Abstract
Primary effusion lymphoma (PEL) is an aggressive form of non-Hodgkin's B cell lymphoma associated with infection by Kaposi's sarcoma associated herpesvirus (KSHV). (+)-JQ1 and I-BET151 are two recently described novel small molecule inhibitors of BET bromodomain chromatin-associated proteins that have shown impressive preclinical activity in cancers in which MYC is over-expressed at the transcriptional level due to chromosomal translocations that bring the MYC gene under the control of a super-enhancer. PEL cells, in contrast, lack structural alterations in the MYC gene, but have deregulated Myc protein due to the activity of KSHV-encoded latent proteins. We report that PEL cell lines are highly sensitive to BET bromodomain inhibitors-induced growth inhibition and undergo G0/G1 cell-cycle arrest, apoptosis and cellular senescence, but without the induction of lytic reactivation, upon treatment with these drugs. Treatment of PEL cell lines with BET inhibitors suppressed the expression of MYC and resulted in a genome-wide perturbation of MYC-dependent genes. Silencing of BRD4 and MYC expression blocked cell proliferation and cell-cycle progression, while ectopic expression of MYC from a retroviral promoter rescued cells from (+)-JQ1-induced growth arrest. In a xenograft model of PEL, (+)-JQ1 significantly reduced tumor growth and improved survival. Taken collectively, our results demonstrate that the utility of BET inhibitors may not be limited to cancers in which genomic alterations result in extremely high expression of MYC and they may have equal or perhaps greater activity against cancers in which the MYC genomic locus is structurally intact and c-Myc protein is deregulated at the post-translational level and is only modestly over-expressed.
Keywords: Myc, PEL, BET, Bromodomain, KSHV
Introduction
Infection with KSHV is the commonest cause of malignancy among patients with AIDS and one of the commonest causes of malignancies among young adults in many parts of the world (1-2). The prognosis of KSHV associated PEL is extremely poor with a median survival of less than 3 months in most reported studies (3-5). Thus, there is an urgent need for novel molecularly targeted therapies for PEL.
Bromodomains are small evolutionary conserved domains present in many chromatin associated proteins and almost all nuclear histone acetyltransferases (6-7). These domains recognize acetylated-lysine residues in histones and regulate protein–protein interactions involved in chromatin-mediated cellular gene transcription, cell proliferation, and viral transcriptional activation (6-7). The human genome contains 41 bromodomain proteins that have been implicated in a wide range of diseases including cancer, inflammation, obesity, diabetes, infectious diseases and cardiovascular disorders (7). The bromodomain and extra-terminal (BET) subfamily is represented by BRD2, BRD3, BRD4, and BRDT and its members share similar domain structure consisting of two highly conserved amino-terminal bromodomains. Several recent studies have shown that BRD4 plays a key role in hematological malignancies (8-11). These studies have been facilitated by the recent development of small molecule compounds, (+)-JQ1 and I-BET151, which displace BET bromodomain proteins from chromatin by competitively binding to the acetyl-lysine recognition pocket in histones (12-13). Thus, pharmacological inhibition of BRD4 by (+)-JQ1 and I-BET151 in a mouse model of MLL-fusion induced AML was shown to block disease progression in vitro and in vivo (9-10). Treatment with BET inhibitors was also shown to have activity in preclinical models of multiple myeloma and Burktitt's lymphoma (8, 14). The anti-proliferative effects of BET inhibitors in the above disease models were associated with a block in the transcription of key oncogenes, most notably MYC, followed by genome-wide suppression of c-Myc-dependent target genes (8-9, 14). However, since BRD4 is ubiquitously expressed, it was unclear why its inhibition would result in specific down-regulation of c-Myc. A recent study addressed this question using the multiple myeloma cell line MM1.S and reported that BRD4, along with Mediator coactivator complex, is present at extremely high levels at a small set of exceptionally large enhancers, called super-enhancers (15). The MM1.S cell line carries an IgH-MYC rearrangement that places the MYC gene under the control of IgH super-enhancer (15). Treatment of MM1.S cells with (+)-JQ1 was found to lead to preferential loss of BRD4 and its associated co-factors at super-enhancers and caused preferential loss of transcription at genes associated with super-enhancers, including the MYC oncogene (15). Based on these results, BET inhibitors would be expected to have activity primarily against cancers in which the MYC gene comes under the control of a super-enhancer and is highly over-expressed at the transcriptional level.
c-Myc has also been shown to be required for proliferation of PEL cells and for maintenance of KSHV latency (16). However, while MYC is frequently deregulated at the genomic/transcriptional level in human cancers, including cancers against which BET inhibitors have shown activity, the MYC genomic locus is structurally intact in PEL (3). Instead, c-Myc is deregulated in PEL at the post-translational level due to the activity of two KSHV latent proteins, LANA and vIRF3/LANA2, which enhance the stability of c-Myc and stimulate its transcriptional activity (17-19). To examine whether BET inhibitors may also have activity against cancers in which MYC is not up-regulated at the transcriptional level, we examined their activity against PEL cells. We demonstrate that the utility of BET inhibitors is not limited to cancers in which genomic alternations bring the MYC genes under the control of a super-enhancer and these compounds may have equal or greater activity against cancers in which the MYC genomic locus is structurally intact and c-Myc protein is deregulated at the post-translational level.
Results
Anti-proliferative effects of (+)-JQ1 on PEL cells lines
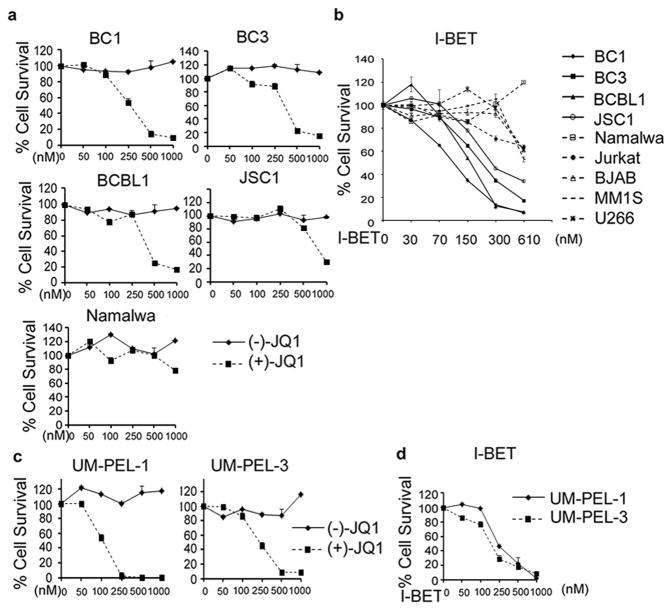
To explore the effect of BRD4 inhibitors on the survival and proliferation of PEL cells, we treated four PEL-derived cell lines, BC1 (KSHV+/EBV+), BC3 (KSHV+/EBV-), BCBL1 (KSHV+/EBV-), and JSC1 (KSHV+/EBV+) with increasing doses of (+)-JQ1. As shown in Figure 1a, treatment with increasing doses of (+)-JQ1 for a period of 5 days strongly reduced the survival of BC1, BC3 and BCBL1 in a dose-dependent manner as measured by an MTS assay (IC50 = 250 nM, 380 nM, and 380 nM for BC1, BC3, BCBL1, respectively). (+)-JQ1 also blocked the proliferation of JSC1 cells, albeit at slightly higher doses (IC50 = 790 nM). In contrast, Burkitt's lymphoma-derived Namalwa (KSHV-/EBV+) cells were relatively resistant to (+)-JQ1 (IC50 = 1130 nM). Treatment with (-)-JQ1, an inactive enantiomer of (+)-JQ1 (12), had no significant growth inhibitory effect on any of the tested cell lines (Figure 1a). To further demonstrate the sensitivity of PEL cells to (+)-JQ1, we next examined its effect on a panel of leukemia and lymphoma cell lines of diverse lineages. The IC50 of (+)-JQ1 for the non-PEL cell lines in this panel ranged from 820 nM to >5 μM, which were considerably higher than its IC50 for the PEL cell lines (Table 1). Collectively, the above results demonstrate that the PEL-derived cell lines are remarkably sensitive to (+)-JQ1-induced growth inhibition.
