Abstract
Zebrafish (Danio rerio) as a model research organism continues to expand its relevance and role in multiple research disciplines, with recent work directed toward models of metabolism, nutrition, and energetics. Multiple technologies exist to assess body composition in animal research models at various levels of detail (tissues/organs, body regions, and whole organism). The development and/or validation of body composition assessment tools can open new areas of research questions for a given organism. Using fish from a comparative nutrition study, quantitative magnetic resonance (QMR) assessment of whole body fat and fat-free mass (FFM) in live fish was performed. QMR measures from two cohorts (n = 26 and n = 27) were compared with chemical carcass analysis (CCA) of FM and FFM. QMR was significantly correlated with chemical carcass values (fat, p < 0.001; lean, p = 0.002), although QMR significantly overestimated fat mass (FM) (0.011 g; p < 0.0001) and underestimated FFM (−0.024 g; p < 0.0001) relative to CCA. In a separate cross-validation group of fish, prediction equations corrected carcass values for FM (p = 0.121) and FFM (p = 0.753). These results support the utilization of QMR—a nonlethal nondestructive method—for cross-sectional or longitudinal body composition assessment outcomes in zebrafish.
Introduction
Aquatic research models have largely been developed for increased understanding of general aquaculture and ecology applications.1 However, particular aquatic models, such as the zebrafish (Danio rerio), have played significant roles in multiple areas of developmental biology and, more recently, molecular biology research. As a model research organism, the zebrafish has recently been used to test questions related to metabolism, nutrition, and energetics.2–6 With the development of improved dietary standards, husbandry/aquaculture conditions, and genetic mutant models, the need for additional technologies to accurately and noninvasively monitor body composition (particularly fat, lean, and bone) has increased. Bridging across multiple research disciplines, body composition influences outcomes related to culture production, adaptive ecological responses, aging, nutrition, and obesity-related outcomes. For traditional laboratory model organisms (particularly rodents and other mammals), multiple technologies exist to assess body composition at various levels of detail ranging from tissues/organs, body regions (e.g., peripheral vs. trunk), and whole body measures. These technologies include bioelectrical impedance, dual-energy X-ray absorptiometry, QMR, computed tomography, and magnetic resonance imaging.7–10 However, both the unique characteristics of many types of aquatic species (e.g., scales, shells, and skeleton) and their requirement for a water environment limit the number of technologies that can be adapted to accommodate these conditions. Additionally, the potential interactions that water may have on the technology and methodologies underlying the body composition measurement result in few options that are currently applied for in vivo body composition assessment in aquatic models.
The current “gold standard” method for analyzing body composition in zebrafish and other animal models is chemical carcass analysis (CCA).11 In this method, animal carcasses undergo dessication either in an oven or by freeze-drying to determine total body water (TBW). Lipid is then extracted from the dried carcass with organic solvents, such as petroleum ether or chloroform/methanol, and determined gravimetrically. Lean mass is generally calculated as the fat-free dry mass remaining after lipid extraction.8 Although this technique has been used for decades, the obvious limitations regarding its destructive nature and inability to target fat deposition/distribution prevent application to long-term studies.11 Studies using this technique require the destruction of multiple live animals to measure changes in response over time, or when controlling for body composition differences at baseline.
While each body composition method has its advantages and disadvantages, the development of body composition assessment tools for alternative animal models can open new areas of research to address questions that were previously inaccessible for these organisms. To this end, determination of fat mass (FM) and fat-free mass (FFM) with QMR has been applied to the zebrafish model in the context of a dietary protein-feeding study.12 The current study includes a validation and cross-validation comparison of QMR with CCA in the zebrafish.
Materials and Methods
Zebrafish, Danio rerio (AB strain), were obtained from the Aquatic Animal Research Core at the UAB and were part of a nutrition study reported previously (Table 1).12 Fish included both sexes and were ∼4 months of age at measurement. All procedures were approved by the UAB IACUC and adhered to standard zebrafish husbandry requirements for housing, anesthesia, and euthanasia.
Table 1.
