancer blocking
The Drosophila Bithorax complex (BX-C) has been intensively studied as a paradigm of gene cluster architecture. It is responsible for the specification of parasegment (PS) identity in the posterior 2/3rds of the fly, from parasegment 5 (PS5) to parasegment 14 (PS14) (Duncan, 1987; Maeda and Karch, 2009, 2006; Peifer et al., 1987). Though the BX-C must orchestrate the differentiation of ten morphologically distinct parasegments, it contains only three homeotic genes Ultrabithorax (Ubx), abdominal-A (abd-A) and Abdominal-B (Abd-B) (Bender et al., 1983; Boulet et al., 1991; Celniker et al., 1990; Karch et al., 1985; Sánchez-Herrero, 1991; Sánchez-Herrero et al., 1985; Simon et al., 1990). The specification of distinct parasegment identities by these three genes depends upon a series of regulatory domains, each directing a parasegment-specific spatial/temporal pattern of expression of one of the three homeotic genes (Karch et al., 1990, 1985; Lewis, 1978; Maeda and Karch, 2009, 2006; Peifer et al., 1987; White and Wilcox, 1985). As illustrated in Figure 1, these parasegment-specific regulatory domains are arranged along the chromosome in the same order as the body segments that they specify. The set of regulatory domains controlling the expression of a single homeotic gene comprise a “Transcription Associated Regulatory Domain” (TARD) (Chetverina et al., 2014). Expression of the Ubx gene is controlled by a TARD composed of two regulatory domains; abx/bx, which is located within the Ubx transcription unit, and bxd/pbx, which is located upstream of the Ubx promoter (Beachy et al., 1985; Little et al., 1990; White and Wilcox, 1985). The abx/bx domain directs Ubx expression in a pattern specifying PS5 identity, while bxd/pbx generates a pattern of expression specifying PS6 identity (Bender et al., 1983; Duncan, 1987; Peifer et al., 1987).
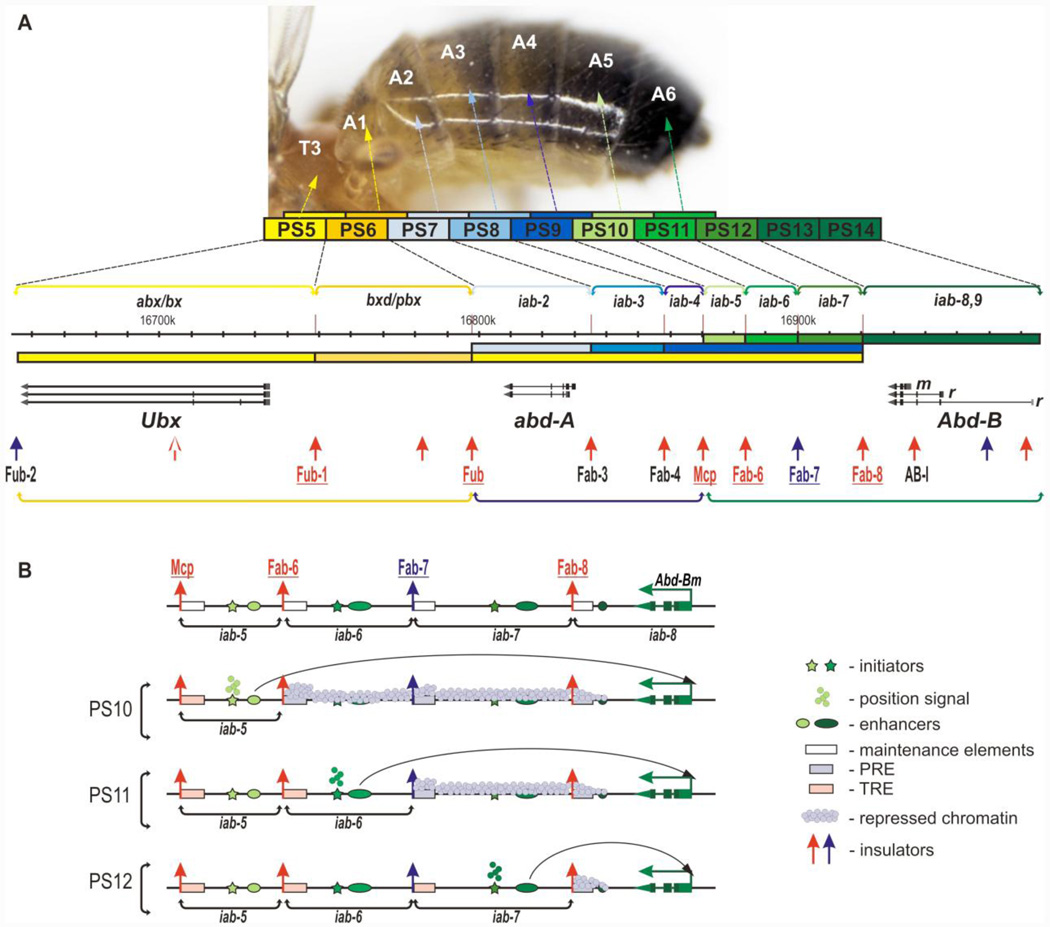
Figure 1. The Bithorax Complex.
(A) Map and coordinates of the BX-C are taken from the FlyBase genome browser D. melanogaster (R6.04). Multicolored boxes above illustrate the embryo parasegments (PS) corresponding to the segments of the adult fly. The same color brackets indicate localization of regulatory domains that control expression of the BX-C genes Ubx, abd-A, and Abd-B (shown as black horizontal arrows). Abd-B has different promoters designated as m and r. The expression patterns of each BX-C gene are shown and the colored bar indicates the regulatory domains according to Maeda and Karch (2006) (darker shades of color indicate higher expression levels). The Ubx, abd-A, and Abd-B regulatory domains are organized into the three transcriptionally associated regulatory domains (TARDs), and the three TARDs are designated by yellow, blue, and green brackets, respectively. Insulators identified in BX-C in ChIP experiments are shown as upward arrows: red – for those that are bound by dCTCF and blue – for the one CTCF-independent insulator, Fab-7. Insulators whose identity has been confirmed by mutations or functional assays are underlined and labeled by color font. Insulators identified only on the basis of ChIP experiments are labeled with black font. Abd-BI (AB-I) is an insulator-like element located upstream of the Abd-Bm promoter. In addition to being associated with known insulator proteins, it functions in bypass assays with other boundaries from the Abd-B TARD (Kyrchanova et al., 2011). The Fub-2 insulator is located downstream of the Ubx transcription unit and is identified on the basis of its association with known insulator proteins. (B) A model for the sequential activation of BX-C regulatory domains going from more anterior to more posterior parasegments (Peifer et al., 1987; Mihaly et al., 2006; Bowman et al., 2014). The model shows the sequential activation of the regulatory domains in the Abd-B TARD. Stars symbolize initiators, the ovals – tissue specific enhancers (for simplicity only one enhancer is shown; however, there may be several stage/tissue specific enhancers in each domain). Activation of successive initiators by position dependent gap and pair rule genes is indicated by the green (light, intermediate and dark) parasegment “position signals.” Each domain has a maintenance element (rectangles on the left side of the regulatory domain). These maintenance elements can be set in the repressive state by the parasegment-specific initiation. In this case the Polycomb complexes assemble on the maintenance element (grey rectangles) and silence the domain (indicated by light grey “repressive chromatin” across the domain). Alternatively, the maintenance elements can be set in the active state (pink rectangles). In this case, TrxG proteins bind to the maintenance element (pink rectangles) and keep the domain in an active state. In the active state the various enhancers in the domain can regulate the target Abd-B promoter. Domains are insulated from each other by boundary elements (upward arrows).
The remaining regulatory domains are responsible for the development of the abdominal segments and are called infra-abdominal (iab) (Celniker et al., 1990; Duncan, 1987; Karch et al., 1990, 1985; Lewis, 1978; Mihaly et al., 2006; Peifer et al., 1987; Sánchez-Herrero, 1991). Three of these, iab-2, iab-3, and iab-4 control the expression of abd-A and they function to specify PS7, PS8, and PS9 respectively (Figure 1A). The Abd-B gene has several alternative promoters, but the morphological features of parasegments PS10-PS13 depend on transcripts produced by the most proximal promoter, Abd-Bm (Figure 1A). The activity of this promoter is regulated by a TARD consisting of four regulatory domains, iab-5, iab-6, iab-7, and iab-8 (Maeda and Karch, 2009, 2006). These domains direct Abd-B expression in patterns appropriate for PS10, PS11, PS12, and PS13 identity, respectively. Abd-B expression from the other more distal promoters is thought to be responsible for PS14 identity (Figure 1A).
