Highlight
The Arabidopsis homolog of human U11-48K protein, a small nuclear ribonucleoprotein, is necessary for correct splicing of minor U12 introns, which is crucial for normal plant growth and development.
Key words: Arabidopsis thaliana, minor spliceosome, plant development, splicing, U12 intron, U11-48K.
Abstract
The minor U12 introns are removed from precursor mRNAs by the U12 intron-specific minor spliceosome. Among the seven ribonucleoproteins unique to the minor spliceosome, denoted as U11/U12-20K, U11/U12-25K, U11/U12-31K, U11/U12-65K, U11-35K, U11-48K, and U11-59K, the roles of only U11/U12-31K and U11/U12-65K have been demonstrated in U12 intron splicing and plant development. Here, the functional role of the Arabidopsis homolog of human U11-48K in U12 intron splicing and the development of Arabidopsis thaliana was examined using transgenic knockdown plants. The u11-48k mutants exhibited several defects in growth and development, such as severely arrested primary inflorescence stems, formation of serrated leaves, production of many rosette leaves after bolting, and delayed senescence. The splicing of most U12 introns analyzed was impaired in the u11-48k mutants. Comparative analysis of the splicing defects and phenotypes among the u11/u12-31k, u11-48k, and u11/12-65k mutants showed that the severity of abnormal development was closely correlated with the degree of impairment in U12 intron splicing. Taken together, these results provide compelling evidence that the Arabidopsis homolog of human U11-48K protein, as well as U11/U12-31K and U11/U12-65K proteins, is necessary for correct splicing of U12 introns and normal plant growth and development.
Introduction
In higher eukaryotes, two distinct spliceosomes catalyze the splicing of introns from precursor mRNAs (pre-mRNAs), which is crucial for regulation of gene expression in cells (Burge et al., 1999). The splicing of major U2-type introns is catalyzed by the major spliceosome (Moore et al., 1993; Staley and Guthrie, 1998), while the splicing of minor U12-type introns is catalyzed by the minor spliceosome (Tarn and Steitz, 1997; Patel and Steitz, 2003; Zhu and Brendel, 2003; Will and Lührmann, 2005; Turunen et al., 2013). Although the minor U12 introns constitute <1% of all introns found in eukaryotes (Burge et al., 1998; Levine and Durbin, 2001; Zhu and Brendel, 2003; Sheth et al., 2006; Alioto, 2007), the importance of U12 intron splicing has been demonstrated in diverse cellular processes in many organisms, including embryonic development in Drosophila and zebra fish (Otake et al., 2002; König et al., 2007; Markmiller et al., 2014), a developmental disorder known as microcephalic osteodysplastic primordial dwarfism 1 and myelodysplastic syndrome in humans (Jafarifar et al., 2014; Madan et al., 2015), isolated familial growth hormone deficiency in humans (Argente et al., 2014), and nonsense-mediated mRNA decay (Zhu and Brendel, 2003; Hirose et al., 2004).
Assembly of the spliceosome during the splicing process requires the formation of protein–RNA, protein–protein, and RNA–RNA complexes. The major spliceosome consists of U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins (snRNPs), while the minor spliceosome consists of U11, U12, U5, U4atac, and U6atac snRNPs (Hall and Padgett, 1996; Tarn and Steitz, 1996a, b; Kolossova and Padgett, 1997; Incorvaia and Padgett, 1998; Will and Lührmann, 2005). More than 200 proteins are found in both the major and minor spliceosome complexes. In addition to the many proteins present in both spliceosomes, seven proteins unique to the minor spliceosome, denoted as U11/U12-20K, U11/U12-25K, U11/U12-31K, U11/U12-65K, U11-35K, U11-48K, and U11-59K, have been identified in animals and plants (Will et al., 2004; Lorković et al., 2005). In plants, these proteins were found to be highly conserved in dicotyledons and monocotyledons (Will et al., 2004; Lorković et al., 2005; Russell et al., 2006). These minor spliceosome proteins are known to associate with the small nuclear RNAs (snRNAs), denoted as U11, U12, and U4atac/U6atac snRNAs (Will et al., 2004), a process which is necessary for recognition of the 5' splice site, 3' splice site, and branch-point site during spliceosome formation (Tarn and Steitz, 1996b; Frilander and Steitz, 1999; Turunen et al., 2013). It was previously demonstrated that human U11/U12-65K protein interacts with U12 snRNA and recognizes branch-point sites (Benecke et al., 2005). A recent study also demonstrated that Arabidopsis (Arabidopsis thaliana) U11/U12-65K interacts with U12 snRNA, which is necessary for U12 intron splicing and plant development (Jung and Kang, 2014). Through functional analysis in Arabidopsis and rice (Oryza sativa), U11/U12-31K was found to be indispensable for U12 intron splicing, and crucial for the normal development of dicot and monocot plants (Kim et al., 2010; Kwak et al., 2012). All of these studies clearly demonstrate that the proteins unique to the minor spliceosome complex are necessary for U12 intron splicing and development in both animals and plants.
Although the aforementioned studies clearly indicate the importance of minor spliceosome proteins in U12 intron splicing and development of plants, the significance of most minor spliceosome proteins has not yet been proven experimentally. The Arabidopsis homolog of human U11-48K (At3g04160), investigated in the present study, is one of the proteins unique to the minor spliceosome complex. It was previously demonstrated that human U11-48K protein interacts with the U11 snRNA and recognizes the 5' splice site of U12 introns, which is important for U12-type splicing and cell growth (Turunen et al., 2008; Tidow et al., 2009). However, the cellular role of the Arabidopsis homolog of human U11-48K in the splicing of U12 introns, and the consequences of U12 intron splicing in plant growth and development, has not yet been determined. In this study, we provide evidence that the Arabidopsis homolog of human U11-48K is necessary for the correct splicing of U12 introns, which is crucial for normal growth and development of plants.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana Columbia-0 ecotype, u11/u12-31k mutant (Kim et al., 2010), u11/u12-65k mutant (Jung and Kang, 2014), and u11-48k mutant (this study) plants were grown at 23 °C under long-day conditions (16h light/8h dark cycle) either in soil or in half-strength Murashige and Skoog medium containing 1% sucrose. For the construction of artificial miRNA (amiRNA)-mediated mutant plants, two amiRNAs with different target sites (Fig. 1A; Supplementary Fig. S3A at JXB online) were designed using the Web MicroRNA Designer (http://wmd3.weigelworld.org/) as previously described (Kim et al., 2010). The amiRNAs targeting the U11-48K gene were cloned into the pBI121 vector that expresses amiRNA under the control of the Cauliflower mosaic virus 35S promoter. The pBI121 vector was transformed into Arabidopsis by vacuum infiltration (Bechtold and Pelletier, 1998) using Agrobacterium tumefaciens GV3101. The T3 homozygous lines were selected and used for phenotype analysis.
