Abstract
Bacillus cereus isolates have been described harboring Bacillus anthracis toxin genes, most notably B. cereus G9241, and capable of causing severe and fatal pneumonias. This report describes the characterization of a B. cereus isolate, BcFL2013, associated with a naturally occurring cutaneous lesion resembling an anthrax eschar. Similar to G9241, BcFL2013 is positive for the B. anthracis pXO1 toxin genes, has a multi-locus sequence type of 78, and a pagA sequence type of 9. Whole genome sequencing confirms the similarity to G9241. In addition to the chromosome having an average nucleotide identity of 99.98% when compared to G9241, BcFL2013 harbors three plasmids with varying homology to the G9241 plasmids (pBCXO1, pBC210 and pBFH_1). This is also the first report to include serologic testing of patient specimens associated with this type of B. cereus infection which resulted in the detection of anthrax lethal factor toxemia, a quantifiable serum antibody response to protective antigen (PA), and lethal toxin neutralization activity.
Introduction
Bacillus cereus infections are typically associated with foodborne illnesses, periodontal diseases, and other opportunistic diseases [1]. However, B. cereus can be associated with more severe and even fatal infections. Within the last decade, B. cereus isolates associated with severe infections have been described harboring B. anthracis toxin genes and/or capsule biosynthesis genes. Hoffmaster et al. described the first B. cereus with B. anthracis toxin genes, B. cereus G9241, to cause a severe pneumonia in a metal worker from Louisiana [2]. Since that report, there have been several additional accounts of infection associated B. cereus resembling G9241 isolated from severe pneumonia cases in metal workers in Texas [3, 4, 5]. With the exception of the initial case from Louisiana, these infections were fatal. In addition to these isolates, Klee et al. characterized two Bacillus isolates harboring B. anthracis virulence genes, now termed B. cereus biovar anthracis, cultured from deceased great apes in Cote d’Ivoire and Cameroon. [6].
In this report, we characterize a B. cereus isolate, BcFL2013, cultured from a swab of a facial lesion resembling an anthrax eschar from a 70-year-old Florida resident. (Fig 1). The patient was hospitalized and received antibiotic treatment but fully recovered. The Florida Department of Health Laboratory performed the B. anthracis-specific polymerase chain reaction assay (LRN PCR) on the isolate from the skin lesion and detected one of the B. anthracis plasmids, pXO1. The isolate was then forwarded to the Centers for Disease Control and Prevention (CDC) for further characterization. In addition to isolate characterization, plasma and serum samples from the patient were collected and forwarded to the CDC for serological testing.
Fig 1. Anthrax-like cutaneous lesion on left check of 70-year-old male Florida resident.
Materials and Methods
The BcFL2013 isolate was inoculated on trypticase soy agar with 5% sheep blood (BD Diagnostics Systems, Franklin Lakes, NJ) and tested to determine susceptibility to gamma phage as previously described [7]. Capsule staining and visualization was performed by incubating cells overnight at 37°C in defibrinated horse blood (Remel, Lenexa, KS). Capsules were visualized using India Ink (Remel, Lenexa, KS) under a 100X oil immersion objective. PCR assays for B. anthracis virulence genes (pagA, lef, cya and capA) were performed as previously described [8, 9]. The pBC210 (previously called pBC218) PCR assay was performed as described by Hoffmaster et al. [3].
The isolate was also molecularly characterized by multilocus sequence typing (MLST) and protective antigen gene (pagA) sequencing prior to performing whole genome sequencing (WGS). MLST and pagA sequence typing was performed as previously described [10, 11]. Briefly, MLST was based on the analysis of seven housekeeping gene partial sequences (glpF, gmk, ilvD, pta, pur, pycA, and tpi) and is described online at http://pubmlst.org/bcereus. For WGS, a 101 X 101 paired-end run was performed in an Illumina GAIIx using TruSeq chemistry and yielded 10,182,400 reads. A de novo assembly was performed using CLC Genomics Workbench 6.0.4 (CLC Inc., Aarhus, Denmark) which yielded 84 contigs consisting of 5.46 Mb, with an N50 of 139,961 bp. [12]. The reads were also mapped against reference sequences for G9241 (CP009590.1) and plasmids associated with G9241, pBC210 (CP009591.1), pBCXO1 (CP009592.1) and pBFH_1 (CP009589.1), to extract homologous sequences from BcFL2013 using CLC Genomics Workbench 8.0.3. The average nucleotide identity (ANI) was calculated using the online calculator at http://enve-omics.ce.gatech.edu/ani/index [13].
