Summary
Anaplasma phagocytophilum is an emerging human pathogen and obligate intracellular bacterium. It inhabits a host cell-derived vacuole and cycles between replicative reticulate cell (RC) and infectious dense-cored (DC) morphotypes. Host–pathogen interactions that are critical for RC-to-DC conversion are undefined. We previously reported that A. phagocytophilum recruits green fluorescent protein (GFP)-tagged Rab10, a GTPase that directs exocytic traffic from the sphingolipid-rich trans-Golgi network (TGN) to its vacuole in a guanine nucleotide-independent manner. Here, we demonstrate that endogenous Rab10-positive TGN vesicles are not only routed to but also delivered into the A. phagocytophilum-occupied vacuole (ApV). Consistent with this finding, A. phagocytophilum incorporates sphingolipids while intracellular and retains them when naturally released from host cells. TGN vesicle delivery into the ApV is Rab10 dependent, up-regulates expression of the DC-specific marker, APH1235, and is critical for the production of infectious progeny. The A. phagocytophilum surface protein, uridine monophosphate kinase, was identified as a guanine nucleotide-independent, Rab10-specific ligand. These data delineate why Rab10 is important for the A. phagocytophilum infection cycle and expand the understanding of the benefits that exploiting host cell membrane traffic affords intracellular bacterial pathogens.
Introduction
Anaplasma phagocytophilum is a tick-transmitted, obligatory intracellular bacterium that proliferates in membrane-bound inclusions of granulocytes and bone marrow progenitor cells. A. phagocytophilum infection in humans, termed human granulocytic anaplasmosis (HGA), is an emerging zoonosis in the USA, Europe and Asia (reviewed by Truchan et al., 2013). HGA presents as an acute febrile illness characterized by chills, headache, malaise, leucopenia, thrombocytopenia and elevated hepatic aminotransferases. More severe complications may include rhabdomyolysis, haemorrhage, shock, seizures, pneumonitis, increased susceptibility to secondary infections and death (Truchan et al., 2013). Promyelocytic HL-60, RF/6A endothelial and human embryonic kidney (HEK-293T) cells are useful in vitro models for studying A. phagocytophilum infection (Klein et al., 1997; Munderloh et al., 2004; Beyer et al., 2014). The bacterium undergoes a biphasic developmental cycle in which an infectious dense-cored (DC) organism invades to reside within a host cell-derived vacuole. Between 4 and 8 h post-infection, the DC develops into the non-infectious reticulate cell (RC) form, which replicates to yield a bacteria-filled inclusion called the A. phagocytophilum-occupied vacuole (ApV). From 8 to 20 h, the intravacuolar population consists exclusively of replicating RC bacteria. Conversion back to the DC form occurs between 28 and 32 h. DC organisms exit host cells between 28 and 36 h to initiate the next round of infection (Troese and Carlyon, 2009). Host–pathogen interactions that are critical for RC-to-DC transition are unknown.
Microbial acquisition of host nutrients is essential for the development of infectious disease. This is especially important for obligate intracellular pathogens whose reduced genomes render them auxotrophic for many metabolites and imposes a reliance on host cell nutrients. A. phagocytophilum satisfies this need, at least in part, by redirecting vesicular traffic to the ApV (Xiong et al., 2009; Huang et al., 2010a; Niu et al., 2012; Xiong and Rikihisa, 2012). Rab GTPases regulate membrane dynamics and route vesicular traffic in eukaryotic cells. Rabs cycle between two distinct states: a cytosolic, inactive guanosine diphosphate (GDP)-bound form and a membrane-associated, active guanosine triphosphate (GTP)-bound form that directs vesicular trafficking and membrane fusion events (Stenmark, 2009). We previously demonstrated that several green fluorescent protein (GFP)-tagged Rabs are specifically recruited to the ApV. Of these, GFP-Rab10 localizes to the ApV in an atypical, guanine nucleotide-independent manner, suggesting that the bacterium actively targets Rab10-positive vesicles (Huang et al., 2010a). Rab10 directs exocytic traffic from the trans-Golgi network (TGN) (Liu and Storrie, 2012). The significance of Rab10 to A. phagocytophilum pathobiology is unknown.
Herein, we provide evidence that Rab10-positive TGN vesicles are routed to and delivered into the ApV in a Rab10-dependent manner. In agreement with this observation, lipidomic analysis revealed that the pathogen incorporates sphingolipids. Rab10-dependent TGN vesicle import into the ApV is critical for bacterial transcriptional up-regulation of a marker of RC-to-DC transition and infectious progeny generation. The A. phagocytophilum surface protein, uridine monophosphate kinase (UMPK), was identified as a guanine nucleotide-independent Rab10-specific ligand. Overall, this report demonstrates that Rab10 is important for A. phagocytophilum TGN parasitism and completion of the pathogen’s biphasic infection cycle and advances understanding of the diverse ways by which intracellular bacteria exploit Rab GTPases and membrane traffic.
Results
Endogenous and ectopically expressed Rab10 localize to the A. phagocytophilum-occupied vacuolar membrane and with intravacuolar A. phagocytophilum organisms
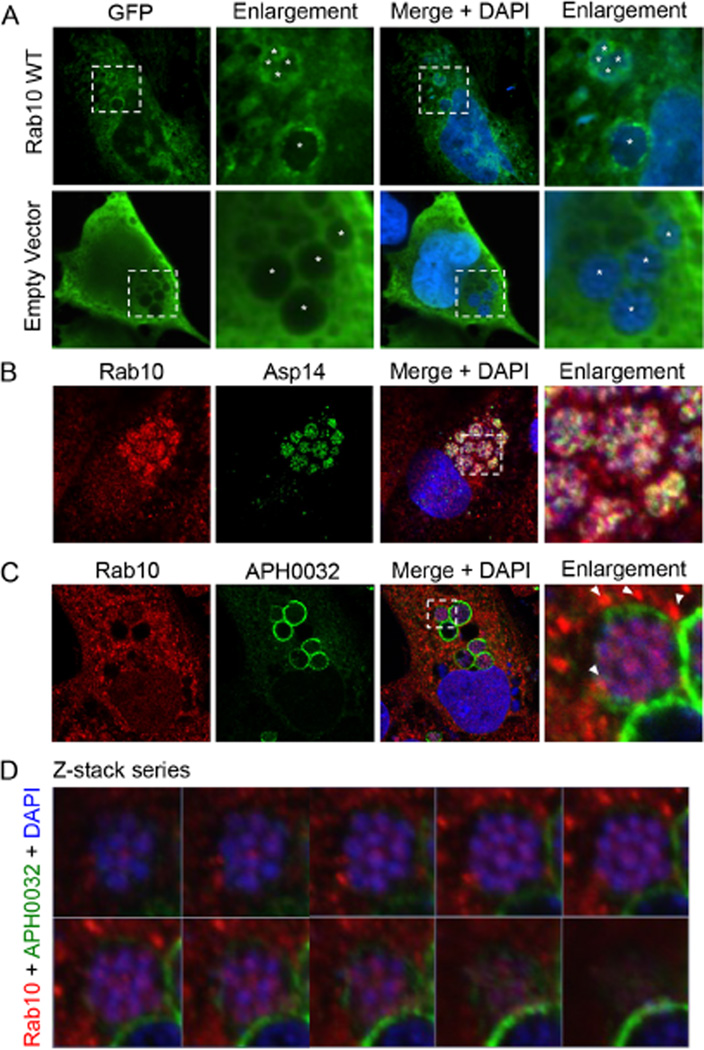
To determine if GFP-Rab10 localizes to the ApV in a host cell type besides myeloid cells (Huang et al., 2010a), we assessed for the phenomenon in infected RF/6A endothelial cells using laser scanning confocal microscopy (LSCM). RF/6A cells were also selected for this purpose because, in addition to being permissive for A. phagocytophilum infection, they are large and flat, which makes them ideal for imaging the ApV (Munderloh et al., 2004; Sukumaran et al., 2011; Beyer et al., 2014). Similar to that previously observed for infected HL-60 cells (Huang et al., 2010a), GFP-Rab10-positive vesicles surrounded ApVs (Fig. 1A). Additionally, GFP-Rab10 was observed within ApVs encircling A. phagocytophilum organisms. GFP alone remained diffuse in the cytosol of infected cells. Given these observations for GFP-Rab10, we hypothesized that endogenous Rab10-positive vesicles are delivered into the ApV. Accordingly, we screened infected RF/6A cells with antibodies against endogenous Rab10 and the bacterial outer membrane protein (OMP), Asp14 (14 kDa A. phagocytophilum surface protein) (Kahlon et al., 2013) or APH0032, which is a bacterial effector that is pronouncedly expressed late (24 to 32 h) during the infection cycle and localizes to the A. phagocytophilum occupied vacuolar membrane (AVM) (Huang et al., 2010b). In addition to producing a vesicular labelling pattern in the cytosol and at the peripheries of ApVs, endogenous Rab10 signal surrounded individual intravacuolar bacteria in ring-like labelling patterns and partially colocalized with Asp14 signal (Fig. 1B and C). Z-stack analysis and three-dimensional (3D) rendering of a representative ApV confirmed the presence of Rab10-positive vesicles within the vacuole associated with intravacuolar bacteria (Fig. 1D and Movie S1). Thus, Rab10-positive vesicles are recruited to the ApV and delivered into its lumen where they associate with A. phagocytophilum organisms. Notably, a minor portion of APH0032-positive ApVs lacked lumenal Rab10 signal, as exemplified by the ApV in the lower-right corners of the images presented in Fig. 1C and D. This observation suggests that delivery of Rab10-positive vesicles into the ApV may be a temporal event that occurs during the period of APH0032 expression.
Fig. 1.
Endogenous and ectopically expressed Rab10 localize to the AVM and with intravacuolar A. phagocytophilum organisms.
A. GFP-Rab10 localizes to the AVM and with intravacuolar bacteria. RF/6A cells expressing GFP-Rab10 were infected with A. phagocytophilum and visualized by LSCM. Asterisks denote bacterial vacuoles.
B. Endogenous Rab10 localizes with A. phagocytophilum surface protein Asp14. A. phagocytophilum-infected RF/6A cells that had been screened with antibodies against Rab10 and Asp14 were examined by confocal microscopy.
C, D. Z-stack series and 3D rendering show endogenous Rab10 associated with A. phagocytophilum organisms within the ApV. A. phagocytophilum-infected RF/6A cells were screened with antibodies against Rab10 and AVM protein APH0032 and visualized by confocal microscopy. (C) Representative infected host cell that was used for Z-stack analysis. Note the localization of punctate Rab10 labelling at the AVM (arrow heads) and within the ApV associated with A. phagocytophilum organisms. (D) Z-stack series showing individual, successive focal planes. (A–C) The regions in the merge + DAPI panels that are demarcated by hatched-line boxes indicate the regions that are magnified in the enlargement panels. The enlargement panel in C was analysed by Z-stack in (D) (A–D) Host cell nuclei and bacterial DNA were stained with DAPI (blue). Results shown are representative of three experiments with similar results.
The ApV localizes adjacent to the Golgi apparatus and TGN-derived vesicles are delivered into its lumen where they associate with A. phagocytophilum organisms
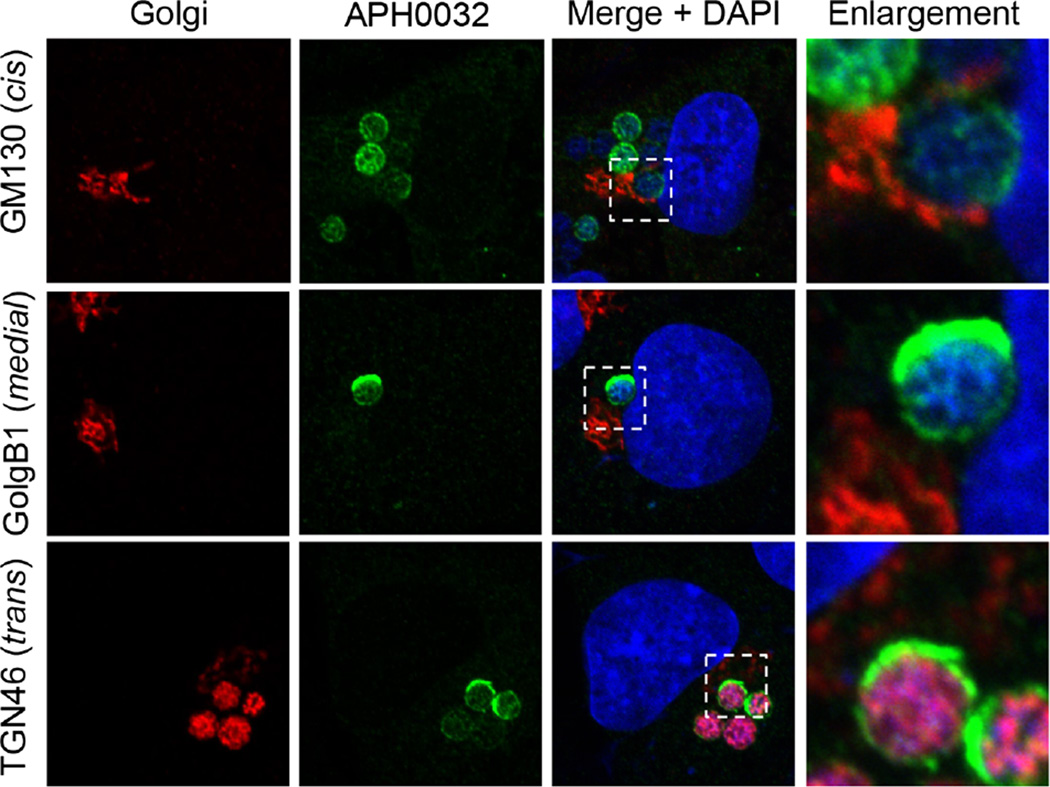
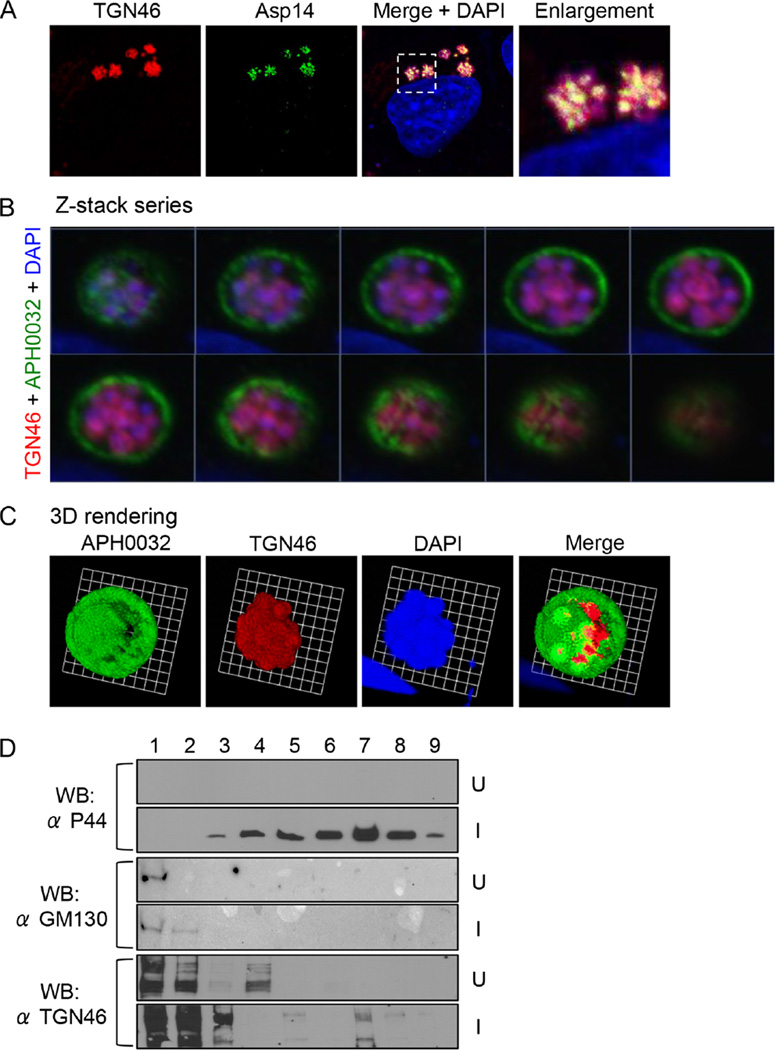
Given that Rab10 directs exocytic traffic from the TGN, we investigated if the ApV associates with the Golgi apparatus and if the Rab10-positive vesicles delivered into its lumen are TGN derived. Screening A. phagocytophilum infected RF/6A cells with antibodies against markers for the cis-Golgi (GM130 and GolgB1), medial-Golgi (GolgB1) and trans-Golgi (TGN46) revealed ApVs closely apposed to and in many instances wrapped by the Golgi (Fig. 2). Signal for TGN46 but not GM130 or GolgB1 was detected within ApV lumens, indicating specific delivery of TGN-derived vesicles or fragments. TGN46 labelling inside ApVs partially colocalized with Asp14 signal (Fig. 3A). Z-stack imaging and 3D rendering of a representative ApV further verified that the presence of TGN46-positive vesicles closely associated with intravacuolar A. phagocytophilum organisms (Fig. 3B and C and Movie S2) and was highly similar to the observed proximal association of Rab10-positive vesicles near the surfaces of the bacteria (Fig. 1B–D and Movie S1). As a complementary approach, organelles from uninfected and A. phagocytophilum-infected HL-60 cells were fractionated using a density gradient centrifugation method that keeps the ApV intact (Niu et al., 2012). Immunoblot analysis revealed altered distribution of TGN46 in the fractions of infected versus uninfected cells (Fig. 3D). Specifically, TGN46 co-migrated to the fraction in which the A. phagocytophilum OMP, P44 (Truchan et al., 2013), was most abundant.