Figure 1. BRD4 inhibitors reduce cell viability in PEL cells lines in a dose-dependent manner.
(a) PEL cell lines (BC1, BC3, BCBL1 and JSC1) were treated with indicated doses of (-)-JQ1 or (+)-JQ1 for 5 days and cell viability measured using an MTS assay (refer to Materials and methods). Namalwa, a Burkitt's lymphoma cell line, was used as a control. (b) PEL cell lines (solid lines) and non-PEL cells lines (dotted lines) were treated in triplicate with the indicated concentrations of I-BET151 and cell viability measured after 5 days using the MTS assay. (c-d) Patient-derived UMP-EL-1 and UM-PEL-3 cells were treated with indicated doses of (-)-JQ1, (+)-JQ1 (c) or I-BET151 (d) for 5 days and cell viability measured using an MTS assay. The values shown are mean ± SD of a representative of two independent experiments performed in triplicate.
Table 1. IC50 values I for (+)-JQ1 (top panel) and I-BET (bottom panel) in PEL and non-PEL derived lymphoma and leukemia cells.
| Cell line | (+)-JQ1 IC50 (nM) |
| BC1 (PEL) | 250 |
| BC3 (PEL) | 380 |
| BCBL1 (PEL) | 380 |
| JSC1 (PEL) | 790 |
| UM-PEL-1 (patient-derived PEL) | 130 |
| UM-PEL-3 (patient-derived PEL) | 220 |
| Namalwa (Burkitt's lymphoma) | 1130 |
| Karpass-299 (lymphoma) | 820 |
| H9 (lymphoma) | 1260 |
| Jurkat (T-cell leukemia) | 1340 |
| MOLT-4 (T-cell leukemia) | 1320 |
| HPB-ALL (T-cell leukemia) | >5000 |
| K562 (chronic myelogenous leukemia) | >5000 |
|
| |
| Cell line | I-BET IC50 (nM) |
|
| |
| BC1 (PEL) | 220 |
| BC3 (PEL) | 460 |
| BCBL1 (PEL) | 330 |
| JSC1 (PEL) | 680 |
| UM-PEL-1 (patient-derived PEL) | 210 |
| UM-PEL-3 (patient-derived PEL) | 180 |
| Namalwa (Burkitt's lymphoma) | 970 |
| Karpass-299 (lymphoma) | 820 |
| BJAB (B lymphoma) | 970 |
| Jurkat (T-cell leukemia) | 1220 |
| MM1S (multiple myeloma) | 760 |
| U266 (multiple myeloma) | 950 |
IC50 values were calculated using Graphpad Prism 5 software.
Anti-proliferative effect of I-BET151 on PEL cell lines
To next demonstrate that the observed growth inhibitory effect of (+)-JQ1 on PEL cell lines is a class-effect, we took advantage of I-BET151, a structural analog of JQ1 (10). Treatment with I-BET151 resulted in a dose-dependent reduction in the proliferation of all tested cell lines. However, similar to the results with (+)-JQ1, the IC50 values of I-BET151 for PEL cell lines (220 nM, 460 nM and 330 nM for BC1, BC3 and BCBL1 cells) were significantly lower than their IC50 for non-PEL cells (970 nM, 970 nM, 1220 nM, 760 nM and 950 nM for BJAB, Namalwa, Jurkat, MM1S and U266 cell lines, respectively) (Figure 1b and Table 1). Taken collectively, the above results confirm that PEL cell lines are remarkably sensitive to BET inhibitors.
Anti-proliferative effects of (+)-JQ1and I-BET151 on UM-PEL-1 and UM-PEL-3 cells
Due to its rarity, many studies on PEL are limited to PEL-derived cell lines that may deviate from the original tumors seen in the patient due to culture-induced adaptations. To circumvent this problem, we tested the effect of BET inhibitors on UM-PEL-1 and UM-PEL-3 cell models (20-21). These models were established by transferring freshly isolated human PEL cells into the peritoneal cavities of NOD/SCID mice and have been subsequently maintained by serial propagation in vivo without in vitro cell growth to avoid the changes in KSHV gene expression evident in cultured cells (20-21). Treatment with (+)-JQ1 inhibited the proliferation of UM-PEL-1 and UM-PEL-3 cells with IC50 values of 130 nM and 220 nM, respectively, while (-)-JQ1 had no inhibitory effect (Figure 1c). Similarly, I-BET151 blocked the proliferation of UM-PEL-1 and UM-PEL-3 cells with IC50 values of 210 nM and 180 nM, respectively (Figure 1d).
BET inhibitors induce cell cycle arrest in PEL cells
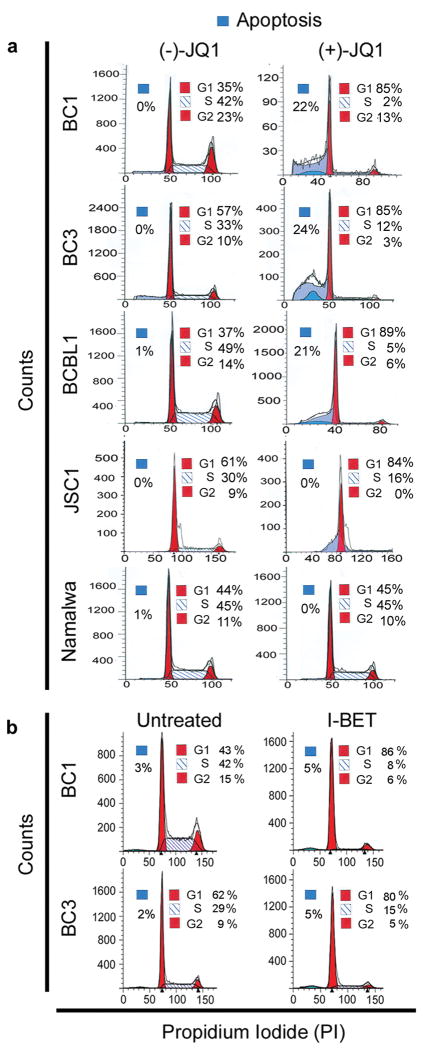
To study the mechanism by which BET inhibitors reduce the proliferation of PEL cells, we examined their effect on cell cycle progression using flow cytometry. Treatment of PEL cells with (+)-JQ1 for 48 hours resulted in a pronounced reduction in cells in the S phase as compared to (-)-JQ1-treated cells (Figure 2a and Supplementary Table 3). This was accompanied by a concomitant increase in cells arrested in the G0/G1 phase (Figure 2a and Supplementary Table 3). Essentially similar results were obtained upon treatment of BC1 and BC3 cells with I-BET151 (Figure 2b and Supplementary Table 3). In contrast, (+)-JQ1 had no major effect on cell-cycle progression in Namalwa cells (Figure 2a). Thus, BET inhibitors appear to delay progression of PEL cells into the S phase of the cell cycle, resulting in G0/G1-growth arrest.