Composition of Diets
| Amount included (g/100 g total) | |||||
|---|---|---|---|---|---|
| Ingredient | MIX | FPH | CAS | SOY | WG |
| Fish protein hydrolysate (82%)a,b | 18.20 | 59.00 | |||
| Casein (vita free) (96%)a,c | 22.75 | 51.00 | |||
| Soy protein isolate (92%)a,d | 4.55 | 52.00 | |||
| Wheat gluten (80%)a,e | 9.10 | 60.00 | |||
| Dextrin | 18.25 | 13.85 | 21.85 | 20.85 | 12.85 |
| Base mix | 20.15 | 20.15 | 20.15 | 20.15 | 20.15 |
| Menhaden oil | 4.67 | 4.67 | 4.67 | 4.67 | 4.67 |
| Corn oil | 2.33 | 2.33 | 2.33 | 2.33 | 2.33 |
| Validation group (n = 26) | 7 | 7 | 2 | 8 | 2 |
| Cross-validation group (n = 27) | 6 | 6 | 7 | 5 | 3 |
Protein content by percentage.
The Scoular Company, Sopropeche-C.P.S.P. 90.
MP Biomedicals, catalog no. 904798.
MP Biomedicals, catalog no. 905456.
Sigma-Aldrich, catalog no. G5004.
CAS, casein; FPH, fish protein hydrolysate; MIX, mixed; SOY, soy protein isolate; WG, wheat gluten.
Body composition assessment
Body composition measurement (FM and FFM) was performed by QMR using the EchoMRI 3-in-1 system (Houston, TX, software v. 2.0) and the tissue probe holder. Measures were taken during the middle of the 14-h light phase, and while not intentionally fasted, animals were last fed 4 h before measurement. QMR has been generally practiced in unanesthetized free-moving animals.8 Initial tests assessing the precision of QMR revealed a high level of variance in free-swimming unanesthetized fish. Considering the reliance of QMR on the proton (1H) environment for the estimation of body components (fat-free water associated vs. fat), nonbuffered tricaine methansulfonate (MS-222, 300 mg/L) was used to rapidly immobilize the fish in a small volume of water (also containing MS-222, 300 mg/L) for the duration of the QMR measure and reduce water movement across the gills within the body cavity. Anesthetic induction was individually performed on single fish to monitor not only swimming motion, loss of equilibrium, and cessation of opercular movement but also maintenance of heart contractions (requiring approximately 2–5 min depending on the individual fish characteristics, e.g., body size).13 Fish were quickly blotted dry and weighed to the nearest 0.001 g and placed in the QMR tissue sample holder (∼15 mL total volume space) with a very small volume of MS-222 containing water (∼200 μL). The duration of the scan is dependent on the precision setting with the high-precision tissue holder scan requiring ∼3 min per scan. Body composition measures, including FFM (nonlipid) and FM (total lipid), are reported to the nearest 0.001 g. Following the scans, fish were subsequently revived by submersion in fresh system water and gentle perfusion of the gills using a disposable pipette. Survival of the fish was >95% using this approach.
Euthanasia
At study completion, fish were euthanized by rapid submersion in ice-cold water with MS-222 (300 mg/L),13 and the carcass was stored at −80°C until analysis.
Chemical carcass analysis
To determine the total lipid content by CCA (modified from Folch et al.),14 whole fish were first verified for biological sex by dissection and visual confirmation of ovaries or testes and then placed into individual aluminum weigh pans and dried at 50°C for 72 h (until attainment of stable dry mass weight). Individual fish were minced with a razor blade to enhance chemical extraction, and carcasses weighing between 25 and 200 mg dry weight were placed in 40-mL glass vials (Fisher #06-406-9, 24-400 Glass Packaging Institute [GPI] thread finish). To each sample, 25 mL of 2:1 (v:v) chloroform:methanol was added (>100:1 solvent:sample), after which vials were capped loosely (Fisher #02-984-23, 24-400 GPI thread finish caps with low-density polyethylene Poly-Seal cone liner) and refrigerated for 24 h.
To extract lipids, samples were heated in a 60°C water bath for 30 min, after which they were left to stand at room temperature for 10 min. Solvent level was returned to the 25-mL mark, after which samples were filtered through 12.5-cm Whatman 541 filter paper, and 20 mL of the filtrate was collected into a clean 40-mL vial. Four milliliters of Milli-Q water was added to each vial, after which samples were capped and shaken for 5 min. Samples were centrifuged in a Beckman model TJ-6 tabletop centrifuge at 850 g for 30 min, after which the upper water–methanol phase was removed by aspiration. The lower phase was evaporated by placing vials in a 50°C water bath under a stream of nitrogen gas. Once dry, lipid samples were transferred to preweighed ¼-dram glass shell vials by three separate rinses of chloroform and dried under air to evaporate the chloroform. Dry shell vials were placed in a vacuum desiccator (Drierite® desiccant) overnight, after which final weights were obtained. The percentage of lipid was calculated as follows:
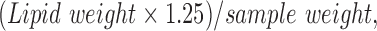 |
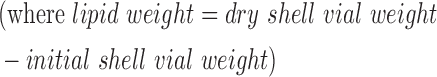 |
with necessary adjustments for sample recovery and transfer between subsequent steps.