In addition, testing of an acute-stage plasma sample (collected on 03/02/2013) and two subsequent convalescent serum samples (collected on 03/14/2013 and 04/04/2013) taken from the patient was performed to detect lethal factor (LF), anti-protective antigen IgG, and anthrax lethal toxin neutralizing activity (TNA) immune response. All serological assays were performed as previously described [14, 15, 16].
Because the samples were collected in the course of clinical care and management, and sent to CDC for routine diagnostic purposes, informed consent was not obtained, and IRB approval was not sought. However, we received express written permission from the patient to publish the photograph presented in this report.
Results
BcFL2013 was beta-hemolytic on sheep blood agar and resistant to gamma phage lysis (Table 1). The colony morphology was typical for B. cereus: large, umbonate (raised center), dark tan and granular. This isolate also produced a capsule (Table 1, Fig 2). Initial analysis of BcFL2013 by PCR revealed it was similar to G9241: positive for the B. anthracis pXO1-encoded toxin genes (pagA, lef, and cya) and negative for the pXO2-encoded capsule (capA) gene. However, unlike G9241, this isolate was negative by the pBC210 plasmid assay. Molecular subtyping showed the isolate had an identical MLST and pagA sequence type as G9241 (ST 78 and genotype 9, respectively).
Table 1. Summary of results for testing of B. cereus BcFL2013 compared to B. cereus G9241 and B. anthracis Ames.
| Assay | B. cereus BcFL2013 | B. cereus G9241 | B. anthracis Ames |
|---|---|---|---|
| Susceptibility to gamma phage | Resistant | Resistant | Susceptible |
| Demonstration of capsule | Positive | Positive | Positive |
| MLST | 78 | 78 | 1 |
| pagA sequence type | 9 | 9 | 6 |
| pXO1 or homolog | Yes | Yes | Yes |
| pXO2 or homolog | No | No | Yes |
| pBC210 or homolog | Partial* | Yes | No |
* Sequence with homology to approximately half of pBC210 was detected (108,352 bp).
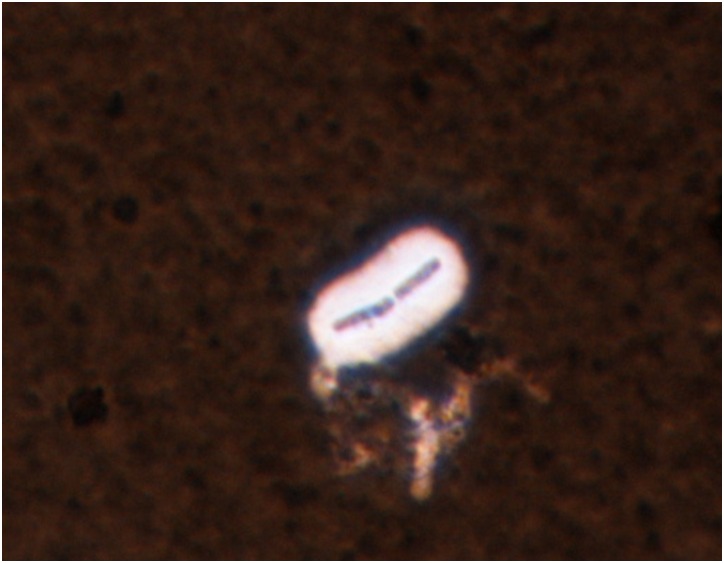
Fig 2. B. cereus BcFL2013 capsule. India ink stain of B. cereus BcFL2013 after overnight growth in defibrinated horse blood.
We performed WGS to further characterize and determine the presence of virulence genes and plasmids in this isolate. Whole genome sequence Illumina reads for BcFL2013 were mapped to that of plasmid pBCXO1 of G9241, which indicated that a homolog of this plasmid is present, although with a ~ 2.5 kb deletion. Comparison of this homolog with pBCXO1 yielded an ANI of >99.98%. This included the anthrax toxin genes (pagA, cya, and lef) and hyaluronan capsule synthase genes (hasACB) which were determined to be 100% identical to those found in pBCXO1. Mapping the Illumina reads of BcFL2013 to plasmid pBC210 of G9241 indicated a homolog of the plasmid was present but had only approximately half the sequence of pBC210 and did not include the bpsXABCDEFGH operon. The 108,352 kb of homologous pBC210 sequence had an ANI of 99.84% when compared to G9241.