Figure 2. BET bromodomain inhibitors induce cell cycle arrest in PEL cells.
(a, b) Cell-cycle analysis of (-/+)-JQ1-(500 nM for 48 hours) and I-BET151 (500 nM for 48 hours)-treated PEL cell lines showing significant G0/G1 arrest. A non-PEL cell line, Namalwa, was used as a control. Cells were stained with propidium iodide (PI) and analyzed by flow cytometry. Results are representative of two independent experiments.
Induction of apoptosis and cellular senescence by (+)-JQ1 in PEL cells
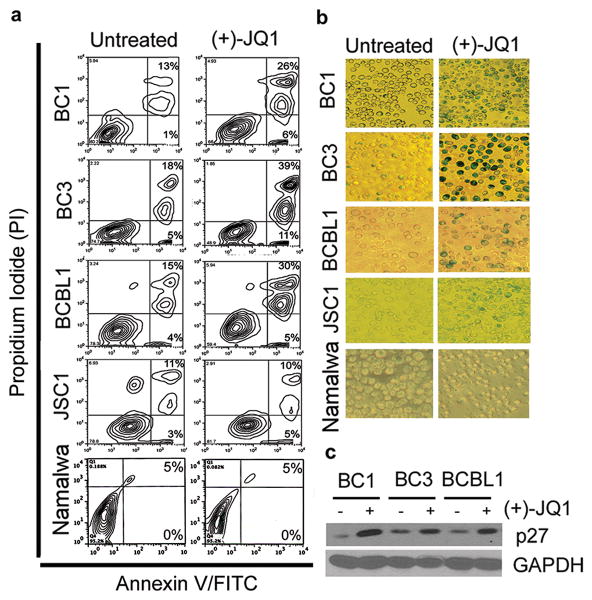
The cell-cycle analysis also suggested an increase in the proportion of apoptotic cells upon (+)-JQ1 treatment, indicating induction of cell death via apoptosis (Figure 2a). Consistent with it, treatment with (+)-JQ1 for 48 hours resulted in a modest increase in Annexin V+/PI+ cells (Figure 3a). In contrast, no increase in the Annexin V+/PI+ fraction was seen upon treatment of JSC1 and Namalwa cells with (+)-JQ1 (Figure 3a). A dose-response experiment demonstrated the dose-dependency of apoptosis induction in response to (+)-JQ1-treatment in BC1 and BCBL1 cells, but not in Namalwa cells (Figure S1a). We also evaluated the effect of increasing doses of (+)-JQ1 on the alterations in the expression of pro- and anti-apoptotic proteins. We did not observe a significant increase in the cleavage of PARP or down-regulation of Mcl-1 upon treatment with (+)-JQ1 in BC1, BCBL1 and Namalwa cells (Figure S1b). A modest reduction in Bcl-xL expression was seen upon treatment of BC1 cells with increasing doses of (+)-JQ1, but was not seen in BCBL1 and Namalwa cells (Figure S1b). We also evaluated the effect of JQ1 on cellular senescence. PEL cells were treated with (+)-JQ1 and subsequently stained with β-galactosidase, a marker for cellular senescence. Consistent with their relative sensitivity to (+)-JQ1-mediated growth inhibition, BC1, BC3 and BCBL1 cells showed strong β-galactosidase staining following (+)-JQ1 treatment, while JSC1 and Namalwa cells showed weak to no increase in staining, respectively (Figure 3b). The increase in β-galactosidase staining in BC1, BC3 and BCBL1 cells was accompanied by up-regulation of p27 (Figure 3c), a known marker of cellular senescence (22). As a control, we also evaluated the effect of (-)-JQ1 on induction of apoptosis and cellular senescence in PEL cells and observed no significant effect (Figure S2). Taken together, treatment of PEL cells with BET inhibitors results in a phenotype of reduced proliferation, G0/G1 cell cycle arrest, mild increase in apoptosis and induction of cellular senescence.
Figure 3. Induction of apoptosis and cellular senescence by (+)-JQ1 in PEL cells.
(a) PEL cells were exposed to 500 nM (+)-JQ1 for 48 hours, stained with Annexin-V-FITC/PI and analyzed for apoptosis by flow cytometry. (b) PEL cells were treated with 500 nM (+)-JQ1 for 72 hours and its effect on cellular senescence detected by β-galactosidase staining. Results are representative of two independent experiments. (c) Western blot analysis showing induction of p27 upon 48 hours of treatment with (+)-JQ1. GAPDH serves as a loading control.
Analysis of differential gene expression in PEL following JQ1 treatment
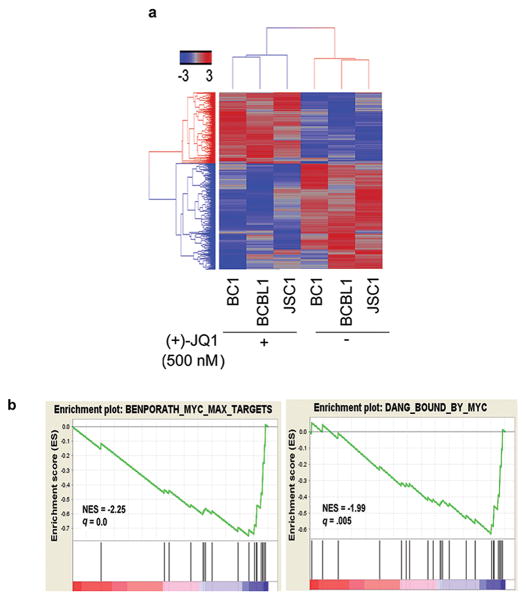
To test the effect of BET inhibition on gene transcription, BC1, BCBL1 and JSC1 cells were treated with (+)-JQ1 (500 nM) for 8 hours followed by genome-wide transcriptome analysis using Illumina's RNA-Seq platform. Unsupervised hierarchical clustering separated samples according to their treatment group, indicating a common transcriptional response to treatment with (+)-JQ1 (Figure 4a). Rather than inducing non-specific changes in gene expression, (+)-JQ1 induced significant changes in a limited number of genes. Thus, there were 350 genes (229 down- and 129 up-regulated genes) whose expression was changed ≥2 fold in all three cell lines. We used a GSEA program to identify functional gene sets whose expression changed significantly with (+)-JQ1 treatment in PEL cells (23). For this purpose, we focused on gene sets containing genes co-regulated in response to genetic and chemical perturbations. Among the top gene signatures identified by this analysis were two gene sets (BENPORATH_MYC_MAX_TARGETS and DANG_BOUND_BY_MYC) containing genes that are known targets of Myc (Figure 4b).
Figure 4. (+)-JQ1 treatment affects expression of MYC-dependent genes.
