Abstract
Background
Nasopharyngeal carcinoma (NPC) is a peculiar Epstein Barr virus (EBV)-associated malignancy that is prevalent in South-East Asia. Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (PIN1) isomerizes specific phosphorylated amino acid residues, which makes it an important regulator in cell survival and apoptosis. In this study, we investigated the contribution made by PIN1 in NPC tumorigenesis and PIN1’s potential role as a therapeutic target.
Methods
The expression of PIN1 was examined in a panel of NPC cell lines, xenografts and primary tumors. The functional roles of PIN1 in NPC cells were elucidated by the knockdown and overexpression of PIN1 in in vitro and in vivo nude mice models by siRNA and lenti-viral transfection, respectively. The antitumor effects of the PIN1 inhibitor Juglone in NPC cells were also evaluated.
Results
We revealed the consistent overexpression of PIN1 in almost all EBV-associated NPC cell lines, xenografts and primary tumors. PIN1 suppression was capable of inhibiting cyclin D1 expression and activating caspase-3 in NPC cells. It positively regulated NPC cell proliferation, colony formation and anchorage-independent growth. The inhibition of PIN1 suppressed tumor growth in vitro and in vivo.
Conclusions
This study demonstrates the oncogenic role of PIN1 in NPC tumorigenesis, and shows that its overexpression can enhance tumor cell growth via the upregulation of cyclinD1. Our findings inform the development of novel treatments targeting PIN1 for NPC patients.
Introduction
Nasopharyngeal carcinoma (NPC) is a distinctive type of head and neck carcinoma that arises from the epithelial cells covering the surface and lining of the nasopharynx. This malignancy exhibits a distinct ethnic and geographical distribution, and is particularly prevalent in Southern China, where almost all cases are nonkeratinizing carcinomas associated with EBV infection [1, 2]. This association with EBV, in addition to the existence of multiple genetic aberrations [3], increases the complexity of NPC research. The poor results achieved by conventional chemo-radiotherapy for patients with advanced loco-regional diseases and distant metastases represent a therapeutic challenge [4]. Given the high relapse and metastasis rates, it is of utmost importance that alternative therapeutic approaches be sought out and developed.
Cancer development involves a complex array of aberrant signaling pathways that fundamentally results in uncontrolled cell proliferation controlled by proline-directed phosphorylation [5]. PIN1 is a highly conserved enzyme that binds to and isomerizes specific phosphorylated serine or threonine residues preceding proline (Ser/Thr-Pro) bonds. It induces conformational changes in certain proteins that prompt changes to their properties, including catalytic activities, subcellular localization, protein–protein interactions and turnover rate [5, 6]. Thus, PIN1 is considered an important regulator in cellular processes, such as cell cycle regulation, cell signaling, transcription and splicing, DNA-damage responses, cell survival and drug resistance [5, 7, 8].
Apart from the importance of PIN1 in modulating the activation of various signaling molecules, it has also been shown to stabilize viral oncoproteins such as the human T-cell leukemia virus type I Tax protein [9]. Moreover, PIN1 has been reported to be involved in the functioning of several viruses, including Kaposi’s sarcoma-associated herpes virus [10], human immunodeficiency virus type I [11, 12] and the hepatitis C virus [13]. Interestingly, it has also been demonstrated to interact with EBV-encode protein [14], although information on its role in EBV-associated malignancy has proven scarce.
This study aimed to elucidate the role of PIN1 in the development of NPC which is consistently associated with EBV infection. By investigating the effects of PIN1 expression on EBV-associated NPC, we revealed its contribution to tumor cell growth and tumorigenesis.
Materials and Methods
Cell lines, xenografts and primary tumors
Three NPC cell lines C666-1 (a naturally EBV-associated, undifferentiated type of NPC) [15], HK1 (an EBV-negative, well-differentiated type of NPC) [16], HK1-EBV (HK1 infected with EBV) [17], immortalized normal nasopharynx epithelial cell lines (NP69 and NP460) [18] established in our laboratories were used in this study. The cervical cancer cell line HeLa was obtained from American Type Culture Collection (ATCC). Two EBV-positive NPC xenografts—xeno-666 [15], xeno-2117 [19], C15 and C17 [20] were also included. Xen-666 and xeno-2117 were established by our NPC group as described previously [15, 19]. C15 and C17 were obtained from Prof. Pierre Busson who established these xenografts [20]. For the immunohistochemical (IHC) staining study, 70 archival formalin-fixed paraffin embedded (FFPE) primary NPC biopsies were recruited from the tissue bank of Department of Anatomical and Cellular Pathology at the Prince of Wales Hospital, The Chinese University of Hong Kong (CUHK). The study protocol was approved by the Clinical Research Ethics Committee of the CUHK. All of the specimens were histologically evaluated as undifferentiated or poorly differentiated carcinomas and confirmed to be EBV-positive, as determined by EBER in situ hybridization [21]. The patients’ characteristics are presented in Table 1.
Table 1. Characteristics of NPC patient specimens recruited for IHC study.
| Variables | No. of patients |
|---|---|
| Age (years) | |
| ≤ 50 | 41 |
| > 50 | 29 |
| Gender | |
| Male | 57 |
| Female | 13 |
| Clinical stage | |
| Early (Stages 1 and 2) | 21 |
| Late (Stages 3 and 4) | 49 |
| RecurrenceRecurrence/distant metastasis | |
| Absent | 53 |
| Present | 17 |
Transfection and drug treatments
HK1, HK1-EBV, C666-1 and HeLa cells were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum and 1% L-glutamine (Life Technologies). The NP69 cells were cultured in keratinocyte serum-free medium (KSFM) with bovine pituitary extracts and recombinant epidermal growth factor (Invitrogen). All of the cells were incubated in a humidified atmosphere with 5% CO2 at 37°C.
Two Silencer Select validated siRNAs targeting PIN1 (siPin1 544 and siPin1 545) and universal negative control siRNA (Life Technologies) were transiently transfected into C666-1 using LipofectamineTM 2000 Transfection Reagent (Invitrogen) according to the manufacturer’s protocols. Cells were collected at indicated time points for further analysis. MG132 (Calbiochem) was applied to HK1 and NP69 cells (at 10 and 15 μM and 5 and 10 μM, respectively) to study the proteasome degradation of PIN1. The PIN1 inhibitor Juglone (Calbiochem) was used to study the effect of PIN1 inhibition on C666-1 cells (at 2, 4, 6 and 8 μM). Treated cells were collected and stored at -80°C until use. To determine the IC50 of Juglone, the cells were seeded into 96-well plates and treated with different Juglone dosages (0.03–20 μM) for 24 hours. Cell viability was then determined by a WST-1 assay.
Establishing stable PIN1-overexpressing cell lines
The pCDH and pCDH-PIN1 plasmids were a gift, generously provided by Dr. Roberta Pang, Department of Surgery, The University of Hong Kong. A lentiviral system was used to establish the stable PIN1-overexpressed NP69 cell line. The pCDH lentiviral vector was packed using the third-generation lentivirus system, according to the Lenti Starter kit manufacturer’s protocols (System Biosciences). Viral supernatant (culture media) was harvested from 293TN producer cells 48 hours after transfection. The virus (with a pCDH or pCDH-PIN1 vector and 10 μg pPACKH1-plasmid mix) was collected and transduced with TransDuxand into NP69 cells. PIN1 expression was confirmed by quantitative polymerase chain reaction (qPCR) and Western blotting. Green fluorescence protein (GFP) reporter expression was visualized under fluorescence microscope to ensure the presence of the transduced DNA.