Regulatory elements in the Bithorax Complex
All the regulatory domains in the BX-C appear to share a similar set of individual regulatory elements (Maeda and Karch, 2011, 2009, 2006). Each domain has an “initiator” element (* in Figure 1B) that is responsible for establishing its activity state, either on or off. The choice of activity state is made at the end of the blastoderm stage and depends upon the action of transcription factors encoded by maternal effect genes and the zygotically active gap and pair-rule genes (Busturia and Bienz, 1993; Ingham and Martinez-Arias, 1986; Irish et al., 1989; Müller and Bienz, 1992; Qian et al., 1991; Shimell et al., 2000; Simon et al., 1990; White and Lehmann, 1986). These transcription factors are differentially expressed along the anterior-posterior (AP) axis and this provides a combinatorial code that sequentially activates the BX-C regulatory domains and defines the parasegmental identity of cells in the posterior 2/3rds of the embryo (Howard, 1990; Pankratz and Jäckle, 1990; Small and Levine, 1991). For example, the combination of maternal and zygotic transcription factors in cells in the PS10 region of the embryo activates the iab-5 initiator setting the iab-5 domain in the on state (Figure 1B) (Iampietro et al., 2010; Mihaly et al., 2006). This combination does not, however, activate the initiator elements in the three other Abd-Bm regulatory domains, iab-6, iab-7, and iab-8, and these domains are set in the off state in PS10 (Figure 1B). The remaining Abd-Bm regulatory domains are activated in a step-wise fashion in the more posterior parasegments, PS11, PS12, and PS13 (Figure 1B). In the PS11 region of the embryo, the iab-6 regulatory domain is set in the on state by the iab-6 initiator, while the iab-7 and iab-8 regulatory domains are in the off state. In PS12, the iab-7 but not the iab-8 regulatory domain is activated (see Figure 1B), while finally, in PS13, all four of the Abd-Bm regulatory domains are activated.
In addition to the parasegment-specific initiator, each regulatory domain has a set of maintenance elements (open rectangle in Figure 1B) (Maeda and Karch, 2006; Schwartz and Pirrotta, 2013; Steffen and Ringrose, 2014). The positional information provided by maternal and zygotic transcription factors is present only transiently in the early embryo and when these factors disappear shortly after gastrulation, the activity state of the regulatory domains, and thus parasegment identity, is remembered by the maintenance elements (Struhl and Akam, 1985). The off state is maintained by Polycomb Response Elements (PREs) that function to recruit Polycomb group proteins (Bantignies et al., 2003; Busturia et al., 1997; Chan et al., 1994; Hagstrom et al., 1997; Mihaly et al., 1997; Müller and Bienz, 1991; Simon et al., 1993). These proteins provide an inheritable memory mechanism that keeps the domain in the off state as development proceeds. Trithorax group proteins are responsible for keeping an activated domain in the on state (Kassis and Brown, 2013; Schwartz and Pirrotta, 2013; Steffen and Ringrose, 2014). Sequences capable of recruiting Trithorax proteins (Trithorax Response Elements, TRE) and maintaining the active state are much less well defined; however, current studies suggest that they are embedded in, but are distinct from, the BX-C PREs (Steffen and Ringrose, 2014).
While the initiators and the maintenance elements are responsible for selecting and remembering, respectively, the activity state of the regulatory domain, the differentiation of parasegment-specific morphological traits depends upon a collection of spatial, tissue and stage-specific enhancers (Barges et al., 2000; Busturia and Bienz, 1993; Mihaly et al., 2006; Pirrotta et al., 1995; Simon et al., 1990). The enhancers in each regulatory domain direct the expression of one of the three BX-C homeotic genes in a spatio-temporal pattern that orchestrates the proper development of the parasegment. That parasegment differentiation depends upon these spatial, tissue and stage-specific enhancers and not the parasegment-specific initiation element is illustrated by an experiment in which the iab-6 (PS11) initiator was replaced by the initiator from iab-5 (PS10) (Iampietro et al., 2010). In these flies, the combination of maternal and zygotic transcription factors in PS10 turns on the iab-5 initiator not only in the iab-5 domain but also in iab-6. The ectopically activated iab-6 regulatory domain enables the enhancers in the iab-6 domain to direct Abd-B expression in PS10 in a pattern appropriate for cells in PS11, not PS10, and as a result PS10 is transformed into a duplicate copy of PS11.
Boundary elements mark the borders of the regulatory domains and ensure their functional independence
In order to properly specify parasegment identity, the initiators and maintenance elements in each of the BX-C regulatory domains must be able to function autonomously, without interference from initiators and maintenance elements in the neighboring regulatory domains (Celniker et al., 1990; Galloni et al., 1993; Gyurkovics et al., 1990; Karch et al., 1994; Lewis, 1978; McCall et al., 1994). Independence is conferred by special elements called chromatin domain boundaries or insulators that are located at the borders of the BX-C regulatory domains (Chetverina et al., 2014; Maeda and Karch, 2009, 2006). The known or suspected boundaries in BX-C include Fub-2, Fub-1, Fub, Fab-3, Fab-4, Mcp, Fab-6, Fab-7, Fab-8 and Abd-BI (see Figure 1A). Five of these boundaries, Fub, Mcp, Fab-6, Fab-7 and Fab-8, are defined by mutations that disrupt their function (Barges et al., 2000; Bender and Lucas, 2013; Celniker et al., 1990; Gyurkovics et al., 1990; Iampietro et al., 2010; Karch et al., 1994; Mihaly et al., 1997). With the exception of Fub, their ability to function as boundary elements or insulators in a heterologous context has been confirmed by a variety of transgene assays (Barges et al., 2000; Ciavatta et al., 2007; Gruzdeva et al., 2005; Hagstrom et al., 1996; Maksimenko et al., 2015, 2014; Pérez-Lluch et al., 2008; Schweinsberg et al., 2004; Schweinsberg and Schedl, 2004; Zhou et al., 1999, 1996). The remaining boundaries have been identified based on their association with known insulator proteins in ChIP experiments and in some cases by their activity in transgene assays (Holohan et al., 2007; Kyrchanova et al., 2011; Smith et al., 2009).
Mcp1 and Fab-71 were the first BX-C boundary mutations isolated (Celniker et al., 1990; Gyurkovics et al., 1990; Karch et al., 1994; Maeda and Karch, 2007; Mihaly et al., 1997; Sipos and Gyurkovics, 2005). Mcp marks the TARD border between the sets of regulatory domains that control the abd-A and Abd-B homeotic genes. The iab-4 regulatory domain is on the proximal side of the Mcp boundary, and it directs abd-A expression in PS9. iab-5 is on the distal side of the boundary and it regulates Abd-B in PS10. The Fab-7 boundary is located within the Abd-B TARD and it separates iab-6, which specifies PS11, and iab-7, which specifies PS12. The Mcp1 and Fab-71 deletions have a gain-of-function phenotype in which the parasegment specified by the regulatory domain proximal to the boundary, PS9 for Mcp1 and PS11 for Fab-71, is transformed into a duplicate copy of the more posterior parasegment, PS10 for Mcp1 and PS12 for Fab-71. This gain-of-function phenotype is due to the misregulation of Abd-B in the affected parasegment. For example, in Fab-71, Abd-B expression in PS11 is controlled by the iab-7 regulatory domain rather than iab-6.
Subsequent studies on Fab-7 revealed that the original Fab-71 allele removes not only the Fab-7 boundary but also the nearby iab-7 PRE (Hagstrom et al., 1997; Mihaly et al., 1997; Mishra et al., 2001). Deletion of just the Fab-7 boundary results in a mixed gain- and loss-of-function phenotypic transformation of PS11 (Mihaly et al., 1997). The mixed phenotype is thought to be due to a competition between the iab-6 and the iab-7 initiators in the fused regulatory domain (Mihaly et al., 1997). The gain-of-function phenotype is due to the activation of the iab-7 regulatory domain in PS11 by the iab-6 initiator. The loss-of-function phenotype (in which PS11 cells assume a PS10 identity) arises because the iab-7 initiator inactivates the iab-6 regulatory domain and as a result Abd-B expression is regulated by iab-5. It is thought that the exclusively gain-of-function phenotype evident in the original Fab-71 mutation arises because the deletion also removes the iab-7 PRE, making it impossible to maintain the fused iab-6:iab-7 domain in the inactive state. As the iab-5 PRE is also deleted in the Mcp mutant, a similar mechanism may account for its gain-of-function phenotype. Mutations in the other BX-C boundaries (Fub, Fab-6 and Fab-8) have similar phenotypic effects (either gain-of-function, or mixed gain- and loss-of-function) on the affected parasegments (invariably the parasegment specified by the regulatory domain proximal to the mutant boundary).
In addition to blocking crosstalk between the adjacent regulatory domains, the BX-C boundaries have insulator functions that closely resemble boundary elements found elsewhere in the fly genome (Chetverina et al., 2014; Maeda and Karch, 2007). In transgene assays, the BX-C insulators can block enhancers from activating transcription, and prevent PREs from silencing transcription (Barges et al., 2000; Ciavatta et al., 2007; Gruzdeva et al., 2005; Hagstrom et al., 1996; Maksimenko et al., 2015, 2014; Pérez-Lluch et al., 2008; Schweinsberg et al., 2004; Schweinsberg and Schedl, 2004; Smith et al., 2009; Zhou et al., 1996). Like other insulators, the BX-C boundaries must be interposed between the enhancer/PRE silencer and the promoter of the reporter gene in order to block regulatory interactions. Moreover, the insulating activity detected in transgene assays is “constitutive”—it is observed throughout development with no apparent tissue or cell type specificity. Though not their normal function in their endogenous context, the BX-C boundaries can also insulate reporters carried by transgenes that have been inserted into a BX-C regulatory domain from the action of enhancers and silencers in the adjacent regulatory domains (Bender and Hudson, 2000; Galloni et al., 1993; McCall et al., 1994; Mihaly et al., 1997). This protection is lost when the intervening boundary is removed.
