Abstract
The cost, prevalence and pain associated with endodontic disease necessitate an understanding of the fundamental molecular aspects of its pathogenesis. This study was aimed to identify the genetic contributors to pulpal pain and inflammation. Inflamed pulps were collected from patients diagnosed with irreversible pulpitis (n=20). Normal pulps from teeth extracted for various reasons served as controls (n=20). Pain level was assessed using a visual analog scale (VAS). Genome-wide microarray analysis was performed using Affymetrix GeneTitan Multichannel Instrument. The difference in gene expression levels were determined by the Significance Analysis of Microarray program using a false discovery rate (q-value) of 5%. Genes involved in immune response, cytokine-cytokine receptor interaction and signaling, integrin cell surface interactions, and others were expressed at relatively higher levels in the in the pulpitis group. Moreover, several genes known to modulate pain and inflammation showed differential expression in asymptomatic and mild pain patients (≥30mm on VAS) compared to those with moderate to severe pain. This exploratory study provides a molecular basis for the clinical diagnosis of pulpitis. With an enhanced understanding of pulpal inflammation, future studies on treatment and management of pulpitis and on pain associated with it can have a biological reference to bridge treatment strategies with pulpal biology.
Introduction
Inflammation of the dental pulp (pulpitis) can be a progressive and devastating pain experience characterized by spontaneous or provoked pain, hyperalgesia, allodynia and difficulty in achieving adequate local anesthesia 1. The activation of dental pulp or peridental nociceptors during endodontic inflammation elicits a pain response that contributes to approximately 90% of dental emergency visits in both private dental clinics and in hospitals 2-5. The economic implication of these emergency visits is reported to cost almost US$1 billion per year 4. However, despite the cost and the prevalence of endodontic disease and the great discomfort associated with it, the fundamental molecular aspects of its pathogenesis are still not fully understood. The current literature on pulpal immune response to microbial infection continues to expand but very little is known on the regulatory mechanisms behind pulpal disease.
Cells comprising the human dental pulp trigger immune responses to a complex array of microorganisms that invade dental tissues 6-8. These immunocompetent cells also form mechanical barriers (i.e. odontoblasts), detect and transmit sensations (nerve fibers) or differentiate (i.e. dental pulp stem cells) to limit infection, signal injury and promote repair, respectively. These cascades of events resulting from dental pulp stimulation by microorganisms result in the release of a plethora of immune mediators that trigger pulpal or odontogenic pain, inflammation or in advanced stages, pulpal necrosis. In addition, several studies have suggested that pulpal events can be reflected in the gingival crevicular fluid (GCF) through measurable levels of protein markers that correlated with pulpal symptoms 9, 10. This shows that the dental pulp is not an isolated environment but rather a vital, reactive tissue that communicates with the outside environment.
Several studies have identified the biological differences between healthy and inflamed dental pulp. Cytokines, cell surface receptors and other protein markers are shown to be either highly increased or decreased in inflamed dental pulp.7, 11-21. A limited number of studies have examined gene expression in inflamed human pulps 22-24. These studies, however, did not explore the differences in gene expression between normal and inflamed dental pulp and the clinical presentation of donor patients (i.e. pain and swelling), which may provide biologic explanation on the variability of clinical signs and symptoms of pulpal inflammation that confound diagnosis.
Although the clinical value of a molecular diagnostic marker may at first appear limited in scope, the emerging correlation of gene expression with clinical signs and symptoms will enhance our understanding on the development of pulpal inflammation - this will add not only to the knowledge base but it will also provide a biological basis for the varying clinical presentations of pulpitis. Prior studies that focused on histological findings have shown a wide variation – from poor to strong - in correlating clinical signs and symptoms with histological findings 25-27. In this study, data from the full genome scan will be utilized to determine if an association exists between gene expression and clinical presentation (i.e. pain) of pulpitis patients.
Results and Discussion
Normal and pulpitis samples exhibited differentially expressed genes
The SAM software generated GSEA data which showed a significantly higher expression of various gene sets that are associated with immune response activation, maintaining cellular function and cell-to-cell interaction, among others (Figure 1) in pulpitis samples. This underscores the utility of both the subjective (patient-derived history) and objective (endodontist-performed testing) diagnostic techniques that clinically delineate a normal from an inflamed dental pulp. Furthermore, the results above re-establish the immunocompetency of the dental pulp that has been shown to carry Toll-like receptor (TLR) -2/4+ cells 7, 28.
Figure 1.
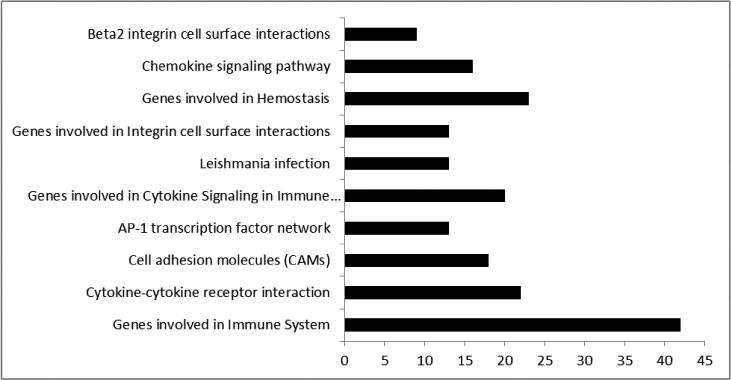
Gene Set Enrichment Analysis (GSEA) results between pulpitis and normal samples. Each bar represents the functional categories and the number of significantly regulated genes between pulpitis and normal groups (q<0.05).
