Abstract
Primary cancer cell dissemination is a key event during the metastatic cascade, but context-specific determinants of this process remain largely undefined. Multiple reports have suggested that the p53 (TP53) family member p63 (TP63) plays an anti-metastatic role through its minor epithelial isoform containing the N-terminal transactivation domain (TAp63). However, the role and contribution of the major p63 isoform lacking this domain, ΔNp63α, remain largely undefined. Here, we report a distinct and TAp63-independent mechanism by which ΔNp63α-expressing cells within a TGFβ-rich microenvironment become positively selected for metastatic dissemination. Orthotopic transplantation of ΔNp63α-expressing human osteosarcoma cells into athymic mice resulted in larger and more frequent lung metastases than transplantation of control cells. Mechanistic investigations revealed that ΔNp63α repressed miR-527 and miR-665, leading to the upregulation of two TGFβ effectors, SMAD4 and TβRII (TGFBR2). Furthermore, we provide evidence that this mechanism reflects a fundamental role for ΔNp63α in the normal wound healing response. We show that ΔNp63α-mediated repression of miR-527/665 controls a TGFβ-dependent signaling node that switches off anti-migratory miR-198 by suppressing the expression of the regulatory factor, KSRP (KHSRP). Collectively, these findings reveal that a novel microRNA network involved in the regulation of physiological wound healing responses is hijacked and suppressed by tumor cells to promote metastatic dissemination.
Introduction
The p53 family of transcription factors, including p53 (TP53), p63 (TP63) and p73 (TP73), are key players in tumor development (1). The p53 gene is the prototypical human tumor suppressor and is mutated or lost in the majority of human cancers (1). Unlike p53, neither p63 nor p73 exhibits frequent somatic mutation in cancer (2,3). Nevertheless, a tumor suppressor role for p73 is suggested by genetic and biochemical studies, while p63 has been linked to tumorigenesis rather than tumor suppression (4). In the normal stratified epithelia p63 functions to maintain cellular regenerative proliferation and survival, and overexpression and/or genomic amplification of p63 is observed in a subset of human cancers (5).
All three p53 family members encode proteins with highly homologous DNA binding domains, through which they regulate both shared and distinct subsets of transcriptional targets (6). Both p63 and p73 are expressed from two distinct promoters, thereby producing isoforms that either contain or lack the N-terminal transactivation domain (TAp63/p73 and ΔNp63/p73, respectively) (3). Differential mRNA splicing also gives rise to multiple C-terminal variants of both p63 and p73. In both normal epithelia and in epithelial cancers including head and neck squamous cell carcinoma (HNSCC), prostate and breast cancer the predominant p63 isoform expressed is ΔNp63α (7,8). Supporting the central role of this isoform, selective knockout of ΔNp63 but not TAp63 in vivo recapitulates the phenotype of complete p63 knockout(9-11).
Regulation by MicoRNAs (miRs) is known to contribute to diverse cellular processes in both normal and cancer cells. While p53 is known to regulate miRs that contribute to its function, the identity and contribution of p63 and p73-regulated miRs are poorly understood (12).
We recently demonstrated that p63 itself directly regulates a subset of miRs that function in a feed-forward circuit to regulate p73 levels and activity (13). This finding is in keeping with an emerging consensus that miRs are particularly prominent within regulatory circuits controlling transcription factor functions. Emerging data suggest that such circuits may be particularly important for regulation of cellular functions associated with tumor metastasis (14). In the last decade, several miRs have been identified as promoters or suppressors of metastatic dissemination. These miRs regulate metastasis through divergent or convergent regulation of metastatic genes within specific pathways. The consequences of this regulation can be the modification of the cancer cell phenotype itself or a modification of the metastatic microenvironment composition (15).
One such metastatic pathway potentially under the control of miRs is the TGFβ pathway. TGFβ is a ubiquitously expressed cytokine that regulates numerous biological activities in a wide range of tissues. In addition to its role in regulating cell development, differentiation, and survival, TGFβ inhibits the proliferation of epithelial, endothelial, and hematopoietic cell lineages (16). A hallmark of neoplastic transformation is the resistance to TGFβ-mediated cytostasis, which ultimately converts the signals produced by this cytokine into oncogenic activities, and particularly enhanced cancer cell invasion and metastasis. This dramatic modification of TGFβ function underlies the adverse prognosis associated with elevated TGFβ levels in developing carcinomas, involving their acquisition of epithelial-mesenchymal transition (EMT), and metastatic chemoresistant phenotypes (17). The molecular mechanisms whereby TGFβ promotes the progression of late stage carcinomas as well as cancer cell invasion and metastasis remain to be elucidated, highlighting the need to understand the seemingly paradoxical activities of TGFβ in normal and malignant cells.
The potential role of p63 in metastasis regulation remains poorly understood. The TAp63 isoform was identified as a metastasis suppressor by decreasing mobility and invasion (18). Additionally, Adorno M et al, have described a mechanism whereby TGFβ functions upstream to abrogate TAp63-mediated metastasis suppression through formation of a ternary complex involving mutant p53 and SMAD2 (19). In contrast the relationship, if any, of the major ΔNp63α isoform to metastasis and TGFβ remain controversial. One study described ΔNp63α as a pro-metastatic protein, while two recent contradictory studies describe ΔNp63 isoform as pro- or anti-EMT, in a TGFβ-dependent manner (18,20-22).
Here we report a systemic analysis of p63-regulated miRs, which uncovered a prominent and selective contribution of ΔNp63α to miR-dependent control of the TGFβ pathway. We show that ΔNp63 and TAp73, but not TAp63, control the expression of miR-527 and miR-665, two miRs that repress the central TGFβ regulators SMAD4 and TGFBR2. We provide evidence that this p63-mediated pathway is linked to control of normal wound healing through TGFβ-dependent regulation of the KSRP/ miR-198 axis. In tumors, we show that this regulatory network orchestrates the metastatic dissemination of tumor cells within the TGFβ-containing microenvironment. Taken together, our findings implicate a miR-dependent circuit involved in normal wound healing via ΔNp63α as a key mediator of TGFβ -induced metastatic dissemination.
Materials and Methods
Tumor cell line and patient tumor material
The cell lines JHU-029, was the generous gifts of David Sidransky (Johns Hopkins University). This line was maintained by the MGH Center for Molecular Therapeutics cell line bank and underwent high-density single nucleotide polymorphism (SNP) typing, revealing that each was unique compared with > 800 other banked lines. Stocks of JHU-029 used in this study express no wild-type p53 due to a frameshift mutation in p53 codon 108 that results in premature truncation. Tetracycline-inducible HA-TAp73β cells were established in JHU-029 using the TREx plasmid system (Invitrogen). Cells were maintained at 37°C with 5% CO2 in either RPMI (Lonza) supplemented with 10% FBS, penicillin, and streptomycin.
The human osteosarcoma cell lines MNNG/HOS (young female high grade osteosarcoma from femur origin transformed in vitro by N-methyl-N′-nitro-N-nitrosoguanidine treatment), were purchased from the American Type Culture Collection and maintained in RPMI 1640 (Lonza) supplemented with 10% fetal bovine serum and 2mmol L−1 L-glutamine. All cell lines were cultured in a humidified 5% CO2/air atmosphere at 37°C. The cell line was maintained by our University cell facility. Genetic characteristics were determined by PCR-single-locus-technology by Eurofins compagny using Promega, PowerPlex 21 PCR Kit. All cell lines were passaged for less than 3 months and tested in July 2015.
Osteosarcoma cell lines subpopulations were obtained at the time of diagnostic biopsy (B) or after surgical resection of lung metastasis (M), in patients diagnosed with osteosarcoma at the Hospital of the University of Navarra (Clínica Universidad de Navarra, CUN, Pamplona, Spain). The clinical features of clinical samples are available in supplemental Table 1. Samples were obtained following patient informed consent, and after ethical approval by the Navarra University Hospital Ethics Committee. All subpopulations were thoroughly characterized as previously described (Patiño-García et al. Clin Cancer Res).
Micro-RNA micro Array analysis
For each sample, 2μg of Trizol extracted total RNA were purified using the RNeasy Mini Cleanup Kit (Qiagen). After having passed sample quality control on the Bioanalyser2100, the samples were labelled using the miRCURY™ Hy3™/Hy5™ power labelling kit and hybridized on the miRCURY™ LNA Array (v.10.0) (Exiqon). Spike-in controls were added in various concentrations covering the full signal range, in both Hy3 and Hy5 labelling reactions giving the opportunity to evaluate the labelling reaction, hybridization and the performance of the array experiment in general. The quantified signals (background correction) were normalized using the global Lowess (LOcallyWEighted Scatterplot Smoothing) regression algorithm. The diagram only represent the microRNAs that passed the filtering criteria across samples; Fold Change > 1.50. The microarray data discussed in this publication have been deposited in NCBI Gene Expression Omnibus (GEO Series accession number GSE25524).
Transient transfection of pre-miR™ and anti-miR™
All the pre-miR™ and anti-miR™ were obtained from Ambion and tranfected at a final concentration of 30nM using the siPORTNeoFX Transfection Agent (Ambion) according to the manufacturer's protocol. The pre-miR™ and anti-miR™ Negative Controls used are random sequences that have been tested in human cell lines and tissues and validated to not produce identifiable effects.
Transient transfection of TAp73b and ΔNp63α
Both TAp73β and ΔNp63α sequences were inserted in pcDNA3 vector (Invitrogen). 2μg of DNA were transfected using FuGENE® HD Transfection Reagent (Promega) according to the manufacturer's protocol. We used pcDNA3 empty vector as a control. These assays were all performed as described (13)
Cell proliferation
JHU-029 cells (2 × 103/well) were plated in 0.1 mL RPMI 1640 with 10% FBS and treated with pre-miRs and anti-miRs at indicated concentration. 72h after transfection a solution of WST-1 was added to the wells. Cell proliferation was assessed by optical density measurement at 470nm with Victor2 1420 (MultilabelCounter, Perkin Elmer). The viability percentage is calculated by: OD at 470nm with indicated pre-miR/ OD at 470nm with pre-miR control × 100. Each assay was repeated in triplicate.