Reverse transcription (RT) and quantitative polymerase chain reaction (qPCR)
RNA samples prepared by TRIZOL and phenol-chloroform extraction were subjected to reverse-transcription using MultiScribe Reverse Transcriptase (Invitrogen). The cDNA obtained was used for real-time qPCR via SYBR Green Master Mix (Applied Biosystems), according to the manufacturer’s protocols. The primers used in this study are listed in Table 2. The PCR reactions were performed using the 7500 Fast Real-Time PCR system (Applied Biosystems). Each sample was analyzed in triplicate. The expression of target genes was normalized against the housekeeping gene β-actin using the 2[-ΔΔCT] method.
Table 2. Quantitative PCR primer sequences.
| Gene | Forward primer (5’→ 3’) | Reverse primer (5’→ 3’) |
|---|---|---|
| PIN1 | GGAGGCCCTGGAGCTGAT | AACTGTGAGGCCAGAGACTCAAA |
| Actin | GTCTTCCCCTCCATCGTG | AGGGGTGAGGATGCCTCTCTT |
Western blotting
The protein samples were separated according to their sizes by electrophoresis, and then electro-transferred to a nitrocellulose membrane using a Bio-Rad Trans-Blot cell. The nitrocellulose membrane was incubated with primary antibodies overnight in 5% non-fat milk or 5% bovine serum albumin. The membrane was then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies. The target proteins were detected by chemiluminescent substrates (GE Life Science) and the emitted signal was detected on X-ray films (Kodak). The membranes were probed with antibodies against human PIN1 (Calbiochem), ACTIN (Santa Cruz), CYCLIN D1 (Lab Vision), β-catenin, c-JUN, c-JUN (Ser73) and c-JUN (Ser63) (Cell Signaling).
Immunohistochemical staining
The FFPE specimens were sectioned (4 μm), de-paraffinized and rehydrated for subsequent immunostaining. Following antigen retrieval, endogenous biotin activity was blocked by normal bovine serum and the sections were incubated in primary antibodies (anti-PIN1, Calbiochem) in a moist chamber. The HRP-labeled secondary antibody was applied to the sections and, finally, the 3,3’-diamino-benzidine (DAB) substrate was applied for color development. PIN1 expression and localization was visualized in red and nucleuses were counterstained with hematoxylin, which appeared blue. The slides were then dehydrated and mounted with Permount mounting medium (Fisher Scientific).
Caspase-3 activity assay
The cell apoptosis of treated and non-treated cells was detected using the CaspACEAssayTM System according to the manufacturer’s protocols (Promega). Wells in duplicates containing blank (no cell extract), negative control (extract from untreated cells), induced apoptosis (extracts from PIN1 inhibitor-treated cells) and cells treated with caspase inhibitor (extracts from PIN1 inhibitor- and Z-VAD-FMK the caspase inhibitor-treated cells) were included in the experiments. The experiments were performed in triplicate and the absorbance of the colors developed was measured at a wavelength of 405 nm by a Perkin Elmer 1420 Multilabel Counter Victor 3 (Perkin Elmer). The caspase-specific activity was calculated according to the manufacturer’s guidelines.
WST-1 and BrdU assays
The cell proliferation reagent WST-1 (Roche) was used to determine the rate of cell proliferation in treated- and non-treated cells. The treated cells were seeded into a 96-well plate at a density of 3000–8000 cells per well. Cell viability was estimated every 24 hours by WST-1, continuously, for 5 days. The absorbance in each well was measured at a wavelength of 450 nm and normalized using a wavelength of 690 nm, detected by the Victor 3 (Perkin Elmer). Cell proliferation was also measured in terms of DNA incorporation within proliferating cells using the BrdU assay (Cell Proliferation ELISA, BrdU colorimetric Kit, Roche).
Anchorage-independent growth assay
The treated and non-treated cells were plated in soft agar (base agarose: 1.8% agarose in KSFM; top agarose: 0.9% agarose pre-mixed with cells; 2 mL overlaying KSFM medium) to evaluate their growth in an anchorage-independent manner. The plates were incubated at 5% CO2 at 37°C for a month. The colonies were visualized using 0.1% p-iodonitro tetrazolium violet (INT) stain (Sigma-Aldrich), and then counted.
Colony formation assay
The treated and non-treated cells were seeded at 8X103 cells per well (C666-1) or 1X103 cells per well (NP69) into 6-well plates. The cells were kept at 37°C in an incubator with 5% CO2 for 21–28 days. The colonies formed were fixed with methanol and visualized in blue using Giemsa (Sigma-Aldrich) staining. The colonies containing over 50 cells were counted.
In vivo tumorigenicity
To elucidate the in vivo tumorigenic effect of PIN1, NP69 cells (treated with siRNAs of PIN1 or with vector control) were subcutaneously inoculated into the flanks of female BALB/c nude mice (nu/nu) (4 mice per group). All mice were anesthesized by 2,2,2-tribromoethanol (Avertin) prior to inoculation. Matrigel (BD Bioscience) was used in the inoculation process for NP69. The mice were inspected daily for tumor formation and the sizes of the tumors formed were recorded. To determine the anti-tumor effect of PIN1 inhibitor in vivo, Juglone (0 mg/kg, 0.5 mg/kg, 1.5 mg/kg) was injected intra-peritoneally into nude mice implanted with C666-1 luciferase cells when the tumors had reached their appropriate sizes. Tumor size was recorded daily and the luciferase salt was injected into the nude mice on days 0, 6 and 8 to monitor the changes in tumor size via an IVISTM 100 Imaging system. All of the mice were sacrificed at the end of the experiments by cervical dislocation and the tumors were preserved for further analysis. No mice were ill, died or required early termination during this study. Ethical approval was obtained from the University Animal Experimentation Ethics Committee (AEEC), CUHK, and the animal license was approved by the Hong Kong Government, Department of Health.
Luciferase reporter assay
The C666-1 cells at 60% confluence in the 96-well plate were co-transfected with FR-TK-Luc plasmid, PIN1 siRNA and reporter plasmid. After 48 hours of transfection, the cells were lysed with a 1X passive lysis buffer (Promega). The lysates were then transferred to a 96-well plate and luciferase activities were assayed using the Dual Luciferase Reporter Kit (Promega) according to the manufacturer’s protocols. The luciferase signal was measured by Victor 3 (Perkin Elmer) with or without automatic injections. The results were expressed as the ratio of Firefly luciferase activity to Renilla luciferase activity.
Signaling pathway array
A Cancer 10-Pathway Reporter Luciferase Kit (SA Biosciences) was used to determine signaling pathway aberrations in the treated cells. In brief, reporter plasmids on the Cignal Finder Array Plate were resuspended and mixed with 0.8 μL Fugene HD reagent (Roche) in 100 μL OPTI-MEM medium (Invitrogen). The transfection complex was allowed to form at room temperature for 20 minutes. The treated cells were then collected and reverse-transfected with reporter plasmid at a density of 1.5X104 cells per well. The dual-luciferase reporter assay was conducted after 24 hours of incubation.
Statistical analysis
All of the experiments were conducted in triplicate and the results were presented as mean with standard error mean (SEM). Student t-test was used to determine statistical significance, with a P-value of less than 0.05 considered significant.
