Publisher's Note: There is an Inside Blood Commentary on this article in this issue.
Key Points
B-cell–specific expression of Myd88p.L252P leads to the development of DLBCL in mice.
The Myd88p.L252P mutation cooperates with BCL2 amplifications in ABC-DLBCL lymphomagenesis in vivo.
Abstract
The adaptor protein MYD88 is critical for relaying activation of Toll-like receptor signaling to NF-κB activation. MYD88 mutations, particularly the p.L265P mutation, have been described in numerous distinct B-cell malignancies, including diffuse large B-cell lymphoma (DLBCL). Twenty-nine percent of activated B-cell–type DLBCL (ABC-DLBCL), which is characterized by constitutive activation of the NF-κB pathway, carry the p.L265P mutation. In addition, ABC-DLBCL frequently displays focal copy number gains affecting BCL2. Here, we generated a novel mouse model in which Cre-mediated recombination, specifically in B cells, leads to the conditional expression of Myd88p.L252P (the orthologous position of the human MYD88p.L265P mutation) from the endogenous locus. These mice develop a lymphoproliferative disease and occasional transformation into clonal lymphomas. The clonal disease displays the morphologic and immunophenotypical characteristics of ABC-DLBCL. Lymphomagenesis can be accelerated by crossing in a further novel allele, which mediates conditional overexpression of BCL2. Cross-validation experiments in human DLBCL samples revealed that both MYD88 and CD79B mutations are substantially enriched in ABC-DLBCL compared with germinal center B-cell DLBCL. Furthermore, analyses of human DLBCL genome sequencing data confirmed that BCL2 amplifications frequently co-occurred with MYD88 mutations, further validating our approach. Finally, in silico experiments revealed that MYD88-mutant ABC-DLBCL cells in particular display an actionable addiction to BCL2. Altogether, we generated a novel autochthonous mouse model of ABC-DLBCL that could be used as a preclinical platform for the development and validation of novel therapeutic approaches for the treatment of ABC-DLBCL.
Introduction
Toll-like receptors (TLRs) belong to a class of pattern-recognition receptors.1 Ten distinct human TLRs have been described.1 Except for TLR3, all TLRs require the adaptor protein MYD88 to initiate downstream signaling.1,2 Upon TLR activation, MYD88 is recruited to the Toll/interleukin-1 receptor (TIR) domain of the activated TLR via its own TIR domain.3 MYD88 recruits IRAK1, -2, and -4 to ultimately form the myddosome.4 IRAK4-mediated phosphorylation of IRAK1 and IRAK2 promotes TRAF6 recruitment,5 which subsequently ubiquitylates and activates TAK1,6 leading to NF-κB activation.2
Inappropriate TLR signaling through somatic MYD88 mutations has been described in numerous hematologic malignancies, such as chronic lymphocytic leukemia (CLL),7 Waldenström macroglobulinemia,8 and diffuse large B-cell lymphoma (DLBCL).9 Specifically, 29% of activated B-cell–type DLBCLs (ABC-DLBCLs), which typically display constitutive NF-κB activation, carry the p.L265P mutation (position according to the protein accession NP_002459) in the hydrophobic core of the MYD88 TIR domain.9 ABC-DLBCL was shown to display MYD88p.L265P-dependent NF-κB activation and STAT3 phosphorylation.9 The MYD88p.L265P mutation is exceedingly rare in non-ABC-DLBCLs, such as germinal center B-cell (GCB) and primary mediastinal B-cell lymphoma.9 Moreover, MYD88p.L265P-mutant ABC-DLBCL appears to be dependent on continued MYD88p.L265P expression.9 Together, these data implicate MYD88-dependent oncogenic NF-κB signaling as an integral contributor to ABC-DLBCL pathogenesis and as a potential therapeutic target. However, despite the clinical relevance of the MYD88p.L265P mutation, no autochthonous mouse model that faithfully mimics this critical genomic aberration has yet been generated.
Materials and methods
Experimental mice
B-cell–specific Cre expression was achieved by using Cd19Cre,10 AidCre,11 and Cd21Cre12 deleter mouse strains (The Jackson Laboratory). The targeting vector depicted in Figure 1A was constructed by using standard techniques. The gene targeting strategy was based on the National Center for Biotechnology Information transcript NM_010851.2 in which exon 1 contains the translation initiation codon. Wild-type (wt) exons 2-6, including the entire 3′ untranslated region were flanked with LoxP sites. An additional polyadenylation signal (human growth hormone polyadenylation [hGHpA] signal) has been inserted between the 3′ untranslated region and the distal LoxP site to prevent downstream transcription of the mutated exon 5. The size of the LoxP-flanked region is 4.7 kb. Exons 2-6, including the splice acceptor site of intron 1, have been duplicated and inserted downstream of the distal LoxP site. The p.L252P mutation has been introduced into the duplicated exon 5. Positive selection markers were flanked by Frt (neomycin resistance; NeoR) and F3 (puromycin resistance; PuroR) sites and inserted into intron 1 and downstream of the hGHpA, respectively. The targeting vector was generated by using BAC clones from the C57BL/6J RPCIB-731 BAC library and was transfected into the C57BL/6N Tac embryonic stem (ES) cell line. Targeted clones were isolated by using double positive (NeoR and PuroR) selection, and correct integration was verified by Southern blotting. Upon blastocyst injection, germ line–transmitting transgenic mice were received. The conditional Myd88c-p.L252P allele was obtained after in vivo Flp-mediated removal of the selection markers. This allele expresses the wild-type Myd88 protein, because the presence of the hGHpA cassette downstream of wild-type exon 6 prevents transcription of the mutant exons 2-6. The constitutive Myd88p.L252P allele is obtained after in vivo Cre-mediated deletion of wild-type exons 2-6 and hGHpA. This allele expresses the mutant Myd88L252P protein. The remaining recombination sites are located in nonconserved regions of the genome. To generate conditional BCL2 knockin mice (Rosa26LSL.BCL2.IRES.GFP), a Rosa26 locus-targeting vector was used in which CAGS (cytomegalovirus early enhancer/chicken β actin) promoter-driven expression of the transgene (BCL2) and downstream expression of an internal ribosomal entry site–driven green fluorescent protein (IRES-GFP, as a reporter for Cre-mediated recombination) is prevented by a LoxP-flanked STOP cassette (supplemental Figure 5C, available on the Blood Web site). The transgenic mice expressed the transgenes only after Cre-mediated excision of the LoxP-flanked STOP cassette. The targeting vector was electroporated into ES cells (BRUCE4), which were screened for correct integration by standard Southern blot methods. Correctly targeted ES cells were used to generate chimeras, which were backcrossed onto a pure C57BL/6N background and examined for germ line transmission.
