Highlight
The transcription factor WRKY22 increases susceptibility to aphids in Arabidopsis via the suppression of salicylic acid signalling.
Keywords: Arabidopsis thaliana, mechanostimulation, Myzus persicae, plant–insect interaction, plant resistance to aphids, touch, transcription factors.
Abstract
Aphids induce many transcriptional perturbations in their host plants, but the signalling cascades responsible and the effects on plant resistance are largely unknown. Through a genome-wide association (GWA) mapping study in Arabidopsis thaliana, we identified WRKY22 as a candidate gene associated with feeding behaviour of the green peach aphid, Myzus persicae. The transcription factor WRKY22 is known to be involved in pathogen-triggered immunity, and WRKY22 gene expression has been shown to be induced by aphids. Assessment of aphid population development and feeding behaviour on knockout mutants and overexpression lines showed that WRKY22 increases susceptibility to M. persicae via a mesophyll-located mechanism. mRNA sequencing analysis of aphid-infested wrky22 knockout plants revealed the up-regulation of genes involved in salicylic acid (SA) signalling and down-regulation of genes involved in plant growth and cell-wall loosening. In addition, mechanostimulation of knockout plants by clip cages up-regulated jasmonic acid (JA)-responsive genes, resulting in substantial negative JA–SA crosstalk. Based on this and previous studies, WRKY22 is considered to modulate the interplay between the SA and JA pathways in response to a wide range of biotic and abiotic stimuli. Its induction by aphids and its role in suppressing SA and JA signalling make WRKY22 a potential target for aphids to manipulate host plant defences.
Introduction
As plants are sessile organisms in often dynamically changing environments, plasticity is fundamental to survival. Transcriptional regulation plays an important role in how plants cope with environmental stimuli. In Arabidopsis approximately 50 transcription factor families have been identified, accounting for approximately 2000 genes (Guo et al., 2005; Mitsuda and Ohme-Takagi, 2009). Together with signal perception and transduction elements, these transcription factors participate in complex and dynamic networks that regulate developmental processes and responses to (a)biotic stress. Insect infestations are typical situations that require quick transcriptional reprogramming in order to mount an effective defence response. Aphids are phloem-feeding insects that manoeuvre their piercing–sucking mouthparts between cells and reach the vascular bundle without inflicting major physical damage (Minks and Harrewijn, 1989). They are vectors of many plant viruses and deprive the plant of photoassimilates. Aphids cause strong transcriptional perturbations in plants, inducing or repressing up to several thousand genes, whereas other insects such as caterpillars and cell-content feeders alter the expression of only up to several hundred genes (De Vos et al., 2005; Kusnierczyk et al., 2007; Barah et al., 2013; Dubey et al., 2013; Kerchev et al., 2013; Appel et al., 2014; Foyer et al., 2015). An open question is, however, whether these transcriptional changes lead to enhanced resistance to aphids or whether they are unsuccessful or even counter-effective modulations. Aphids are known to secrete effectors via their saliva into the apoplast and the vascular bundle (Rodriguez and Bos, 2012), and might be able to manipulate the host plant physiology for their own benefit. In this study, genome-wide association (GWA) mapping revealed WRKY22 (At4g01250) as one of the candidate genes for affecting feeding behaviour of the generalist aphid Myzus persicae (Sulzer) on Arabidopsis thaliana. WRKY22 is a member of the WRKY transcription factor family, which was discovered in the 1990s and named after its binding affinity to the W-box promoter motif (Eulgem et al., 2000). WRKY22 and its homologue WRKY29 are part of group IIe WRKYs and are both established markers of pathogen-triggered immunity (PTI). Pathogen-associated molecular patterns (PAMPs) such as flagellin, chitin and cellulysin are recognized elicitors of the mitogen-activated protein kinase (MAPK) cascade that induce WRKY22 and WRKY29 within 30min post-inoculation (Asai et al., 2002; Dong et al., 2003; Navarro et al., 2004; Mészáros et al., 2006; Thilmony et al., 2006; Schikora et al., 2011; González-Lamothe et al., 2012; Shi et al., 2015). In general, PTI results in the accumulation of reactive oxygen species and callose deposition and involves salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) signalling (Yi et al., 2014). Although the exact role of WRKY22 and WRKY29 in PTI is unknown, WRKY22 has been shown to be required for resistance to the hemibiotrophic pathogen Pseudomonas syringae (Hsu et al., 2013) and WRKY29 has been described as conferring resistance to P. syringae as well as to the necrotrophic pathogen Botrytis cinerea (Asai et al., 2002). In this study, we assessed the involvement of WRKY22 in plant resistance to M. persicae aphids and its downstream transcriptional effects.
Materials and methods
Plants and insects
A collection of 344 natural accessions of A. thaliana was obtained from the ABRC Stock Center (Baxter et al., 2010). This set was selected in a previous study to represent most intraspecific genetic variation and minimal redundancy (Platt et al., 2010), and was genotyped for 214000 single nucleotide polymorphism (SNPs) with AtSNPtile1 arrays (Atwell et al., 2010; Li et al., 2010; Horton et al., 2012). Transfer (T)-DNA lines SALK_094892 (wrky22-3) and SALK_098205 (wrky22-4), and TRANSPLANTA-inducible overexpression lines TPT_4.01250.1C and TPT_4.01250.1E were obtained from NASC (Coego et al., 2014). Seeds were cold stratified for 72h at 4 °C before they were sown in pots (5cm diameter) with pasteurized (4h at 80 °C) Arabidopsis potting soil (Lentse Potgrond, Lent, The Netherlands) in a climate room at 24±1 °C, 50–70% relative humidity, 8 h–16h light–dark photoperiod, and a light intensity of 200 μmol m−2 s−1. Homozygous T-DNA plants were selected based on PCR and harvested for seeds for subsequent experiments. The location of the T-DNA insertion was confirmed via sequencing, and abolition of WRKY22 expression was tested with RT-qPCR (Supplementary Table S1 at JXB online). Expression of WRKY22 in the TRANSPLANTA-inducible overexpression lines (Coego et al., 2014) was measured with RT-qPCR 24h after application of 10 μM oestradiol in water to the plant trays (Supplementary Table S1). Green peach aphids, M. persicae, were reared on radish, Raphanus sativus (L.), at 19 °C, 50–70% relative humidity and a 16 h–8h light–dark photoperiod.
Automated video tracking
Aphid behaviour was tracked on 344 natural accessions of Arabidopsis (n=5–6 per accession) according to the methodology of Kloth et al. (2015). One adult, wingless aphid was introduced into a well of a 96-well plate containing a leaf disc of 6mm diameter, abaxial side up, on 1% agar substrate. Wells were covered with cling film to avoid aphid escape, and 20 aphids were recorded on 20 different accessions simultaneously with a camera mounted above the plate, at 22±1 °C. EthoVision® XT 8.5 video tracking and analysis software (Noldus Information Technology bv, Wageningen, The Netherlands) was used for automated acquisition of aphid position and velocity. The number and duration of probes were subsequently calculated with the statistical computing program R (R Core Team, 2013). Leaf discs were made of intermediately aged leaves of 4- to 5-week-old Arabidopsis plants, one disc per plant. Aphid behaviour was recorded for 85min, starting at 4.5h after inoculation of the aphids. The video-tracking assay was performed in an incomplete block design with each complete replicate consisting of 18 blocks of 20 accessions. Sixty plants were screened each day across three blocks, and one replicate of the complete Hapmap collection was acquired in 6 days. An alpha design was generated with Gendex (http://designcomputing.net/gendex/) to assign accessions to each block. Five to six replicates were acquired per accession.
GWA mapping and haplotype analysis
GWA mapping was performed on the proportion of aphids making long probes (> 25min) with scan_GLS (Kruijer et al., 2015), using a kinship matrix based on all SNPs to account for population structure. SNPs with a minor allele frequency <0.05 were excluded from analysis. Block and replicate were included in the model as covariates. SNPs with −log10(P) value larger than 4 were taken as candidate loci. Generalized heritability was estimated as in Oakey et al. (2006). For haplotype analysis, SNPs with a minor allele frequency above 5% were retrieved from the Arabidopsis 1001 genomes browser for 173 accessions (Cao et al., 2011). For each domain, haplotypes were defined as unique SNP combinations with a frequency above 5%. For exons, only non-synonymous SNPs were included. A promoter region of 1000kb was used and gene domains were obtained from Interpro (Mitchell et al., 2015). Promoter motifs were retrieved from Athamap (Hehl and Bülow, 2014).
RT-qPCR
For each sample, two intermediately aged leaves per 4- to 5-week-old Arabidopsis plant were harvested between 12.00 and 15.00h. Samples were immediately frozen in liquid nitrogen, and stored at −80 °C until processing. RNA was isolated from homogenized leaf material with an InviTrap® Spin Plant RNA kit, and treated with Ambion® TURBO DNA-freeTM according to the manufacturer’s instructions. RNA was quantified with a NanoDrop® ND-1 000 spectrophotometer, and integrity was assessed with gel electrophoresis. DNA-free RNA was converted into cDNA using the Bio-Rad iScriptTM cDNA synthesis kit. Quantitative reverse transcription PCR was carried out on a Bio-Rad IQTM5 system using SYBR Green. For each primer combination (Supplementary Table S1), RT-qPCR products were sequenced to validate the region of amplification. To test aphid induction of WRKY22, PR1, VSP2, and PDF1.2, plants were treated with and without aphids (n=4). For infested samples, a Petri dish with indentation for the petiole was used to contain 15 adult M. persicae aphids on the leaf, to inflict as little mechanostimulation as possible. Four biological replicates were collected for three treatments: (1) an empty Petri dish for 48h, (2) a Petri dish for 48h with addition of aphids in the last 6h, and (3) a Petri dish with aphids for 48h.
