Highlight
The C-terminus of SiAGO1b is an essential motif for the interaction between SiAGO1b and SiHYL1, and plays a key role in regulating growth, development and stress responses in Setaria italic.
Key words: AGO1, development, EMS mutant, foxtail millet, growth, HYL1, map-based cloning, RNA-seq.
Abstract
Foxtail millet (Setaria italica (L.) P. Beauv), which belongs to the Panicoideae tribe of the Poaceae, is an important grain crop widely grown in Northern China and India. It is currently developing into a novel model species for functional genomics of the Panicoideae as a result of its fully available reference genome sequence, small diploid genome (2n=18, ~510Mb), short life cycle, small stature and prolific seed production. Argonaute 1 (AGO1), belonging to the argonaute (AGO) protein family, recruits small RNAs and regulates plant growth and development. Here, we characterized an AGO1 mutant (siago1b) in foxtail millet, which was induced by ethyl methanesulfonate treatment. The mutant exhibited pleiotropic developmental defects, including dwarfing stem, narrow and rolled leaves, smaller panicles and lower rates of seed setting. Map-based cloning analysis demonstrated that these phenotypic variations were attributed to a C–A transversion, and a 7-bp deletion in the C-terminus of the SiAGO1b gene in siago1b. Yeast two-hybrid assays and BiFC experiments revealed that the mutated region was an essential functional motif for the interaction between SiAGO1b and SiHYL1. Furthermore, 1598 differentially expressed genes were detected via RNA-seq-based comparison of SiAGO1b and wild-type plants, which revealed that SiAGO1b mutation influenced multiple biological processes, including energy metabolism, cell growth, programmed death and abiotic stress responses in foxtail millet. This study may provide a better understanding of the mechanisms by which SiAGO1b regulates the growth and development of crops.
Introduction
Foxtail millet (Setaria italica (L.) P. Beauv), which belongs to the Panicoideae subfamily, was domesticated from the wild species, green foxtail (Setaria viridis (L.) P. Beauv) more than 8000 years ago in Northern China (Zohary et al., 2012). It remains an important cereal crop in arid and semi-arid regions of China and India. Reference genomes of two different foxtail millet accessions are available (Bennetzen et al., 2012; Zhang et al., 2012). Comparative genome analysis revealed a high level of collinearity between foxtail millet and rice (Oryza sativa) (Devos and Gale, 1997), indicating a promising future for comparative functional genomics. In addition, foxtail millet has been proposed recently as a novel model species for functional genomics studies of the Panicoideae because of its small diploid genome (2n=18, ~510Mb), short life cycle, small stature, prolific seed production and C4 photosynthesis (Diao et al., 2014; Muthamilarasan and Prasad, 2015).
RNA interference (RNAi) is a conserved mechanism that acts as both a defense mechanism against viruses and transposon blooms, and a method of gene regulation that can influence either transcription rate or mRNA stability (Baulcombe, 2004; Vaucheret, 2006). Both transcriptional and post-transcriptional RNAi mechanisms depend on short noncoding RNAs such as small interfering RNAs (siRNAs) and microRNAs (miRNAs). To date, this mechanism has been shown to regulate various biological processes including development, metabolism, and immunity in both plants and animals (Zhang et al., 2013). RNAi-mediated gene silencing typically caused the destruction of specific mRNA molecules (Finnegan and Matzke, 2003). Dicer-like protein (DCL) and Argonaute (AGO) are two vital proteins in the plant RNAi process. DCL proteins contain two domains that possess endonuclease function. DCL slices mRNA into 21–25 nt small RNAs (sRNAs). The sRNAs are then captured by AGO to form the core of the RNA-induced silencing complex (RISC). The sRNAs unwind into single strands and lead the RISC to target mRNA. The RISC then captures the target mRNA and cleaves it into segments. Thus, the target genes are silenced post-transcriptionally (Baulcombe, 2004).
The AGO family contains ten members in Arabidopsis thaliana (Vaucheret, 2008), 19 in rice (Kapoor et al., 2008) and 17 in foxtail millet (Luo et al., 2013; Bennetzen et al., 2012). These members can be divided into four subfamilies: MEL1, AGO4, AGO7, and AGO1. MEL1 is involved in premeiotic mitosis and meiosis during sporophyte development (Nonomura et al., 2007). The AGO4 subfamily combines with siRNA to form complexes that then recruit DNA methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) and other proteins to mediate methylation modification in DNA fragments containing sequences complementary to siRNA sequences (Ye et al., 2012). AGO7 participates in the trans-acting small interfering RNA (ta-siRNA) pathway (Nagasaki et al., 2007). AGO1 is the core element of the RISC complex. AGO1 combines with 5′-U miRNAs and siRNAs (Takeda et al., 2008) and slices target mRNA under the guidance of miRNAs and siRNAs (Qi et al., 2005). Disruption of AGO1 function in different species generally results in phenotypes including dwarfed stems, narrow leaves, and sterile inflorescences in plants (Wu et al., 2009). Previous research on Arabidopsis showed that AGO1 can interact with HYPONASTIC LEAVES 1 (HYL1), an important protein that plays a role in the correct recognition of slice sites in target mRNAs (Fang and Spector, 2007; Yang et al., 2014). hyl1 mutants show similar phenotypes to ago1 mutants and exhibit greater sensitivity to abscisic acid (ABA) (Lu and Fedoroff, 2000).
The reference genome for foxtail millet included five genes belonging to the AGO1 subfamily (Bennetzen et al., 2012); however, the specific functions of these genes are uncharacterized. AGO proteins contain three characteristic domains: PAZ, MID, and PIWI (Song and Joshua-Tor, 2006). The PAZ domain binds to the 3′ ends of sRNAs (Mi et al., 2008). The MID domain binds to the 5′ ends of sRNAs (Ma et al., 2005). The PIWI domain has an RNase H function that provides the mRNA slicer activity (Liu et al., 2004; Rivas et al., 2005; Song et al., 2004). In this study, we employed a forward genetics approach to map and characterize an ethyl methanesulfonate (EMS)-induced foxtail millet mutant that exhibited pleiotropic defects in plant growth and development, as well as hypersensitivity to ABA and drought stress. Map-based cloning identified the candidate gene as SiAGO1b, which encodes an argonaute protein, an important component of the RNA-induced silencing complex. The siago1b mutant allele identified in this study does not appear to contain any polymorphisms in these three conserved domains; however, it does encode a protein that lacks a C-terminal region of SiAGO1b. We show that this region, not previously believed to be essential for AGO1 function, influences the protein’s interaction with SiHYL1, which affects growth, development and drought tolerance in foxtail millet. Transcriptome analysis revealed that the SiAGO1b mutation strongly influenced transcriptional regulation in foxtail millet. These results demonstrate the functional role of SiAGO1b in foxtail millet and support its importance in plant growth and development.
Materials and methods
Plant materials and growth conditions
The siago1b mutant was derived by EMS treatment of the foxtail millet variety Yugu1 (the accession used for the creation of the reference genome sequence). Yugu1 seeds were mutagenized with 0.5% (v/v) EMS solution overnight. One M2 line was identified that exhibited the phenotype of dwarfing, narrow and rolled leaves, and lower seed setting rate. For morphological analysis, the mutant line was backcrossed to Yugu1 and selfed to clean the background mutations. The segregation ratio of normal and mutant phenotypes was recorded. Ten individuals of the siago1b mutants and wild-type plants were selected to measure the agronomic traits.