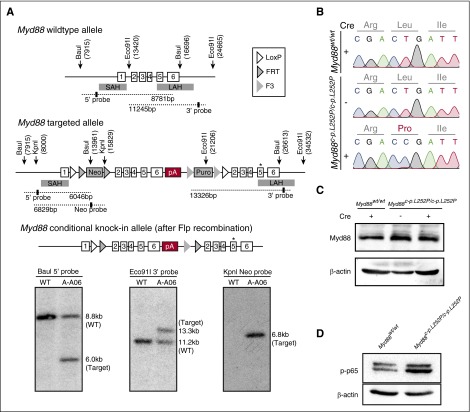
Figure 1.
Construction of a conditional Myd88p.L252P allele. (A) Targeting of the Myd88 locus in C57BL/6N Tac ES cells. The endogenous Myd88 locus was targeted with the linearized vector described in the supplemental Data. The targeted allele before (middle panel) and after Flp-mediated recombination of FRT and F3 sites (bottom panel) is schematically depicted. The Southern blots of BauI, Eco91I, and KpnI digested genomic DNA probed with a 5′, a 3′, and a Neo probe, respectively, are shown below the schematic illustration of the targeting strategy. Positions of restriction sites and probes are shown in the schematic drawing above. (B) Myd88p.L252P mRNA is expressed upon Cre-mediated recombination in MEFs. Myd88wt/wt and Myd88c-p.L252P/c-p.L252P MEFs were isolated. RNA was isolated from both cell lines before LentiCre application (Sanger sequencing chromatograms, top and middle panels) and the Myd88 mRNA sequence was determined after reverse transcription. The wild-type sequence was recovered from both cell lines. After LentiCre application and puromycin selection, only the p.L252P sequence could be recovered from Myd88c-p.L252P/c-p.L252P MEFs (Sanger sequencing chromatogram, bottom panel). (C) The Myd88p.L252P isoform is expressed in Myd88c-p.L252P/c-p.L252P MEFs after LentiCre-mediated recombination. Myd88wt/wt and Myd88c-p.L252P/c-p.L252P MEFs were LentiCre exposed and puromycin selected, as in (B). Whole-cell lysates were separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto polyvinylidene difluoride membranes before Myd88 and β-actin, which served as loading controls, and were visualized by immunoblotting. Both Myd88wt and Myd88p.L252P proteins were expressed at equal levels. (D) Conditional LentiCre-mediated Myd88p.L252P expression leads to p65 Ser-536 phosphorylation. Myd88wt/wt and Myd88c-p.L252P/c-p.L252P MEFs were transduced with LentiCre and puromycin selected, as in (B). Upon selection, cells were lysed, proteins were separated on SDS-PAGE, and pSer-536 p65 was visualized by immunoblot. SAH, short arm of homology; LAH, long arm of homology; pA, polyadenylation signal sequence.
The Eµ:Myc model has been described earlier and was used, as previously described.13 The Eµ:Tcl1 model of CLL was described earlier.14 All the experiments that involved the breeding and/or treatment of rodents were approved by the local animal care committee and the relevant authorities (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen, AZ: 84-02.04.2014.A146/A083).
Transplantation experiments
Animal protocols based on the Eµ:Myc transgenic mouse model as used in this study were approved by the local governmental review board (Landesamt Berlin) and conformed to regulatory standards. Isolation, retroviral infection, transplantation of Eµ:Myc transgenic fetal liver cells (ie, hematopoietic stem cells), and subsequent monitoring of the recipient mice regarding lymphoma onset were performed as previously described.15 Specifically, numerous independently isolated fetal liver cell populations were stably transduced with a murine stem cell virus-based Myd88p.L252P-IRES-GFP retrovirus or a murine stem cell virus–empty GFP construct as a control before their propagation in recipient mice.
Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) murine samples were sliced at 2 to 4 μm. Sections were stained with hematoxylin and eosin (H&E) and antibodies against Ki-67 (Cell Marque), B220 (RA3-6B2BD,), p65 (C-20; Santa Cruz,), Irf4 (M-17; Santa Cruz Biotechnology,), Bcl6 (C-19; Santa Cruz Biotechnology), and Cd138 (553712; BD). Staining intensities were scored by 2 independent observers who used a 4-tier scale. FFPE human samples were sliced at 2 to 4 µm. Sections were stained with H&E and antibodies against CD10 (NCL-L-CD10-270; Novocastra), BCL6 (M7211; Dako), IRF4 (M7259; Dako), BCL2 (M0877; Dako), and CD138 (138M-16; Cell Marque).
Sequence analysis of immunoglobulin genes and cloning of polymerase chain reaction products
RNA was extracted from sections of frozen biopsies with TRI Reagent (Sigma, Taufkirchen, Germany). One microgram of RNA was converted to complementary DNA with QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). Mouse IgH rearrangements were analyzed by using previously published methods.16 Each polymerase chain reaction (PCR) was analyzed on a Qiaxcel Advanced Instrument (Qiagen) by using ScreenGel Software v1.2. Direct sequencing was performed with the ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit v3.1 (Life Technologies, Darmstadt, Germany) on an ABI 3130 sequencer (Applied Biosystems, Foster City, CA). Sequences were analyzed by using 4Peaks Software v1.7.2 (The Netherlands Cancer Institute, Amsterdam, The Netherlands), Lasergene software (DNAStar, Madison, WI), and by manual review. Sequences were compared with mouse germ line Ig gene sequences with International ImMunoGeneTics database.17 In addition, the PCR products of 1 case were cloned with the TOPO TA Cloning Kit (Life Technologies), which resulted in 32 sequences. To generate genealogic trees, sequence analysis focused on the IGHV segment, starting within FR1. To exclude possible Taq errors, only mutations shown more than once were considered for analysis (see supplemental Data).
Results
To assess the role of MYD88p.L265P in B-cell lymphomagenesis, we generated a conditional Myd88p.L252P allele (Myd88c-p.L252P) that is expressed from the endogenous locus upon Cre-mediated recombination (Figure 1A). Murine Myd88p.L252P is at the orthologous position of human Myd88p.L265P. To verify Cre-inducible Myd88p.L252P expression, we derived murine embryonic fibroblasts (MEFs) from wild-type and Myd88c-p.L252P/c-p.L252P mice. By using Sanger sequencing, we detected expression of Myd88wt messenger RNA (mRNA) in both Myd88wt/wt and Myd88c-p.L252P/c-p.L252P MEFs, indicating lack of background expression of Myd88p.L252P in the absence of Cre recombinase. In contrast, cytomegalovirus promoter-driven Cre expression mediated by lentiviral transduction of Myd88c-p.L252P/c-p.L252P MEFs induced recombination in the Myd88 locus, leading to expression of Myd88p.L252P-mutant mRNA (Figure 1B). Myd88 protein abundance was unaffected by stable Cre expression, indicating that the Myd88p.L252P isoform is expressed at endogenous levels (Figure 1C). This Myd88p.L252P expression translated into robust p65 Ser-536 phosphorylation, indicating activation of NF-κB signaling (Figure 1D).