Electrical penetration graph recording
Feeding behaviour of M. persicae aphids was investigated with electrical penetration graph (EPG) recording on 4- to 5-week-old Arabidopsis plants, using direct current (DC) according to the methodology of ten Broeke et al. (2013). To adjust the radish-reared aphids to Arabidopsis, aphids were transferred to Col-0 Arabidopsis plants 24h before the experiments. EPG recording was performed at 22±2 °C and light intensity of 120 μmol m−2 s−1, using clean plants and one aphid per plant. An electrode was inserted in the potting soil and a thin gold wire of 1.5cm was gently attached to the dorsum of an adult, wingless aphid with silver glue. The electrical circuit was completed when the aphid’s piercing–sucking stylet mouthparts penetrated the plant cuticle. Electrical signals associated with stylet activities were recorded and annotated with EPG Stylet+ software (http://www.epgsystems.eu) and further processed in R (R Core Team, 2013; Tjallingii, 1988). Between 20 and 24 biological replicates were measured on T-DNA lines (Col-0: n=24; wrky22-3: n=22; wrky22-4: n=20) and between 15 and 19 on overexpression lines (Col-0: n=15; OE.c: n=19; OE.e: n=19). WRKY22 overexpression was induced by supplying 10 μM oestradiol solution to the plants 24h before the experiment. To correct for potential side-effects of oestradiol, the wild-type plants received the same oestradiol treatment as the overexpression lines.
Aphid population development
To assess aphid developmental rate and population size, 2.5-week-old Arabidopsis plants were infested with one M. persicae neonate of age 0–24h and placed in a climate room at 24±1 °C, 50–70% relative humidity, 8 h–16h light–dark photoperiod, 200 μmol m−2 s−1 light intensity. A soap-diluted water barrier prevented aphids from moving between plants. None of the aphids developed wings. From day 7 onwards, occurrence of the first offspring was checked twice per day using 5× magnification glasses (Col-0: n=18; wrky22-3: n=19; wrky22-4: n=22). The number of aphids per plant was counted at 14 days after infestation. Plants without an adult aphid 8 days after introduction and plants without any adults or neonates 14 days after introduction were excluded from the analysis.
Statistics
Data were tested for a normal distribution and homogeneity of variances using Shapiro’s test and Levene’s test. Non-parametric data sets were assessed with the Mann–Whitney U-test (two groups) or Kruskall–Wallis test (more than two groups). Data sets with a normal distribution were tested with Student’s t-test (two groups) or a one-way ANOVA (more than two groups).
RNA-seq analysis
RNA-seq analysis was conducted on leaves of Col-0 and wrky22-3 with three biological replicates per treatment. Five leaves of five different 4- to 5-week-old plants were pooled per sample. Plants had been exposed to one of three treatments: (1) an empty clip cage for 48h, (2) a clip cage for 48h with addition of 15 aphids in the last 6h, and (3) a clip cage with 15 aphids for 48h. Only fourth-instar nymphs and adult M. persicae aphids were used. Experiments were conducted simultaneously in a climate chamber (24±1 °C, 50–70% relative humidity, 8 h–16h light–dark photoperiod, and a light intensity of 120 μmol m−2 s−1), but in separate cages with an air circulation system that prevented contamination of plant volatiles between treatments (Menzel et al., 2014). Samples were harvested in two batches between 13.00 and 16.00h, immediately frozen in liquid nitrogen, and stored at −80 °C until processing. RNA was isolated and checked according to the description above (260/280 OD range: 2.0–2.2, 260/230 OD range: 1.9–2.3). Library preparation was performed with a TruSeq™ RNA Sample Prep Kit (Illumina®) and between 11 million and 24 million single-end 50-bp reads were sequenced per sample with Illumina® HiSeqTM 2000 in three lanes, multiplexed with 12 samples per lane. Reads were cleaned from adaptors and trimmed to 51bp using the program Trimmomatic version 0.32 (Bolger et al., 2014). Quality control was performed with FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). Reads were mapped to the TAIR10 Arabidopsis reference genome (https://www.arabidopsis.org/) with Tophat version 2.0.13, intron length 20–2000 (Trapnell et al., 2012; Trapnell et al., 2013). An index file was built with Bowtie 2 (Langmead et al., 2009). Transcript assembly, quantification, normalization and differential expression analysis were performed with Cufflinks, using the bias detection and correction algorithm, multi-read correction for reads mapping to multiple locations, and a minimum alignment count of 10. Treatments were compared both between plant lines (Col-0 versus mutant) and within plant line (empty clip cage versus 6h post-inoculation (hpi), empty clip cage versus 48 hpi, and 6 hpi versus 48 hpi). Only differentially expressed genes (false discovery rate Q-value<0.05) with an absolute fold change ≥2 (log2≥1) were taken into account. Differentially expressed genes were tested for overrepresentation of biological processes against a reference set including all transcripts in the complete data set with at least 1 count, using the application BiNGO in Cytoscape (Maere et al., 2005; Cline et al., 2007). Genes associated with cell-wall processes were selected and classified based upon their TAIR description, and the heatmap was constructed with the R package ‘gplots’ (Warnes et al., 2009).
Results
GWA mapping
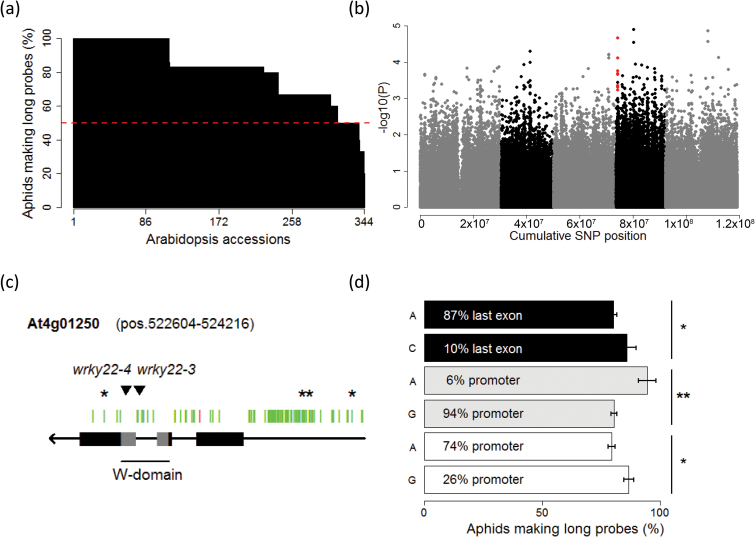
To identify genes involved in resistance to the green peach aphid, M. persicae, GWA mapping was performed on 344 natural accessions of Arabidopsis, using a selection of approximately 214 000 SNPs (Atwell et al., 2010; Li et al., 2010; Horton et al., 2012). The behaviour of aphids was screened on these accessions with an automated video-tracking platform (Kloth et al., 2015). The number and duration of plant penetrations was estimated by analysing the location and movement of aphids on single leaf discs. It is known that aphids need on average 25min to penetrate the epidermis and mesophyll before they reach the vascular bundle (van Helden and Tjallingii, 1993; Tjallingii, 1994; Prado and Tjallingii, 2007). Therefore, we used the proportion of aphids making long probes (>25min) as a proxy for the success rate of phloem ingestion. The majority of the Arabidopsis accessions did not show indications of resistance to aphids, but on 10% of the accessions at least half of the aphids were unsuccessful in feeding after 4.5h of infestation (Fig. 1A and Supplementary Table S2). GWA mapping of aphid feeding behaviour revealed seven genomic regions with a −log10(P) value above 4 and a heritability of 10% (Fig. 1B and Table 1). WRKY22 (At4g01250) was identified as a candidate gene in a 40kb region around a polymorphism with a −log10(P) value of 4.7 (chromosome 4 position 543516). Other candidates in the region included a gene with unknown function (At4g01290), a methyltransferase and a gene (At4g01240) with an MYB-like domain (At4g01280). Resequenced data of 173 accessions (Cao et al., 2011) showed that WRKY22 contained one non-synonymous SNP in its coding region, and that most of the polymorphisms were confined to the introns and the promoter region (Fig. 1C). A silent SNP in the last exon and two SNPs in the promoter were correlated with aphid feeding behaviour (Fig. 1D). Both polymorphisms in the promoter coincided with an AT-hook DNA-binding motif of AHL20, a transcription factor involved in plant defence to bacteria (Lu et al., 2010). Because WRKY22 is involved in PAMP-triggered immune responses (Asai et al., 2002; Navarro et al., 2004) and its expression is induced by M. persicae and Brevicoryne brassicae aphids (De Vos et al., 2005; Barah et al., 2013), we conducted further experiments to assess whether WRKY22 is involved in resistance to aphids.
Fig. 1.
Genome-wide association mapping of aphid feeding behaviour. (A) Phenotypic distribution of the proportion of M. persicae aphids making long probes (>25min) during a 1.5h recording on plants from 344 natural Arabidopsis accessions 4.5h post-inoculation. For accessions for which the percentage of long probes was below the dotted line, at least half of the aphids were unsuccessful in feeding. (B) Genome-wide associations with 214 000 SNPs. SNPs in red are positioned in a 40kb region around WRKY22 (highest −log10(P)=4.7). (C) All SNPs in WRKY22 and its 1000kb promoter region according to 173 resequenced Arabidopsis accessions (green: silent; red: non-synonymous). Predicted gene domains are shown in grey, unknown domains in black. Triangles represent T-DNA insertions. (D) One synonymous SNP in the last exon and two SNPs in the promoter had an effect on aphid feeding behaviour (*P<0.05, **P<0.01, Student’s t-test, chromosome 4, positions 523037, 524726 and 525079).
Table 1.