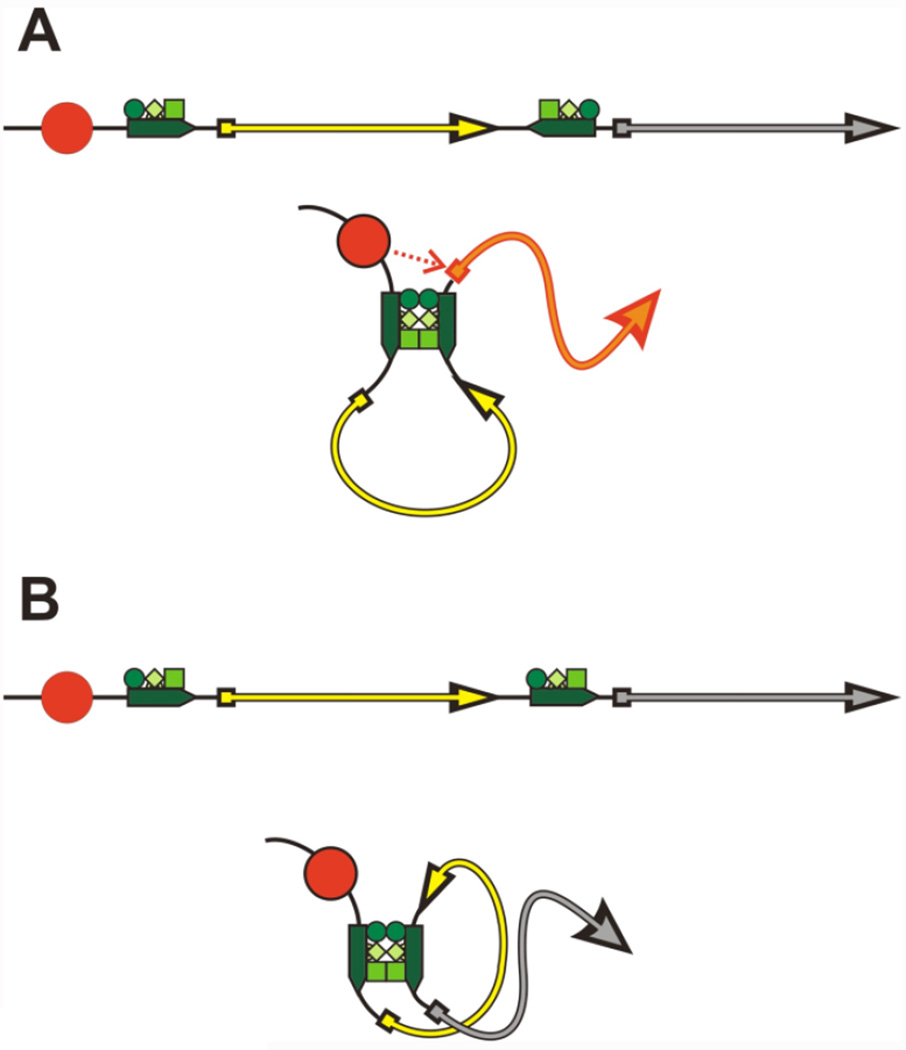
The BX-C boundaries also resemble other insulator elements in their architectural activities— insulator bypass and long-range interactions (Chetverina et al., 2014; Maeda and Karch, 2007). Insulator bypass takes place when two copies of an insulator are placed between the regulatory element (enhancer/silencer) and a reporter gene (Cai and Shen, 2001; Muravyova et al. 2001; Kyrchanova and Georgiev, 2014). In the example shown in Figure 2A, when two copies of the Fab-8 boundary are placed on either side of a yellow reporter, they block an upstream enhancer from activating a yellow expression (Kyrchanova et al., 2007). However, the upstream enhancers are able to bypass the two Fab-8 boundaries and activate a white reporter located downstream of the distal Fab-8 boundary. It is thought that the two insulators pair with each other bringing sequences on the far sides of the two insulators in close proximity. Insulator bypass typically depends upon the relative orientation of the insulators with respect to each other and requires either two copies of the same insulator or the appropriately matched insulators that can pair with each other (Kyrchanova et al., 2011, 2008a, 2007). If the orientation of one of the insulators is reversed, the insulators can still pair, but because of the topology of the loop that is formed, strong activation of the distant gene is not observed (Figure 2B). One of the known exceptions to this orientation dependence “rule” is Fab-7. Two Fab-7 insulators can mediate enhancer activation of the white reporter in the bypass assay independently of their relative orientation (Rodin et al., 2007). The second architectural function is in long distance interactions. Both Mcp and Fab-7 have been shown to mediate long distance regulatory interactions between transgenes inserted at great distances from each other (Bantignies et al., 2003; Li et al., 2011; Muller et al., 1999; Vazquez et al., 2006). These long distance regulatory interactions are dependent on the formation of stable and direct insulator:insulator physical connections. Long distance interactions have also been found for several non-BX-C boundaries, including the homie insulator from the even-skipped locus and the gypsy transposon su(Hw) insulator (Fujioka et al., 2013, 2009).
Figure 2. Insulator-insulator interactions and insulator bypass.
This diagram illustrates a transgene assay for insulator bypass. The enhancer is shown as a red circle. The two reporter genes in the transgene are indicated by yellow and grey lines with arrowheads. The two insulators in the transgene are indicated by thick green arrows, while the factors that associate with the insulators and mediate pairing interactions are indicated by colored circles and boxes. Note that the orientation of the insulators in A and B is different. The insulators in A are arranged in opposite orientations, while the insulators in B are in the same orientation. Interactions between insulators in the A and B transgenes generate loops with different topological configurations. (A) Head-to tail pairing of insulators in the opposite orientation configures the intervening loop in a manner that brings the enhancer (red circle) into a close proximity to the promoter driving expression of the grey reporter. Activation of the grey reporter is illustrated by the change in color to red. (B) In this transgene, head-to-tail pairing of insulators in the same orientation generates a loop in which the enhancer is far from the promoter of the grey reporter. In this case, the enhancer doesn’t activate the reporter expression. Note that in both the A and B configurations, the yellow reporter is not activated by the upstream enhancer.
The boundary paradox
The positioning of the BX-C homeotic genes and their respective regulatory domains relative to the boundary elements that insulate each domain poses a regulatory paradox (Maeda and Karch, 2007). As illustrated for the Abd-B TARD in Figure 1B, three of the four Abd-B regulatory domains are located downstream of the Abd-Bm transcription unit. For each of these regulatory domains there is at least one boundary element located between it and the Abd-Bm promoter. For example, in PS10, the iab-5 regulatory domain drives Abd-Bm expression; however, it is separated from its target promoter by three boundaries, Fab-6, Fab-7, and Fab-8 which might be expected to block regulatory interactions. A similar problem of intervening boundary elements exists for iab-6 and iab-7, as well as elsewhere in the BX-C. For example, iab-3 and iab-4, control abd-A expression in PS8 and PS9 (Figure 1A). These domains are located upstream of the abd-A promoter and are separated from the abd-A promoter by either one boundary element (Fab-3 for iab-3) or two boundary elements (Fab-3 and Fab-4 for iab-4). A similar configuration of regulatory domains is present in the Ubx TARD. While the Ubx transcription unit is located within the abx/bx domain, the bxd/pbx regulatory domain is located upstream of the transcription unit and is separated from the Ubx promoter by the Fub-1 boundary element (see Figure 1A).
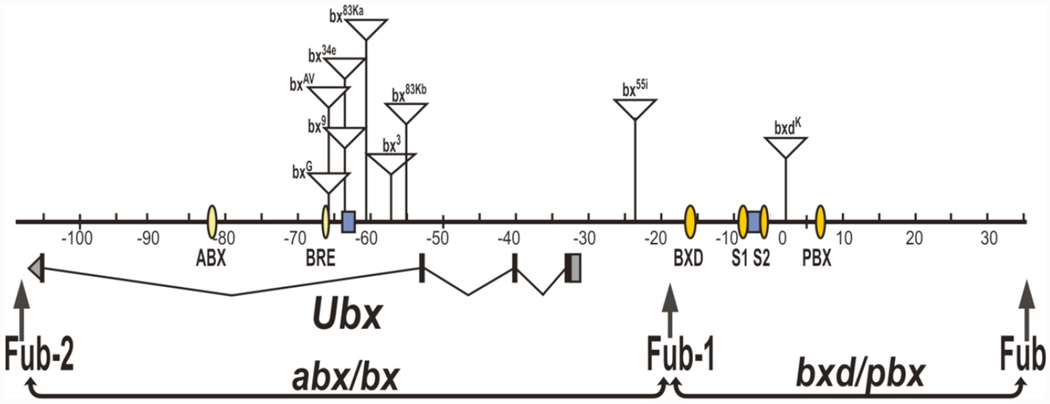
There are good reasons to believe that the presence of intervening boundary elements should disrupt interactions between the regulatory elements in the regulatory domains and their target promoters. To begin with, much of the early genetic analysis of the BX-C, and in particular the Ubx gene, relied on spontaneous mutations. A substantial fraction of these spontaneous mutations were gypsy transposon insertions into the Ubx region of the complex (Bender et al., 1983; Duncan, 1987; Peifer and Bender, 1986). bx mutations disrupt Ubx regulation in PS5, transforming the cells in this parasegment towards a PS4 identity. Seven of the ten spontaneous bx mutations have a gypsy transposon inserted within the Ubx transcription unit between the Ubx promoter and the abx enhancer (and bx PRE) (see Figure 3). In these mutants, the su(Hw) insulator carried by the gypsy transposon blocks the abx enhancer from activating a Ubx expression in PS5. The disruption of Ubx expression by the bx gypsy insertions can be suppressed by mutations in the su(Hw) gene or by excising the su(Hw) insulator. Similarly, gypsy transposons inserted into the bxd/pbx region block the enhancers in this upstream regulatory domain from activating the Ubx expression in PS6, transforming the parasegment towards a PS5 identity (Figure 3).