Validation study
To assess precision from QMR for FM and FFM, three separate readings were recorded for each fish in the study, and the coefficient of variation (CV) was calculated for each fish.
Twenty-six zebrafish were used for the validation study. To determine accuracy, measured FM and FFM values were compared with CCA values in the validation group using the same data acquired for measuring precision. Using the linear regressions, prediction equations were generated to correct QMR estimated fat and lean mass for an independent sample of fish (cross-validation, see below).
Cross-validation study
A second group of zebrafish (n = 27) was randomly selected to assess whether prediction equations generated from the validation group accurately predicted chemical composition. The fish were scanned using the same techniques as the validation group and subsequently euthanized and processed for CCA.
Data analyses
Data were analyzed using R Statistical Software Package (v. 2.15.2, R Foundation for Statistical Computing, Vienna, Austria). Precision was determined using the coefficient of variation (CV = standard deviation [SD]/mean) ×100) for all fish in the study (n = 53). QMR and CCA were compared using linear regression analysis and paired t-tests, with significance denoted at α < 0.05. Measurement bias for FM and FFM was compared with residual differences between QMR and CCA values, with the CCA values considered as the gold standard. Prediction equations generated from the validation set of animals were applied to QMR-measured FM and FFM for the cross-validation cohort to predict FM and FFM. Body weight, FM, and FFM are reported as mean and SD of the mean.
Results
Validation–precision
Using three measures of the same fish with identical QMR system settings, the average CV measured by QMR was 10.8% ± 8.4% for FM and 3.3% ± 3.2% for FFM for all fish in the study (n = 53) (Fig. 1). Precision for FM was significantly less than that for FFM (p < 0.0001). CV values significantly decreased as both QMR FM and QMR FFM increased (FM, p = 0.01; FFM, p < 0.001) across a range of masses from 0.009 to 0.029 g for FM and 0.034–0.223 g for FFM.
FIG. 1.
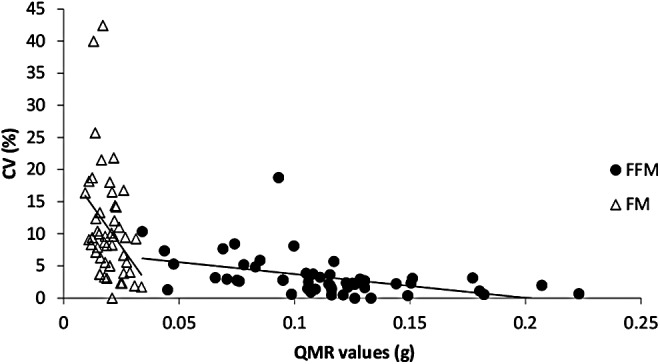
CV as a function of body compartment mass measured by QMR for FFM and FM (n = 53). CV, coefficient of variation; FFM, fat-free mass; FM, fat mass; QMR, quantitative magnetic resonance.
Validation–accuracy
Using approximately half of the total sample size representing both sexes and all diet groups (Table 1), significant correlations between QMR readings and CCA values were observed for both FFM (p = 0.002) and FM (p < 0.001) (Fig. 2). Despite the correlations, QMR significantly overestimated FM by ∼92% (0.011 g; p < 0.0001) and underestimated lean mass by ∼15% (−0.024 g; p < 0.0001) (Fig. 3). There was also a significant bias in the measurement of FFM, with underestimation increasing as FFM increased (p < 0.001) (Fig. 3). Based on the linear regressions derived from these observed relationships, prediction equations were generated to correct for QMR–CCA differences (Table 2).
FIG. 2.