Western blotting analysis
Samples containing equal amounts of protein (depending on the antibody, 20-50μg) from lysates of cultured cell lines underwent electrophoresis on SDS-polyacrylamide gel and were transferred to PVDF membranes. The membranes were blocked in 3% BSA-PBS-0.05% Tween at room temperature for 1h and blots were probed overnight at 4°C with primary antibodies (TβRII, 1:1,000; Abcam), (SMAD4, 1:500; Santa Cruz Biotechnologies), (SMAD3 and P-SMAD3, 1:1,000; Cell Signaling Technologies), (p63, 1:1,000; Sigma-Aldrich), (p73, 1:500; Calbiochem) or GAPDH (1:2,000; Cell signaling) to detect proteins of interest. After incubation, the membranes were washed 3 times with washing buffer (PBS containing 0.05% Tween) for 5min. Membranes were then incubated for 1h with 1:10,000 diluted secondary antibodies (Santa Cruz Biotechnologies, Santa Cruz, CA) at room temperature. Specific proteins were detected using G-Box (Syngene, Cambridge, UK) after washing.
Quantitative Reverse Transcription-PCR
Total RNA was extracted from cultured cells, human biopsies or xenografts using TRIreagent (Invitrogen Life Technologies). Total RNA was reversed transcribed using the ThermoScript RT-PCR System (Life Technologies). Real-time monitoring of PCR amplification of complementary DNA (cDNA) was performed using DNA primers (primers sequences are available upon request) on CFX96 real-time PCR detector system (Bio-Rad, Marnes la Coquette, France) with SYBR PCR Master Mix buffer (Bio-Rad). Target gene expression was normalized to the average B2M and GAPDH levels in respective samples as an internal standard, and the comparative cycle threshold (Ct) method was used to calculate relative quantification of target mRNAs. Each assay was performed in triplicate.
Quantitative Reverse Transcription-PCR miR
A specific RT was performed for each miR from 100 ng of total RNA, using a specific stem-loop RT primer (50 nM) and the MultiScribe Reverse transcriptase (Applied Biosystems). The RT conditions were as follows: 30 minutes at 16°C followed by 30 seconds at 20°C, 30 seconds at 42°C, 1 second at 50°C for 60 cycles, and finally 5 minutes at 85°C. Mature miRs’ expression levels were measured by real-time qRT-PCR using the SYBR PCR Master Mix buffer (Bio-Rad) and a CFX96 real-time PCR detector system (Bio-Rad). The expression of each gene was normalized to the small nuclear U6B RNA as a reference. All experiments were performed in triplicate. Primer sequences for the RT and qPCR are available in Supplementary Table 3 and 4 respectively.
Lentiviral and retroviral production and mRNA QRT-PCR
P63 shRNA lentiviral particles were produced by transfection of required viral plasmids (4μg of each: RSV-RRE, RGR, VSV-G and 4 μg of p63 All isoforms shRNA; p63Allsi; or 4μg of GFP shRNA; GFPsi) following the manufacturer's protocol (CalPhos Transfection Kit; Clontech) in HEK 293 T cell line. HEK 293T were cultured in DMEM medium enriched with 10%FBS and 1X non essential amino acids (Lonza). JHU-029 cells were infected with HEK 293T supernatants (with GFPsi as a control or p63Allsi) and selected by antibiotic resistance (puromycin, 1μg/mL) for several weeks. P63 All isoforms-shRNA targeted sequence is 5’-GGGTGAGCGTGTTATTGATGCT-3’. P63-directed shRNA was validated not to cross react with p53 or p73 (13). Primer sequences for mRNA QRT-PCR validation are available upon request.
3’UTR Reporter assays
50,000 cells were seeded in 24-well plates and were transfected with the indicated reporter constructs depending on the different conditions. Lysates were prepared at 48 hours after transfection, and luciferase activity was measured using the Dual Luciferase Reporter Assay system (Promega) and a luminometer (MicroLumat Plus, EG&G Berthold).
For PAI-1 reporter assay, cells were transfected with the reporter vector ((CAGA)9-lux) and pRL SV40 (containing Renilla luciferase, Promega) together with TAp73β, ΔNp63α or pre-miRs following manufacturer's recommendation (siPORT™ NeoFX™ Transfection Agent, Invitrogen). (CAGA)9-lux reporter vector was generated using pGL3-basic vector (Promega) where 9 repetitions of the CAGA box sequence were inserted between BglII and HindIII restriction sites.
From the MNNG/HOS cell line cDNA, two fragments of the SMAD4 3’UTR (90bp-1900bp and 1900bp-4055bp; primers sequences are in supplemental table 5) were separately cloned into pMIR-REPORT™ Luciferase vectors (Ambion) using the Platinum® Taq DNA Polymerase High Fidelity (Life technologies). Each fragment was cloned downstream of the Firefly Luciferase between HindIII and SpeI restriction sites.
For Smad4 3’UTR reporter assays, cotransfection with 10ng of either control reporter vector (pMIR control, Ambion), UTR-reporter vector (pMIR SMAD4 3’UTR) or mutated UTR-reporter vector (pMIR mutated SMAD4 3’UTR) all together with pre/anti-miRs control, 527 or 371-5p (15μM) and 25ng of pRL SV40 (Promega) was performed.
For psi-check2 TβRII 3’UTR reporter assays, cells were cotransfected with control reporter vector (psi-ckeck2 control, Promega), UTR-reporter vector (psi-check2 TβRII 3’UTR) or mutated UTR-reporter vector (psi-check2 mutated TβRII 3’UTR) together with pre/anti-miRs control, 527, 665 and 371-5p (15μM) was done.
Psi-check2 TβRII 3’UTR was a kind gift from Dr. Ioanna Keklikoglou (German Cancer Research Center (DKFZ), Heidelberg, Germany) where 3’UTR of the TβRII gene was inserted between XhoI and NotI sites of the psi-check2 vector.
TβRII and SMAD4 3’UTR site directed mutagenesis
According to the TargetScan database miR-527 recognizes two putative sequences within the SMAD4 3’UTR and miR-665 recognizes two putative sequences within the TβRII 3’UTR (supplemental figure 2).
Mutant seed binding sequences were created with Quickchange lightning Multi site-Directed Mutagenesis kit (Agilent technologies, Inc) according to the manufacturer's indications. Mutagenic primers were randomly designed with the web-based Quickchange Primer Design Program (supplemental table 5). For 3’UTR SMAD4 fragment 1 and fragment 2 a single seed sequence was mutated. For 3’UTR TβRII 2 seed sequences were mutated (supplemental figure 2).
Migration and invasion tests
To evaluate migration ability, JHU-029 or MNNG/HOS cell lines were transfected with Tap73β, ΔNp63α and control vectors or indicated pre-miRs/anti-miRs. After 24h of transfection cells were starved during 5 hours with RPMI 1640 supplemented with 1% FBS and then pre-treated or not with 5ng/mL TGFβ (R&D systems). All different conditions were trypsinized and treated or not with 5ng/mL TGFβ before being seeded on the upper side of a Transwell chamber (Falcon) on a porous transparent polyethylene terephthalate (PET) membrane (8-μm pore size). Different cell densities were used depending on the experiment to increase or decrease cell migration. Indeed, to be able to observe an increased cell migration in our experimental conditions, we should not already have a saturated migration in our control condition. The percentage migration increase variation between the different control conditions (pre-miR Ct and anti-miR Ct) is voluntarily set up (by cell confluence variation) to facilitate the observation of either an increase or a decrease cell migration in presence of TGFβ. The lower chamber of the transwell was filled with growth medium containing 10% FBS. After 24 hours of incubation, cells on the upper side of the filters were mechanically removed and cells migrated to the lower side were fixed (with glutaraldehyde 10%), stained (with violet crystal 0.1%), and counted. Five random fields were analyzed for each chamber.
Matrigel-coated transwell were used for invasion assay (50ng Matrigel/well). After 24 h incubation, cells that had invaded to the lower surface of membrane were fixed, stained, and counted. Five random fields were analyzed for each chamber.
Wound-healing test
50×103 JHU-029 cells were seeded in 24 well-plate and transfected with Tap73β, ΔNp63α and control vectors or indicated pre-miRs/anti-miRs. 24h after transfection cells were starved during 5 hours with RPMI 1640 supplemented with 1% FBS and then treated or not with 5ng/mL TGFβ. 24h later wells were confluent and scratches were performed. Cell migration was recorded during 24h every 10 minutes at fixed points and not recovered areas were calculated.
Zymography
The presence of MMP-2 and MMP-9 in serum-free conditioned media was analyzed by zymography in 10% sodium dodecyl sulfate–polyacrylamide gels containing 1 mg/mL gelatin (Sigma-Aldrich). After removal of sodium dodecyl sulfate with Triton X-100, neutral protease activity was revealed by incubation of the gels for 48 hours at 37°C in 50mM Tris buffer (pH 7.4) containing 2 mM CaCI2, followed by staining in 0.05% Coomassie blue in an aqueous solution containing 20% isopropanol and 10% acetic acid. Pictures were taken using G-Box (Syngene, Cambridge, UK).
Animal Treatment
All procedures involving mice [their housing in the Experimental Therapeutic Unit at the Faculty of Medicine of Nantes (France) and care, the method by which they were anesthetized and killed, and all experimental protocols] were conducted in accordance with the institutional guidelines of the French Ethical Committee (CEEA.PdL.06). Mice were anesthetized by inhalation of of isoflurane/air (1.5%, 1 Lmin−1). Tumor volume was measured 3 times weekly and tumor volume was calculated by using the formula: length × (thickness2) / 2. Data points were expressed as average tumor volume ± SEM.
ΔNp63α-MNNG/HOS: MNNG/HOS cells were transiently transfected with either pcDNA3 empty vector or pcDNA3-ΔNp63α (Invitrogen) following the manufacturer's protocol (FuGENE® HD Transfection Reagent, Promega). Cells expressing the insert were selected by antibiotic pressure (2mg/mL geneticin) for several weeks. MNNG/HOS osteolytic xenograft model was induced by paratibial injection of 1×106 of either control or ΔNp63α overpressing human MNNG/HOS cells in five weeks-old female athymic nude mice (Harlan Sprague-Dawley, Inc.). Tumor volumes were monitored until a maximum of 2500 mm3. Tumor and metastasis samples were collected to analyze gene expression by RT-qPCR.