Johnson et al. reported on the complete sequence of the third G9241 plasmid, originally named pBClin29 and noted as ~29-kb long, which they renamed pBFH_1 [17]. The complete pBFH_1 plasmid is 52,166 bp and encodes hypothetical phage proteins as previously described [2, 17]. Our sequencing of the BcFL2013 isolate revealed a 48-kb contig homologous to pBFH_1 (ANI of 98.74%). Analysis of the remaining reads (those not mapping to the chromosome or the three previously mentioned plasmids) did not indicate the presence of any additional plasmids.
The unmapped reads remaining after mapping to the three plasmids were used for a de novo assembly which yielded a putative chromosomal sequence ~ 5.14 MB. When compared to the sequence for the G9241 chromosome (CP009590.1), the putative BcFL2013 chromosomal sequence yielded an ANI of 99.98%. The shotgun sequence for BcFL2013 was deposited into DDBJ/EMBL/GenBank under accession no. JHQN01000000.
Serological testing of acute-stage plasma and two subsequent convalescent serum samples taken from the patient was performed to detect anthrax toxin lethal factor (LF), anti-protective antigen (PA) IgG, and anthrax lethal toxin neutralizing activity (TNA) immune response (Table 2). LF was detected in the acute sample (0.819 ng/mL) but not in the convalescent samples. These results were similar to levels observed for cutaneous anthrax cases described previously [18]. Anti-PA IgG was detected only in the convalescent samples (18.2 and 48.0 μg/mL, respectively). Similarly, anthrax lethal toxin neutralization activity was detected in the convalescent samples (ED50 = 156.5 and ED50 = 194.5, respectively) but was not tested for in the acute plasma sample.
Table 2. Results of serological testing of specimens from 2013 Florida patient with anthrax-like cutaneous lesion.
| Assay results | |||
|---|---|---|---|
| Specimen and date collected | Lethal factor detection | Anti-PA ELISA | Toxin neutralization assay |
| Plasma (03/02/2013) | 0.819 ng/mL | <LLOQb | Not tested |
| Serum (03/14/2013) | <LODa | 18.2 μg/mL | 156.5 (ED50) |
| Serum (04/04/2013) | <LOD | 48.0 μg/mL | 194.5 (ED50) |
aLOD, limit of detection
bLLOQ, lower limit of quantification
Discussion
To date, six B. cereus isolates, associated with human infections, have been described containing B. anthracis toxin genes: four isolates (G9241 from LA, 03BB87, 03BB102, Elc2) from metal workers in Texas and Louisiana with respiratory infections and two isolates (laboratory-acquired G9241 infection from Illinois and BcFL2013) resulting in cutaneous infections [2–5, 19]. In addition to these, the CDC received a B. cereus isolate harboring B. anthracis toxin genes (LA4726) from Louisiana in 2007 from a pneumonia patient who was also a metal worker (data not shown). Four of these isolates (03BB87, LA4726, G9241, and BcFL2013) had identical subtypes (ST 78) by MLST. The two remaining metal worker isolates (03BB102 and Elc2) were MLST subtypes ST 11 and ST 108, respectively. All seven metal worker isolates had sequence homology to B. anthracis toxin genes (pagA, lef, and cya) on pXO1. Only one isolate, 03BB102, had homology to the B. anthracis capsule genes (cap operon) on the pXO2 plasmid; however, it was not observed to produce a polyglutamate capsule like B. anthracis [3]. Phenotypically, all of the B. cereus isolates from the metal workers produced a capsule (either hyaluronic acid and/or exo-polysaccaride) although they differed in composition from the B. anthracis capsule. The BcFL2013 isolate described here has one of the capsule operons associated with G9241, hasACB. This would suggest that BcFL2013 produced, at very least, a hyaluronic acid capsule, but further studies would be required to determine the exact capsule composition.
In addition to these isolates, two B. cereus isolates have been isolated from non-human primates in Cote d’Ivoire and Cameroon (CI and CA) which harbor B. anthracis virulence genes and have been described as B. cereus biovar anthracis [6]. Unlike the strains from the metal workers, the B. cereus biovar anthracis isolates harbor both B. anthracis virulence plasmids (99–100% identity) [20]. Additionally, the CA and CI isolates have a non-functional plcR gene, a transcriptional regulator, due to an insertion at the 3’ end of the gene [6]. B. anthracis also has a non-functional plcR gene resulting from of a nonsense mutation in the gene [21]. Conversely, BcFL2013 has a predicted full length plcR protein with 100% identity to that of G9241.