Assessment of drought tolerance and ABA response
To investigate variation in drought tolerance of the siago1b mutant, well-watered mutant and wild-type plants were subjected to drought treatment at either the seeding stage (6 days after germination) or four leaves stage (3 weeks after germination). Water was withheld for 12 days, and plants were then re-watered for 5 days. In addition, variation in water loss rates of fresh leaves between wild-type and siago1b mutant plants were monitored as described previously (Han et al., 2013) for leaves excised from wild-type and siago1b mutant plants grown for 25 d in a culture room at 28 °C under 16/8h light/dark cycles. Ten independent biological replicates were used for each measurement.
To assess ABA responses, foxtail millet seeds of siago1b and Yugu1 were germinated on moist filter paper containing 0, 2, 5 and 10 μM concentrations of ABA. Germination rates were recorded after 10 days in the growth chamber. Fifty seeds were used for each ABA treatment and three independent replicates were carried out for each combination of genotype and treatment. After germination, the lengths of cotyledons and roots where measured for 10 individuals from each ABA treatment.
Mapping and cloning of SiAGO1b
For map-based cloning, an F2 mapping population derived from a cross between the siago1b mutant and the foxtail millet variety Liaogu1 was constructed and grown from June to September at the Shunyi Station of the Chinese Academy of Agricultural Sciences in Beijing, China. Liaogu1 is a foxtail millet cultivar that flowers at approximately the same time as Yugu1 but shows a high density of genetic polymorphisms relative to Yugu1. Genomic DNA from F2 plants was extracted for segregation analysis using available simple sequence repeat (SSR) markers (Zhang et al., 2014). New SSR markers were developed based on the foxtail millet genome sequence information from the S. italica genome project V2.2 (http://www.phytozome.net) database if necessary. Single-nucleotide polymorphism (SNP) markers were developed based on SNP comparison data between Yugu1 and Liaogu1 (Jia et al., 2013). The SSR marker primer sequences and SNP marker loci are listed in Supplementary Table S1 at JXB online.
Sequencing and phylogenetic analysis of candidate proteins
Reference sequences of the candidate genes located in the mapping region were retrieved from the S. italica genome project V2.2. Genes in the mapped region were PCR amplified and the PCR products were sequenced using an Applied Biosystems 3730 sequencer (Applied Biosystems, Foster City, CA, USA) and analysed by DNAMAN8 software (Lynnon Biosoft, Quebec, Canada). Alignments of full-length candidate protein sequences used for phylogenetic analysis were produced by CLUSTALW (Thompson et al., 2002). The phylogenetic tree was constructed using MEGA5.0 software (Tamura et al., 2011) and the neighbor-joining method, with 1000 bootstrap value trials. The alignment file is included in Supplementary Table S2.
Yeast two-hybrid analysis
A yeast two-hybrid assay was performed using the Matchmaker Gold Yeast Two-Hybrid System (Cat no. 630489, Clontech, Mountain View, CA, USA). The full-length coding regions of SiAGO1b/siago1b and SiHYL1 were fused in frame to pGBKT7 and pGADT7, separately, to construct pGBKT7–SiAGO1b, pGBKT7–ΔSiAGO1b and pGADT7–SiHYL1 vectors. Test vectors were co-transformed into the yeast strain Gold Saccharomyces cerevisiae, and interactions were tested by SD/–Ade/–His/–Leu/–Trp plate selection, following the manufacturer’s instructions.
Bimolecular fluorescence complementation assay in foxtail millet protoplasts
For the bimolecular fluorescence complementation (BiFC) assay, SiAGO1b and ΔSiAGO1b were each cloned into the pSPYNE vector and fused to the N-terminus of the yellow fluorescent protein (YFP). The coding sequence of SiHYL1 was cloned into the pSPYCE vector, resulting in a fusion open reading frame (ORF) that also contained the C-terminus of the YFP. Protoplasts were isolated from fresh leaves of 7d-old foxtail millet seedling. Both protoplast isolation and transfection followed a protocol described previously (Kim et al., 2015). To investigate the expression and subcellular localization of the mutated gene, ΔSiAGO1b was recombined into p16318:GFP vector, and introduced into foxtail millet protoplasts by PEG-mediated transfection. YFP and green fluorescent protein (GFP) fluorescence was detected and captured by confocal microscopy (LSM700, Carl Zeiss, Germany).
Transcriptome sequencing and quantitative real-time reverse transcription PCR analysis
Mutant siago1b and wild-type (WT) Yugu1 plants were grown in a growth chamber with 16h of light at 28 °C and 8h of dark at 25 °C each day for 3 weeks. The aboveground parts of siago1b and WT plants were harvested and total RNA was extracted for transcriptome sequencing. RNA quality and purity were examined using an Agilent Bioanalyzer 2100 (Agilent Technologies, Waldbronn, Germany). The cDNA library was constructed following the Illumina sequencing manual. The cDNA libraries of mutant siago1b and the WT were sequenced on an Illumina HiSeq 2000 Genome Analyzer (Illumina, San Diego, CA, USA) with three independent biological replicates for each genotype. Raw sequencing data obtained in this study have been deposited at EMBL-EBI in the European Nucleotide Archive database under the accession number ERP014695. For the quantitative real-time reverse transcription PCR (qRT-PCR) assay, RNA was extracted from the leaves, panicles, and stems of siago1b and WT plants that had developed to the heading stage using Trizol (Cat no. 15596-026, Invitrogen, Paisley, UK). After removing contaminating DNAs with a Purelink RNA Kit (Cat no. 12183018, Invitrogen, UK), the RNAs were reverse transcribed using a PrimeScript II 1st Strand cDNA Synthesis Kit (Cat no. 6210A, Takara, Otsu Shiga, Japan). The cDNAs were then used as templates for qRT-PCR. Quantitative PCR was performed using a FastStart Universal SYBR Green Master kit (Cat no. 04913914001, Roche, Mannheim, Germany) on an Applied Biosystems 7300 Analyzer (Applied Biosystems, Foster City, CA, USA). The S. italica Actin gene (primer pairs: 5′-GTGCTTTCCCTCTACGCCAGTG-3′, 5′-ACCGCTGAGCACAATGTTACCA-3′) was used as the internal control. The primers used for qRT-PCR are listed in Supplementary Table S3. Each qRT-PCR assay was carried out with three independent replicates and each replicate corresponded to three technical repeats.
Analysis of the transcriptome data
The 100-bp paired-end reads generated from the siago1b and WT plants were processed by removing contaminants (reads containing adapters, unknown or low-quality bases) using in-house Perl scripts, and then trimmed using SolexaQA (Hiremath et al., 2011). Clean reads were aligned to the foxtail millet genome database (S. italica v2.2, DOE-JGI, www.phytozome.net) using Bowtie2 and TopHat (Langdon, 2015). Differentially expressed genes (DEGs) and transcript expression analysis were performed using Cufflinks (Trapnell et al., 2012). Genes with a false discovery rate ≤0.001 and an absolute log2-fold change value ≥1 were identified as DEGs. To obtain functional annotation and classification for DEGs, we used Blast2GO to perform gene ontology (GO) annotations with regard to biological process, molecular function and cellular component (Conesa and Gotz, 2008). AgriGO was used to perform GO functional enrichment analysis with default parameters (Du et al., 2010). Enriched GO terms were visualized by ReviGO (Supek et al., 2011) and Cytoscape software (Shannon et al., 2003). For pathway analysis, all DEGs were mapped to terms in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. KOBAS 2.0 was employed to identify statistically significantly enriched metabolic pathways (Xie et al., 2011). Twenty-nine genes were selected to validate the gene expression in the Illumina data using qRT-PCR.