To determine whether B-cell–specific Myd88p.L252P expression promotes lymphomagenesis, we compared Cd19Cre/wt;Myd88c-p.L252P/wt (hereafter M-Cd19) and Cd19Cre/wt (hereafter Cd19) mice. The mice were longitudinally monitored by magnetic resonance imaging (MRI). Although no gross lymphoproliferative disease (LPD) was detectable in Cd19 mice, M-Cd19 mice developed splenomegaly and occasional lymphadenopathy starting at the age of ∼60 weeks (Figure 2B). Similar MRI morphologic signs of LPD were observed when B-cell–specific Myd88p.L252P expression was induced by Cre expression that was driven off the Aid promoter (hereafter M-Aid), which is active in GCB cells, during both T cell–dependent and –independent immune responses (supplemental Figure 1A).11,18 Furthermore, Cd21 promoter-driven Cre, which is expressed when immature transitional B cells differentiate into mature long-lived peripheral B cells,12 induced an LPD in Cd21Cre/wt;Myd88c-p.L252P/wt mice (hereafter M-Cd21) that was indistinguishable from that observed in M-Cd19 mice (supplemental Figure 1A).
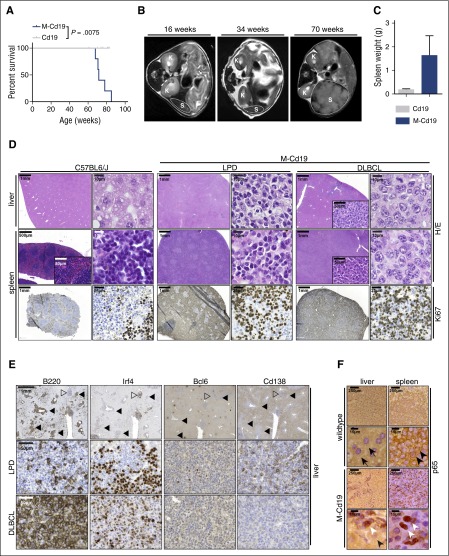
Figure 2.
B-cell–specific Myd88p.L252P expression drives lymphoproliferation and lymphomagenesis in vivo. (A) B-cell–specific expression of Myd88p.L252P significantly reduces overall survival in vivo. Kaplan-Meier curves illustrate the overall survival of M-Cd19 mice. M-Cd19 mice display a significantly reduced survival compared with the respective controls (log-rank test). (B) Serial MRI scans in 16-, 34-, and 70-week-old M-Cd19 mice revealed the occurrence of splenomegaly in 70-week-old M-Cd19 mice. (C) M-Cd19 mice display splenomegaly at the time of death. Preterminal M-Cd19 and Cd19 mice were sacrificed and spleen weight was recorded. Bars represent the average (n = 3); error bars represent standard deviations. (D) M-Cd19 mice develop lymphoproliferative disease and occasional lymphoma. The top panels show H&E staining of spleens and livers isolated from C57BL/6 and M-Cd19 mice at the time of death. Although the organ architecture appeared normal in C57BL/6 wild-type mice, the architecture of spleens isolated from M-Cd19 mice was largely disrupted by infiltration of small mature lymphocytes (LPD columns) or large blastoid cells (DLBCL columns). The bottom panel shows the partial and complete disruption of the spleen by infiltrates with high proliferative indices. (E) Immunohistochemical characterization of the liver infiltrates of M-Cd19 mice. Areas of infiltrates morphologically resembling DLBCL (marked with solid triangles) showed a homogeneous staining pattern of B220 and Irf4 positivity, whereas they were negative for Bcl6 and Cd138 (bottom panel). Infiltrated areas of small, mature lymphocytes (marked with open triangles) displayed a more heterogeneous staining pattern of positive and negative cells for B220, Irf4, and Cd138, whereas staining was largely negative for Bcl6 (middle panel). (F) The lymphoma cells infiltrating spleens and livers of M-Cd19 mice displayed a largely nuclear localization of p65, indicating NF-κB activation. Black arrows indicate hepatocytes, black arrowheads indicate cytoplasmic p65 staining in splenic lymphocytes in C57BL/6 wild-type mice, and white arrowheads indicate nuclear p65 staining in lymphoma cells in M-Cd19 mice.
The MRI morphologic occurrence of lymphoproliferative lesions translated into a significantly (P = .0075) reduced overall survival of M-Cd19 mice (median survival, 501 days) compared with Cd19 mice (median survival, not reached in 10 mice observed for 427 to 581 days [average, 539 days]) (Figure 2A). Similar data were obtained when Cre expression was driven off the Aid or Cd21 promoter in M-Aid and M-Cd21 mice (median survival, 574 and 610 days, respectively) (supplemental Figure 1B). We noted that the majority of M-Cd19, M-Aid, and M-Cd21 mice included in this study displayed splenomegaly (4 of 4, M-Cd19; 2 of 3, M-Aid; and 2 of 3, M-Cd21), occasionally accompanied by macroscopic lymphadenopathy (1 of 4, M-Cd19; 2 of 3, M-Aid; and 1 of 3, M-Cd21). The strong oncogenic potential of the Myd88p.L252P mutation was also verified in an alternative mouse model of aggressive B-cell lymphoma in which Myd88p.L252P dramatically shortened tumor onset driven by oncogenic Myc (Eµ:Myc transgenic fetal liver cells transplanted into lethally irradiated recipient mice, stably transduced with either empty vector [n = 40; median onset not reached] or Myd88p.L252P [n = 12; median onset, 36 days]; log-rank Mantel-Cox P < .0001) (supplemental Figure 2).
