Abstract
Endogenous retroviruses (ERVs) undergo de novo DNA methylation during the first few days of mammalian embryogenesis, although the factors that control the targeting of this process are largely unknown. We asked if KAP1 (KRAB-associated protein 1) is involved in this mechanism because of its previously defined role in maintaining ERVs silent through the histone methyltransferase ESET and histone H3 lysine 9 trimethylation. Here, we demonstrate that introduced ERV sequences are sufficient to direct rapid de novo methylation of a flanked promoter in embryonic stem (ES) cells. This mechanism requires the presence of an ERV sequence-recognizing KRAB-zinc finger protein (ZFP) and both KAP1 and ESET. Furthermore, this process can also take place on a strong cellular promoter and leads to methylation signatures that are subsequently maintained in vivo throughout embryogenesis. Finally, we show that methylation of ERVs residing in the genome is affected by knockout of KAP1 in early embryos. KRAB-ZFPs, KAP1 and ESET are thus likely responsible for the early embryonic instatement of stable epigenetic marks at ERV-containing loci.
Keywords: de novo DNA methylation, endogenous retroviral silencing, KRAB-associated protein 1 (KAP1), TRIM28, TIF1β, KRAB-zinc finger protein (KRAB-ZFP), ESET, SETDB1
Introduction
Endogenous retroviruses (ERVs) account for close to ten percent of mammalian genomes (Waterston et al., 2002) and are both drivers of evolution and threats to genetic integrity, because of their ability first to retrotranspose and second to alter the expression of neighboring genes through cis-acting transcriptional influences. In mice, ERVs display residual retrotransposition activity, leading to polymorphic integrations and differential gene regulation between mouse strains (Qin et al., 2010; Takabatake et al., 2008; Zhang et al., 2008). Moreover, around ten percent of spontaneous mutations in inbred mice are linked to ERVs (Maksakova et al., 2006), for instance dactylaplasia that results from a MusD neo-insertion (Friedli et al., 2008; Kano et al., 2007). In humans, the non-LTR retroelement LINE1 (L1) retains some activity and, remarkably, patients with Rett syndrome due to mutations in the DNA methylation binding protein MeCP2 are more prone to L1 retrotransposition (Muotri et al., 2010).
ERVs are duly inactivated during early embryogenesis by histone modifications and de novo DNA methylation (reviewed in (Rowe and Trono, 2011)). This process counteracts the prior genome-wide erasure of DNA methylation that begins at the zygote phase, leaving transposons in a lowly methylated state including L1s and to a lesser extent intracisternal A-type particles (IAPs), (Feng et al., 2010; Lane et al., 2003). De novo DNA methylation is key to ERV repression from plants to mammals, and involves methylation of cytosine residues at CpG dinucleotides by the enzymes DNMT3A and DNMT3B, acting in conjunction with their catalytically inactive cofactor DNMT3L (Chedin et al., 2002; Okano et al., 1999; Suetake et al., 2004). Later in development and in adult tissues, levels of de novo methyltransferases are reduced compared to preimplantation embryos (Carlson et al., 1992), yet pre-established DNA methylation patterns are perpetuated during DNA replication by the maintainance DNA methyltransferase DNMT1, recruited to hemimethylated DNA by UHRF1 (Bestor et al., 1988; Bostick et al., 2007; Sharif et al., 2007). DNA methylation is particularly crucial to constrain the transcription of some ERVs including IAPs, the expression of which become uncontrolled following inactivation of DNA methylation machinery (Bourc’his and Bestor, 2004; Chen et al., 2003; Gaudet et al., 2004; Walsh et al., 1998), although not in DNMT1 knockout ES cells unless they are first differentiated (Hutnick et al., 2010), highlighting the particular importance for DNA methylation later in development. Interestingly, alterations in ERV DNA methylation patterns impact on the expression of neighbouring genes (Duhl et al., 1994; Macfarlan et al., 2011; Macfarlan et al., 2012; Michaud et al., 1994; Rebollo et al., 2011).
DNA methylation is conditioned by the density and spacing of CpG dinucleotides, can be affected by DNA-binding proteins such as CTCF and REST, and is influenced by the histone code, with for instance methylation of histone 3 at lysine 4 (H3K4) preventing the docking of the DNMT3A-DNMT3L complex (Glass et al., 2009; Jia et al., 2007; Lienert et al., 2011; Okitsu and Hsieh, 2007; Ooi et al., 2007; Stadler et al., 2011; Weber et al., 2007). The factors and pathways that recruit de novo DNA methylases to specific genomic targets, notably ERVs, remain largely undefined, although it was recently shown that DNMT3L and the lysine methyltransferase G9a are factors required for the initiation of proviral de novo DNA methylation (Leung et al., 2011; Ooi et al., 2010).
Here, we investigate the role played in this process by KAP1 (KRAB-associated protein 1, also known as tripartite motif protein 28 –TRIM28- or TIF1b), a cofactor to the sequence-specific DNA-binding KRAB-zinc finger proteins (ZFPs), a large family of tetrapod-restricted transcriptional factors that co-evolved with ERVs (Emerson and Thomas, 2009; Friedman et al., 1996; Thomas and Schneider, 2011; Urrutia, 2003). KAP1 was previously demonstrated to maintain ERVs in a silent state in embryonic stem (ES) cells via the histone methyltransferase ESET (also known as SETDB1) and secondary H3K9 trimethylation (Matsui et al., 2010; Rowe et al., 2010). It is also known that the artificial tethering of a KRAB domain to the vicinity of a promoter can lead to its DNA methylation in transgenic mice but not in ES cells (Wiznerowicz et al., 2007), and that maintenance of imprinting marks depends on ZFP57 and KAP1 (Li et al., 2008; Messerschmidt et al., 2012; Quenneville et al., 2011; Zuo et al., 2012). Of relevance here, the retrovirus murine leukemia virus (MLV) is known to be recognized by the mouse-specific KRAB-ZFP ZFP809 through its proline primer binding site (PBS Pro) sequence and undergos KAP1-dependent silencing in embryonic carcinoma (EC) cells (Wolf et al., 2008; Wolf and Goff, 2007; Wolf and Goff, 2009), although it is not known if these factors are sufficient to promote DNA methylation. Of note, MLV-based vectors are themselves prone to DNA methylation weeks after their integration in ES or EC cells, through a process that is independent of the presence of a PBS Pro sequence (Leung et al., 2011; Niwa et al., 1983; Wang et al., 1998), as also documented for the cytomegalovirus (CMV) promoter (Meilinger et al., 2009).