Results
The siago1b mutant displays pleiotropic developmental defects
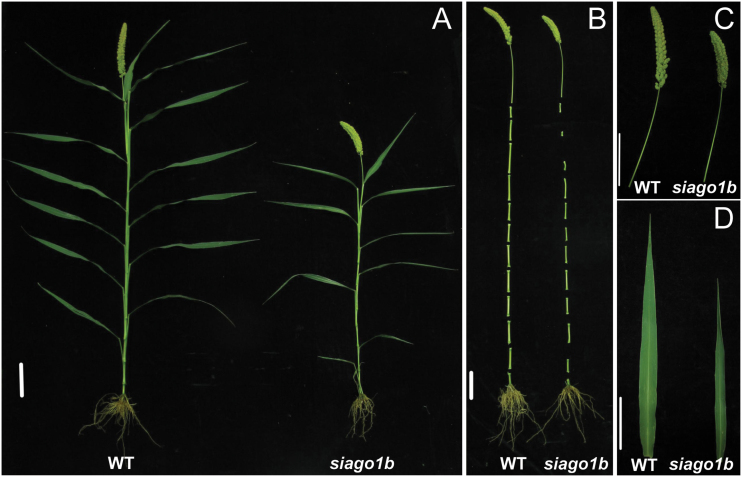
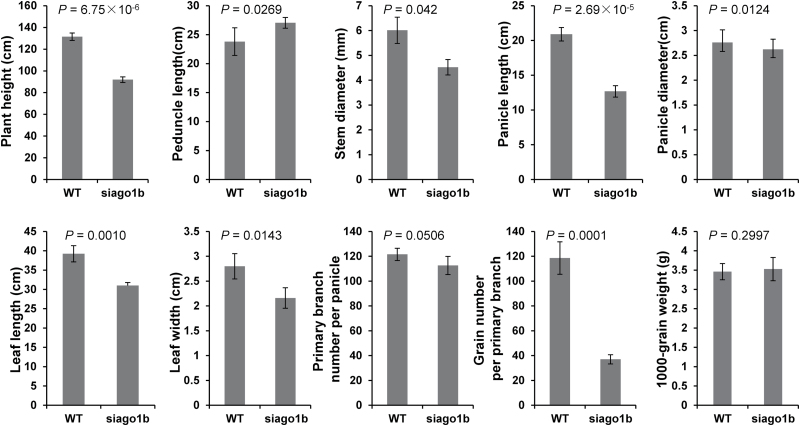
At maturity, siago1b plants were ~70% of the height of WT plants (Fig. 1A). The siago1b internodes from the top to the bottom were shorter and narrower than wild-type plants (Fig. 1B). The peduncle length, leaf length, leaf width, panicle length, and panicle diameter were diminished significantly in siago1b plants (Figs. 1C, D). Grain number per branch also varied between siago1b and wild-type plants with the WT averaging 118 grains per branch, but siago1b only 37 grains per branch (Fig. 2). However, no significant variation between the two was observed for the number of primary branches per panicle or 1000-grain weight (Fig. 2). These phenotypes were consistent with the ago1b mutant in rice (Wu et al., 2009).
Fig. 1.
The phenotypes of the wild-type (WT) and siago1b. (A) The gross morphologies of the WT and siago1b. (B) The panicles and internodes of the WT and siago1b. (C) The panicles and peduncles of the WT and siago1b. (D) The second upper leaves of the WT and siago1b. Scale bar: 10cm.
Fig. 2.
Phenotype statistics of siago1b and the wild-type (WT). The statistics of ten S. italica agronomic traits of the WT and siago1b. Data are the means of ten independent biological replicates and the P value of Welch’s two-sample t test are shown.
Drought and ABA response in seedling growth of siago1b
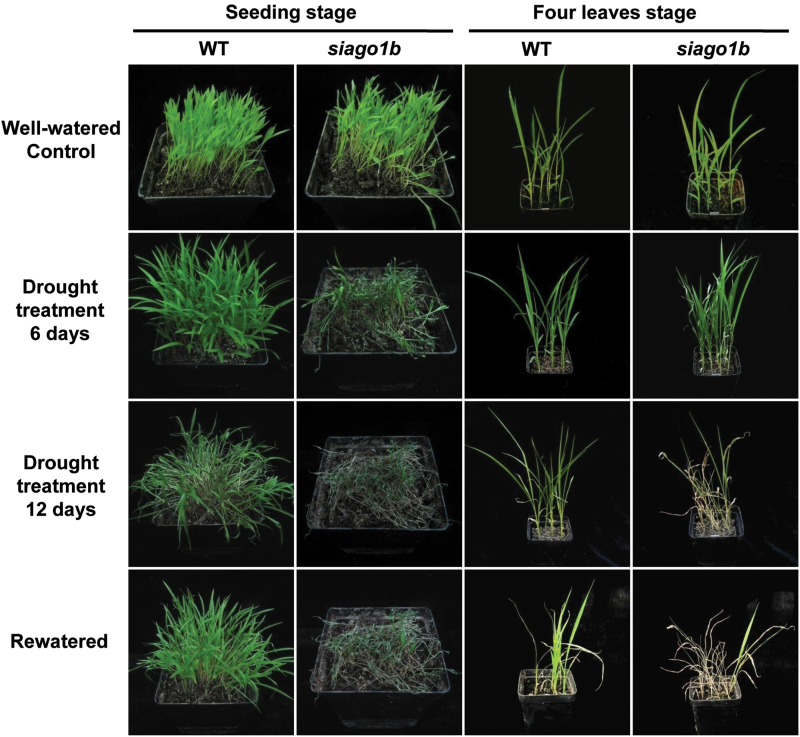
Both wild-type and siago1b seedlings were subjected to a 2-week drought treatment at either the emergence or four leaf stage. During water deprivation, the siago1b mutant plants withered and showed more severe wilting than the WT plants. WT seedlings showed obvious wilting on day 12, while the siago1b mutant seedlings exhibited obvious wilting by day 6 and most siago1b individuals were dead and desiccated by day 12 (Fig. 3). Additionally, siago1b seedlings lost water more quickly than WT seedlings did (Fig. 4A).
Fig. 3.
Morphological differences in the drought tolerance of siago1b and the wild-type (WT). Seeding stage refers to plants grown in soil for 10 days after sowing under well-watered conditions. Four leaves stage refers to plants grown for 3 weeks before drought treatment. Water was withheld for 12 days, after which the plants were rewatered for 5 days.
Fig. 4.
Siago1b mutant response to dehydration and ABA treatment. (A) Water loss from whole seedling of siago1b mutant and the WT. Water loss is expressed as the percentage of initial fresh weight of seedlings. (B, C, D) Difference in germination rates, cotyledon length, and primary root length between siago1b mutant and the WT in response to exogenous ABA. Data are means from ten individuals. Asterisks indicate a significant difference between siago1b and WT plants (n=10, Welch’s two-sample t test, P<0.001).