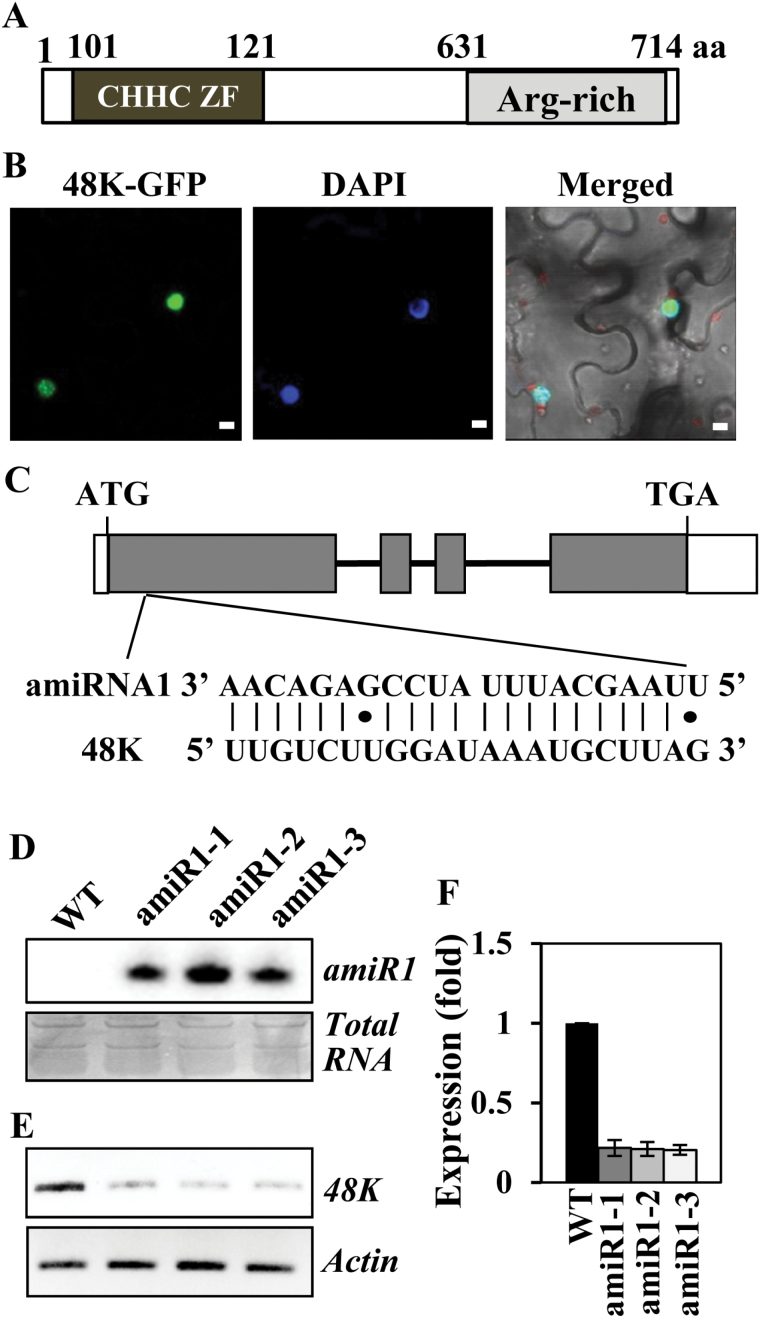
Fig. 1.
Domain structure and cellular localization of the Arabidopsis homolog of human U11-48K protein and generation of artificial miRNA-mediated knockdown plants. (A) Schematic representation of the domain structure of the Arabidopsis homolog of human U11-48K. The conserved CHHC-type zinc finger (ZF) motif and arginine (Arg)-rich region are shown. (B) GFP signals from the 48K–GFP-expressing tobacco plant were observed using a confocal microscope. DAPI was used to stain the nucleus. Scale bar=10 μm. (C) Position of the artificial miRNA1 (amiR1) target site and the sequences of amiR1, along with its target, U11-48K (48K). Exons and introns are represented as gray boxes and thick lines, respectively, and the untranslated regions are represented as white boxes. (D) Confirmation of mature amiR1 generation. Total RNA extracted from each transgenic line (amiR1-1, amiR1-2, and amiR1-3) was separated via denaturing 12% PAGE, and the expression of 21 nucleotide long mature amiR1 in each line was confirmed by northern blotting. (E, F) Down-regulation of U11-48K in the transgenic plants. The levels of U11-48K in each transgenic plant were confirmed by (E) RT–PCR and (F) real-time RT–PCR analysis. The numbers 1, 2, and 3 in (F) indicate amiR1-1, amiR1-2, and amiR1-3, respectively. Values are means ±SE obtained from three independent biological replicates. (This figure is available in colour at JXB online.)
Analysis of cellular localization of the Arabidopsis homolog of human U11-48K
To determine the cellular localization of the Arabidopsis homolog of human U11-48K protein, the cDNA encoding a full-length U11-48K was amplified with gene-specific primers (forward primer, 5' ATGGATCGACCACCGTCGTTG 3'; and reverse primer, 5' TCACTCTTTTTCTGTTGGTATG 3'), and ligated in front of a green fluorescent protein (GFP) gene in the Cassava vein mosaic virus CsV-GFP3-PA vector, which expresses the fusion protein under the control of the CsV promoter, and the resulting vector was infiltrated into tobacco leaves using A. tumefaciens GV3101. The GFP signals in the leaves of the tobacco plants were observed using a Zeiss LSM510 laser scanning confocal microscope (Carl Zeiss, http://www.zeiss.com/microscopy/) with the excitation and emission wavelengths of 488nm and 505nm, respectively. The nuclei were stained with DAPI (4',6-diamidino-2-phenylindole), and the signals were observed with the excitation and emission wavelengths of 360nm and 460nm, respectively.
Reverse transcription–PCR (RT–PCR) and northern blot analysis
To determine the expression levels of target genes, total RNA was extracted from the frozen tissues using the Plant RNeasy extraction kit (Qiagen, http://www.qiagen.com), and 200ng of RNA was reverse transcribed and amplified using the One-step RT-PCR kit (Qiagen) with the gene-specific primers listed in Supplementary Table S1. To detect 21-mer mature amiRNAs in the transgenic plants, 20 μg of total RNA was separated via denaturing 12% PAGE and transferred to a nylon membrane, as previously described (Kim et al., 2010; Jung and Kang, 2014). RNA blots were hybridized with a radiolabeled probe complementary to the amiRNA, and the signals were detected using a FLA7000 Phosphorimager (GE Healthcare Life Sciences, http://www.gelifesciences.com).