Determining how particular ERVs are targeted for de novo DNA methylation is technically challenging, owing to their genomic representation in multiple copies of often differentially methylated status in ES cells and early embryos. To circumvent this difficulty, we inserted previously identified genomic targets of KAP1 derived either from the proximal region of IAPs or from PBS Pro (present in some 146 endogenous MLVs in the C57Black6 mouse genome) into lentiviral vectors, either upstream or downstream of a promoter driving a GFP reporter. In both configurations, these introduced retroviral elements were sufficient to induce not only the silencing but, in parallel, de novo DNA methylation of the flanking promoter. Methylation was remarkably rapid with an adjacent retroviral promoter (MND), occuring within 48 hours, but slower with a cellular promoter (PGK), occuring within six days. For PBS Pro, we could demonstrate that DNA methylation was conditioned by the expression of ZFP809. We then determined that de novo DNA methylation of introduced ERVs in ES cells was dependent on both KAP1 and ESET, and that sequence-specific ERV methylation could be recapitulated in vivo and maintained through development, with ERV sequences directing DNA methylation of an adjacent retroviral or cellular promoter. Finally, we documented that methylation footprints of genomic ERVs are affected by KAP1 loss in early embryos. These data reveal that KAP1 shapes DNA methylation signatures at ERV-containing loci in early mouse development.
Materials and Methods
Lentiviral vectors
All MND vectors are described before (Rowe et al., 2010). For ERV-directed repression of a cellular promoter, the transfer vector pRRLSIN.cPPT.PGK-GFP.WPRE (available from Addgene) was employed that contains a human phosphoglycerate kinase-1 (PGK) promoter to distinguish it from the endogenous mouse promoter during methylation analysis. Pro and B2 sequences were cloned into this vector by annealing primers upstream (into the XhoI site) or downstream (between the BamHI and AgeI sites) of the promoter in the antisense orientation. Or IAP1 or IAP4 (described before, (Rowe et al., 2010)) were cloned upstream of the promoter into the XhoI site in the antisense orientation. Cloning was verified by sequencing (Microsynth). Vectors were produced by transient transfection of 293T cells with the transfer vector, packaging and VSVG envelope plasmids (Barde et al., 2010) and titrated on 3T3 fibroblasts.
Cell culture and flow cytometry
ES cells were cultured in Glasgow Minimum Essential Media (GMEM, Sigma: G5154) with sodium pyruvate (used at 1mM, Sigma: S8636), MEM non-essential amino acids (used at 1x, Gibco: 11140-035), L-glutamine (used at 2mM, Gibco: 25030-024), 2-mercaptoethanol (used at 0.1mM, Sigma), ES cell tested FBS (Gibco: 16141-079) and leukaemia inhibitory factor (LIF, used at 1,000 units/ml, Chemicon: ESG1107). Cells were grown on 0.2% gelatin (Sigma: 48723-500G-F)-coated plates and split every two days. ES cell lines used were two KAP1loxP/loxP lines called ES3 and ES6 and their derived KAP1-conditional knockout cell lines that are transduced with tamoxifen (4-0HT)-inducible cre (Rowe et al., 2010). Knockout cells were collected 3-4 days after treatment with 4-0HT (overnight at 1mM, Sigma: H7904) as stated. G9a parental or stable knockout ES cells and Eset-inducible knockout ES cells were from Yoichi Shinkai (Dong et al., 2008; Matsui et al., 2010; Tachibana et al., 2008) (The RIKEN Institute, Japan). DNMT1-/-, DNMT3A-/-, DNMT3B-/- (triple knockout or TKO) ES cells were from Masaki Okano (Tsumura et al., 2006) (The RIKEN Institute, Japan). F9 EC (embryonic carcinoma) cells, primary mouse embryonic fibroblasts (MEFs), 3T3 fibroblasts and 293T cells were also used where stated. Vector titers and GFP repression were measured by flow cytometry and for KAP1 and ESET knockout experiments, cells were stained with an anti-SSEA-1 PE- conjugated antibody or isotype control (BD Pharmingen: 560142 and 555584) and SSEA-1 high-expressing cells gated for undifferentiated cells.
Quantitative bisulfite pyrosequencing
Genomic DNA was converted (200ng/sample) using an EpiTect Bisulfite kit (Qiagen: 59104) and used for PCR (primers, one tagged with biotin, were designed using PyroMark Assay Design Software 2.0). PCR products were checked by Sanger sequencing and verified on agarose gels for each experiment before immobilizing on 96 well plates using a Vacuum Prep Workstation and pyrosequencing using PyroMark Gold Reagents (Qiagen: 972804). We thank Alex Reymond and Jacqueline Chrast for kind use of their pyrosequencer (at the Center for Integrative Genomics, University of Lausanne, Switzerland). Results were analysed using Pyro Q-CpG Software. See Supplementary Table I for primer sequences.
Immunoblotting
Cells were washed with cold PBS and resuspended in radioimmunoprecipitation buffer to prepare total cell extracts. Protein was quantified by BCA Protein Assay Reagents (Pierce) and normalized for loading on a 10% denaturing SDS-polyacrylamide gel. Wet transfer was performed and primary antibodies used were: anti-KAP1 (Mouse mAb, MAB3662, Chemicon), anti-IAP GAG (kind gift from Bryan Cullen, Duke University, U.S.A., see (Bogerd et al., 2006)) and anti-PCNA (mouse mAb, clone PC10, Cat. No. NA03, Calbiochem).
293T cell transfection
Cells were seeded at 1x10e5 per well in a 12 well plate and transfected with 100ul transfection mix per well, containing 6ul FuGene 6 (Promega) and 1ug DNA in serum free media, after incubating this mixture for 15 minutes at room temperature.
Quantification of RNA and DNA copy number
RNA was extracted and qRT-PCR performed as before (Rowe et al., 2010). Primer specificity was confirmed by dissociation curves and samples normalized to Titin, Gapdh, or Eef1a1 in Fig. 2D because it reacts with both human and mouse Eef1a1. For in vivo experiments, genomic DNA was extracted from embryos to measure DNA copy numbers by performing Taqman qPCR for HIV Gag, GFP and Titin as a normalizer, in comparison to a titration curve of plasmid containing sequences of HIV Gag, GFP and Titin. A cell line with a known number of vector copies was used as control. Results presented are means of values obtained for HIV Gag and GFP primers. See Supplementary Table I for primer sequences.
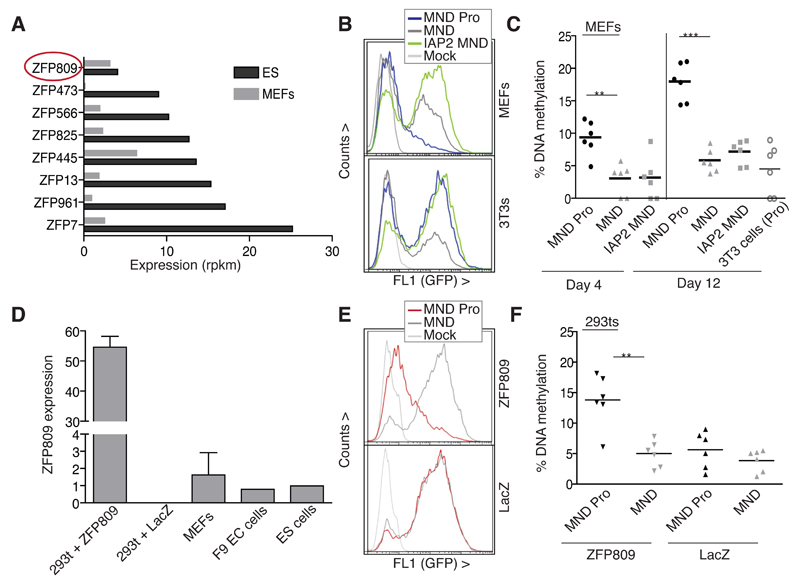
Fig. 2. ERV DNA methylation patterns are conditioned by KRAB-ZFP expression profiles.