SNPs and corresponding genes associated with the proportion of aphids making long probes (>25min, −log 10(P) value>4)
Only the highest scoring SNP is shown per gene. Genes were grouped in one linkage disequilibrium (LD) region, if they were located within 20kb from each other. Chr.: chromosome.
| LD region | Chr. | Position | −log10(P) | AGI code | Description |
|---|---|---|---|---|---|
| 1 | 1 | 28995670 | 6.6 | At1g77160 | Protein of unknown function (DUF506) |
| 2 | 2 | 10866313 | 4.3 | At2g25530 | AFG1-like ATPase family protein |
| 3 | 3 | 20709836 | 4.2 | At3g55800 | Chloroplast enzyme sedoheptulose-1,7-bisphosphatase (SBPase) |
| 4 | 4 | 519513 | 4.1 | At4g01240 | S-Adenosyl-l-methionine-dependent methyltransferase superfamily protein |
| 4 | 4 | 536493 | 4.1 | At4g01280 | Homeodomain-like superfamily protein, SANT DNA-binding MYB-like domain |
| 4 | 4 | 543516 | 4.7 | At4g01290 | Unknown protein |
| 5 | 4 | 6641192 | 4.5 | At4g10790 | UBX domain-containing protein |
| 5 | 4 | 6644022 | 4.9 | At4g10800 | BTB/POZ domain-containing protein |
| 6 | 5 | 15927540 | 4.9 | At5g39770 | Pseudogene homologous to AtMSU81, restriction Endonuclease |
| 7 | 5 | 19854700 | 4.1 | At5g48965 | Mutator-like transposase family |
| 7 | 5 | 19858466 | 4.1 | At5g48970 | Mitochondrial substrate carrier family protein |
Mesophyll-located susceptibility to aphids
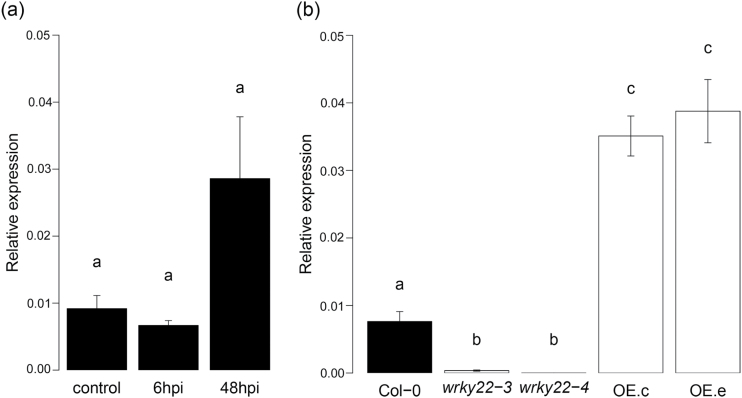
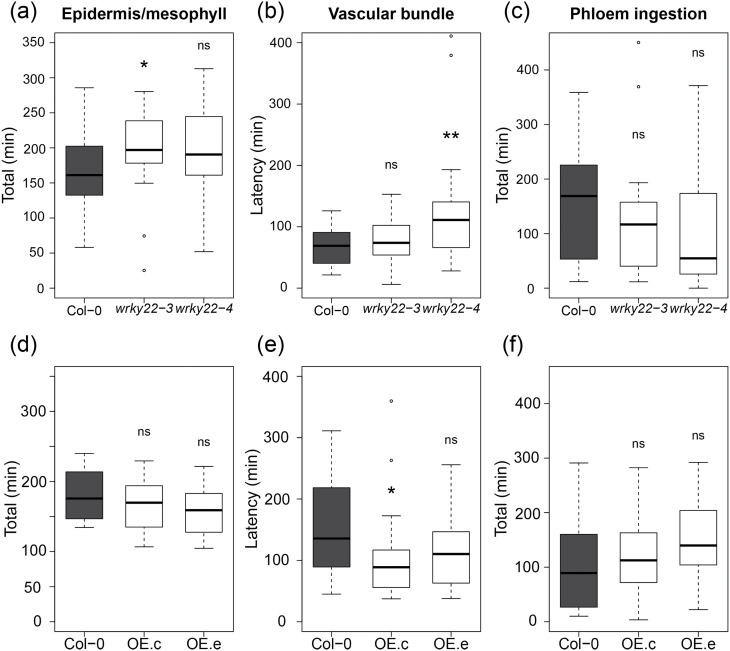
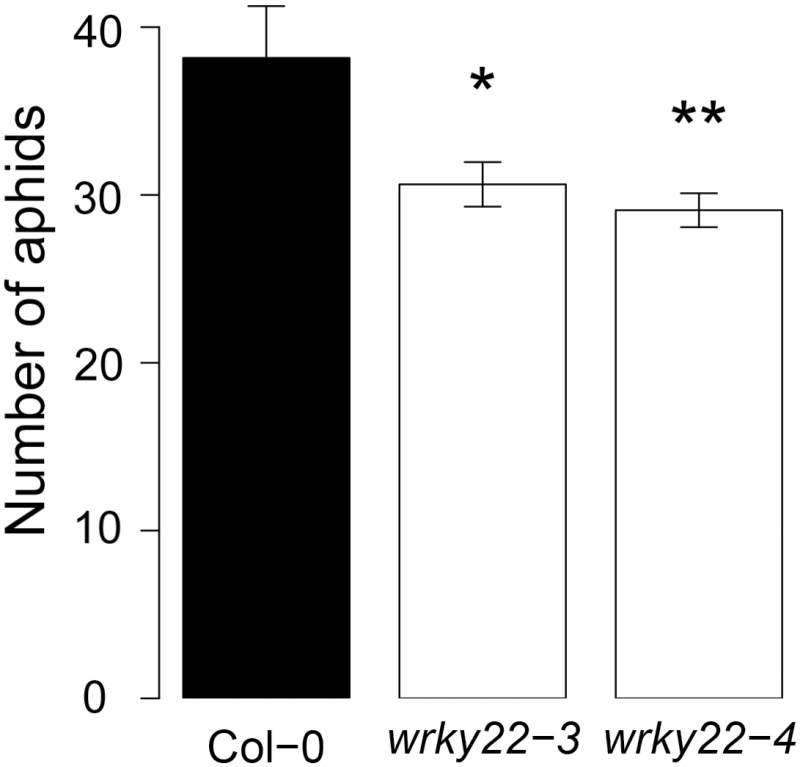
To validate the previously reported induction of WRKY22 by aphid infestation (De Vos et al., 2005; Barah et al., 2013), RT-qPCR was performed on wild-type plants with aphids and without aphids. WRKY22 expression was unaffected at 6h post-infestation (hpi), and showed a non-significant increase at 48 hpi (Fig. 2A). Two wrky22 transfer (T)-DNA insertion lines and two WRKY22-inducible overexpression lines (Coego et al., 2014) were selected for further experiments. RT-qPCR confirmed that both T-DNA lines were true knockouts, and that the overexpression lines showed a 3- to 5-fold up-regulation of WRKY22 at 24h after induction with oestradiol (Fig. 2B). For a detailed insight into aphid feeding behaviour on knockout and overexpression lines, we used electrical penetration graph (EPG) recordings (McLean and Kinsey, 1964; Tjallingii, 1988). Aphid feeding behaviour was affected on both wrky22 knockout lines; on wrky22-3 aphids spent almost 20% more time on penetrating the epidermis and mesophyll, and on wrky22-4 aphids showed an hour’s delay in reaching the vascular bundle compared with the wild-type (Col-0) (Fig. 3A–C and Supplementary Table S3). One of the WRKY22-inducible overexpression lines showed the opposite trend, with aphids arriving almost an hour earlier at the vascular bundle compared with the wild-type (Fig. 3D–F and Supplementary Table S4). The other WRKY22 overexpression line did not show any differences compared with the wild-type. The total time of phloem ingestion was not affected in any of the (mutant) lines, suggesting that in the first 8h of infestation, the overall effects are small and confined to activities in the epidermis and/or mesophyll. An aphid population development assay on wrky22-3 and wrky22-4 showed that after 2 weeks of infestation, aphid populations were approximately 20% smaller on the knockouts compared with the wild-type (Fig. 4). Both behavioural experiments and population assays indicate that WRKY22 increases susceptibility to M. persicae aphids.
Fig. 2.
WRKY22 expression. (A) WRKY22 expression in the wild-type without M. persicae aphids (control) and after 6 and 48h of aphid infestation. (B) Expression in the wild-type (Col-0), wrky22-3 and wrky22-4 knockout lines, and WRKY22-inducible overexpression lines OE.c and OE.e. Overexpression lines were induced with oestradiol 24h before sampling (one-way ANOVA and Student’s t-test; different letters refer to significant differences).
Fig. 3.
Aphid behaviour on wrky22 knockout lines (upper panels) and WRKY22 overexpression lines (lower panels). (A, D) The total time M. persicae aphids were penetrating the epidermis and mesophyll during 8-h recordings on knockout lines wrky22-3 and wrky22-4 (A), and overexpression lines OE.c and OE.e (D). (B, E) Time between the start of the recording and the first contact with either a phloem or xylem bundle measured on knockout (B), and overexpression lines (E). (C, F) The total time aphids were ingesting phloem on knockout (C), and overexpression lines (F); knockout and overexpression lines were compared with the wild-type with Mann–Whitney U-test (*P<0.05; **P<0.01). To test the effect of overexpression, all plants were induced with oestradiol 24h before the assay.
Fig. 4.
Aphid population size on wild-type and knockout plants. The total number of M. persicae aphids per plant was counted 2 weeks after infestation with one neonate aphid. Mutant lines were compared with the wild-type with Student’s t-test (*P<0.05; **P<0.01).