Fig. 2.
The ApV localizes adjacent to the Golgi apparatus, and TGN-derived vesicles are delivered into its lumen. Anaplasma phagocytophilum infected RF/6A cells were screened with antibodies against AVM marker, APH0032, and GM130 (cis-Golgi), GolgB1 (cis-Golgi and medial-Golgi) or TGN46 (trans-Golgi) and examined using LSCM. Results shown are representative of three experiments with similar results.
Fig. 3.
The ApV localizes adjacent to the Golgi apparatus, and TGN-derived vesicles are delivered into the ApV lumen where they associate with A. phagocytophilum organisms.
A. TGN46 signal colocalizes with intravacuolar bacteria. A. phagocytophilum-infected cells were screened with antibodies against TGN46 and Asp14 and examined by LSCM. The region that is demarcated by a hatched-line box is magnified in the enlargement panel.
B. Z-stack series shows TGN46 within the ApV. A. phagocytophilum-infected RF/6A cells were screened with antibodies against TGN46 and APH0032. Successive focal planes of a representative A. phagocytophilum vacuole are presented.
C. 3D rendering of the Z-stack series presented in B shows TGN46-positive vesicles encasing individual A. phagocytophilum organisms within the vacuole. These 3D images are also presented in Movie S2.
D. TGN46 cofractionates with A. phagocytophilum organisms. Uninfected (U) or A. phagocytophilum-infected HL-60 cells (I) were homogenized, and the post-nuclear supernatants were subjected to density gradient centrifugation. Successive 1 ml aliquots were analysed by Western blot (WB) using antibodies against TGN46, GM130 and A. phagocytophilum major surface protein, P44. Results shown are representative of three experiments with similar results.
Given the abundance of TGN vesicles/fragments delivered into the ApV, we examined Golgi integrity during A. phagocytophilum infection. ApVs were demarcated using an antibody against the cytoskeletal protein, vimentin, which localizes to the AVM (Sukumaran et al., 2011). In uninfected host cells and host cells with low A. phagocytophilum loads (one to three ApVs per cell), the Golgi remained intact, and intravacuolar bacteria were TGN46 positive (Fig. S1A and B). However, bacterial load-dependent fragmentation of the Golgi was observed for cells having medium (4–10 ApVs per cell) and high bacterial loads (≥ 11 ApVs per cell) (Fig. S1B). Additionally, even though the cis-Golgi, medial-Golgi and trans-Golgi were each fragmented and dispersed throughout host cells with high bacterial loads, only a TGN46 signal was detected within ApVs (Fig. S1B and C). This finding reinforces that A. phagocytophilum selectively targets TGN-derived exocytic traffic. Furthermore, in cells with high A. phagocytophilum loads, not all of the ApV lumens were visually TGN46 positive (Fig. S1B), which is likely due to the bacterial demand for TGN vesicles exceeding the supply. Collectively, these data support the findings that the ApV interacts with the Golgi apparatus and intercepts TGN-derived traffic and that TGN-derived vesicles or fragments are delivered into the ApV lumen.
Structured illumination microscopic analyses confirm that Rab10-positive and TGN46-positive vesicles are present in the ApV associated with intravacuolar A. phagocytophilum organisms
The partial colocalization of Rab10 and TGN46 signals with Asp14-associated and 4′,6-diamidino-2-phenylindole (DAPI)-associated signals of A. phagocytophilum observed using LSCM suggested that Rab10-positive trans-Golgi-derived vesicles are present within the ApV lumen in close proximity to intravacuolar A. phagocytophilum bacteria. LSCM has a lateral resolution of approximately 200 nm (Allen et al., 2014). Within cells, and presumably within the ApV, macro-molecular associations occur in spatial distances less than 200 nm. Therefore, structured illumination microscopy, a form of super-resolution microscopy, which has a lateral resolution of approximately 100 nm (Allen et al., 2014), was employed to more accurately gauge whether Rab10 and TGN46 signals were associated with intravacuolar A. phagocytophilum organisms. Supporting the phenomena observed via LSCM, SIM imaging revealed the Rab10 signal to exhibit a vesicle-like pattern that was diffuse in the host cell cytosol and a more clustered pattern that partially localized with Asp14-positive and DAPI-positive A. phagocytophilum organisms (Fig. 4A). Likewise, the TGN46 signal pronouncedly colocalized with bacterial-associated Asp14 and DAPI signals (Fig. 4B). These data indicate that Rab10 and TGN46 are within 100 nm of the surfaces of A. phagocytophilum organisms.
Fig. 4.
Structured illumination microscopic analyses confirm that Rab10-positive and TGN46-positive vesicles are present in the ApV proximally associated with intravacuolar A. phagocytophilum organisms.
A, B. A. phagocytophilum-infected RF/6A cells that had been screened with antibodies against (A) Rab10 and Asp14 or (B) TGN46 and Asp14 were examined by SIM. The regions in the merge + DAPI panels that are demarcated by hatched-line boxes indicate the regions that are magnified in the enlargement panels. Host cell nuclei and bacterial DNA were stained with DAPI (blue). Results shown are representative of three experiments with similar results. Nuc, host cell nucleus.
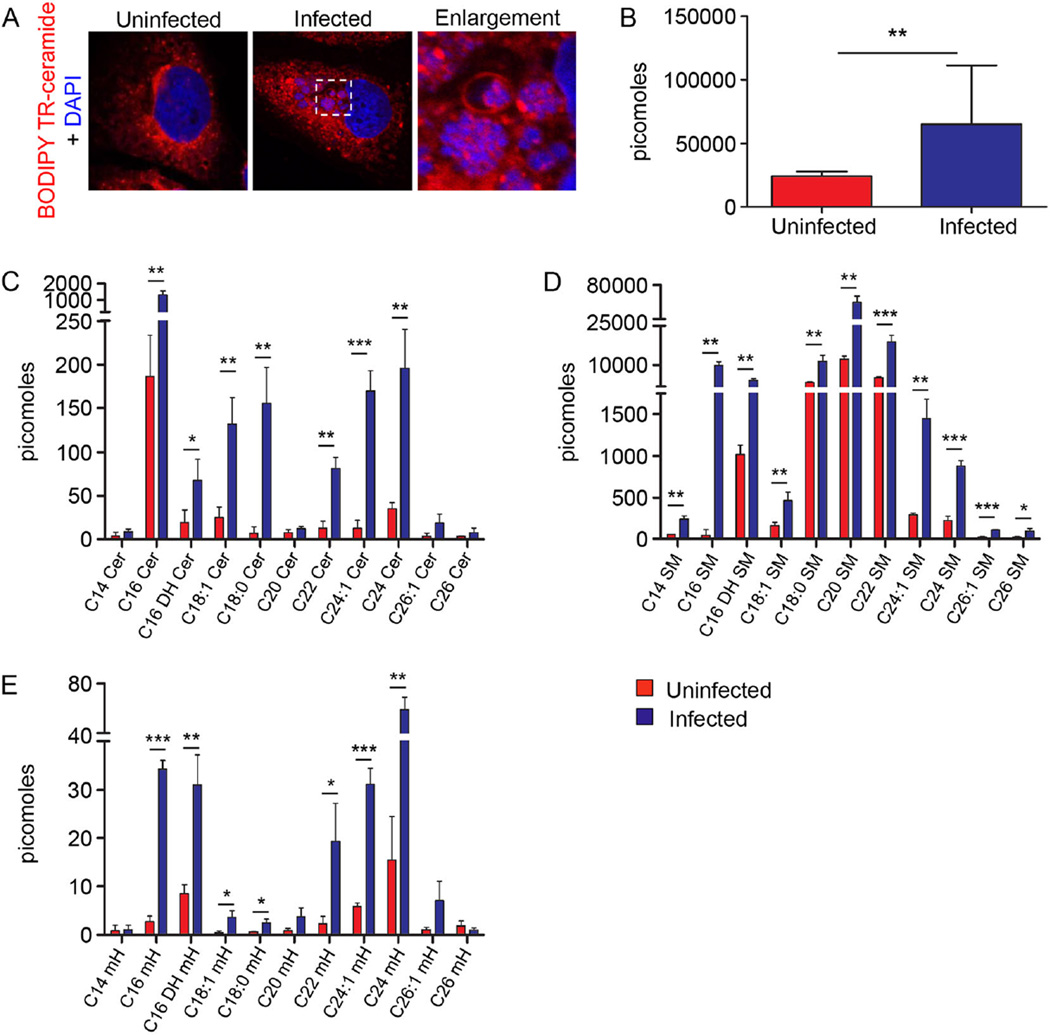
Sphingolipids are delivered into the ApV and incorporated by A. phagocytophilum
Anaplasma phagocytophilum lacks genes that are necessary for sphingolipid synthesis (Dunning Hotopp et al., 2006). Given that the TGN is enriched in sphingolipids (van Meer, 1998) and TGN vesicles accumulate in the ApV, we examined for the delivery of sphingolipids into the ApV lumen using BODIPY Texas Red (TR) ceramide. This fluorescently labelled lipid accumulates in the Golgi, where it is modified into fluorescently labelled sphingolipids that subsequently traffic to the plasma membrane via TGN-derived secretory vesicles (Pagano et al., 1991). Infected host cells were incubated with BODIPY TR ceramide and examined using LSCM. BODIPY TR ceramide-positive vesicles were observed surrounding ApVs and within ApV lumen associating with bacteria in ring-like labelling patterns (Fig. 5A). Because the infection was asynchronous and all A. phagocytophilum bacteria were TR positive, we inferred that BODIPY TR-derived sphingolipids associated with both RC and DC forms. Furthermore, ultra performance liquid chromatography–electrospray ionization–tandem mass spectrometry (UPLC-ESI-MS/MS) analyses detected the presence of multiple ceramide, sphingomyelin and monohexyl sphingolipid subspecies in A. phagocytophilum DC organisms that had been naturally released from infected HL-60 cells, but not in media from uninfected control HL-60 cells (Fig. 5B–E). These results substantiate that host cell sphingolipids that are typically found in the TGN are delivered into the ApV lumen and are incorporated by A. phagocytophilum.
Fig. 5.
Sphingolipids are delivered into the ApV and incorporated by A. phagocytophilum.
A. BODIPY TR-labelled sphingolipids localize with intravacuolar A. phagocytophilum organisms. BODIPY TR ceramide was added to uninfected and A. phagocytophilum-infected cells and examined by LSCM. Host cell nuclei and bacterial DNA were stained with DAPI (blue). Results are representative of three experiments.
B–E. A. phagocytophilum incorporates host cell sphingolipids. Host cell-free A. phagocytophilum DC organisms recovered from the media of infected HL-60 cells were subjected to UPLC-ESI-MS/MS to detect total sphingolipids (B), ceramides (Cer) (C), sphingomyelins (SM) (D) and monohexosyl sphingolipids (mH) (E). Media from uninfected HL-60 cells was processed and examined in parallel. Results are the mean ± standard deviation of triplicate samples and are representative of three biological replicates with similar results. Statistically significant (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001) values are indicated.