Figure 3. gypsy transposon insertions in the Ubx region of BX-C.
This figure shows a map of the Ubx region of BX-C and includes the Ubx transcription unit and the two Ubx regulatory domains, abx/bx and bxd/pbx. The numbered line represents the BX-C DNA walk in kilobases from its starting point as in Bender et al., 1983a. Triangles indicate the sites of insertion of mutations that are associated with gypsy transposon insertions. Vertical small yellow ovals designate enhancers, blue rectangles – PRE elements.
The effects of “ectopic” insulator elements, carried by transposons, on the functioning of the Ubx TARD have been duplicated in the Abd-B TARD in boundary replacement experiments. In these experiments, the Fab-7 boundary was replaced either by the gypsy transposon su(Hw) insulator, or the scs insulator from the 87A7 heat shock locus (Hogga et al., 2001; Hogga and Karch, 2002). Both of these boundaries were able to recapitulate the insulating activity of Fab-7 and block crosstalk between the iab-6 and iab-7 regulatory domains. However, they also disrupted Abd-B regulation in a complicated way. In both replacements, segment A6 (PS11) in the adult cuticle is transformed into a duplicate copy of A5 (PS10). This would imply that iab-5 is regulating Abd-B expression in both PS10 and PS11. Surprisingly, however, Abd-B protein doesn’t seem to be expressed in the embryonic epidermis in either PS11 or PS10. This would imply that the su(Hw) and scs boundaries not only prevent iab-6 from regulating Abd-B in PS11, but also block the more proximal domain, iab-5, from regulating Abd-B. The fact that both PS10 and PS11 develop with a PS10 (A5)-like identity even though Abd-B isn’t expressed in these parasegments is thought to be due to an unusual property of iab-5. In chromosomal rearrangements that separate iab-5 and iab-6 from Abd-B, iab-5 is able to induce a pattern of abd-A expression appropriate for specifying PS10 identity (Hogga et al., 2001). This is not the only unexpected observation. While the su(Hw) element has insulator activity in the epidermis, this activity is lost in the CNS. Abd-B expression in PS10 in the CNS resembles wild type, while in PS11 the pattern resembles that in PS12. In the former case, su(Hw) is not able to block iab-5 from regulating Abd-B, while in the latter case, it is unable to prevent crosstalk between iab-6 and iab-7.
While heterologous insulators disrupt critical regulatory interactions when inserted into BX-C regulatory domains, the endogenous boundaries do not. Instead, they are permissive for contacts between active regulatory domains and their regulatory targets. There are reasons to believe that this phenomenon is not unique to the regulatory domains of the Drosophila BX-C. Hi-C experiments have defined the topologically associated domains (TADs) in which interactions between regulatory elements and genes are seemingly able to bypass (putative) intervening insulators (Ciabrelli and Cavalli, 2015; Dixon et al., 2012; Ghavi-Helm et al., 2014; Hou et al., 2012; Sexton et al., 2012; Zhang et al., 2013). Thus, understanding how this might occur in a context in which the key regulatory interactions are well defined should help to illuminate how the topology of the chromatin fiber can be manipulated to impact gene regulation.
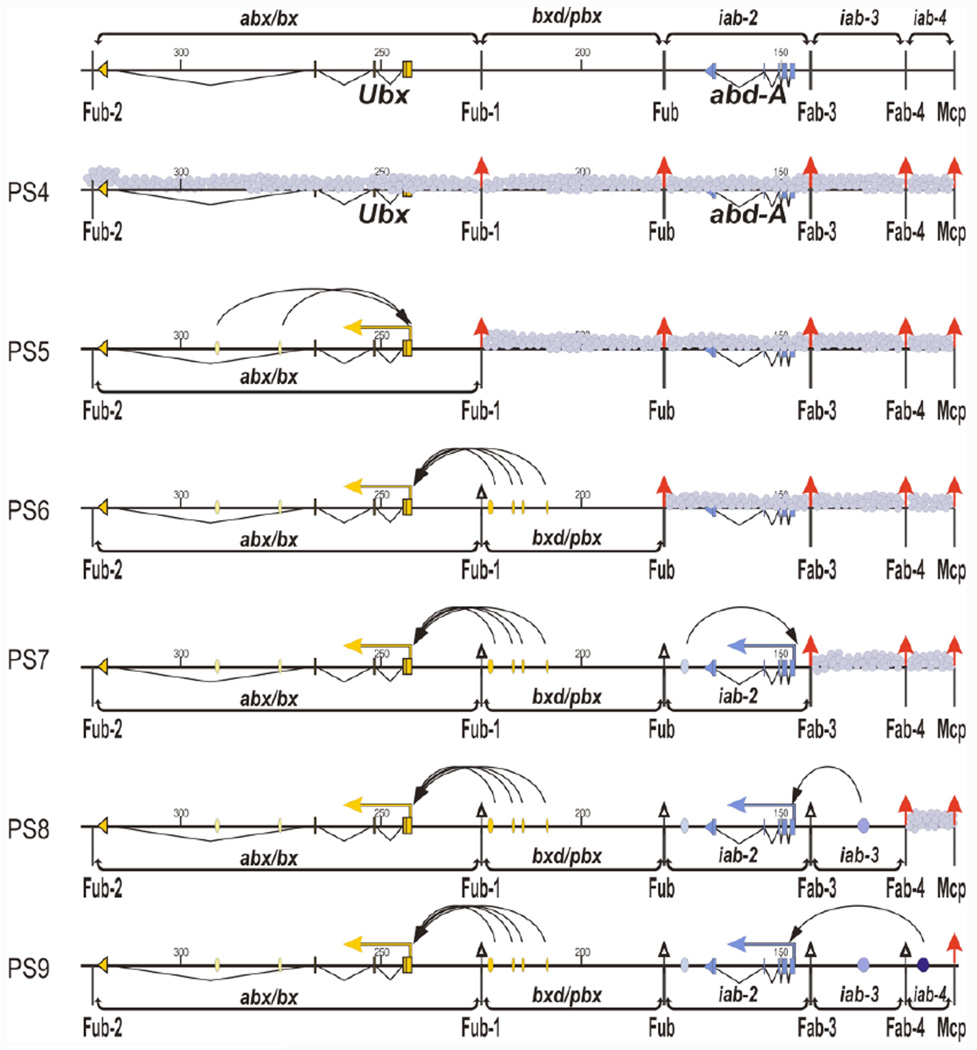
Opening the gate: Sequential domain activation and insulator inactivation
One model couples insulator activity to the successive activation of the BX-C regulatory domains moving from anterior to posterior parasegments (c.f., Peifer et al., 1987). In PS5, only the most proximal BX-C regulatory domain, abx/bx, is activated, while all of the other domains are turned-off. Fub-1, which defines the distal endpoint of the abx/bx regulatory domain, is functional and it prevents the inactive bxd/pbx regulatory domain from interfering with Ubx regulation by the abx/bx enhancers (Figure 4). In these PS5 cells all of the more distal regulatory domains are off and the insulators are fully functional. In the adjacent more posterior parasegment, PS6, the proximal abx/bx and the PS6 specific regulatory domain bxd/pbx are both activated, while all of the more distal BX-C regulatory domains are in the off state. Ubx expression in these PS6 cells is directed by the bxd/pbx domain. However, this requires the neutralization of the intervening Fub-1 insulator so that the spatial/tissue/stage-specific regulatory elements in bxd/pbx can interact with the Ubx promoter (Figure 4). By contrast, the boundary that defines the distal edge of bxd/pbx, Fub, must remain active in this model in PS6 cells so that it can block crosstalk with the adjacent iab-2 domain. Consistent with this idea, when the Fub boundary is deleted the bxd/pbx initiators activate iab-2 inappropriately in PS6 (Bender and Lucas, 2013). A similar scenario would apply for the sequential activation of the three abd-A regulatory domains (Figure 4). In PS7, three regulatory domains would be opened and activated, abx/bx, bxd/pbx, and iab-2, while the remaining regulatory domains would be shut down. The activation of iab-2 would be accompanied by the neutralization of the Fub boundary; however, the Fab-3 boundary on the distal side of iab-2 would need to remain active in order to prevent crosstalk with the silenced iab-3 regulatory domain. Regulation of abd-A by iab-3 in PS8, and by iab-4 in PS9 would require the sequential inactivation of the intervening boundaries. In PS8, Fab-3 would be neutralized so that the iab-3 regulatory domain could control abd-A expression, while Fab-4 would remain active to insulate iab-3 from iab-4. In PS9, both the Fab-3 and Fab-4 would be neutralized so that the iab-4 regulatory domain can interact with the abd-A promoter.
Figure 4. The open-gate model.