To investigate ABA responses of the mutants, we quantified the seed germination rate (SGR) of siago1b and wild-type plants under different ABA concentrations. The SGR of WT was slightly affected by exogenous ABA, whereas for the siago1b mutant, the SGR decreased significantly in response to exogenous ABA. Under 10 μM ABA treatment, none of the mutant seeds germinated (150 seeds, 3 independent replicates, 10 days after sowing), while the SGR of WT seeds was above 70% under the same treatment conditions (Fig. 4B). Growth of siago1b mutant seedlings was also severely affected by exogenous ABA treatment, as evidenced by shorter primary root and cotyledon (Fig. 4C, D, Supplementary Fig. S1) than WT plants when treated with equal concentrations of exogenous ABA.
Map-based cloning of the SiAGO1b gene
The SiAGO1b gene was isolated using a map-based cloning approach and an F2 population derived from a cross of mutant siago1b and wild-type foxtail millet plants of the variety Liaogu1. In the F2 generation, a total 780 individuals were phenotypically scored, of which 595 were wild-type and 185 exhibited a dwarf phenotype, with narrow and rolled leaves, which was consistent with a mendelian ratio of 3:1 for normal phenotype to mutant phenotype offspring (χ2=0.62<χ2 0.05=3.84). This suggested that a single recessive gene controlled the multiple phenotypes observed for siago1b. For map-based cloning, more than 800 F2 homozygous recessive individuals were used. Bulked segregation analysis showed that the SiAGO1b gene was on chromosome 7 and was genetically linked with SSR markers CAAS7027 and CAAS7029. Additional SSR and SNP markers were employed to fine-map SiAGO1b to a 46.3-kb region between SNP markers SNP027326466 and SNP27372797 on chromosome 7, with two and four recombinants, respectively (Fig. 5).
Fig. 5.
Map-based cloning of the SiAGO1b gene. SiAGO1b was mapped in the interval between molecular markers SNP027326466 and SNP 27372797 on chromosome 7 using 780 recessive individual plants showing a mutant-like phenotype from an F2 population. Numbers under the markers indicate recombinants. Numbers between markers indicate the physical distance. The white arrows indicate ORFs. The orange arrow stands for the candidate gene.
SiAGO1b encodes an argonaute protein
Using the S. italica genome database V2.2, five ORFs were identified in the mapping interval (Table 1). Sequencing of genomic DNA from the target region revealed a 7-bp deletion and a 1-bp shift in the 22nd exon of Seita.7G201100 (Fig. 6A). Seita.7G201100 encodes a protein containing the two characteristic domains of argonaute (AGO) proteins: PAZ and PIWI (Fig. 6B). Phylogenetic analysis and protein sequence alignment showed that the Seita.7G201100 was most closely related to OsAGO1b, which belongs to subfamily AGO1 (Fig. 6C). Therefore, the target gene was named SiAGO1b. The siago1b mutant allele was predicted to encode a protein (ΔSiAGO1b) with a frame shift mutation after amino acid 1068 and early termination at amino acid 1073 (Fig. 6B). Multiple sequence alignment of the SiAGO1b protein and its homologous proteins in soybean (Glycine max), maize (Zea mays), rice, Brachypodium distachyon and wheat (Triticum aestivum) revealed that the C-terminal motif of the SiAGO1b (–PLPALKENVKRVMFYC) protein is highly conserved among these organisms. However, ΔSiAGO1 has a mutation in this region (–QLSRRT) (Fig. 6D). The alignment result indicated that the mutated region of SiAGO1b protein is probably a functional motif.
Table 1.
Gene IDs, locations and functional annotations in the mapped region
| Gene ID | Location | Functional annotation |
|---|---|---|
| Seita.7G201100 | scaffold_7: 27330567 - 27344429 | Eukaryotic translation initiation factor 2C |
| Seita.7G201200 | scaffold_7: 27338776 - 27340117 | There are no functional annotations for this locus |
| Seita.7G201300 | scaffold_7: 27340287- 27342020 | There are no functional annotations for this locus |
| Seita.7G201400 | scaffold_7: 27354369 - 27356190 | Eukaryotic translation initiation factor 3 |
| Seita.7G201500 | scaffold_7: 27365483 - 27367466 | Protein of unknown function (DUF1618) |
Fig. 6.
The structure and phylogenetic analysis of target gene SiAGO1b. (A) Gene structure of SiAGO1b. The mutation site is indicated by a red arrow. (B) Protein structure of the wild-type (WT) SiAGO1b and mutant ΔSiAGO1b. The mutant site is indicated by a red arrow in WT SiAGO1b. (C) Phylogenetic relationships of AGO family proteins of foxtail millet, Arabidopsis and rice. SiAGO1b was most closely related to OsAGO1b, which belongs to subfamily AGO1. A red arrow indicates the position of SiAGO1b. (D) The multiple alignments of SiAGO1b homologous proteins in different organisms. The organism name and gene locus name are shown before protein sequences. ΔSiAGO1 indicates the mutant protein. A red box indicates the C-terminal conserved region. A red line indicates the mutant protein sequence in the siago1b mutant.
SiAGO1b mutation influenced its interaction with SiHYL1 and transcript accumulation level in leaf and panicle
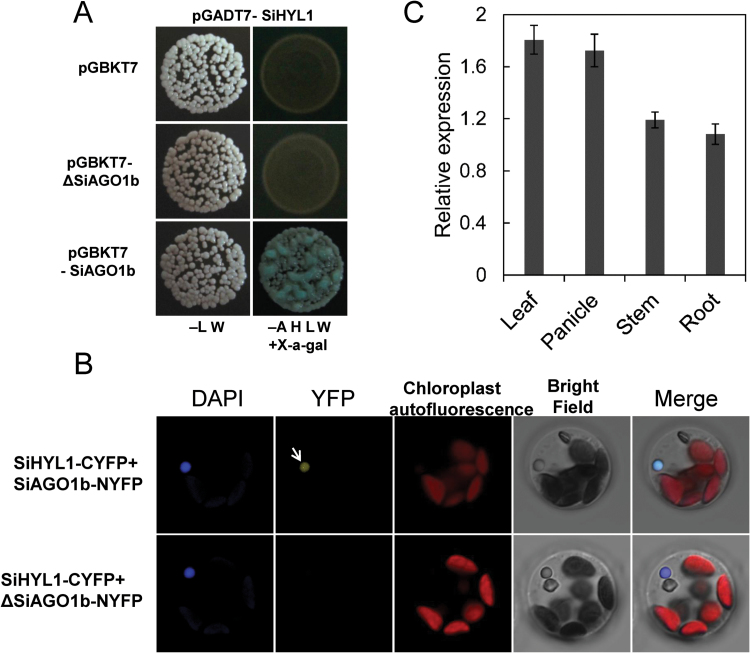
The Arabidopsis homologous protein of SiAGO1b, AtAGO1, interacts with the HYL1 protein (Fang and Spector, 2007). In foxtail millet, Seita.7G329000 is the homolog of HYL1, which was named SiHYL1. The yeast strain (Gold Saccharomyces cerevisiae) carrying BD-SiAGO1b+AD-SiHYL1 grew well on SD/–Ade/–His/–Leu/–Trp yeast growth medium. However, the yeast strain carrying BD-ΔSiAGO1b+AD-SiHYL1 could not grow on SD/–Ade/–His/–Leu/–Trp yeast growth medium (Fig. 7A).
Fig. 7.