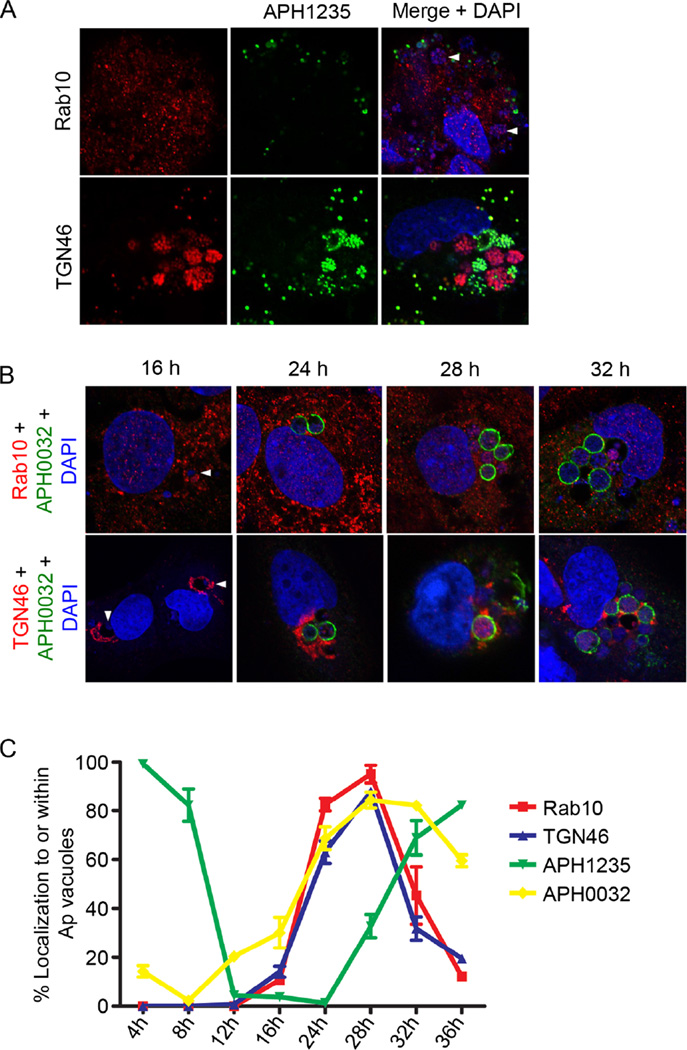
Anaplasma phagocytophilum acquisition of Rab10-positive TGN vesicles precedes conversion to the infectious DC form
To better understand the relevance of Rab10-positive TGN vesicle delivery to A. phagocytophilum intracellular development, we examined if the vesicles specifically associate with the DC or RC form. Infected RF/6A cells were screened with Rab10 or TGN46 antibody together with an antibody against the A. phagocytophilum DC-specific protein, APH1235 (Troese et al., 2011; Mastronunzio et al., 2012). An RC-specific marker has yet to be identified. As previously reported (Troese et al., 2011; Mastronunzio et al., 2012), APH1235-positive A. phagocytophilum organisms were detected bound to host cell surfaces and as intracellular clusters, presumably within vacuoles (Fig. 6A). Strikingly, Rab10 and TGN46 were nearly undetectable on APH1235-positive bacteria. This observation indirectly suggests that RC but not DC organisms predominantly associate with TGN vesicles.
Fig. 6.
A. phagocytophilum acquisition of Rab10-positive TGN vesicles precedes conversion to the APH1235-positive infectious DC form.
A. Rab10 and TGN46 colocalize with A. phagocytophilum organisms during the replicative phase of bacterial development. A. phagocytophilum infected RF/6A cells were screened with antibodies against Rab10 or TGN46 and the A. phagocytophilum DC marker, APH1235.
B, C. RF/6A cells were synchronously infected with A. phagocytophilum, stained with DAPI, screened with antibodies against A. phagocytophilum Rab10, TGN46, APH1235 and APH0032 and examined by confocal microscopy at 4, 8, 12, 16, 24, 28 and 32 h post-infection. Results shown are representative of three experiments. (B) Representative confocal micrograph images of A. phagocytophilum-infected cells at 16, 24, 28 and 32 h screened with APH0032 and either Rab10 or TGN46 antibodies. Data are representative of three experiments with similar results.
A and B. Arrowheads denote ApVs in (A) and (B) because the antibody against the AVM marker, APH0032, was not used in (A) and A. phagocytophilum does not abundantly express APH0032 at 16 h. (C) Acquisition of TGN46 and Rab10 by A. phagocytophilum precedes bacterial expression of APH1235. Percentages of ApVs that were positive for Rab10, TGN46, APH135 and APH0032 over the time course are presented. Data are the mean (± standard deviation) of triplicate samples and are representative of two experiments with similar results.
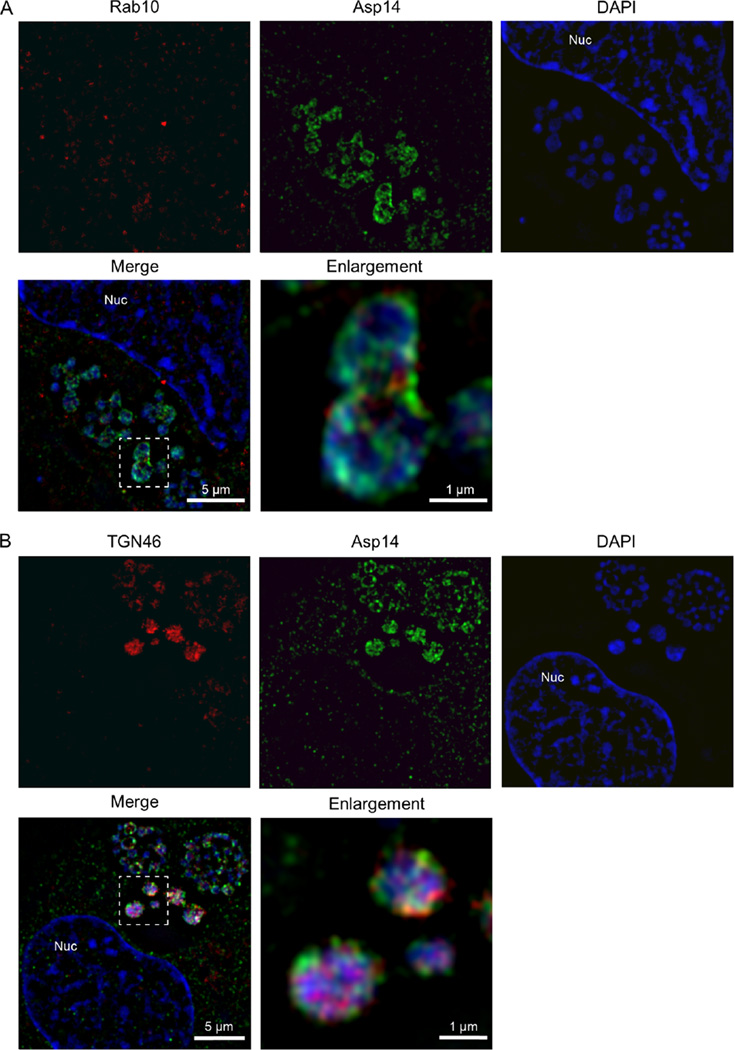
To further investigate this phenomenon, we assessed Rab10 and TGN46 localization within infected cells by LSCM over a 36 h synchronized infection time course. We used LSCM to examine APH0032 and APH1235 expression kinetics to verify that the infection cycle proceeded as expected. APH0032 expression/AVM localization was examined for the additional reason that one of our initial experiments suggested that Rab10 vesicle import into the ApV is a temporal event that occurs coincident with APH0032 expression (Fig. 1C and D). Consistent with previous observations (Huang et al., 2010b), the percentages of APH0032-positive AVMs pronouncedly rose from 8 to 28 h (Fig. 6B and C). Also as previously reported (Troese et al., 2011; Mastronunzio et al., 2012), the percentages of A. phagocytophilum organisms expressing APH1235 dropped from 100% at 4 h to nearly 0% between 12 and 24 h and then pronouncedly increased again to more than 80% by 36 h (Fig. 6C). The localization of TGN46 and Rab10 around and within ApVs occurred synchronously. Rab10-positive vesicles and TGN vesicles were detected surrounding ApVs beginning at 16 h (Fig. 6B and C). Both markers were observed within ApVs beginning at 24 h and peaked at 28 h, a time point at which nearly 100% of ApVs were Rab10 and TGN46 positive. After 28 h, intravacuolar Rab10 and TGN46 signals waned considerably. Notably, the time points at which Rab10 and TGN46 signals associated with the ApV and A. phagocytophilum organisms preceded those when the bacteria were APH1235 positive. The coincident and temporal associations of Rab10 and TGN46 signals with A. phagocytophilum organisms indirectly suggested that Rab10 and TGN46 antibodies did not non-specifically label the bacteria. To verify that this was the case, host cell-free RC and DC organisms were screened via LSCM. Both RCs and DCs were stained by DAPI and labelled by P44 antibody (Fig. S2). Rab10 and TGN46 antibodies labelled only the RC morphotype. Collectively, these data demonstrate that delivery of Rab10-positive TGN vesicles into the ApV and their association with intravacuolar bacteria precedes A. phagocytophilum conversion from the replicative RC form to the infectious DC form.
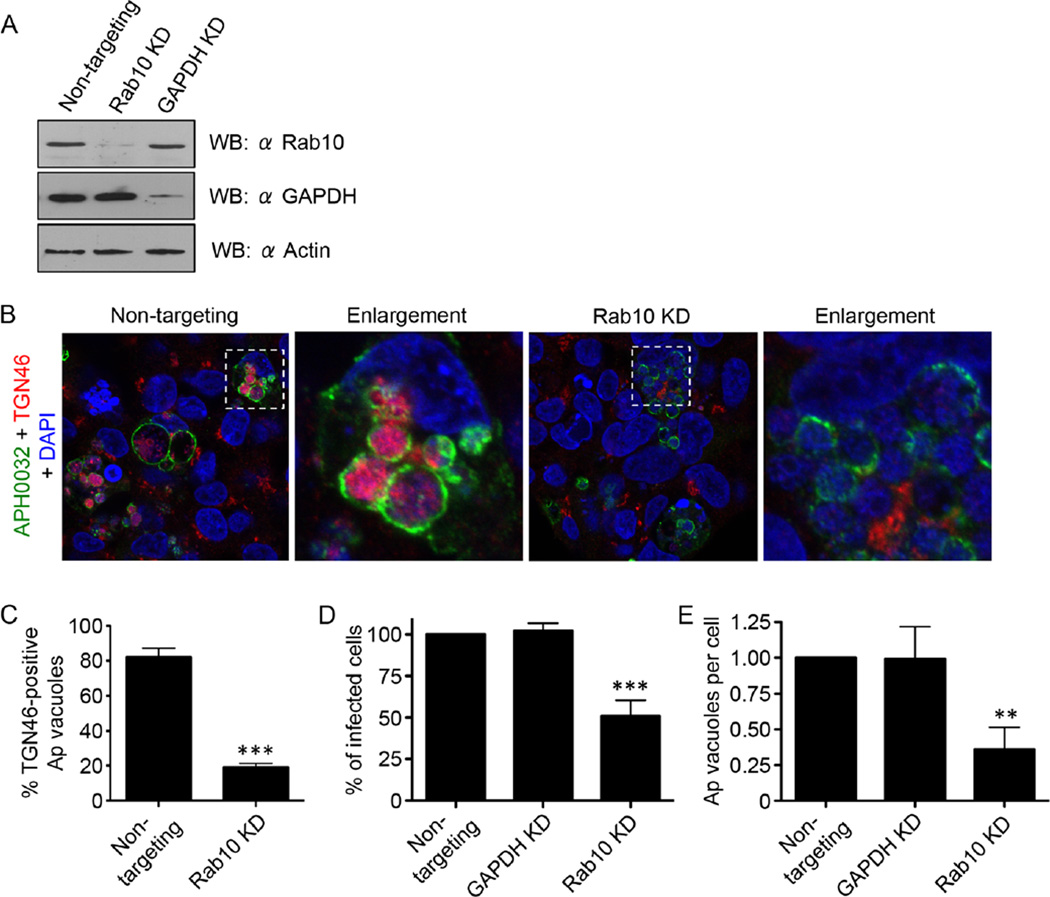
Knockdown of Rab10 markedly reduces both TGN vesicle delivery into the ApV and infection
To define the significance of Rab10 to A. phagocytophilum TGN parasitism, we assayed for TGN vesicle recruitment to and delivery into the ApV in infected host cells in which Rab10 had been knocked down. For this experiment, we used HEK-293T cells because they are susceptible to A. phagocytophilum infection and have a high transfection efficiency (≥ 75%) relative to RF/6A and HL-60 cells (<20%) (Beyer et al., 2014). Furthermore, GFPR-ab10 and TGN46 localize to and within the ApV, respectively, in these cells (Fig. S3A and B). HEK-293T cells were treated with small interfering RNA (siRNA) targeting Rab10, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or non-targeting siRNA. Knockdown of target protein expression was confirmed by Western blot (Fig. 7A). Rab10 knockdown had no effect on Golgi apparatus integrity, as TGN46 labelling revealed the organelle to be perinuclear, condensed and structurally intact (Fig. S3C). Treated cells were infected with A. phagocytophilum followed by LSCM examination. The percentage of ApVs with lumenal TGN46 signal was reduced by approximately fourfold in Rab10 siRNA-targeting versus non-targeting siRNA-treated HEK-293T cells (Fig. 7C). Also, whereas the Golgi apparatus was fragmented and barely detectable in infected control cells, it remained intact in infected cells in which Rab10 had been knocked down (Fig. 7B). Furthermore, the percentage of infected cells and the number of ApVs per cell were reduced by approximately twofold and threefold, respectively, in Rab10-knockdown cells compared with control cells (Fig. 7D and E). Taken together, these data suggest that Rab10 is critical for TGN recruitment to the ApV, for TGN vesicle uptake by the ApV and for productive A. phagocytophilum infection.
Fig. 7.
Knockdown of Rab10 markedly reduces both TGN vesicle delivery into the ApV and A. phagocytophilum infection. HEK-293T cells were treated with Rab10-targeting, non-targeting or GAPDH-targeting siRNA for 72 h.
A. Western blot showing knockdown of Rab10 or GAPDH after appropriate siRNA treatment.
B–D. After siRNA treatment, the cells were infected with A. phagocytophilum (Ap) for 48 h and processed for confocal microscopy. (B and C) Rab10 knockdown markedly inhibits translocation of TGN vesicles into the ApV. Rab10-targeting and non-targeting siRNA-treated infected cells were screened with antibodies against APH0032 and TGN46 and stained with DAPI (blue). (B) Representative confocal micrographs. Regions that are demarcated by hatched-line boxes are magnified in the corresponding enlargement panels. (C) Percentage ± standard deviation (SD) of TGN46-positive ApVs in non-targeting siRNA-treated or Rab10-targeting siRNA-treated infected cells. (D and E) Rab10 knockdown (KD) significantly reduces A. phagocytophilum infection. siRNA-treated infected cells were screened with an antibody against A. phagocytophilum P44 to denote bacteria and examined by immunofluorescence microscopy to determine the percentage ± SD of infected cells (D) and mean number ± SD of ApVs per cell (E). Statistically significant (**P ≤ 0.01; ***P ≤ 0.001) values are indicated. Data presented in each panel are representative of three experiments with similar results.