To characterize the disease occurring in M-Cd19 mice, we initially performed histologic examinations. H&E staining of spleens and livers of M-Cd19, M-Aid, and M-Cd21 mice revealed infiltrates of lymphoid cells in these organs. In particular, the architecture of the spleens of these mice was largely disrupted by these infiltrates (4 of 4, M-Cd19; 3 of 3, M-Aid; 3 of 3, M-Cd21). Of note, we did not detect any infiltration of the bone marrow (n = 3 per genotype; supplemental Figure 3). Further analyses of the splenic and hepatic infiltrates revealed that these lesions constituted a largely monomorphic lymphoid cell population with indolent appearance, consistent with an LPD. Intriguingly, in a subset of mice (1 of 4, M-Cd19; 1 of 3, M-Aid; and 1 of 3, M-Cd21), diffuse large lymphoid infiltrates resembling DLBCL were detectable (Figure 2D; supplemental Figure 1D-E). Of note, the Ki-67 indices did not substantially differ between areas with LPD and DLBCL morphology (Figure 2D). To determine the clonality of these infiltrates, we next performed Southern blot analyses to detect clonal immunoglobulin (Ig) rearrangements. We were not able to detect clonal Ig rearrangements in genomic DNA isolated from areas with LPD-like appearance that were detected in M-Cd19, M-Aid, and M-Cd21 mice (supplemental Figure 4A). Strikingly, oligoclonal Ig rearrangement patterns were detected in DNA isolated from infiltrates that displayed DLBCL-like morphology in M-Cd19, M-Aid, and M-Cd21 mice (indicated by white triangles in supplemental Figure 4A). DNA isolated from spleens of an Eµ:Tcl1-driven CLL mouse model served as a positive control for the development of a clonal lymphoid neoplasm.14 Altogether, our data strongly indicate that B-cell–specific Myd88c-p.L252P expression largely leads to an LPD, with only occasional development of mono-/oligoclonal lymphoma.
To further characterize the LPD and lymphomas, we next performed immunohistochemistry in liver infiltrates of M-Cd19 mice. As shown in Figure 2E, the areas with DLBCL-like morphology displayed a strikingly uniform staining pattern. These lesions were B220 and Irf4 positive and were Bcl6 and Cd138 negative, resembling the immunophenotype of a post-GCB lymphoid malignancy.19 In contrast, the areas of LPD displayed a rather heterogeneous staining pattern for B220, Irf4, and Cd138 and were largely negative for Bcl6, further underscoring the nonclonal characteristics of these lesions. We noted that areas of LPD and DLBCL-like morphology in M-Aid and M-Cd21 mice displayed similar staining patterns (supplemental Figure 1F-G).
To further interrogate the nature of the DLBCL-like lesions, we next used immunohistochemistry to assess subcellular NF-κB localization, because constitutive NF-κB activation, indicated by nuclear enrichment, is a hallmark feature of ABC-DLBCL.9 These experiments revealed a uniform nuclear staining of the NF-κB subunit p65 in the DLBCL-like lesions detected in spleens and livers of M-Cd19, M-Aid, and M-Cd21 mice (Figure 2F; supplemental Figure 1C). Given that ABC-DLBCL, which is characterized by constitutive NF-κB activation, carries the p.L265P mutation in 29% of the cases,9 and given that the clonal lymphoproliferation we observed in M-Cd19 mice displayed morphologic features of DLBCL, we hypothesized that the B-cell–specific Myd88c-p.L252P expression drives ABC-DLBCL development in our model.
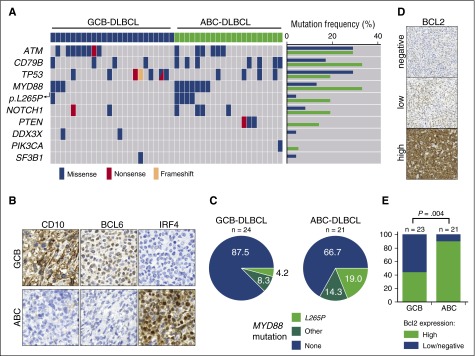
To cross-validate our observations, we analyzed MYD88 mutation frequency in human DLBCLs (n = 24, GCB-DLBCL; n = 21, ABC-DLBCL). We performed targeted sequencing of FFPE-extracted DNA by multiplex PCR covering the MYD88, ATM, BTK, CD79B, DDX3X, FBXW7, MAPK1, NOTCH1, PIK3CA, PIK3CD, PTEN, PTPN6, SF3B1, TP53, and XPO1 genes (Figure 3A). DLBCL subtypes were determined based on immunohistochemical stains for CD10, BCL6, and IRF4 following the Hans algorithm (Figure 3B).19 The non-GCB of Hans algorithm was designated as “ABC” in our study. Confirming previously published data, we found a substantial overrepresentation of MYD88 and CD79B mutations in ABC-DLBCLs compared with GCB-DLBCLs (Figure 3A,C; supplemental Table 1).9,20,21 Furthermore, the anti-apoptotic oncogene BCL2 is frequently deregulated in human DLBCL.22,23 Whereas the chromosomal translocation t(14;18)(q32;q21), which juxtaposes BCL2 and the IGHV enhancer, is commonly observed in GCB-DLBCL,23 focal copy number gains are more commonly observed in ABC-DLBCL (∼30% to 40%).22-25 Indeed, mining of the whole-genome sequencing data on DLBCL from the German International Cancer Genome Consortium Molecular Mechanisms in Malignant Lymphoma by Sequencing Project (https://icgc.org/ and C.L., M.S., and R.S., manuscript in preparation) revealed that 6 DLBCLs carry the MYD88p.L265P mutation. Remarkably, all of these samples belonged to the ABC-DLBCL subtype and carried a copy number gain of 18q detected by both fluorescent in situ hybridization with a BCL2-specific probe and somatic copy number variant mapping.26,27 A critical role for BCL2, particularly in activated B cells, is further corroborated by published transcriptome analyses. Specifically, BCL2 mRNA was not expressed in GCB cells but was massively induced during activation of peripheral blood B cells.28 Consistent with this, the majority of ABC-DLBCLs displayed BCL2 mRNA levels more than fourfold higher than GCB cells.28 Furthermore, exogenous expression of MYD88p.L265P in B cells was recently shown to induce the accumulation of self-reactive B cells in vivo only when apoptosis was opposed by Bcl2 overexpression.29 Additional circumstantial evidence suggesting that ABC-DLBCL in particular might depend on functional BCL2 activity could be derived from recently published, large-scale in vitro drug sensitivity profiling experiments. Specifically, re-assessment of a data set published by Stegmeier and colleagues revealed that MYD88-mutant DLBCLs in particular display an actionable BCL2 addiction, which could be exploited by the targeted high-affinity BCL2 inhibitor ABT-263 (supplemental Figure 5A-B).30 The BTK inhibitor ibrutinib was recently shown to synergistically interact with the BCL2 family inhibitor navitoclax (ABT-263) in killing ABC-DLBCL cells.31 We validated these observations in our cohort of DLBCL samples by using immunohistochemistry to assess BCL2 expression. As shown in Figure 3D-E, high-level BCL2 expression was significantly enriched in ABC-DLBCL patients compared with patients who had GCB-DLBCL. Altogether, these genomic, transcriptomic, immunohistochemical, and functional data strongly suggest that enhanced BCL2 activity plays a central pathomechanistic role in the development of ABC-DLBCL. On the basis of these considerations, we next asked whether lymphoma development in M-Cd19 mice could be accelerated by increased BCL2 gene dosage in vivo. To mimic BCL2 amplification in vivo, we generated a novel conditional allele in which BCL2.IRES.GFP was targeted into the Rosa26 locus. BCL2 expression was prevented by the insertion of a LoxP.STOP.LoxP cassette upstream of the translation-initiating codon (supplemental Figure 5C). By using this allele, we generated Cd19Cre/wt;Rosa26LSL.BCL2.IRES.GFP/wt;Myd88c-p.L252P/wt mice (hereafter M-B-Cd19), in which Cd19-driven Cre mediates the excision of the LoxP.STOP.LoxP cassette from the Rosa26 locus, leading to CAGS-promoter-driven BCL2 overexpression in addition to Myd88p.L252P expression. MRI scans revealed that M-B-Cd19 mice developed splenomegaly substantially earlier than M-Cd19, M-Aid, or M-Cd21 mice (Figures 2B and 4B; supplemental Figure 1A). Furthermore, M-B-Cd19 mice displayed a significantly reduced overall survival (median survival, 179 days) compared with M-Cd19, M-Aid, and M-Cd21 mice (long-rank P = .0008, P = .0001, and P < .0001, respectively) (Figure 4A). Of note, all of the M-B-Cd19 mice displayed excessive lymphadenopathy and/or splenomegaly at the time of death (splenomegaly in 7 of 8 mice; lymphadenopathy in 6 of 8 mice).