The protein interaction and gene expression analysis of SiAGO1b. (A) Result of yeast two-hybrid assay. Yeast two-hybrid assays showing that SiHYL1 interacts with SiAGO1b, but not with mutant protein ΔSiAGO1b. –LW indicates yeast medium SD/–Leu/–Trp, –AHLW indicates yeast medium SD/–Ade/–His/–Leu/–Trp. 5-Bromo-4-chloro-3-indolyl α-d-galactopyranoside (X-α-gal) was added to the solid yeast medium, and the same amount of yeast was used in each assay. The interaction was judged from the blue color and yeast growth density. (B) The relative expression of ΔSiAGO1b gene in mutant leaf, panicle, stem, and root. Total RNA was isolated from various tissues of WT and siago1 seedlings grown in culture. qPCR was conducted with three biological replicates. (C) BiFC experiments between SiAGO1b, mutant protein ΔSiAGO1b and SiHYL1. Protein partners was fused to an N-terminal fragment or C-terminal fragment of YFP, respectively, and co-infiltrated into foxtail millet protoplasts. DAPI was used to label the nucleus. BiFC signals between SiAGO1b and SiHYL1 were observed in nucleus region. No BiFC signals were observed between mutant protein ΔSiAGO1b and SiHYL1. Negative and positive control test is shown in Supplementary Figs S2 and S3.
To further confirm the interaction between SiAGO1b (ΔSiAGO1b) and SiHYL1, we employed BiFC assays with SiAGO1b tagged with the N-terminal domain of YFP and SiHYL1 fused into the C-terminal domain of YFP. A YFP fluorescence signal was detected in the nucleus, indicating that SiAGO1b interacts with SiHYL1 (Fig. 7B, Supplementary Fig. S2). The result is consistent with a previous report from Arabidopsis (Fang and Spector, 2007). However, no BiFC signal was detected between the mutated protein ΔSiAGO1b and SiHYL1. Simultaneously, we determined the subcellular localization of ΔSiAGO1b. A fluorescence signal from a ΔSiAGO1b-GFP fusion protein can be clearly detected in the nucleus, indicating that loss of C-terminal motif in SiAGO1b does not affect its translation or subcellular localization (see Supplementary Fig. S3). Together, these results suggest that the C-terminal polypeptide of SiAGO1b is necessary for protein–protein interaction between SiAGO1b and SiHYL1.
qRT-PCR was used to assess the expression of SiAGO1b in different tissues. The relative expression level of SiAGO1b was higher in siago1b mutant panicles and leaves than wild-type, but expression in the stem was not significantly different between the two genotypes (Fig. 7C). This suggests that there may be a feedback mechanism to increase the expression of SiAGO1b in siago1b mutant panicles and leaves in response to the loss of the functional SiAGO1b protein activity.
DEG analysis of siago1b mutant by transcriptome sequencing
Argonaute protein is a key component of the RISC complex that regulates gene expression in a range of biological processes (Mallory and Hervé, 2010). Therefore, mutations of AGO1 are likely to produce both direct and indirect changes in the abundance of the downstream target genes. Transcriptome sequencing was employed to compare the expression profiles of WT and siago1b mutant plants, resulting in the identification of 1598 differentially expressed genes (see Supplementary Table S4). GO enrichment analysis for the up- and down-regulated genes in siago1b was performed to identify the major biological processes and molecular functions regulated by SiAGO1b. Thirty-nine biological processes (P<0.05, Supplementary Table S5) were enriched among genes up-regulated in siago1b, and 22 for the down-regulated genes (P<0.05, Supplementary Table S6). GO terms involved in stress responses and oxidation–reduction were enriched among both up- and down-regulated genes. Interestingly, the majority of all genes annotated as participated in transcriptional regulation, protein metabolism, and programmed cell death were up-regulated in the mutant (Fig. 8A). GO terms associated with energy metabolism (e.g. carbohydrate metabolism and lipid metabolism) were enriched specifically among the genes down-regulated in siago1b (Fig. 8B). Supplementary Fig. S4 shows the DEGs distributed among the seven most enriched biological and 15 molecular GO terms. Supplementary Table S7 lists the 37 up-regulated genes and 34 down-regulated genes, which showed the greatest change in expression between mutant and wild-type plants. These genes were mainly distributed in three functional pathways: genes related to abscisic acid (ABA) signaling and stress responses, transcription factors controlling organ development, and genes regulating floral development (Fig. 8C). Other genes controlling plant normal growth and development showed significant changes in expression. Notably, two DEGs had no homologous genes in Arabidopsis and rice. These may be foxtail millet-specific genes that possess unique functions (Supplementary Table S8). Among these 71 genes with the greatest difference in expression between mutant and wild-type plants, 27 had homologs in Arabidopsis that have already been annotated (Table 2). These 29 genes were selected to validate the RNA-seq gene expression analysis through the use of qRT-PCR (Supplementary Fig. S5).
Fig. 8.
Enriched biological processes and candidate differentially expressed genes (DEGs) of the siago1b mutant. (A, B) Functional enrichment analysis of up- and down-regulated genes. Each circle represent a gene ontology (GO) term in red, as shown in the color bar ranging from 1.0 to 1×10–11 (P value); P<0.05 was used as the threshold. (C) Expression patterns of DEGs previously characterized in Arabidopsis or rice. Clustering based on average log2 FPKM of genes involved in phytohormone signal transduction, transcription regulation and stress responses.
Table 2.