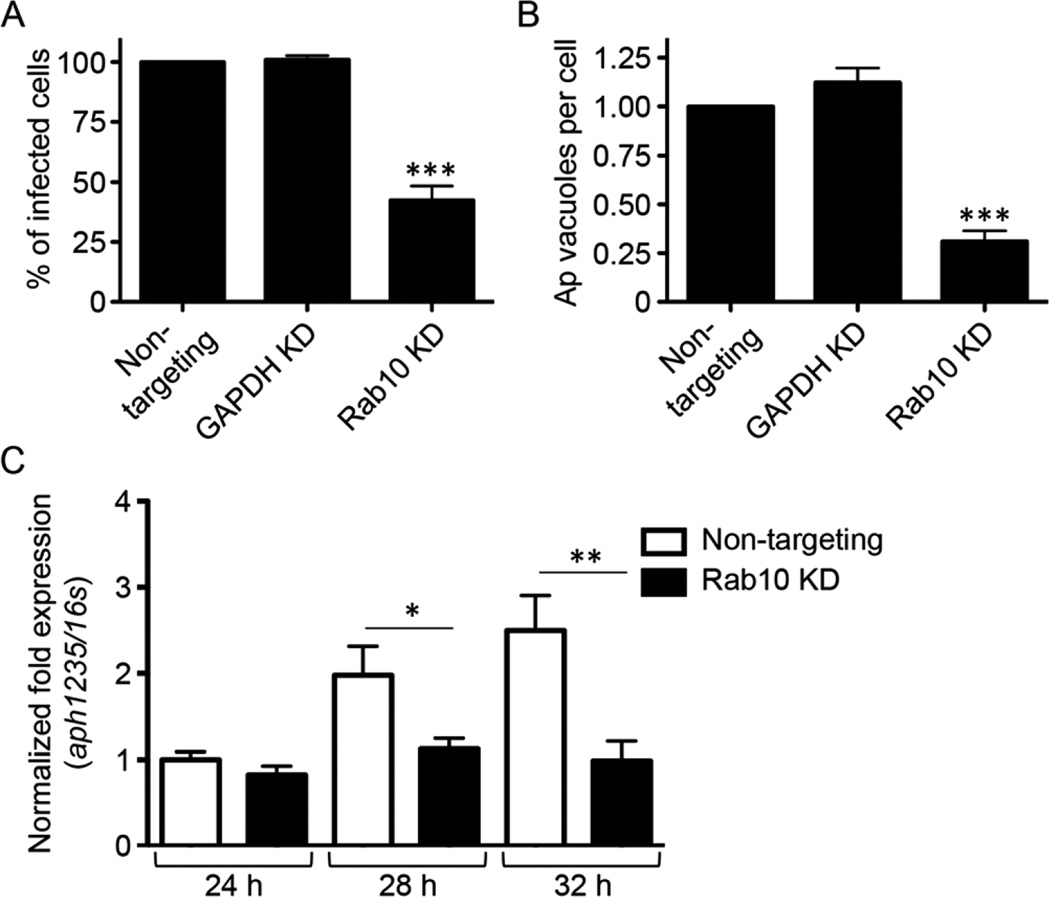
Rab10-dependent acquisition of TGN vesicles is important for the generation of infectious DC progeny
Because the A. phagocytophilum load was pronouncedly reduced in host cells in which Rab10 had been knocked down (Fig. 7D and E), yet many of these cells were observed to harbour numerous late-stage ApVs (Fig. 7B), we reasoned that acquisition of Rab10-positive TGN vesicles was not important for bacterial replication but was instead critical for the production of infectious progeny. To test this hypothesis, media from A. phagocytophilum-infected HEK-293T cells that had been treated with Rab10, GAPDH or non-targeting siRNA was collected at 48 h post-infection and added to naïve cells. In untreated cells, this media contains naturally liberated, infectious A. phagocytophilum DC organisms (Beyer et al., 2014). If siRNA treatment inhibited RC-to-DC conversion, then fewer DC bacteria would be released into the media and, consequently, would produce a lower level of infection in recipient cells. At 24 h, the percentage of infected host cells and the number of ApVs per cell was more than twofold and nearly fourfold lower, respectively, in cells that had been incubated with media from infected Rab10-knockdown cells relative to those that had received media from infected non-targeting siRNA-treated control cells (Fig. 8A and B). To confirm that Rab10 knockdown inhibited RC-to-DC transition, the experiment was repeated and quantitative reverse transcription (qRT)-PCR was performed on RNA collected at 24, 28 and 32 h post-inoculation to assess for aph1235 expression. The selected time points are those during which A. phagocytophilum TGN acquisition, aph1235 up-regulation (Troese et al., 2011;Mastronunzio et al., 2012) and RC-to-DC transition (Troese and Carlyon, 2009) would normally occur. As expected (Troese et al., 2011; Mastronunzio et al., 2012), aph1235 expression rose over the time course in host cells that had been treated with non-targeting siRNA (Fig. 8C). In contrast, aph1235 transcript levels did not increase at any time point in host cells in which Rab10 had been knocked down. Thus, Rab10-dependent acquisition of TGN vesicles is critical for aph1235 up-regulation, RC-to-DC conversion and the production of A. phagocytophilum infectious progeny.
Fig. 8.
Knockdown of Rab10 markedly reduces generation of infectious progeny and aph1235 transcription.
A. Rab10 knockdown (KD) reduces generation of infectious progeny. HEK-293T cells were treated with Rab10-targeting, GAPDH-targeting or non-targeting siRNA for 72 h and subsequently infected with A. phagocytophilum (Ap). At 48 h post-infection, a time point at which infectious DC organisms would be present in the media if the bacterial developmental cycle had proceeded normally, media from the infected siRNA-treated cells was added to naïve HEK-293T cells. At 24 h, the recipient cells were screened with antibody against Ap P44 and examined by immunofluorescence microscopy to determine the percentages of infected cells (A) and the mean number (± standard deviation) of ApVs per cell (B). Data presented are representative of three experiments with similar results.
C. Knocking down Rab10 inhibits transcription of the Ap DC marker, aph1235. Total RNAs isolated from non-targeting siRNA-treated or Rab10-targeting siRNA-treated, Ap-infected cells at 24, 28 and 32 h were subjected to qRT-PCR using primers specific for aph1235. Relative aph1235 transcript levels were normalized to Ap 16S rRNA gene transcript levels using the 2−ΔΔCT method. Data presented are representative of two experiments with similar results. Statistically significant (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001) values are indicated.
Identification of an A. phagocytophilum Rab10-specific ligand
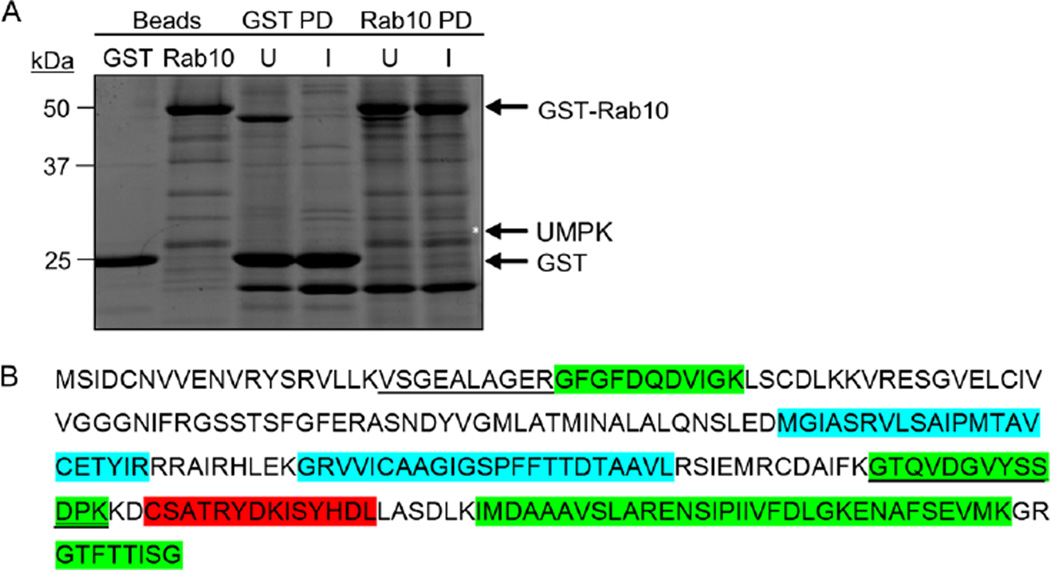
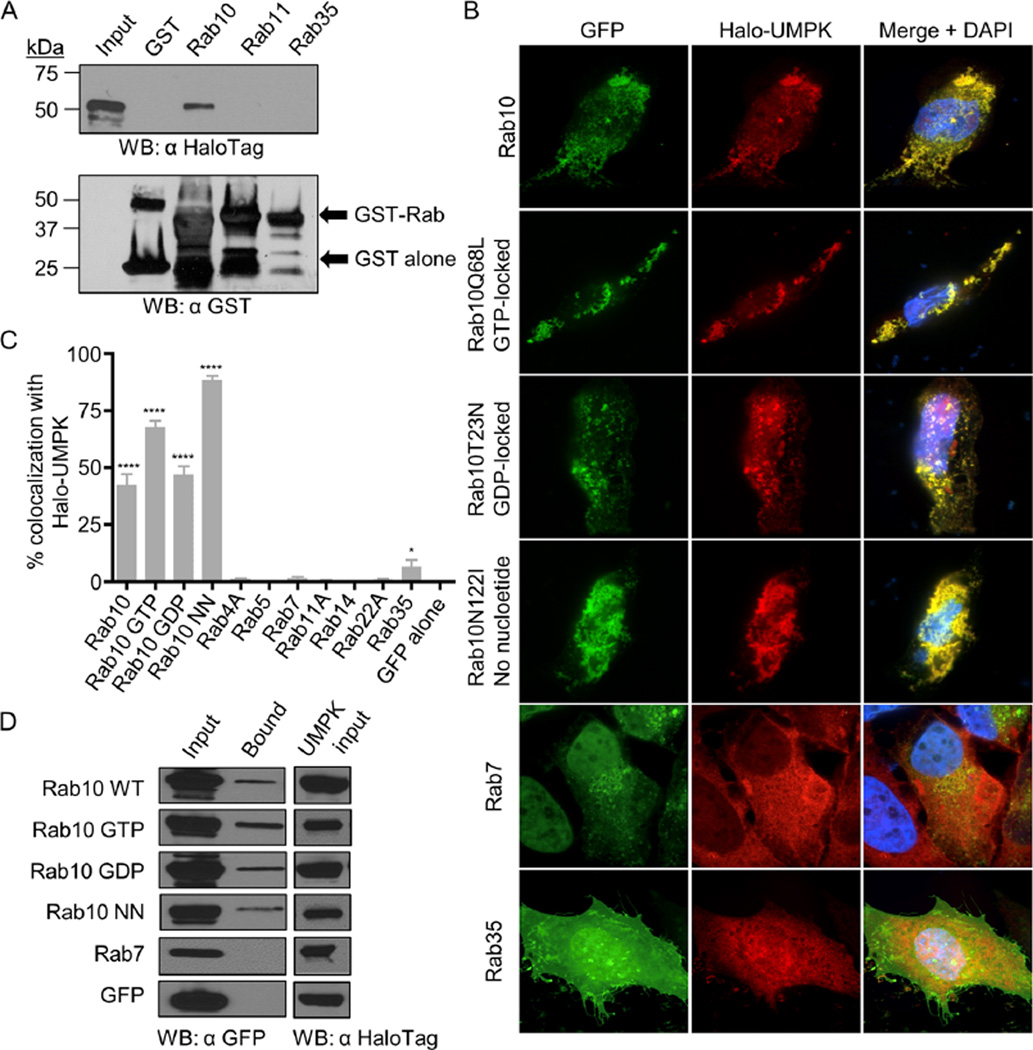
Because Rab10-positive vesicles are recruited to the ApV in a guanine nucleotide-independent, tetracycline-sensitive manner (Huang et al., 2010a) and are delivered into the lumen where they associate with A. phagocytophilum organisms, we reasoned that at least one pathogen-encoded protein contributes to these processes. To identify potential A. phagocytophilum Rab10-interacting proteins, we incubated whole-cell lysates of infected and uninfected HL-60 cells with glutathione sepharose beads coated with glutathione-S-transferase (GST)-tagged Rab10 or GST alone. A 26 kDa protein that was specifically isolated from infected cells by the GST-Rab10 beads (Fig. 9A) was identified by mass spectrometry as APH0111 (Fig. 9B). This protein is annotated as UMPK (Dunning Hotopp et al., 2006), a nucleotide scavenger enzyme that catalyses the conversion of uridine monophosphate to uridine diphosphate (Yan and Tsai, 1999). To validate that UMPK and Rab10 interact and to evaluate the specificity of the interaction, whole-cell lysates of Escherichia coli expressing Halo-tagged UMPK were incubated with beads coated with GST alone, GST-Rab10, GST-Rab11 or GST-Rab35. Rab11 and Rab35 were included because they also localize to the AVM (Huang et al., 2010a). Only GST-Rab10-coupled beads precipitated recombinant UMPK (Fig. 10A).
Fig. 9.
Identification of an A. phagocytophilum Rab10-specific ligand.
A. GST-Rab10 beads pull down A. phagocytophilum UMPK (APH0111). Glutathione sepharose beads coupled with GST-Rab10 or GST alone were incubated with whole-cell lysates of uninfected or A. phagocytophilum-infected HL-60 cells. Eluates from pulldown beads and beads alone were resolved by SDS-PAGE and stained with Sypro Ruby. The band of approximately 26 kDa (denoted by asterisk) that was unique to the eluate from GST-Rab10 beads incubated with A. phagocytophilum-infected cell lysate was excised and analysed by mass spectroscopy. PD, pulldown. U, uninfected. I, infected.
B. A. phagocytophilum UMPK amino acid sequence. Underlined peptide sequences are those that were identified by LC-MS/MS following precipitation of UMPK by GST-Rab10. Peptides that were identified as being on the A. phagocytophilum surface in our prior report (Kahlon et al., 2013) are denoted by green highlighting. Blue highlighting demarcates predicted transmembrane domains. Red highlighting indicates the peptide encompassing amino acids 182–195 against which antiserum was raised.