Phenotypically, the CA and CI isolates differ from B. anthracis as they are motile, gamma-phage resistant, and, in some cases, penicillin-resistant and, unlike most B. cereus, are non-hemolytic on blood agar. [6]. The CA and CI isolates differ from the other B. cereus isolates in that they express both a hyaluronic acid and polyglutamate capsules [22]. By MLST, the CA and CI isolates, along with two metal worker isolates (03BB102 and Elc2), are more closely related to B. anthracis [3, 20]. It is of interest that, to date, the B. cereus biovar anthracis isolates have not been cultured from humans. Whether this is the result of virulence differences, decreased human exposure, or an artifact of limited diagnostic capacity in regions where these strains have been detected remains to be determined.
This is the first report of a B. cereus isolate harboring B. anthracis toxin genes associated with a naturally occurring cutaneous infection. However, as previously mentioned, there was a laboratory-acquired cutaneous infection associated with B. cereus G9241 in 2011 [19]. Unlike the previous cases associated with these types of B. cereus isolates, the Florida patient did not have any previous history of metal work. The patient had recent contact with apparently healthy horses but this is not a known risk factor for infection. Previous cases caused by these types of isolates occurred in Texas and Louisiana, neither of which were visited by this patient recently. It remains unknown how the Florida patient became infected.
In addition to being the first report of a naturally acquired B. cereus infection resembling cutaneous anthrax, this is the first report of demonstrated LF toxemia and a detectable humoral response to the toxin with in vitro toxin neutralization activity from an infection caused by a B. cereus. However, Brezillon et al. showed that the B. cereus biovar anthracis isolates expressed both B. anthracis toxin and capsule genes in animal models [22]. Previous reports of serious and fatal B. cereus infections did not include serologic testing of patients to determine if the toxin genes were expressed in vivo resulting in toxemia. These methods have been used successfully to detect toxin and antibody response in systemic and cutaneous anthrax cases [18, 23]. This case demonstrates that B. anthracis toxin genes are not only present but are also expressed in vivo and, in this case, associated with the clinical presentation resembling an anthrax eschar.
Acknowledgments
This publication made use of the Bacillus cereus Multi Locus Sequence Typing website (http://pubmlst.org/bcereus/) developed by Keith Jolley and sited at the University of Oxford (Jolley & Maiden 2010, BMC Bioinformatics, 11:595). The development of this site has been funded by the Wellcome Trust.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Turenne CY, Snyder JW, Alexander DC. Bacillus and other aerobic endospore-forming bacteria In: Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, et al. , editors. Manual of Clinical Microbiology. Sterling: ASM Press; 2015. pp. 441–461. [Google Scholar]
- 2.Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, et al. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc Natl Acad Sci USA. 2004;101:8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmaster AR, Hill KK, Gee JE, Marston CK, De BK, Popovic T, et al. Characterization of Bacillus cereus isolates associated with fatal pneumonias: strains are closely related to Bacillus anthracis and harbor B. anthracis virulence genes. J Clin Micro. 2006;44:3352–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avashia SB, Riggins WB, Lindley C, Hoffmaster A, Drumgoole R, Nekomoto T, et al. Fatal pneumonia among metalworkers due to inhalation exposure to Bacillus cereus containing Bacillus anthracis toxin genes. Clin Infect Dis. 2007;44:414–416. [DOI] [PubMed] [Google Scholar]
- 5.Wright AM, Beres SB, Consamus EN, Long SW, Flores AR, Barrios R, et al. Rapidly progressive, fatal, inhalation anthrax-like infection in a human. Arch Pathol Lab Med. 2011;135:1447–1459. 10.5858/2011-0362-SAIR.1 [DOI] [PubMed] [Google Scholar]