Pre-miR injection: HOS osteolytic xenograft model was induced by an i.m. injection of 1.5 × 106 human HOS LucF GFP cells next to the tibia of five week-old female athymic nude mice (Harlan Sprague-Dawley, Inc.), leading to a rapidly growing tumor in soft tissue with secondary contiguous bone invasion. Once palpable tumors, mice were randomly assigned to pre-miR control or pre-miR-527 or pre-miR-665. When tumors reached 200mm3, pre-miRs were injected at a 50nM concentration every 2 days. The pre-miRs are injected directly into the tumor volume. A total of 20μl at 50nM were injected in 4 different locations for each tumor. Each experimental group consisted of 8 mice. It is important to note that in our model the baseline metastatic rate depends on the number of cells initially used for para-tibial injection. Thus, different cell numbers injected accounts for the different baseline metastatic rates shown in Figure 5 and Figure 6.
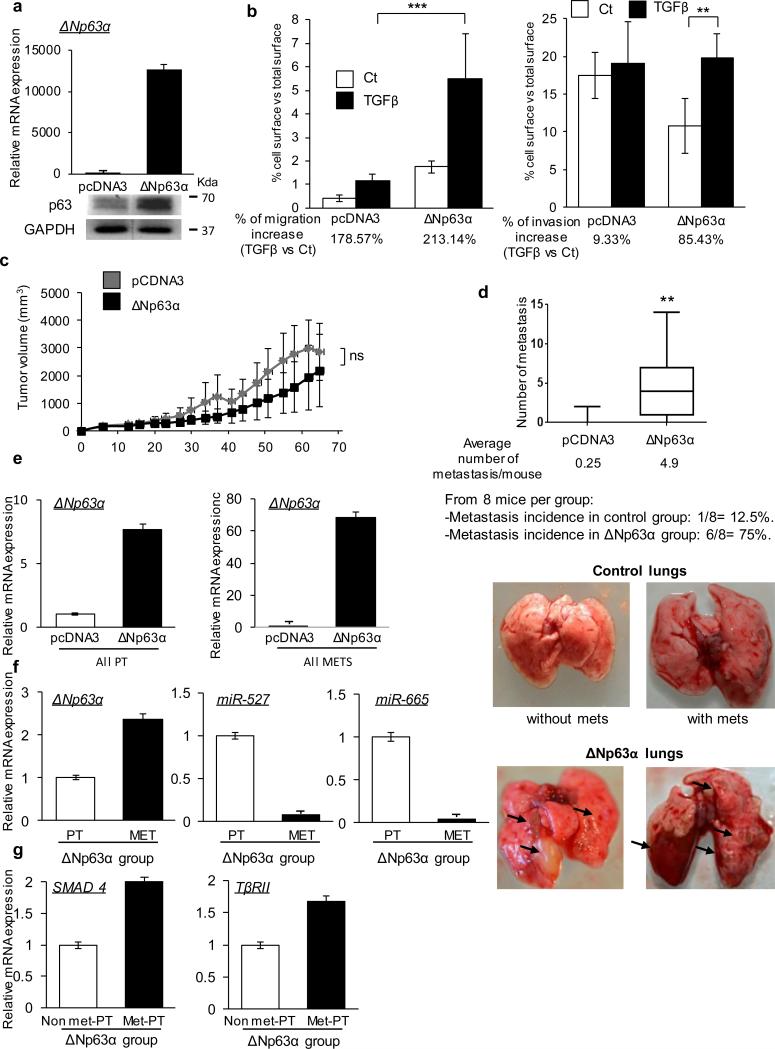
Figure 5. p63 induces TGFβ-mediated metastasis in vivo.
(a) Overexpression of ΔNp63α mRNA (top) and protein (bottom) in ΔNp63α-HOS group vs control group. (b) HOS cells were transfected with ΔNp63α or control vectors and treated or not with TGFβ (5ng/mL) after 24h. Cell migration (left) and invasion (right) were analyzed after 24h. Results are expressed in terms of cell-occupied area vs total area. Two-way ANOVA followed by Tukey's multiple comparison tests were used (*** p< 0.001). (c) Tumor progression was calculated with the formula (t2 × l)/2 where t is the tumor thickness and l is the tumor length. Average tumor volumes for ΔNp63α or control groups over time are represented in the graph. Mann-Whitney statistical test was used. (d) Number of metastasis found at tumor volume endpoint (2500mm3) for each mouse and average for ΔNp63α or control groups. Lung images were obtained after sacrifice showing the difference in appearance of the ΔNp63α metastasis vs the control group. Arrows indicate metastatic foci. Mann-Whitney statistical test was used. (e) ΔNp63α, (f) miR-527, miR-665, (g) TβRII and SMAD4 expression levels were analyzed by qRT-PCR in the indicated groups (primary tumors PT, metastasis MET, primary tumors that did not produce metastasis (Non met-PT) and that did (Met-PT)). All error bars show s.e.m. for triplicate measurements from representative experiments.
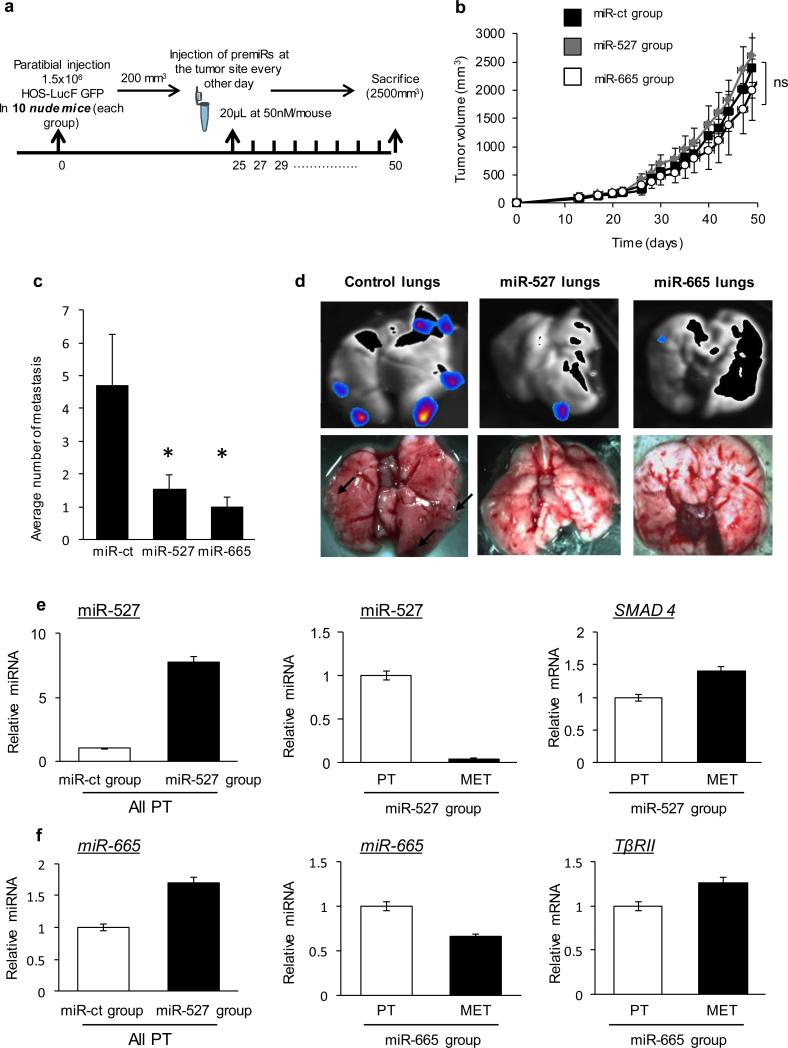
Figure 6. miR-527 and miR-665 regulate metastasis dissemination in vivo.
(a) Experimental schema. 1.5M HOS LucF GFP were injected paratibially in nude mice. At 200mm3 tumor volume mice were treated every 48h with 50nM pre-miR control (miR ct-group), pre-miR-527 (miR-527 group) or pre-miR-665 (miR-665 group). (b) Tumor progression was calculated as in Fig. 5. Average tumor volumes for miR-527, miR-665 or control groups over time are represented in the graph. Kruskal-Wallis test followed by Dunnett's multiple comparison test were used. (c) Average number of metastasis found at tumor volume endpoint (2500mm3) for miR-527, miR-665 or control groups. Mann-Whitney statistical test was used. * p< 0.05. (d) Bioluminescence imaging of lungs after sacrifice for miR-527, miR-665 and control groups. (e) miR-527 and SMAD4 expression levels were analyzed by qRT-PCR. (f) miR-665 and TβRII expression levels were analyzed by qRT-PCR. All error bars show s.e.m. for triplicate measurements from representative experiments.
When tumor volume reached ≥10% of body weight, mice were sacrificed prior to an intraperitoneal injection of luciferine substrate (0.12 mg D-luciferine/mg mouse; Interchim) to evaluate lung metastasis by bioluminescence (NightOwl BERTHOLD LB981). Tumor and metastasis samples were collected to analyze gene expression by RT-qPCR.
Statistical analysis
For each experimental datapoint, the SEM from replicate experiments was calculated as noted in the legends and is shown as error bars. All error bars show SEM for at least triplicate measurement from representative experiments. The mean ± SEM was calculated for all groups and compared by two-tailed paired Students t test or by ANOVA, with the Bonferroni multiple comparisons test for post-hoc analysis. Prism 3.0 software was used for all statistical analysis.
Results
A subset of microRNAs regulated selectively by ΔNp63 target the TGFβ pathway
Expression of the different isoforms of p63 seems to be correlated with the metastatic potential of human cancers including squamous cell carcinoma (SCC) (23-26), prostate cancer (27), bladder cancer (28) and some breast cancers (29). To identify miRs regulated by p63 we ablated endogenous p63 expression by lentiviral RNA interference, followed by global miR expression profiling using the Exiqon Locked Nucleic Acid (LNA) Platform (Figures 1A and 1B). We performed this initial experiment in JHU-029 squamous cell carcinoma (SCC) cells, a human tumor-derived line that expresses high levels of endogenous ΔNp63α, which is the major p63 isoform expressed in both normal basal epithelial cells and the majority of human SCCs (8,30). At 48 hours a significant fraction of miRs identified as reproducibly regulated by p63 were upregulated following p63 knockdown, consistent with the established ability of the major enodogenous ΔNp63α isoform to function as a transcriptional repressor (Figure 1B; the full list of significantly regulated miRs is provided in Supplemental Figure 1A) (8,31).