(a) Heat map representation of 1,443 genes that are up- or down-regulated ≥1.5 fold in two of the three PEL cell lines following 8 hour treatment with 500 nM (+)-JQ1. (b) Gene Set Enrichment Analysis showing enrichment of gene sets containing target genes of MYC among genes affected by (+)-JQ1 treatment in PEL cells; NES=normalized enrichment score, q= false discovery rate.
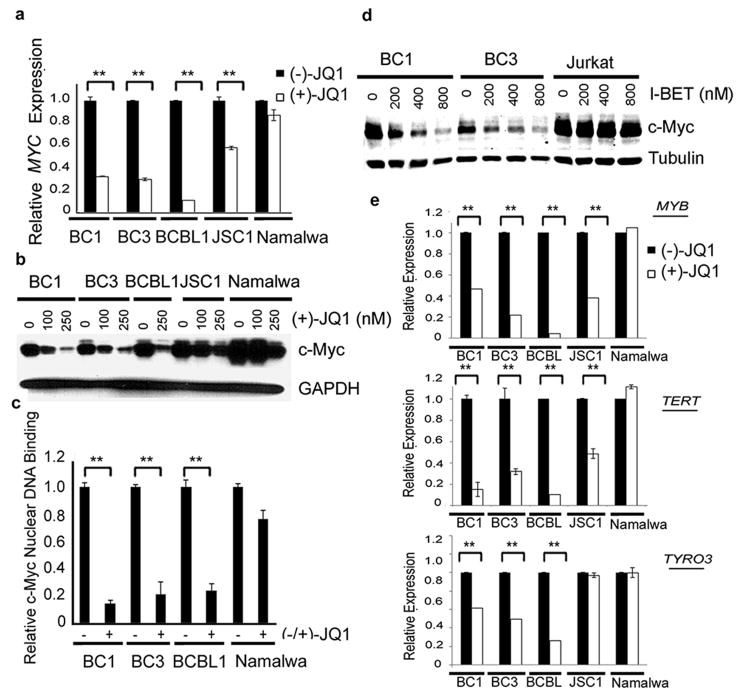
(+)-JQ1 inhibits Myc transcription, expression, Myc downstream target genes and Myc Nuclear Binding
We used qRT-PCR to confirm down-regulation of MYC by (+)-JQ1 in PEL cell lines. we observed significant reduction in MYC expression in BC1, BC3, BCBL1 and JSC1 cells following exposure to (+)-JQ1 as compared to the (-)-JQ1-treated cells (Figure 5a). In contrast, (+)-JQ1 had no significant inhibitory effect on MYC expression in the Namalwa cells (Figure 5a). We also observed a dose-dependent decline in c-Myc protein expression in BC1, BC3 and BCBL1 cells (Figure 5b), which was associated with depletion of chromatin-bound c-Myc, as measured by an ELISA-based DNA-binding assay (Figure 5c). The baseline level of c-Myc protein was higher in JSC1 cells as compared to the other PEL cell lines and its significant residual expression was observed in the (+)-JQ1-treated cells (Figure 5b), which probably contributed to their reduced sensitivity to the drug in the MTS assay (Table 1). Additional support for the hypothesis that the level of c-Myc is a key determinant of response to (+)-JQ1 was provided by studies on Namalwa cells, which are relatively resistant to (+)-JQ1 (IC50=1130 nM) (Table 1). The basal level of c-Myc was significantly higher in Namalwa cell as compared to the PEL cell lines and remained higher following treatment with (+)-JQ1 (Figure 5b). A close correlation between the response to BET inhibitors and c-Myc level was also observed in experiments involving treatment with I-BET151. As shown in Figure 5d, I-BET151 resulted in a dose-dependent decline in c-Myc protein levels in the BC1 and BC3 cell lines. However, similar to Namalwa cells, the basal level of c-Myc in the I-BET151-resistant Jurkat cells was relatively high as compared to the PEL cell lines and was not significantly affected by treatment with the drug (Figure 5d). Collectively, the above results support the hypothesis that the level of c-Myc is a key determinant of response to BET inhibitors.
Figure 5. BET inhibitors block MYC transcription programs in PEL.
(a) Quantitative RT-PCR analysis of MYC levels in PEL and non-PEL (Namalwa) cells treated with 250 nM of (-)- or (+)-JQ1 for 48 hours. Real-time PCR reactions were performed in triplicate and the data presented as fold change in target gene expression (Mean ± SD) after normalization with GAPDH as a housekeeping gene. The results shown are representative of two independent experiments. (b) PEL and non-PEL (Namalwa) cells treated with indicated doses of (+)-JQ1 for 48 hours were lysed and analyzed by Western blot for the expression of c-Myc. GAPDH was used as a loading control. (c) PEL cell lines (BC1, BC3 and BCBL1) and non-PEL (Namalwa) cells were treated with 250 nM of (-)- or (+)-JQ1 for 72 hours and nuclear extracts were isolated to study the effect of (+)-JQ1 on nuclear c-Myc protein level as measured by an ELISA-based DNA-binding assay. The results shown are representative of two independent experiments performed in triplicate. ** indicates p > 0.01 calculated using a two-tailed Student's t-test. (d) Western blot analysis of c-Myc upon 48 hours of indicated concentrations of I-BET151 treatment with Tubulin as a loading control. (e) Quantitative RT-PCR analysis of MYC downstream target genes – MYB and TERT, and TYRO3 in the indicated cell lines treated with 250 nM of (-)- or (+)-JQ1 for 48 hours. GAPDH was used as a housekeeping gene to normalize gene expression data. The results shown are a representative of two independent experiments performed in triplicate. ** indicates p > 0.01 calculated using a two-tailed Student's t-test.
Finally, we used qRT-PCR analysis to examine the effect of (+)-JQ1 on the expression of two known downstream MYC target genes, MYB and TERT. We also included TYRO3, a gene known to be down-regulated by (+)-JQ1, in the analysis (8). (+)-JQ1 resulted in significant reduction in the expression of MYB and TERT in BC1, BC3, BCBL1 and JSC1 cell lines and significant reduction in TYRO3 expression in BC1, BC3, and BCBL1 cell lines as compared to (-)-JQ1-treated cells (Figure 5e). (+)-JQ1 also reduced the expression of two additional MYC target genes PMM2 (24) and SLC19A1 (25) in BC1 and BCBL1 cells (Figure S3). In contrast, (+)-JQ1 treatment had no significant impact on the expression of MYB, TERT and TYRO3 in the Namalwa cells (Figure 5e). Thus, sensitivity to (+)-JQ1 correlates closely with the degree of down-regulation of MYC-target genes.