Figure 3.
MYD88 mutations and high expression of BCL2 are enriched in ABC-DLBCL. (A) MYD88 mutations are substantially enriched in human ABC-DLBCL. Human DLBCLs were stratified as ABC- or GCB-DLBCL following the Hans algorithm, as depicted in (B). After immunohistochemistry-based stratification, DNA was isolated from tissue sections and subjected to targeted deep sequencing by a multiplex PCR, which covered the ATM, BTK, CD79B, DDX3X, FBXW7, MAPK1, MYD88, NOTCH1, PIK3CA, PIK3CD, PTEN, PTPN6, SF3B1, TP53, and XPO1 genes. MYD88 mutations per se, and particularly the MYD88p.L265P mutation were substantially enriched in ABC-DLBCL. Similarly, CD79B and PTEN mutations were enriched in ABC-DLBCL. (C) Distribution of MYD88 mutations detected in human DLBCL samples is shown in pie charts. The samples were classified into high and low or negative expression of BCL2, as shown in (D). (E) High protein expression levels of BCL2 are significantly enriched in ABC-DLBCL (Fisher’s exact test).
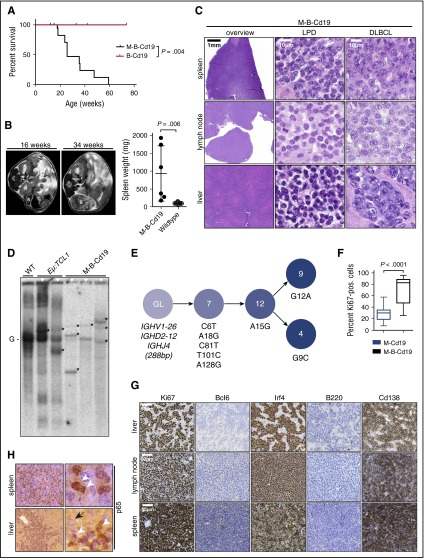
Figure 4.
Combination of B-cell–specific BCL2 overexpression and Myd88p.L252P expression drives ABC-DLBCL development in vivo. (A) Kaplan-Meier curve illustrates overall survival of M-B-Cd19 mice. M-B-Cd19 mice display a significantly reduced survival compared with the respective controls (log-rank test). (B) M-B-Cd19 mice develop splenomegaly at 25 to 35 weeks. MRI scans and quantification of spleen weights at autopsy are shown (two-tailed Student t test). (C) The architecture of spleens and lymph nodes isolated from M-B-Cd19 mice was completely disrupted by a homogeneous infiltrating population of large lymphoblastoid cells (DLBCL column). Small areas of more heterogeneous lymphoid cell populations consisting of small lymphoid cells were occasionally detectable (LPD column). The livers of M-B-Cd19 mice were diffusely infiltrated by large blastoid cells with DLBCL-like morphology. (D) Variable-diversity-joining–recombination analysis by Southern blot analysis revealed the presence of clonal populations in tumors of M-B-Cd19 mice. Tumors from leukemic Eµ:TCL1 mice were used as oligoclonal controls. The germ line configuration is also present in the WT controls and is depicted as “G.” Asterisks indicate clonal rearrangements. Samples of mice with lymphoma detected by histologic analyses are marked with white triangles (top). (E) Genealogic trees of murine DLBCL isolated from M-B-Cd19 mice. Igh rearrangements were cloned and individual clones were sequenced. Analysis focused on the Ighv segments starting with FR1. Genealogic tree was derived from 32 sequences. The tree-like structures demonstrate ongoing somatic hypermutation. Numbers in circles show number of sequence reads; numbering of mutations is in regard to the germ line sequence of Ighv1-26. (F) The Ki-67 index of DLBCL lesions from M-B-Cd19 mice was significantly higher than that in M-Cd19 mice (P = 2.27 × 10−12, two-tailed Student t test; n = 3 mice per genotype; 20 fields of view per lesion). (G) Lymphoma infiltrates of M-B-Cd19 mice were immunohistochemically analyzed and stained positive for Irf4 and Cd138 and negative for B220 and Bcl6. The Ki-67 stainings are quantified in (F).
To further characterize the nature of the lymphadenopathy that we observed in M-B-Cd19 mice by MRI, we next performed histologic examination of infiltrated organs (livers, lymph nodes, and spleens). Morphologic assessment of the splenic, hepatic, and lymph node lesions detected in M-B-Cd19 mice revealed that these infiltrates consisted almost entirely of diffuse large lymphoid cells with blastoid to plasmablastoid appearance (Figure 4C). Intriguingly, plasmablasts have been implicated as the cell of origin of ABC-DLBCL.24 In marked contrast to the phenotype observed in M-Cd19 mice, in which the bulk of the lesions consisted of LPD-like infiltrates (Figure 2D), areas of LPD were only rarely detectable in M-B-Cd19 mice. Furthermore, the Ki-67 index of the lymphoid lesions in M-B-Cd19 mice was significantly higher compared with lesions that emerged in M-Cd19 mice (median, 71.57% ± 3.65% in M-B-Cd19 mice vs 28.27% ± 2.40% in M-Cd19 mice) (Figure 4F). The Ki-67 indices observed in M-B-Cd19 mice matched those typically detected in aggressive human lymphoma.32 Leukemic effusion was detectable only in M-B-Cd19 mice (supplemental Figure 6).