Results
Overexpression of PIN1 in NPC primary tumors and tumor lines
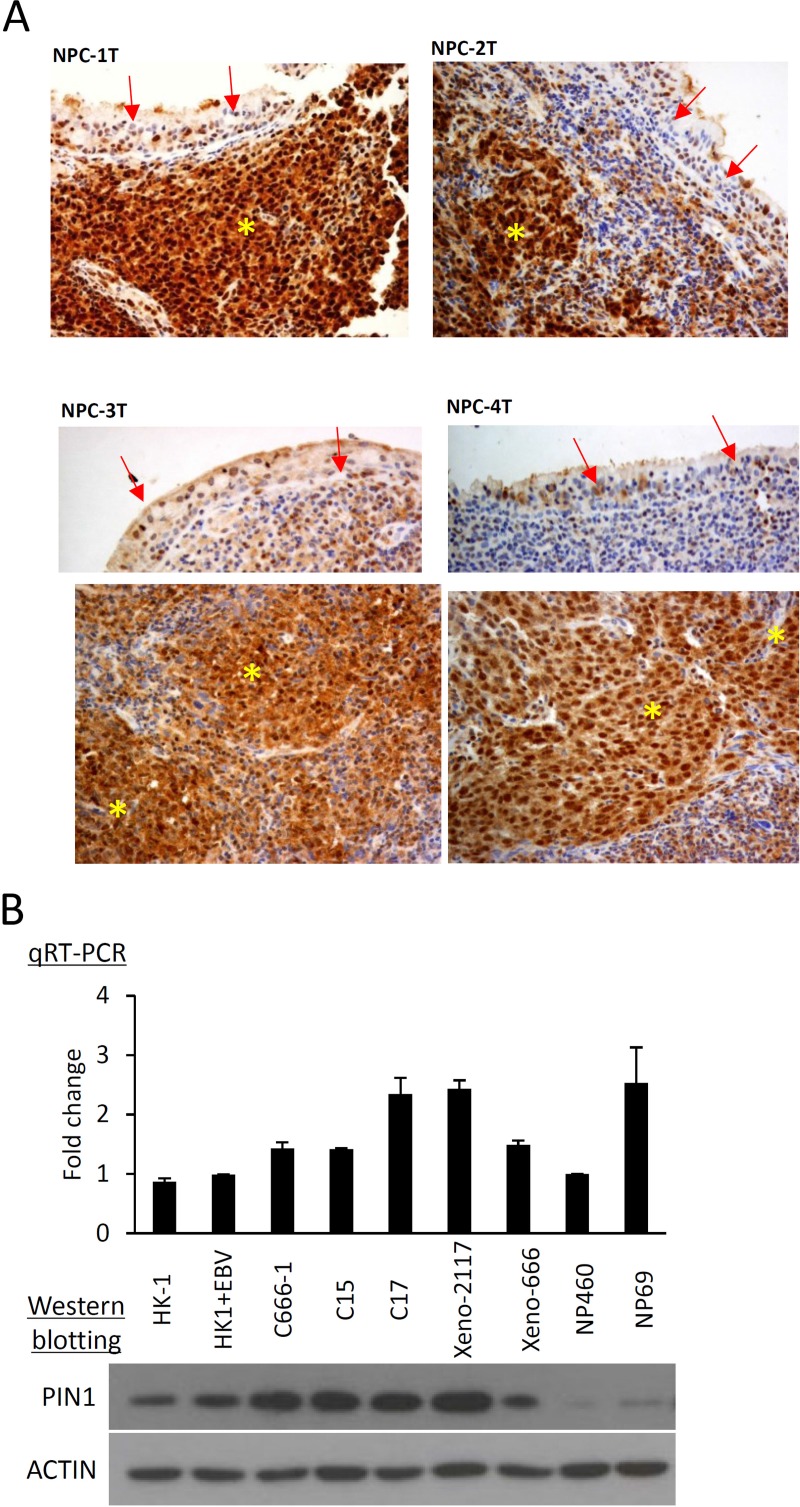
Using immunohistochemical staining, we detected the overexpression of PIN1 in 70/70 (100%) NPC primary tumors, compared with the adjacent normal nasopharyngeal epithelium (Fig 1A). PIN1 overexpression was also demonstrated in a panel of EBV-positive NPC cell line (C666-1) and xenografts (C15, C17, xeno-2117 and xeno-666) by Western blotting, whereas only weak PIN expression was detected in the immortalized normal nasopharyngeal epithelial cell lines (NP460 and NP69) (Fig 1B). Despite the dramatic reduction in PIN1 protein expression in the immortalized normal NPC cells, no obvious differences in PIN1 mRNA transcriptions were observed between the normal NP cell lines and the NPC tumors. This finding suggests a post-translational regulation of PIN1 in normal nasopharyngeal cells.
Fig 1. Overexpression of PIN1 in EBV-associated NPC.
(A) IHC staining was used to illustrate the overexpression of PIN1 in representative NPC primary tumors (NPC1-NPC4). All of the cases were EBV-positive undifferentiated carcinomas. NPC1, NPC3 and NPC4 are from the patients with stage 3 disease. NPC2 is from a patient with stage 2 NPC. Strong PIN signals were found in the tumor cells, which are indicated by yellow “*”. The adjacent normal epithelium served as the control in which weak PIN1 signals were observed. The red arrows indicate normal nasopharyngeal epithelium. (B) Expression of PIN1 in NPC tumor lines, xenografts and immortalized normal NP cells, detected by qRT-PCR (upper panel) and Western blot (lower panel). For qRT-PCR, the PIN1 transcription in NP460 was used as a reference. The relative expression of PIN1 transcripts was indicated as a fold difference over the reference. β-actin was used for loading normalization. Similarly, ACTIN was used as an internal loading control in the Western blot analysis. The weak PIN1 expression in NP69 and NP460 suggested that the downregulation of PIN1 involved post-translational regulation.
To determine the mechanism of post-translational regulation on PIN1 proteins, HK-1 and NP69 were treated with the proteasome inhibitor MG132 (Carbobenzoxy-Leu-Leu-leucinal). The Western blot results revealed increased PIN1 protein expression in both HK1 and NP69 cells after MG132 treatment (S1 Fig). This indicates that PIN1 protein expression is regulated by proteasome degradation.
The consistent overexpression of PIN1 suggests its potential oncogenic role in NPC development. To investigate its oncogenic function, siRNAs and the PIN1 inhibitor Juglone were used to determine the effect of PIN1 suppression on NPC cell growth. Subsequently, the effects of PIN1 expression on cell growth and tumorigenesis were examined in normal nasopharyngeal epithelial cells (NP69) transfected with a PIN1 expressing vector.
PIN1 regulates NPC cell growth and cyclin D1 expression
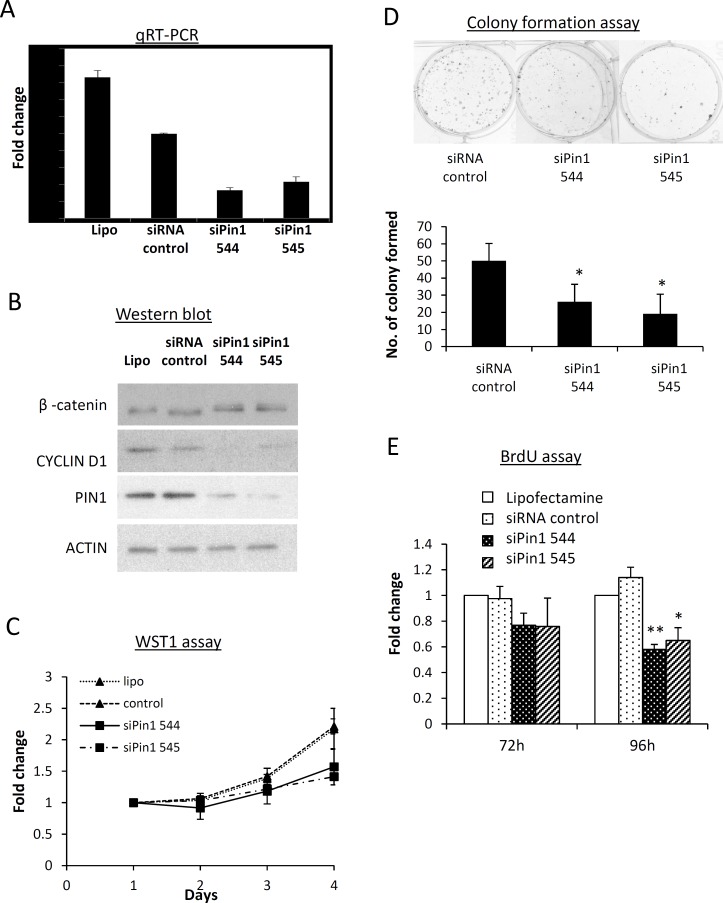
The effect of PIN1 knockdown on NPC cell growth was investigated by transfecting C666-1 cells with PIN1-specific siRNAs (siPin1 544 and siPin1 545). Fig 2A and 2B show the marked downregulation of PIN1 mRNA and protein in the C666-1cells transfected with siPIN1. The results of the WST-1 assay demonstrated observable growth inhibition in the siPIN1-transfected C666-1 cells, compared with the control (Fig 2C). Furthermore, the colony formation ability was significantly reduced in the siPIN1-transfected C666-1 (Fig 2D). These findings indicate that PIN1 expression regulates NPC cell growth. The results of the BrdU assay revealed significant suppression of DNA synthesis in the PIN1 knockdown NPC cells (Fig 2E). Importantly, the expression of cell cycle regulator cyclin D1 was repressed in the PIN1 knockdown C666-1 cells (Fig 2B). The co-transfection of pCMV-cyclinD1, a cyclin D1-expressing vector with siPIN1, restored the cyclin D1 expression, cell proliferation, DNA synthesis and colony formation ability in C666-1 cells compared with those of siRNA-treated cells (S2 Fig). This suggests that PIN1 regulates NPC cell growth by modulating cyclin D1 expression.