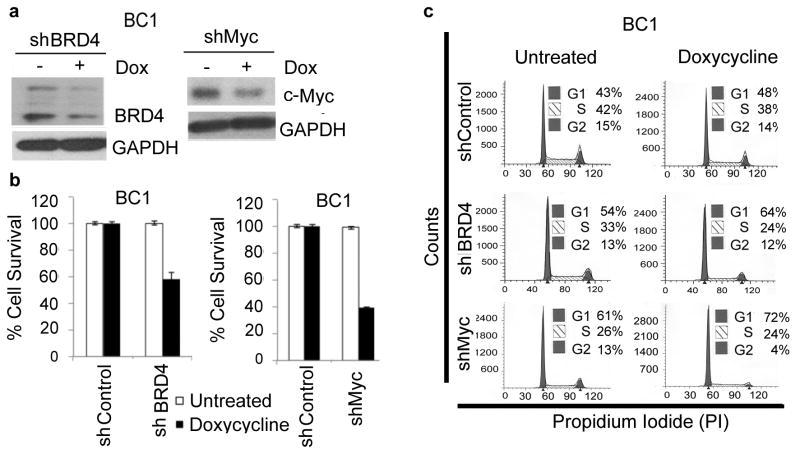
Suppression of BRD4 and c-Myc expression reduces PEL cell viability
In the case (+)-JQ1 exerts its effect in PEL cells by inhibiting BRD4 and downregulating c-Myc, then silencing of BRD4 and MYC by shRNAs would be expected to resemble the effect of (+)-JQ1 treatment. To test this hypothesis, we generated stable clones of BC1 cells expressing tetracycline-inducible H1 promoter (H1/TO)-driven shRNAs targeting BRD4 and MYC, respectively. Treatment of BC1-H1/TO-BRD4-shRNA and BC1-H1/TO-MYC-shRNA cells with doxycycline resulted in downregulation of BRD4 and c-Myc, respectively, as determined by Western blotting (Figure 6a). More importantly, doxycycline treatment of BC1-H1/TO-BRD4-shRNA and BC1-H1/TO-MYC-shRNA cells was accompanied by approximately 40% and 60% reduction in cellular proliferation (Figure 6b and Supplemtary Table 4), respectively, and G0/G1 cell-cycle arrest (Figure 6c), thus resembling the phenotype of (+)-JQ1-treated cells. In contrast, doxycycline treatment had no significant effect on cell proliferation or cell-cycle progression in BC1 cells expressing a scrambled shRNA as a control (Figure 6b and 6c).
Figure 6. Silencing of BRD4 and MYC blocks proliferation of PEL cells.
(a) Western blot of whole cell lysates from BC1 cells stably expressing tetracycline-inducible shRNAs targeting BRD4 and MYC. Cells were grown in the absence and presence of doxycycline (500 ng/mL) for 11 days. GAPDH was used as a loading control. (b) BC1 cells stably transduced with control, BRD4 or MYC shRNAs were treated with doxycycline (500 ng/mL, 5 days) and cell survival was measured by MTS assay. The values shown are mean ± S.D of a representative of two independent experiments performed in triplicate. (c) BC1 cells stably transduced with control, BRD4 or MYC shRNAs were treated with doxycycline (500 ng/mL, 5 days) and stained with propidium iodide for cell-cycle analysis. The data represents the mean ± S.D of two independent experiments.
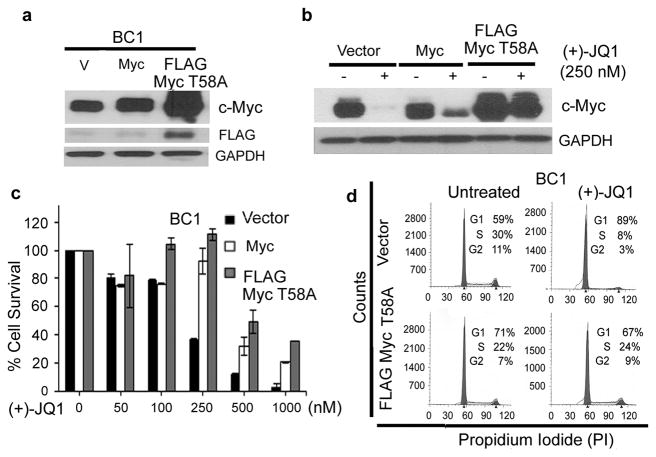
Reconstitution of MYC rescues (+)-JQ1-treated cells from growth-suppression
To confirm the functional involvement of c-Myc down-regulation in the anti-proliferative effect of (+)-JQ1, we studied whether ectopic expression of c-Myc from a retroviral-derived promoter would rescue them from JQ1-induced growth inhibition. For this purpose, we generated BC1 cells stably expressing either wild-type c-Myc or its FLAG-tagged T58A mutant, which is resistant to phosphorylation-induced degradation. Consistent with the increased stability of the T58A mutant, c-Myc protein expression was modestly increased in the wild-type c-Myc-transduced cells but strongly increased in the c-Myc-T58A mutant transduced cells (Figure 7a). We next compared the response of the resulting cells to (+)-JQ1. While (+)-JQ1 treatment led to near complete disappearance of c-Myc expression in the control cells, modest and strong residual c-Myc expression was seen in the wild-type Myc and T58A-Myc transduced cells, respectively (Figure 7b). Both Myc constructs conferred significant protection against (+)-JQ1-induced growth inhibition, with the T58A-Myc expressing cells showing slightly higher protection (Figure 7c). The addition of ectopic Myc to BC1 cells also rescued them from (+)-JQ1-induced cell-cycle arrest as determined by flow cytometry (Figure 7d and Supplementary Table 5). Taken collectively, these results demonstrate that down-regulation of Myc plays a major role in the growth-inhibitory effect of BET inhibitors in PEL cells.
Figure 7. Reconstitution of MYC rescues (+)-JQ1-treated cells from growth-suppression.
(a) BC1 cells infected with retroviral vectors expressing an empty vector, Myc or FLAG-tagged Myc T58A were analyzed for expression of the transduced proteins by Western blotting using antibodies against c-Myc and FLAG. GAPDH was used as a loading control. (b) BC1 cells stably expressing Myc or Myc T58A were treated with 250 nM of (+)-JQ1 for 48 hours and the effect of JQ1 on c-Myc expression was examined by Western blotting. GAPDH was used to show equal protein loading. (c) PEL cells expressing vector control or exogenous Myc or Myc T58A were treated with 500 nM of (+)-JQ1 for 5 days and the cell survival was measured by an MTS assay. The values shown are mean ± SD of three independent experiments performed in triplicate. (d) BC1 cells stably expressing vector control or exogenous Myc or Myc T58A were treated with 250 nM of (+)-JQ1 for 48 hours and stained with propidium-iodide for cell cycle analysis. The data represents one of three independent experiments.
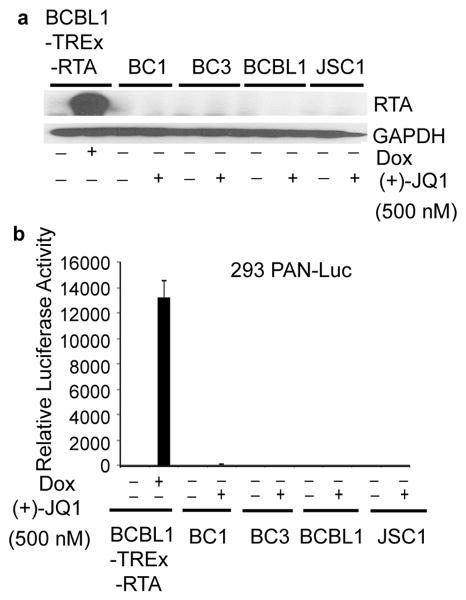
Lack of induction of KSHV lytic replication by (+)-JQ1 in PEL
Myc is required for maintenance of KSHV latency and its shRNA-mediated silencing results in the induction of KSHV Replication and Transcription Activator (RTA), the master regulator of KSHV lytic replication (16). Therefore, we next examined whether treatment with (+)-JQ1 results in lytic reactivation of the virus, as measured by expression of RTA using immunoblotting. As a positive control, we used BCBL1-TREx-RTA cells, which express RTA from a tetracycline-inducible promoter and undergo a complete cycle of viral replication upon treatment with doxycycline (26). While treatment of BCBL1-TREx-RTA cells with doxycycline resulted in robust induction of RTA, treatment of PEL cell lines with (+)-JQ1 failed to do so (Figure 8a). We have previously generated a reporter cell line, designated 293-PAN-Luc, which expresses the firefly luciferase gene under the control of KSHV PAN promoter and responds to infection with KSHV by increasing luciferase activity in a sensitive and quantitative manner (27). To confirm the lack of lytic replication induction by (+)-JQ1, cell-free supernatants from (+)-JQ1-treated PEL cells were applied to 293PAN-Luc cells and assessed for increase in luciferase activity. As shown in Figure 8b, exposure of 293PAN-Luc cells to supernatant from doxycycline-treated BCBL1-TREx-RTA cells led to a significant increase in luciferase activity while supernatant from (+)-JQ1-treated PEL cells failed to do so. Thus, (+)-JQ1 induced growth inhibition and apoptosis is not accompanied by production of infectious virions.