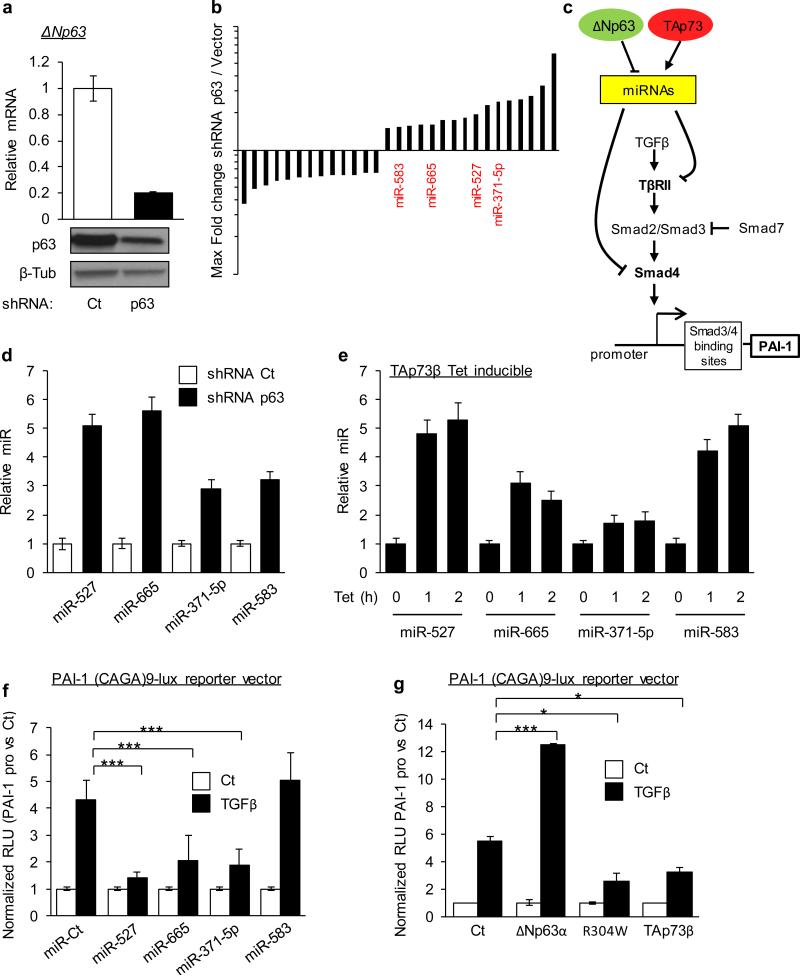
Figure 1. p63-regulated microRNAs target the TGFβ pathway.
(a) Knockdown of endogenous p63 RNA (top) and protein (bottom) by p63-directed or control (Ct) lentiviral shRNA in JHU-029 human SCC cells at 48h. Beta-tubulin (β-Tub) serves as a loading control. (b) Array analysis showing the fold-change and direction of change for all miRs regulated ≥ 1.5-fold in p63-ablated versus control samples shown in (a). miRs predicted to target members of the TGFβ pathway are indicated in red. (c) Graphical abstract of our working hypothesis. (d) Validation of p63 repression of miRs targeting the TGFβ pathway, by real-time quantitative RT-PCR (qRT-PCR) at 72 hours post lentiviral p63-directed or control shRNA expression in JHU-029 cells. (e) Validation of p73 activation of miRs targeting the TGFβ pathway, by qRT-PCR at 1 and 2 hours post tretracycline treatment using a tetracycline-inducible TAp73β JHU-029 human SCC cells. (f) p63/p73 - regulated miRs regulate the TGFβ pathway. A reporter construct containing the SMAD4 biding motif of PAI-1 promoter fused to luciferase was co-transfected with the indicated synthetic miR duplexes. Results show relative luciferase units (RLU) normalized to the control reporter lacking the SMAD4 biding motif. (g) p63/p73 regulates the TGFβ pathway. A reporter construct containing the SMAD4 biding motif of PAI-1 promoter fused to luciferase was co-transfected with the indicated p63 and p73 isoforms. Results show relative luciferase units (RLU) normalized to the control reporter lacking the SMAD4 binding motif. All error bars show s.e.m. for triplicate measurements from representative experiments. * p< 0.05, ** p< 0.01, *** p< 0.001. Two-way ANOVA followed by Tukey's multiple comparison tests were used.
Remarkably, examination of predicted target genes of these p63-regulated miRs using a combination of in silico resources (TargetScan, MicroCosm, DianaLab and miRANDA) revealed that four of the sixteen significantly upregulated miRs (miR-527, miR-665, miR-371-5p and miR-583) were all predicted to have targets in the TGFβ pathway including TGFBR2 and SMAD4 (Figure 1C and Supplemental Figure 1A) (32,33). Because p63 and p73 are known to regulate a common subset of genes, we performed direct validation of p63 and p73-dependent regulation of these four endogenous miRs (Figure 1D and 1E) (8,34). For p63 we used the same lentiviral RNA interference approach employed for the array analysis, then validated directly by qPCR the four miRs as repressed by endogenous ΔNp63. In order to determine whether TAp63 isoforms regulated these endogenous miRs, we used cells expressing tetracycline (tet)-inducible TAp63α; no miR regulation was observed (Supplemental Figure 1B). In order to test p73-dependent miR regulation we performed the same experiment using tet-inducible TAp73β, a major p73 isoform expressed in epithelial cells (35) (ΔNp73 being not expressed in our SCC model, it appeared not relevant to study its implication in this network). Induction of TAp73β mRNA is detectable within one hour following addition of tetracycline in these cells (Supplemental Figure 1C), and induction of each of the four miRs was also observed at this early time point. As a control we tested up-regulation of the direct p73 transcriptional target gene PUMA, which was coincident with p73-dependent miR regulation (Figure 1E and Supplemental Figure 1C). Thus, each of these miRs is selectively regulated by ΔNp63 and TAp73β, but not by TAp63 isoforms.
We then tested the ability of p63/p73 and their regulated miRs to control the TGFβ pathway (Figure 1C), employing the Smad3/4-specific reporter construct (CAGA)9-lux (36). This reporter includes 9 copies of the SMAD3/4 biding motif fused to luciferase. Co-transfection of the reporter with the corresponding synthetic miR duplexes in each case, except for miR-583, inhibited TGFβ-induced luciferase expression relative to the control reporter lacking the SMAD3/4 biding motif. Since the array-based analysis was not validated experimentally for miR-583, this miR will no longer be part of the study (Figure 1F). Consistent with our proposed model for p63/p73-dependent regulation of this pathway (Figure 1C), ectopic ΔNp63α expression, which decreases expression of these endogenous miRs, stimulated the TGF-β induced luciferase reporter expression. This effect was abolished by the introduction of a single point mutation in ΔNp63α, R304W, which abrogates the ability of p63 to bind to DNA, emphasizing the role of direct DNA binding in this regulatory process (Figure 1G)(37). The role of endogenous ΔNp63α in this pathway was shown through knockdown experiments, which led to a decrease in TGFβ-dependent luciferase reporter induction (Supplemental Figure 1D). Finally, ectopic expression of TAp73β, which increases the expression of these endogenous miRs, led to an inhibition of TGFβ-induced luciferase expression compared to control (Figure 1G).
miR-527 and miR-665 repress Smad4 and TβRII, respectively
Two predicted seed binding sequences for miR-665 and one for miR-371-5p are present within the TβRII 3’UTR, and two predicted seed sequences for miR-527 are present within the SMAD4 3’UTR (Supplemental Figure 2A, B). Thus, in order to validate the predicted TGFβ pathway proteins targeted by miR-527, miR-665 and miR-371-5p, we first performed 3’UTR reporter assays. Using a TβRII 3’UTR reporter construct, we validated the miR-665 as a robust repressor of TβRII, and eliminated miR-371-5p from the potential candidates (Supplemental Figure 2C). To address direct SMAD4 and TβRII regulation by miR-527 and miR-665 respectively, we introduced point mutations within these 3’UTR reporter constructs, then co-transfected these constructs with miR mimics (also called pre-miRs) (Supplemental Figure 2A, B). As predicted, in both cases miR-mediated down regulation of the 3’UTR reporter was completely abolished when a reporter containing the mutant UTR was used instead (Figure 2A). Of note, the long size of the SMAD4 3’UTR forced us to divide it in two smaller segments and to make two reporter constructs; this revealed to us that only the second binding site for miR-527 within the SMAD4 3’UTR was active (Figure 2A). To test the endogenous miR regulation, we co-transfected anti-miRs into SCC cells with either the wild type or mutant UTR reporter. Consistent with our model, 527 and 665 anti-miRs mediated up-regulation of the SMAD4 and TβRII 3’UTR constructs, respectively (Supplemental Figure 2D).