A) Comparison of expression levels of selected KRAB-ZFPs in ES cells and MEFs based on mRNA-sequencing data. Rpkm: The number of sequencing reads normalized by gene length and to the total read number. B) The Pro sequence is repressed in MEFs where ZFP809 expression is retained. Top: The MND Pro vector is repressed in MEFs compared to the empty (MND) or IAP2 MND vector, whereas all vectors are highly expressed in 3T3 cells (bottom). Mock: nontransduced. C) The Pro sequence directs de novo promoter methylation in MEFs. DNA methylation was measured at days 4 and 12 post transduction with the stated vectors. Promoter methylation of the MND Pro vector was also measured in 3T3 cells at day 12 as a control. P values: p=0.0052 (day 4) and p=0.0001 (day 12). D) 293t cells were complemented with ZFP809 (or LacZ as a control) and expression compared to endogenous levels in the stated cells by qRT-PCR. Bars show means and s.e.m. of triplicate transfections (293t cells) or 2 independent cell lines (MEFs) or individual cell lines (EC and ES cells). E) and F) ZFP809 is responsible for Pro-sequence directed repression and DNA methylation. 293t cells were transduced with the stated vectors (or nontransduced: Mock) and then transfected with either ZFP809 or LacZ. GFP repression (E) and promoter methylation (F) were measured 5 days post transfection. P=0.0042.
Lentiviral transgenesis
Lentiviral vectors for transgenesis were prepared using Episerf medium (Invitrogen: 10732022), the particle concentration obtained by p24 ELISA (Perkin-Elmer: NEK050B001KT) and the infectious titer determined on HCT116 cells by GFP flow cytometry. Vectors for each transgenesis experiment were produced and titered in parallel in order to be in the same range. Fig. 4 titers: 2x10e9 (IAP2 MND) and 4x10e9 (MND). Fig. 5 titers: 1.4x10e9 (PGK Pro) and 1.2x10e9 (PGK). Fig. S4B titers: 2.3x10e9 (IAP4 PGK) and 2.4x10e9 (IAP1 PGK). Transgenesis was performed by perivitelline injection of vectors into fertilized oocytes that were transferred to foster mothers (strain B6D2F1/J) and then recovered at E13. Photographs were taken using the same saturation, gain and exposure settings for all embryos, and GFP results displayed all represent the same image processing settings.
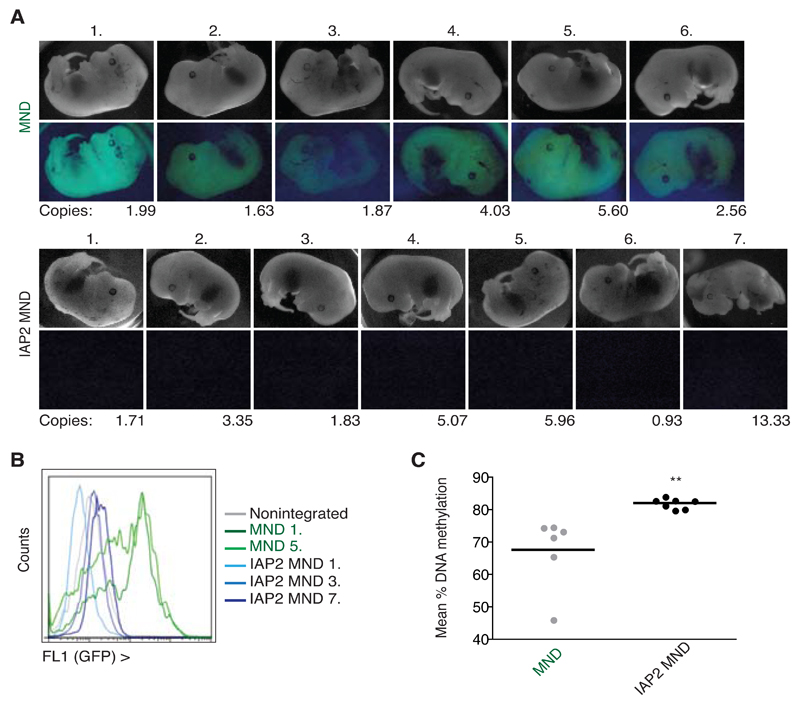
Fig. 4. ERV-guided KAP1-induced DNA methylation is recapitulated in embryogenesis.
A) The MND and IAP2 MND vectors were used for lentiviral transgenesis with scoring of embryos for vector integration and GFP expression at E13. For the MND group, 6 embryos were integrated with copy numbers stated (top) and for the IAP2 MND group, 10 embryos were integrated, none of which were green with 7 shown (bottom). B) MEFs were cultured from sample embryos (stated on the right) or a non-integrated embryo as a control and GFP measured by flow cytometry. C) Embryos from A) (N=6 for MND and N=7 for IAP2 MND) were assessed for methylation of the MND promoter (means are shown per embryo over 10 CpGs). P=0.0057 (unpaired two-tailed t test).
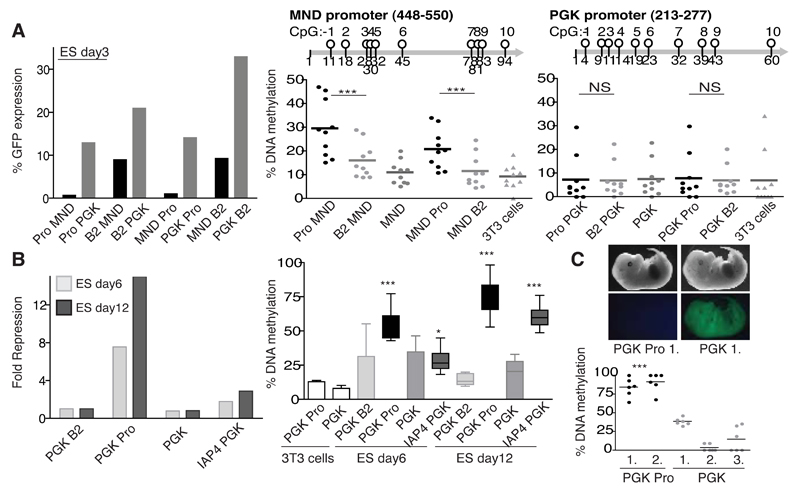
Fig. 5. ERV sequences can induce repression and DNA methylation of a cellular promoter.