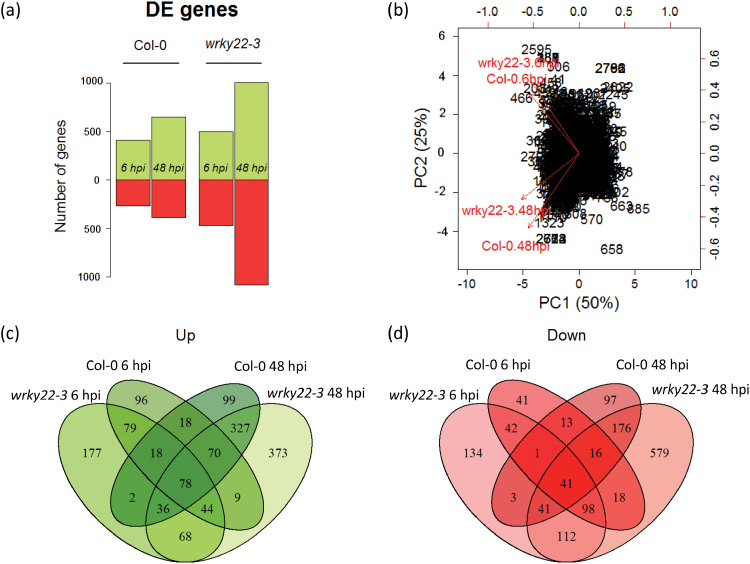
Transcriptomic signature of wrky22-3
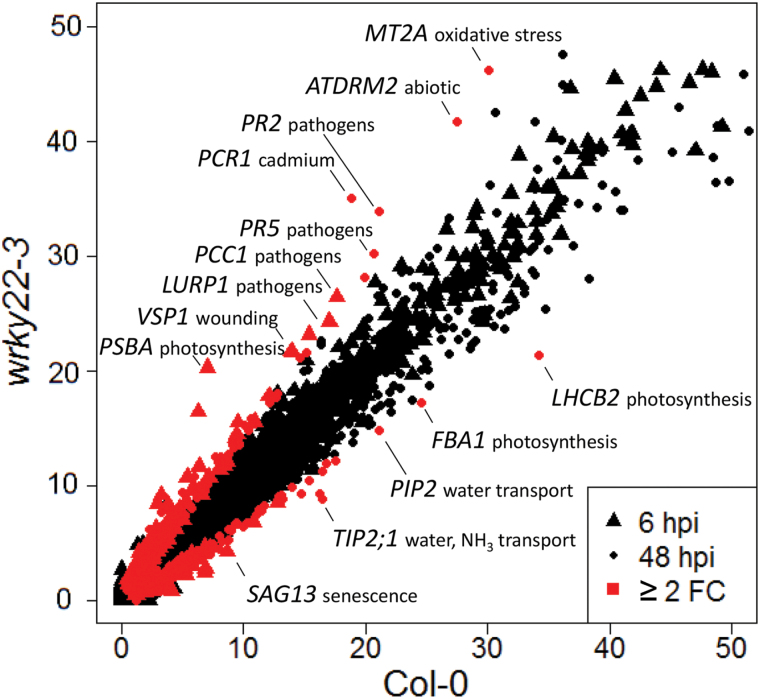
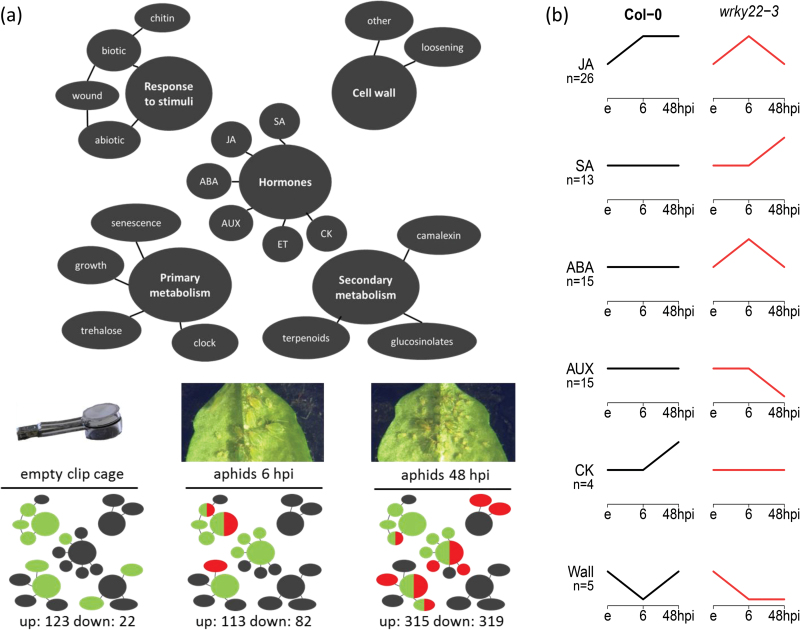
To study the role of WRKY22 in resistance to aphids, mRNA sequencing (RNA-seq) analysis was performed on wrky22-3 and wild-type plants that had received one of three treatments: (1) an empty clip cage for 48h, (2) a clip cage for 48h with addition of aphids in the last 6h, and (3) a clip cage with aphids for 48h. For each treatment three biological replicates were sampled, each consisting of a pool of five leaves from different plants. Samples were sequenced for single end 50-bp reads, and for each sample at least 9.5 million reads mapped to unique loci on the Arabidopsis reference genome (Supplementary data file S1). The total number of differentially expressed (DE) genes increased with the duration of infestation, from approximately 700 at 6 hpi to 1000 at 48 hpi in the wild-type. In both treatments, wrky22-3 contained twice as many up- and down-regulated genes as the wild-type (Fig. 5A). Principal component analysis showed that the duration of infestation was the major factor explaining differential expression (Fig. 5B–D). Highly abundant transcripts that were up-regulated in wrky22-3 compared with the wild-type included the JA reporter VSP1 (6 hpi) and pathogenesis-related genes such as PR2 and PR5 (48 hpi) (Fig. 6). Photosynthesis- and water-transport-related genes were down-regulated in wrky22-3 at 48 hpi (Fig. 6). Gene ontology (GO) enrichment analysis of the total set of DE genes revealed an overrepresentation of up-regulated JA-, SA- and abscisic acid (ABA)-responsive genes in wrky22-3 at both 6 and 48 hpi (Fig. 7A). The JA pathway was mainly characterized by up-regulation of genes of the ethylene response factor (ERF) branch (Vos et al., 2015), e.g. the AP2/ERF transcription factor RAP2.6, and PDF1.2 (Table 2). The majority of the DE genes associated with JA and ABA showed a peak at 6 hpi in wrky22-3, whereas most SA-responsive genes reached their highest level at 48 hpi (Fig. 7B). The induction of SA-responsive genes in the T-DNA line at 48 hpi coincided with a suppression of genes associated with auxin (AUX) responsiveness, plant growth and cell wall loosening (Fig. 7A, B).
Fig. 5.
Differentially expressed (DE) genes between treatments with and without aphids in the wild-type and wrky22-3. (A) The number of DE genes between control and infestation treatments (green bars: up-regulated, red bars: down-regulated). (B) Biplot of the two first principal components of differentially expressed genes between control and infestation treatments (DE genes ≥2-fold). (C) Overlap in up-regulated genes, and (D) down-regulated genes.
Fig. 6.
Gene transcripts of aphid-infested wild-type and wrky22-3 plants. Differentially expressed genes between wild-type and knockout plants (≥2-fold change) are shown in red. Axes depict the square-root transformation of the normalized number of transcripts (the number of fragments per kilobase of transcript per million reads mapped (FPKM)); genes ≤2500 FPKM are shown (including all DE genes in the dataset). Annotations include gene name and biological process; wounding: wound responsive; pathogens: pathogen responsive; cadmium: responsive to cadmium; abiotic: responsive to several abiotic stresses.
Fig. 7.
Enriched biological processes in wrky22-3. (A) Over-representation of biological processes in the knockout relative to the wild-type. Balloons refer to a process, or to the biosynthesis of, or responsiveness to the respective compound (SA: salicylic acid; JA: jasmonic acid; ABA: abscisic acid; AUX: auxin; ET: ethylene; CK: cytokinin; clock: circadian clock). Balloon colour indicates enrichment in the knockout (green: up-regulated; red: down-regulated; green/red: both up- and down-regulated; the total number of DE genes is depicted below the charts). (B) Relative expression patterns between treatments within each plant line. Only the dominant pattern (≥50% of the genes) of significant perturbations (≥2-fold, q-value <0.05) between treatments with and without aphids is shown. (Wall: cell wall loosening; n: number of genes associated with the biological process; e: empty clip cage; 6: 6 hpi.)
Table 2.
Differentially expressed genes (≥2-fold) of over-represented biological processes in wrky22-3 relative to the wild type
GO enrichment and gene classification are according to the BiNGO Cytoscape app (SA: salicylic acid; JA: jasmonic acid; ABA: abscisic acid; AUX: auxin; ET: ethylene; CK: cytokinin) (Cline et al., 2007; Maere et al., 2005).