Fig 2.
PIN1 suppression inhibits cell growth, DNA synthesis, colony formation ability and cyclin D1 expression in NPC cells (A) qRT-PCR and (B) Western blot were used in the downregulation of PIN1 transcription and protein expression in PIN1 siRNAs (siPIN1 544 and siPIN1 545)-treated NPC cells and C666-1, respectively. In these PIN1 knockdown cells, reduced cyclin D1 expression was observed. ACTIN was used as the loading control in the Western blot analysis. (C) WST-1 assay revealed growth inhibition in the NPC cells transfected with siPin1 544 and siPin1 545. (D) Colony formation ability was significantly suppressed in PIN1-silenced C666-1 cells. Representative photos of colonies formed by siRNA-treated and control cells are shown. Statistical significance was determined by Student t-test, where a P-value of less than 0.05 was considered significant (*P < 0.05, **P < 0.01). (E) A BrdU assay was used to reveal the significant inhibition of DNA synthesis in the PIN knockdown NPC cells.
PIN1 inhibitor induces caspase-3 and reduces tumor size
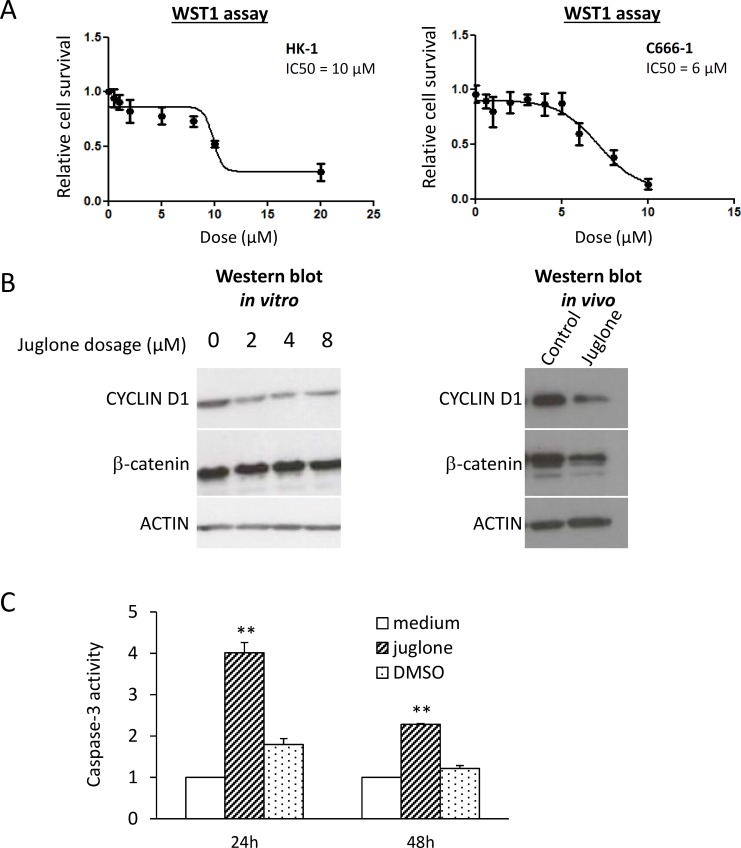
A PIN1 inhibitor, Juglone, was used to elucidate the effects of PIN inhibition in NPC cells in vitro and in vivo. A WST-1 assay showed that the half maximal inhibitory concentration (IC50) of Juglone for C666-1 and HK1 was 6 and 10 μM, respectively (Fig 3A). This finding indicates that C666-1 cells with high PIN1 expression are more susceptible to Juglone treatment. The PIN1 inhibitor Juglone suppressed the cyclin D1 expression in C666-1 cells (Fig 3B, left panel). The in vivo effect of the PIN1 inhibitor on cyclin D1 repression was further confirmed by the intra-tumor injection of Juglone into C666-1 xenografts in nude mice models (Fig 3B, right panel). Juglone treatment also significantly enhanced caspase-3 activity in C666-1 cells, compared with the controls (Fig 3C). The results suggest that Juglone inhibits cell growth and induces apoptosis in NPC cells.
Fig 3. PIN1 inhibitor Juglone suppresses tumor cell growth and induces caspase-3 activity.
(A) HK-1 (left panel) and C666-1 (right panel) were treated with the PIN1 inhibitor Juglone for 24 hours to determine the drug dose sensitivity. The IC50 values of the C666-1 and HK1 cells were 6 μM and 10 μM, respectively. (B) The expression of cyclin D1 was suppressed by Juglone in a dose-dependent manner, but no significant changes in β-catenin expression were detected. The expression of cyclin D1 and β-catenin in the Juglone-treated C-666 xenograft model was examined by Western blot. Reduced expression of cyclin D1 was observed in the tumor that had received Juglone treatment. PIN1 inhibition resulted in suppression of cyclin D1 both in vitro and in vivo, and there was a modest reduction in the β-catenin level in the in vivo model. (C) The C666-1 cells exhibited significantly higher caspase-3 activity after Juglone treatment, compared with their untreated or DMSO-treated counterparts. This indicates that PIN1 inhibition can activate the caspase apoptotic pathway in NPC cells. Statistical significance was determined by Student t-test, P-value of less than 0.05 was considered significant (*P < 0.05, **P < 0.01).
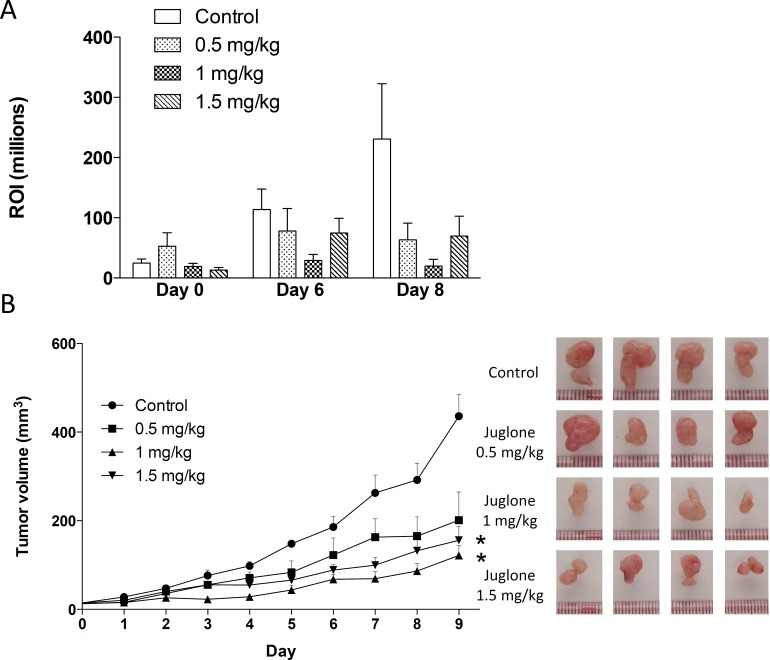
Fig 4A and 4B demonstrate the in vivo antitumor effect of Juglone in NPC cells. The ROI count was highly reduced in the mice treated with Juglone, compared with that of the controls (Fig 4A). As Fig 4B shows, the tumor sizes in the mice treated with Juglone were significantly reduced, compared with the control (Fig 4B). A significant inhibition of tumor growth was observed in mice treated with 1 mg/kg and 1.5 mg/kg Juglone.