This figure shows the coupling of insulator inactivation with the successive activation of BX-C regulatory domains going from anterior to posterior parasegments. The top track shows the Ubx and abd-A genes and their associated regulatory domains with coordinates according to Martin et al., 1995. Enhancers are designated as ovals with different shades: yellow and dark-yellow represent enhancers in abx/bx and bxd/pbx, respectively, that direct Ubx expression in PS5 and PS6; pale-blue, blue and deep blue represent enhancers in iab-2, iab-3, and iab-4, respectively, that direct abd-A expression in PS7-9. The location of insulators between the regulatory domains is indicated by arrows. Active insulators in each parasegment are shown by arrows that are colored red, while insulators that have been neutralized are indicated by unfilled black arrows. Small grey circles designate repressive chromatin status.
The open-gate model requires a parasegment-specific regulation of the insulator activity. Since transgene assays indicate that BX-C boundaries are active irrespective of the parasegment, tissue or developmental stage, parasegment specificity can’t be an intrinsic property of the BX-C boundary elements (Maeda and Karch, 2007; Chetverina et al., 2014). Rather, there would have to be a mechanism that couples the sequential activation of each BX-C regulatory domain by the parasegment-specific initiator with the neutralization of the boundary element on the centromere proximal side of that domain (Peifer et al., 1987). In fact, a mechanism of this sort exists for coordinating the activation of the initiator element in a regulatory domain with the inactivation of the PRE (Bowman et al., 2014). However, unlike the BX-C PREs, which are thought to be functionally equivalent and compatible with all of the BX-C initiators, there also must be a precise matching of initiator and boundary, as the initiator must not be able to neutralize the boundary on the distal side of the domain (or any other more distal boundaries). In this case, the constitutive activity of BX-C boundary elements in transgene assays would be explained by the failure to include the matching parasegment-specific initiator in the reporter construct. Likewise, since heterologous boundaries, like su(Hw) and scs, are not neutralized in the appropriate fashion (see above), they would presumably lack the elements that mediate initiator-dependent regulation of boundary activity.
While the open-gate model fits nicely with the sequential proximal-to-distal activation of the Ubx and abd-A regulatory domains in successive parasegments, it doesn’t explain how insulator activity is neutralized in the Abd-B TARD. Like the Ubx and abd-A TARDs, the Abd-B regulatory domains are sequentially activated in a proximal-to-distal order in successively more posterior parasegments. However, since all four of the Abd-B regulatory domains are downstream (centromere proximal) of the Abd-Bm promoter (Figure 1B), only one domain, iab-8, does not have a boundary element between it and the promoter. In fact, in PS10 where iab-5 is turned on, there would be three functional boundary elements, Fab-6, Fab-7, and Fab-8, interposed between it and the Abd-Bm promoter (Figure 1B). The open-gate model also doesn’t explain the results of an initiator swap experiment, in which the iab-6 initiator in the iab-6 regulatory domain is replaced by the initiator element from iab-5 (Iampietro et al., 2010). If inactivation of the boundary element on the proximal side of the regulatory domain is a critical function for the initiator element (as is postulated in the open-gate model) then the neutralization target for the iab-5 initiator would be Mcp. On the other hand, it should not be able to neutralize the distal boundary element Fab-6. However, the gain-of-function phenotype of the iab-5 replacement would require a change in insulator matching specificity when the initiator is placed in the iab-6 regulatory domain as it would have to neutralize the Fab-6, not the Mcp boundary element.
Insulator bypass by matched boundaries
A second model attributes the permissiveness of boundary elements in BX-C to insulator bypass (Cai and Shen, 2001; Muravyova et al., 2001; Kyrchanova et al 2011). In this model, the BX-C boundaries would have two distinct functions. The first is to prevent adventitious interactions between adjacent regulatory domains. The second is an architectural function that brings active regulatory domains into close proximity with their target homeotic genes (Cai and Shen, 2001; Muravyova et al., 2001). Unlike the open-gate model, which ascribes special regulatory properties to the boundary elements from the BX-C, the blocking and architectural activities of the BX-C boundaries would be essentially the same as those of boundary elements found elsewhere in the fly genome.
The architectural functions evident in the bypass assay are (like enhancer blocking activity) non-autonomous and require that the interacting partners be appropriately matched. The “ideal” matching pair appears to be two copies of the same boundary element, e.g., su(Hw), scs, Mcp, Fab-7 or Fab-8. In contrast, heterologous boundary combinations, e.g., su(Hw) with scs or Mcp with su(Hw) often don’t function or function only poorly in the bypass assay (Kuhn et al., 2003; Kyrchanova et al., 2008a, 2007; Majumder and Cai, 2003; Maksimenko et al., 2008). This incompatibility likely arises because the boundaries utilize different sets and arrangements of proteins, and this generates a high degree of specificity in the matching process. Consistent with this idea, multimerized binding sites for either the Su(Hw) or the Zw5 (scs) proteins support bypass, while pairing multimerized Su(Hw) binding sites with multimerized Zw5 binding sites doesn’t support bypass (Kyrchanova et al., 2008a). On the other hand, bypass is observed when a composite Zw5/Su(Hw) multimer is paired with itself in the correct (opposite) orientation.
The bypass model makes two predictions. The first is that boundaries from the BX-C will form compatible matching pairs, while heterologous boundaries (e.g., su(Hw) and scs) from elsewhere in the genome will not generally be compatible with BX-C boundaries. Bypass assays indicate that the first prediction is correct. Pairwise combinations of Fab-3, Fab-4, Mcp, Fab-6, and Fab-8 with themselves and with each other indicate that all of these boundaries form matching pairs that are able to mediate the bypass (Kyrchanova et al., 2011, 2008b). One reason they might do so is that they all utilize the insulator protein dCTCF (Holohan et al., 2007; Kyrchanova et al., 2011; Moon et al., 2005; Moshkovich et al., 2011; Smith et al., 2009). The exception in this group is Fab-7. Unlike the other insulators, it doesn’t bind the dCTCF protein (Holohan et al., 2007). Moreover, it is selective in its interactions with other BX-C boundaries. Bypass is observed for Fab-7 in combination with either of the two other boundaries that are also part of the Abd-B TARD, Fab-6 and Fab-8, while bypass is not observed for Fab-3, Fab-4, and Mcp (Kyrchanova et al., 2011). Fab-3 and Fab-4 are included in the abd-A TARD, while Mcp separates the abd-A TARD from the Abd-B TARD.
The second prediction is that there must be mechanisms that help direct the individual regulatory domains to their appropriate homeotic gene targets. One of these would presumably be the pairing specificity exhibited by the Fab-7 boundary. Another, likely more important mechanism could involve interactions between the boundaries that comprise each BX-C TARD and sequences in the vicinity of the promoter of the corresponding homeotic gene. Such interactions have been observed within the Abd-B TARD. The region upstream of the Abd-Bm promoter (Abd-BI in Figure 1) binds several known insulator proteins including dCTCF and in bypass assays behaves like a boundary element (Kyrchanova et al., 2011, 2008b). Moreover, like Fab-7, Abd-BI is specific in its interactions with other BX-C boundaries. Thus, bypass is observed when it is paired with Fab-6, Fab-7, and Fab-8, which are all part of the Abd-B TARD. In contrast, bypass is not observed when the Abd-BI element is paired with boundaries from the abd-A TARD.
While this model would seemingly account for both the bypass of insulators and the specificity in regulatory domain:BX-C homeotic gene interactions, there are a number of findings that don’t seem to fit. The first is the apparent failure of the Fab-8 boundary to fully substitute for Fab-7 (Iampietro et al., 2008). Like Fab-7, the Fab-8 replacement boundary blocks adventitious regulatory interactions between iab-6 and iab-7. However, the Fab-8 replacement is not permissive for regulatory interactions between iab-6 and the Abd-Bm promoter and PS11 is transformed into a duplicate copy of PS10. Unlike the gypsy and scs replacements discussed above, Abd-Bm is expressed in a PS10 like pattern in both PS10 and PS11 indicating that iab-5:Abd-Bm interactions are not disrupted. While these results would be inconsistent with the expectations of the bypass model, there is a plausible explanation why the Fab-8 replacement failed to support bypass. The Fab-8 sequence used in the replacement experiment lack sequences from the proximal side of the Fab-8 boundary. The missing sequences include a promoter targeting sequence called the “PTS” (Lin et al., 2007, 2004, 2003; Zhou and Levine, 1999). This sequence appears to be an integral part of the intact Fab-8 boundary element. When the PTS sequence is deleted from the Fab-8 boundary, pairing interactions with the Abd-BI element are lost (Kyrchanova et al., 2011, 2008b). The PTS sequence is not, however, sufficient for pairing with Abd-BI on its own as an intact boundary element is required for the Fab-8:Abd-BI bypass.
A second finding that is inconsistent with the predictions of the bypass model comes from experiments in which a bacterial Dam methylase-Gal4 fusion protein was tethered via Gal4 DNA binding sequences to the (centromere) proximal side of the Fab-7 boundary (Cléard et al., 2006). As expected, the tethered Dam methylase modified GATC sequences in close proximity to the multimerized Gal4 binding sequences. Consistent with the predictions of the bypass model, a peak of GATC methylation was also observed in the Abd-BI element. Also as predicted, this methylation is dependent upon the presence of the Fab-7 boundary element. When the boundary was deleted, the methylation peak in the Abd-BI element disappeared. However, contrary to the expectations of the bypass model the frequency of methylation of GATC sequences in Abd-BI was also inversely dependent upon the transcriptional activity of the Abd-Bm promoter. The methylation frequency was highest in adult heads, where the entire BX-C is shut off. In contrast, in the adult abdomen where Abd-Bm is actively transcribed and contact would be predicted in the bypass model, the extent of methylation of GATC sequences in Abd-BI was not much different from the background levels observed when the Fab-7 boundary was deleted.