| Process | Treatment | Direction | Name | AGI code | Description |
|---|---|---|---|---|---|
| ABA | Empty cage, 6 hpi | Up | ANNAT4 | At2g38750 | Annexin, Golgi-mediated secretion |
| 6 hpi | Up | HAI1 | At5g59220 | HIGHLY ABA-INDUCED PP2C gene 1 | |
| 6 hpi | Up | HD-Zip-I | At3g61890 | Homeodomain leucine zipper class I | |
| 48 hpi | Up | ACR8 | At1g12420 | ACT DOMAIN REPEAT 8 | |
| 48 hpi | Up | AMY1 | At4g25 000 | ALPHA-AMYLASE-LIKE 1, starch mobilization | |
| 48 hpi | Up | Dehydrins | At3g50970, At1g20440 | Membrane located, freeze tolerance | |
| 48 hpi | Up | ERF48 | At2g40340 | ABA responsive AP2/ERF transcription factor | |
| 48 hpi | Up | LTI78 | At5g52310 | LOW-TEMPERATURE-INDUCED 78 | |
| 48 hpi | Up | WRKY63 | At1g66600 | ABA responsive WRKY transcription factor | |
| AUX | 6, 48 hpi | Down | CCA1 | At2g46830 | Negative regulator of circadian rhythm |
| 48 hpi | Down | AXR3 | At1g04250 | AUXIN RESISTANT 3 | |
| 48 hpi | Down | GH3s | At2g47750, At5g13360 | GH3 auxin responsive gene family | |
| 48 hpi | Down | SAURs | At1g20470, At1g29500, At1g29510, At3g03820, At4g22620, At4g38840, At4g38850, At4g38860, At5g18020, At5g18030, At5g18050 | SAUR(-like) auxin-responsive proteins | |
| CK | 48 hpi | Down | ARRs | At1g19050, At1g74890, At3g57040, At5g62920 | Arabidopsis response regulator (ARR) family |
| JA | Empty cage, 6, 48 hpi | Up | JAZs | At2g34600, At5g13220, At1g17380, At1g19180 | JAZ7, JAZ10, JAZ5, JAZ1, Jasmonate-Zim- domain proteins |
| Empty cage, 6, 48 hpi | Up | MDHAR4 | At3g09940 | Monodehydroascorbate reductase | |
| Empty cage, 6, 48 hpi | Up | MYB47 | At1g18710 | JA-responsive MYB transcription factor | |
| Empty cage, 6, 48 hpi | Up | TAT3 | At2g24850 | Tyrosine aminotransferase, JA responsive | |
| Empty cage | Up | VSP2 | At5g24770 | VEGETATIVE STORAGE PROTEIN 2 | |
| Empty cage, 6, 48 hpi | Up | VSP1 | At5g24780 | VEGETATIVE STORAGE PROTEIN 1 | |
| Empty cage, 6 hpi | Up | AOCs | At3g25760, At3g25780 | Allene Oxide Cyclase family, JA biosynthesis | |
| Empty cage, 6 hpi | Up | OPR3 | At2g06050 | OXOPHYTODIENOATE-REDUCTASE 3, JA biosynthesis | |
| Empty cage | Up | EXT4 | At1g76930 | Extensin | |
| Empty cage | Up | JR1 | At3g16470 | JASMONAtE RESPONSIVE 1 | |
| 6, 48 hpi | Up | PDF1.2 | At5g44420 | PLANT DEFENSIN 1.2 | |
| 6 hpi | Up | DAD1 | At2g44810 | DEFECTIVE ANTHER DEHISCENCE 1, JA biosynthesis | |
| 6 hpi | Up | JAR1 | At2g46370 | Jasmonate-amido synthetase | |
| 6 hpi | Up | LOX3 | At1g17420 | LIPOXYGENASE 3 | |
| 6 hpi | Up | RAP2.6 | At1g43160 | AP2/ERF transcription factor | |
| SA | 6, 48 hpi | Up | GRX480 | At1g28480 | Glutaredoxin family, suppresses PDF1.2 |
| 6, 48 hpi | Up | LURP1 | At2g14560 | Resistance to Hyaloperonospora parasitica | |
| 6, 48 hpi | Up | WRKY18 | At4g31800 | WRKY18 | |
| 48 hpi | Up | WRKYs | At5g01900, At5g22570 | WRKY38, WRKY62 | |
| 48 hpi | Up | MYB77 | At3g50060 | MYB77 | |
| 48 hpi | Up | WAK1 | At1g21250 | CELL WALL-ASSOCIAtED KINASE 1 | |
| JA, SA, ABA | 6 hpi | Up | CIR1 | At5g37260 | MYB transcription factor |
| 48 hpi | Up | MYBs | At1g06180, At1g57560, At5g67300, At2g16720 | MYB13, MYB50, MYB44, MYB7 | |
| 48 hpi | Up | MPK11 | At1g01560 | MAP KINASE 11 | |
| 48 hpi | Up | PDR12 | At1g15520 | ABC transporter family, MAPK cascade | |
| Camalexin | Empty cage, 48 hpi | Up | PAD3 | At3g26830 | PHYTOALEXIN DEFICIENT 3, camalexin biosynthesis |
| 48 hpi | Up | P450 | At4g39950 | Cytochrome P450, indo-3-acetaldoxime (IAOx) biosynthesis | |
| Terpenoids | Empty cage, 6, 48 hpi | Up | TSP4 | At1g61120 | TERPENE SYNTHASE 4 |
| Empty cage | Up | TPS10 | At2g24210 | TERPENE SYNTHASE 10 | |
| Cell wall | 48 hpi | Down | Expansins | At1g20190, At1g26770, At1g69530, At2g20750, At2g40610 | Expansin family, cell wall loosening and multidimensional cell growth |
Enhanced negative JA–SA crosstalk in wrky22-3
Upon aphid infestation, the wrky22-3 transcriptome showed evidence of initial suppression, but eventual up-regulation of SA signalling. The expression of PR1, a robust SA-reporter gene (Pieterse et al., 2012), was 2-fold down-regulated at 6 hpi, but 3.5-fold up-regulated at 48 hpi in wrky22-3 compared with the wild-type. Transcript levels of JA-reporter genes PDF1.2 and VSP1 were consistently more abundant in wrky22-3 (Table 3), suggesting a possible role of negative JA–SA crosstalk (Spoel and Dong, 2008; Pieterse et al., 2012). A potential antagonizing candidate is NIMIN-2, encoding an SA-suppressing protein (Weigel et al., 2005), which was up-regulated in wrky22-3 6 hpi (Table 3). Apart from SA antagonism, there were also signs of JA antagonism. Several up-regulated genes in wrky22-3, i.e. GRX480, WRKY51, and WRKY62 (Table 3), have previously been implicated as potential suppressors of JA signalling (Mao et al., 2007; Ndamukong et al., 2007; Gao et al., 2011).
Table 3.
JA- and SA-signalling-related gene expression in wrky22-3 plants compared with wild-type plants with and without aphids
Differentially expressed genes with at least 2-fold absolute change are shown (ns: not significant; emp: empty clip cage; SA/JA sig: SA/JA signalling; SA/JA suppr: suppression of SA/JA signalling).
| Fold change | ||||||
|---|---|---|---|---|---|---|
| Gene | Name | Role | Emp | 6 hpi | 48 hpi | Reference |
| VSP1 | VEGETATIVE STORAGE PROTEIN 1 | JA sig | 13.0 | 2.5 | 2.8 | (Anderson et al., 2004; Lorenzo et al., 2004) |
| VSP2 | VEGETATIVE STORAGE PROTEIN 2 | JA sig | 7.5 | ns | ns | (Anderson et al., 2004; Lorenzo et al., 2004) |
| PDF1.2, 1.2C | PLANT DEFENSIN 1.2A, 1.2C | JA sig | ns | 2.3 | 3.5 | (Lorenzo et al., 2003; Penninckx et al., 1998) |
| PR1 | PATHOGENESIS-RELATED GENE 1 | SA sig | ns | 0.4 | 3.5 | (van Loon et al., 2006) |
| NIMIN-1 | NIM1-INTERACTING 1 | SA suppr | ns | ns | 2.5 | (Weigel et al., 2001; Weigel et al., 2005) |
| NIMIN-2 | NIM1-INTERACTING 2 | SA suppr | ns | 2.0 | 2.5 | (Weigel et al., 2001; Weigel et al., 2005) |
| GRX480 | Glutaredoxin | JA suppr | ns | 2.8 | 2.8 | (Ndamukong et al., 2007) |
| WRKY51 | WRKY DNA-BINDING PROTEIN 51 | JA suppr | ns | ns | 4.3 | (Gao et al., 2011) |
| WRKY62 | WRKY DNA-BINDING PROTEIN 62 | JA suppr | ns | ns | 2.8 | (Mao et al., 2007) |
JA induction by mechanostimulation in wrky22-3
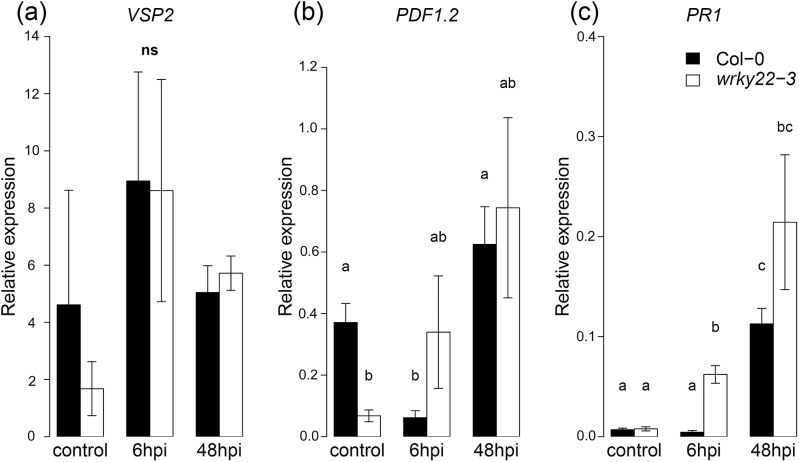
Remarkably, the treatment with empty clip cages changed the expression of almost 150 genes in the knockout relative to the wild-type. Most of them were up-regulated in wrky22-3 and showed a significant overrepresentation of JA-responsive genes, including VSP2 of the touch- and wound-responsive MYC-branch of the JA signalling pathway (Lange and Lange, 2015) (Fig. 7 and Table 2). In order to see if mechanostimulation by clip cages had affected the plant’s response to aphids, RT-qPCR was conducted on aphid-infested leaves without clip cages (see Materials and methods). We found that in clean wrky22-3 plants, PDF1.2 expression was lower compared with wild-type plants (Fig. 8). After 6h of aphid infestation, PR1 was up-regulated in wrky22-3 while PDF1.2 only showed a non-significant increase (Fig. 8). These results suggest that, in the RNA-seq analysis, the over-represented JA response may have been clip cage-induced and was likely involved in the suppression of the SA response to aphids at 6 hpi.