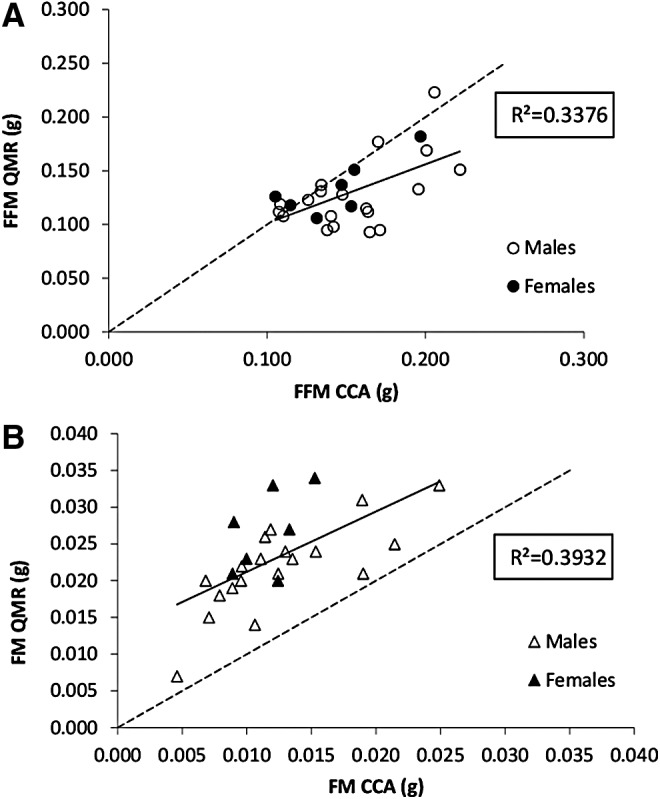
Relationships between QMR and CCA for (A) FFM and (B) FM for the validation group (n = 26). The line of regression is represented by the solid line, while the line of identity, provided for reference, is represented by the dashed line. CCA, chemical carcass analysis.
FIG. 3.
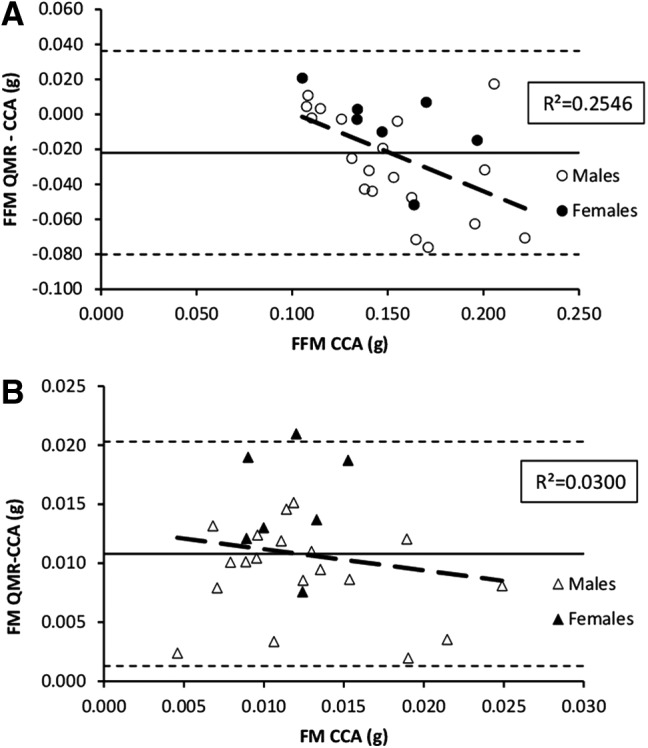
Residual plots for (QMR–CCA) body composition compartments for the validation group (n = 26) for (A) FFM and (B) FM. Group mean differences (solid line) ± 2 SD (dotted line) shown. Regression line (dashed line) demonstrates a significant bias in FFM and FM. SD, standard deviation.
Table 2.
Prediction Equations for FFM and FM (in Grams)
| Dependent variable | Prediction equations | Model r2 | p |
|---|---|---|---|
| CCA FFM | FFM = 0.6141 × QMR FFM +0.0723 | 0.3099 | 0.0020 |
| CCA FM | FM = 0.4630 × QMR FM +0.0016 | 0.3561 | 0.0008 |
Equations were generated from QMR values for the validation group.
CCA, chemical carcass analysis; FFM, fat-free mass; FM, fat mass; QMR, quantitative magnetic resonance.