Figure 8. Lack of induction of KSHV lytic replication by (+)-JQ1 in PEL cells.
(a) Immunoblot analysis of RTA in PEL cells treated with (+)-JQ1 (48 hours, 500 nM). RTA expression in BCBL1-TREx-RTA cells upon treatment with doxycycline (48 hours, 100 ng/mL) serves as a positive control for lytic induction. GAPDH is used as a loading control. (b) (+)-JQ1-treatment (72 hours, 500 nM) of PEL cells does not induce production of infectious virions. Cell-free supernatants were collected from PEL cells treated with 500 nM (+)-JQ1 for 72 hours and used to infect 293PAN-Luc cells containing a luciferase reporter construct driven by PAN promoter. Supernatant from doxycycline-treated BCBL-Trex-RTA cells was used a positive control. Data represents one of two experiments performed in duplicate (mean +/- SD, n=2).
(+)-JQ1 exhibits in vivo growth inhibitory activity against PEL in mouse xenografts
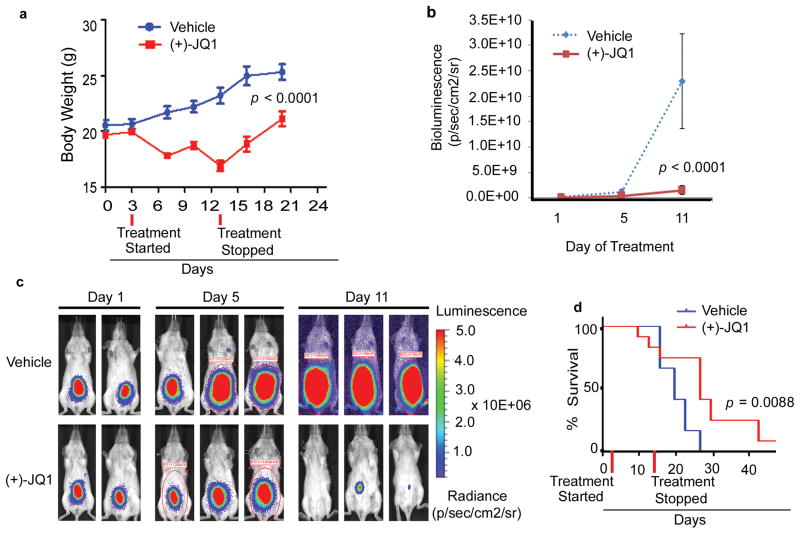
To check the in vivo efficacy of (+)-JQ1 against PEL, we injected the BC1 cells expressing an RSV-promoter driven firefly luciferase cDNA (BC1-Luc) intra-peritoneally into NOD/SCID mice. Animals with established tumors, as detected by bioluminescence imaging on day 3, were randomly assigned to vehicle control (n= 12) and (+)-JQ1 (50 mg/kg twice daily for 11 days; n = 12). Animals treated with (+)-JQ1 showed reduced tumor growth as determined by bioluminescence imaging and measurement of body weight, a surrogate for tumor-induced ascites (Figures 9a-c). In addition, the median survival of (+)-JQ1-treated animals was significantly improved as compared to the control vehicle-treated animals (27 days vs. 20 days; p=.0088) (Figures 9d).
Figure 9. (+)-JQ1 impairs in vivo growth of PEL in a mouse xenograft model.
(a) 5 week old female NOD/SCID mice were injected intra-peritoneally (IP) with 2 × 107 BC1-Luc cells. Upon establishment of tumors, mice were randomly segregated into vehicle control or (+)-JQ1 (50 mg/kg IP twice daily × 11 days) groups. Change in body weight of mice assigned to vehicle control or (+)-JQ1 (p <0.0001) are shown. (b) Tumor burden of vehicle control or (+)-JQ1-treated mice as measured by bioluminescence imaging (p<0.0001, day 11). (c) Representative whole body bioluminescence images of mice injected with BC1-Luc cells 1-, 5- and 11-days post vehicle control or (+)-JQ1 treatment. (d) Survival curves (Kaplan-Meier) of mice injected with PEL cells showing an increase in survival of (+)-JQ1 treated mice when compared with vehicle control (p = 0.008, log-rank test).
Discussion
As a master regulator of cell proliferation, c-Myc contributes to the disease progression in a number of human cancers, making it a prime therapeutic target (28). However, conventional approaches to directly block c-Myc function using small molecule compounds have proven to be largely ineffective. Using a first in class inhibitor of BET bromodomains, which block Myc transcription and function indirectly through displacement of chromatin bound co-activator proteins, we provide evidence that PEL cell lines are sensitive to BET inhibitors in vitro and in vivo. Indeed, the involvement of c-Myc in the inhibitory effects of BET inhibitors in PEL cell lines is supported by the following lines of evidence. First, we observed significant down-regulation of c-Myc transcript and protein levels in PEL cell lines following treatment with BET inhibitors, which was accompanied by depletion of nuclear c-Myc. Second, we observed a marked enrichment of down-stream target genes of c-Myc among the genes perturbed in PEL cell lines following treatment with BET inhibitors. Third, there was a good correlation between the sensitivity to BET inhibitors and degree of down-regulation of MYC and its downstream target genes. Fourth, shRNA-mediated silencing of MYC and BRD4 in BC1 cells resulted in cell-cycle arrest, thus resembling the cell-cycle effects observed in cells treated with the BET inhibitors. Consistent with our results in BC1 cells, knock-down of MYC has been also reported to induce cell-cycle arrest in the PEL-derived BC3 cell line (16). Finally, over-expression of c-Myc via an ectopic retroviral promoter rescued PEL cells from (+)-JQ1-induced growth inhibition.