Consistent with the monomorphic appearance of the blastoid lesions detected in M-B-Cd19 mice, clonality analyses using Southern blot to detect Ig rearrangements revealed that these infiltrates were of clonal or oligoclonal origin (3 of 3 lesions isolated from independent mice) (Figure 4D). Expression of the mutant Myd88 allele in the lymphomas was verified by reverse transcriptase PCR sequencing analysis (supplemental Figure 7). To ask whether the oligoclonal lymphomas underwent somatic hypermutation, as would be expected in the case of ABC-DLBCL, we analyzed a clonal lesion for Igh rearrangements by direct sequencing. As shown in Figure 4E, we could detect a clonal Ighv1-26, Ighd2-12, Ighj4 rearrangement. The sequence shows an in-frame rearrangement with mutations but no stop-codons. Thus, this rearrangement is potentially functional. Next, we cloned the amplified Ighv gene rearrangements from this sample and sequenced 32 subclones to investigate intraclonal diversity. All analyzed sequences shared 5 mutations compared with the most homologous germ line sequence (Ighv1-26). Further mutations could be detected in several subclones (Figure 4E), demonstrating intraclonal diversity for this analyzed case.
Next, we used immunohistochemistry to further characterize the lymphomas isolated from M-B-Cd19 mice. As shown in Figure 4G and supplemental Figure 8, the lesions stained uniformly positive for Irf4 and were entirely negative for Bcl6, consistent with the somatic hypermutation detected in Figure 4E and further corroborating the post-germinal center (GC) nature of the infiltrates.24 Furthermore, the lymphomas lost B220 expression and stained strongly positive for Cd138, indicating ABC origin of these lesions.33-36 This is in line with analyses of human DLBCL, which indicate that GC-experienced Cd138-positive non-GC-DLBCLs are typically of plasmablastoid differentiation.37-39 In agreement with constitutive NF-κB activation, which is typically observed in human ABC-DLBCL, we found almost exclusive nuclear p65 staining in the lymphomas detected in M-B-Cd19 mice (Figure 4H).
Although MYD88 mutations have not been detected in human multiple myeloma,8,40-42 the strong Cd138 positivity of the lymphomas growing in M-B-Cd19 mice suggested the possibility that these lesions might be of plasmacellular origin.8,40-42 However, despite considerable effort, we were unable to detect a plasma cell expansion in the bone marrow of M-Cd19, M-Aid, M-Cd21, and M-B-Cd19 mice. More importantly, we did not find lymphoma infiltration of the bone marrow in M-Cd19, M-Aid, and M-Cd21 mice, and only 1 of 4 M-B-Cd19 mice showed an isolated lymphoma infiltrate that did not display signs of plasmacellular differentiation (supplemental Figures 3 and 9). In line with a lack of a consistent bone marrow infiltration, which is an obligatory feature of Waldenström macroglobulinemia and is frequently observed in multiple myeloma, we did not detect mono- or oligoclonal gammopathy in either M-Cd19 or M-B-Cd19 mice (supplemental Figure 10). However, we did detect an increased abundance of the Ig fraction in M-Cd19 mice compared with wild-type controls by using serum electrophoresis (upper and middle panels of supplemental Figure 10). This gammopathy was even more prominent in M-B-Cd19 mice (lower panel of supplemental Figure 10). The overall polyclonality strongly suggests a nonneoplastic origin of the gammopathy observed in M-Cd19 and M-B-Cd19 mice. Thus in summary, the morphologic appearance, as well as the immunophenotype of the lesions observed in M-B-Cd19 mice strongly suggests that these lymphomas represent ABC-DLBCL.
Discussion
Here, we generated a novel conditional Myd88p.L252P allele, which is at the orthologous position of the human Myd88p.L265P mutation and is expressed from the endogenous locus upon Cre-mediated recombination. We chose to design an allele that allows expression outside the natural genomic context to most faithfully mimic the situation in human malignancies. B cell–specific expression of this mutant leads to an LPD and occasional (∼30%) transformation into an aggressive lymphoma that morphologically and immunophenotypically resembles human ABC-DLBCL. Interestingly, development of LPD and lymphoma occurred at a similar rate, regardless of the Cre allele. We specifically used a pan B-cell Cre allele (Cd19Cre), a Cre allele that is active in GCB cells (AidCre), and a Cre allele that is expressed when immature transitional B cells differentiate into mature peripheral B cells (Cd21Cre). These data might indicate that Myd88p.L252P expression leads to a block in B-cell differentiation after the GC reaction. Consistent with this postgerminal nature of Myd88p.L252P-expressing cells, we find that the resulting lymphomas are consistently Bcl6 negative and express Irf4 (Figure 2E; supplemental Figure 1F-G). These observations are in line with data from human lymphoma patients, which suggests that oncogenic MYD88 mutations are enriched in post-GC malignancies, such as ABC-DLBCL, Waldenström macroglobulinemia, and the IGHV-mutant GC-experienced subset of CLL patients.7-9,24,43-45 Furthermore, crossing in an additional allele that allows conditional BCL2 expression from the Rosa26 locus leads to 100%-penetrant development of ABC-DLBCL.
Both MYD8846-48 and BCL224,49-51 have been shown to be critical for transition through the GC. Thus, it is conceivable that mice displaying B-cell–specific Myd88p.L252P expression (M-Cd19) or Myd88p.L252P expression in combination with BCL2 overexpression (M-B-Cd19) might accumulate post-GCB cells. Consistent with this, we find a polyclonal gammopathy in both M-Cd19 and M-B-Cd19 mice (supplemental Figure 10).
MYD88 is frequently mutated in ∼29% of ABC-DLBCL cases.9 However, MYD88 mutations are even more common in Waldenström macroglobulinemia, in which these alterations are detected in ∼90% of the cases.8,52 Thus, it is somewhat surprising that our Myd88p.L252P-expressing mice develop clonal ABC-DLBCL rather than Waldenström macroglobulinemia. One possible explanation might be the different genomic landscapes of these distinct B-cell neoplasias. For instance, mutations in the carboxyl terminal domain of the chemokine receptor CXCR4 have been detected in a substantial portion of patients with Waldenström macroglobulinemia (∼30% of the patients), whereas they are absent in ABC-DLBCL.2,9,53 Furthermore, somatic mutations affecting CD79A and CD79B are present in ∼20% of human ABC-DLBCLs, whereas these alterations are rare or absent in other lymphoma entities.20,24 Thus, it will be interesting to combine our novel Myd88p.L252P allele with additional B-cell–specific genetic alterations to determine whether further manipulation might enable a plasmacytoid differentiation.