A) Side-by-side comparison in ES cells of repression and DNA methylation of the MND vs. PGK promoter using vectors with either the B2 or Pro sequence cloned upstream (written before) or downstream (written after) of each promoter at day 3. Vectors were normalized on 3T3 cells, which were analysed as a control for DNA methylation (the PGK Pro and MND Pro samples). One representative experiment of two is shown. P values are <0.0001 (Pro MND vs. B2 MND and MND Pro vs. MND B2). B) Fold repression (left, normalized to the PGK B2 vector and to expression in 3T3 cells) and DNA methylation (right) of the PGK promoter within the stated vectors at 6 and 12 days post transduction. 3T3 cells (day6) were analysed as a negative control for DNA methylation. P values over 10 CpGs for day6: PGK B2 vs. PGK Pro, p=<0.0001 or vs. IAP4 PGK, p=0.0227. For day12: PGK B2 vs. PGK Pro, p=<0.0001 or vs. IAP4 PGK, p=<0.0001. C) The Pro sequence directs methylation of the PGK promoter in vivo. 0/5 integrated embryos were green for the PGK Pro vector vs. 4/9 green for the empty (PGK) vector. One embryo is shown for each (top) where the copy numbers were 2.4 (PGK Pro) and 2.8 (PGK). Bottom: DNA methylation results for the top embryos and others (numbered). PGK Pro 1. and 2. are significantly more methylated than PGK 1., p=0.0003 and p=0.0004 respectively. One representative experiment of three is shown here.
Statistics
GraphPad Prism version 4.00 (www.graphpad.com) was used for all statistical analyses. For all DNA methylation analyses, two groups (as detailed in the legends) were compared across multiple CpG positions using paired two-tailed t tests (except where stated). Any additional statistical analyses are specified in the legends.
Results
Introduced ERV sequences can induce rapid de novo DNA methylation in ES cells
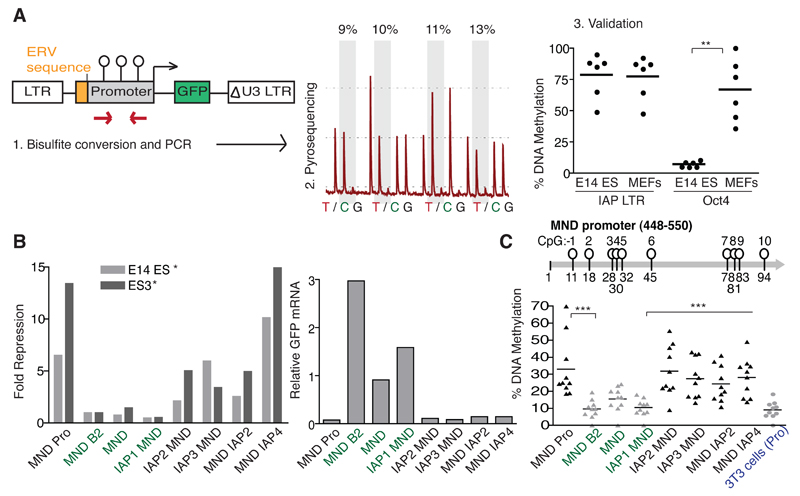
Since repeat-derived sequences are largely methylated in ES cells, we set up a method to measure the de novo DNA methylation directed at ERV-specific elements by cloning these into lentiviral vectors, a gene transfer system itself not prone to spontaneous methylation, compared to MLV-based vectors (Wang et al., 1998). To quantify levels of de novo DNA methylation, we used bisulfite pyrosequencing and verified this technique with primers for endogenous ERVs and the Oct4 promoter confirming the latter to be methylated in mouse embryonic fibroblasts (MEFs) but not ES cells, whereas the bulk of ERVs harbored this modification in both cell types (Fig. 1A). In ES cells transduced with lentiviral vectors, we confirmed that PBS Pro and previously described KAP1-sensitive IAP elements mediated repression of an adjacent MND retroviral promoter, whether placed upstream or downstream (Fig. 1B and Fig. S1A) as previously reported (Rowe et al., 2010). Bisulfite analyses then further revealed that de novo DNA methylation systematically correlated with repression, and that it surprisingly was present at four days post-transduction (Fig. 1C). A time-course study showed that this process was already under way at day 3 and then progressively increased, with mean levels across the promoter reaching around 75% at day 7 (Fig. S1B). Control vectors, whether containing no KAP1-targeted retroviral sequence, the point-mutated B2 PBS Pro derivative, or the previously identified KAP1-insensitive IAP1 element (Rowe et al., 2010), underwent neither repression nor DNA methylation. Moreover, in 3T3 fibroblasts, no ERV-directed promoter methylation was detected, suggesting this process may be restricted to ES cells (Fig. 1C).
Fig. 1. ERV sequences can induce rapid de novo DNA methylation in ES cells.
A) Technique to measure repression and DNA methylation. Left: In a lentiviral vector, ERV sequences are cloned either immediately upstream (shown here) or downstream (as stated per experiment) of an internal promoter and repression measured by GFP. Bisulfite quantitative pyrosequencing of the promoter (middle) reveals the ratio of thymine (T) vs. cytosine (C) at each CpG position (shaded in grey) to determine the % DNA methylation. Right: Validation with primers for global IAP LTRs and the Oct4 promoter, p=0.0022. Points show individual CpG positions. B) ERV-directed repression of a GFP reporter in ES cells. Left: Flow cytometry 4 days post transduction. All vectors had an internal “MND” promoter (Rowe et al., 2010), which was empty or contained an ERV sequence (Pro, B2, IAP1, IAP2, IAP3 or IAP4) either upstream (when written before MND) or downstream (when written after MND) of the promoter. Both E14 ES and ES3 ES cell lines showed significant ERV repression (p=0.0260 and p= 0.0311 respectively, unpaired one-tailed t tests) compared to control vectors (shown in green). Repression is normalized to the MND B2 control vector and to expression in 3T3 control cells. Right: GFP mRNA quantification (normalized to titin) for one representative cell line 6 days post transduction; repressed vectors showed significantly less mRNA than controls (shown in green), p=0.0042 (unpaired one-tailed t test). C) Day 4 analysis of de novo methylation at the MND promoter. N=10 (CpG positions – see map). Here the methylation profile is also shown for the MND Pro vector in 3T3 control cells. Significant differences in ES cells: The MND Pro vs. MND B2 vector (p=0.0006); IAP2 MND (p=0.001) or IAP3 MND or MND IAP2 or MND IAP4 (p=<0.0001 for all) all vs. the IAP1 MND control vector.
ERV DNA methylation patterns are conditioned by KRAB-ZFP expression profiles
KAP1 has previously been shown to repress ERVs in ES cells and early embryos (Rowe et al., 2010) and is known to be recruited to specific DNA sequences through KRAB-ZFPs (Friedman et al., 1996; Thomas and Schneider, 2011). One characterized KRAB-ZFP is ZFP809, which targets KAP1 to the PBS Pro sequence (Wolf and Goff, 2007; Wolf and Goff, 2009). Since we previously found ERVs to be regulated by KAP1 in mouse ES cells but not in MEFs (Rowe et al., 2010), we hypothesized that this difference may correlate with the expression profiles of KRAB-ZFPs, especially since KAP1 is itself still present in MEFs. We therefore used mRNA-sequencing data from ES cells and MEFs to compare expression levels of KRAB-ZFPs between these cell types. While most KRAB-ZFPs were enriched in ES cells compared to MEFs (although there is no evidence that any of them repress ERVs), with mean expression levels at 3.421 and 1.4242 respectively, ZFP809 displayed similar levels in the two cell types (Fig. 2A and data not shown). Correspondingly, when we compared silencing of the PBS Pro and IAP vectors in MEFs, we found the former but not the latter to undergo repression in these cells (Fig. 2B), consistent with the lack of activation of endogenous IAPs in MEFs upon KAP1 removal (Rowe et al., 2010). Indeed, we propose that IAPs are already stably methylated in MEFs so it is not necessary for their cognate KRAB-ZFP to be maintained. PBS Proinduced repression was accompanied by de novo DNA methylation in MEFs, albeit at lower levels than in ES cells (Fig. 2C). In order to demonstrate this mechanism to be determined by the presence of the KRAB-ZFP, we complemented human 293T cells with mouse ZFP809, leading to repression but also significant PBS Pro-dependent DNA methylation of the adjacent promoter (Fig. 2D-F). This level of DNA methylation, however, is not sufficient to maintain silencing, which is reversible (Fig. S2).