Analysis of the splicing patterns of U12 intron-containing genes
Total RNA was extracted from 3-week-old wild-type and amiRNA mutant plants. The RNA samples were treated with RQ1 DNase (Promega, http://www.promega.com) and further purified using an RNeasy clean-up kit (Qiagen). RT–PCR analysis of the splicing patterns was conducted essentially as described previously (Kim et al., 2010; Jung and Kang, 2014). Briefly, 200ng of RNA was amplified using a One-step RT-PCR kit (Qiagen) with gene-specific primers that were designed to form base pairs with the 5' end of the first exon and the 3' end of the last exon in each gene (Kim et al, 2010; Supplementary Table S1). The PCR products were separated on a 1–2% agarose gel and visualized under UV light. The identities of the PCR products were verified by cloning and sequencing.
Quantitative analysis of splicing efficiency
The splicing efficiency of U12 introns was analyzed by quantitative real-time RT–PCR as described (Koprivova et al., 2010; Gu et al., 2014). The gene-specific oligonucleotide primers corresponding to the exon/exon regions were designed to amplify the spliced (mature) transcripts, and the primers corresponding to the intron/exon regions were designed to amplify the unspliced (precursor) transcripts (Supplementary Table S2). A 100ng aliquot of total RNA was analyzed using a QuantiTect SYBR RT-PCR kit (Qiagen) with the gene-specific primers in a Rotor-Gene Q real-time thermal cycling system (Qiagen).
Results
Structural features and characterization of Arabidopsis homolog of human U11-48K
U11-48K proteins harbor a CHHC-type zinc finger motif at the N-terminal half and an arginine-rich region at the C-terminal half (Fig. 1A). Arabidopsis homolog of human U11-48K protein is comprised of 714 amino acids, in contrast to the human U11-48K protein which contains only 339 amino acids (Supplementary Table S3). A large difference in the length of the U11-48K proteins in human and Arabidopsis has already been documented, which demonstrates that the difference is mainly due to a long amino acid extension at the N-terminus of the Arabidopsis protein (Lorkovic et al., 2005). The previous report also demonstrated that although low sequence homology was observed between the 48K proteins, conserved amino acids were found throughout the entire length of the protein, indicating that they are true orthologs of the U11-48K proteins (Lorkovic et al., 2005). Here, we analyzed in more detail structural features of plant U11-48K proteins. Unlike animal U11-48K proteins that are comprised of 330–340 amino acids and share ~90% amino acid sequence similarity with each other, the length and amino acid sequences of plant U11-48K proteins tend to be more diverse (Supplementary Table S3). Analysis of the amino acid sequences revealed that U11-48K proteins in dicotyledonous plants contained ~700 amino acids, whereas those in monocotyledonous plants were comprised of ~400–600 amino acids (Supplementary Table S3). Sequence comparison indicated that the sequence similarity of U11-48K proteins among dicot plants was ~30–80%, whereas monocot plants share ~50–90% similarity (Supplementary Table S3). Comparison between dicots and monocots revealed amino acid sequence similarity of ~30%, while the similarities between plants and humans were <25% (Supplementary Table S3). Notably, the length and amino acid sequences of U11-48K proteins were more diverse than those of other plant U11/U12 proteins, which share ~60% sequence similarity among dicot and monocot plant species (Supplementary Table S3). Although diverse sequence similarity was observed for the entire peptide of the U11-48K proteins, all harbored a well-conserved CX5HX9HX3C-type zinc finger motif at the N-terminal half and an arginine-rich region at the C-terminal half (Supplementary Fig. S1). Importantly, the human U11-48K protein also harbors a CX5HX9HX3C-type zinc finger motif at the N-terminal half, which is necessary for the specific binding to the 5' splice site of U12-type introns (Tidow et al., 2009). Our present analysis and previous reports indicate that plant U11-48K proteins are true orthologs of the U11-48K proteins. However, considering the lack of direct experimental evidence to support the binding of plant U11-48K protein to the 5' splice site of U12 introns, we named it Arabidopsis homolog of human U11-48K. To determine the subcellular localization of the Arabidopsis homolog of human U11-48K, the vector expressing the 48K–GFP fusion protein was transiently expressed in tobacco leaves, and the cellular expression of the 48K–GFP fusion protein was examined via confocal analysis. The results showed that the GFP signals were exclusively observed in the nucleus (Fig. 1B), indicating that Arabidopsis homolog of human U11-48K protein is localized to the nucleus.
Arabidopsis homolog of human U11-48K plays an essential role in the normal growth and development of Arabidopsis
To determine the functional roles of Arabidopsis homolog of human U11-48K during plant growth and development, we first tried to obtain homozygous T-DNA insertion mutant lines. Two mutant lines, SALK_142557C and SALK_106445C in which the T-DNA was inserted into the first exon and the fourth exon of the U11-48K gene, respectively, were obtained from the Arabidopsis Biological Resource Center and were analyzed. However, we were not able to select the homozygous allele of the mutants, suggesting that Arabidopsis homolog of human U11-48K may play an essential role in plant development. We therefore generated u11-48k knockdown mutant plants using the amiRNA-mediated knockdown method, as described previously (Schwab et al., 2006; Kim et al., 2010). Three amiRNA lines targeted to the first exon in the U11-48K gene were generated (Fig. 1C) and used for phenotype analysis. Expression of the mature 21 nucleotide long amiRNA and down-regulation of U11-48K in the mutant plants were confirmed by northern blot and RT–PCR analyses (Fig. 1D–F).
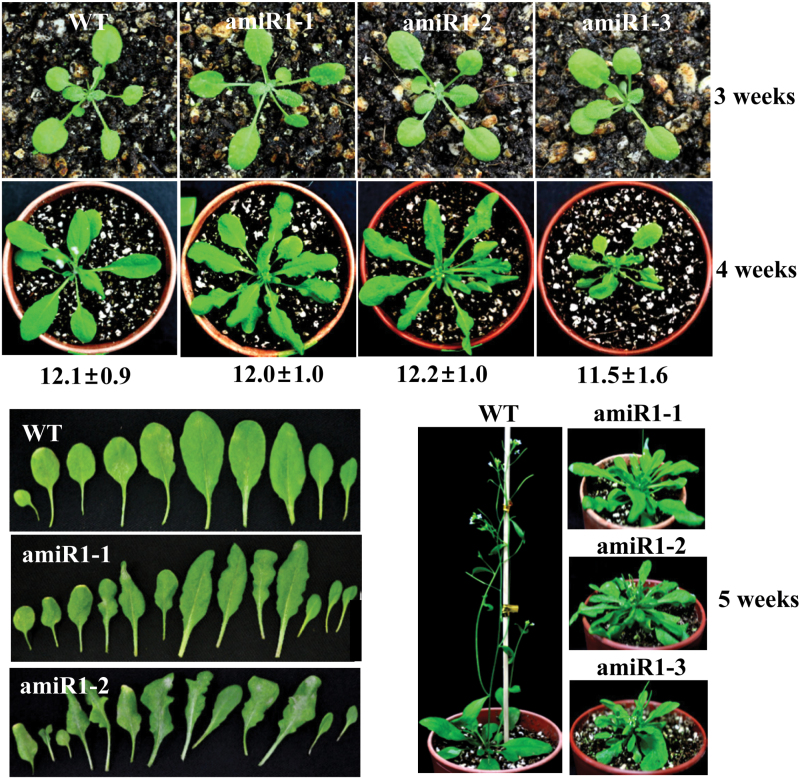
The wild-type and u11-48k mutant plants displayed similar growth with no noticeable differences during the first 3 weeks (Fig. 2). However, at a later stage of growth, the u11-48k mutant plants exhibited several defects in growth and development, such as severely arrested primary inflorescence stems, formation of serrated leaves, and production of many rosette leaves after bolting (Fig. 2). Since the u11-48k mutants exhibited severe defects in the formation of primary inflorescence stems, we carefully examined whether Arabidopsis homolog of human U11-48K is involved in flowering time control. However, no differences in leaf numbers were observed between the wild type and u11-48k mutants at the time of flowering (Fig. 2), indicating that flowering time is not affected by Arabidopsis homolog of human U11-48K. To confirm further whether the developmental-defect phenotypes observed in the u11-48k mutants resulted from the specific knockdown of the U11-48K gene, another amiRNA line targeted to a different position in the first exon of the U11-48K gene was generated, and their phenotypes were analyzed. Identical developmental-defect phenotypes were observed in the second amiR2 mutant line, comparable with the first amiR1 mutant line (Supplementary Fig. S2). Altogether, these results clearly indicate that Arabidopsis homolog of human U11-48K plays an essential role in the normal growth and development of Arabidopsis.