Cross-validation
The cross-validation group was similar to the validation group in all aspects regarding body weight, FM, and FFM (Fig. 4 and Table 3), with both sexes and all diet treatment groups represented (Table 1). Similar to the validation group, QMR significantly overestimated FM by ∼83% (0.010 g; p < 0.0001), while significantly underestimating FFM by ∼12% (−0.020 g; p < 0.001) (Fig. 5). After prediction equation correction in the cross-validation group, corrected QMR measures of FM and FFM were significantly correlated with CCA and not significantly different from CCA values (Table 4). However, tests for residual bias with the corrected QMR values indicated significance with both FM (p < 0.0001) and FFM (p < 0.0001), which persisted with combined models of free waste (FW), FFM, and FM prediction (R2 = 0.8232, p < 0.0001).
FIG. 4.
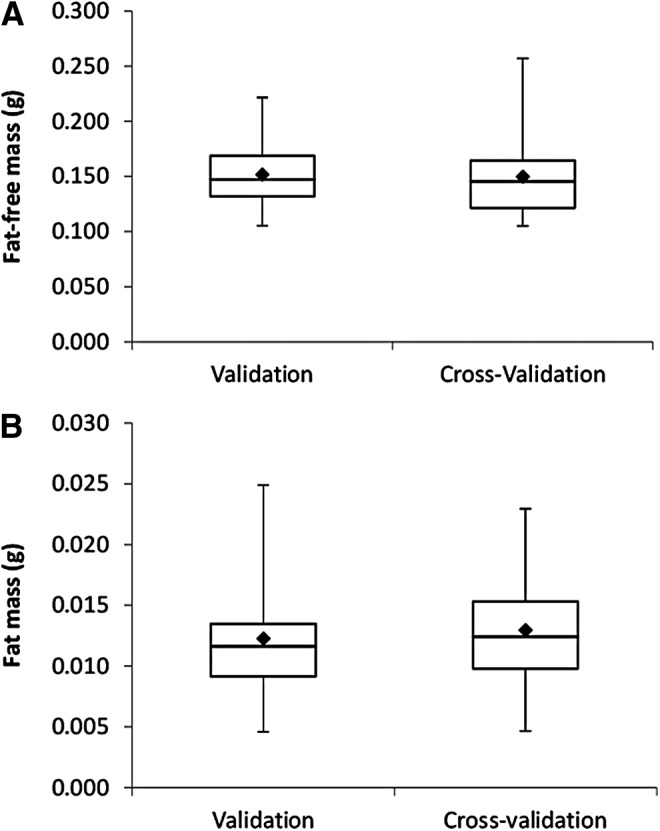
Box plots for (A) CCA FFM and (B) CCA FM for the validation (n = 26) and cross-validation (n = 27) groups, with the 25th and 75th percentiles shown as the upper and lower halves of each box plot, along with the 10th and 90th percentiles (error bars).
Table 3.
Population Statistics for the Validation (n = 26) and Cross-Validation (n = 27) Groups
| Validation | Cross-validation | ||||
|---|---|---|---|---|---|
| Males (n = 19) | Females (n = 7) | Males (n = 17) | Females (n = 10) | p | |
| Body weight (g) | 0.17 ± 0.04 | 0.16 ± 0.03 | 0.18 ± 0.05 | 0.15 ± 0.05 | 0.381 |
| Standard length (mm) | 26.22 ± 2.21 | 23.76 ± 1.54 | 26.86 ± 1.78 | 24.88 ± 2.66 | 0.375 |
| CCA FM (g) | 0.01 ± 0.005 | 0.01 ± 0.002 | 0.01 ± 0.005 | 0.01 ± 0.005 | 0.836 |
| CCA FFM (g) | 0.15 ± 0.03 | 0.15 ± 0.03 | 0.17 ± 0.04 | 0.14 ± 0.04 | 0.351 |
Values are mean ± SD.
SD, standard deviation.
FIG. 5.