Our novel models represent a preclinical platform that could be exploited for the in vivo development and validation of novel therapeutic approaches to treat ABC-DLBCL, which remains a difficult-to-treat clinical entity.54,55 For instance, it will be interesting to see whether combined targeting of the apoptotic machinery and the myddosome, through BCL2 inhibition and IRAK4 repression, will result in synergistic toxicity in Myd88-driven ABC-DLBCL.55 Our models might also serve to delineate the potential role of NF-κB and MAPKK6 signaling as actionable pathways downstream of the myddosome complex in ABC-DLBCL.56 For this purpose, it will be interesting to assess the cytotoxic potential of TAK1, IkB kinase, and MAPKK6 inhibitors in our Myd88-driven models of ABC-DLBCL. Altogether, we demonstrated the oncogenic role of a novel Myd88p.L252P allele in B-cell neoplastic disease in vivo. The resulting autochthonous mouse models of ABC-DLBCL represent useful preclinical tools for the development, validation, and refinement of novel treatment strategies for ABC-DLBCL in the clinical arena.
Acknowledgments
The authors are indebted to their patients, who provided primary material. The authors thank Alexandra Florin, Marion Müller, and Ursula Rommerscheidt-Fuss (Institute of Pathology, University Hospital Cologne) for their outstanding technical support.
This work was supported by the Volkswagenstiftung (Lichtenberg Program [H.C.R.]), the Deutsche Forschungsgemeinschaft (KFO-286 [H.C.R., H.K., F.T.W., and L.P.F.], KA 2853/4-1 [H.K.], and SFB-685 [A.N.R.W., O.-O.W.]), the Deutsche Jose Carreras Leukämie Stiftung (R12/26 [H.C.R. and L.P.F.]), the Marga und Walter Boll Stiftung (210-05-13 [L.P.F.]), the Helmholtz-Gemeinschaft (Preclinical Comprehensive Cancer Center [H.C.R. and C.A.S.]), the Else Kröner-Fresenius Stiftung (EKFS-2014-A06 [H.C.R.]), the Marlene Porsche Foundation for Cancer Prevention (S.B.), the Else-Übelmesser Stiftung (Juniorprofessoren-Programn [A.N.R.W]), the Deutsche Krebshilfe (111724 [H.C.R.]), and the German Ministry of Science and Education in the framework of the International Cancer Genome Consortium Molecular Mechanisms in Malignant Lymphoma by Sequencing Project (01KU1002A-J [C.L., P.M., L.T., and R.S.]).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: H.C.R. conceived and supervised the experiments and wrote the manuscript; L.P.F., H.K., M.A.-M., J.M.S., R.B., and F.T.W. provided mouse strains and materials; T.P. and Y.A.-B. performed magnetic resonance imaging scans and volumetric analyses; G.K., S.C.S., K.V., M.P., S.B., P.L., P.M., M. Ortmann, and R.B. performed immunohistochemistry and pathologic analyses; and L.M.S., A.N.R.W., O.-O.W., A.H.S., A.R.K., G.K., P.L., D.K., M. Odenthal, C.F., C.A.S., M.M.-R., M.R., M.A.-M., M.S., C.L., L.T., and R.S. performed experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A full list of members and affiliations of the German International Cancer Genome Consortium Molecular Mechanisms in Malignant Lymphoma by Sequencing Project Consortium appears in the supplemental Data.
Correspondence: H. Christian Reinhardt, Department I of Internal Medicine, University Hospital of Cologne, Weyertal 115B, 50931 Cologne, Germany; e-mail: christian.reinhardt@uk-koeln.de.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loiarro M, Volpe E, Ruggiero V, et al. Mutational analysis identifies residues crucial for homodimerization of myeloid differentiation factor 88 (MyD88) and for its function in immune cells. J Biol Chem. 2013;288(42):30210–30222. doi: 10.1074/jbc.M113.490946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465(7300):885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye H, Arron JR, Lamothe B, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418(6896):443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 6.Xia ZP, Sun L, Chen X, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461(7260):114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Treon SP, Xu L, Yang G, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N Engl J Med. 2012;367(9):826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 9.Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robbiani DF, Bothmer A, Callen E, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135(6):1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117(6):787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Riabinska A, Daheim M, Herter-Sprie GS, et al. Therapeutic targeting of a robust non-oncogene addiction to PRKDC in ATM-defective tumors. Sci Transl Med. 2013;5(189):189ra78. doi: 10.1126/scitranslmed.3005814. [DOI] [PubMed] [Google Scholar]
- 14.Bichi R, Shinton SA, Martin ES, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci USA. 2002;99(10):6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1(3):289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 16.Montesinos-Rongen M, Sánchez-Ruiz M, Brunn A, et al. Mechanisms of intracerebral lymphoma growth delineated in a syngeneic mouse model of central nervous system lymphoma. J Neuropathol Exp Neurol. 2013;72(4):325–336. doi: 10.1097/NEN.0b013e31828b7a98. [DOI] [PubMed] [Google Scholar]
- 17.Lefranc MP, Giudicelli V, Ginestoux C, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37(Database issue):D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crouch EE, Li Z, Takizawa M, et al. Regulation of AID expression in the immune response. J Exp Med. 2007;204(5):1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 20.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, Ju H, Kim DH, et al. CD79B and MYD88 mutations in diffuse large B-cell lymphoma. Hum Pathol. 2014;45(3):556–564. doi: 10.1016/j.humpath.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 22.Dierlamm J, Murga Penas EM, Bentink S, et al. Deutsche Krebshilfe Network Project “Molecular Mechanisms in Malignant Lymphomas”. Gain of chromosome region 18q21 including the MALT1 gene is associated with the activated B-cell-like gene expression subtype and increased BCL2 gene dosage and protein expression in diffuse large B-cell lymphoma. Haematologica. 2008;93(5):688–696. doi: 10.3324/haematol.12057. [DOI] [PubMed] [Google Scholar]
- 23.Testoni M, Zucca E, Young KH, Bertoni F. Genetic lesions in diffuse large B-cell lymphomas. Ann Oncol. 2015;26(6):1069–1080. doi: 10.1093/annonc/mdv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rui L, Schmitz R, Ceribelli M, Staudt LM. Malignant pirates of the immune system. Nat Immunol. 2011;12(10):933–940. doi: 10.1038/ni.2094. [DOI] [PubMed] [Google Scholar]