Twenty-seven genes whose expression was significantly altered between the wild-type and the siago1b mutant that have homologous genes studied in Arabidopsis
| Gene ID | Homologous gene in Arabidopsis | Fold change in siago1b | Gene name in Arabidopsis | Gene annotation |
|---|---|---|---|---|
| Seita.1G334700 | AT1G77760.1 | 5.711 | GNR1, NIA1, NR1 | Nitrate reductase 1 |
| Seita.9G440900 | AT5G65010.1 | 5.205 | ASN2 | Asparagine synthetase 2 |
| Seita.6G178500 | AT1G77760.1 | 3.957 | GNR1, NIA1, NR1 | Nitrate reductase 1 |
| Seita.7G150500 | AT3G54420.1 | 3.492 | ATCHITIV, ATEP3, CHIV, EP3 | Homolog of carrot EP3-3 chitinase |
| Seita.8G132600 | AT3G48280.1 | 3.237 | CYP71A25 | Cytochrome P450, family 71, subfamily A, polypeptide 25 |
| Seita.2G270400 | AT1G02850.2 | 3.141 | BGLU11 | β-Glucosidase 11 |
| Seita.8G008100 | AT1G69490.1 | 3.067 | ANAC029, ATNAP, NAP | NAC-like, activated by AP3/PI |
| Seita.9G379000 | AT1G78290.2 | 2.800 | SNRK2-8, SNRK2.8, SRK2C | Protein kinase superfamily protein |
| Seita.5G455700 | AT3G56970.1 | 2.709 | BHLH038, ORG2 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
| Seita.2G368800 | AT3G18830.1 | 2.427 | ATPLT5, ATPMT5, PMT5 | Polyol/monosaccharide transporter 5 |
| Seita.3G386200 | AT1G56010.2 | 2.305 | anac021, ANAC022, NAC1 | NAC domain containing protein 1 |
| Seita.7G059700 | AT5G45890.1 | −11.917 | SAG12 | Senescence-associated gene 12 |
| Seita.6G048800 | AT5G23960.1 | −4.917 | ATTPS21, TPS21 | Terpene synthase 21 |
| Seita.9G011100 | AT3G26300.1 | −4.516 | CYP71B34 | Cytochrome P450, family 71, subfamily B, polypeptide 34 |
| Seita.8G200200 | AT3G07990.1 | −3.120 | SCPL27 | Serine carboxypeptidase-like 27 |
| Seita.8G247500 | AT4G37050.1 | −2.916 | AtPLAIVC, PLA V, PLP4 | PATATIN-like protein 4 |
| Seita.6G233600 | AT1G32640.1 | −2.804 | ATMYC2, JAI1, JIN1, MYC2, RD22BP1, ZBF1 | Basic helix–loop–helix (bHLH) DNA-binding family protein |
| Seita.5G311800 | AT2G19770.1 | −2.463 | PRF5 | Profilin 5 |
| Seita.3G067200 | AT5G23960.1 | −2.446 | ATTPS21, TPS21 | Terpene synthase 21 |
| Seita.1G332100 | AT5G08640.1 | −2.228 | ATFLS1, FLS, FLS1 | Flavonol synthase 1 |
| Seita.2G134400 | AT1G19670.1 | −2.214 | ATCLH1,ATHCOR1,CLH1,CORI1 | Chlorophyllase 1 |
| Seita.1G207000 | AT2G02860.1 | −2.201 | ATSUC3,ATSUT2,SUC3,SUT2 | Sucrose transporter 2 |
| Seita.9G471200 | AT2G21140.1 | −2.171 | ATPRP2,PRP2 | Proline-rich protein 2 |
| Seita.8G139500 | AT5G13930.1 | −2.157 | ATCHS,CHS,TT4 | Chalcone and stilbene synthase family protein |
| Seita.5G469800 | AT4G01470.1 | −1.948 | ATTIP1.3,GAMMA-TIP3,TIP1;3 | Tonoplast intrinsic protein 1;3 |
| Seita.8G211600 | AT3G09220.1 | −1.946 | LAC7 | Laccase 7 |
| Seita.8G212000 | AT3G09220.1 | −1.910 | LAC7 | Laccase 7 |
Discussion
The C-terminus of SiAGO1b is an essential motif for the interaction between SiAGO1b and SiHYL1, which plays an important role in plant growth and development
To maintain normal growth and development, plant gene expression must be under strict control. AGO proteins mediate target cleavage under the guidance of sRNAs, such as miRNAs. Most miRNAs are incorporated into AGO1-associated silencing complexes in plants. AGO1 is considered the most important slicer protein for sRNA-mediated target-RNA cleavage (Voinnet, 2009). AtAGO1 was the first reported member of the AGO gene family, so named because the leaves of the atago1 mutant showed an Argonauta squid tentacles-like character (Bohmert et al., 1998). Rice has four AGO1 homologs. Rice AGO1 homolog knockdown mutants showed pleiotropic developmental phenotypes. The rice AGO1 mutants exhibited severe dwarfing, narrow and rolled leaves, and a lower seed setting rate (Wu et al., 2009). The foxtail millet siago1b mutant showed many of the same phenotypes observed in rice. In addition, the peduncle length, panicle length and panicle diameter were diminished significantly in the siago1b mutant. The HYL1 protein was previously shown to interact with AGO1 in Arabidopsis (Fang and Spector, 2007). Like the ago1 mutant, the hyl1 mutant exhibited dwarf, narrow and rolled leaves and a lower seed setting rate. Two ABA-inducible genes, KIN2 and COR47 (Gilmour et al., 1992; Kurkela and Borg-Franck, 1992), exhibited increased transcript levels in the hyl1 mutant. This suggested that the HYL1 is sensitive to ABA (Lu and Fedoroff, 2000).
Sequencing of the siago1b allele did not identify any mutations in the characteristic domains of AGO1 protein: PAZ, MID and PIWI (Song and Joshua-Tor, 2006). However, a 7-bp deletion and 1-bp shift were identified in the last exon of SiAGO1b. To investigate whether the mutated region is a functional element in foxtail millet, the foxtail millet homolog of HYL1 (SiHYL1) was cloned. Yeast two-hybrid assays and BiFC experiments revealed that WT SiAGO1b protein interacts with SiHYL1 protein but the mutant SiAGO1b protein does not (Fig. 7A, 7B). Thus, the C-terminus of SiAGO1 is essential for its interaction with SiHYL. The SiAGO1 C-terminus has a highly conserved motif across different plant species (Fig. 6D). The siago1b mutant lacks this motif, which may result in a structural alteration that abolishes the ability to interact with SiHYL1. The loss of function in the C-terminus of SiAGO1b made the mutant more sensitive to ABA and drought stress, and in addition led to the serious growth and developmental phenotypes.
The siago1b mutants are more sensitive to ABA and drought stress
Through expression analysis of the 27 genes whose homologs have already been studied in Arabidopsis with the most significant changes in expression in the siago1b mutant, several genes related to stress and abscisic acid (ABA) signal response were identified whose expressions changed markedly in siago1b. GNR1 (AT1G77760.1), which was up-regulated in the mutant, encodes a cytosolic minor isoform of nitrate reductase, which is involved in the first step of nitrate assimilation and contributes about 15% of the nitrate reductase activity in shoots. The stomata of the mutant are less sensitive to ABA, and the mutant plants exhibit very poor growth on nitrate medium (Desikan et al., 2002). The up-regulated gene SNRK2-8 (AT1G78290.2) encodes a member of the SNF1-related protein kinase (SnRK2) family. SNRK2-8 plays an important role in the ABA pathway, osmotic stress and drought stress signaling in Arabidopsis (Mizoguchi et al., 2010). Up-regulated gene ATCHITIV (AT3G54420.1) encodes an EP3 chitinase. The expression of ATCHITIV responds to environmental stresses in Arabidopsis. For instance, ATCHITIV responds to cold, light intensity, wounding, salt stress, and water deprivation (Takenaka et al., 2009). The down-regulated gene AtPLAIVC (AT4G37050.1) encodes a patatin-related phospholipase A. AtPLAIVC is expressed in the gynoecium and is induced by ABA or phosphate deficiency in Arabidopsis roots. A loss-of-function mutant exhibited an impaired response to phosphate deficiency during root development. In addition, a novel function of AtPLAIVC was reported: in root development it has a function at the interface between phosphate deficiency and auxin signaling (Rietz et al., 2010). The down-regulated gene AtFLS1 (AT1G32640.1) encodes an MYC-related transcriptional activator with a DNA-binding domain (a basic helix–loop–helix leucine zipper motif). The transcription of this gene is induced by dehydration stress and ABA treatment (Carvalhais et al., 2015). AtSUC3 (AT2G02860.1) encodes a sucrose transporter in sieve elements and some sink tissues, and is also down-regulated in the mutant. The loss-of-function mutant of AtSUC3 reduced the expression of genes AtSUC2 and AtSUC4, which respond to abiotic stresses and ABA. Thus, AtSUC3 is an important regulator in plant abiotic stress tolerance via the ABA signal pathway (Gong et al., 2015).