Fig. 2.
Development-defect phenotypes of the u11-48k mutant plants. The wild-type (WT) and artificial miRNA-mediated mutant (amiR1-1, amiR1-2, and amiR1-3) plants were grown in soil under long-day conditions, and photographs were taken on the indicated weeks. Identical results were obtained from three independent biological replicates, for which a representative example is shown. The numbers below the 4 week panel indicate the number of leaves present when floral buds appeared. Values are means ±SE obtained from three independent biological replicates, five plants per replicate. (This figure is available in colour at JXB online.)
Arabidopsis homolog of human U11-48K plays a role in senescence
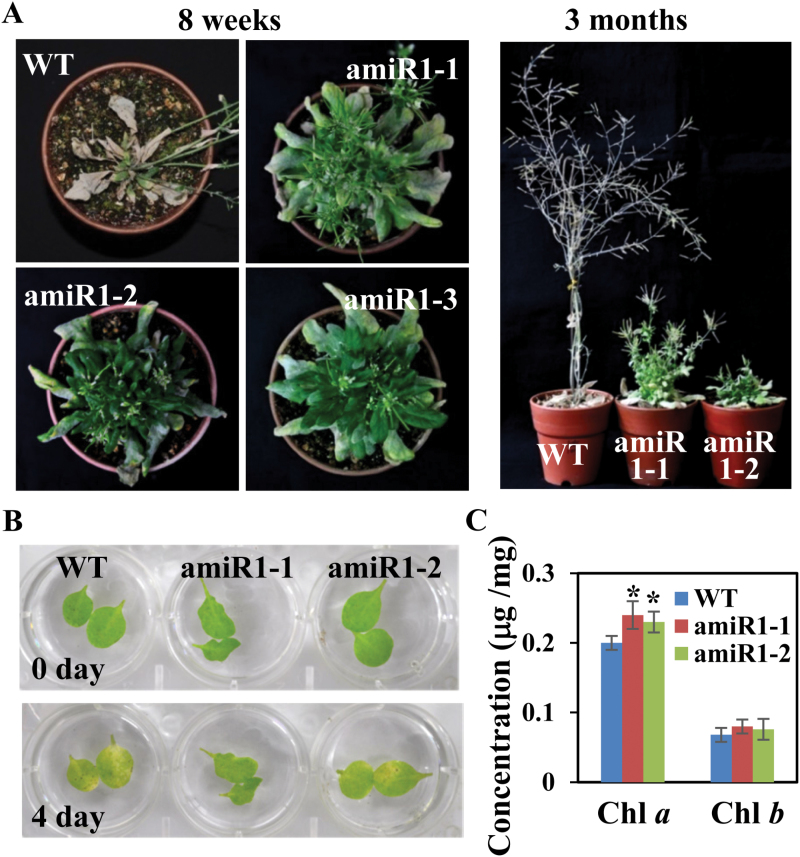
As it was evident that Arabidopsis homolog of human U11-48K is involved in the growth and development of Arabidopsis, we subsequently examined whether it is involved in senescence. When the plants were grown in soil for >8 weeks, the u11-48k mutants survived beyond the death of the wild-type plants (Fig. 3A; Supplementary Fig. S2). To examine further the possible involvement of Arabidopsis homolog of human U11-48K in senescence, the dark-induced senescence assay was performed on the leaves of 4-week-old plants. The results showed that greening of the leaves of u11-48k mutant plants was maintained for a much longer period than in the wild-type plants when incubated under dark conditions (Fig. 3B). Consequently, the Chl a contents in the u11-48k mutant plants were higher than those in the wild-type plants (Fig. 3C). These results suggest that Arabidopsis homolog of human U11-48K is positively involved in senescence.
Fig. 3.
Delayed senescence of the u11-48k mutant plants. (A) Prolonged survival of the u11-48k mutant plants (amiR1-1, amiR1-2, and amiR1-3). Identical results were obtained from three independent experiments, for which a representative example is shown. (B) Dark-induced senescence. Rosette leaves from 4-week-old wild type (WT) and u11-48k mutants were floated on water in the dark for 4 d. (C) Chlorophyll content in the leaves of each plant was measured 4 d after dark-induced senescence. Values are means ±SE obtained from three independent biological replicates, two leaves per replicate, and asterisks above the columns indicate statistically different values between WT and mutant plants (P≤0.05). (This figure is available in colour at JXB online.)