Induction of new architectural elements
The Dam methylation experiments indicate that the topological configuration of boundary and regulatory elements in BX-C varies depending on transcriptional activity (Cléard et al., 2006). In the inactive state the Fab-7 boundary is in close proximity with the Abd-BI insulator-like element upstream of the Abd-Bm promoter. Presumably this is also true for the Fab-6 and Fab-8 boundaries. However, in tissues in which the Abd-B promoter is turned on this topological configuration is altered, presumably in a manner that enables the enhancers in the distal-most active regulatory domain to contact and regulate the Abd-Bm promoter. For example, in PS12, three regulatory domains, iab-5, iab-6, and iab-7 are turned on; however, since Abd-B expression in this parasegment is controlled by iab-7, the regulatory elements in this domain must be brought into proximity with the Abd-Bm promoter. One mechanism that would generate a parasegment-specific change in the topological configuration of the Abd-B TARD would be to induce a new architectural element in the iab-7 domain when this domain is activated in PS12. This architectural element would interact with Abd-BI, presumably displacing Fab-7 (as well as the architectural elements in the more proximal regulatory domains, iab-6 and iab-5) and bring the iab-7 enhancers in close proximity to the Abd-Bm promoter. In this “topological reconfiguration” model, equivalent parasegmentally regulated architectural elements would be present in the other Abd-B TARD regulatory domains, as well as in the regulatory domains controlling Ubx and abd-A expression.
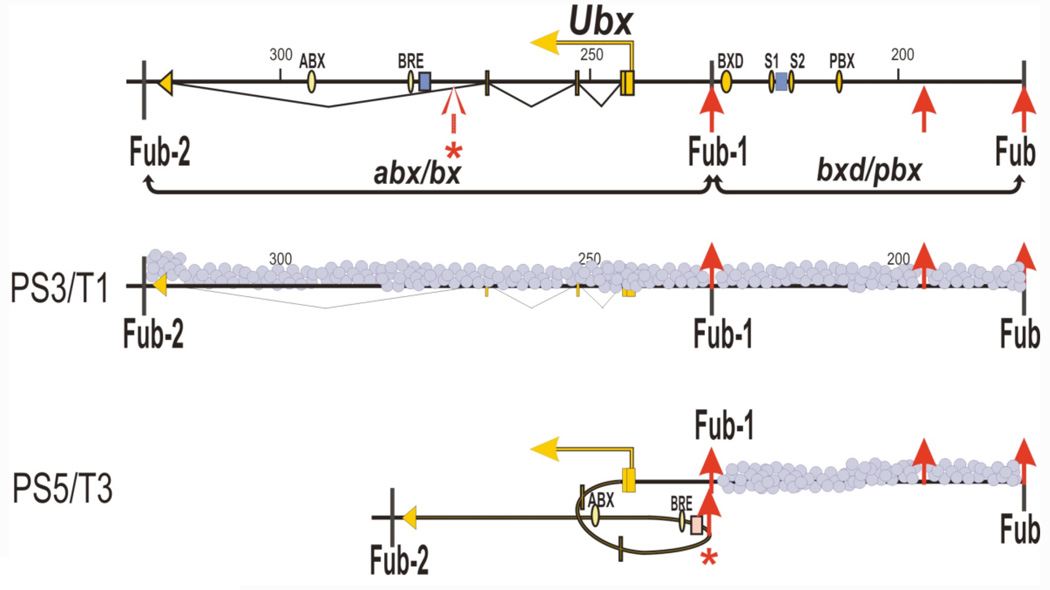
Although parasegment-specific architectural elements have not been identified in iab-7 or in any other Abd-B regulatory domain, an element of this type has been found in the Ubx regulatory domain abx/bx. This developmentally regulated architectural element is located 30 kb downstream of the Ubx promoter, and just upstream of the enhancers and other regulatory elements in abx that control Ubx expression in PS5 (Figure 6). The element is marked by a tissue specific site for the dCTCF insulator protein (Magbanua et al., 2015). dCTCF is not bound to the architectural element in tissues (the first thoracic leg imaginal disc) in which the Ubx gene is not expressed. It is, however, bound in tissues where the Ubx gene is active (the third thoracic leg imaginal disc). Moreover, dCTCF binding to the architectural element is correlated with a change in the topology of the abx/bx regulatory domain. In tissues where Ubx is turned off, the regulated architectural element shows little association with the Ubx promoter (Figure 6). However, in tissues where the Ubx gene is turned on, the architectural element is in close proximity to the Ubx promoter. In addition to interacting with the Ubx promoter, the regulated architectural element also interacts with sequences in the abx enhancer. Moreover, this interaction is also tissue specific and seen only in tissues in which the Ubx promoter is active. These findings would suggest a model in which contacts between the abx enhancer and the Ubx promoter in T3 are facilitated by the activation of the dCTCF architectural element. Further support for a model of this type comes from an analysis of one of the bx mutations induced by the insertion of a gypsy transposon. The transposon insertion is located between the abx enhancers and the developmentally regulated dCTCF architectural element. While the transposon doesn’t interfere with interactions between the architectural element and the Ubx promoter or with dCTCF binding, it appears to destabilize interactions between the architectural element and the abx enhancers.
Figure 6. The parasegment-specific architectural element in abx/bx.
The top track shows the map of the Ubx regulatory domain with coordinates according to Martin et al., 1995. dCTCF binding sites are marked as red upward arrows. The parasegmentally regulated dCTCF binding site is marked by an open arrow with an asterisk underneath; dCTCF does not bind to this site in PS3 where Ubx is turned off. However, in PS5 where the Ubx gene is turned on, dCTCF binds to the parasegmentally regulated site and can mediate interactions with architectural elements in a close proximity to the Ubx promoter. This interaction brings the abx enhancers in contact with the Ubx promoter.
Conclusion
The recent discovery of TADs has revealed the global domain organization of the genome and has emphasized the general role of boundaries in genome architecture (Dixon et al. 2012; Sexton et al. 2012). However, how boundaries function to organize genome topology remains unclear. Insights into boundary function are likely to come from both genome-wide studies of chromatin architecture and the in-depth analysis of specific genomic loci. In particular, the BX-C in Drosophila has contributed greatly to our understanding of the role of boundaries in the organization of functional regulatory domains and in this review we have presented our current understanding of how these elements function in the context BX-C. It is clear that many aspects remain unresolved and we have focused on the striking paradox that the organization of homeotic genes and regulatory domains in BX-C places boundaries/insulators in between regulatory domains (specifically their enhancers/silencers) and their target promoters. Yet, while boundary elements block adventitious interactions between adjacent regulatory domains, they do not prevent the regulatory domains from interacting with their target promoters. This paradox points to the intriguing possibility of a topological code that we have yet to decipher. We have outlined three models that have been proposed to explain this paradox, the open-gate model, the bypass model and the architectural element induction model, however none of these currently appear able to satisfactorily explain all the experimental results. Indeed, each model highlights specific areas that require further investigation.
First, the open-gate model requires a regulatory interaction between the parasegment specific initiator (either direct or indirect) and one of the two boundaries that bracket the regulatory domain. However, how the insulator activity of a boundary might be switched off in one parasegment, but remain on in neighboring parasegments is unknown. Nor is obvious how only one of the boundaries in the domain might be targeted for inactivation. It is striking that boundaries and PREs are often positioned close together in the BX-C. Perhaps being juxataposed with a Polycomb-silenced chromatin may affect the enhancer-blocking function of the insulators. Indeed, there are examples in vertebrates of chromatin effects on the insulator function (Lefevre et al. 2008; Kanduri et al. 2000). Post-translational modification of CTCF has been reported and the sumoylation of CTCF has been linked to the SUMO-E3 ligase activity of the Polycomb group protein Pc2 (MacPerson et al., 2009).
Second, the bypass model is based on transgene assays for boundary pairing that reveal a language of boundary element communication that we do not yet understand. Both the specificity of boundary pairing in insulator bypass assays and genomic data on insulator protein occupancy at boundaries indicate that different boundaries have distinct properties. We need to understand more about how boundary diversity impacts function. What determines the specificity and orientation of boundary interactions? How does orientation affect influence regulatory interactions. Also, these relationships may be dependent on the specific architecture within the BX-C and more approaches that investigate insulator function within the endogenous context are required; such analysis will be facilitated by CRISPR genome editing as in the recent study on the role of CTCF sites at the mouse HoxA locus (Narendra et al., 2015).
Third, the architectural element induction model suggests that we need to know more about the modulation of insulator function. The BX-C presents a powerful system for the analysis of how the functioning of architectural elements might be modulated as the parasegmental activity of the complex provides a set of distinct states for comparison. However, it has been difficult to access parasegment-specific tissue for analysis. Recently, an approach has been developed through the parasegment-specific expression of a fluorescently tagged nuclear protein that enables the preparation of tissue from individual parasegments by the isolation of fluorescent nuclei in a fluorescence-activated cell sorter (Bowman et al., 2014). This procedure opens up the potential for the comparative molecular analysis of insulator complexes across the different parasegmental regulatory states, enabling systematic study of how insulator complexes may be modulated through the effects on the binding of individual components, the composition of complexes and the post-transcriptional modification of the insulator complex components.