KAP1 and ESET are required for de novo DNA methylation of ERVs
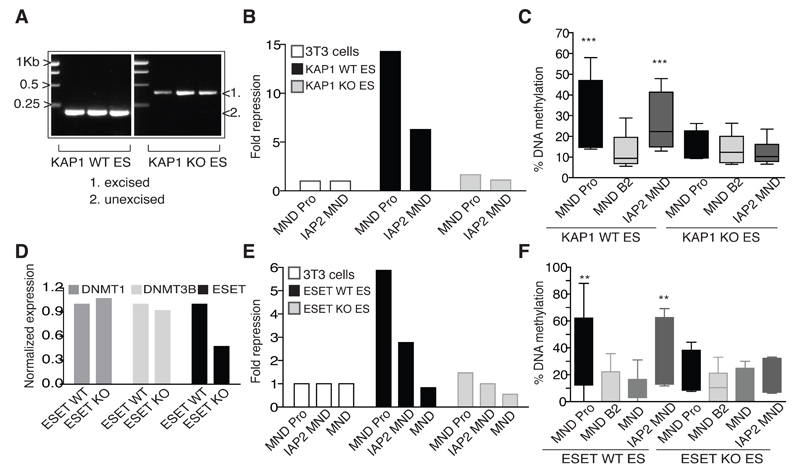
In order to determine the role of KAP1 in ERV de novo DNA methylation, we used a previously described conditional knockout ES cell line (Rowe et al., 2010). Since Kap1 deletion is ultimately lethal in early embryos and ES cells ((Rowe et al., 2010) and see Fig. S3), experiments were performed within three days of inducing this process (a time-point established from Fig. S1B). At that point, both repression and DNA methylation of the lentivirally introduced, IAP- or Pro PBS-flanked promoter could be documented in control cells, while both processes were abrogated by KAP1 removal (Fig. 3A-C). Similar experiments in Eset-conditional knockout ES cells demonstrated this histone methyltransferase to also be required for ERV-directed repression and de novo DNA methylation (Fig. 3D-F). Both KAP1 and ESET are thus necessary not only for the maintenance of proviral silencing (Leung et al., 2011; Matsui et al., 2010; Rowe et al., 2010), but also required initially for de novo DNA methylation of introduced ERV sequences.
Fig. 3. KAP1 and ESET are required for de novo DNA methylation of ERVs.
A) Kap1-inducible knockout (KO) ES cells were genotyped 3 days post excision by PCR. Triplicate samples of independently excised cells are shown. 171- and 390-bp products represent loxP-flanked or excised Kap1, respectively. B) Fold repression 48 hours post vector transduction (and 3 days post Kap1 excision). Vectors were titered on 3T3 cells first so that equal infectious units between vectors could be used for transduction of ES cells and 3T3 cells in parallel. Results were then normalized to the MND B2 control vector and to 3T3 cell expression. One representative experiment of two is shown. C) The same samples from B) were used to assess de novo methylation of the MND promoter over 10 CpG positions (see Fig. 1C for a CpG map). One representative experiment of two is shown. P values: MND Pro vs. MND B2, p=0.0001; IAP2 MND vs. MND B2, p=<0.0001. D) Eset-inducible knockout (KO) ES cells were assessed for a reduction of ESET (or DNMT mRNAs as a control) 3 days post excision by qRT-PCR. E) Fold repression 48 hours post vector transduction (and 3 days post Eset excision) normalized as in B). One representative experiment of two is shown. F) The same samples from E) were used to assess de novo methylation of the MND promoter over 10 CpG positions. One representative experiment of two is shown. P values: MND Pro vs. MND B2, p=0.0085; IAP2 MND vs. MND B2, p=0.0014.
ERV-guided KAP1-induced DNA methylation is recapitulated in embryogenesis
De novo DNA methylation patterns established early in embryogenesis are maintained later in development through the action of DNMT1 (Gaudet et al., 2004; Walsh et al., 1998). We asked if DNA methylation marks promoted by KAP1 in ES cells could also be induced in vivo, and whether in this setting they were perpetuated until late in embryogenesis. To this end, control and KAP1-sensitive IAP sequence-containing MND-GFP lentiviral vectors were injected into fertilized mouse oocytes, and the resulting embryos were collected at E13 to assess integration, repression and promoter methylation. While all embryos containing the control vector were bright or dull green, none of the IAP LV-harbouring embryos expressed GFP to detectable levels, whether examined by microscopy or by flow cytometry (Fig. 4A,B). Of note, MEFs transgenic for the non-repressed promoter displayed variability in GFP intensity within the population likely due to KAP1-independent silencing mechanisms, including relating to integration site differences (Barklis et al., 1986; Ellis, 2005). Strikingly, DNA methylation at E13 mirrored data obtained in ES cells, with high promoter methylation for all IAP-vector-harboring embryos, whereas DNA methylation of the lentivirally introduced promoter was lower and more variable in the absence of an adjacent KAP1-tethering element (Fig. 4C). These results suggest that KAP1 machinery acts in early embryos as a lock to ensure active DNA methylation and thus robust ERV silencing throughout development.
ERV sequences can induce repression and DNA methylation of a cellular promoter
Since cellular promoters can lie in close proximity to ERVs in the genome and ERV methylation status can influence cellular gene expression (Duhl et al., 1994; Macfarlan et al., 2011; Macfarlan et al., 2012; Michaud et al., 1994; Rebollo et al., 2011), we assessed whether our KAP1-dependent ERV sequences could also direct de novo DNA methylation of a cellular promoter. PBS Pro could induce some repression of a juxtaposed PGK (phosphogylcerate) promoter, albeit less efficiently than for MND, and with only minimal if any DNA methylation at day 3 post-transduction (Fig. 5A). However, by six days, repression was clear (around 7-fold, Fig. S4A) so we used this time-point to demonstrate that both PBS Pro and IAP sequences could direct de novo DNA methylation of the cellular promoter with levels further increasing after twelve days (Fig. 5B). Furthermore, PBS Pro or a KAP1-sensitive IAP sequence induced repression and DNA methylation of the adjacent PGK promoter when lentiviral vectors containing these elements were used to generate transgenic embryos (Fig. 5C and Fig. S4B).