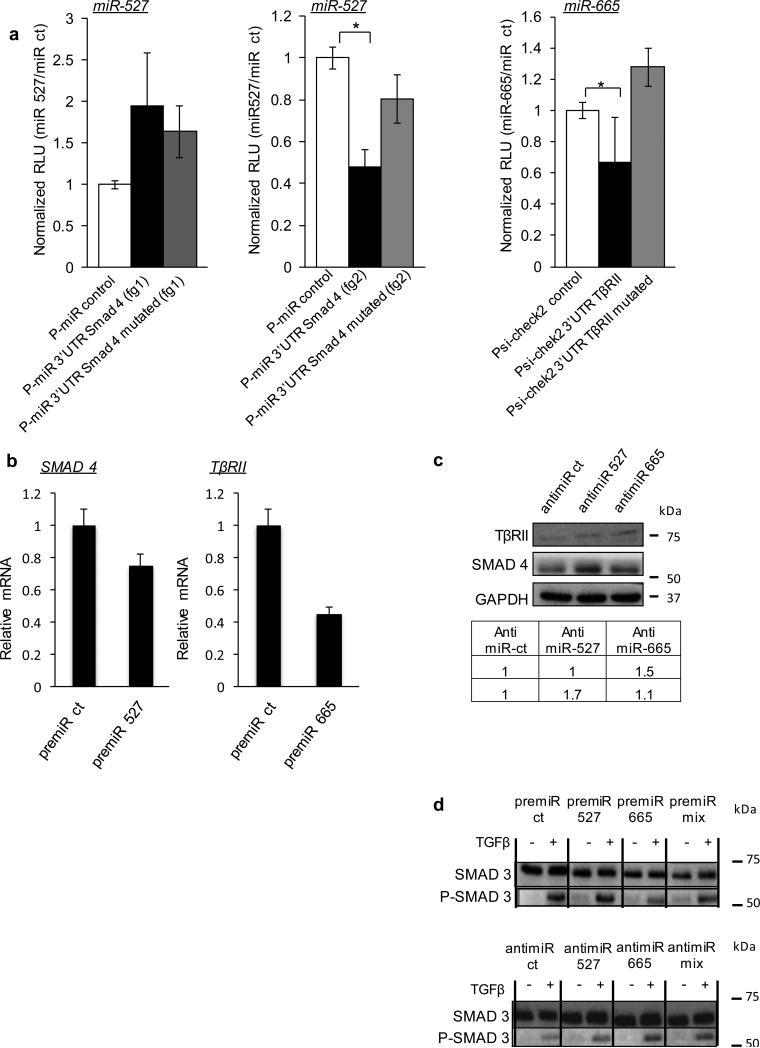
Figure 2. miR-527 and miR-665 repress Smad4 and TβRII, respectively.
(a) miRs 527 and 665 repress gene expression via SMAD4 and TβRII 3′ UTRs, respectively. This repression is lost when mutant seed sequences are introduced. Co-transfection of the indicated miR mimics or control (ct) miR, together with the UTR-reporter, mutated UTR-reporter or control reporter; results show relative luciferase units (RLU) normalized to the control miR. (fg indicates fragment; SMAD4 3’UTR was too large to be cloned in a single construct). One-way ANOVA followed by Dunnett's multiple comparison tests were used, * p< 0.05. (b) qRT–PCR for SMAD4 and TβRII RNA levels in 48h miR-transfected JHU-029 cell line. (c) JHU-029 cells were transfected with anti-miR ct (controls) or anti-miR candidates (anti-miR 527 and anti-miR 665) and SMAD4 and TβRII expression levels were evaluated by western blotting 48h after transfection. Densitometry measurements are indicated. (d) Immunoblotting for P-SMAD3 and SMAD3 from JHU-029 cells transfected with pre/anti miR ct (controls) or pre/anti miRs candidates (pre/anti-miR 527 and pre/anti-miR 665). Cells were treated or not with 5ng/mL TGFβ for 15 minutes. All error bars show s.e.m. for triplicate measurements from representative experiments.
We next tested effects on the endogenous mRNA levels for SMAD4 and TGFBR2, performing ectopic expression of microRNAs with pre-miRs. In addition, we tested the contribution of the endogenous miRs using an anti-miR strategy. As predicted, transfection of the two miR mimics suppressed the endogenous SMAD4 mRNA level (Figure 2B), while expression of specific anti-miRs caused a significant increase in endogenous SMAD4 mRNA for miR-527 and miR-665 but not for miR-371 (Supplemental Figure 2E). For TβRII, only the miR mimic for miR-665 was able to repress endogenous TβRII mRNA level (Figure 2B) and only the anti-miR-665 increased the endogenous TβRII mRNA level (Supplemental Figure 2E). Of note, these experiments confirmed again that miR-371-5p cannot regulate either SMAD4 or TβRII, so we discarded it from our list of potential miRs regulating the TGFβ pathway. Because miRs can also be translational regulators, we next evaluated the effects of the endogenous miRs using anti-miRs followed by western blot. These experiments confirmed miR-527 as a regulator of SMAD4 and miR-665 as a regulator of TβRII at the protein level (Figure 2C). As a validation of our model and of the TGFβ pathway regulation, we performed western blots for the endogenous phosphorylated form of SMAD3 (P-SMAD3) after treatment with TGFβ. Indeed, in control conditions, we observed an increase in P-SMAD3 as a result of TGFβ treatment (Figure 2D). After transfection of the respective pre-miRs and anti-miRs, only those corresponding to miR-665 were able to regulate the phosphorylation of SMAD3 following TGFβ treatment. This finding is expected given that only this miR has a target upstream of SMAD3 in the TGFβ pathway, namely TβRII (Figure 2D). This result has been confirmed in MNNG-HOS cell line (Supplemental Figure 2F).
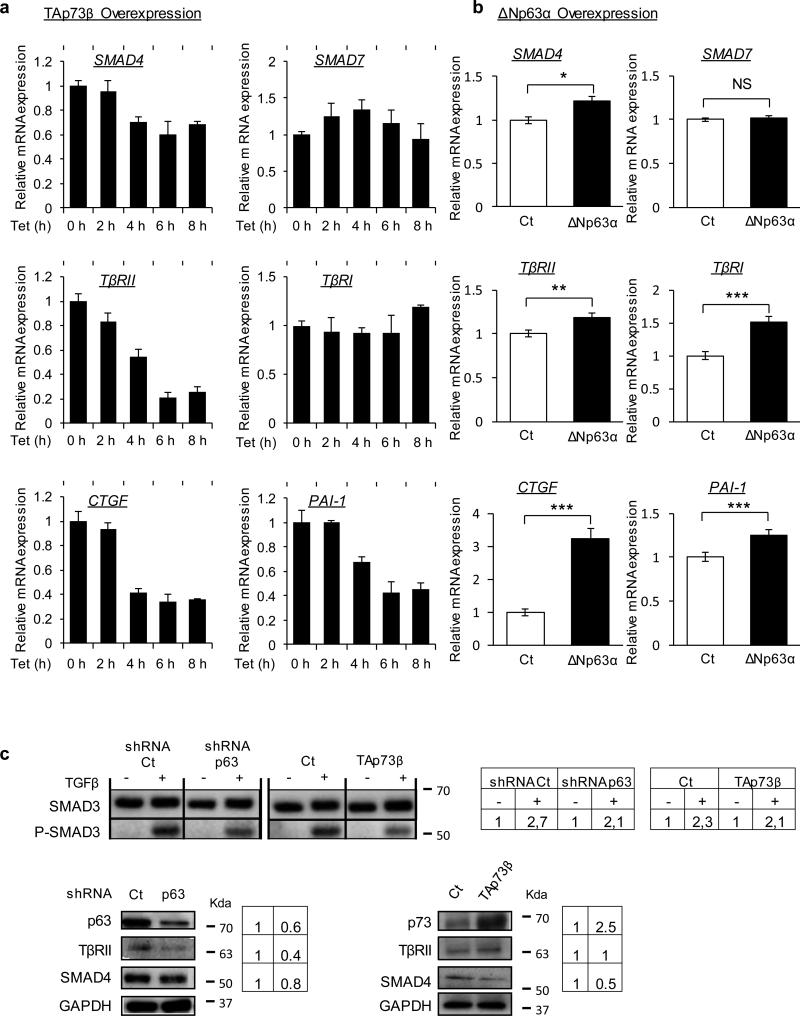
ΔNp63α and TAp73β regulate Smad4 and TβRII
According to our model, if ΔNp63α and TAp73β regulate miR-527 and miR-665 controlling SMAD4 and TβRII, then the modulation of these factors should impact the level of SMAD4 and TβRII and consequently the TGFβ response in the cells. This hypothesis was first tested by qPCR, which showed that SMAD4 but not SMAD7 expression was specifically repressed by ectopic expression of TAp73β or knock-down of endogenous p63 (Figure 3A and Supplemental Figure 3A). The same regulation was observed for TβRII (Figure 3A and Supplemental Figure 3). As predicted by our model, the principal final targets of the pathway, CTGF and PAI-1 were also regulated in the same way (Figure 3A and Supplemental Figure 3). The effect on PAI-1 was also confirmed in the MNNG-HOS cell line (Supplemental Figure 3B). The ectopic expression of ΔNp63α, described as a repressor of our miRs of interest, induced as expected the expression level of SMAD4, TβRII, CTGF and PAI-1 (Figure 3B). Of note, we also observed the induction of TβRI, which is not a predicted target of these miRs but which supports functionally the over-expression of TβRII. Furthermore, the protein levels of SMAD4 and TβRII analyzed by western blot were also consistent with our model. Indeed, ablation of endogenous p63 and p73 overexpression both repressed SMAD4 and TβRII protein levels and also repressed the phosphorylation of SMAD3 following TGFβ treatment (Figure 3C). The effect of ΔNp63α overexpression was then evaluated in the MNNG-HOS osteosarcoma cell line model lacking this protein isoform. As expected, we observed an increase of the SMAD4 and TβRII protein levels and also a strong increase of the phosphorylation of SMAD3 following TGFβ treatment (Supplemental Figure 3C).
Figure 3. p63 and p73 regulate SMAD4 and TβRII.
(a) mRNA expression of indicated key members of the TGFβ pathway analyzed over time by qRT-PCR after induction of TAp73β by a tetracycline inducible system in the JHU-029 cell line. (b) qRT-PCR for the indicated key members of the TGFβ pathway in JHU-029 after 48h of transfection with pcDNA3 (control) or pcDNA3 ΔNp63α. * p< 0.05, ** p< 0.01, *** p< 0.001. Two-way ANOVA followed by Tukey's multiple comparison tests were used. (c) Immunoblotting for SMAD3 and P-SMAD3 in JHU-029 after 48h of transfection with TAp73β and shRNA for p63 with their respective controls. Cells were treated or not with 5ng/mL TGFβ for 15 minutes to show lower activation of the TGFβ pathway. p63, p73, SMAD4 and TβRII protein levels were analyzed in JHU-029 cells after 48h of transient overexpression of TAp73β or knockdown of p63 using shRNA. Densitometry measurements are indicated. All error bars show s.e.m. for triplicate measurements from representative experiments.