Fig. 10.
Validation of A. phagocytophilum UMPK-Rab10 interactions.
A. GST-Rab10 but not other GST-tagged Rabs or GST alone pull down UMPK. A lysate of E. coli expressing Halo-UMPK was divided equally and incubated with GST, GST-Rab10, GST-Rab11 and GST-Rab35 coated beads. Input E. coli lysate and bead eluates were analysed by immunoblot using HaloTag and GST antibodies.
B. Halo-UMPK colocalizes specifically with vesicles that are positive for GFP-Rab10 isoforms in an aggregative pattern. HeLa cells were cotransfected to express Halo-UMPK together with GFP-Rab10 isoforms or other GFP-tagged Rab GTPases and analysed by confocal microscopy. Rab10Q68L (GTP locked); Rab10T32N (GDP locked); Rab10N122I [incapable of binding any GTP or GDP (no nucleotide)].
C. Vesicles that are positive for GFP-Rab10 isoforms differentially colocalize with UMPK. One hundred cells coexpressing Halo-tagged A. phagocytophilum UMPK and GFP-tagged Rab GTPases or Halo-tagged E. coli UMPK and GFP-Rab10 were assessed per condition for Halo and GFP signal colocalization. Statistically significant (****P ≤ 0.0001; *P ≤ 0.05) values are indicated.
D. Halo-UMPK interacts with all Rab10 isoforms in living cells. HeLa cells were cotransfected to express Halo-UMPK together with GFP-Rab10 isoforms, GFP-Rab7 or GFP alone. Whole-cell lysates of the transfected cells (Input) were mixed with Halo-Link resin, and the resin eluate (Bound) was analysed by Western blot (WB) with GFP antibody. Portions of each transfection lysate were analysed with HaloTag antibody to confirm Halo-UMPK bait expression (UMPK input). Data presented in A and D are representative of two experiments with similar results. Data presented in B and C are representative of three experiments with similar results.
To assess if UMPK colocalizes with Rab10 in living cells and in a guanine nucleotide-independent manner, HeLa cells were cotransfected to express Halo-UMPK and GFP-tagged wild-type Rab10, GTP-locked Rab10Q68L, GDP-locked Rab10T23N or guanine nucleotide binding defective Rab10N122I and examined by LSCM. HeLa cells coexpressing Halo-UMPK and GFP alone or one of several additional GFP-tagged Rabs that are known to be recruited to the ApV (Rab1A, Rab4A, Rab11A, Rab14, Rab22A and Rab35) or not (Rab5 and Rab7) (Huang et al., 2010a) served as controls. Consistent with the guanine nucleotide-independent specificity of UMPK for Rab10, coexpression of Halo-UMPK with each GFP-tagged Rab10 isoform yielded aggregative vesicular labelling patterns where Halo and GFP signals completely overlapped (Fig. 10B). Of the other Rab GTPases, Halo-UMPK poorly localized with Rab35 and did not colocalize with any other Rab (Fig. 10B and C). Halo and GFP signals did not colocalize in cells that expressed either Halo alone and Halo-UMPK and GFP alone (Fig. S4A) or GFP-Rab10 (Fig. S4B). Halo-tagged E. coli UMPK and GFP-Rab10 also did not colocalize in transfected cells (Fig. S4C), further indicating the specificity of the A. phagocytophilum UMPK-Rab10 interaction. To confirm that UMPK is capable of binding Rab10 in living cells, Halo-UMPK was coexpressed with and assessed for the ability to precipitate GFP-tagged Rab10 isoforms. All GFP-tagged Rab10 isoforms, but neither GFP nor GFP-Rab7, coprecipitated with Halo-UMPK (Fig. 10D). Taken together, these data identify A. phagocytophilum UMPK as a Rab10-specific ligand that binds the GTPase in a guanine nucleotide-independent manner.
UMPK is an A. phagocytophilum surface protein
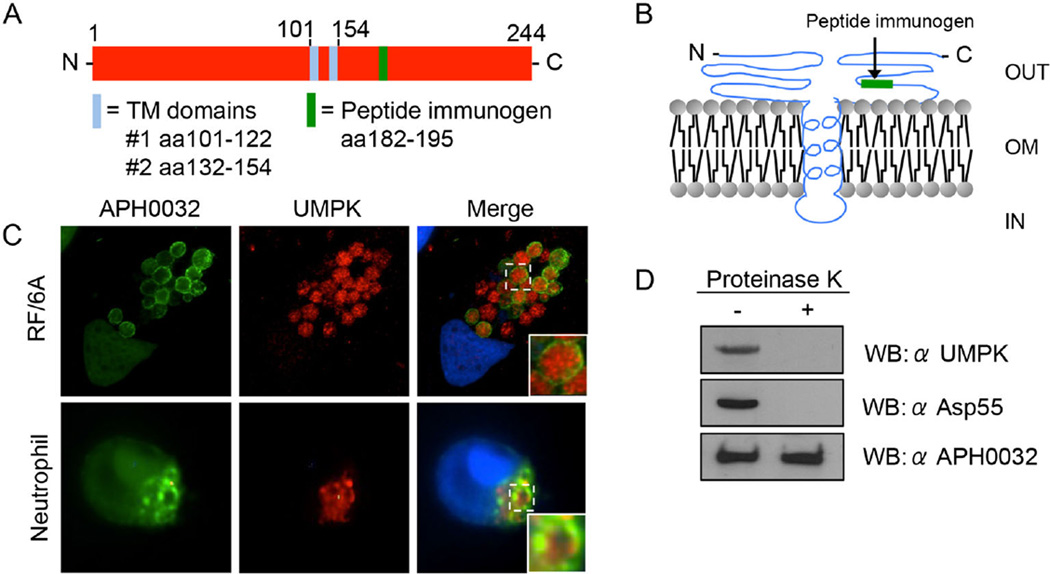
Uridine monophosphate kinase is a 244-amino-acid protein with a predicted molecular mass of 26 kDa and two transmembrane domains that putatively present amino acids 1 to 100 and 155 to 244 on the bacterial surface (Figs 9B and 11A and B). Our previous evaluation of the A. phagocytophilum surface proteome identified UMPK as a likely surface protein (Kahlon et al., 2013). Mining the UMPK tryptic peptides recovered in that study (Kahlon et al., 2013) revealed that they mapped to the protein’s two predicted surface-exposed regions (Fig. 9B). To verify that UMPK is a surface protein, A. phagocytophilum-infected RF/6A cells and human neutrophils were screened with antibody specific for a peptide corresponding to predicted surface-exposed UMPK residues, 189–195 (Figs 9B, 11A and B and S5A), together with APH0032 antibody. In both infected host cell types, UMPK182–195 antibody labelled intravacuolar bacteria, but not the APH0032-positive AVM (Fig. 11C). Next, to determine if UMPK residues 182–195 are an immunoaccessible domain that is exposed on the bacterial outer membrane, the surfaces of intact host cell-free A. phagocytophilum organisms were treated with proteinase K, a proven approach for verifying surface presentation of A. phagocytophilum and Chlamydia trachomatis OMPs (Wang et al., 2006; Ojogun et al., 2012; Kahlon et al., 2013; Seidman et al., 2014). Western-blotted lysates of proteinase K-treated and vehicle-treated DC bacteria were screened with UMPK182–195 antiserum. Positive control antiserum targeted a confirmed surface-exposed epitope of Asp55 (55 kDa A. phagocytophilum surface protein) (Ge and Rikihisa, 2007), while negative control antiserum targeted APH0032, which is not an OMP (Huang et al., 2010b). Following surface proteolysis, neither UMPK182–195 nor Asp55 was detected (Fig. 11D). APH0032 was amply detected in lysates of both treated and vehicle control-treated bacteria. Immunofluorescence examination of A. phagocytophilum-infected host cells at multiple time points post-infection revealed that the bacterium constitutively expressed UMPK (Fig. S5B). Thus, UMPK is an A. phagocytophilum surface-localized Rab10-specific ligand that is expressed throughout the course of infection.
Fig. 11.
UMPK is an A. phagocytophilum surface protein.
A. UMPK is a 244-amino-acid protein with two predicted TM domains at residues 101–122 and 132–154. Antiserum was raised against a peptide that corresponds to amino acids 182–195, which is predicted to be surface exposed.
B. Predicted outer membrane (OM) topology of UMPK. OUT, extracellular. IN, interior to the OM.
C. UMPK localizes within the ApV. A. phagocytophilum-infected RF/6A cells, and human neutrophils were screened with antibodies against UMPK and APH0032 and visualized by confocal microscopy. The insets denoted by solid boxes are enlarged versions of the vacuoles demarcated by hatched-line boxes. Host cell nuclei and bacterial DNA were stained with DAPI (blue).
D. Proteinase K removes a surface-exposed UMPK epitope. Intact A. phagocytophilum organisms were incubated with proteinase K (+) or vehicle control (−) followed by washing to remove cleaved proteins. The bacteria were lysed and analysed by Western blot (WB) using antibodies against UMPK residues 182–195, a known-surface exposed epitope of the A. phagocytophilum surface protein, Asp55, and APH0032, which is not a surface protein. Results presented are representative of two experiments with similar results.
Discussion
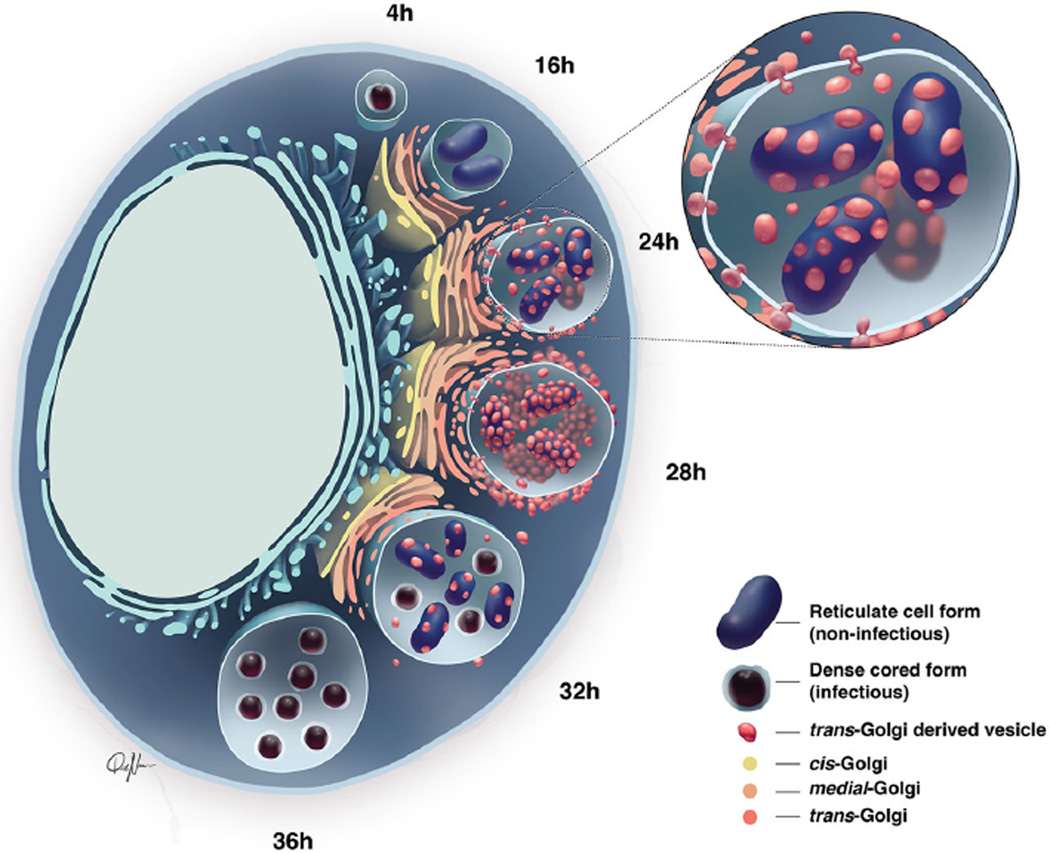
Abu Kwaik and Bumann (2013) coined the term, ‘nutritional virulence’ to describe the phenomenon that, without access to host nutrients, microbial pathogens cannot survive and cause disease. This study provides important insights into how A. phagocytophilum satisfies its nutritional virulence requirements, specifically host–pathogen interactions that are critical for it to complete its biphasic developmental cycle and yield infectious progeny. Figure 12 presents our hypothetical model for these findings as follows. After A. phagocytophilum has entered its host cell, transitioned from the DC to the RC form and begun dividing, the ApV localizes to and is wrapped by the Golgi apparatus. During the period of expansive bacterial replication, which occurs between 12 and 28 h, A. phagocytophilum targets TGN exocytic traffic in a Rab10-dependent manner. Hijacked Rab10-positive TGN vesicles localize to and within the ApV. Within the ApV lumen, the bacterium acquires sphingolipid-rich contents of the imported TGN vesicles and reconverts to its infectious DC form.
Fig. 12.
Model: Rab10-positive TGN vesicles are delivered into the ApV lumen and their association with replicative A. phagocytophilum RC organisms is a precursor for the bacteria to develop into the infectious DC form to complete the infection cycle. Between 12 and 16 h post-infection, after A. phagocytophilum has converted from the DC to RC form, the ApV has positioned itself in close proximity to the Golgi apparatus. By 24 h, the ApV has begun to intercept TGN-derived vesicles, which cross the AVM into the ApV lumen and associate with intravacuolar A. phagocytophilum RC organisms. TGN vesicle delivery into the ApV lumen and A. phagocytophilum association with TGN vesicles occurs around 28 h. Between 28 and 32 h, Rab10-positive TGN vesicle delivery into the ApV and association with intralumenal bacteria has waned considerably. Coincident with this phenomenon, TGN vesicle-negative A. phagocytophilum organisms have transitioned from the RC to DC form.
Rab10 is important for RC-to-DC conversion. APH1235 contributes to invasion of mammalian host cells (Mastronunzio et al., 2012), is up-regulated during RC-to-DC transition (Troese et al., 2011) and is therefore a marker for the expression profile associated with the generation of infectious progeny. Rab10 knockdown in host cells did not affect bacterial replication but impeded TGN vesicle import into the ApV, aph1235 expression and the generation of DC bacteria. In addition to TGN vesicles, A. phagocytophilum also promotes delivery of autophagic bodies into the ApV lumen as a major amino acid source (Niu et al., 2012). However, autophagy does not involve Rab10, and autophagosome recruitment would likely not be affected in Rab10 siRNA-treated cells. Thus, while autophagy is important for intracellular growth, it may not be involved in generating infectious progeny. A. phagocytophilum organisms that are positive for Rab10 and TGN46 are APH1235 negative and vice versa, suggesting that the bacterium incorporates/utilizes TGN vesicle contents before RC-to-DC conversion occurs. The pathogen may use TGN vesicle contents to compensate for its auxotrophies and as carbon sources to fuel the energy demand associated with the RC-to-DC switch.