KAP1 shapes DNA methylation of endogenous retroviruses in ES cells and embryos
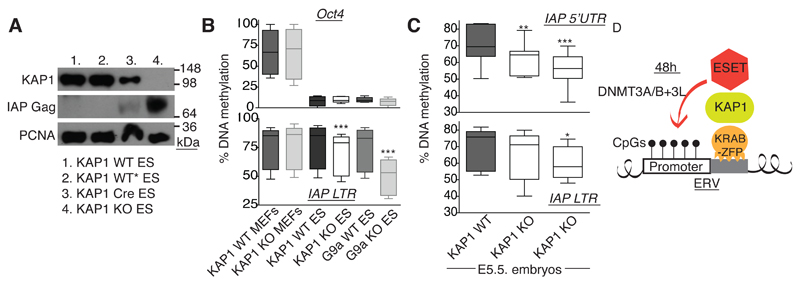
Our Southern blot analyses previously failed to detect significant global loss of DNA methylation at ERVs upon Kap1 deletion (Rowe et al., 2010) but here, we revisited this issue using the more sensitive bisulfite pyrosequencing technique to examine ERVs in ES cells and early embryos. As expected, IAPs became highly overexpressed following KAP1 removal in ES cells, which here we record to translate in a marked accumulation of IAP GAG protein (Fig. 6A). We then measured IAP methylation, using the Oct4 promoter as a control (Fig. 6B): Whereas global IAP LTRs were unaffected in Kap1-knockout MEFs, they displayed a mild but significant decrease in knockout ES cells in line with the minimal decrease previously observed at ERVs in Eset-knockout ES cells (Matsui et al., 2010). A comparatively greater loss of IAP DNA methylation was measured in an ES cell line stably deleted for the the G9a histone methyltransferase (Dong et al., 2008; Tachibana et al., 2008). However, this cell line grows continuously, allowing cumulative decreases in DNA methylation, whereas Kap1-deleted cells only divide a few times before dying. The overexpression of IAPs in Kap1-deleted cells is most likely a consequence of the removal of histone repressive marks such as H3K9me3 (Matsui et al., 2010; Rowe et al., 2010), especially as we confirmed with conventional bisulphite sequencing at one IAP locus that no molecules are completely or even largely demethylated (Fig. S5). However, it is possible that such a small decrease in DNA methylation contributes to the phenotype, particularly as we find that 5-Aza treatment of MEFs induces fairly conservative decreases in IAP DNA methylation (Fig. S6), yet a massive upregulation of their transcription (Fig. 7B). Interestingly, we also found using two different primer sets, that DNA methylation levels at ERVs were mildly but significantly lower in E5.5 Kap1 knockout embryos compared to wild type littermates (Fig. 6C and Fig. S7). Although only 1/23 further knockout embryos was obtained (at E4.5), likely due to the high lethality, this embryo also displayed less DNA methylation at IAPs than its two control littermates (p=0.0002 and 0.0014 respectively, data not shown). Altogether, these data support a role for KAP1 in genome-wide ERV DNA methylation during early embryogenesis.
Fig. 6. KAP1 shapes DNA methylation of endogenous retroviruses in ES cells and embryos.
A) KAP1 knockout in ES cells leads to an accumulation of IAP GAG p73 expression. ES cells with loxP-flanked Kap1 untreated (1.) or treated with 4-OHT (2.) or transduced with 4-OHT-inducible Cre, without (3.) or with 4-OHT (4.). Cells were harvested 4 days post 4-OHT treatment. B) KAP1 WT and KO ES cells were analysed for DNA methylation of global IAP LTRs (bottom). Methylation of the Oct4 promoter (top), G9a KO ES cells and KAP1 KO MEFs were used as controls. P values for the IAP LTR: KO MEFs were not different to WT MEFs (p=0.2186); KAP1 KO ES cells were significantly different to WT (p=0.0008) and G9a KO ES were significantly different to WT (p=0.0002). C) Kap1 heterozygous mice (C57BL/6) were crossed and embryos dissected at E5.5 to measure DNA methylation of endogenous IAPs using primers either in the 5’UTR (top) or LTR (bottom). Here two knockouts and one WT embryo were analysed. Top: p=0.0059 and 0.0006 respectively. Bottom: p=0.1613 (not significant) and 0.0493 respectively (paired one-tailed t tests). D) Summary model: KRAB-ZFPs, KAP1 and ESET are necessary for de novo methylation of ERVs, occurring within 48 hours. This process is sequence-specific and takes place in ES cells and embryogenesis.
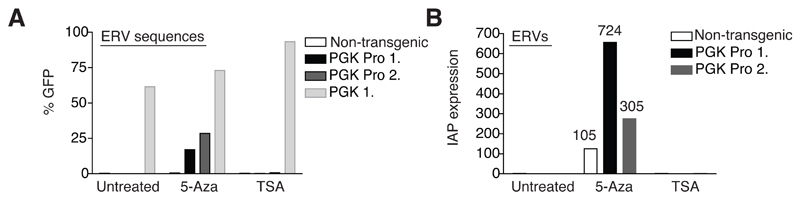
Fig. 7. DNA methylation of introduced ERV sequences and resident ERVs becomes critical late in development.
A) Transgenic MEFs from Fig. 5C and additionally, a MEF cell line not containing any vector (non-transgenic) were treated with 5-Aza (48 hours) or TSA (24 hours) or left untreated and GFP measured the next day. While TSA could increase the % GFP of MEFs transgenic for the empty vector (PGK 1.), GFP was only resurrected in Pro vector-transgenic MEFs upon 5-Aza addition. B) In the same experiment as shown in A), endogenous IAP transcripts were also measured in the stated MEF lines by qRT-PCR. Fold upregulation is shown normalized to untreated controls for each cell line.
DNA methylation becomes critical for ERV silencing later in development
Finally, in order to address the relevance of DNA methylation of ERV sequences and endogenous ERVs in development, we derived MEFs from transgenic embryos in which our ERV-derived proviruses were integrated. Treatment with the DNA methyltransferase inhibitor 5-aza (but not the HDAC inhibitor, TSA) was sufficient to relieve silencing and this was true for both ERV sequences newly introduced within the context of lentiviral vectors and endogenous IAPs that were upregulated by up to around 700 fold (Fig. 7). To ask if DNA methylation already played a role in silencing ERVs at an earlier developmental stage, we also examined DNMT triple knockout (TKO) ES cells (Tsumura et al., 2006), in which all three DNA methyltransferases are deleted (Fig. S8). Levels of IAP GAG protein and of IAP and MERV-L RNA were significantly higher in these than in control ES (Fig. S8A,B), consistent with a previous study (Matsui et al., 2010). Of note, it was recently reported that in DNMT1 KO ES cells and in another ES cell line where all DNMT proteins were depleted (by knockout of DNMT3A/3B and knockdown of DNMT1) (Meissner et al., 2005), IAP protein was only detected upon LIF (leukemia inhibitory factor) withdrawal, hence induction of differentiation for six or more days, a time point at which OCT4 protein was undetected in differentiated cultures by immunofluoresence (Hutnick et al., 2010). However, the TKO ES cells used here were maintained in standard ES cell conditions (see methods) and were largely undifferentiated as indicated by comparable levels of OCT4 protein to WT (Fig. S8A) as well as high surface expression of SSEA1 (Fig. S8C), in contrast to ES cells cultured without LIF, which downregulated this marker (Fig. S8D). In addition, these cells have previously been found to conserve their self-renewal ability (Tsumura et al., 2006). Still, we cannot exclude that some differentiating cells contributed more prominently to the observed IAP upregulation. Importantly, the overexpression of these elements in ES cells (around 40 fold) is modest compared to that seen in MEFs (Fig. 7) supporting a critical role of DNA methylation in ERV silencing later in development (Gaudet et al., 2004; Walsh et al., 1998) in contrast to in ES cells where KAP1-dependent histone methylation is present (Hutnick et al., 2010; Karimi et al., 2011; Matsui et al., 2010; Rowe et al., 2010) and reviewed in (Leung and Lorincz, 2011; Rowe and Trono, 2011). Indeed both 5-aza treatment and KAP1 knockout need to be performed to induce such a dramatic upregulation of IAPs in ES cells (to around 400 fold, (Rowe et al., 2010)).