Overall, the boundary paradox in the BX-C highlights the importance of chromatin topology in gene regulation and presents the BX-C as a powerful system to further investigate the molecular mechanisms and topological grammar that underlie the regulatory architecture of the genome.
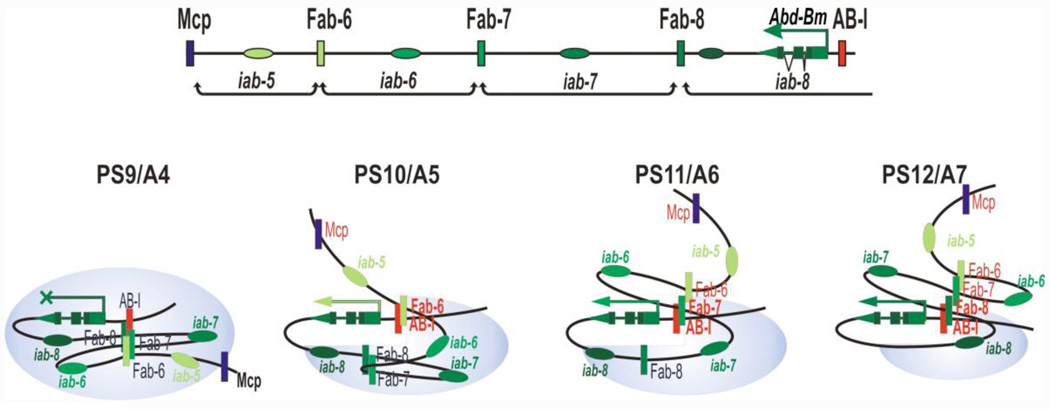
Figure 5. The insulator bypass model.
This figure depicts insulator bypass in the Abd-B TARD. The top track shows the four regulatory domains in the Abd-B TARD and Abd-Bm promoter. Insulators are shown as vertical rectangles with the same color as the enhancers (ovals) whose function they facilitate. In PS10, where the iab-5 regulatory domain is active, the Fab-6 insulator interacts with the insulator-like element, Abd-BI (AB-I), located upstream of the Abd-Bm promoter. This brings iab-5 enhancers to the promoter. In PS11, interactions between the Fab-7 insulator and Abd-BI brings the iab-6 enhancers to the Abd-Bm promoter. Insulators of active domains are marked by red, and insulators that interact with Abd-BI marked by bold. The blue zone symbolizes the repressive domain. All elements outside of the blue zone are active.
HIGHLIGHT.
-
-
Boundary elements ensure the functional autonomy of BX-C regulatory domains.
-
-
Boundaries function by blocking regulatory interactions.
-
-
BX-C regulatory domains are separated from their target genes by boundaries.
-
-
Regulation of target gene transcription requires a bypass mechanism.
-
-
We discuss several possible mechanisms for boundary bypass.
Acknowledgments
This work was supported by the Ministry of Education and Science of the Russian Federation (project no. 14.B25.31.0022) and a grant from NIH GM043432 to PS, by Russian Science Foundation N 14-24-00166 to PG and by Wellcome Trust grant 089834/Z/09/Z to RW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–2420. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barges S, Mihaly J, Galloni M, Hagstrom K, Müller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Dev. Camb. Engl. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- Beachy PA, Helfand SL, Hogness DS. Segmental distribution of bithorax complex proteins during Drosophila development. Nature. 1985;313:545–551. doi: 10.1038/313545a0. [DOI] [PubMed] [Google Scholar]
- Bender W, Akam M, Karch F, Beachy PA, Peifer M, Spierer P, Lewis EB, Hogness DS. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Bender W, Hudson A. P element homing to the Drosophila bithorax complex. Dev. Camb. Engl. 2000;127:3981–3992. doi: 10.1242/dev.127.18.3981. [DOI] [PubMed] [Google Scholar]
- Bender W, Lucas M. The border between the ultrabithorax and abdominal-A regulatory domains in the Drosophila bithorax complex. Genetics. 2013;193:1135–1147. doi: 10.1534/genetics.112.146340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet AM, Lloyd A, Sakonju S. Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Dev. Camb. Engl. 1991;111:393–405. doi: 10.1242/dev.111.2.393. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Deaton AM, Domingues H, Wang PI, Sadreyev RI, Kingston RE, Bender W. H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife. 2014;3:e02833. doi: 10.7554/eLife.02833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busturia A, Bienz M. Silencers in abdominal-B, a homeotic Drosophila gene. EMBO J. 1993;12:1415–1425. doi: 10.1002/j.1460-2075.1993.tb05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busturia A, Wightman CD, Sakonju S. A silencer is required for maintenance of transcriptional repression throughout Drosophila development. Dev. Camb. Engl. 1997;124:4343–4350. doi: 10.1242/dev.124.21.4343. [DOI] [PubMed] [Google Scholar]
- Cai HN, Shen P. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science. 2001;291:493–495. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Sharma S, Keelan DJ, Lewis EB. The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 1990;9:4277–4286. doi: 10.1002/j.1460-2075.1990.tb07876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays News Rev. Mol. Cell. Dev. Biol. 2014;36:163–172. doi: 10.1002/bies.201300125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciabrelli F, Cavalli G. Chromatin-Driven Behavior of Topologically Associating Domains. J. Mol. Biol. 2015;427:608–625. doi: 10.1016/j.jmb.2014.09.013. [DOI] [PubMed] [Google Scholar]
- Ciavatta D, Rogers S, Magnuson T. Drosophila CTCF is required for Fab-8 enhancer blocking activity in S2 cells. J. Mol. Biol. 2007;373:233–239. doi: 10.1016/j.jmb.2007.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat. Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan I. The bithorax complex. Annu. Rev. Genet. 1987;21:285–319. doi: 10.1146/annurev.ge.21.120187.001441. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Sun G, Jaynes JB. The Drosophila eve insulator Homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading. PLoS Genet. 2013;9:e1003883. doi: 10.1371/journal.pgen.1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Wu X, Jaynes JB. A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Dev. Camb. Engl. 2009;136:3077–3087. doi: 10.1242/dev.036467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni M, Gyurkovics H, Schedl P, Karch F. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 1993;12:1087–1097. doi: 10.1002/j.1460-2075.1993.tb05750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EEM. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol. Cell. Biol. 2005;25:3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurkovics H, Gausz J, Kummer J, Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990;9:2579–2585. doi: 10.1002/j.1460-2075.1990.tb07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- Hogga I, Karch F. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Dev. Camb. Engl. 2002;129:4915–4922. doi: 10.1242/dev.129.21.4915. [DOI] [PubMed] [Google Scholar]
- Hogga I, Mihaly J, Barges S, Karch F. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol. Cell. 2001;8:1145–1151. doi: 10.1016/s1097-2765(01)00377-x. [DOI] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol. Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K. The blastoderm prepattern. Semin. Cell Biol. 1990;1:161–172. [PubMed] [Google Scholar]
- Iampietro C, Cléard F, Gyurkovics H, Maeda RK, Karch F. Boundary swapping in the Drosophila Bithorax complex. Dev. Camb. Engl. 2008;135:3983–3987. doi: 10.1242/dev.025700. [DOI] [PubMed] [Google Scholar]
- Iampietro C, Gummalla M, Mutero A, Karch F, Maeda RK. Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet. 2010;6:e1001260. doi: 10.1371/journal.pgen.1001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Martinez-Arias A. The correct activation of Antennapedia and bithorax complex genes requires the fushi tarazu gene. Nature. 1986;324:592–597. doi: 10.1038/324592a0. [DOI] [PubMed] [Google Scholar]
- Irish VF, Martinez-Arias A, Akam M. Spatial regulation of the Antennapedia and Ultrabithorax homeotic genes during Drosophila early development. EMBO J. 1989;8:1527–1537. doi: 10.1002/j.1460-2075.1989.tb03537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Karch F, Bender W, Weiffenbach B. abdA expression in Drosophila embryos. Genes Dev. 1990;4:1573–1587. doi: 10.1101/gad.4.9.1573. [DOI] [PubMed] [Google Scholar]
- Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 1994;22:3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F, Weiffenbach B, Peifer M, Bender W, Duncan I, Celniker S, Crosby M, Lewis EB. The abdominal region of the bithorax complex. Cell. 1985;43:81–96. doi: 10.1016/0092-8674(85)90014-5. [DOI] [PubMed] [Google Scholar]
- Kassis JA, Brown JL. Polycomb group response elements in Drosophila and vertebrates. Adv. Genet. 2013;81:83–118. doi: 10.1016/B978-0-12-407677-8.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn EJ, Viering MM, Rhodes KM, Geyer PK. A test of insulator interactions in Drosophila. EMBO J. 2003;22:2463–2471. doi: 10.1093/emboj/cdg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Chetverina D, Maksimenko O, Kullyev A, Georgiev P. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res. 2008a;36:7019–7028. doi: 10.1093/nar/gkn781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Georgiev P. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 2014;588:8–14. doi: 10.1016/j.febslet.2013.10.039. [DOI] [PubMed] [Google Scholar]
- Kyrchanova O, Ivlieva T, Toshchakov S, Parshikov A, Maksimenko O, Georgiev P. Selective interactions of boundaries with upstream region of Abd-B promoter in Drosophila bithorax complex and role of dCTCF in this process. Nucleic Acids Res. 2011;39:3042–3052. doi: 10.1093/nar/gkq1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol. Cell. Biol. 2007;27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O, Toshchakov S, Podstreshnaya Y, Parshikov A, Georgiev P. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol. Cell. Biol. 2008b;28:4188–4195. doi: 10.1128/MCB.00229-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre P, Witham J, Lacroix CE, Cockerill PN, Bonifer C. The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol. Cell. 2008;32:129–139. doi: 10.1016/j.molcel.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Li H-B, Müller M, Bahechar IA, Kyrchanova O, Ohno K, Georgiev P, Pirrotta V. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol. Cell. Biol. 2011;31:616–625. doi: 10.1128/MCB.00849-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Chen Q, Lin L, Smith S, Zhou J. Promoter targeting sequence mediates enhancer interference in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3237–3242. doi: 10.1073/pnas.0605730104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Chen Q, Lin L, Zhou J. The Promoter Targeting Sequence mediates epigenetically heritable transcription memory. Genes Dev. 2004;18:2639–2651. doi: 10.1101/gad.1230004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Wu D, Zhou J. The promoter targeting sequence facilitates and restricts a distant enhancer to a single promoter in the Drosophila embryo. Dev. Camb. Engl. 2003;130:519–526. doi: 10.1242/dev.00227. [DOI] [PubMed] [Google Scholar]