Many intracellular bacteria parasitize host cell lipids (reviewed by Vromman and Subtil, 2014). C. trachomatis was the first bacterium shown to recruit the Golgi apparatus and to deliver both ceramide and cholesterol into the lumen of its inclusion (Hackstadt et al., 1995, 1996; Scidmore et al., 1996; Wolf and Hackstadt, 2001; Carabeo et al., 2003; Heuer et al., 2009). C. trachomatis hijacks the TGN by targeting Rab6, Rab11 and Rab14 (Rzomp et al., 2003; Lipinski et al., 2009; Capmany and Damiani, 2010), whereas A. phagocytophilum does so by co-opting Rab10. A. phagocytophilum acquires endogenous host cell and BODIPY TR-labelled sphingolipids, the latter of which would have been derived from precursor BODIPY TR ceramide and produced in the TGN. Because both RC and DC forms were BODIPY TR positive and lipidomics revealed that naturally liberated DC bacteria were enriched in sphingolipids, we infer that DC organisms retain TGN-derived sphingolipids assimilated during the RC stage. How this occurs is unknown. The close apposition of TGN vesicles with intravacuolar A. phagocytophilum organisms could allow for lipid transfer proteins, which are present in TGN vesicle membranes and mediate lipid exchange between donor and acceptor membranes (Lev, 2010; Blom et al., 2011), to deliver host-derived sphingolipids to the bacterium. Such sphingolipids may help stabilize the A. phagocytophilum peptidoglycan-deficient cell wall, as a similar role has been established for cholesterol (Lin and Rikihisa, 2003).
The Golgi is destabilized to the point that it is barely detectable in host cells with high A. phagocytophilum burdens. This alters neither Rab10 ApV localization nor bacterial replication, as both proceed equally as well in host cells treated with the Golgi destabilizing agent, brefeldin A, or vehicle control (Mott et al., 1999; Huang et al., 2010a). Golgi fragmentation does not occur in cells with lower loads likely because the bacterial demand does not exceed the rate at which the organelle is replenished. Under high A. phagocytophilum load, however, the demand is too high to allow for Golgi replenishment, which results in destabilization of the organelle. Based on GM130 (cis-Golgi marker) labelling, a previous report noted that Golgi destabilization occurred in host cells heavily infected with A. phagocytophilum (Xiong and Rikihisa, 2012). Because this study did not label for the TGN (Xiong and Rikihisa, 2012), it could not determine the fate of TGN-derived vesicles and their relevance to A. phagocytophilum infection.
In terms of pathogen–Rab interactions, it is well documented that bacterial manipulation of Rabs can culminate in SNARE-mediated fusion of the routed vesicles with the pathogen-occupied vacuolar membrane. This, in turn, transports vesicle contents into the lumen for bacterial use (Arasaki et al., 2012; Sherwood and Roy, 2013). A lesser-understood phenomenon that also occurs and for which evidence has only recently begun to accumulate is the transport of entire vesicles into pathogen-occupied vacuoles (POVs) in the absence of membrane fusion. Indeed, intact lipid droplets, peroxisomes, multivesicular bodies and endoplasmic reticulum-derived vesicles are delivered into the C. trachomatis inclusion (Beatty, 2006; Cocchiaro et al., 2008; Dumoux et al., 2012; Boncompain et al., 2014). Also, Rab14, which like Rab10 directs TGN exocytic traffic (Junutula et al., 2004), was detected within the C. trachomatis inclusion (Capmany and Damiani, 2010). Until this report, import of host cell-derived vesicles into a POV had not been observed for another vacuole-adapted intracellular bacterium besides C. trachomatis. How intracellular bacteria orchestrate membrane fusion-independent delivery of Rab-decorated vesicles into the POV lumen is undefined. Notably, it was recently demonstrated that two secreted proteins of the protozoan pathogen, Toxoplasma gondii, localize to and function synergistically to mediate passive transfer of small molecules across the parasitophorous vacuole membrane (Gold et al., 2015).
Anaplasma phagocytophilum UMPK is a novel bacterial Rab ligand that specifically binds Rab10 in a guanine nucleotide-independent manner, which suggests that it interacts with a region of Rab10 that is unaltered by its guanine nucleotide-bound status. UMPK’s annotated function in pyrimidine biosynthesis (Yan and Tsai, 1999) and its role in engaging Rab10 demonstrated herein categorize it as a moonlighting protein, or a protein that performs multiple autonomous, unrelated functions (Henderson and Martin, 2011, 2013). There are over 200 examples of bacterial metabolic proteins and chaperones functioning as virulence factors (Henderson and Martin, 2011, 2013). As typified by A. phagocytophilum UMPK, which is present on the bacterium’s surface, these proteins often execute their virulence functions at sites that are topologically distinct from their housekeeping roles (Henderson and Martin, 2011, 2013). E. coli UMPK is a moonlighting protein that localizes not only to the cytoplasm where it functions as both a nucleotide scavenger enzyme and a transcriptional regulator but also to the outer membrane where it performs an unknown function (Kholti et al., 1998; Landais et al., 1999). Also, Yersinia pestis cytidine monophosphate kinase, a UMPK homolog, makes an undefined contribution to virulence (Walker et al., 2012).
Based on UMPK’s surface presentation and the proximity of Rab10-positive TGN vesicles to bacteria in the ApV lumen, it will be important to investigate if UMPK binds Rab10 as a receptor on the newly imported vesicles to promote utilization of their contents. However, because a reliable system for performing knockout complementation studies in A. phagocytophilum is currently lacking, this hypothesis cannot yet be evaluated. As important progress towards effective systems for assessing bona fide gene function in obligate intracellular bacteria continues to be made (Mishra et al., 2012; Cheng et al., 2013; Beare and Heinzen, 2014; Wood et al., 2014), we anticipate that it will be possible to validate A. phagocytophilum UMPK’s role during infection in the near future.
Obligate intracellular bacteria cause infections that can be debilitating, chronic and even fatal. Because they must parasitize nutrients from their hosts to survive, understanding their nutritional virulence strategies may elucidate new treatment options. This study indicates A. phagocytophilum Rab10-dependent TGN parasitism could potentially be targeted as a novel therapeutic. Accordingly, it will be important to assess for translocation of intact vesicles into the POVs of other intracellular bacterial species and to ultimately determine if UMPK or other bacterial surface proteins mediate interactions with the delivered vesicular cargo.
Experimental procedures
Cultivation of uninfected and A. phagocytophilum-infected host cell lines and neutrophils
Uninfected and A. phagocytophilum (NCH-1 strain)-infected human promyelocytic HL-60 cells [CCL-240; American Type Culture Collections (ATCC), Manassas, VA] and RF/6A rhesus monkey choroidal endothelial cells (CRL-1780; ATCC) were cultured as described previously (Huang et al., 2010b). HeLa 229 epithelial cells (CCL-1.2; ATCC) were grown in Roswell Park Memorial Institute 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% foetal bovine serum (FBS; Gemini Bio-Products, Sacramento, CA) at 37°C with 5% CO2. HEK-293T cells were cultured in Dulbecco’s modified Eagle’s medium with l-glutamine, 4.5 g l−1 of d-glucose and 100 mg l−1 of sodium pyruvate (Invitrogen) supplemented with 10% FBS, 1× minimal essential medium non-essential amino acids (Invitrogen) and 15 mM of HEPES (Affymetrix, Cleveland, OH) at 37°C with 5% CO2. Human neutrophils were isolated from peripheral blood from healthy donors using Polymorphprep (Axis-Shield; Greiner Bio-One, Frickenhausen, Germany) as described previously (Carlyon et al., 2004). The protocol (HM11407) for obtaining donor blood for the purpose of isolating neutrophils has been reviewed and approved by the Virginia Commonwealth University Institutional Review Board with respect to scientific content and compliance with applicable research and human subjects regulations.
Plasmids
Previously described constructs encoding GFP-tagged and GST-tagged Rab proteins were kind gifts from Marci Scidmore (Cornell University) and have been described previously (Rzomp et al., 2003; Cortes et al., 2007; Huang et al., 2010a). APH0111 (UMPK) DNA was synthetically generated and cloned into the SgfI and PmeI sites of the pFN21K vector mammalian expression vector by Promega Corporation (Madison, WI). The insert was subcloned into the SgfI and PmeI sites of the E. coli expression vector pFN18A (Promega) using the Flexi system transfer kit (Promega). The pFN21K control vector that was used for expressing the Halo tag alone was constructed by digesting the original pFN21K vector with SgfI and PmeI to remove the barnase gene. The digested vector was religated with a double-stranded DNA bridge that was generated using oligonucleotides 5′-CGCGTAAGGGTAGGTTT-3′ (sense strand) and 5′-AAACGTACCCTTACGCGAT-3′ (antisense strand). DNA encoding the E. coli UMPK gene was amplified from Alpha-Select Gold Efficiency E. coli (Bioline, Taunton, MA) using primers 5′-ATATGCGATCGCCGCTACCAATGCAAAACCCGTCTATAAACGCATTC-3′ (sense strand; spacer nucleotides are italicized; SgfI site is underlined) and 5′-AACTGTTTAAACTTATTCCGTGATTAAGTCCCTTCTTTTTCACC-3′ (antisense strand; spacer nucleotides are italicized; PmeI site is underlined) and cloned into the SgfI and PmeI sites of the pFN21K. DNA encoding the A. phagocytophilum UMPK gene was amplified from NCH-1 DNA using primers 5′-GACCGCGATCGCCGATTGTAATGTTGTGGAGAATGTGCGTTATTCTA-3′ (sense strand; spacer nucleotides are italicized; SgfI site is underlined) and 5′-GTCGGTTTAAACCTATCCAGATATAGTAGTAAACGTACCCC-3′ (antisense strand; spacer nucleotides are italicized; PmeI site is underlined) and ligated into pFN21K or pFN18A. All inserts were amplified with Platinum Pfx DNA polymerase (Invitrogen, Grand Island, NY), and the sequences verified by DNA sequencing.
GST-Rab10-mediated precipitation of endogenous UMPK
BL21 E. coli (Novagen, EMD Millipore, Billerica, MA) transformed with a plasmid encoding GST-Rab10 were grown to an optical density at 600 nm of 0.4 and induced with 100 µM of isopropyl β-d-1-thiogalactopyranoside at 16°C for 16 h. GST-Rab10 was solubilized from inclusion bodies using 1% sarkosyl in Brymora bacterial lysis buffer [10 mM Tris–HCl (pH 7.4), 250 mM NaCl, 1% (vol/vol) Triton x-100, 2.5 mM MgCl2] and combined with the soluble fraction. GST-Rab10 was immobilized on glutathione sepharose beads (GE Healthcare Life Sciences, Little Chalfont, UK) with the addition of 2% Triton X-100 and 20 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate. Fifty million uninfected or A. phagocytophilum-infected HL-60 cells were lysed in radioimmunoprecipitation assay buffer [10 mM Tris-Cl (pH 8.0), 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate and 140 mM NaCl]. The lysates were pre-cleared with glutathione beads to eliminate non-specific binding and then mixed with 5 mg of GST or GST-Rab10 glutathione beads for 2 h at 4°C. The beads were washed and boiled, and the eluates were resolved on a 20 cm acrylamide tris-glycine 10–20% gradient gel (Jule Inc., Milford, CT) using the Protean II XL Cell electrophoresis rig (BioRad, Hercules, CA). The gel was stained with SYPRO Ruby (Invitrogen). The approximately 26 kDa band of interest that was exclusively recovered from the A. phagocytophilum-infected host cell lysate was excised and submitted to the W. M. Keck Facility at Yale University (New Haven, CT) for mass spectroscopy and database searching.
Immunofluorescence assays and microscopy
Cells for immunofluorescence assays were grown and infected on #1½ 12 × 12 mm glass coverslips for LSCM (Electron Microscopy Sciences, Hatfield, PA) or #1½ HP (i.e. 0.17 ± 0.005 mm thick) high-performance glass coverslips (Zeiss, Thornwood, NY) for SIM. They were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) for 30 min followed by permeabilization with 0.5% Triton X-100 for 10 min and washed in 1× phosphate-buffered saline (PBS). In some cases, host cell-free A. phagocytophilum RC and/or DC organisms were recovered from infected HL-60 cells as described (Carlyon, 2005) and fixed to glass slides. The cells or host cell-free bacteria were screened with antibodies for immunofluorescence microscopy as previously described (Huang et al., 2010c). Commercial primary antibodies were against TGN46 (Sigma-Aldrich, St. Louis, MO), GM130 (BD Biosciences, San Jose, CA), GolgB1 (Sigma-Aldrich), HaloTag (Promega), GFP (Invitrogen), Rab10 (Sigma-Aldrich) and vimentin (Abcam, Cambridge, MA). Antibodies against Asp14, APH1235, APH0032 and P44 were described previously (Huang et al., 2010b; Troese et al., 2011; Kahlon et al., 2013). Affinity-purified rabbit polyclonal antiserum targeting a peptide corresponding to A. phagocytophilum UMPK amino acids 182 to 195 was generated by New England Peptide (Gardner, MA). Secondary antibodies conjugated to Alexa Fluor fluorochromes were obtained from Invitrogen. BODIPY ceramide/bovine serum albumin complex (Invitrogen) was added to infected RF/6A cells per manufacturer’s instructions. Coverslips were mounted with Prolong Gold Anti-fade reagent with or without DAPI (Invitrogen). Images were obtained using an Olympus BX51 microscope affixed with a disc-spinning unit (Olympus, Center Valley, PA) or a Zeiss LSM 700 laser scanning confocal microscope. For SIM, coverslips were stained with 1 µg ml−1 of DAPI in 1× PBS for 5 min and mounted with Prolong Gold Anti-fade reagent without DAPI (Invitrogen). Images were obtained using a Nikon N-SIM superresolution microscope. LSCM and SIM were performed at the VCU Microscopy Facility, which is supported, in part, by funding from NIH-NINDS Center core grant (5P30NS047463) and NIH-NCI Cancer Center Support Grant (P30 CA016059). 3D movies were generated using Volocity Image Analysis Software (Perkin Elmer, Waltham, MA).