The ΔNp63α and TAp73β -regulated miR network controls TGFβ-induced migration
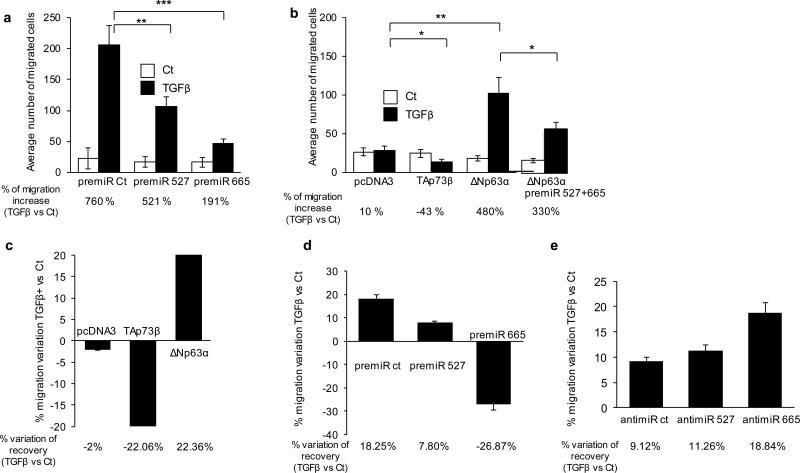
We first tested the functional relevance of our model using multiple independent migration assays. The pro-migration properties of TGFβ were assessed using a boyden chamber test.
We first transfected pre-miRs for our two miRs of interest. Both pre-miRs corresponding to miR-527 and miR-665 decreased migration induced by TGFβ, from 760% to 521% and from 760% to 191%, respectively (Figure 4A) while those pre-miRs do not affect cell viability at all under the same experimental conditions (Supplemental Figure 4A).
Figure 4. The p63 and p73-controlled miR network regulates TGFβ induced migration.
(a) JHU-029 cells were transfected with indicated miR mimics or with control. 24h later cells were treated or not with TGFβ (5ng/mL) and were re-treated before the migration test. (b) JHU-029 cells were transfected with TAp73β, ΔNp63α or pcDNA3 (control). Cells were treated or not with TGFβ (5ng/mL) 24h later and re-treated before the migration test. Migrated cells were counted after 24h of migration in boyden chambers. Migrated cells were counted after 24h of migration in boyden chambers. * p< 0.05, ** p< 0.01, *** p< 0.001. Two-way ANOVA followed by Tukey's multiple comparison tests were used. All error bars show s.e.m. for triplicate measurements from representative experiments. (c) Scratch assay was performed 48h after transfection of JHU-029 cells with TAp73β, ΔNp63α or pcDNA3 (control), prior to a 24h TGFβ treatment (or not) at 5ng/mL. (d, e) The same scratch assay was performed 48h after transfection of the indicated miR mimics/anti-miRs or with controls.
We then performed the same experiment in presence of ΔNp63α and TAp73β. After 24 hours of TGFβ treatment at 5ng/ml concentration, we observed a 10% increase in migration compared to control conditions. As predicted by our molecular analysis, ectopic expression of TAp73β decreased global migration by 43% (Figure 4B). In contrast, the expression of ΔNp63α increased strongly the effects of TGFβ as indicated by an increase in migration from 10 to 480% of the untreated conditions. Interestingly, without TGFβ treatment the expression of ΔNp63α seems to slightly decrease the basal migratory potential of the cells, as previously noted (Figure 4B) (18). Direct evidence showing that ΔNp63α regulates TGFβ signaling via downregulation of miR-527 and miR-665 is provided by the abrogation of the pro-migration potential of ΔNp63α following overexpression of miR-527 and miR-665 Figure 4B). Accordingly, we also observed by Western Blot that overexpressing these miRs abrogated the effects on SMAD4 caused by ΔNp63α expression (Supplemental Figure 4B). Direct evidence showing that TAp73β regulates TGFβ signaling via upregulation of miR-527 and miR-665 is provided by the abrogation of the anti-migration potential of TAp73β following anti-miR-527 and 665 transfection (Supplemental Figure 4C). Finally, we validated the pro-migration potential of TβR2 in the presence of TGFβ and the inhibition of this effect by the miR-665 (Supplemental Figure 4D).
Of note, the difference in the response to TGFβ treatment between the respective controls of Figure 4A and 4B is due to the intentional difference in the cell number plated, which depended on the experimental hypothesis (increased or decreased migration).
The migration potential of the cells in the presence of TGFβ was also tested using an in vitro wound-healing assay, also called a scratch assay. Ectopic expression of TAp73β decreased the cell migration potential by about 22% compared to control, while expression of ΔNp63α increased it by more than 22% (Figure 4C). We then tested the contribution of these miRs directly using pre-miRs and anti-miRs. Overexpression of pre-miR 527 and 665 decreased the cell migration potential by about 10% and 45%, respectively, compared to the anti-miR control, while overexpression of anti-miR 527 and 665 increased it by more than 2% and 10%, respectively (Figure 4D and E 4). Globally, those data support the pro-migratory potential of endogenous p63 and the anti-migratory effect of endogenous miR-527 and miR-665, in a TGFβ-rich micro-environment context.
ΔNp63α induces TGFβ-mediated metastasis in vivo
Because we demonstrated in vitro that ΔNp63α increases cell migration in presence of TGFβ, we wished to verify whether ΔNp63α could increase in vivo the metastatic potential of tumor cells in a physiological context where TGFβ is both present and important. One such in vivo model is osteosarcoma, a primary bone tumor. This type of tumor recruits osteoclasts and thereby induces abnormal bone resorption, which in turn releases growth factors including TGFβ from the bone matrix and stimulates tumor proliferation and potentially metastasis dissemination (38-40). In this way, osteosarcoma cells establish a vicious cycle of bone resorption and tumor growth in which TGFβ has been shown to play a role (38).
To study the influence of ΔNp63α on tumor cell metastatic potential in presence of TGFβ, we used the human osteosarcoma cell line MNNG/HOS. An important characteristic of the MNNG/HOS cells for these experiments is their very low ΔNp63α expression level, providing us the opportunity to directly assess the contribution of increased ΔNp63α through stable ectopic expression of this protein (Figure 5A). We first validated in vitro that repression of miRs 527 and 665 was observed following overexpression of ΔNp63α (Supplemental Figure 5A). Indeed, we observed an increased sensitivity of the p63-expressing MNNG/HOS to the pro-migratory potential of TGFβ (Figure 5B). The same result was observed in an experiment-assessing invasion through matrigel at the bottom of a boyden chamber. In presence of TGFβ, the invasion of the p63-expressing MNNG/HOS increased by 85% whereas in the same conditions, the control cells increased their invasion by only 9% (Figure 5B). The matrix metalloproteinase (MMPs) are proteases, activated by the TGFβ pathway, capable of degrading multiple extracellular matrix proteins. The pro-invasive potential of ΔNp63α in presence of TGFβ could be partially explained by an increased activity of MMP-9 as demonstrated by our zymography experiment (Supplemental Figure 5B). Taken together, these in vitro observations validate our hypothesis that ΔNp63α increases TGFβ-dependent migration and invasion.
An in vivo evaluation of our hypothesis was then performed using a human xenograft model involving MNNG/HOS cells that recapitulates features of the human disease (40). We injected control MNNG/HOS cells or their ΔNp63α-expressing counterparts (1 to 1.5 million cells) paratibially into athymic mice. Importantly, no significant difference in orthotopic tumor growth was observed between control and p63-expressing MNNG/HOS following injection (Figure 5C). At 65 days following injection, the animals were sacrificed, the lungs dissected and the number of metastasic nodules evaluated using an optical microscope. We observed a remarkable increase in the number of lung metastatic nodules in mice injected with ΔNp63α-expressing MNNG/HOS compared to the control counterparts (an average of 4.9 vs 0.25 metastatic nodules/mouse respectively) (Figure 5D). Moreover, the metastatic nodules from ΔNp63α-expressing tumors were not only more numerous but also larger (Figure 5D). Notably, the analysis of the orthotopic tumors at the end of the experiment confirmed that after 65 days the transfected MNNG/HOS were still overexpressing ΔNp63α relative to control cells by 7-fold (Figure 5E). Even more dramatic was the expression of ΔNp63α in the metastatic lesions, which was more than 60-fold compared to the control metastases. This finding suggests strong selection for ΔNp63α expression during the metastatic process. Similarly, a higher level of ΔNp63α was also observed in metastases than in the respective orthotopic tumors (Figure 5F). Moreover, as predicted by our in vitro data, the metastases showed a strong decrease in endogenous miR-527 and miR-665 expression levels (Figure 5F). Finally, in this same group of mice harboring a ΔNp63α-overexpressing tumor, the orthotopic tumors that gave rise to metastases expressed a higher level of both SMAD4 and TβRII compared to orthotopic tumors that did not metastasize (Figure 5G). The protein level of SMAD4 and P-SMAD3 (as a readout of TβR2 activity) were evaluated by immunohistochemistry. We observed in the tumors that gave rise to metastasis both a larger area of cells expressing SMAD4 and the presence of proliferative tumor cell follicles harboring nuclear P-SMAD3 (Supplemental Figure 5C).