Fig 4. PIN1 inhibitor Juglone suppresses tumor growth in vivo.
(A) Using in vivo imaging, the tumor growth of luciferase-tagged C666-1 was revealed after treatment with different dosages of Juglone (ROI counts per million photons per second). The luciferase signal was increased in the control untreated tumors during the Juglone treatment. In the mice treated with Juglone in different doses (0.5, 1 and 1.5 mg/kg), consistently lower luciferase signals were observed during the 8-day treatment periods. (B) Significant inhibition of tumor growth was shown in the mice treated with 0.5, 1 and 1.5 mg/kg Juglone. Four nude mice were used in each study group. Statistical significance was determined by Student t-test, where a P-value of less than 0.05 was considered significant (*P < 0.05, **P < 0.01).
PIN1 induces anchorage-independent growth of nasopharyngeal epithelial cells
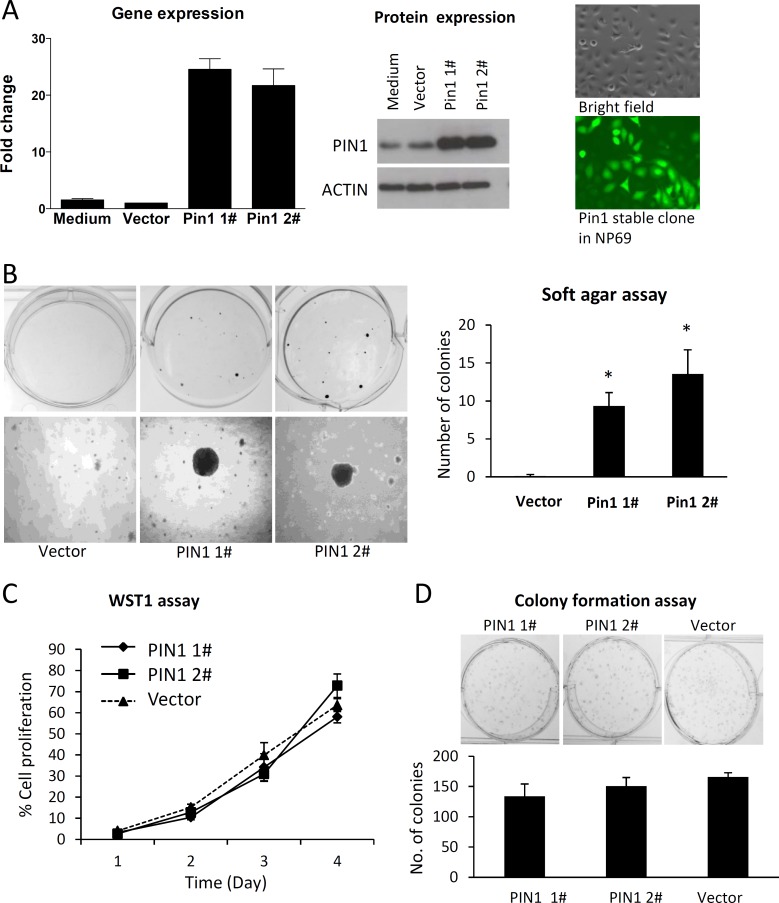
The tumorigenic effect of PIN1 overexpression was further studied in an immortalized nasopharyngeal epithelial cell line (NP69) transfected with two PIN1 mimics (pCDH-CMV-MCS-EF1-copGFP-PIN1/“Pin1 1#” and “Pin1 2#”). The overexpression of PIN1 transcripts and proteins was observed in the PIN1-transfected NP69 cells (Fig 5A). Fig 5A shows the high transfection efficiency of PIN1-transfected cells expressing GFP.
Fig 5. PIN1 overexpression enhances anchorage-independent growth in nasopharyngeal epithelial cells.
(A) The overexpression of PIN1 was detected in NP69 cells transfected with Pin1-expressing lenti-viral vectors (Pin1 1# and Pin1 2#) by qPCR (left panel) and Western blot (middle panel). Representative photos of PIN1-transfected cells by bright field (right upper panel) and fluorescence field (right lower panel) are shown. The transfection efficiency was monitored by GFP expression. (B) Enhanced anchorage-independent growth on soft agar in NP69 cells with PIN1-overexpression, compared with control cells (left upper panel, colonies in 6-well plates; left lower panel, colonies under microscope; right panel, average data in histogram). (C-D) No significant effect of PIN1 overexpression on cell growth and colony formation ability was detected in the immortalized nasopharyngeal epithelial cells NP69, by WST-1 and colony formation assay, respectively. Statistical significance was determined by Student t-test, where a P-value of less than 0.05 was considered significant (*P < 0.05).
The overexpression of PIN1 in N69 cells significantly induced anchorage-independent cell growth in the soft agar assays (Fig 5B). The number of colonies formed in soft agar by PIN1-expressing NP69 cells was significantly higher than those of vector control. However, PIN1 overexpression did not induce cell proliferation and colony formation ability in NP69 cells (Fig 5C and 5D).
PIN1 overexpression activates MAPK/JNK pathway
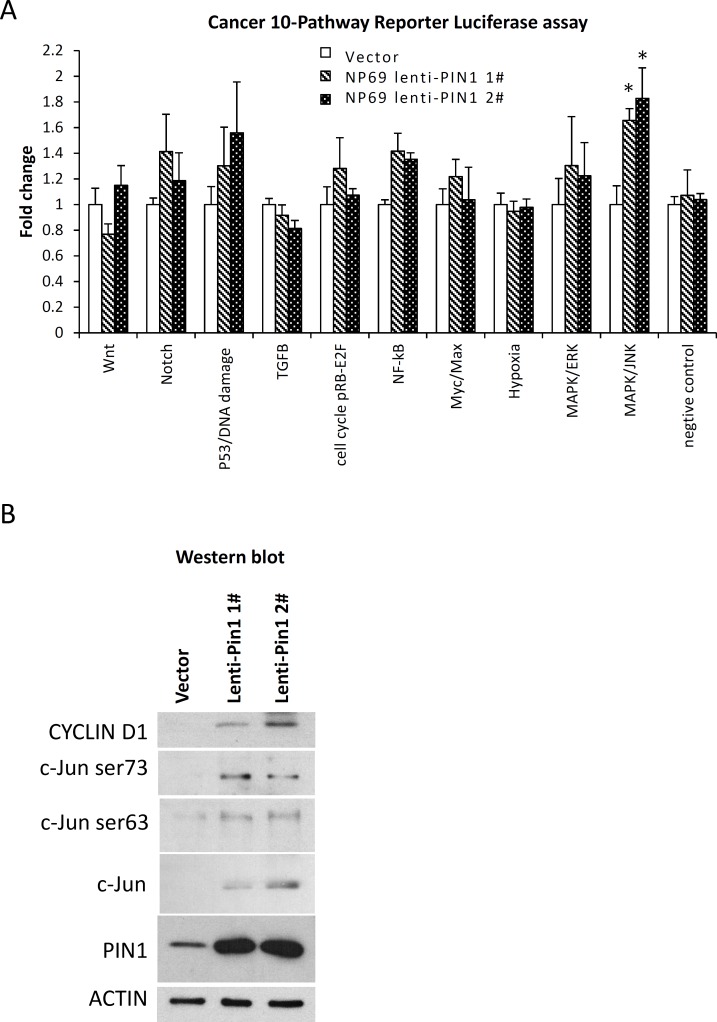
A reporter luciferase assay revealed that the MAPK/JNK signaling pathway was significantly activated in NP69 cells stably expressing PIN1, compared with those transfected with a vector (Fig 6A). The activation of the NOTCH, p53 and NF-κB pathways was also observed in PIN1-expressed NP69 cells, although it did not reach statistical significance. Aside from cyclin D1, Western blotting confirmed the upregulation of both c-Jun and phosphoryl-c-Jun (Ser63 and Ser73) in NP69 cells stably transfected with PIN1 (Fig 6B). This finding suggests that PIN1 induces the growth of nasopharyngeal epithelial cells by activating the MAPK/JNK pathway.