- 25.Lenz G, Wright GW, Emre NC, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA. 2008;105(36):13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richter J, Schlesner M, Hoffmann S, et al. ICGC MMML-Seq Project. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316–1320. doi: 10.1038/ng.2469. [DOI] [PubMed] [Google Scholar]
- 27.Kretzmer H, Bernhart SH, Wang W, et al. ICGC MMML-Seq project; BLUEPRINT project. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat Genet. 2015;47(11):1316–1325. doi: 10.1038/ng.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 29.Wang JQ, Jeelall YS, Beutler B, Horikawa K, Goodnow CC. Consequences of the recurrent MYD88(L265P) somatic mutation for B cell tolerance. J Exp Med. 2014;211(3):413–426. doi: 10.1084/jem.20131424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahal R, Frick M, Romero R, et al. Pharmacological and genomic profiling identifies NF-κB-targeted treatment strategies for mantle cell lymphoma. Nat Med. 2014;20(1):87–92. doi: 10.1038/nm.3435. [DOI] [PubMed] [Google Scholar]
- 31.Mathews Griner LA, Guha R, Shinn P, et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci USA. 2014;111(6):2349–2354. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salles G, de Jong D, Xie W, et al. Prognostic significance of immunohistochemical biomarkers in diffuse large B-cell lymphoma: a study from the Lunenburg Lymphoma Biomarker Consortium. Blood. 2011;117(26):7070–7078. doi: 10.1182/blood-2011-04-345256. [DOI] [PubMed] [Google Scholar]
- 33.Tan LH. A practical approach to the understanding and diagnosis of lymphoma: an assessment of the WHO classification based on immunoarchitecture and immuno-ontogenic principles. Pathology. 2009;41(4):305–326. doi: 10.1080/00313020902884501. [DOI] [PubMed] [Google Scholar]
- 34.Bai M, Skyrlas A, Agnantis NJ, et al. B-cell differentiation, apoptosis and proliferation in diffuse large B-cell lymphomas. Anticancer Res. 2005;25(1A):347–362. [PubMed] [Google Scholar]
- 35.Fournier EM, Velez MG, Leahy K, et al. Dual-reactive B cells are autoreactive and highly enriched in the plasmablast and memory B cell subsets of autoimmune mice. J Exp Med. 2012;209(10):1797–1812. doi: 10.1084/jem.20120332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coro ES, Chang WL, Baumgarth N. Type I IFN receptor signals directly stimulate local B cells early following influenza virus infection. J Immunol. 2006;176(7):4343–4351. doi: 10.4049/jimmunol.176.7.4343. [DOI] [PubMed] [Google Scholar]
- 37.Chang CC, McClintock S, Cleveland RP, et al. Immunohistochemical expression patterns of germinal center and activation B-cell markers correlate with prognosis in diffuse large B-cell lymphoma. Am J Surg Pathol. 2004;28(4):464–470. doi: 10.1097/00000478-200404000-00005. [DOI] [PubMed] [Google Scholar]
- 38.Colomo L, López-Guillermo A, Perales M, et al. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood. 2003;101(1):78–84. doi: 10.1182/blood-2002-04-1286. [DOI] [PubMed] [Google Scholar]
- 39.Said JW. Aggressive B-cell lymphomas: how many categories do we need? Mod Pathol. 2013;26(Suppl 1):S42–S56. doi: 10.1038/modpathol.2012.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mori N, Ohwashi M, Yoshinaga K, et al. L265P mutation of the MYD88 gene is frequent in Waldenström’s macroglobulinemia and its absence in myeloma. PLoS One. 2013;8(11):e80088. doi: 10.1371/journal.pone.0080088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu L, Hunter ZR, Yang G, et al. MYD88 L265P in Waldenström macroglobulinemia, immunoglobulin M monoclonal gammopathy, and other B-cell lymphoproliferative disorders using conventional and quantitative allele-specific polymerase chain reaction. Blood. 2013;121(11):2051–2058. doi: 10.1182/blood-2012-09-454355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiménez C, Sebastián E, Chillón MC, et al. MYD88 L265P is a marker highly characteristic of, but not restricted to, Waldenström’s macroglobulinemia. Leukemia. 2013;27(8):1722–1728. doi: 10.1038/leu.2013.62. [DOI] [PubMed] [Google Scholar]
- 43.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44(1):47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 46.Hua Z, Gross AJ, Lamagna C, et al. Requirement for MyD88 signaling in B cells and dendritic cells for germinal center anti-nuclear antibody production in Lyn-deficient mice. J Immunol. 2014;192(3):875–885. doi: 10.4049/jimmunol.1300683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hou B, Saudan P, Ott G, et al. Selective utilization of Toll-like receptor and MyD88 signaling in B cells for enhancement of the antiviral germinal center response. Immunity. 2011;34(3):375–384. doi: 10.1016/j.immuni.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer-Bahlburg A, Khim S, Rawlings DJ. B cell intrinsic TLR signals amplify but are not required for humoral immunity. J Exp Med. 2007;204(13):3095–3101. doi: 10.1084/jem.20071250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito M, Novak U, Piovan E, et al. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2009;106(27):11294–11299. doi: 10.1073/pnas.0903854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ci W, Polo JM, Cerchietti L, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113(22):5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith KG, Light A, O’Reilly LA, Ang SM, Strasser A, Tarlinton D. bcl-2 transgene expression inhibits apoptosis in the germinal center and reveals differences in the selection of memory B cells and bone marrow antibody-forming cells. J Exp Med. 2000;191(3):475–484. doi: 10.1084/jem.191.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kapoor P, Paludo J, Vallumsetla N, Greipp PR. Waldenström macroglobulinemia: What a hematologist needs to know. Blood Rev. 2015;29(5):301–319. doi: 10.1016/j.blre.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 53.Hunter ZR, Xu L, Yang G, et al. The genomic landscape of Waldenstrom macroglobulinemia is characterized by highly recurring MYD88 and WHIM-like CXCR4 mutations, and small somatic deletions associated with B-cell lymphomagenesis. Blood. 2014;123(11):1637–1646. doi: 10.1182/blood-2013-09-525808. [DOI] [PubMed] [Google Scholar]
- 54.Tilly H, Gomes da Silva M, Vitolo U, et al. ESMO Guidelines Committee. Diffuse large B-cell lymphoma (DLBCL): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v116–v125. doi: 10.1093/annonc/mdv304. [DOI] [PubMed] [Google Scholar]
- 55.Kelly PN, Romero DL, Yang Y, et al. Selective interleukin-1 receptor-associated kinase 4 inhibitors for the treatment of autoimmune disorders and lymphoid malignancy. J Exp Med. 2015;212(13):2189–2201. doi: 10.1084/jem.20151074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]