Discussion
The present work demonstrates the involvement of sequence-specific KRAB-ZFPs, KAP1, and ESET in establishing early embryonic DNA methylation patterns at ERV sequences that are subsequently maintained through embryogenesis. This suggests these factors to be required during early development first for the histone-based silencing and then for the de novo methylation of genomic ERVs, even though the evidence provided here is limited to newly introduced ERV sequences. Our results support a model (Fig. 6D) whereby sequences located in the 5’ region of ERV genomes recruit specific KRAB-ZFPs, which brings about KAP1, ESET and most likely other chromatin modifiers, the conjugated action of which leads to transcriptional repression and de novo promoter methylation. This phenomenon implies the secondary recruitment of de novo DNA methyltransferases, consistent with the documented immunoprecipitation of KAP1 with DNMT3A and DNMT3B (Li et al., 2008; Quenneville et al., 2011; Zuo et al., 2012), and the reported interaction of DNMT3A with the B-BOX coiled coil domain of KAP1 (Zuo et al., 2012). Since ERV DNA methylation requires ESET, it could be that this enzyme directly recruits DNMT3AB and 3L. In that respect, histone methyltransferases have previously been revealed to act as docking sites for DNA methylation machinery (Dong et al., 2008; Tachibana et al., 2008) and, importantly, ESET has been proposed to bind directly to DNMT3A and tether it to promoters, leading to their DNA methylation-mediated silencing (Li et al., 2006). Alternatively, de novo DNA methylation may result indirectly from a lack of transcription and in turn active marks such as H3K4me3 (Jia et al., 2007; Ooi et al., 2007), due to the recruitment of H3K9me3 and histone deacetylation at ERVs.
We reveal here that KAP1-recruiting ERV sequences are able to trigger repression and de novo DNA methylation of a strong cellular promoter as well as of a retroviral promoter, at least within the context of newly introduced lentiviral vectors. The slower kinetics observed with the PGK promoter, compared with its MND counterpart, may be due to partial protection of its CpG-dense core by transcription factors, as recently proposed (Lienert et al., 2011; Stadler et al., 2011). These data indicate that KAP1-mediated silencing of ERVs may dampen expression from nearby cellular transcription units by inducing promoter CpG methylation. This is supported by our observation that the methylation status of CpG islands correlating with their immediate proximity to KAP1-recruiting sites in ES cells (Quenneville et al., 2012). Relevant to ERVs, in a recent study, polymorphic copies of ERVs across mouse strains were employed to determine whether histone and DNA methylation marks could spread from ERVs to cellular promoters. Results showed that, while H3K9me3 marks could spread several kilobases, it was infrequent for DNA methylation to spread from an IAP to neighbouring gene promoters, except where genes lie within 500bp of an IAP (Rebollo et al., 2011). Our results now implicate a KAP1/ESET complex in this mechanism. Since DNA methylation rarely spreads from ERVs as far as nearby promoters in the genome, in future work it will be interesting to investigate the possible presence of barrier elements protecting cellular genes from the spread of repressive marks including DNA methylation, nucleated at ERV sequences.
Here, we observed significant DNA methylation of a PBS Pro-, retroviral promoter-containing lentiviral vector in MEFs, which express similar levels of ZFP809 to ES cells, as well as in 293T cells engineered to express high levels of this KRAB-ZFP. In contrast, a KAP1-restricted IAP sequence induced neither repression nor de novo DNA methylation of the MND promoter in MEFs, suggesting that the cognate KRAB-ZFP is not expressed in these targets. The matching KRAB-ZFP(s) could be any of those enriched in ES cells except ZFP568, which has already been shown not to be responsible for repressing IAPs in vivo (Shibata et al., 2011). That ZFP809 induced only low methylation in MEFs and 293T cells, compared to ES cells, explains why the repression is reversible, as previously noted when KRAB is artificially tethered to a promoter in cell lines (Barde et al., 2009; Groner et al., 2010; Quenneville et al., 2012; Wiznerowicz and Trono, 2003).
Surprisingly, we found very little decrease in DNA methylation levels at resident ERVs in KAP1 knockout embryos despite the hereby-demonstrated role of KAP1 in shaping DNA methylation patterns at ERV sequences in embryogenesis. It could be that loss of DNA methylation that follows loss of repressive histone marks can not be measured accurately due to the earlier lethality of knockout embryos (at or before E5.5) (Cammas et al., 2000) or that the majority of DNA methylation at genomic ERVs is KAP1-independent at this stage. Along these lines, since IAPs are relatively resistant to genome-wide DNA demethylation in pre-implantation embryos (Lane et al., 2003), methylation may be maintained without requiring de novo methyltransferase activity. Importantly, this and previous studies (Matsui et al., 2010; Rowe et al., 2010) indicate that histone modifications, rather than DNA methylation, are primarily responsible for silencing ERVs in ES cells, as IAPs get immediately and markedly upregulated upon KAP1 or ESET removal in ES cells, in the absence of any spectacular change in their DNA methylation status. It is noteworthy that ERVs exhibit of range of DNA methylation rates in ES cells, suggesting that some, including many IAPs, may escape demethylation, while others may get demethylated but rapidly re-methylated following their KRAB/KAP1 recognition. Moreover, the impact of DNA methylation per se on transcription, at least during this period, needs to be further investigated, as blocking histone-based repression of IAPs suffices to induce their upregulation. It may be that subtle alterations at specific positions, critical to block transcription, have so far escaped analyses, or that DNA methylation does not have the same consequences in ES and somatic cells, for instance if some factors important for repressing transcription at DNA methylated loci are expressed in differentiated but not in undifferentiated cells.
Our in vivo experiments with vectors reveal that ERV methylation patterns are faithfully copied late into development leading to extremely high DNA methylation levels (upwards of 80%) that mimic natural levels at IAP LTRS resident in the genome. The self-perpetuating nature of DNA methylation alleviates the need for the continuous expression of cognate ERV-binding KRAB-ZFP repressors. It also explains why the maintenance of DNA methylation at ERVs throughout development is critical to suppress their transcription, notably for IAPs (Walsh et al., 1998). In sum, the present work implies that ERV sequence recognition by KRAB-ZFPs/KAP1 and the lysine methyltransferase ESET is necessary to target ERV-containing loci for rapid de novo DNA methylation in early development, leading to stable site-specific DNA methylation signatures across the genome.