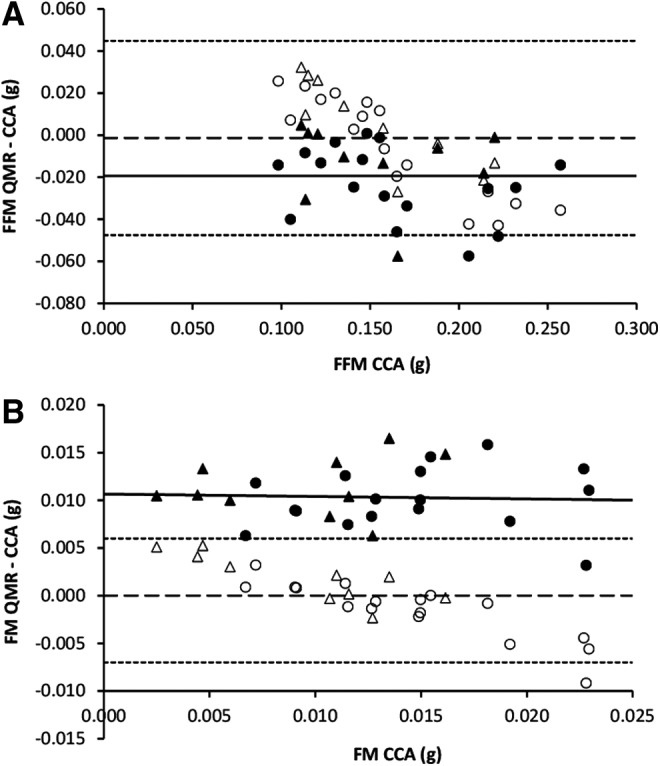
Residual plots for QMR–CCA for the cross-validation group (n = 27) before and after correction by derived prediction equations for (A) FFM and (B) FM. Uncorrected values (closed markers) with group mean (solid line) and prediction-corrected values (open markers) with mean (dashed line) ± 2 SD (dotted lines) shown for each body compartment. Males are represented by circles, while females are represented by triangles.
Table 4.
QMR Values for the Cross-Validation Group
| Mean uncorrected QMR | Mean predicted QMR | Mean CCA | p | r2 | |
|---|---|---|---|---|---|
| FFM | 0.141 ± 0.043 | 0.159 ± 0.027 | 0.160 ± 0.045 | 0.7528 | 0.826 |
| FM | 0.023 ± 0.006 | 0.012 ± 0.003 | 0.013 ± 0.005 | 0.1209 | 0.749 |
Values are mean ± SD (in grams) for QMR FFM and FM before (uncorrected) and after (predicted) prediction equation corrections with CCA FFM and FM. Paired t-test significance (p values) and r2 values for predicted QMR versus CCA.
Discussion
Carcass analysis is considered the gold standard in determining FM and FFM in research organisms, including zebrafish, but is not compatible with repeated measures in longitudinal studies due to the technique's destructive nature. With the zebrafish emerging as an increasingly popular model in biomedical and nutrition research, validation of more noninvasive methods for measuring body composition is needed. In this study, we assessed the precision and accuracy of QMR to measure FM and FFM in live anesthetized zebrafish. In zebrafish, lipids associated with FM are primarily stored as TAGS (triacylglycerols), and the main storage sites include visceral, intramuscular, and subcutaneous adipocyte depots.15,16 QMR estimates total lipid for FM, which includes both storage (nonpolar) and structural (polar) lipids. Therefore, an overestimation of FM using QMR measures can be expected, which is comparable to our technique for CCA.17–19 CV values improved significantly as mass in both body composition measurements (FM and FFM) increased (Fig. 1). It should also be noted that FM, which is normally present in proportionally smaller quantities in a nondiseased state, was significantly less than FFM in this study using young healthy fish. FM estimates ranged from 0.007 to 0.036 g, while lean mass ranged from 0.065 to 0.243 g. When validating in vivo methods of body composition analysis, the precision for FM is often less than that for FFM.7,8,20 CV is a function of body mass such that with a fixed measurement error or uncertainty, there is more variance with a smaller body compartment mass compared with larger body composition compartment masses.8,20 Therefore, the smaller amounts of FM present compared to FFM in the current study may contribute to differences in precision between the body compartments. However, it should also follow that as FM increases in the fish, QMR measurements for that component will be more precise.
In both the validation and cross-validation groups, QMR was found to overestimate FM to a much greater degree (proportionally) than FFM was underestimated; similar results have been observed in QMR validation studies for rodents, crustaceans, piglets, and bats compared with CCA.8,17–20 However, several studies also showed that QMR underestimated FM when using deuterium dilution and a four-component model as the gold standard.21–26 Values for both FM and FFM, while significantly different from CCA values, were still significantly correlated for each body compartment measure. For FFM, residual bias increased significantly with increasing CCA mass, while residual bias did not occur for FM. The derived prediction equations were able to correct for errors in both measured body composition values for the cross-validation study group (Table 3). However, neither prediction equation was able to correct for residual bias in FM or FFM (Fig. 5). Applying a multiple regression model using all three components (FM, FFM, and FW) as predictor variables was also unable to improve the residual bias in either FM or FFM, suggesting this was not simply due to misassignment of one body component compartment to the other. It is not completely clear why bias was not corrected in the cross-validation group by the prediction equations, an issue that needs consideration in future studies.