We then validated the relevance of this endogenous miR/TGFβ pathway in osteosarcoma patient samples. We obtained from the University of Navarra 6 pairs of osteosarcoma primary tumors and their respective lung metastasis biopsy specimens. qPCR analysis revealed a decreased level of miR-527 and miR-665 in the metastases and a concomitant increase of SMAD4 and TβRII (Supplemental Figure 5D). Collectively, these data support the pro-migratory potential of ΔNp63α through its regulation of the endogenous miR-527/miR-665/Smad4/TβRII network in an in vivo TGFβ-rich microenvironmental context.
miR-527 and miR-665 regulate metastatic dissemination in vivo
We next evaluated directly the metastasis regulation potential of miR-527 and miR-665 in vivo through injection of the respective pre-miR constructs into osteosarcoma orthotopic tumors. As noted, this type of tumor was selected for its metastatic dissemination sensitivity to TGFβ (40). Orthotopic tumors were generated by paratibial injection of 1.5 million MNNG/HOS cells expressing both luciferase and GFP (41). At 200mm3 of tumor volume, mice were treated every 48h with 50nM pre-miR control (miRct-group) or pre-miR-527 (miR-527 group) (Figure 6A). No differences in primary orthotopic tumor growth were observed between miR-527 and miR-665-treated tumors versus the control group (Figure 6B). At the tumor volume endpoint (2500mm3), the average number of lung metastases for miR-527, miR-665 or the control group were evaluated both visually and using bioluminescence imaging of lungs after sacrifice (Figure 6C, D). Mice injected with miR-527 and miR-665 demonstrated a significant reduction of lung metastatic nodules. To credential the mechanism of this effect a gene expression analysis was then performed on biopsies from the control and the miR-injected groups. We first validated the efficiency of the miR injection into the orthotopic tumor. Indeed, qPCR analysis revealed a seven and a two-fold increase in miR expression in the orthotopic tumor from the miR-527 and miR-665-treated group, respectively (Figure 6E-F). In agreement with our model attributing anti-metastatic potential to miR-527 and miR-665, we observed a lower level of miR-527 or miR-665 in the metastatic nodules compared to the orthotopic tumor, and this difference was reflected functionally in a higher level of SMAD4 or TβRII, respectively, in the metastatic lesions (Figure 6E-F). These findings were further confirmed at the protein level through immunohistochemistry for SMAD4 and P-SMAD3 (Supplemental figure 6A). Notably, the few metastases observed in the miR-527 and miR-665 groups express a lower level of SMAD4 and TβRII respectively compared to the miR-ct group, validating their inhibition by miR-527 and miR-665. Correspondingly, immunohistochemistry showed decreased SMAD4 and P-SMAD3 in these specimens (Supplemental Figure 6B).
In keeping with a contribution of this pathway in carcinomas as well as mesenchymal tumors, we found a weak but statistically significant inverse correlation between ΔNp63 and miR-527 in a small cohort of breast tumor specimens (Supplemental figure 6C), and the analysis of a larger cohort from GEO reveled again a weak but statistically significant inverse correlation between miR-527 and SMAD4 (Supplemental figure 6D).
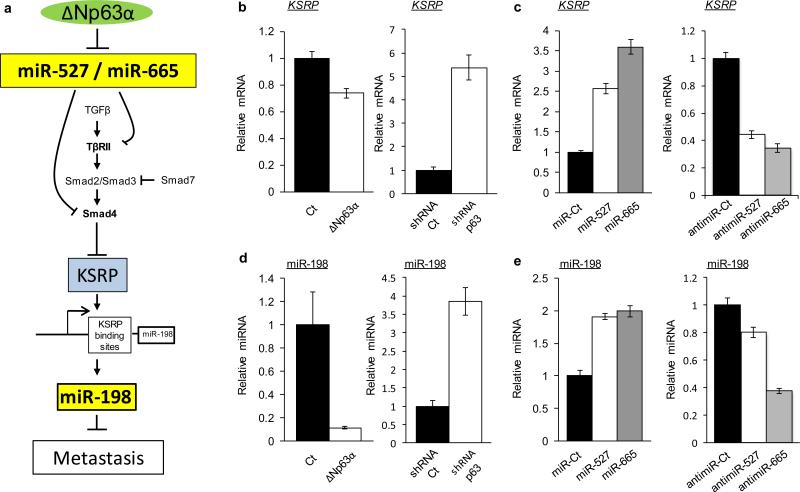
p63, miR-527 and miR-665 regulate a wound healing pathway
Finally, we sought to discover the normal physiological context for the regulatory network we had uncovered. Of immediate interest was wound healing, a biological process involving complex regulation of various pathways including cell migration, proliferation and extracellular matrix remodeling, generally stimulated by TGFβ and recently shown to involve ΔNp63α (42-45). Thus we asked whether the endogenous ΔNp63α/miR network regulating the TGFβ pathway we identified was associated with a newly identified pathway controlling the normal wound healing response. Recently, TGFβ signaling has been described as switching off anti-migratory miR-198 expression by downregulating the RNA-binding protein KSRP (46). Indeed, KSRP promotes the biogenesis of a subset of microRNAs harboring the GUG motif as in miR-198 within its terminal loop (47). Moreover it has been demonstrated that phosphorylated R-SMAD proteins, the transducers of TGFβ signaling, activate the expression of a miR targeting KSRP (46). Supporting the link between our model and the wound healing KSRP/miR-198 pathway (Figure 7A), we noted that in our initial microarray analysis, shRNA-mediated inhibition of endogenous p63 resulted in a nearly 6-fold upregulation of miR-198 (Supplemental Figure 1A). Further supporting this link, we found that the expression of KSRP is repressed by the expression of ΔNp63α and increased following the expression of both miR-527 and miR-665. In keeping with regulation of this wound-healing pathway by the endogenous p63/miR-527/665 circuit, we observed that KSRP is up regulated by repression of endogenous p63, while repression of either endogenous miR-527 or miR-665 with anti-miRs leads to KSRP repression (Figure 7C).
Figure 7. p63, miR-527 and miR-665 regulate a wound-healing pathway.
(a) Schematic representation of the interplay between the p63/miR-527/miR-665 pathway and the wound healing KSRP/miR-198 pathway. (b) KSRP expression level analyzed by qRT-PCR after 48h overexpression of ΔNp63α or shRNA against p63 and their respective controls in HOS cells and 029 cells, respectively. (c) Expression of the pre-miR-527, pre-miR-665, anti-miR-527, anti-miR-665 and their controls. (d) miR-198 expression level analyzed by qRT-PCR after 48h overexpression of ΔNp63α or shRNA against p63 and their respective controls in HOS cells and 029 cells, respectively. (e) Expression of the pre miR-527, pre-miR-665, anti-miR-527, anti-miR-665 and their respective controls. All error bars show s.e.m. for triplicate measurements from representative experiments.
We then validated directly the predicted consequences of these effects on miR-198 expression. As anticipated, miR-198 is repressed by the expression of ΔNp63α and activated after repression of endogenous p63 (Figure 7D). Conversely the overexpression of both miR-527 and miR-665 leads to expression of miR-198, while repression of these endogenous miRs with anti-miRs led to miR-198 repression (Figure 7E). Thus, the p63/miR-527/665 circuit we have discovered is an endogenous regulator of the key wound-healing factors KSRP and miR-198.
Discussion
Metastatic dissemination of cancer is a complex and context-dependent process that is responsible for the vast majority of cancer lethality. Understanding the molecular details of this process will thus be critical for developing more effective therapeutics. Here we demonstrate that ΔNp63α, the major p63 isoform expressed in the epithelium and squamous malignancies including HNSCC, enhances TGFβ-dependent metastatic dissemination. This effect is mediated through direct inhibition of a miR transcriptional network involving miR-527 and miR-665. A key novel feature of our work is demonstrating how ΔNp63α functions to control cellular microenvironmental cues via TGFβ. In contrast to TAp63, which is proposed to be controlled by upstream TGFβ/SMAD activation (19), we reveal a distinct pathway whereby ΔNp63α controls downstream TGFβ/SMAD activity. Supporting a selective role for ΔNp63α in this mechanism, we provide evidence that this pathway is involved in the normal wound healing response, which itself has been linked to ΔNp63α (44,45). Indeed, in the context of our orthotopic model we observed substantially increased ΔNp63α expression in the metastatic lesions relative to the respective primary tumors, and in advanced squamous carcinomas we have shown that ΔNp63α mRNA levels exceed those of TAp63 by up to 200-fold (48). While multiple mechanisms including clonal selection or epigenetic regulation are likely to contribute to selection for ΔNp63α in these distinct contexts, these findings collectively point to ΔNp63α as a driver of tumor progression and metastasis.
Our data suggest that in a TGFβ-rich microenvironment, ΔNp63α is a pro-metastatic transcription factor. This context and isoform-specific effect of ΔNp63α may explain the lack of agreement in the literature on the contribution of this p63 isoform to cancer dissemination. One study showed that ΔNp63α promotes tumor formation and metastasis through the regulation of brachyury expression, an important gene for limb development which is also implicated in both epithelial to mesenchymal transition (EMT) and mesenchymal to epithelial transition (MET)(20). Related to the EMT process, ΔNp63α also promotes the expression of at least one important mesenchymal marker in bladder cancer as well as accelerates EMT in normal human keratinocytes (28). In contrast, other studies have reported p63 and/or ΔNp63α as an anti-metastatic factor, through the upregulation of caspase-1 and CD82 for example (49,50). Additionally, ΔNp63α was shown to negatively regulate Erk signaling for further inhibition of cell migration and invasion in breast cancer (29).
Our study resolves some of these conflicting data by emphasizing the contribution of the tumor microenvironment. This is a parameter not accounted for in many studies in the field, yet so important for understanding the complexity of metastatic dissemination. Including TGFβ in the equation describing the role of p63 in metastatic dissemination helps clarify this role. Accordingly, we performed in vivo validation of our hypothesis in an orthotopic bone tumor model for two essential reasons. First, these cells express little or no ΔNp63α, providing us opportunity to experimentally introduce this protein in order to study its effects. Second and even more importantly, osteosarcoma develops in a bone microenvironment, the largest reservoir for TGFβ (51). When tumor cells reach bony sites, a ‘vicious cycle’ between tumor cells and local bony cells is established resulting in a loss of equilibrium between bone resorption and bone matrix deposition. In the case of osteolytic metastatic lesions, the balance will bend towards the osteoclastic population leading to a high bone resorption. Degradation of bone matrix releases high quantities of cytokines (TGFβ and IGF-1), which further enhance tumor cell proliferation that in turn will cause more osteolysis and liberation of TGFβ, both locally and in the general circulation (40). Thus, our findings demonstrate a general mechanism whereby ΔNp63α-expressing cells, in a TGFβ-rich microenvironment, are positively selected during metastatic dissemination owing to the repression of miR-527 and miR-665 and the subsequent upregulation of two major proteins of the TGFβ pathway: SMAD4 and TβRII. To demonstrate in vivo that the pro metastatic activity of ΔNp63α is mediated by the repression of miR-527 and miR-665, another control in vivo experiment might be performed where a group of mice injected with ΔNp63α overexpressing cells are also treated with miR-527 and miR-665.