- Little JW, Byrd CA, Brower DL. Effect of abx, bx and pbx mutations on expression of homeotic genes in Drosophila larvae. Genetics. 1990;124:899–908. doi: 10.1093/genetics/124.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. The CTCF insulator protein is posttranslationally modified by SUMO. Mol. Cell. Biol. 2009;29:714–725. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CH, Mayeda CA, Davis CA, Ericsson CL, Knafels JD, Mathog DR, Celniker SE, Lewis EB, Palazzolo MJ. Complete sequence of the bithorax complex of Drosophila. Proc Natl Acad Sci U S A. 1995;92:8398–402. doi: 10.1073/pnas.92.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda RK, Karch F. Gene expression in time and space: additive vs hierarchical organization of cis-regulatory regions. Curr. Opin. Genet. Dev. 2011;21:187–193. doi: 10.1016/j.gde.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The bithorax complex of Drosophila an exceptional Hox cluster. Curr. Top. Dev. Biol. 2009;88:1–33. doi: 10.1016/S0070-2153(09)88001-0. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. Making connections: boundaries and insulators in Drosophila. Curr. Opin. Genet. Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Dev. Camb. Engl. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- Magbanua JP, Runneburger E, Russell S, White R. A variably occupied CTCF binding site in the ultrabithorax gene in the Drosophila bithorax complex. Mol. Cell. Biol. 2015;35:318–330. doi: 10.1128/MCB.01061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Cai HN. The functional analysis of insulator interactions in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5223–5228. doi: 10.1073/pnas.0830190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimenko O, Bartkuhn M, Stakhov V, Herold M, Zolotarev N, Jox T, Buxa MK, Kirsch R, Bonchuk A, Fedotova A, Kyrchanova O, Renkawitz R, Georgiev P. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 2015;25:89–99. doi: 10.1101/gr.174169.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimenko O, Golovnin A, Georgiev P. Enhancer-promoter communication is regulated by insulator pairing in a Drosophila model bigenic locus. Mol. Cell. Biol. 2008;28:5469–5477. doi: 10.1128/MCB.00461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimenko O, Kyrchanova O, Bonchuk A, Stakhov V, Parshikov A, Georgiev P. Highly conserved ENY2/Sus1 protein binds to Drosophila CTCF and is required for barrier activity. Epigenetics Off. J. DNA Methylation Soc. 2014;9:1261–1270. doi: 10.4161/epi.32086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall K, O’Connor MB, Bender W. Enhancer traps in the Drosophila bithorax complex mark parasegmental domains. Genetics. 1994;138:387–399. doi: 10.1093/genetics/138.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J, Barges S, Sipos L, Maeda R, Cléard F, Hogga I, Bender W, Gyurkovics H, Karch F. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Dev. Camb. Engl. 2006;133:2983–2993. doi: 10.1242/dev.02451. [DOI] [PubMed] [Google Scholar]
- Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Dev. Camb. Engl. 1997;124:1809–1820. doi: 10.1242/dev.124.9.1809. [DOI] [PubMed] [Google Scholar]
- Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 2001;21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, Burke LJ, Renkawitz-Pohl R, Ohlsson R, Zhou J, Renkawitz R, Lobanenkov V. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–1701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Bienz M. Sharp anterior boundary of homeotic gene expression conferred by the fushi tarazu protein. EMBO J. 1992;11:3653–3661. doi: 10.1002/j.1460-2075.1992.tb05450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Bienz M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 1991;10:3147–3155. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Hagstrom K, Gyurkovics H, Pirrotta V, Schedl P. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics. 1999;153:1333–1356. doi: 10.1093/genetics/153.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–498. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. Transcription. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz MJ, Jäckle H. Making stripes in the Drosophila embryo. Trends Genet. TIG. 1990;6:287–292. doi: 10.1016/0168-9525(90)90234-w. [DOI] [PubMed] [Google Scholar]
- Peifer M, Bender W. The anterobithorax and bithorax mutations of the bithorax complex. EMBO J. 1986;5:2293–2303. doi: 10.1002/j.1460-2075.1986.tb04497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Karch F, Bender W. The bithorax complex: control of segmental identity. Genes Dev. 1987;1:891–898. doi: 10.1101/gad.1.9.891. [DOI] [PubMed] [Google Scholar]
- Pérez-Lluch S, Cuartero S, Azorín F, Espinàs ML. Characterization of new regulatory elements within the Drosophila bithorax complex. Nucleic Acids Res. 2008;36:6926–6933. doi: 10.1093/nar/gkn818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V, Chan CS, McCabe D, Qian S. Distinct parasegmental and imaginal enhancers and the establishment of the expression pattern of the Ubx gene. Genetics. 1995;141:1439–1450. doi: 10.1093/genetics/141.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Capovilla M, Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991;10:1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S, Kyrchanova O, Pomerantseva E, Parshikov A, Georgiev P. New properties of Drosophila fab-7 insulator. Genetics. 2007;177:113–121. doi: 10.1534/genetics.107.075887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Herrero E. Control of the expression of the bithorax complex genes abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Dev. Camb. Engl. 1991;111:437–449. doi: 10.1242/dev.111.2.437. [DOI] [PubMed] [Google Scholar]
- Sánchez-Herrero E, Vernós I, Marco R, Morata G. Genetic organization of Drosophila bithorax complex. Nature. 1985;313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat. Rev. Genet. 2013;14:853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- Schweinsberg SE, Schedl P. Developmental modulation of Fab-7 boundary function. Dev. Camb. Engl. 2004;131:4743–4749. doi: 10.1242/dev.01343. [DOI] [PubMed] [Google Scholar]
- Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shimell MJ, Peterson AJ, Burr J, Simon JA, O’Connor MB. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev. Biol. 2000;218:38–52. doi: 10.1006/dbio.1999.9576. [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W, Shimell MJ, O’Connor M. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev. Biol. 1993;158:131–144. doi: 10.1006/dbio.1993.1174. [DOI] [PubMed] [Google Scholar]
- Simon J, Peifer M, Bender W, O’Connor M. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 1990;9:3945–3956. doi: 10.1002/j.1460-2075.1990.tb07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos L, Gyurkovics H. Long-distance interactions between enhancers and promoters. FEBS J. 2005;272:3253–3259. doi: 10.1111/j.1742-4658.2005.04757.x. [DOI] [PubMed] [Google Scholar]
- Small S, Levine M. The initiation of pair-rule stripes in the Drosophila blastoderm. Curr. Opin. Genet. Dev. 1991;1:255–260. doi: 10.1016/s0959-437x(05)80079-6. [DOI] [PubMed] [Google Scholar]
- Smith ST, Wickramasinghe P, Olson A, Loukinov D, Lin L, Deng J, Xiong Y, Rux J, Sachidanandam R, Sun H, Lobanenkov V, Zhou J. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev. Biol. 2009;328:518–528. doi: 10.1016/j.ydbio.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat. Rev. Mol. Cell Biol. 2014;15:340–356. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- Struhl G, Akam M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 1985;4:3259–3264. doi: 10.1002/j.1460-2075.1985.tb04075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J, Müller M, Pirrotta V, Sedat JW. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol. Biol. Cell. 2006;17:2158–2165. doi: 10.1091/mbc.E06-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RAH, Wilcox M. Regulation of the distribution of Ultrabithorax proteins in Drosophila. Nature. 1985;318:563–567. doi: 10.1002/j.1460-2075.1985.tb03889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, Lehmann R. A gap gene, hunchback, regulates the spatial expression of Ultrabithorax. Cell. 1986;47:311–321. doi: 10.1016/0092-8674(86)90453-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wong C-H, Birnbaum RY, Li G, Favaro R, Ngan CY, Lim J, Tai E, Poh HM, Wong E, Mulawadi FH, Sung W-K, Nicolis S, Ahituv N, Ruan Y, Wei C-L. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Ashe H, Burks C, Levine M. Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Dev. Camb. Engl. 1999;126:3057–3065. doi: 10.1242/dev.126.14.3057. [DOI] [PubMed] [Google Scholar]
- Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]
- Zhou J, Levine M. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell. 1999;99:567–575. doi: 10.1016/s0092-8674(00)81546-9. [DOI] [PubMed] [Google Scholar]