The distribution of males and females, while not differing significantly between the validation and cross-validation groups, was heavily skewed toward males in both groups in this sample. Although the derived prediction equations were able to correct measured QMR values to CCA values, it is currently unknown whether these equations could be improved for single sex body composition determination in zebrafish. The accuracy of the prediction equations slightly improved when derived separately for males, but due to the limited number in our sample, we were unable to derive separate prediction equations for females (Table 5). This may be particularly important with older fish at varying levels of sexual maturity and reproductive stages. Additionally, between-study differences in zebrafish strains, diets, husbandry conditions, and so forth, should be considered to develop and compare prediction equations in both male and female populations if absolute (e.g., tissue mass) rather than relative body composition assessments are required.
Table 5.
Prediction Equations and QMR Values for the Cross-Validation Group (in Males Only)
| Dependent variable | Prediction equations | Mean uncorrected QMR | Mean predicted QMR | Mean CCA | p | r2 |
|---|---|---|---|---|---|---|
| FFM | FFM = 0.6545 × QMR FFM +0.0710 | 0.151 ± 0.044 | 0.170 ± 0.0284 | 0.169 ± 0.044 | 0.9488 | 0.835 |
| FM | FM = 0.6798 × QMR FM − 0.0023 | 0.025 ± 0.006 | 0.015 ± 0.004 | 0.015 ± 0.005 | 0.9518 | 0.736 |
Values are mean ± SD (in grams) for QMR FFM and FM before (uncorrected) and after (predicted) prediction equation corrections with CCA FFM and FM. Paired t-test significance (p values) and r2 values for predicted QMR versus CCA.
One body compartment measure in QMR that has not yet been validated in zebrafish is TBW. At the time of this study, a hydration constant for freshwater fish was unavailable, limiting the ability to validate QMR measurement of this component. Determination of the hydration constant may extend validation of QMR measurements to TBW across a range of body sizes, although challenges arising from the water environment used for measurement may be expected.
In conclusion, we found QMR to be a valid noninvasive technique for measuring FM and FFM in the zebrafish. Despite the small residual bias that remained after applying prediction equations for FM and FFM, there were no significant differences between the CCA values and corrected QMR values in the cross-validation group. Applying this technique in future studies will allow researchers to track changes in zebrafish body composition over time, potentially during both pre- and posttreatment periods (with one or more successful interventions) at both the individual fish and population levels to determine changes in body composition. This may further contribute to the value of this model in areas of biomedical and nutrition research.
Acknowledgments
We acknowledge the Small Animal Phenotyping Core for assistance with body composition measures using QMR and the Nutrition Obesity Research Center Aquatic Animal Models Core for assistance with zebrafish diet manufacture and husbandry.
Funding
This work was supported in part by a pilot/feasibility grant from the UAB Nutrition Obesity Research Center to DLS (P30DK056336). This work was also supported by the UAB Small Animal Phenotyping Core (P60DK079626 and P30DK056336).
Disclosure Statement
Research reported in this publication was supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health. The content and opinions expressed herein are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or any other organization with which the authors are affiliated.