Exogenously applied gibberellic acid partially recovers the stunted stem growth in the u11-48k mutant plants
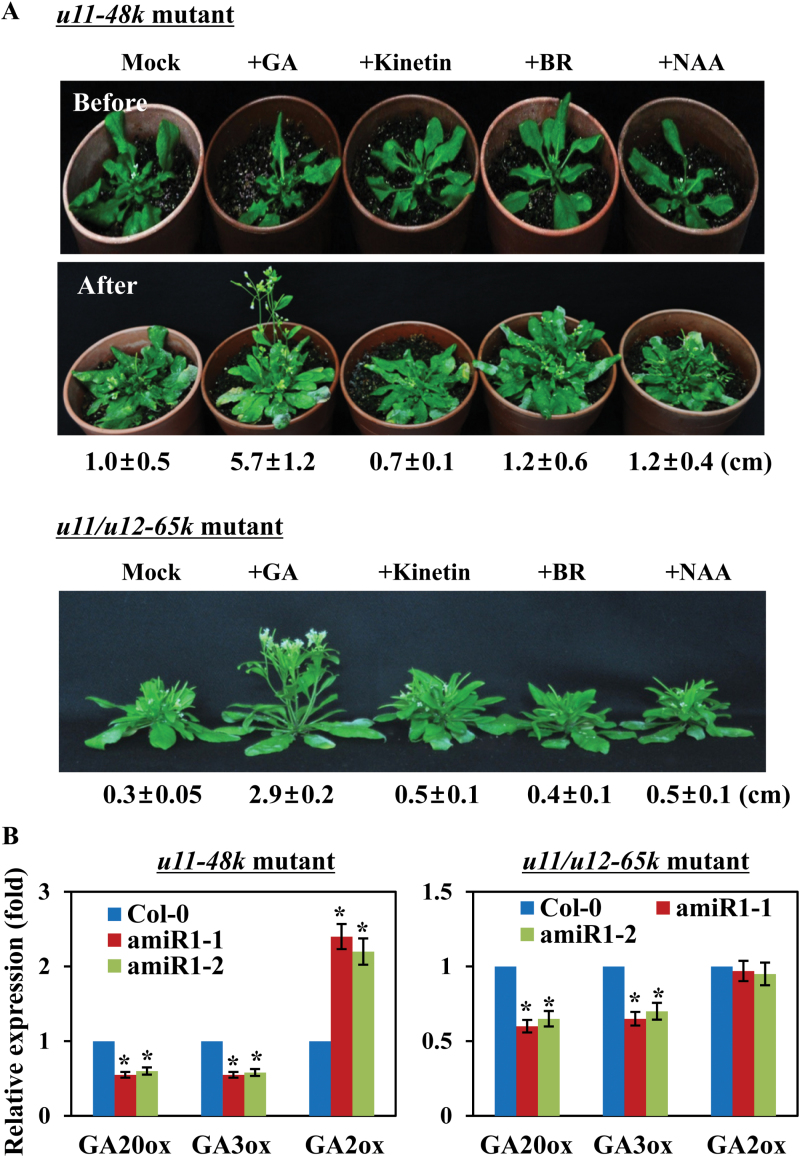
A previous study demonstrated that the stunted inflorescence stems of u11/u12-31k mutants were recovered by the application of gibberellic acid (GA) (Kim et al., 2010). To determine whether the phenotype of stunted primary inflorescence stems was related to hormone functions, the effect of exogenously applied hormones on the growth of the u11-48k mutant and u11/u12-65k mutant plants (Jung and Kang, 2014) was next examined. To accomplish this, the amiR1 mutant plants were treated with different hormones, including GA3 (100 µM), cytokinin (kinetin, 50 µM), brassinosteroid [BR (24-epibrassinolide, 5 µM], and auxin [α-naphthalene acetic acid (NAA), 0.5 µg ml–1], all of which are known to influence stem growth and development, and then the growth of primary inflorescence stems was examined. The results showed that the length of the primary inflorescence stems of the u11-48k and u11/u12-65k mutant plants had increased by ~6-fold (from 1.0cm to 5.7cm) and 10-fold (from 0.3cm to 2.9cm) 2 weeks after GA application, respectively, whereas none of the other hormones affected the growth of the primary inflorescence stems of the mutant plant (Fig. 4A). To support that the GA-mediated recovery of the stem growth is specific to the u11-48k and u11/u12-65k mutant plants, the growth of the primary inflorescence stems of the wild-type plants was examined after application of each hormone. Notably, none of the hormones affected the growth of the primary inflorescence stems of the wild-type plants (Supplementary Fig. S3). These results suggest that the abnormal growth and stunted inflorescence stems of the u11-48k and u11/u12-65k mutants as well as the u11/u12-31k mutants are partially related to the defect in GA biosynthesis.
Fig. 4.
Effect of exogenously applied hormones on the stem length of the u11-48k and u11/u12-65k mutant plants. (A) Four-week-old mutant plants were treated with 100 µM GA, 50 µM kinetin, 5 µM BR, or 0.5 µg ml–1 NAA, and stem lengths of the plants were measured 2 weeks after the application of each hormone. Values are means ±SE obtained from three independent experiments, five plants per experiment. (B) The transcript levels of the genes involved in GA metabolism were determined in the meristemic region of the mutant plants. Values are means ±SE obtained from three independent biological replicates, and asterisks above the columns indicate statistically different values between wild-type and mutant plants (P≤0.05). (This figure is available in colour at JXB online.)
Because the inflorescence stem growth of the u11-48k and u11/u12-65k mutants is partially recovered by exogenously applied GA, we next determined whether the genes involved in GA metabolism are disturbed in the knockdown plants. We investigated, via real-time RT–PCR analysis using the primers listed in Supplementary Table S2, the expression levels of the genes encoding key enzymes involved in GA metabolism: GA20-oxidase (GA20ox) and GA3-oxidase (GA3ox) involved in GA biosynthesis, and GA2-oxidase (GA2ox) involved in GA catabolism. Because exogenously applied GA influences the growth of primary inflorescence stems, we specifically wanted to determine the expression levels of these genes in meristemic regions of the plants. When the u11-48k and u11/u12-65k mutant plants began to bolt, the stems, leaves, and roots of the plants were removed and only the meristem-containing regions were collected for RNA extraction and subsequent analysis. Notably, the transcript levels of the genes involved in GA biosynthesis (GA20ox and GA3ox) were significantly down-regulated in the u11-48k and u11/u12-65k mutants compared with the wild-type plants (Fig. 4B). The transcript level of the gene involved in GA catabolism (GA2ox) was markedly up-regulated in the u11-48k mutants, but was not noticeably reduced in the u11/u12-65k mutants (Fig. 4B). A large decrease in the transcript levels of GA biosynthesis-related genes together with a significant increase in the transcript level of the GA catabolism-related gene in the u11-48k mutants and a marked decrease in the transcript levels of GA biosynthesis-related genes in the u11/u12-65k mutants suggest that the amounts of GA in both mutants may be reduced compared with those in the wild-type plants, which is consistent with the observation that exogenously applied GA partially recovers the stunted stem growth in both mutant plants (Fig. 4A). To examine whether the genes encoding GA20ox, GA3ox, and GA2ox contain U12 introns, and if their splicing patterns are altered in the mutant plants, the intron sequences in these GA metabolism-related genes were analyzed. None of the GA20ox, GA3ox, and GA2ox genes contained U12 introns (the U12-type intron database; http://genome.crg.es/cgi-bin/u12db/u12db.cgi; Supplementary Fig. S4), and splicing patterns of GA20ox, GA3ox, and GA2ox genes were not altered in the mutants compared with those of the wild-type plants (Supplementary Fig. S4). These results suggest that the defect in GA biosynthesis is responsible, at least in part, for abnormal growth and stunted inflorescence stems of the u11-48k and u11/u12-65k knockdown plants.