- 6.Klee SR, Ozel M, Appel B, Boesch C, Ellerbrok H, Jacob D, et al. Characterization of Bacillus anthracis-like bacteria isolated from wild great apes from Cote d’Ivoire and Cameroon. J Bacteriol. 2006;188:5333–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marston CK, Gee JE, Popovic T, Hoffmaster AR. Molecular approaches to identify and differentiate Bacillus anthracis from phenotypically similar Bacillus species isolates. BMC Microbiol. 2006;6:22 10.1186/1471-2180-6-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmaster AR, Meyer RF, Bowen MP, Marston CK, Weyant RS, et al. Evaluation and validation of a real-time polymerase chain reaction assay for rapid identification of Bacillus anthracis. Emerg Infect Dis. 2002;8:1178–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson PJ, Hugh-Jones ME, Adair DM, Green G, Hill KK, Kuske CR, et al. PCR analysis of tissue samples from the 1979 Sverdlovsk anthrax victims: The presence of multiple Bacillus anthracis strains in different victims. Proc Natl Acad Sci USA. 1998;95:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Priest FG, Barker M, Baillie LW, Holmes EC, Maiden MCJ. Population structure and evolution of the Bacillus cereus group. J Bacteriol. 2004;186:7959–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmaster AR, Fitzgerald CC, Ribot E, Mayer LW, Popovic T. Molecular subtyping of Bacillus anthracis and the 2001 bioterrorism-associated anthrax outbreak, United States. Emerg Infect Dis. 2002;8:1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gee JE, Marston CK, Sammons SA, Burroughs MA, Hoffmaster AR. Draft genome of Bacillus cereus strain BcFL2013, a clinical isolate similar to G9241. Genome Announc. 2014;2:e00469–14. 10.1128/genomeA.00469-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. [DOI] [PubMed] [Google Scholar]
- 14.Boyer AE, Quinn CP, Woolfitt AR, Pirkle JL, McWilliams LG, Stamey KL, et al. Detection and quantification of anthrax lethal factor in serum by mass spectrometry. Anal Chem. 2007;79:8463–8470. [DOI] [PubMed] [Google Scholar]
- 15.Semenova VA, Schiffer J, Steward-Clark E, Soroka S, Schmidt DS, Brawner MM, et al. Validation and long term performance characteristics of a quantitative enzyme linked immunosorbent assay (ELISA) for human anti-PA IgG. J Immunol Methods. 2012;376:97–107. 10.1016/j.jim.2011.12.002 [DOI] [PubMed] [Google Scholar]
- 16.Li H, Soroka SD, Taylor TH, Stamey KL, Stinson KW, Freeman AE, et al. Standardized, mathematical model-based and validated in vitro analysis of anthrax lethal toxin neutralization. J Immunol Methods. 2008;333:89–106. 10.1016/j.jim.2008.01.007 [DOI] [PubMed] [Google Scholar]
- 17.Johnson SL, Daligault HE, Davenport KW, Jaissle J, Frey KG, Ladner JT, et al. Complete genomes sequences for 35 biothreat assay-relevent Bacillus species. Genome Announc. 2015;3:e00151–15. 10.1128/genomeA.00151-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyer AE, Quinn CP, Beesley CA, Gallegos-Candela M, Marston CK, Cronin LX, et al. Lethal factor toxemia and anti-protective antigen antibody activity in naturally acquired cutaneous anthrax. J Infect Dis. 2011;204:1321–1327. 10.1093/infdis/jir543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser J. UPDATED: University of Chicago microbiologist infected from possible lab accident. Science. 2011. Available: http://news.sciencemag.org/2011/09/updated-university-chicago-microbiologist-infected-possible-lab-accident.
- 20.Klee SR, Brzuszkiewicz EB, Nattermann H, Bruggemann H, Dupke S, Wollher A, et al. The genome of a Bacillus isolate causing anthrax in chimpanzees combines chromosomal properties of B. cereus with B. anthracis virulence plasmids. Plos One. 2010;5:e10986 10.1371/journal.pone.0010986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agaisse H, Gominet M, Okstad OA, Kolsto AB, Lereclus D. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol Microbiol 1999;32: 1043–1053. [DOI] [PubMed] [Google Scholar]
- 22.Brezillon C, Haustant M, Dupke S, Corre J-P, Lander A, Franz T, et al. Capsule, toxins, and atxA as virulence factors of emerging Bacillus cereus biovar anthracis. PloS Negl Trop Dis. 9:e0003455 10.1371/journal.pntd.0003455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinn CP, Semenova VA, Elie CM, Romero-Steiner S, Greene C, Li H, et al. Specific, sensitive, and quantitative enzyme-linked immunosorbent assay for human Immunoglobulin G antibodies to anthrax protective antigen. Emerg Infect Dis. 2002;8:1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.