Colocalization studies
HeLa cells were seeded onto 12 × 12 mm coverslips (Electron Microscopy Sciences) and cotransfected with plasmid DNA encoding Halo alone, Halo-A. phagocytophilum UMPK, Halo-E. coli UMPK or GFP-UMPK, and various GFP-Rabs or GFP alone was processed and labelled using Lipofectamine 2000 (Invitrogen). HaloTag TMR Direct ligand (Promega) was added to wells containing cells transfected to express Halo-tagged protein. After 24 h, the cells were processed and labelled, where applicable, as described earlier, and examined by immunofluorescence microscopy for colocalization of Halo and GFP signals.
Infection assays
Neutrophils and HL-60 cells were infected with A. phagocytophilum DC organisms released from infected HL-60 cells by sonication as previously described (Troese and Carlyon, 2009). RF/6A or HEK-293T cells were infected with A. phagocytophilum DC bacteria that had been naturally released from infected RF/6A cells into the culture media as follows. Adherent host cells to be infected were seeded onto 12 mm coverslips (Electron Microscopy Sciences) and overlain with 200 µl of A. phagocytophilum-laden media from heavily infected RF/6A cells. The supernatant was spun down on top of the cells at 1000 × g for 3 min followed by a 1 h incubation at 37°C with 5% CO2. The cells were washed with 1× PBS to remove unbound bacteria, and fresh media was added. The cells were returned to 37°C with 5% CO2 for 24 or 48 h or specific time points, after which the cells were fixed in 4% paraformaldehyde and examined by immunofluorescence microscopy.
Density gradient centrifugation
Uninfected or infected HL-60 cells of 2 × 107 were washed with ice-cold 1× PBS two times followed by one wash in cold homogenization buffer (HB; 250 mM sucrose, 10 mM Tris–HCl, pH 7.4,1 mM EDTA). The cells were suspended in 1 ml of cold HB with protease inhibitors (Roche) and transferred to a type B dounce homogenizer (Gerresheimer Kimble Chase LLC, Vineland, NJ). The cells were homogenized for 20 to 30 strokes until ≥ 90% of the host cells were lysed, as verified by the trypan blue exclusion assay. The homogenate was centrifuged at 500 × g for 5 min to pellet the nuclei and unbroken cells. The supernatant was overlain on top of a 5%, 15% or 25% continuous Opti-prep (Sigma-Aldrich) gradient. The gradient was centrifuged at 200,000 × g for 3 h in a Beckman Coulter (Indianapolis, IN) Optima XE-100 ultracentrifuge. Twelve 1 ml fractions were collected from the top of the gradient. Protein was concentrated from the fractions by trichloroacetic acid precipitation. Equal volumes of fractions 1 to 9 were immunoblotted using TGN46, GM130 and P44 antibodies.
Western blot analyses
Cell lysates or gradient centrifugation fractions were analysed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot as previously described (Troese et al., 2011). Commercially available primary antibodies targeted β-actin (Santa Cruz Biotechnology, Santa Cruz, CA), GAPDH (Sigma-Aldrich), GFP (Invitrogen), HaloTag (Promega), TGN46 (Sigma-Aldrich) and Rab10 (Sigma-Aldrich). Rabbit antisera against A. phagocytophilum proteins P44 (IJdo et al., 1999) and APH0032 (Huang et al., 2010b) were described previously. Rabbit antibody targeting a surface-exposed epitope of Asp55 (Ge and Rikihisa, 2007) was a kind gift from Yasuko Rikihisa (Ohio State University, Columbus, OH).
UPLC-ESI-MS/MS lipidomics of A. phagocytophilum DC organisms
Media from 3 × 107 A. phagocytophilum-infected (≥ 90%) HL-60 cells, which contained DC bacteria that had been naturally released, was spun at 300 × g twice to pellet host cells. The supernatant was spun at 1000 × g to remove host cellular debris. Next, the supernatant was spun at 5200 × g for 10 min to pellet DC organisms. The resulting pellet was washed twice with ice-cold PBS. All spins were performed at 4°C. As a background control, the media from 3 × 107 uninfected HL-60 cells was processed and analysed by UPLC-ESI-MS/MS in parallel. The A. phagocytophilum DC and control samples were examined for the presence of sphingolipids using UPLC-ESI-MS/MS as previously described (Simanshu et al., 2013; Wijesinghe et al., 2014). Three separate biological replicates were examined in triplicate.
Rab10 siRNA knockdown
Seeded onto 12 mm glass coverslips (Electron Microscopy Sciences) were 4 × 105 HEK-293T cells. After 16 to 20 h, 80 µl of 5 µM ON-TARGETplus human Rab10 siRNA SMARTpool (GE Dharmacon, Lafayette, CO) was mixed with 320 µl of media and added to the wells. Non-targeting or GAPDH-targeting siRNA (GE Dharmacon) was added to control wells. After 72 h, 200 µl of media containing A. phagocytophilum DC organisms that had been released from infected RF/6A cells was added, and the bacteria were spun on the cells as described earlier. Additionally, cells from one well of each siRNA treatment were harvested for Western blot analysis to confirm knockdown. At 48 h post-infection, cells were harvested for quantitative PCR, processed for microscopy analyses, or the DC-laden media was added to naïve HEK-293T cells to detect the presence of infectious progeny.
Quantitative PCR and qRT-PCR
To analyse A. phagocytophilum load after siRNA treatment, DNA was isolated from host cells using the DNeasy Blood and Tissue Kit (Qiagen, Boston, MA). Fifty nanograms of DNA was analysed with primers specific for A. phagocytophilum 16S rDNA and host cell β-actin using SsoFast EvaGreen Supermix (BioRad) as previously described (Ojogun et al., 2012). To detect APH1235 transcript at 24, 28 and 32 h post-infection, cDNA was generated and analysed by qRT-PCR as previously described (Kahlon et al., 2013).
Precipitation of GFP-Rab10 proteins using Halo-UMPK
HeLa cells were seeded into a T-75 flask per pulldown and cotransfected with 12 µg each of Halo-UMPK (pFN21K) and GFP-Rab constructs using Lipofectamine 2000 (Invitrogen) per the manufacturer’s instructions. After 24 h, the cells were trypsinized from the flask, spun down at 300 × g and subjected to one freeze–thaw cycle before complete lysis with Mammalian lysis buffer containing protease inhibitors (Promega). The pulldown was performed according to Promega’s HaloTag Mammalian Pull-Down and Labeling Systems Technical Manual (TM#342) with the addition of 2.5 mM MgCl2 in the binding and wash buffers. The eluates were resolved by SDS-PAGE, transferred to nitrocellulose and probed with GFP antibody (Invitrogen) as described (Troese et al., 2011).
Precipitation of Halo-UMPK by GST-Rab10 beads
BL21(DE3) E. coli (Novagen, EMD Millipore) transformed with the pFN18A Halo-UMPK construct were grown to an optical density at 600 nm of 0.4 and induced with 100 µM isopropyl β-d-1-thiogalactopyranoside at 25°C for 4 h. A culture of 50 ml culture was pelleted and lysed with Bugbuster (Novagen, EMD Millipore) containing EDTA-free Complete Protease Inhibitor (Roche, Indianapolis, IN). The lysate was divided equally, and each aliquot was incubated with 5 mg each of GST, GST-Rab10, GST-Rab11 or GST-Rab35 glutathione beads overnight on a rotator at 4°C. The beads were washed and boiled, and the eluates were resolved by SDS-PAGE and immunoblotted with HaloTag antibody (Promega).
Identification of UMPK as an A. phagocytophilum surface protein
Putative transmembrane domains in A. phagocytophilum UMPK were identified using TMPred (http://www.ch.embnet.org/software/TMPRED_form.html). Surface-exposed proteins of A. phagocytophilum DC organisms were removed by proteolysis as previously described (Ojogun et al., 2012), except that the bacteria were treated with 4 mg ml−1 of proteinase K for 45 min at 25°C. We previously reported recovering UMPK as an A. phagocytophilum surface protein using a surface biotinylation-affinity chromatography approach (Kahlon et al., 2013). To provide the previously unpublished sequences of the peptides recovered in this study, we mined the peptide spectra from this analysis and denoted them in Fig. S5.
Statistical analyses
One-way analysis of variance or Student’s t-test was performed using the Prism 5.0 software package (GraphPad; San Diego, CA) to assess statistical significance as described. Statistical significance was set at P ≤ 0.05.
Supplementary Material
Acknowledgments
We thank Yasuko Rikihisa (Ohio State University, Columbus, OH) for Asp55 antiserum and her and Qingming Xiong (Ohio State) for helpful advice on density gradient purification. We thank C. Dan Nacu of the Virginia Commonwealth University Department of Communication Arts and Design for producing the model in Fig. 12. We thank Bob Heinzen and Charles Larson for critical review of this manuscript, Scott Henderson for helpful discussions regarding SIM and Christine Van Duyn for technical assistance. This study was supported by funding from NIH grants AI072683 (to J. A. C.), HL125353 (to C. E. C.) and AI109068 (to C. E. C); National Center for Advancing Translational Sciences grant UL1TR000058 (to J. A. C.); the Veteran’s Administration (VA Merit Review I BX001792 to C. E. C. and a Research Career Scientist Award 13F-RCS-002 to C. E. C.); the Israel Binational Science Foundation (BSF2011360 to C. E. C.); the Center for Clinical and Translational Research Endowment Fund of Virginia Commonwealth University (VCU) (to J. A. C.). LSCM and SIM were performed at the VCU Microscopy Facility, which is supported, in part, with funding from NIH-NINDS Center core grant 5P30NS047463 and NIH-NCI Cancer Center Support grant (P30 CA016059). VCU Lipidomics/Metabolomics Core services and products in support of the research project were generated, in part, by the VCU Massey Cancer Center with funding from NIH-NCI Cancer Center Support grant, P30 CA016059.
Footnotes
Author contributions
H. K. T. and J. A. C. designed the experiments, analysed results and wrote the paper. H. K. T., L. V., C. L. C. and N. O. performed the experiments. B. P. G., D. S. W. and C. E. C. performed and analysed data generated by UPLC-ESI-MS/MS.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher’s web-site:
Fig. S1. The Golgi apparatus is fragmented in Anaplasma phagocytophilum-infected host cells with high bacterial burdens. (A) The Golgi apparatus is intact, compact and perinuclear in uninfected host cells. RF/6A cells were screened with antibodies against GM130, GolgB1 or TGN46. (B) The trans-Golgi network (TGN) is fragmented in host cells having high A. phagocytophilum burdens. RF/6A cells were infected with different loads of A. phagocytophilum organisms for 28–32 h and processed for laser scanning confocal microscopy (LSCM). A. phagocytophilum-occupied vacuoles (ApVs) were visualized with antibody against the intermediate filament protein, vimentin, which cages ApVs. The TGN and TGN-derived vesicles delivered into the ApV were visualized with TGN46 antibody. (C) The cis-Golgi and medial-Golgi are fragmented in host cells having high A. phagocytophilum burdens. Heavily infected RF/6A cells were screened with antibodies targeting the cis-Golgi (GM130 and GolgB1) and medial-Golgi (GolgB1). A. phagocytophilum-occupied vacuolar membranes and intravacuolar bacteria were visualized with APH0032 antibody. The cells were examined by confocal microscopy. Host cell nuclei and bacterial DNA were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Hatched-line boxes indicate regions that were magnified in the enlargement panels. Data are representative of two experiments with similar results.
Fig. S2. Host cell-free A. phagocytophilum reticulate cells (RCs) are Rab10 and TGN46 positive, while host cell-free dense-core morphotypes (DCs) are Rab10 and TGN46 negative. A. phagocytophilum RC and DC organisms were isolated from infected HL-60 cells, fixed to glass slides, screened with antibodies against (A) Rab10, (B) TGN46 or (C) P44 and examined by LSCM. Bacterial DNA was stained with DAPI (blue). Results shown are representative of three experiments with similar results.
Fig. S3. HEK-293T cells are appropriate for assessing the effects of Rab10 knockdown on A. phagocytophilum infection and Golgi recruitment to the ApV. (A) Green fluorescent protein (GFP)-Rab10 localizes to ApVs in HEK-293T cells. HEK-293T cells were transfected to express GFP-Rab10 for 24 h and subsequently incubated with A. phagocytophilum. After 48 h, the cells were processed for LSCM. Bacteria were visualized with antibody against major surface protein P44. Arrow points to an ApV surrounded by a GFP-Rab10 signal. An arrow denotes pronounced GFP-Rab10 signal localized to an ApV. (B) TGN vesicles are delivered into ApVs in HEK-293T cells. A. phagocytophilum-infected HEK-293T cells were screened with antibodies against TGN46 and APH0032 and examined by LSCM. (C) The Golgi is intact following Rab10 knockdown in HEK-293T cells. HEK-293T cells were treated with non-targeting or Rab10-targeting small interfering RNA. After 72 h, the cells were fixed and processed for confocal microscopy. The Golgi was visualized with TGN46 antibody. (A–C) Host cell nuclei and bacterial DNA were visualized with DAPI (blue). Data are representative of two experiments with similar results.
Fig. S4. Halo-uridine monophosphate kinase (UMPK) and GFP, Halo and GFP-Rab10 and Halo-tagged Escherichia coli UMPK and GFP-Rab10 fail to colocalize when coexpressed in HeLa cells. (A–C) Transfected HeLa cells expressing Halo-UMPK and GFP (A), Halo alone and GFP-Rab10 (B) or Halo-tagged E. coli UMPK and GFP-Rab10 (C) were examined using confocal microscopy. Host cell nuclei were stained with DAPI (blue). Data are representative of two experiments with similar results.
Fig. S5. Validation of antibody specific for UMPK amino acids 182–195. (A) Western blot of uninfected or A. phagocytophilum-infected RF/6A cell lysates probed with UMPK antibody detects a band having an apparent molecular weight of approximately 26 kDa, which corresponds to the expected size of UMPK, in the infected cell lysate only. Data are representative of two experiments with similar results. (B) UMPK is constitutively expressed throughout infection. RF/6A cells were synchronously infected with A. phagocytophilum for 4, 8, 12, 16, 24, 28 or 32 h, after which, the cells were fixed, screened with UMPK antibody, stained with DAPI and examined by confocal microscopy. Results shown are representative of three experiments. Statistically significant (***P ≤ 0.001; *P ≤ 0.05) values are indicated.
Movie S1. Endogenous Rab10-positive vesicles colocalize with intravacuolar A. phagocytophilum organisms. A. phagocytophilum infected RF/6A cells were screened with antibodies against Rab10 (red) and APH0032 (green), stained with DAPI (blue) and visualized by LSCM. Z-stack images obtained for a representative ApV in Fig. 1D were used for three-dimensional rendering.