Importantly, we provide evidence for the endogenous physiological context in which this ΔNp63α-mediated pathway normally functions. ΔNp63α is an established mediator of epithelial regenerative proliferation that has been, not surprisingly, implicated in the wound healing process (44,45). Additionally, it is well established that wound healing involves complex regulation of similar pathways as metastatic dissemination, including cell migration and extracellular matrix remodeling, generally stimulated by TGFβ (42,43). The physiological relevance of our findings is therefore strongly supported by our demonstration of the connection between the ΔNp63α/miRs network regulating the TGFβ pathway on the one hand, and TGFβ-dependent regulation of wound healing through KSRP/miR-198 on the other. By demonstrating the link between this metastatic network and the normal wound healing response, this study reveals a new mechanism through which deregulation of p63 may contribute to human cancer.
In conclusion, we have uncovered a new microRNAs network regulated by ΔNp63α in the context of a TGFβ-rich microenvironment. This network is involved in the physiological regulation of wound healing, but is hijacked by the tumor to disseminate, making our discovery relevant in both physiological and pathological contexts. Our study opens some potential therapeutic possibilities. For example, miR-527 and miR-665 along with SMAD4 and TβRII expression levels might be used clinically as biomarkers to determine whether or not patients are likely to develop metastasis. Ultimately, understanding such metastatic regulatory networks should also provide the opportunity for effective therapeutic intervention.
Supplementary Material
Acknowledgments
This paper was written as a part of research project which received funding from the Seventh Framework Programme ([FP7/2007-2013]) under grant agreement no264817 – BONE-NET. LWE is funded by RO1 DE-015945. Camille Jacques is funded by INSERM and Région Pays de la loire. Philippe Hulin, Nantes University, PICell for time-lapse microscopy.
Footnotes
Conflict of interest: The authors declare no conflict of interest
Author Contributions
L.R.C, B.O. and L.W.E, designed research aims, designed and performed experiments, analyzed data and wrote the paper., C.J., F.L, M.T.G, M.B, D.S, T.Q, performed experiments and analyzed data. F.Le.,provided tissues. F.V, D.H., contributed to manuscript preparation. B.O. and L.W.E contributed equally to the present work.
References
- 1.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137(3):413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Moll UM, Slade N. p63 and p73: roles in development and tumor formation. Molecular cancer research : MCR. 2004;2(7):371–86. [PubMed] [Google Scholar]
- 3.Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26(36):5169–83. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 4.Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes & development. 2008;22(19):2677–91. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–8. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 6.Leong CO, Vidnovic N, DeYoung MP, Sgroi D, Ellisen LW. The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest. 2007;117(5):1370–80. doi: 10.1172/JCI30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sniezek JC, Matheny KE, Westfall MD, Pietenpol JA. Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope. 2004;114(12):2063–72. doi: 10.1097/01.mlg.0000149437.35855.4b. [DOI] [PubMed] [Google Scholar]
- 8.Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer cell. 2006;9(1):45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 9.Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell death and differentiation. 2006;13(6):1037–47. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- 10.Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, et al. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139(4):772–82. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell stem cell. 2009;5(1):64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ory B, Ramsey MR, Wilson C, Vadysirisack DD, Forster N, Rocco JW, et al. A microRNA-dependent program controls p53-independent survival and chemosensitivity in human and murine squamous cell carcinoma. J Clin Invest. 121(2):809–20. doi: 10.1172/JCI43897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nature cell biology. 2013;15(6):546–54. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481(7380):190–4. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 16.Massague J. TGFbeta in Cancer. Cell. 2008;134(2):215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor MA, Parvani JG, Schiemann WP. The pathophysiology of epithelial-mesenchymal transition induced by transforming growth factor-beta in normal and malignant mammary epithelial cells. J Mammary Gland Biol Neoplasia. 15(2):169–90. doi: 10.1007/s10911-010-9181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 467(7318):986–90. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137(1):87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 20.Cho MS, Chan IL, Flores ER. DeltaNp63 transcriptionally regulates brachyury, a gene with diverse roles in limb development, tumorigenesis and metastasis. Cell Cycle. 9(12):2434–41. doi: 10.4161/cc.9.12.12051. [DOI] [PubMed] [Google Scholar]
- 21.Danilov AV, Neupane D, Nagaraja AS, Feofanova EV, Humphries LA, Direnzo J, et al. DeltaNp63alpha-Mediated Induction of Epidermal Growth Factor Receptor Promotes Pancreatic Cancer Cell Growth and Chemoresistance. PloS one. 2011;6(10):e26815. doi: 10.1371/journal.pone.0026815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh JE, Kim RH, Shin KH, Park NH, Kang MK. {Delta}Np63alpha protein triggers epithelialmesenchymal transition and confers stem cell properties in normal human keratinocytes. J Biol Chem. 286(44):38757–67. doi: 10.1074/jbc.M111.244939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell death and differentiation. 2011;18(9):1487–99. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu X, Coates PJ, Boldrup L, Nylander K. p63 contributes to cell invasion and migration in squamous cell carcinoma of the head and neck. Cancer letters. 2008;263(1):26–34. doi: 10.1016/j.canlet.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Barbieri CE, Tang LJ, Brown KA, Pietenpol JA. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 2006;66(15):7589–97. doi: 10.1158/0008-5472.CAN-06-2020. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Lu H, Yan B, Romano RA, Bian Y, Friedman J, et al. DeltaNp63 versatilely regulates a Broad NF-kappaB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res. 2011;71(10):3688–700. doi: 10.1158/0008-5472.CAN-10-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(38):15312–7. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran MN, Choi W, Wszolek MF, Navai N, Lee IL, Nitti G, et al. The p63 protein isoform DeltaNp63alpha inhibits epithelial-mesenchymal transition in human bladder cancer cells: role of MIR-205. J Biol Chem. 2013;288(5):3275–88. doi: 10.1074/jbc.M112.408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergholz J, Zhang Y, Wu J, Meng L, Walsh EM, Rai A, et al. DeltaNp63alpha regulates Erk signaling via MKP3 to inhibit cancer metastasis. Oncogene. 2014;33(2):212–24. doi: 10.1038/onc.2012.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thurfjell N, Coates PJ, Vojtesek B, Benham-Motlagh P, Eisold M, Nylander K. Endogenous p63 acts as a survival factor for tumour cells of SCCHN origin. International journal of molecular medicine. 2005;16(6):1065–70. [PubMed] [Google Scholar]
- 31.Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Molecular and cellular biology. 2003;23(7):2264–76. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic acids research. 2008;36(Database issue):D149–53. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic acids research. 2008;36(Database issue):D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cellular and molecular life sciences : CMLS. 2004;61(7-8):822–42. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeYoung MP, Johannessen CM, Leong CO, Faquin W, Rocco JW, Ellisen LW. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006;66(19):9362–8. doi: 10.1158/0008-5472.CAN-06-1619. [DOI] [PubMed] [Google Scholar]
- 36.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. The EMBO journal. 1998;17(11):3091–100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Celli J, Duijf P, Hamel BC, Bamshad M, Kramer B, Smits AP, et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99(2):143–53. doi: 10.1016/s0092-8674(00)81646-3. [DOI] [PubMed] [Google Scholar]
- 38.Kozlow W, Guise TA. Breast cancer metastasis to bone: mechanisms of osteolysis and implications for therapy. J Mammary Gland Biol Neoplasia. 2005;10(2):169–80. doi: 10.1007/s10911-005-5399-8. [DOI] [PubMed] [Google Scholar]
- 39.Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22(5):571–84. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamora A, Talbot J, Bougras G, Amiaud J, Leduc M, Chesneau J, et al. Overexpression of Smad7 Blocks Primary Tumor Growth and Lung Metastasis Development in Osteosarcoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014 doi: 10.1158/1078-0432.CCR-13-3191. [DOI] [PubMed] [Google Scholar]
- 41.Rousseau J, Escriou V, Perrot P, Picarda G, Charrier C, Scherman D, et al. Advantages of bioluminescence imaging to follow siRNA or chemotherapeutic treatments in osteosarcoma preclinical models. Cancer gene therapy. 2010;17(6):387–97. doi: 10.1038/cgt.2009.89. [DOI] [PubMed] [Google Scholar]
- 42.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453(7193):314–21. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 43.Hameedaldeen A, Liu J, Batres A, Graves GS, Graves DT. FOXO1, TGF-beta regulation and wound healing. International journal of molecular sciences. 2014;15(9):16257–69. doi: 10.3390/ijms150916257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ichikawa T, Suenaga Y, Koda T, Ozaki T, Nakagawara A. DeltaNp63/BMP-7-dependent expression of matrilin-2 is involved in keratinocyte migration in response to wounding. Biochemical and biophysical research communications. 2008;369(4):994–1000. doi: 10.1016/j.bbrc.2008.02.128. [DOI] [PubMed] [Google Scholar]
- 45.Noszczyk BH, Majewski ST. p63 expression during normal cutaneous wound healing in humans. Plastic and reconstructive surgery. 2001;108(5):1242–7. doi: 10.1097/00006534-200110000-00022. discussion 48-50. [DOI] [PubMed] [Google Scholar]
- 46.Sundaram GM, Common JE, Gopal FE, Srikanta S, Lakshman K, Lunny DP, et al. 'See-saw' expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature. 2013;495(7439):103–6. doi: 10.1038/nature11890. [DOI] [PubMed] [Google Scholar]
- 47.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459(7249):1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramsey MR, Wilson C, Ory B, Rothenberg SM, Faquin W, Mills AA, et al. FGFR2 signaling underlies p63 oncogenic function in squamous cell carcinoma. J Clin Invest. 2013;123(8):3525–38. doi: 10.1172/JCI68899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Celardo I, Grespi F, Antonov A, Bernassola F, Garabadgiu AV, Melino G, et al. Caspase-1 is a novel target of p63 in tumor suppression. Cell death & disease. 2013;4:e645. doi: 10.1038/cddis.2013.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu J, Liang S, Bergholz J, He H, Walsh EM, Zhang Y, et al. DeltaNp63alpha activates CD82 metastasis suppressor to inhibit cancer cell invasion. Cell death & disease. 2014;5:e1280. doi: 10.1038/cddis.2014.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dallas SL, Rosser JL, Mundy GR, Bonewald LF. Proteolysis of latent transforming growth factor-beta (TGF-beta )-binding protein-1 by osteoclasts. A cellular mechanism for release of TGF-beta from bone matrix. The Journal of biological chemistry. 2002;277(24):21352–60. doi: 10.1074/jbc.M111663200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.