A genome-wide identification and functional characterization of the argonaute gene family in foxtail millet was performed by Yadav et al. (2015). The study found that SiAGO1b had highest mRNA accumulation in all tissues compared with other AGO family members. They also reported that members of the S. italica AGO1 subfamily showed substantial inductions of expression in response to ABA (12.4-fold up-regulation at 1h ABA treatment). In our study, the germination rate and seedling growth of siago1b were significantly repressed by exogenous ABA (Fig. 4, Supplementary Fig. S1). The seedlings of siago1b lost water more quickly than those of the WT. In addition, the drought tolerance ability of the siago1b mutant decreased obviously. It is probable that these phenomena were caused by disordered expression of ABA-related genes in the siago1b mutant.
The mutation of siago1b affected genes related to plant growth and development
The same analysis of genes with the greatest changes in expression in siago1b and homologs studied in Arabidopsis also identified several transcription factors. ANAC029 (AT1G69490.1), up-regulated, encodes a NAC transcription factor. ANAC029 plays an important role in leaf senescence in Arabidopsis and other plant species (Guo and Gan, 2006). ANAC029 also plays a role in petal senescence, independent of endogenous ethylene control (Shinozaki et al., 2014). Up-regulated gene BHLH038 (AT3G56970.1) encodes a member of the basic helix–loop–helix transcription factor protein family. These transcription factor genes are up-regulated strongly from cell proliferation to expansion in young developing leaves of Arabidopsis. The mutant plants developed smaller rosettes than WT plants (Andriankaja et al., 2014). Up-regulated gene ANAC022 (AT1G56010) encodes a transcription factor involved in the formation of the shoot apical meristem and auxin-mediated lateral roots. The results of previous studies indicated that ANAC022 responds to plant hormones. ANAC022 might be involved in auxin and gibberellin signaling pathways in promoting the development of lateral roots (Wang et al., 2006). These transcription factors were all up-regulated and are involved in the regulation of development and senescence of flowers, leaves, and roots. The expression changes of these transcription factors could lead to pleiotropic developmental defects in plants. The blocked growth and development phenotypes of the siago1b mutant were likely caused by the expression changes of these transcription factors.
Genes controlling the reproductive process were severely repressed
One of the notable phenotypes associated with the siago1b mutant was its significant decrease in grain yield without a decrease in thousand-grain weight. This indicated that the development process of flowers was likely defective in the siago1b mutant. There were DEGs whose homologs in Arabidopsis control flowering development. In addition to ANAC029, which plays a role in petal senescence, AtTPS21 (AT5G23960.1), down-regulated in siago1b, encodes a sesquiterpene synthase involved in generating group A sesquiterpenes. The flowering stage is controlled by the expression of AtTPS21 in Arabidopsis (Yu et al., 2015). The down-regulated gene PRF5 (AT2G19770.1) encodes a profilin 5 protein, which is an actin monomer-binding protein that regulates actin cytoskeleton organization. It is expressed predominantly in mature pollen and growing pollen tubes (Wang et al., 2008). AtTIP1.3 (AT4G01470.1), down-regulated, encodes a tonoplast intrinsic protein, belonging to a subfamily of aquaporins. It functions as a water and urea channel in pollen and contributes to normal male fertility in adverse environmental conditions (Wudick et al., 2014). In addition, two genes associated with the formation of flavonoids in mutant siago1 were significantly down-regulated. Flavonols are important compounds for conditional male fertility in plants. AtFLS1 (AT5G08640.1) encodes a flavonol synthase that catalyses the formation of flavonols from dihydroflavonols (Falcone et al., 2010). AtCHS (AT5G13930.1) encodes chalcone synthase (CHS), which also plays an essential role in the biosynthesis of flavonoid (Sun et al., 2015). The defective development of siago1b flowers may result from the repression of certain genes controlling the reproductive process.
The expression of other genes controlling plant normal growth and development was also significantly altered. For example, ASN2 (AT5G65010.1) encodes an asparagine synthetase and was up-regulated. Mutations disrupting the function of this gene exhibit defects in development. This gene is essential for nitrogen assimilation, distribution, and remobilization in plants (Gaufichon et al., 2013).
In summary, map-based cloning of an EMS-induced pleiotropic mutant in foxtail millet identified the causal gene SiAGO1b. Initial characterization of the mutant was carried out at the molecular level. Protein interaction and RNA-seq analysis provided some clues to the function and pathways of SiAGO1b in foxtail millet. These results revealed that a motif in the C-terminus of SiAGO1b is vitally important to maintain normal growth and drought stress tolerance. The findings of this study may help to promote further studies of how the molecular mechanism of AGO1 is either varied or conserved among different plant species.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Morphological differences in the ABA treatment.
Figure S2. Negative control of BiFC assays.
Figure S3. Subcellular localization of ΔSiAGO1b.
Figure S4. Differentially expressed gene (DEG) distribution in the most enriched gene ontology (GO) terms.
Figure S5. Twenty-nine differentially expressed genes (DEGs) selected for validation of the Illumina data using quantitative real-time reverse transcription PCR (qRT-PCR).
Table S1. Simple sequence repeat (SSR) primer sequences and single-nucleotide polymorphism (SNP) loci.
Table S2. Multiple sequence alignment for phylogenetic analysis.
Table S3. Primers used for qRT-PCR.
Table S4. Genes that were differentially expressed in siago1b compared with the wild-type.
Table S5. Thirty-nine biological processes that were enriched for the up-regulated genes.
Table S6. Twenty-two biological processes that were enriched for the down-regulated genes.
Table S7. The top 37 up-regulated genes and 34 down-regulated genes that were most differentially expressed.
Table S8. Significant differentially expressed genes between wild-type and siago1b mutant which have no homologous genes in Arabidopsis and rice.
Acknowledgements
This work was supported by the National High Technology Research and Development Program of China (863 Program) (grant number 2013AA102603), National Natural Science Foundation of China (grant numbers 31301328, 31522040), Beijing Natural Science Foundation (6142019), Fundamental Research Funds of ICS-CAAS (Grant to G.J., 2013007), China Agricultural Research System (grant number CARS07-13.5-A02), and The Agricultural Science and Technology Innovation Program of CAAS.