Fig. 8.
The effect of aphid infestation without clip cage on the expression of JA and SA reporter genes. RT-qPCR measurements of expression of the JA reporters VSP2 (A) and PDF1.2 (B), and the SA reporter PR1 (C) in wild-type and wrky22-3 plants (Student’s t-test and the Mann–Whitney U-test; different letters denote significant differences). Aphids were contained on the leaves without inflicting major mechanical stimulation (see Materials and methods).
Known resistance genes
Several genes described in earlier studies as being involved in enhancing resistance to M. persicae were up-regulated in wrky22-3. Expression of the MYB-domain-containing protein MYBR1, found to affect M. persicae reproduction under the influence of harpin proteins (Liu et al., 2010), was more than 2-fold induced at 48 hpi (Supplementary data file S1). Also several other genes with affiliation to known resistance factors for M. persicae were up-regulated, such as the phloem protein PP2-A12 at 48 hpi, the myrosinase-binding protein MBP1 at 48 hpi, and several xyloglucan endotransglucosylases/hydrolases (XTHs) at 6 and 48 hpi (Mewis et al., 2005; Divol et al., 2007; Zhang et al., 2011; Louis and Shah, 2013).
Differential expression of cell wall-related genes
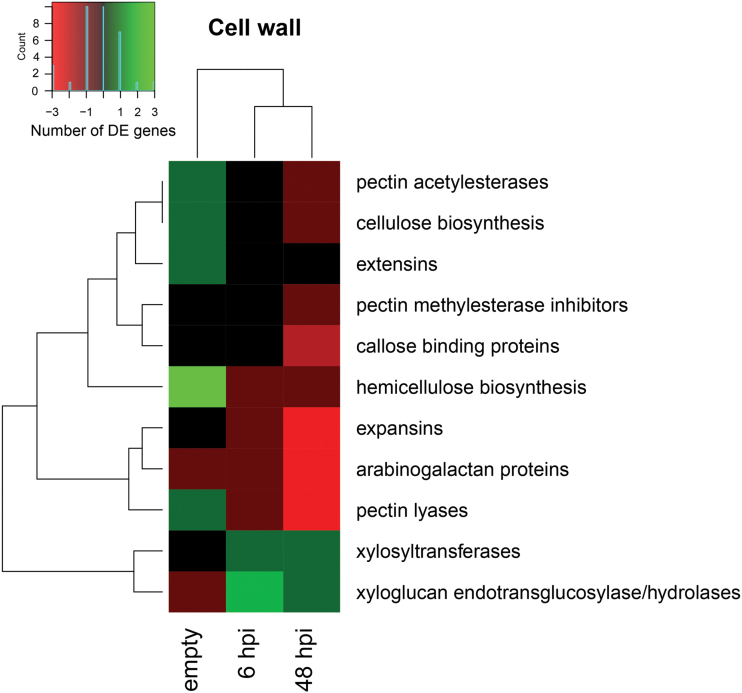
After 48h of aphid infestation, the wrky22-3 transcriptome was characterized by down-regulation of genes associated with cell wall loosening (Fig. 7 and Table 2). To assess all cell wall-related processes, DE genes with cell wall annotation were selected and grouped into categories based on their name and function (Fig. 9). We did not observe an up-regulation of touch-responsive xyloglucan endotransglucosylases/hydrolases (XTHs) by the empty clip cage. The empty clip cage did, however, cause a 3- to 5-fold up-regulation of the cellulose synthase-like genes CSLA1, CSLA10, and CSLA15, involved in hemicellulose biosynthesis (Liepman et al., 2005). Aphids up-regulated XTHs and down-regulated, for example, expansins, involved in cell-wall loosening, and pectin lyases, involved in pectin breakdown. While cell-wall loosening is a prerequisite for cell elongation, a process mainly regulated by CK and AUX (Taiz, 1984; Yadav et al., 2009; Albersheim et al., 2011), the transcriptomic patterns indicate an aphid-induced arrest of symplastic cell growth in wrky22-3.
Fig. 9.
Expression of cell-wall related genes in wrky22-3 and the effect of mechanostimulation by empty clip cages and aphid infestation. Genes and treatments are clustered according to the number of differentially expressed genes (≥2-fold change), using Ward’s minimum variance method (red: down-regulated; green: up-regulated in wrky22-3 compared with the wild-type).
Discussion
The effect of WRKY22 on M. persicae aphids
Plant responses to aphids are known to involve many transcriptional perturbations including multiple phytohormonal pathways (De Vos et al., 2005; De Vos et al., 2007; Smith and Boyko, 2007; Foyer et al., 2015). It is, therefore, a challenge to unravel the genetic basis of effective defence mechanisms against aphids. In this study, we explored natural variation in Arabidopsis to find genes related to impaired feeding behaviour of M. persicae. Natural variation in the occurrence of long probes, a proxy for the success rate of phloem ingestion, was associated to several genomic regions, including the WRKY22 locus. Polymorphisms in the WRKY22 promoter and in the last exon most strongly correlated with variation in aphid feeding behaviour. Even though the associations had low statistical power and heritability, knockout lines confirmed an effect of WRKY22 on aphid performance. Without a functional WRKY22 protein, it was more difficult for aphids to penetrate the epidermis and mesophyll and they arrived later at the vascular bundle. One WRKY22 overexpression line showed the opposite trend, although the impact was smaller than in the wrky22 knockouts, most likely due to the moderate extent and short time frame of the overexpression (3- to 5-fold change, induced 24h before the experiments). The effects of WRKY22 on aphid performance were marginal in the first 8h, but more substantial after an infestation period of 2 weeks as reflected by aphid population size. Overall, these assays indicate that WRKY22 promotes susceptibility to M. persicae aphids. Our RNA-seq and RT-qPCR analyses indicate that WRKY22-mediated susceptibility is associated with the suppression of SA signalling. Aphid infestation of wrky22-3 plants resulted in faster and potentially stronger up-regulation of the SA pathway than in wild-type plants. Even though it has been described that JA-induced defences are most effective against aphids (De Vos et al., 2007; Walling, 2008), SA-induced mechanisms have been shown to have a detrimental impact on aphids as well (Li et al., 2006; Moloi and Westhuizen, 2006). The down-regulation of pectin lyases and expansins (Fig. 9) suggests that there is less degradation of pectin and less loosening of the cell wall matrix in the wrky22 mutant. Fortification of the primary cell wall may have hampered the penetration of the mesophyll apoplast by aphids. This would explain why aphids required more time in probing the epidermis and mesophyll and were delayed in reaching the vascular bundle on the wrky22 mutants (Fig. 3 and Supplementary Table S3). We can, however, not exclude the involvement of other resistance factors in the mesophyll, such as the accumulation of reactive oxygen species or secondary metabolites.
Involvement of WRKY22 in biotic and abiotic stress responses
Apart from WRKY22’s responsiveness to aphids, we observed a strong activation of the JA pathway in wrky22-3 as a result of the use of clip cages. JA accumulation is known to be induced by mechanical stimuli (Ichimura et al., 2000; Chehab et al., 2012), wounding, and damage-associated molecular patterns (DAMPs) (Doares et al., 1995; Denoux et al., 2008; Vidhyasekaran, 2014). Since there were no obvious signs of plant damage, the up-regulation of the JA pathway was most likely triggered by touch-induced surface stimulation of our samples. WRKY22 has been shown to be induced in response to touch and wounding (Lee et al., 2004; Kilian et al., 2007). Our data suggest that WRKY22 acts as a suppressor of JA signalling in response to these stimuli. Many other abiotic stimuli have been described to induce WRKY22 as well, such as prolonged darkness, submergence, cold acclimation, light perception, salinity, potassium starvation, and exposure to ozone (Folta et al., 2003; Hampton et al., 2004; Monte et al., 2004; Lee et al., 2005; Tosti et al., 2006; Chawade et al., 2007; Kilian et al., 2007; Zhou et al., 2011; Göhre et al., 2012; Hsu et al., 2013; Kim et al., 2013; Sugimoto et al., 2014). Hsu et al. (2013) identified several potential downstream targets of WRKY22, including genes involved in drought resistance and phosphate starvation. The accumulating evidence for its involvement in abiotic stress responses warrants a change of view, i.e. that WRKY22 is not solely involved in PTI. A parallel can be drawn with WRKY40, previously known as a repressor of PTI but recently also recognized as a central player in ABA inhibition during abiotic stress (Chen et al., 2010; Friedel et al., 2012). Although abiotic and biotic stimuli are most likely perceived via stress-specific mechanisms and require differential plant responses, signal-transduction pathways might converge via common regulators, such as WRKY22, in order to fine-tune the interplay between phytohormones.
SA–JA signal integration
One of the major questions is whether WRKY22 is an activator or repressor of SA and JA signalling. Our transcriptome analysis of wrky22-3 revealed up-regulation of JA signalling upon mechanostimulation and up-regulation of SA signalling upon aphid infestation. This would suggest that in wild-type plants, WRKY22 is a suppressor of JA and SA signalling. From previous studies we know, however, that WRKY22 and WRKY29 confer resistance to (hemi)biotrophic and necrotrophic pathogens (Asai et al., 2002; Hsu et al., 2013), and that they are induced by PAMP-triggered MAPK cascades which result in the activation of SA, JA and ET signalling (Zipfel et al., 2004). There is no direct evidence that WRKY22 and its homologue WRKY29 induce SA, JA and ET signalling, and the possibility exists that they are involved in MAPK-triggered processes independent of SA and JA signalling. Nevertheless, their requirement for PTI makes them unlikely candidates for consistent suppression of plant defence hormones. Rather, WRKY22 could be an integrator of SA and JA signals, inhibiting or enforcing both pathways, depending on their interaction with other transcription factors and signalling pathways (Fig. 10). Similarly, WRKY70 has been proposed to be capable of inducing and inhibiting both SA and JA signalling, depending on the strength of the induction (Li et al., 2004; Ülker et al., 2007). Although many questions remain with regard to the underlying mechanism, our study shows that WRKY22 plays a role in both SA and JA signalling and is involved in transcriptional reprogramming in response to mechanostimulation and aphid infestation. To understand the function of WRKY22, its transcriptional network needs to be further unravelled under multiple biotic and abiotic stress conditions. With respect to aphids, WRKY22 increases susceptibility. Its responsiveness to aphid infestation and its potential to suppress JA and SA signalling would make WRKY22 an excellent target for aphids to manipulate JA- and SA-dependent host plant defences for their own benefit.