Fig 6. Overexpression of PIN1 activates the MAPK/JNK pathway.
(A) Using the Cancer 10-Pathway Reporter Luciferase assay, significant activation of the MAPK/JNK signaling pathway was observed in two stably PIN1-overxpressing NP69 cell lines (NP69lenti-PIN1 1# and NP69lenti-PIN1 2#). For these PIN1-expressing cells, the activity was also moderately activated in the NOTCH, P53/DNA damage and NF-κB pathways. (B) Western blot analysis demonstrated the upregulation of c-Jun, phosphorylated c-Jun (Ser63 and Ser73) and cyclin D1 in the stable PIN1-transfected NP69 cells. Statistical significance was determined by Student t-test, where a P-value of less than 0.05 was considered significant (*P < 0.05).
Discussion
In a recent study, Lu et al. demonstrated the association of PIN1 promoter polymorphisms and risk of NPC [22]. In the current study, we provide the functional evidence on the importance of PIN1 overexpression in NPC tumorigenesis. We show that PIN1 suppression inhibits in vitro and in vivo tumor growth in NPC cells while its overexpression induces the anchorage-independent growth of nasopharyngeal epithelial cells. The findings suggest that PIN1 overexpression contributes to NPC tumorigenesis, and that its inhibition may be a potential therapeutic strategy for EBV-associated NPC.
PIN1 is an important enzyme that binds phosphorylated Ser/Thr-Pro motifs and catalyzes the cis/trans-isomerization of proline-containing peptides [5]. The overexpression of PIN has been reported in various human tumor types, including breast [23], esophageal [24], prostate [25] and colorectal cancer [26, 27]. In this study, constitutive PIN1 overexpression was found in EBV-associated NPC tumor cells, but PIN1 protein was scarcely detected in EBV-negative nasopharyngeal epithelial cells. Since PIN1 transcription is similar in both NPC and normal NP cells, the PIN overexpression in the tumor cells is regulated by post-transcriptional mechanisms. A recent study reported that PIN1 expression is regulated by the tumor suppressive miRNA miR-296-5p in prostate cancer [28]. However, aberrant mir-296-5p expression is rarely detected in NPC [29]. The restoration of PIN expression in the MG132-treated NP cells suggests that the process of proteasomal degradation is an important mechanism in regulating PIN1 function. Basu et al. showed that the proteasomal degradation of PIN1 was crucial in its interaction with BCL2 and tumor cell survival [30]. Together with the current findings, the lack of or inadequate proteasomal degradation may be a potential mechanism in explaining PIN1 accumulation in EBV-associated NPC, which subsequently aids tumor cell survival. In our recent study, we revealed the interaction of EBV latent protein EBNA1 with PIN1 in NPC cells (unpublished data). Nevertheless, the binding did not inhibit the proteasomal degradation PIN1. The mechanisms for PIN1 overexpression in NPC cells needs to further elucidate in future studies.
PIN1 is crucial in tumor cell transformation, as it activates oncogenic pathways and growth enhancers [31] while inactivating tumor suppressors and growth inhibitors [32, 33]. Among the PIN1-activated oncogenes, Cyclin D1 is of particular importance in NPC tumorigenesis. The overexpression of cyclin D1 has been detected in more than 90% of primary NPC tumors. In NPC cells, cyclin D1 not only plays an essential role in cell proliferation but its overexpression can stabilize EBV infection [34]. PIN1 is thought to play an important role in NPC pathogenesis by regulating cyclin D1 expression. Although there is no significant effect of PIN1 overexpression on cell proliferation and colony formation in normal epithelial cells (Fig 5C), cyclin D1 is up-regulated by PIN1 expression. PIN1 overexpression may promote EBV infection in nasopharyngeal epithelial cells via cyclin D1 upregulation as we reported previously [34]. Further studies are needed to determine the role of PIN1 in the enhancement of EBV infection and the maintenance of the stable latent EBV genome in nasopharyngeal epithelial cells—the critical steps in NPC tumorigenesis.
PIN1 has been shown to regulate cyclin D1 via the JNK, WNT or NF-κB pathways. As Fig 6A and 6B show, PIN1 significantly activated the MAPK/JNK pathway and induced cyclin D1 expression in NP69 cells. It is probable that PIN1 modulates cyclin D1 expression by activating the JNK pathway. Cyclin D1 expression can be induced by the interaction between PIN1 and the p-Ser63/73-Pro motifs in Jun, which positively regulates Jun transcriptional activity on its target genes. Our findings concur with those of studies on human breast cancer in which PIN1 overexpression resulted in increased JNK activity [35]. In addition to JNK, PIN1 can also modulate cyclin D1 expression via the β-catenin of the WNT pathway [36]. PIN1 overexpression has also been shown to up-regulate cyclin D1 and β-catenin in hepatocellular carcinomas (HCC) [37]. However, PIN1 expression did not significantly up-regulate the β-catenin expression in NPC cells, and thus the WNT pathway might not be involved in PIN1-modulated cyclin D1 expression. It is well established that the NF-κB and NOTCH pathways are critically activated pathways in NPC [38–40]. PIN1 has been reported to target the pThr254-Pro motif in NFκB1 and p65 [41], which inhibits p65 from binding to its inhibitor IκB, resulting in the protein nuclear accumulation and stability of these proteins. Our study confirms that PIN1 overexpression can up-regulate the NF-κB, NOTCH and p53 pathways. Further studies are required to fully elucidate the role of PIN1 in regulating these cancer-related signaling pathways in NPC. The current study also reveals the role of PIN1 in inducing anchorage-independent cell growth and cyclin D1 expression in normal nasopharyngeal epithelial cells (Fig 5B), suggesting that PIN1 is likely to play an important role in NPC tumorigenesis. The implication of PIN1 in carcinogenesis has also been demonstrated in HCC [37].
Regarding the tumorigenic properties of PIN1, we evaluated the effects of the PIN1 inhibitor Juglone on NPC tumor cell growth, apoptosis and in vivo tumor development. Juglone is a natural component of the Juglans mandshurica Maxim that was previously proven to be a potent cytotoxic agent in human tumor cell lines by exerting multiple anti-tumor activities such as cell apoptosis [42–44]. Apoptosis is a well-controlled type of cell death, and its induction has proven a useful approach in cancer therapies [45–47]. Our study confirmed that Juglone significantly induced caspase-3 activity and inhibited tumor growth in vivo. Concordant with our PIN1 siRNA study, the depletion of PIN1 suppresses tumor cell proliferation, DNA synthesis and colony formation in NPC cells. Previous studies have shown that PIN1 inhibition can suppress the Neu- and Ras-induced transformed phenotypes, in addition to inducing mitotic arrest and apoptosis in breast cancer cells. Moreover, PIN1 knockout mice have demonstrated aberrant cell proliferation that resulted in various abnormalities such as retinal degeneration, neurological abnormality and testicular atrophy [48]. The increase in caspase-3 activity via Juglone suggested that it might induce tumor cell apoptosis and inhibit tumor formation in the nude mice model. Our findings indicate that Juglone might serve as a potential anti-tumor drug for NPC patients.