References
- Andriankaja ME, Danisman S, Mignolet-Spruyt LF, et al. 2014. Transcriptional coordination between leaf cell differentiation and chloroplast development established by TCP20 and the subgroup Ib bHLH transcription factors. Plant Molecular Biology 85, 233–245. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. 2004. RNA silencing in plants. Nature 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, et al. 2012. Reference genome sequence of the model plant Setaria . Nature Biotechnology 30, 555–561. [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. 1998. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO Journal 17, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhais LC, Dennis PG, Badri DV, Kidd BN, Vivanco JM, Schenk PM. 2015. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Molecular Plant-Microbe Interactions 28, 1049–1058. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S. 2008. Blast2GO: A comprehensive suite for functional analysis in plant genomics. International Journal of Plant Genomics 2008, 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Griffiths R, Hancock J, Neill S. 2002. A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America 99, 16314–16318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Gale MD. 1997. Comparative genetics in the grasses. Plant Molecular Biology 35, 3–15. [PubMed] [Google Scholar]
- Diao X, Schnable J, Bennetzen JL, et al. 2014. Initiation of Setaria as a model plant. Frontiers of Agricultural Science and Engineering 1, 16–20. [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone FM, Rius S, Emiliani J, Pourcel L, Feller A, Morohashi K, Casati P, Grotewold E. 2010. Cloning and characterization of a UV-B-inducible maize flavonol synthase. The Plant Journal 62, 77–91. [DOI] [PubMed] [Google Scholar]
- Fang Y, Spector DL. 2007. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Current Biology 17, 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Matzke MA. 2003. The small RNA world. Journal of Cell Science 116, 4689–4693. [DOI] [PubMed] [Google Scholar]
- Gaufichon L, Masclaux-Daubresse C, Tcherkez G, et al. 2013. Arabidopsis thaliana ASN2 encoding asparagine synthetase is involved in the control of nitrogen assimilation and export during vegetative growth. Plant, Cell & Environment 36, 328–342. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Artus NN, Thomashow MF. 1992. cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana . Plant Molecular Biology 18, 13–21. [DOI] [PubMed] [Google Scholar]
- Gong X, Liu M, Zhang L, Ruan Y, Ding R, Ji Y, Zhang N, Zhang S, Farmer J, Wang C. 2015. Arabidopsis AtSUC2 and AtSUC4, encoding sucrose transporters, are required for abiotic stress tolerance in an ABA-dependent pathway. Physiologia Plantarum 153, 119–136. [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S. 2006. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. The Plant Journal 46, 601–612. [DOI] [PubMed] [Google Scholar]
- Han X, Tang S, An Y, et al. 2013. Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis . Journal of Experimental Botany 64, 4589–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiremath PJ, Farmer A, Cannon SB, et al. 2011. Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa. Plant Biotechnology Journal 9, 922–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Huang X, Zhi H, et al. 2013. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nature Genetics 45, 957–961. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Arora R, Lama T, Nijhawan A, Khurana JP, Tyagi AK, Kapoor S. 2008. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics 9, 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Moon SJ, Min MK, et al. 2015. Functional characterization and reconstitution of ABA signaling components using transient gene expression in rice protoplasts. Frontiers in Plant Science 6, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkela S, Borg-Franck M. 1992. Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana . Plant Molecular Biology 19, 689–692. [DOI] [PubMed] [Google Scholar]
- Langdon WB. 2015. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Mining 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441. [DOI] [PubMed] [Google Scholar]
- Lu C, Fedoroff N. 2000. A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. The Plant Cell 12, 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Xu H, Liu Z, et al. 2013. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nature Genetics 45, 573–577. [DOI] [PubMed] [Google Scholar]
- Mallory A, Vaucheret H. 2010. Form, function, and regulation of ARGONAUTE proteins. The Plant Cell 22, 3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. 2005. Structural basis for 5’-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, et al. 2008. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell 133, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi M, Umezawa T, Nakashima K, Kidokoro S, Takasaki H, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. 2010. Two closely related subclass II SnRK2 protein kinases cooperatively regulate drought-inducible gene expression. Plant & Cell Physiology 51, 842–847. [DOI] [PubMed] [Google Scholar]
- Muthamilarasan M, Prasad M. 2015. Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theoretical and Applied Genetics 128, 1–14. [DOI] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, et al. 2007. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proceedings of the National Academy of Sciences of the United States of America 104, 14867–14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K, Morohoshi A, Nakano M, Eiguchi M, Miyao A, Hirochika H, Kurata N. 2007. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. The Plant Cell 19, 2583–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ. 2005. Biochemical specialization within Arabidopsis RNA silencing pathways. Molecular Cell 19, 421–428. [DOI] [PubMed] [Google Scholar]
- Rietz S, Dermendjiev G, Oppermann E, Tafesse FG, Effendi Y, Holk A, Parker JE, Teige M, Scherer GF. 2010. Roles of Arabidopsis patatin-related phospholipases a in root development are related to auxin responses and phosphate deficiency. Molecular Plant 3, 524–538. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nature Structural & Molecular Biology 12, 340–349. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki Y, Tanaka T, Ogiwara I, Kanekatsu M, van Doorn WG, Yamada T. 2014. Expression of an AtNAP gene homolog in senescing morning glory (Ipomoea nil) petals of two cultivars with a different flower life span. Journal of Plant Physiology 171, 633–638. [DOI] [PubMed] [Google Scholar]
- Song JJ, Joshua-Tor L. 2006. Argonaute and RNA--getting into the groove. Current Opinion in Structural Biology 16, 5–11. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437. [DOI] [PubMed] [Google Scholar]
- Sun W, Meng X, Liang L, Jiang W, Huang Y, He J, Hu H, Almqvist J, Gao X, Wang L. 2015. Molecular and biochemical analysis of chalcone synthase from Freesia hybrid in flavonoid biosynthetic pathway. PLoS ONE 10, e119054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, Smuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. 2008. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant & Cell Physiology 49, 493–500. [DOI] [PubMed] [Google Scholar]
- Takenaka Y, Nakano S, Tamoi M, Sakuda S, Fukamizo T. 2009. Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: chitinase inhibitor allosamidin enhances stress tolerance. Bioscience, Biotechnology, and Biochemistry 73, 1066–1071. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Current Protocols in Bioinformatics Chapter 2, Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes & Development 20, 759–771. [DOI] [PubMed] [Google Scholar]
- Vaucheret H. 2008. Plant ARGONAUTES. Trends in Plant Science 13, 350–358. [DOI] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. [DOI] [PubMed] [Google Scholar]
- Wang Y, Duan L, Lu M, Li Z, Wang M, Zhai Z. 2006. Expression of NAC1 up-stream regulatory region and its relationship to the lateral root initiation induced by gibberellins and auxins. Science in China Series C: Life Sciences 49, 429–435. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH. 2008. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis . Plant Physiology 148, 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, Ni F, Wu X, Qi Y. 2009. Rice microRNA effector complexes and targets. The Plant Cell 21, 3421–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudick MM, Luu DT, Tournaire-Roux C, Sakamoto W, Maurel C. 2014. Vegetative and sperm cell-specific aquaporins of Arabidopsis highlight the vacuolar equipment of pollen and contribute to plant reproduction. Plant Physiology 164, 1697–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. 2011. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Research 39, W316–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav C B, Muthamilarasan M, Pandey G, et al. 2015. Identification, characterization and expression profiling of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in foxtail millet. Plant Molecular Biology Reporter 33, 43–55. [Google Scholar]
- Yang X, Ren W, Zhao Q, Zhang P, Wu F, He Y. 2014. Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Research 42, 12224–12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Wang W, Iki T, Liu C, Wu Y, Ishikawa M, Zhou X, Qi Y. 2012. Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Molecular Cell 46, 859–870. [DOI] [PubMed] [Google Scholar]
- Yu ZX, Wang LJ, Zhao B, Shan CM, Zhang YH, Chen DF, Chen XY. 2015. Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and Patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Molecular Plant 8, 98–110. [DOI] [PubMed] [Google Scholar]
- Zhang G, Liu X, Quan Z, et al. 2012. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nature Biotechnology 30, 549–554. [DOI] [PubMed] [Google Scholar]
- Zhang S, Tang C, Zhao Q, et al. 2014. Development of highly polymorphic simple sequence repeat markers using genome-wide microsatellite variant analysis in foxtail millet [Setaria italica (L.) P. Beauv]. BMC Genomics 15, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xie M, Ren G, et al. 2013. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proceedings of the National Academy of Sciences of the United States of America 110, 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary D, Hopf M, Weiss E. 2012. Domestication of Plants in the Old World: The origin and spread of domesticated plants in Southwest Asia, Europe, and the Mediterranean Basin . Oxford: Oxford University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.