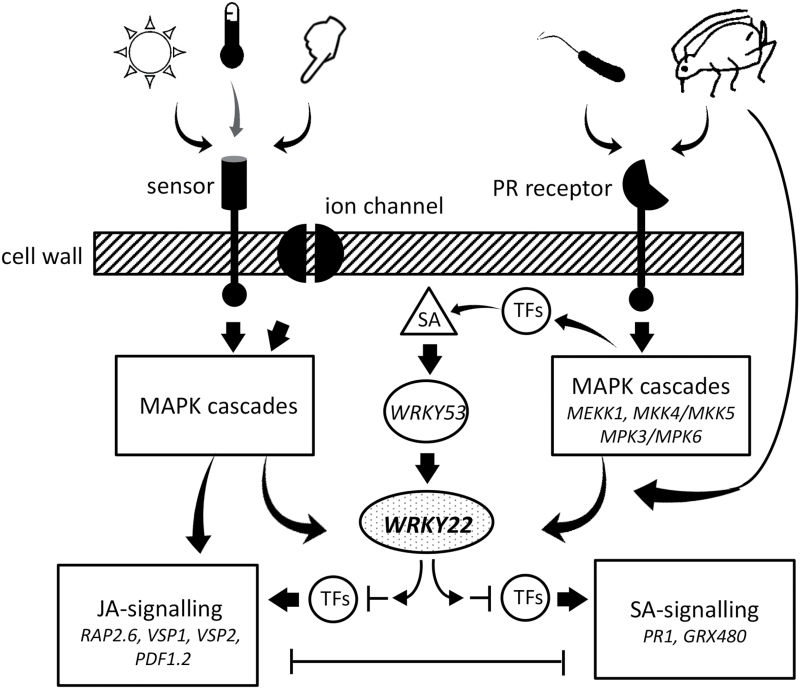
Fig. 10.
Hypothetical model of WRKY22’s role in plant response to abiotic (left) and biotic (right) stresses. Changes in, for example, light, temperature and touch are perceived via sensors and ion channels; plant invasion by organisms such as bacteria and aphids is mainly perceived via pattern-recognition (PR) receptors. These stimuli induce WRKY22 directly via MAPK cascades (Ichimura et al., 2000; Asai et al., 2002), or indirectly via SA accumulation (Miao et al., 2004; Miao and Zentgraf, 2007). Alternatively, aphid effectors secreted via the saliva may induce WRKY22 via PR-receptor-independent routes. WRKY22 subsequently integrates signalling of the JA and SA pathway, by inhibiting or activating specific transcription factors (TFs) and other regulatory genes.
Supplementary data
Supplementary data are available at JXB online.
Data file S1. Differentially expressed genes between wrky22-3 and wild-type rosette leaves with and without aphid infestation.
Table S1. Primers used for PCR and RT-qPCR.
Table S2. Percentage of aphids making long probes (>25min) 4.5 hour after inoculation on 344 natural Arabidopsis accessions.
Table S3. Aphid feeding behaviour, measured by 8-hour EPG recordings on wild-type (Col-0) and wrky22 T-DNA lines (wrky22-3 and wrky22-4).
Table S4. Aphid feeding behaviour, measured by 8-hour EPG recordings on wild-type (Col-0) and wrky22-inducible overexpression lines (OE.c and OE.e).
Acknowledgements
This work was supported by The Netherlands Organization for Scientific Research (NWO) through the Technology Foundation Perspective Programme ‘Learning from Nature’ (STW10989) and the ZonMw programme ‘Enabling Technologies’ (435000017). We would like to thank Johan Bucher of the Department of Plant Breeding, Wageningen University, for providing the clip cages.
References
- Albersheim P, Darvill A, Roberts K, Sederoff R, Staehelin A. 2011. Plant Cell Walls . New York: Garland Science, Taylor & Francis Group. [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. 2004. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. The Plant Cell 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel HM, Fescemyer H, Ehlting J, Weston D, Rehrig E, Joshi T, Xu D, Bohlmann J, Schultz J. 2014. Transcriptional responses of Arabidopsis thaliana to chewing and sucking insect herbivores. Frontiers in Plant Science 5, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu W, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, et al. 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barah P, Winge P, Kusnierczyk A, Tran DH, Bones AM. 2013. Molecular signatures in Arabidopsis thaliana in response to insect attack and bacterial infection. PLoS ONE 8, e58987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I, Brazelton JN, Yu D, et al. 2010. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genetics 6, e1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114 – 2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, et al. 2011. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nature Genetics 43, 956–965. [DOI] [PubMed] [Google Scholar]
- Chawade A, Brautigam M, Lindlof A, Olsson O, Olsson B. 2007. Putative cold acclimation pathways in Arabidopsis thaliana identified by a combined analysis of mRNA co-expression patterns, promoter motifs and transcription factors. BMC Genomics 8, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab EW, Yao C, Henderson Z, Kim S, Braam J. 2012. Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Current Biology 22, 701–706. [DOI] [PubMed] [Google Scholar]
- Chen H, Lai Z, Shi J, Chen Z, Xu X. 2010. Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biology 10, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, et al. 2007. Integration of biological networks and gene expression data using Cytoscape. Nature Protocols 2, 2366–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coego A, Brizuela E, Castillejo P, Ruíz S, Koncz C, del Pozo JC, Piñeiro M, Jarillo JA, Paz-Ares J, León J. 2014. The TRANSPLANTA collection of Arabidopsis lines: a resource for functional analysis of transcription factors based on their conditional overexpression. The Plant Journal 77, 944–953. [DOI] [PubMed] [Google Scholar]
- De Vos M, Kim JH, Jander G. 2007. Biochemistry and molecular biology of Arabidopsis-aphid interactions. Bioessays 29, 871–883. [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RMP, et al. 2005. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Molecular Plant-Microbe Interactions 18, 923–937. [DOI] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. 2008. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Molecular Plant 1, 423–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divol F, Vilaine F, Thibivilliers S, Kusiak C, Sauge MH, Dinant S. 2007. Involvement of the xyloglucan endotransglycosylase/hydrolases encoded by celery XTH1 and Arabidopsis XTH33 in the phloem response to aphids. Plant, Cell and Environment 30, 187–201. [DOI] [PubMed] [Google Scholar]
- Doares SH, Syrovets T, Weiler EW, Ryan CA. 1995. Oligogalacturonides and chitosan activate plant defensive genes through the octadecanoid pathway. Proceedings of the National Academy of Sciences of the United States of America 92, 4095–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Chen C, Chen Z. 2003. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Molecular Biology 51, 21–37. [DOI] [PubMed] [Google Scholar]
- Dubey NK, Goel R, Ranjan A, Idris A, Singh SK, Bag SK, Chandrashekar K, Pandey KD, Singh PK, Sawant SV. 2013. Comparative transcriptome analysis of Gossypium hirsutum L. in response to sap sucking insects: aphid and whitefly. BMC Genomics 14, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. 2000. The WRKY superfamily of plant transcription factors. Trends in Plant Science 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Folta K, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP. 2003. Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. The Plant Journal 36, 203–214. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Verrall SR, Hancock RD. 2015. Systematic analysis of phloem-feeding insect-induced transcriptional reprogramming in Arabidopsis highlights common features and reveals distinct responses to specialist and generalist insects. Journal of Experimental Botany 66, 495–512. [DOI] [PubMed] [Google Scholar]
- Friedel S, Usadel B, von Wirén N, Sreenivasulu N. 2012. Reverse engineering: a key component of systems biology to unravel global abiotic stress cross-talk. Frontiers in Plant Science 3, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q-M, Venugopal S, Navarre D, Kachroo A. 2011. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiology 155, 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göhre V, Jones AME, Sklenář J, Robatzek S, Weber APM. 2012. Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Molecular Plant-Microbe Interactions 25, 1083–1092. [DOI] [PubMed] [Google Scholar]
- González-Lamothe R, El Oirdi M, Brisson N, Bouarab K. 2012. The conjugated auxin indole-3-acetic acid–aspartic acid promotes plant disease development. The Plant Cell 24, 762–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A, He K, Liu D, Bai S, Gu X, Wei L, Luo J. 2005. DATF: a database of Arabidopsis transcription factors. Bioinformatics 21, 2568–2569. [DOI] [PubMed] [Google Scholar]
- Hampton CR, Bowen HC, Broadley MR, Hammond JP, Mead A, Payne KA, Pritchard J, White PJ. 2004. Cesium toxicity in Arabidopsis. Plant Physiology 136, 3824–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehl R, Bülow L. 2014. Athamap web tools for the analysis of transcriptional and posttranscriptional regulation of gene expression in Arabidopsis thaliana . In: Staiger D, ed. Plant circadian networks , Vol. 1158 New York: Springer Science+Business Media, 139–156. [DOI] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, et al. 2012. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nature Genetics 44, 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F-C, Chou M-Y, Chou S-J, Li Y-R, Peng H-P, Shih M-C. 2013. Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. The Plant Cell 25, 2699–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K. 2000. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. The Plant Journal 24, 655–665. [DOI] [PubMed] [Google Scholar]
- Kerchev PI, Karpinska B, Morris JA, Hussain A, Verrall SR, Hedley PE, Fenton B, Foyer CH, Hancock RD. 2013. Vitamin C and the abscisic acid-insensitive 4 transcription factor are important determinants of aphid resistance in Arabidopsis. Antioxidants & Redox Signaling 18, 2091–2105. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. 2007. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal 50, 347–363. [DOI] [PubMed] [Google Scholar]
- Kim Y-S, Sakuraba Y, Han S-H, Yoo S-C, Paek N-C. 2013. Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant and Cell Physiology 54, 1660–1672. [DOI] [PubMed] [Google Scholar]
- Kloth KJ, ten Broeke CJM, Thoen MPM, Hanhart-van den Brink M, Wiegers GL, Krips OE, Noldus LPJJ, Dicke M, Jongsma MA. 2015. High-throughput phenotyping of plant resistance to aphids by automated video tracking. Plant Methods 11, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W, Boer MP, Malosetti M, Flood PJ, Engel B, Kooke R, Keurentjes JJB, van Eeuwijk FA. 2015. Marker-based estimation of heritability in immortal populations. Genetics 199, 379–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusnierczyk A, Winge P, Midelfart H, Armbruster WS, Rossiter JT, Bones AM. 2007. Transcriptional responses of Arabidopsis thaliana ecotypes with different glucosinolate profiles after attack by polyphagous Myzus persicae and oligophagous Brevicoryne brassicae . Journal of Experimental Botany 58, 2537–2552. [DOI] [PubMed] [Google Scholar]