Arabidopsis homolog of human U11-48K is essential for correct splicing of U12 introns
To ascertain whether the abnormal growth and development of the u11-48k mutant plants were due to defects in the splicing of U12-type introns, the splicing patterns of U12-type introns were investigated in the u11-48k mutants at the time of bolting. Arabidopsis harbors ~230 U12-type intron-containing genes (the U12-type intron database; http://genome.crg.es/cgi-bin/u12db/u12db.cgi; Jung and Kang, 2014). Among them, those known to be conserved across organisms and whose putative functions have been clearly identified were chosen, and their splicing patterns were analyzed by RT–PCR as previously described (Kim et al., 2010; Jung and Kang, 2014). The splicing of all U12 introns examined in this study, including HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE 2 (HSI2), Quartre-QuarT 1 (QQT1), NHX5, NHX6, GSH2, RGTB2, and drought induced 19-2 (Di19-2), was found to be defective in the u11-48k mutant plants (Fig. 5; Supplementary Fig. S5). These results indicate that the developmental defects observed in u11-48k mutant plants were due to impairment in U12 intron splicing.
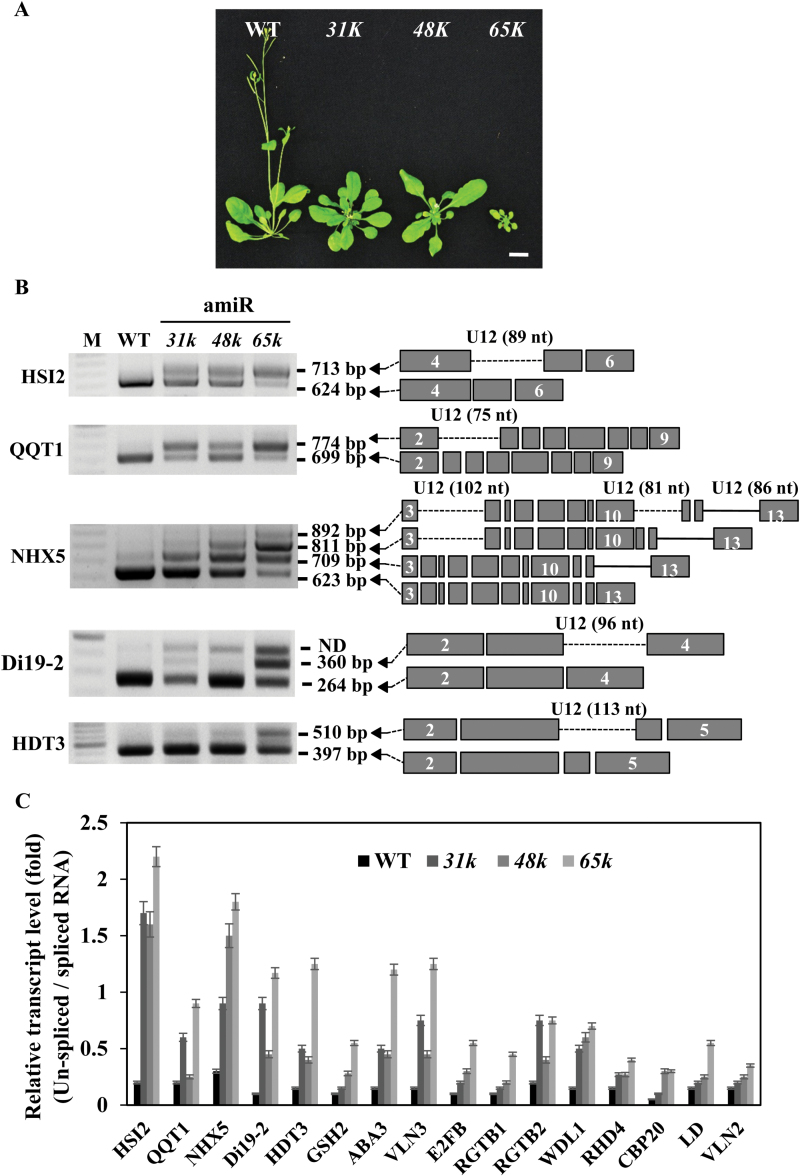
Fig. 5.
Comparison of phenotype and abnormal splicing of U12 introns in the u11/u12-31k, u11-48k, and u11/u12-65k mutant plants. (A) Growth-defect phenotypes of the u11/u12-31k (31k), u11-48k (48k), and u11/u12-65k (65k) mutant plants. Scale bar=1cm. (B) The splicing patterns of selected U12 intron-containing transcripts were analyzed by RT–PCR in wild type (WT), 31k, 48k, and 65k mutant plants. All primers used were designed to form base pairs with the 5' end of the first exon and the 3' end of the last exon in each gene. Identical results were obtained from three independent experiments, for which a representative example is shown. (C) Relative levels of spliced and unspliced transcripts of U12 intron-containing genes in the u11/u12-31k, u11-48k, and u11/u12-65k mutant plants were analyzed by quantitative real-time RT–PCR, and the levels of the unspliced forms relative to those of the spliced forms are expressed in each genotype. Data are the means ±SE obtained from three independent biological replicates. (This figure is available in colour at JXB online.)
The severity of developmental-defect phenotypes of the mutants is closely related to the degree of impairment in U12 intron splicing
Given the essential roles of U11/U12-31K (Kim et al., 2010), U11/U12-65K (Jung and Kang, 2014), and Arabidopsis homolog of human U11-48K (this study) in U12 intron splicing as well as normal growth and development of plants, it would be interesting to determine whether the severity of developmental-defect phenotypes of each mutant is related to the degree of impairment in U12 intron splicing. To address this question, the splicing patterns of the U12 introns in u11/u12-31k, u11-48k, and u11/u12-65k mutant plants were analyzed in parallel. Growth of the u11/u12-65k mutant was observed to be more severely retarded than that of u11/u12-31 and u11-48k mutants (Fig. 5A). Notably, splicing of all U12 introns analyzed was impaired only in the u11/u12-65k mutant, whereas splicing of several U12 introns examined in this study was not affected in u11/u12-31k and u11-48k mutants. Moreover, the degree of impairment in U12 intron splicing was indeed related to the severity of the developmental-defect phenotypes. The intensities of pre-mRNAs were much stronger in the u11/u12-65k mutant than in both the u11/u12-31 and u11-48k mutants. Consequently, the intensities of mature mRNAs were much weaker in the u11/u12-65k mutant (Fig. 5B; Supplementary Fig. S6), demonstrating that the defects in U12 intron splicing were much more significant in the u11/u12-65k mutant than in the u11/u12-31 and u11-48k mutants. To confirm further the differences in the defects in U12 intron splicing among different mutant plants, the levels of mature (spliced) and precursor (unspliced) RNAs were quantitatively determined by real-time RT–PCR analysis. The results showed that the relative levels of unspliced RNAs in the u11/u12-65k mutants were much higher than those in the u11/u12-31 and u11-48k mutants (Fig. 5C). To verify that the mutation of U11/U12-31K, U11-48K, or U11/U12-65K does not affect the splicing of U2-type introns, the splicing patterns of selected U2 intron-containing transcripts were analyzed by RT–PCR in the mutant plants. Clearly, splicing of all U2 intron genes tested was not altered in the mutant plants (Supplementary Fig. S7), confirming that splicing of only U12-type introns was affected in the mutant plants. Altogether, these results indicate that the severity of developmental defects in the mutants is indeed closely correlated with the degree of impairment in U12 intron splicing, reinforcing the conclusion that the correct splicing of U12 introns is essential for the normal growth and development of plants.