Movie S2. TGN-derived vesicles colocalize with intravacuolar A. phagocytophilum organisms. A. phagocytophilum-infected RF/6A cells were screened with antibodies against TGN46 (red) and APH0032 (green), stained with DAPI (blue) and visualized by LSCM. Z-stack images obtained for a representative ApV in Fig. 3B were used for three-dimensional rendering.
References
- Abu Kwaik Y, Bumann D. Microbial quest for food in vivo: ‘nutritional virulence’ as an emerging paradigm. Cell Microbiol. 2013;15:882–890. doi: 10.1111/cmi.12138. [DOI] [PubMed] [Google Scholar]
- Allen JR, Ross ST, Davidson MW. Structured illumination microscopy for superresolution. Chem Phys Chem. 2014;15:566–576. doi: 10.1002/cphc.201301086. [DOI] [PubMed] [Google Scholar]
- Arasaki K, Toomre DK, Roy CR. The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe. 2012;11:46–57. doi: 10.1016/j.chom.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA, Heinzen RA. Gene inactivation in Coxiella burnetii. Methods Mol Biol (Clifton, NJ) 2014;1197:329–345. doi: 10.1007/978-1-4939-1261-2_19. [DOI] [PubMed] [Google Scholar]
- Beatty WL. Trafficking from CD63-positive late endocytic multivesicular bodies is essential for intracellular development of Chlamydia trachomatis. J Cell Sci. 2006;119:350–359. doi: 10.1242/jcs.02733. [DOI] [PubMed] [Google Scholar]
- Beyer AR, Truchan HK, May LJ, Walker NJ, Borjesson DL, Carlyon JA. The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation. Cell Microbiol. 2014;17:504–519. doi: 10.1111/cmi.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom T, Somerharju P, Ikonen E. Synthesis and biosynthetic trafficking of membrane lipids. Cold Spring Harb Perspect Biol. 2011;3:a004713. doi: 10.1101/cshperspect.a004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompain G, Muller C, Meas-Yedid V, Schmitt-Kopplin P, Lazarow PB, Subtil A. The intracellular bacteria Chlamydia hijack peroxisomes and utilize their enzymatic capacity to produce bacteria-specific phospholipids. PLoS One. 2014;9:e86196. doi: 10.1371/journal.pone.0086196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capmany A, Damiani MT. Chlamydia trachomatis intercepts Golgi-derived sphingolipids through a Rab14-mediated transport required for bacterial development and replication. PLoS One. 2010;5:e14084. doi: 10.1371/journal.pone.0014084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabeo RA, Mead DJ, Hackstadt T. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc Natl Acad Sci U S A. 2003;100:6771–6776. doi: 10.1073/pnas.1131289100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlyon JA. Laboratory maintenance of Anaplasma phagocytophilum. Current Protocols in Microbiology. 2005;Chapter 3(Unit 3A):2. doi: 10.1002/9780471729259.mc03a02s00. [DOI] [PubMed] [Google Scholar]
- Carlyon JA, Abdel-Latif D, Pypaert M, Lacy P, Fikrig E. Anaplasma phagocytophilum utilizes multiple host evasion mechanisms to thwart NADPH oxidase-mediated killing during neutrophil infection. Infect Immun. 2004;72:4772–4783. doi: 10.1128/IAI.72.8.4772-4783.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Nair AD, Indukuri VV, Gong S, Felsheim RF, Jaworski D, et al. Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog. 2013;9:e1003171. doi: 10.1371/journal.ppat.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc Natl Acad Sci U S A. 2008;105:9379–9384. doi: 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Rzomp KA, Tvinnereim A, Scidmore MA, Wizel B. Chlamydia pneumoniae inclusion membrane protein Cpn0585 interacts with multiple Rab GTPases. Infect Immun. 2007;75:5586–5596. doi: 10.1128/IAI.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumoux M, Clare DK, Saibil HR, Hayward RD. Chlamydiae assemble a pathogen synapse to hijack the host endoplasmic reticulum. Traffic. 2012;13:1612–1627. doi: 10.1111/tra.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, et al. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006;2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Rikihisa Y. Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. J Bacteriol. 2007;189:7819–7828. doi: 10.1128/JB.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold DA, Kaplan AD, Lis A, Bett GC, Rosowski EE, Cirelli KM, et al. The Toxoplasma dense granule proteins GRA17 and GRA23 mediate the movement of small molecules between the host and the parasitophorous vacuole. Cell Host Microbe. 2015;17:642–652. doi: 10.1016/j.chom.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore MA, Rockey DD. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci U S A. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Rockey DD, Heinzen RA, Scidmore MA. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- Henderson B, Martin A. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun. 2011;79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B, Martin A. Bacterial moonlighting proteins and bacterial virulence. Curr Top Microbiol Immunol. 2013;358:155–213. doi: 10.1007/82_2011_188. [DOI] [PubMed] [Google Scholar]
- Heuer D, Rejman Lipinski A, Machuy N, Karlas A, Wehrens A, Siedler F, et al. Chlamydia causes fragmentation of the Golgi compartment to ensure reproduction. Nature. 2009;457:731–735. doi: 10.1038/nature07578. [DOI] [PubMed] [Google Scholar]
- Huang B, Hubber A, McDonough JA, Roy CR, Scidmore MA, Carlyon JA. The Anaplasma phagocytophilum-occupied vacuole selectively recruits Rab-GTPases that are predominantly associated with recycling endosomes. Cell Microbiol. 2010a;12:1292–1307. doi: 10.1111/j.1462-5822.2010.01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Troese MJ, Howe D, Ye S, Sims JT, Heinzen RA, et al. Anaplasma phagocytophilum APH_0032 is expressed late during infection and localizes to the pathogen-occupied vacuolar membrane. Microb Pathog. 2010b;49:273–284. doi: 10.1016/j.micpath.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Troese MJ, Ye S, Sims JT, Galloway NL, Borjesson DL, Carlyon JA. Anaplasma phagocytophilum APH_1387 is expressed throughout bacterial intracellular development and localizes to the pathogen-occupied vacuolar membrane. Infect Immun. 2010c;78:1864–1873. doi: 10.1128/IAI.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJdo JW, Wu C, Magnarelli LA, Fikrig E. Serodiagnosis of human granulocytic ehrlichiosis by a recombinant HGE-44-based enzyme-linked immunosorbent assay. J Clin Microbiol. 1999;37:3540–3544. doi: 10.1128/jcm.37.11.3540-3544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junutula JR, De Maziere AM, Peden AA, Ervin KE, Advani RJ, van Dijk SM, et al. Rab14 is involved in membrane trafficking between the Golgi complex and endosomes. Mol Biol Cell. 2004;15:2218–2229. doi: 10.1091/mbc.E03-10-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlon A, Ojogun N, Ragland SA, Seidman D, Troese MJ, Ottens AK, et al. Anaplasma phagocytophilum Asp14 is an invasin that interacts with mammalian host cells via its C terminus to facilitate infection. Infect Immun. 2013;81:65–79. doi: 10.1128/IAI.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholti A, Charlier D, Gigot D, Huysveld N, Roovers M, Glansdorff N. pyrH-encoded UMP-kinase directly participates in pyrimidine-specific modulation of promoter activity in Escherichia coli. J Mol Biol. 1998;280:571–582. doi: 10.1006/jmbi.1998.1910. [DOI] [PubMed] [Google Scholar]
- Klein MB, Miller JS, Nelson CM, Goodman JL. Primary bone marrow progenitors of both granulocytic and monocytic lineages are susceptible to infection with the agent of human granulocytic ehrlichiosis. J Infect Dis. 1997;176:1405–1409. doi: 10.1086/517332. [DOI] [PubMed] [Google Scholar]
- Landais S, Gounon P, Laurent-Winter C, Mazie JC, Danchin A, Barzu O, Sakamoto H. Immunochemical analysis of UMP kinase from Escherichia coli. J Bacteriol. 1999;181:833–840. doi: 10.1128/jb.181.3.833-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11:739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- Lin M, Rikihisa Y. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect Immun. 2003;71:5324–5331. doi: 10.1128/IAI.71.9.5324-5331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski AR, Heymann J, Meissner C, Karlas A, Brinkmann V, Meyer TF, Heuer D. Rab6 and Rab11 regulate Chlamydia trachomatis development and Golgin-84-dependent Golgi fragmentation. PLoS Pathog. 2009;5:e1000615. doi: 10.1371/journal.ppat.1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Storrie B. Are Rab proteins the link between Golgi organization and membrane trafficking? Cell Mol Life Sci. 2012;69:4093–4106. doi: 10.1007/s00018-012-1021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronunzio JE, Kurscheid S, Fikrig E. Postgenomic analyses reveal development of infectious Anaplasma phagocytophilum during transmission from ticks to mice. J Bacteriol. 2012;194:2238–2247. doi: 10.1128/JB.06791-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G. Lipids of the Golgi membrane. Trends Cell Biol. 1998;8:29–33. doi: 10.1016/s0962-8924(97)01196-3. [DOI] [PubMed] [Google Scholar]
- Mishra MK, Gerard HC, Whittum-Hudson JA, Hudson AP, Kannan RM. Dendrimer-enabled modulation of gene expression in Chlamydia trachomatis. Mol Pharm. 2012;9:413–421. doi: 10.1021/mp200512f. [DOI] [PubMed] [Google Scholar]
- Mott J, Barnewall RE, Rikihisa Y. Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect Immun. 1999;67:1368–1378. doi: 10.1128/iai.67.3.1368-1378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munderloh UG, Lynch MJ, Herron MJ, Palmer AT, Kurtti TJ, Nelson RD, Goodman JL. Infection of endothelial cells with Anaplasma marginale and A. phagocytophilum. Vet Microbiol. 2004;101:53–64. doi: 10.1016/j.vetmic.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc Natl Acad Sci U S A. 2012;109:20800–20807. doi: 10.1073/pnas.1218674109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojogun N, Kahlon A, Ragland SA, Troese MJ, Mastronunzio JE, Walker NJ, et al. Anaplasma phagocytophilum outer membrane protein A interacts with sialylated glycoproteins to promote infection of mammalian host cells. Infect Immun. 2012;80:3748–3760. doi: 10.1128/IAI.00654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano RE, Martin OC, Kang HC, Haugland RP. A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J Cell Biol. 1991;113:1267–1279. doi: 10.1083/jcb.113.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzomp KA, Scholtes LD, Briggs BJ, Whittaker GR, Scidmore MA. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect Immun. 2003;71:5855–5870. doi: 10.1128/IAI.71.10.5855-5870.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scidmore MA, Fischer ER, Hackstadt T. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J Cell Biol. 1996;134:363–374. doi: 10.1083/jcb.134.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman D, Ojogun N, Walker NJ, Mastronunzio J, Kahlon A, Hebert KS, et al. Anaplasma phagocytophilum surface protein AipA mediates invasion of mammalian host cells. Cell Microbiol. 2014;16:1133–1145. doi: 10.1111/cmi.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RK, Roy CR. A Rab-centric perspective of bacterial pathogen-occupied vacuoles. Cell Host Microbe. 2013;14:256–268. doi: 10.1016/j.chom.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanshu DK, Kamlekar RK, Wijesinghe DS, Zou X, Zhai X, Mishra SK, et al. Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids. Nature. 2013;500:463–467. doi: 10.1038/nature12332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Sukumaran B, Mastronunzio JE, Narasimhan S, Fankhauser S, Uchil PD, Levy R, et al. Anaplasma phagocytophilum AptA modulates Erk1/2 signalling. Cell Microbiol. 2011;13:47–61. doi: 10.1111/j.1462-5822.2010.01516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troese MJ, Carlyon JA. Anaplasma phagocytophilum dense-cored organisms mediate cellular adherence through recognition of human P-selectin glycoprotein ligand 1. Infect Immun. 2009;77:4018–4027. doi: 10.1128/IAI.00527-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troese MJ, Kahlon A, Ragland SA, Ottens AK, Ojogun N, Nelson KT, et al. Proteomic analysis of Anaplasma phagocytophilum during infection of human myeloid cells identifies a protein that is pronouncedly upregulated on the infectious dense-cored cell. Infect Immun. 2011;79:4696–4707. doi: 10.1128/IAI.05658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchan HK, Seidman D, Carlyon JA. Breaking in and grabbing a meal: Anaplasma phagocytophilum cellular invasion, nutrient acquisition, and promising tools for their study. Microbes and Infection/Institut Pasteur. 2013;15:1017–1025. doi: 10.1016/j.micinf.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vromman F, Subtil A. Exploitation of host lipids by bacteria. Curr Opin Microbiol. 2014;17:38–45. doi: 10.1016/j.mib.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Walker NJ, Clark EA, Ford DC, Bullifent HL, McAlister EV, Duffield ML, et al. Structure and function of cytidine monophosphate kinase from Yersinia pseudotuberculosis, essential for virulence but not for survival. Open Biol. 2012;2:120142. doi: 10.1098/rsob.120142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Berg EA, Feng X, Shen L, Smith T, Costello CE, Zhang YX. Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci. 2006;15:122–134. doi: 10.1110/ps.051616206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijesinghe DS, Brentnall M, Mietla JA, Hoeferlin LA, Diegelmann RF, Boise LH, Chalfant CE. Ceramide kinase is required for a normal eicosanoid response and the subsequent orderly migration of fibroblasts. J Lipid Res. 2014;55:1298–1309. doi: 10.1194/jlr.M048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Hackstadt T. Sphingomyelin trafficking in Chlamydia pneumoniae-infected cells. Cell Microbiol. 2001;3:145–152. doi: 10.1046/j.1462-5822.2001.00098.x. [DOI] [PubMed] [Google Scholar]
- Wood DO, Wood RR, Tucker AM. Genetic systems for studying obligate intracellular pathogens: an update. Curr Opin Microbiol. 2014;17:11–16. doi: 10.1016/j.mib.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Rikihisa Y. Subversion of NPC1 pathway of cholesterol transport by Anaplasma phagocytophilum. Cell Microbiol. 2012;14:560–576. doi: 10.1111/j.1462-5822.2011.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Lin M, Rikihisa Y. Cholesterol-dependent Anaplasma phagocytophilum exploits the low-density lipoprotein uptake pathway. PLoS Pathog. 2009;5:e1000329. doi: 10.1371/journal.ppat.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Tsai MD. Nucleoside monophosphate kinases: structure, mechanism, and substrate specificity. Adv Enzymol Relat Areas Mol Biol. 1999;73:103–134. doi: 10.1002/9780470123195.ch4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.