- Lange MJP, Lange T. 2015. Touch-induced changes in Arabidopsis morphology dependent on gibberellin breakdown. Nature Plants 1, 14025. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B-H, Henderson DA, Zhu J-K. 2005. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. The Plant Cell 17, 3155–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Polisensky DH, Braam J. 2004. Genome-wide identification of touch- and darkness-regulated Arabidopsis genes: a focus on calmodulin-like and XTH genes. New Phytologist 165, 429–444. [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. 2004. The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. The Plant Cell 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Xie Q, Smith-Becker J, Navarre D, Kaloshian I. 2006. Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Molecular Plant-Microbe Interactions 19, 655–664. [DOI] [PubMed] [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. 2010. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 107, 21199–21204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman AH, Wilkerson CG, Keegstra K. 2005. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proceedings of the National Academy of Sciences of the United States of America 102, 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Lü B, Wang X, Zhang C, Zhang S, Qian J, Chen L, Shi H, Dong H. 2010. Thirty-seven transcription factor genes differentially respond to a harpin protein and affect resistance to the green peach aphid in Arabidopsis. Journal of Biosciences 35, 435–450. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. 2004. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. The Plant Cell 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. 2003. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. The Plant Cell 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis J, Shah J. 2013. Arabidopsis thaliana–Myzus persicae interaction: shaping the understanding of plant defense against phloem-feeding aphids. Frontiers in Plant Science 4, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zou Y, Feng N. 2010. Overexpression of AHL20 negatively regulates defenses in Arabidopsis. Journal of Integrated Plant Biology 52, 801–808. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. 2005. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. [DOI] [PubMed] [Google Scholar]
- Mao P, Duan M, Wei C, Li Y. 2007. WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant and Cell Physiology 48, 833–842. [DOI] [PubMed] [Google Scholar]
- McLean DL, Kinsey MG. 1964. A technique for electronically recording aphid feeding and salivation. Nature 202, 1358–1359. [Google Scholar]
- Menzel TR, Weldegergis BT, David A, Boland W, Gols R, van Loon JJA, Dicke M. 2014. Synergism in the effect of prior jasmonic acid application on herbivore-induced volatile emission by Lima bean plants: transcription of a monoterpene synthase gene and volatile emission. Journal of Experimental Botany 65, 4821–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros T, Helfer A, Hatzimasoura E, et al. 2006. The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. The Plant Journal 48, 485–498. [DOI] [PubMed] [Google Scholar]
- Mewis I, Appel HM, Hom A, Raina R, Schultz JC. 2005. Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiology 138, 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmerman P, Zentgraf U. 2004. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Molecular Biology 55, 853–867. [DOI] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U. 2007. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. The Plant Cell 19, 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minks AK, Harrewijn P. 1989. World crop pests. Aphids. Their biology, natural enemies and control . Amsterdam: Elsevier Science Publishers. [Google Scholar]
- Mitchell A, Chang H-Y, Daugherty L, et al. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Research 43, D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M. 2009. Functional analysis of transcription factors in Arabidopsis. Plant and Cell Physiology 50, 1232–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloi MJ, van der Westhuizen AJ. 2006. The reactive oxygen species are involved in resistance responses of wheat to the Russian wheat aphid. Journal of Plant Physiology 163, 1118–1125. [DOI] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH. 2004. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proceedings of the National Academy of Sciences of the United States of America 101, 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JDG. 2004. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiology 135, 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel RR, Gatz C. 2007. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. The Plant Journal 50, 128–139. [DOI] [PubMed] [Google Scholar]
- Oakey H, Verbyla A, Pitchford W, Cullis B, Kuchel H. 2006. Joint modeling of additive and non-additive genetic line effects in single field trials. Theoretical and Applied Genetics 113, 809–819. [DOI] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Metraux JP, Broekaert WF. 1998. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. The Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM. 2012. Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Platt A, Horton M, Huang YS, et al. 2010. The scale of population structure in Arabidopsis thaliana . PLoS Genetics 6, e1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado E, Tjallingii WF. 2007. Behavioral evidence for local reduction of aphid-induced resistance. Journal of Insect Science 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2013. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rodriguez PA, Bos JIB. 2012. Toward understanding the role of aphid effectors in plant infestation. Molecular Plant-Microbe Interactions 26, 25–30. [DOI] [PubMed] [Google Scholar]
- Schikora A, Schenk ST, Stein E, Molitor A, Zuccaro A, Kogel K. 2011. N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK61. Plant Physiology 157, 1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Febres VJ, Jones JB, Moore GA. 2015. Responsiveness of different citrus genotypes to the Xanthomonas citri ssp. citri-derived pathogen-associated molecular pattern (PAMP) flg22 correlates with resistance to citrus canker. Molecular Plant Pathology 16, 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Boyko EV. 2007. The molecular bases of plant resistance and defense responses to aphid feeding: Current status. Entomologia Experimentalis et Applicata 122, 1–16. [Google Scholar]
- Spoel SH, Dong X. 2008. Making sense of hormone crosstalk during plant immune responses. Cell Host & Microbe 3, 348–351. [DOI] [PubMed] [Google Scholar]
- Sugimoto M, Oono Y, Gusev O, Matsumoto T, Yazawa T, Levinskikh M, Sychev V, Bingham G, Wheeler R, Hummerick M. 2014. Genome-wide expression analysis of reactive oxygen species gene network in Mizuna plants grown in long-term spaceflight. BMC Plant Biology 14, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L. 1984. Plant cell expansion: Regulation of cell wall mechanical properties. Annual Review of Plant Physiology 35, 585–657. [Google Scholar]
- ten Broeke CJM, Dicke M, van Loon JJA. 2013. Performance and feeding behaviour of two biotypes of the black currant-lettuce aphid, Nasonovia ribisnigri, on resistant and susceptible Lactuca sativa near-isogenic lines. Bulletin of Entomological Research 103, 511–521. [DOI] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SH. 2006. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli . The Plant Journal 46, 43–53. [DOI] [PubMed] [Google Scholar]
- Tjallingii WF. 1988. Electrical recording of stylet penetration activities. In: Minks AK, Harrewijn P, eds. Aphids, their biology, natural enemies and control , Vol. 2B Amsterdam: Elsevier, 95–108. [Google Scholar]
- Tjallingii WF. 1994. Sieve element acceptance by aphids. European Journal of Entomology 91, 47–52. [Google Scholar]
- Tosti N, Pasqualini S, Borgogni A, Ederli L, Falistocco E, Crispi S, Paolocci F. 2006. Gene expression profiles of O3-treated Arabidopsis plants. Plant, Cell and Environment 29, 1686–1702. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnology 31, 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B, Mukhtar MS, Somssich IE. 2007. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226, 125–137. [DOI] [PubMed] [Google Scholar]
- van Helden M, Tjallingii WF. 1993. Tissue localisation of lettuce resistance to the aphid Nasonovia ribisnigri using electrical penetration graphs. Entomologia Experimentalis et Applicata 68, 269–278. [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ. 2006. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Vidhyasekaran P. 2014. PAMP Signals in Plant Innate Immunity: Signal Perception and Transduction . Dordrecht: Springer Science+Business Media. [Google Scholar]
- Vos IA, Moritz L, Pieterse CMJ, Van Wees SCM. 2015. Impact of hormonal crosstalk on resistance and fitness of plants under multi-attacker conditions. Frontiers in Plant Science 6, 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling LL. 2008. Avoiding effective defenses: Strategies employed by phloem-feeding insects. Plant Physiology 146, 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, et al. 2009. gplots: Various R programming tools for plotting data. R package , Version 2.17. https://cran.r-project.org/web/packages/gplots/index.html. [Google Scholar]
- Weigel R, Bäuscher C, Pfitzner AP, Pfitzner U. 2001. NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Molecular Biology 46, 143–160. [DOI] [PubMed] [Google Scholar]
- Weigel RR, Pfitzner UM, Gatz C. 2005. Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. The Plant Cell 17, 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Yadav PK, Yadav D, Yadav KDS. 2009. Pectin lyase: A review. Process Biochemistry 44, 1–10. [Google Scholar]
- Yi SY, Shirasu K, Moon JS, Lee S, Kwon S. 2014. The activated SA and JA signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition. PLoS ONE 9, e88951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, Shi HJ, Chen L, et al. 2011. Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae . BMC Plant Biology 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jiang Y, Yu D. 2011. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Molecules and Cells 31, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. 2004. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.