Discussion
The present study clearly demonstrated that Arabidopsis homolog of human U11-48K protein, the minor spliceosomal protein which interacts with the U11 snRNA and recognizes the 5' splice site of U12 introns in humans (Turunen et al., 2008; Tidow et al., 2009), is indispensable for correct U12 intron splicing, and crucial for the normal growth and development of plants. Similar to U11/U12-31K and U11/U12-65K (Kim et al., 2010; Jung and Kang, 2014), Arabidopsis homolog of human U11-48K was observed to affect plant development after the bolting stage rather than during the vegetative stage (Fig. 2). The present finding of the essential role of Arabidopsis homolog of human U11-48K in plant development further reinforces the previously determined roles of the minor spliceosomal proteins in cell proliferation and viability of plants, as well as in animals. As U12 introns are often found in genes involved in information processes, such as DNA repair, RNA processing, and translation (Burge et al., 1998; Otake et al., 2002; Will et al., 2004; König et al., 2007), it is clear that tight regulation of U12 intron splicing by Arabidopsis homolog of human U11-48K, as well as U11/U12-31K and U11/U12-65K, is essential for the normal development of living organisms which harbor U12-type introns.
Despite the increasing amount of information on the role of Arabidopsis homolog of human U11-48K, U11/U12-31K, and U11/U12-65K in U12 intron splicing and plant growth and development, it remains unknown at present whether the observed developmental defects in these mutant plants were caused by the abortion of splicing of all U12-type introns. It is difficult to interpret the importance and exact role of U12 intron-containing genes in plant growth and development due to the fact that >50 of the putative U12 intron-containing genes encode proteins whose biological and molecular functions are as yet unknown. In addition, as previously observed in u11/u12-65k mutant plants, defects in U12 intron splicing affect diverse alternative splicing, including exon skipping, intron retention, alternative 3' sites, and alternative 5' sites (Jung and Kang, 2014). Although it is still premature to link the observed development-defect phenotypes directly to impairment in the splicing of specific U12 introns, it is likely that cumulative defects in the splicing of growth- and development-related U12 intron-containing genes, such as POD, which regulates early embryo patterning (Li et al., 2011), QQT1, which is associated with microtubules during cell division (Lahmy et al., 2007), HSI2, which is involved in seedling growth (Tsukagoshi et al., 2007), and the Di19 gene family, which is involved in stress and light signaling pathways (Milla et al., 2006), are responsible for the abnormal development phenotypes observed in these mutant plants. Notably, the comparative analysis carried out herein on the splicing defects and abnormal development phenotypes among the u11/u12-31k, u11-48k, and u11/12-65k mutants revealed that the severity of abnormal development was closely correlated with the degree of impairment in U12 intron splicing (Fig. 5). These results reinforce the proposition that cumulative defects in the splicing of U12 intron-containing genes result in abnormal growth and development of the mutant plants.
The process of intron splicing occurs through spliceosome assembly, splicing, and subsequent spliceosome disassembly (Staley and Guthrie, 1998, 1999), during which interactions between spliceosome-associated proteins and snRNAs are required. It has been demonstrated from studies on animal systems that U11-48K protein interacts with the U11 snRNA and recognizes the 5' splice site of U12 introns (Turunen et al., 2008; Tidow et al., 2009). The U11/U12-65K protein was shown to interact with U12 snRNA and recognize the branch-point sites of U12 introns during minor spliceosome assembly in humans and Arabidopsis (Benecke et al., 2005; Jung and Kang, 2014). As a key component of the core complexes between U11/U12-65K, -59K and -48K proteins found in plants and animals (Tidow et al., 2009; Jung and Kang, 2014), the U11-48K protein is necessary for spliceosome assembly and U12 intron splicing in plants, as well as in animals.
In conclusion, our findings clearly demonstrate that Arabidopsis homolog of human U11-48K is an indispensable spliceosomal protein, crucial for U12 intron splicing and the normal growth and development of plants. As evidenced by the close correlation of the severity of abnormal development among the u11/u12-31k, u11-48k, and u11/12-65k mutants with the degree of impairment in U12 intron splicing, it is likely that cumulative defects in the splicing of U12 intron-containing genes resulted in the abnormal growth and development observed in mutant plants. Future research should focus on analyzing whether the observed development-defect phenotypes are due to the impairment in the splicing of specific U12 introns or a subset of U12 introns in cells.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Amino acid sequence comparison of U11-48K proteins in diverse plant species.
Figure S2. Development-defect phenotypes of the u11-48k mutant plants.
Figure S3. Effect of exogenously applied hormones on the stem length of the wild-type plants.
Figure S4. Splicing of GA20ox, GA3ox, and GA2ox genes in the mutant plants.
Figure S5. Abnormal splicing of U12 introns in the u11-48k mutant plants.
Figure S6. Comparison of abnormal splicing of U12 introns in the u11/u12-31k, u11-48k, and u11/u12-65k mutant plants.
Figure S7. Splicing of selected U2 introns in the mutant plants.
Table S1. Gene-specific primer pairs used in RT–PCR experiments.
Table S2. Gene-specific primer pairs used in quantitative analysis of splicing efficiency by real-time RT–PCR.
Table S3. Comparison of U11-48K proteins and other U11/U12 proteins found in diverse plant species and in humans.
Acknowledgements
We thank Dr Detlef Weigel for providing the pRS300 vector. This work was supported by grants from the Mid-career Researcher Program through the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology (2011-0017357), and from the Next-Generation BioGreen21 Program (PJ01105401), Rural Development Administration, Republic of Korea.