Conclusions
Our study provides evidence supporting the oncogenic role of PIN1 in NPC tumorigenesis. The overexpression of PIN1 might enhance tumor cell growth via the upregulation of cyclinD1. Thus, PIN1 inhibition serves as a potential therapeutic approach for NPC patients.
Supporting Information
Using Western blot, elevated PIN1 proteins were observed in the nasopharyngeal epithelial cells, NP69 and HK-1, after treatment with proteasome inhibitor MG132 (0–15 μM). ACTIN was used for loading normalization.
(TIF)
(A) The expression of PIN1 and cyclin D1 in NPC C666-1 cells transfected with PIN1 siRNAs and cyclin D1-expressing vectors was observed via Western blot. Using (B) WST-1, (C) BrdU and (D) colony formation assays, the expression of cyclin D1 was shown to restore the cell growth and DNA synthesis in the PIN1 knockdown NPC cells.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by grant from the Chinese University of Hong Kong (Focus Investigation Scheme-A), and the Hong Kong Research Grants Council – GRF (470413, 470312, 471211), CRF (CUHK8/CRF/11R), AoE NPC (AoE/M-06/08), Theme-Based Research Scheme (T12-403/11 and T12-401/13-R).
References
- 1.Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer cell. 2004;5(5):423–8. Epub 2004/05/18. . [DOI] [PubMed] [Google Scholar]
- 2.Pathmanathan R, Prasad U, Chandrika G, Sadler R, Flynn K, Raab-Traub N. Undifferentiated, nonkeratinizing, and squamous cell carcinoma of the nasopharynx. Variants of Epstein-Barr virus-infected neoplasia. The American journal of pathology. 1995;146(6):1355–67. Epub 1995/06/01. [PMC free article] [PubMed] [Google Scholar]
- 3.Lo KW, Huang DP. Genetic and epigenetic changes in nasopharyngeal carcinoma. Seminars in cancer biology. 2002;12(6):451–62. Epub 2002/11/27. . [DOI] [PubMed] [Google Scholar]
- 4.Chan ATC. Current treatment of nasopharyngeal carcinoma. European Journal of Cancer. 2011;47:S302–S3. 10.1016/s0959-8049(11)70179-4 [DOI] [PubMed] [Google Scholar]
- 5.Lu KP. Prolyl isomerase Pin1 as a molecular target for cancer diagnostics and therapeutics. Cancer cell. 2003;4(3):175–80. Epub 2003/10/03. . [DOI] [PubMed] [Google Scholar]
- 6.Liou YC, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends in biochemical sciences. 2011;36(10):501–14. Epub 2011/08/20. 10.1016/j.tibs.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nature reviews Molecular cell biology. 2007;8(11):904–16. Epub 2007/09/20. 10.1038/nrm2261 . [DOI] [PubMed] [Google Scholar]
- 8.Khanal P, Yun HJ, Lim SC, Ahn SG, Yoon HE, Kang KW, et al. Proyl isomerase Pin1 facilitates ubiquitin-mediated degradation of cyclin-dependent kinase 10 to induce tamoxifen resistance in breast cancer cells. Oncogene. 2012;31(34):3845–56. Epub 2011/12/14. 10.1038/onc.2011.548 . [DOI] [PubMed] [Google Scholar]
- 9.Jeong SJ, Ryo A, Yamamoto N. The prolyl isomerase Pin1 stabilizes the human T-cell leukemia virus type 1 (HTLV-1) Tax oncoprotein and promotes malignant transformation. Biochemical and biophysical research communications. 2009;381(2):294–9. Epub 2009/04/03. 10.1016/j.bbrc.2009.02.024 . [DOI] [PubMed] [Google Scholar]
- 10.Guito J, Gavina A, Palmeri D, Lukac DM. The cellular peptidyl-prolyl cis/trans isomerase Pin1 regulates reactivation of Kaposi's sarcoma-associated herpesvirus from latency. Journal of virology. 2013. Epub 2013/11/01. 10.1128/JVI.02877-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misumi S, Inoue M, Dochi T, Kishimoto N, Hasegawa N, Takamune N, et al. Uncoating of human immunodeficiency virus type 1 requires prolyl isomerase Pin1. The Journal of biological chemistry. 2010;285(33):25185–95. Epub 2010/06/10. 10.1074/jbc.M110.114256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watashi K, Khan M, Yedavalli VR, Yeung ML, Strebel K, Jeang KT. Human immunodeficiency virus type 1 replication and regulation of APOBEC3G by peptidyl prolyl isomerase Pin1. Journal of virology. 2008;82(20):9928–36. Epub 2008/08/08. 10.1128/JVI.01017-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim YS, Tran HT, Park SJ, Yim SA, Hwang SB. Peptidyl-prolyl isomerase Pin1 is a cellular factor required for hepatitis C virus propagation. Journal of virology. 2011;85(17):8777–88. Epub 2011/06/18. 10.1128/JVI.02533-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narita Y, Murata T, Ryo A, Kawashima D, Sugimoto A, Kanda T, et al. Pin1 interacts with the Epstein-Barr virus DNA polymerase catalytic subunit and regulates viral DNA replication. Journal of virology. 2013;87(4):2120–7. Epub 2012/12/12. 10.1128/JVI.02634-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung ST, Huang DP, Hui AB, Lo KW, Ko CW, Tsang YS, et al. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. International journal of cancer Journal international du cancer. 1999;83(1):121–6. Epub 1999/08/17. . [DOI] [PubMed] [Google Scholar]
- 16.Huang DP, Ho JH, Poon YF, Chew EC, Saw D, Lui M, et al. Establishment of a cell line (NPC/HK1) from a differentiated squamous carcinoma of the nasopharynx. International journal of cancer Journal international du cancer. 1980;26(2):127–32. Epub 1980/08/01. . [DOI] [PubMed] [Google Scholar]
- 17.Lo AK, Lo KW, Tsao SW, Wong HL, Hui JW, To KF, et al. Epstein-Barr virus infection alters cellular signal cascades in human nasopharyngeal epithelial cells. Neoplasia. 2006;8(3):173–80. Epub 2006/04/14. 10.1593/neo.05625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsao SW, Wang X, Liu Y, Cheung YC, Feng H, Zheng Z, et al. Establishment of two immortalized nasopharyngeal epithelial cell lines using SV40 large T and HPV16E6/E7 viral oncogenes. Biochimica et biophysica acta. 2002;1590(1–3):150–8. Epub 2002/06/14. . [DOI] [PubMed] [Google Scholar]
- 19.Huang DP, Ho JH, Chan WK, Lau WH, Lui M. Cytogenetics of undifferentiated nasopharyngeal carcinoma xenografts from southern Chinese. International journal of cancer Journal international du cancer. 1989;43(5):936–9. Epub 1989/05/15. . [DOI] [PubMed] [Google Scholar]
- 20.Busson P, Ganem G, Flores P, Mugneret F, Clausse B, Caillou B, et al. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinomas. International journal of cancer Journal international du cancer. 1988;42(4):599–606. Epub 1988/10/15. . [DOI] [PubMed] [Google Scholar]
- 21.Pak MW, To KF, Lo YM, Chan LY, Tong JH, Lo KW, et al. Nasopharyngeal carcinoma in situ (NPCIS)—pathologic and clinical perspectives. Head & neck. 2002;24(11):989–95. Epub 2002/11/01. 10.1002/hed.10161 . [DOI] [PubMed] [Google Scholar]
- 22.Lu Y, Huang GL, Pu XX, He YX, Li BB, Liu XY, et al. Association between PIN1 promoter polymorphisms and risk of nasopharyngeal carcinoma. Mol Biol Rep. 2013;40(5):3777–82. 10.1007/s11033-012-2454-6 . [DOI] [PubMed] [Google Scholar]