References
- 1.Skalski GT, et al. . Variable intake, compensatory growth, and increased growth efficiency in fish: models and mechanisms. Ecology 2005; 86:1452–1462 [Google Scholar]
- 2.Watts SA, Powell M, D'Abramo LR. Fundamental approaches to the study of zebrafish nutrition. ILAR J 2012;53:144–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walters JW, et al. . Visualization of lipid metabolism in the zebrafish intestine reveals a relationship between NPC1L1-mediated cholesterol uptake and dietary fatty aAcid. Chem Biol 2012;19:913–925 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Ulloa PE, et al. . Zebrafish as a model organism for nutrition and growth: towards comparative studies of nutritional genomics applied to aquacultured fishes. Rev Fish Biol and Fish 2011;21:649–666 [Google Scholar]
- 5.Seth A, Stemple DL, Barroso I. The emerging use of zebrafish to model metabolic disease. Dis Model Mech 2013;6:1080–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang L, Miller YI. Emerging applications for zebrafish as a model organism to study oxidative mechanisms and their roles in inflammation and vascular accumulation of oxidized lipids. Free Radic Biol Med 2012;53:1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith D, Johnson M, Nagy T. Precision and accuracy of bioimpedance spectroscopy for determination of in vivo body composition in rats. Int J Body Compos Res 2009;7:21–26 [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson MS, Smith DL, Jr., Nagy TR. Validation of quantitative magnetic resonance (QMR) for determination of body composition in rats. Int J Body Compos Res 2009;7:99–107 [PMC free article] [PubMed] [Google Scholar]
- 9.Rose BS, et al. . Whole body composition of rats determined by dual energy X-ray absorptiometry is correlated with chemical analysis. J Nutr 1998;128:246–250 [DOI] [PubMed] [Google Scholar]
- 10.Judex S, et al. . Quantification of adiposity in small rodents using micro-CT. Methods 2010;50:14–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson MS, Nagy TR: Animal body composition models. In: Human Body Composition. Lohman TG, Heymsfield SB, Wang Z. and Going SB. (eds), pp. 141–150, Human Kinetics, Champaign, IL, 2005 [Google Scholar]
- 12.Smith DL Jr., et al. . Dietary protein source influence on body size and composition in growing zebrafish. Zebrafish 2013;10:439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson JM, Bunte RM, Carty AJ. Evaluation of rapid cooling and tricaine methanesulfonate (MS222) as methods of euthanasia in zebrafish (Danio rerio). J Am Assoc Lab Anim Sci 2009;48:785–789 [PMC free article] [PubMed] [Google Scholar]
- 14.Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957;226:497–509 [PubMed] [Google Scholar]
- 15.Imrie D, Sadler KC. White adipose tissue development in zebrafish is regulated by both developmental time and fish size. Dev Dyn 2010;239:3013–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flynn E, Trent C, Rawls J. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio). J Lipid Res 2009;50:1641–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones AS, Johnson MS, Nagy TR. Validation of quantitative magnetic resonance for the determination of body composition of mice. Int J Body compos Res 2009;7:67–72 [PMC free article] [PubMed] [Google Scholar]
- 18.O'Regan SM, Guglielmo CG, Taylor GM. Measurement of arthropod body composition using quantitative magnetic resonance. Invertebr Biol 2012;131:216–223 [Google Scholar]
- 19.McGuire LP, Guglielmo CG. Quantitative magnetic resonance: a rapid, noninvasive body composition analysis technique for live and salvaged bats. J Mammal 2010; 91:1375–1380 [Google Scholar]
- 20.Mitchell AD. Validation of quantitative magnetic resonance body composition analysis for infants using piglet model. Pediatr Res 2011;69:330–335 [DOI] [PubMed] [Google Scholar]
- 21.Andres A, Mitchell AD, Badger TM. QMR: validation of an infant and children body composition instrument using piglets against chemical analysis. Int J Obes (Lond) 2010;34:775–780 [DOI] [PubMed] [Google Scholar]
- 22.Gallagher D, et al. . Quantitative magnetic resonance fat measurements in humans correlate with established methods but are biased. Obesity (Silver Spring) 2010;18:2047–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zanghi BM, et al. . Noninvasive measurements of body composition and body water via quantitative magnetic resonance, deuterium water, and dual-energy x-ray absorptiometry in cats. Am J Vet Res 2013;74:721–732 [DOI] [PubMed] [Google Scholar]
- 24.Zanghi BM, et al. . Noninvasive measurements of body composition and body water via quantitative magnetic resonance, deuterium water, and dual-energy x-ray absorptiometry in awake and sedated dogs. Am J Vet Res 2013;74:733–743 [DOI] [PubMed] [Google Scholar]
- 25.Tinsley F, Taicher G, Heiman ML. Evaluation of a quantitative magnetic resonance method for mouse whole body composition analysis. Obes Res 2004;12:150–160 [DOI] [PubMed] [Google Scholar]
- 26.Bosaeus M, et al. . Accuracy of quantitative magnetic resonance and eight-electrode bioelectrical impedance analysis in normal weight and obese women. Clin Nutr 2014;33:471–477 [DOI] [PubMed] [Google Scholar]