BET inhibitors have been previously shown to down-regulate MYC and possess selective activity against Burkitt's lymphoma (BL) and multiple myeloma (8, 14), which carry chromosomal translocations involving the MYC locus (29-30). The selective down-regulation of MYC by (+)-JQ1 has been attributed to the preferential loss of BRD4 at the super-enhancers and consequent transcription elongation defects that preferentially impacted genes with super-enhancers, including MYC (15). In contrast, we demonstrate the activity of BET inhibitors against PEL, in which the MYC locus is structurally intact. More importantly, we observed that PEL cells are highly sensitive to BET bromodomain inhibitors. Indeed, the IC50 of both (+)-JQ1 and I-BET151 for PEL cell lines was generally lower than their IC50 for a panel of hematologic cell lines of diverse origins, including cell lines derived from multiple myeloma and BL. Although the precise reason for the remarkable sensitivity of PEL cell lines to BET inhibitors is not known at the present, our results suggest that the relative level of the c-Myc protein may play a role in this process. The basal and the post-treatment levels of the c-Myc protein were significantly higher in the Namalwa and Jurkat cells lines that are relatively resistant to BET inhibitors as compared to the drug-sensitive PEL cell lines. Even among the PEL cell lines, the basal and post-treatment levels of c-Myc protein correlated well with their resistance to BET inhibitors. Thus, a possible explanation for the greater sensitivity of PEL cells to BET inhibitors may lie in their relatively modest expression of the c-Myc protein that is easily suppressed by the drug treatment. The modest elevation of c-Myc in PEL cells may in turn reflect the fact that the PEL cells lack structural alterations in the MYC gene (3) and the elevation of c-Myc activity in these cells is primarily due to post-translational mechanisms involving enhanced stabilization or activity of the protein (17-19). Thus, while the current paradigm suggests that the ideal candidates for BET inhibitors are those cancers in which MYC is highly over-expressed due to the activity of super-enhancers (15), our results suggest that such cancers may not be the most sensitive to treatment with these compound because they will require much higher doses of the inhibitors to completely down-regulate c-Myc expression from the extremely high basal levels. Instead, our study suggests that cancers, such as PEL, which lack structural alterations at the MYC locus and express only modestly high levels of the c-Myc protein, are probably equally good or perhaps better candidates for treatment with the BET inhibitors.
While our results suggest a key role of c-Myc down-regulation in the cellular response to BET inhibitors, perturbation in other cellular and viral genes and signaling pathways is also likely to contribute to this process. Additionally, while ectopic expression of c-Myc successfully protected PEL cells from low doses of (+)-JQ1, it failed to confer protection when exposed to higher doses of the drug. Thus, the effect of BET inhibitors on Myc-independent pathways may also contribute to their growth inhibitory effects against PEL cells. Indeed, I-BET has been shown to block the expression of a number of inflammatory genes that have been implicated in the pathogenesis of KSHV-associated malignancies (13). Additionally, KSHV-encoded LANA tethers viral genomes to host mitotic chromosomes that ensure maintenance of viral episomes in dividing cells (31-32). The interaction between the carboxy-terminal domain of LANA and host chromatin is mediated via BRD4 (33-34). BRD4 was also shown to associate with mitotic chromosomes throughout mitosis and co-localize with LANA and the KSHV episomes on host mitotic chromosomes (33-34). Thus, it is conceivable that the effect of BET inhibitors on LANA-BRD4 interactions and KSHV episome maintenance may also contribute to its growth inhibitory effects in PEL cells.
Finally, induction of lytic replication is increasingly believed to play an important role in KSHV-tumorigenesis (35). Silencing of Myc was recently shown to induce expression of RTA and lytic protein K8 (16). In contrast, we observed that (+)-JQ1 did not induce expression of lytic genes or production of infectious virions, thereby allaying these safety concerns. While the exact mechanism for the lack of lytic reactivation by (+)-JQ1 is not clear at the present, its effects on Myc-independent genes may be responsible for this process.
In summary, we have used first in class BET inhibitors to provide strong in vitro and in vivo pre-clinical rationales for targeting BRD4 and MYC for the treatment of PEL. Epigenetic modulators affect thousands of genes and many targets of BET inhibitors have already been described in the literature (9, 11-12, 15, 36). Although our study does not identify any new target of the BET inhibitors, it addresses a more pressing and clinically relevant issue regarding the therapeutic development of these compounds: namely, the range of cancers against which these compounds are likely to show activity. We demonstrate that the utility of BET inhibitors may not be limited to malignancies in which c-Myc is highly over-expressed due to genomic alterations that bring the MYC gene under the control of a super-enhancer and that they may have equal or even greater activity against cancers in which the MYC genomic locus is structurally intact and c-Myc is only modestly over-expressed. Thus, our results considerably extend the range of cancers that are candidates for clinical testing of BET inhibitors.
Materials and methods
Cell lines and reagents
293FT, BC1, BC3, BCBL1, JSC1, Namalwa, Jurkat, BJAB, H9, K562 cells were obtained from ATCC (Manassas, VA); human multiple myeloma cell lines, MM1S and U266, were from Dr. Ellen Vitetta (UT Southwestern Medical Center) and Karpass-299 and HPB-ALL were from Dr. Alan Epstein (USC). UM-PEL-1 and UM-PEL-3 were provided by Drs. Izidore Lossos and Juan Ramos, respectively (both from U Miami). 293-PAN-Luc and BCBL1-TREx-RTA cells have been described previously (27). BC1-Luc cells were generated by infection with a retroviral vector encoding a Rous Sarcoma Virus promoter driven firefly luciferase construct. (+)-JQ1 and its inactive enantiomer (-)-JQ1 were generously provided by Dr. James Bradner (Dana-Farber Cancer Institute). I-BET151 was from ChemieTek and Doxycycline was purchased from Sigma (St. Louis, MO).
Retroviral and lentiviral constructs
Retroviral constructs containing cMyc-T58A and Flag epitope-tagged wild-type c-Myc were obtained from Addgene. Recombinant retroviruses were generated and used to infect PEL cell lines as described previously (37). For generation of shRNA encoding lentiviral vectors, shRNA oligonucleotides (Supplementary Table 2) directed against human c-Myc and BRD4 mRNAs were annealed and cloned into a modified pENT entry vector containing a tetracycline-inducible H1 promoter (H1/TO). The cassette containing the H1/TO promoter along with the shRNA hairpin was concatenated as described earlier (37) so as to result in eight copies of the H1/TO-shRNA cassettes in each vector. Recombination based sub-cloning was used to transfer the concatenated H1/TO-shRNA cassettes into pSLIK destination vector (38). 293FT cells were used to induce virus production as described previously (37). Packaged virus was then used to infect PEL cells and positive clones were selected with G418. Doxycycline (500 ng/mL) was used to induce the expression of shRNAs.
Animal studies
5 week old female NOD/SCID mice ( NCI) were injected intra-peritoneally (IP) with 2 × 107 BC1-Luc cells. To assess establishment of tumors, mice were imaged 3 days post inoculation using an IVIS Spectrum Imaging system (PerkinElmers, Waltham, MA). Mice were randomly segregated into two treatment groups [vehicle control and (+)-JQ1, n = 12 per group] and were treated IP twice daily with (+)-JQ1 (50 mg/kg) or vehicle control (10% β-cyclodextrin, Sigma). Bioluminescence imaging and body weight (a surrogate for tumor induced ascites) were used to assess tumor burden. All animal procedures were performed under IACUC approved protocols.
Statistical analyses
Two-tailed paired Student's t test was used to test for differences between two groups. Differences with a p≤0.05 were considered as statistically significant. All experiments were repeated a minimum of two times with duplicate/triplicate samples.