- 23.Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, Wang DG. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. The American journal of pathology. 2004;164(5):1727–37. Epub 2004/04/28. 10.1016/S0002-9440(10)63731-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin H, Jiang J, Sun L, Zheng F, Wu C, Peng L, et al. The prolyl isomerase Pin1 is overexpressed in human esophageal cancer. Oncology letters. 2011;2(6):1191–6. Epub 2012/08/01. 10.3892/ol.2011.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayala G, Wang D, Wulf G, Frolov A, Li R, Sowadski J, et al. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer research. 2003;63(19):6244–51. Epub 2003/10/16. . [PubMed] [Google Scholar]
- 26.Kuramochi J, Arai T, Ikeda S, Kumagai J, Uetake H, Sugihara K. High Pin1 expression is associated with tumor progression in colorectal cancer. Journal of surgical oncology. 2006;94(2):155–60. Epub 2006/07/19. 10.1002/jso.20510 . [DOI] [PubMed] [Google Scholar]
- 27.Kim CJ, Cho YG, Park YG, Nam SW, Kim SY, Lee SH, et al. Pin1 overexpression in colorectal cancer and its correlation with aberrant beta-catenin expression. World journal of gastroenterology: WJG. 2005;11(32):5006–9. Epub 2005/08/27. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee KH, Lin FC, Hsu TI, Lin JT, Guo JH, Tsai CH, et al. MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in prostate cancer by directly targeting Pin1. Biochimica et biophysica acta. 2014;1843(9):2055–66. 10.1016/j.bbamcr.2014.06.001 . [DOI] [PubMed] [Google Scholar]
- 29.Spence T, Bruce J, Yip KW, Liu FF. MicroRNAs in nasopharyngeal carcinoma. Chin Clin Oncol. 2016;5(2):17 10.21037/cco.2016.03.09 . [DOI] [PubMed] [Google Scholar]
- 30.Basu A, Das M, Qanungo S, Fan XJ, DuBois G, Haldar S. Proteasomal degradation of human peptidyl prolyl isomerase pin1-pointing phospho Bcl2 toward dephosphorylation. Neoplasia. 2002;4(3):218–27. Epub 2002/05/04. 10.1038/sj/neo/7900233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajbhandari P, Finn G, Solodin NM, Singarapu KK, Sahu SC, Markley JL, et al. Regulation of estrogen receptor alpha N-terminus conformation and function by peptidyl prolyl isomerase Pin1. Molecular and cellular biology. 2012;32(2):445–57. Epub 2011/11/09. 10.1128/MCB.06073-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicole Tsang YH, Wu XW, Lim JS, Wee Ong C, Salto-Tellez M, Ito K, et al. Prolyl isomerase Pin1 downregulates tumor suppressor RUNX3 in breast cancer. Oncogene. 2013;32(12):1488–96. Epub 2012/05/15. 10.1038/onc.2012.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min SH, Lau AW, Lee TH, Inuzuka H, Wei S, Huang P, et al. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Molecular cell. 2012;46(6):771–83. Epub 2012/05/23. 10.1016/j.molcel.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang CM, Yip YL, Lo KW, Deng W, To KF, Hau PM, et al. Cyclin D1 overexpression supports stable EBV infection in nasopharyngeal epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(50):E3473–82. Epub 2012/11/20. 10.1073/pnas.1202637109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. The EMBO journal. 2001;20(13):3459–72. Epub 2001/07/04. 10.1093/emboj/20.13.3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou CX, Gao Y. Aberrant expression of beta-catenin, Pin1 and cylin D1 in salivary adenoid cystic carcinoma: relation to tumor proliferation and metastasis. Oncology reports. 2006;16(3):505–11. Epub 2006/07/26. . [PubMed] [Google Scholar]
- 37.Pang RW, Lee TK, Man K, Poon RT, Fan ST, Kwong YL, et al. PIN1 expression contributes to hepatic carcinogenesis. The Journal of pathology. 2006;210(1):19–25. Epub 2006/07/15. 10.1002/path.2024 . [DOI] [PubMed] [Google Scholar]
- 38.Man CH, Wei-Man Lun S, Wai-Ying Hui J, To KF, Choy KW, Wing-Hung Chan A, et al. Inhibition of NOTCH3 signalling significantly enhances sensitivity to cisplatin in EBV-associated nasopharyngeal carcinoma. The Journal of pathology. 2012;226(3):471–81. Epub 2011/10/20. 10.1002/path.2997 . [DOI] [PubMed] [Google Scholar]
- 39.Chung GT, Lou WP, Chow C, To KF, Choy KW, Leung AW, et al. Constitutive activation of distinct NF-kappaB signals in EBV-associated nasopharyngeal carcinoma. The Journal of pathology. 2013;231(3):311–22. Epub 2013/07/23. 10.1002/path.4239 . [DOI] [PubMed] [Google Scholar]
- 40.Lo KW, Chung GT, To KF. Deciphering the molecular genetic basis of NPC through molecular, cytogenetic, and epigenetic approaches. Seminars in cancer biology. 2012;22(2):79–86. Epub 2012/01/17. 10.1016/j.semcancer.2011.12.011 . [DOI] [PubMed] [Google Scholar]
- 41.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, et al. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Molecular cell. 2003;12(6):1413–26. Epub 2003/12/24. . [DOI] [PubMed] [Google Scholar]
- 42.Kamei H, Koide T, Kojima T, Hashimoto Y, Hasegawa M. Inhibition of cell growth in culture by quinones. Cancer biotherapy & radiopharmaceuticals. 1998;13(3):185–8. Epub 2000/06/13. . [DOI] [PubMed] [Google Scholar]
- 43.Chao SH, Greenleaf AL, Price DH. Juglone, an inhibitor of the peptidyl-prolyl isomerase Pin1, also directly blocks transcription. Nucleic acids research. 2001;29(3):767–73. Epub 2001/02/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paulsen MT, Ljungman M. The natural toxin juglone causes degradation of p53 and induces rapid H2AX phosphorylation and cell death in human fibroblasts. Toxicology and applied pharmacology. 2005;209(1):1–9. Epub 2005/11/08. 10.1016/j.taap.2005.03.005 . [DOI] [PubMed] [Google Scholar]
- 45.Aragane Y, Kulms D, Metze D, Wilkes G, Poppelmann B, Luger TA, et al. Ultraviolet light induces apoptosis via direct activation of CD95 (Fas/APO-1) independently of its ligand CD95L. The Journal of cell biology. 1998;140(1):171–82. Epub 1998/02/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–19. Epub 2004/01/28. . [DOI] [PubMed] [Google Scholar]
- 47.Zhivotovsky B, Kroemer G. Apoptosis and genomic instability. Nature reviews Molecular cell biology. 2004;5(9):752–62. Epub 2004/09/02. 10.1038/nrm1443 . [DOI] [PubMed] [Google Scholar]
- 48.Liou YC, Ryo A, Huang HK, Lu PJ, Bronson R, Fujimori F, et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(3):1335–40. Epub 2002/01/24. 10.1073/pnas.032404099 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Using Western blot, elevated PIN1 proteins were observed in the nasopharyngeal epithelial cells, NP69 and HK-1, after treatment with proteasome inhibitor MG132 (0–15 μM). ACTIN was used for loading normalization.
(TIF)
(A) The expression of PIN1 and cyclin D1 in NPC C666-1 cells transfected with PIN1 siRNAs and cyclin D1-expressing vectors was observed via Western blot. Using (B) WST-1, (C) BrdU and (D) colony formation assays, the expression of cyclin D1 was shown to restore the cell growth and DNA synthesis in the PIN1 knockdown NPC cells.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.