Supplementary Information
Eight Supplementary Figures and their legends
Supplementary Table I: List of primers
Acknowledgments
We thank P.V. Maillard for advice, A. Reymond and J. Chrast (from the Center for Integrative Genomics, University of Lausanne) for use of their pyrosequencer, and Carine Gubelmann and Andrea Corsinotti for the ZFP809 plasmid. We also thank D. Schubeler for supplying us with the G9a knockout and parental ES cells, originally from Y. Shinkai (The Riken Institute, Japan) who provided us with the Eset-inducible knockout ES cells, and B. Cullen (Duke University) for the anti-IAP GAG antibody. We thank F. Spitz, (EMBL, Heidelberg), in whose lab T.A. performed the experiment depicted in Fig. S5. This work was supported by grants from the Swiss National Science Foundation and the European Research Council to D.T.
Footnotes
Authors’ Contributions
H.M.R. conceived the study, designed and performed the experiments, analyzed the data and wrote the manuscript. S.O., S.V., M.F., D.M. and T.A. contributed to experiments. J.M. provided help with pyrosequencing setup and D.T. conceived the study, designed experiments and wrote the manuscript.
Competing Interests
The authors declare that they have no competing financial interests.
References
- Barde I, Laurenti E, Verp S, Groner AC, Towne C, Padrun V, Aebischer P, Trumpp A, Trono D. Regulation of episomal gene expression by KRAB/KAP1-mediated histone modifications. J Virol. 2009;83:5574–80. doi: 10.1128/JVI.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barde I, Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Neurosci. 2010;Chapter 4 doi: 10.1002/0471142301.ns0421s53. Unit 4 21. [DOI] [PubMed] [Google Scholar]
- Barklis E, Mulligan RC, Jaenisch R. Chromosomal position or virus mutation permits retrovirus expression in embryonal carcinoma cells. Cell. 1986;47:391–9. doi: 10.1016/0092-8674(86)90596-9. [DOI] [PubMed] [Google Scholar]
- Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203:971–83. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–9. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development. 2000;127:2955–63. doi: 10.1242/dev.127.13.2955. [DOI] [PubMed] [Google Scholar]
- Carlson LL, Page AW, Bestor TH. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992;6:2536–41. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A. 2002;99:16916–21. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schubeler D, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum Gene Ther. 2005;16:1241–6. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:el000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–7. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedli M, Nikolaev S, Lyle R, Arcangeli M, Duboule D, Spitz F, Antonarakis SE. Characterization of mouse Dactylaplasia mutations: a model for human ectrodactyly SHFM3. Mamm Genome. 2008;19:272–8. doi: 10.1007/s00335-008-9106-0. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–78. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Rideout WM, 3rd, Meissner A, Dausman J, Leonhardt H, Jaenisch R. Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing. Mol Cell Biol. 2004;24:1640–8. doi: 10.1128/MCB.24.4.1640-1648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JL, Fazzari MJ, Ferguson-Smith AC, Greally JM. CG dinucleotide periodicities recognized by the Dnmt3a-Dnmt3L complex are distinctive at retroelements and imprinted domains. Mamm Genome. 2009;20:633–43. doi: 10.1007/s00335-009-9232-3. [DOI] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Denervaud N, Bucher P, Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:el000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285:21082–91. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–51. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H, Kurahashi H, Toda T. Genetically regulated epigenetic transcriptional activation of retrotransposon insertion confers mouse dactylaplasia phenotype. Proc Natl Acad Sci U S A. 2007;104:19034–9. doi: 10.1073/pnas.0705483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–87. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci U S A. 2011;108:5718–23. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DC, Lorincz MC. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci. 2011;37:127–33. doi: 10.1016/j.tibs.2011.11.006. [DOI] [PubMed] [Google Scholar]
- Li H, Rauch T, Chen ZX, Szabo PE, Riggs AD, Pfeifer GP. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem. 2006;281:19489–500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–57. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schubeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–7. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–31. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WE, Spada F, Leonhardt H. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep. 2009;10:1259–64. doi: 10.1038/embor.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–77. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 1994;8:1463–72. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Yokota Y, Ishida H, Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–13. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007;27:2746–57. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Wolf D, Hartung O, Agarwal S, Daley GQ, Goff SP, Bestor TH. Dynamic instability of genomic methylation patterns in pluripotent stem cells. Epigenetics Chromatin. 2010;3:17. doi: 10.1186/1756-8935-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Wang Z, Shang J, Bekkari K, Liu R, Pacchione S, McNulty KA, Ng A, Barnum JE, Storer RD. Intracisternal A particle genes: Distribution in the mouse genome, active subtypes, and potential roles as species-specific mediators of susceptibility to cancer. Mol Carcinog. 2010;49:54–67. doi: 10.1002/mc.20576. [DOI] [PubMed] [Google Scholar]
- Quenneville S, Turelli P, Bojkowska K, Raclot C, Offner S, Kapopoulou A, Trono D. The KRAB-ZFP/KAP1 System Contributes to the Early Embryonic Establishment of Site-Specific DNA Methylation Patterns Maintained during Development. Cell Rep. 2012 doi: 10.1016/j.celrep.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–72. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, Zhang Y, Goyal P, Keane TM, Jones S, Hirst M, et al. Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 2011;7:e1002301. doi: 10.1371/journal.pgen.1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–40. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology. 2011;411:273–87. doi: 10.1016/j.virol.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–12. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Shibata M, Blauvelt KE, Liem KF, Jr, Garcia-Garcia MJ. TRIM28 is required by the mouse KRAB domain protein ZFP568 to control convergent extension and morphogenesis of extra-embryonic tissues. Development. 2011;138:5333–43. doi: 10.1242/dev.072546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–23. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27:2681–90. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takabatake T, Ishihara H, Ohmachi Y, Tanaka I, Nakamura MM, Fujikawa K, Hirouchi T, Kakinuma S, Shimada Y, Oghiso Y, et al. Microarray-based global mapping of integration sites for the retrotransposon, intracisternal A-particle, in the mouse genome. Nucleic Acids Res. 2008;36:e59. doi: 10.1093/nar/gkn235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011;21:1800–12. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–14. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4:231. doi: 10.1186/gb-2003-4-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–7. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Wang L, Robbins PB, Carbonaro DA, Kohn DB. High-resolution analysis of cytosine methylation in the 5long terminal repeat of retroviral vectors. Hum Gene Ther. 1998;9:2321–30. doi: 10.1089/hum.1998.9.16-2321. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Jakobsson J, Szulc J, Liao S, Quazzola A, Beermann F, Aebischer P, Trono D. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J Biol Chem. 2007;282:34535–41. doi: 10.1074/jbc.M705898200. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–61. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Cammas F, Losson R, Goff SP. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J Virol. 2008;82:4675–9. doi: 10.1128/JVI.02445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–4. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 2008;4:e1000007. doi: 10.1371/journal.pgen.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Sheng J, Lau HT, McDonald CM, Andrade M, Cullen DE, Bell FT, Iacovino M, Kyba M, Xu G, et al. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem. 2012;287:2107–18. doi: 10.1074/jbc.M111.322644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.