Supplementary Material
Acknowledgments
The authors thank Dr. James Bradner (Harvard University) for his generous contribution of (+)- JQ1 and (-)- JQ1, Dr. Peter Howley (Harvard University) for BRD4 antibody, Gary Hayward (Johns Hopkins University) for RTA antibody, and Drs. Lossos and Ramos from University of Miami for UM-PEL-1 and UM-PEL-3 cells respectively. This work was supported by grants from the National Institutes of Health (CA139119, DE019811 and P30CA014089) and STOP Cancer Foundations. Flow Cytometry was performed in the USC Flow Cytometry Core Facility that is supported in part by the National Cancer Institute Cancer Center Shared Grant award P30CA014089 and the USC Provost Office Dean's Development Funds.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
Supplementary information: Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Morris K. Cancer? In Africa? Lancet Oncol. 2003 Jan;4(1):5. doi: 10.1016/s1470-2045(03)00969-0. [DOI] [PubMed] [Google Scholar]
- 2.Wabinga HR, Parkin DM, Wabwire-Mangen F, Mugerwa JW. Cancer in Kampala, Uganda, in 1989-91: changes in incidence in the era of AIDS. Int J Cancer. 1993 Apr 22;54(1):26–36. doi: 10.1002/ijc.2910540106. [DOI] [PubMed] [Google Scholar]
- 3.Nador RG, Cesarman E, Chadburn A, Dawson DB, Ansari MQ, Sald J, et al. Primary effusion lymphoma: a distinct clinicopathologic entity associated with the Kaposi's sarcoma-associated herpes virus. Blood. 1996;88(2):645–56. [PubMed] [Google Scholar]
- 4.Boulanger E, Daniel MT, Agbalika F, Oksenhendler E. Combined chemotherapy including high-dose methotrexate in KSHV/HHV8-associated primary effusion lymphoma. Am J Hematol. 2003 Jul;73(3):143–8. doi: 10.1002/ajh.10341. [DOI] [PubMed] [Google Scholar]
- 5.Boulanger E, Hermine O, Fermand JP, Radford-Weiss I, Brousse N, Meignin V, et al. Human herpesvirus 8 (HHV-8)-associated peritoneal primary effusion lymphoma (PEL) in two HIV-negative elderly patients. Am J Hematol. 2004 May;76(1):88–91. doi: 10.1002/ajh.20048. [DOI] [PubMed] [Google Scholar]
- 6.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007 May 4;282(18):13141–5. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 7.Chung CW, Witherington J. Progress in the discovery of small-molecule inhibitors of bromodomain--histone interactions. J Biomol Screen. 2011 Dec;16(10):1170–85. doi: 10.1177/1087057111421372. [DOI] [PubMed] [Google Scholar]
- 8.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011 Sep 16;146(6):904–17. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011 Oct 27;478(7370):524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011 Oct 27;478(7370):529–33. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, et al. BET bromodomain inhibition targets both c-MYC and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012 Aug 17; doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010 Dec 23;468(7327):1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010 Dec 23;468(7327):1119–23. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011 Oct 4;108(40):16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013 Apr 11;153(2):320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Chen S, Feng J, Deng H, Sun R. Myc is required for the maintenance of Kaposi's sarcoma-associated herpesvirus latency. J Virol. 2010 Sep;84(17):8945–8. doi: 10.1128/JVI.00244-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bubman D, Guasparri I, Cesarman E. Deregulation of c-Myc in primary effusion lymphoma by Kaposi's sarcoma herpesvirus latency-associated nuclear antigen. Oncogene. 2007 Jul 26;26(34):4979–86. doi: 10.1038/sj.onc.1210299. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Martin HJ, Liao G, Hayward SD. The Kaposi Sarcoma Associated Herpesvirus LANA Protein Stabilizes and Activates c-Myc. J Virol. 2007 Jul 18; doi: 10.1128/JVI.00804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubyova B, Kellum MJ, Frisancho JA, Pitha PM. Stimulation of c-Myc transcriptional activity by vIRF-3 of Kaposi sarcoma-associated herpesvirus. J Biol Chem. 2007 Nov 2;282(44):31944–53. doi: 10.1074/jbc.M706430200. [DOI] [PubMed] [Google Scholar]
- 20.Bhatt S, Ashlock B, Natkunam Y, Ramos JC, Mesri EA, Lossos IS. Preclinical Activity of Brentuximab Vedotin (SGN-35) in Primary Effusion Lymphoma (PEL). 53rd ASH Annual Meeting and Exposition; San Diego. 2011. [Google Scholar]
- 21.Sarosiek KA, Cavallin LE, Bhatt S, Toomey NL, Natkunam Y, Blasini W, et al. Efficacy of bortezomib in a direct xenograft model of primary effusion lymphoma. Proc Natl Acad Sci U S A. 2010 Jul 20;107(29):13069–74. doi: 10.1073/pnas.1002985107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao H, Darzynkiewicz Z. Biomarkers of cell senescence assessed by imaging cytometry. Methods Mol Biol. 2013;965:83–92. doi: 10.1007/978-1-62703-239-1_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci U S A. 2003 Jul 8;100(14):8164–9. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003 May 1;17(9):1115–29. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura H, Lu M, Gwack Y, Souvlis J, Zeichner SL, Jung JU. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J Virol. 2003 Apr;77(7):4205–20. doi: 10.1128/JVI.77.7.4205-4220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J, Punj V, Matta H, Mazzacurati L, Schamus S, Yang Y, et al. K13 Blocks KSHV Lytic Replication and Deregulates vIL6 and hIL6 Expression: a Model of Lytic Replication Induced Clonal Selection in Viral Oncogenesis. PLoS ONE. 2007 Oct 24;2(10):e1067. doi: 10.1371/journal.pone.0001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009 Nov 1;15(21):6479–83. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002 Oct;2(10):764–76. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 30.Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, et al. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci U S A. 2000 Jan 4;97(1):228–33. doi: 10.1073/pnas.97.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ballestas ME, Kaye KM. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J Virol. 2001 Apr;75(7):3250–8. doi: 10.1128/JVI.75.7.3250-3258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garber AC, Hu J, Renne R. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J Biol Chem. 2002 Jul 26;277(30):27401–11. doi: 10.1074/jbc.M203489200. [DOI] [PubMed] [Google Scholar]
- 33.You J, Srinivasan V, Denis GV, Harrington WJ, Jr, Ballestas ME, Kaye KM, et al. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J Virol. 2006 Sep;80(18):8909–19. doi: 10.1128/JVI.00502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ottinger M, Christalla T, Nathan K, Brinkmann MM, Viejo-Borbolla A, Schulz TF. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J Virol. 2006 Nov;80(21):10772–86. doi: 10.1128/JVI.00804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholas J. Human herpesvirus 8-encoded proteins with potential roles in virus-associated neoplasia. Front Biosci. 2007;12:265–81. doi: 10.2741/2063. [DOI] [PubMed] [Google Scholar]
- 36.Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. BET bromodomain inhibition activates transcription via a transient release of P-TEFb from 7SK snRNP. J Biol Chem. 2012 Sep 5; doi: 10.1074/jbc.M112.410746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X, Santat LA, Chang MS, Liu J, Zavzavadjian JR, Wall EA, et al. A versatile approach to multiple gene RNA interference using microRNA-based short hairpin RNAs. BMC Mol Biol. 2007;8:98. doi: 10.1186/1471-2199-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci U S A. 2006 Sep 12;103(37):13759–64. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.