References
- Alioto TS. 2007. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Research 35, D110–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argente J, Flores R, Gutiérrez-Arumí A, Verma B, Martos-Moreno GÁ, Cuscó I, Oghabian A, Chowen JA, Frilander MJ, Pérez-Jurado LA. 2014. Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency. EMBO Molecular Medicine 6, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. 1998. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods in Molecular Biology 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Benecke H, Lührmann R, Will CL. 2005. The U11/U12 snRNP 65K protein as a molecular bridge, binding the U12 snRNA and U11-59K protein. EMBO Journal 24, 3057–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB, Padgett RA, Sharp PA. 1998. Evolutionary fates and origins of U12-type introns. Molecular Cell 2, 773–785. [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl T, Sharp PA. 1999. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland RF, Cech RT, Atkins C, eds. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 525–560. [Google Scholar]
- Frilander MJ, Steitz JA. 1999. Initial recognition of U12-dependent introns requires both U11/5' splice-site and U12/branchpoint interactions. Genes and Development 13, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Xu T, Lee K, Kang H. 2014. A chloroplast-localized DEAD-box RNA helicase AtRH3 is essential for intron splicing and plays an important role in the growth and stress response in Arabidopsis thaliana . Plant Physiology and Biochemistry 82, 309–318. [DOI] [PubMed] [Google Scholar]
- Hall SL, Padgett RA. 1996. Requirement of U12 snRNA for in vivo slicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 271, 1716–1718. [DOI] [PubMed] [Google Scholar]
- Hirose T, Shu M-D, Steitz JA. 2004. Splicing of U12-type introns deposits an exon junction complex competent to induce nonsense-mediated mRNA decay. Proceedings of the National Academy of Sciences, USA 101, 17976–17981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incorvaia R, Padgett RA. 1998. Base pairing with U6atac snRNA is required for 5' splice site activation of U12-dependdent introns in vivo . RNA 4, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarifar F, Dietrich RC, Hiznay JM, Padgett RA. 2014. Biochemical defects in minor spliceosome function in the developmental disorder MOPD I. RNA 20, 1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HJ, Kang H. 2014. The Arabidopsis U11/U12-65K is an indispensable component of minor spliceosome and plays a crucial role in U12 intron splicing and plant development. The Plant Journal 78, 799–810. [DOI] [PubMed] [Google Scholar]
- Kim WY, Jung HJ, Kwak KJ, Kim MK, Oh SH, Han YS, Kang H. 2010. The Arabidopsis U12-type spliceosomal protein U11/U12-31K is involved in U12 intron splicing via RNA chaperone activity and affects plant development. The Plant Cell 22, 3951–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolossova I, Padgett RA. 1997. U11 snRNA interacts in vivo with the 5' splice site of U12-dependent (AU–AC) pre-mRNA introns. RNA 3, 227–233. [PMC free article] [PubMed] [Google Scholar]
- König H, Matter N, Bader R, Thiele W, Müller F. 2007. Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation. Cell 131, 718–729. [DOI] [PubMed] [Google Scholar]
- Koprivova A, Colas des Francs-Small C, Calder G, Mugford ST, Tanz S, Lee BR, Zechmann B, Small I, Kopriva S. 2010. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. Journal of Biological Chemistry 285, 32192–32199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak KJ, Jung HJ, Lee KH, Kim YS, Kim WY, Ahn SJ, Kang H. 2012. The minor spliceosomal protein U11/U12-31K is an RNA chaperone crucial for U12 intron splicing and the development of dicot and monocot plants. PLoS One 7, e43707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy S, Guilleminot J, Schmit AC, Pelletier G, Chaboute ME, Devic M. 2007. QQT proteins colocalize with microtubules and are essential for early embryo development in Arabidopsis . The Plant Journal 50, 615–626. [DOI] [PubMed] [Google Scholar]
- Levine A, Durbin R. 2001. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Research 29, 4006–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, Xue Y, Jia D-J, Wang T, Hi D-Q, Liu J, Cui F, Xie Q, Ye D, Yang WC. 2011. POD1 regulates pollen tube guidance in response to micropylar female signaling and acts in early embryo patterning in Arabidopsis. The Plant Cell 23, 3288–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ, Lehner R, Forstner R, Barta A. 2005. Evolutionary conservation of minor U12-type spliceosome between plants and humans. RNA 11, 1095–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V, Kanojia D, Li J, et al. 2015. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nature Communications 6, 6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Cloonan N, Lardelli RM, et al. 2014. Minor class splicing shapes the zebrafish transcriptome during development. Proceedings of the National Academy of Sciences, USA 111, 3062–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla M, Townsend J, Chang IF, Cushman JC. 2006. The Arabidopsis AtDi19 gene family encodes a novel type of Cys2/His2 zinc-finger protein implicated in ABA-independent dehydration, high-salinity stress and light signaling pathways. Plant Molecular Biology 61, 13–30. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA. 1993. Splicing of precursors to mRNAs by the spliceosomes. In The RNA world. In: Gesteland RF, Cech RT, Atkins C, eds. The RNA world Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 303–357. [Google Scholar]
- Otake LR, Scamborova P, Hashimoto C, Steitz JA. 2002. The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila . Molecular Cell 9, 439–446. [DOI] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. 2003. Splicing double: insights from the second spliceosome. Nature Reviews Molecular Cell Biology 4, 960–970. [DOI] [PubMed] [Google Scholar]
- Russell AG, Charette JM, Spencer DF, Gray MW. 2006. An early evolutionary origin for the minor spliceosome. Nature 443, 863–866. [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. 2006. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell 18, 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. 2006. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Research 34, 3955–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1999. An RNA switch at the 5' splice site requires ATP and the DEAD box protein Prp28p. Molecular Cell 3, 55–64. [DOI] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. 1996. a. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT–AC) intron in vitro . Cell 84, 801–811. [DOI] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. 1996. b. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT–AC introns. Science 273, 1824–1832. [DOI] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. 1997. Pre-mRNA splicing: the discovery of a new spliceosome doubles the challenge. Trends in Biochemical Sciences 22, 132–137. [DOI] [PubMed] [Google Scholar]
- Tidow H, Andreeva A, Rutherford TJ, Fersht AR. 2009. Solution structure of the U11-48K CHHC zinc-finger domain that specifically binds the 5' splice site of U12-type introns. Structure 17, 294–302. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Morikami A, Nakamura K. 2007. Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proceedings of the National Academy of Sciences, USA 104, 2543–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen JJ, Niemelä EH, Verma B, Frilander MJ. 2013. The significant other: splicing by the minor spliceosome. Wiley Interdisciplinary Reviews. RNA 4, 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen JJ, Will CL, Grote M, Lührmann R, Frilander MJ. 2008. The U11-48K protein contacts the 5' splice site of U12-type introns and the U11-59K protein. Molecular Cell Biology 28, 3548–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2005. Splicing of a rare class of introns by the U12-dependent spliceosome. Journal of Biological Chemistry 386, 713–724. [DOI] [PubMed] [Google Scholar]
- Will CL, Schneider C, Hossbach M, Urlaub H, Rauhut R, Elbashir S, Tuschl T, Lührmann R. 2004. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10, 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Brendel V. 2003. Identification, characterization and molecular phylogeny of U12-dependent introns in the Arabidopsis thaliana genome. Nucleic Acids Research 31, 4561–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.