Differences in gene expression between mild and moderate to severe pain
Among the patients diagnosed with irreversible pulpitis, eight patients reported experiencing zero to mild pain ((≤30mm on VAS) and twelve patient reported moderate to severe pain. The mean VAS scores for patients who reported zero to mild pain and moderate to severe pain was 6.63 (SD = 9.29) and 81.93 (SD = 11.21), respectively (P <0.0001). There were differentially expressed genes between the two groups (Figure 2). Mild pain samples showed a significantly higher expression of genes involved in adaptive immune system, cytokine to cytokine interaction and cytokine signaling, among others. However, when looking at specific genes, several of them that have key roles in inflammation are significantly under-expressed, unchanged or over-expressed in asymptomatic or mild pain patients compared to those with moderate to severe pain (Table 1).
Figure 2.
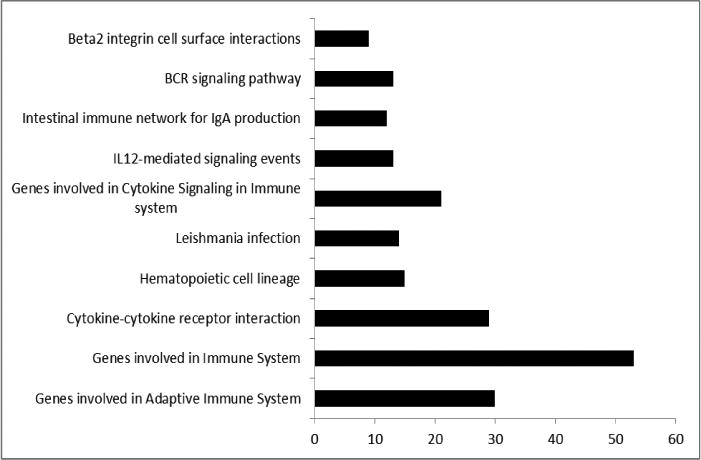
GSEA results between groups that reported none to mild pain and those that reported moderate to severe pain on VAS. Each bar represents the functional categories and the number of significantly regulated genes between none to mild and moderate to severe pain groups (q<0.05).
Table 1. Selected up-regulated or down-regulated genes in pulpitis patients.
| Gene Symbol | Pulpitis vs Normal | None to Mild vs Moderate to Severe Pain | Gene Function (genecards.org) |
|---|---|---|---|
| AMELX | Enamel biomineralization | ||
| CALCRL | Bone metabolism | ||
| CCL20 | Chemotaxis | ||
| CD14 | Innate immune response | ||
| CD163 | Acute phase receptor | ||
| CD79A | B-cell receptor function | ||
| COL10A1 | Collagen formation | ||
| COL11A2 | Collagen formation | ||
| COL12A1 | Collagen formation | ||
| COL14A1 | Collagen formation | ||
| COL15A1 | Collagen formation | ||
| COL15A1 | Collagen formation | ||
| COL18A1 | Collagen formation | ||
| COL1A1 | Collagen formation | ||
| COL1A2 | Collagen formation | ||
| COL21A1 | Collagen formation | ||
| COL4A1 | Collagen formation | ||
| COL4A2 | Collagen formation | ||
| CXCL3 | Chemotaxis | ||
| DEFA1B/1A | Antimicrobial activity | ||
| DEFA3 | Antimicrobial activity | ||
| DSPP | Dentin formation | ||
| IL10RA | IL-10 signaling | ||
| IL1A | Inflammatory response | ||
| IL1B | Inflammatory response | ||
| IL6 | Inflammation, B-cell maturation | ||
| IL8 | Chemotaxis | ||
| LBP | Innate immune response | ||
| MMP13 | Collagen degradation | ||
| MMP20 | Amelogenin degradation | ||
| MMP9 | Collagen degradation | ||
| NOD2 | Innate immune response | ||
| NR5A2 | Antiviral activity | ||
| PTGS2 | Prostaglandin formation | ||
| SCN8A | Sodium ion permeability | ||
| TLR1 | Innate immune response | ||
| TLR2 | Innate immune response | ||
| TLR3 | Innate immune response | ||
| TLR4 | Innate immune response | ||
| TLR6 | Innate immune response | ||
| TLR8 | Innate immune response | ||
| TLR9 | Innate immune response | ||
| TNFA | Inflammatory response |
 Downregulated
Downregulated
 Upregulated
Upregulated
 No difference
No difference
Fold change: Downregulated ≤0.5; Upregulated: ≥1.5; q<0.05
Relief from pain is a very important part in the practice of endodontics. Patients often judge the success of treatment and the efficiency of the dentist based on their pain experience. The decision whether to perform root canal therapy (RCT) to relieve pain or manage infection relies heavily on clinical diagnostic tests which dichotomize the pulpal diagnosis to reversible and irreversible pulpitis. To arrive at a diagnosis, dentists depend mainly on pain history and responses to sensibility tests (namely thermal tests and electric pulp testing,) that have been used for decades; but whether the responses to these clinical tests correlate well with pulpal histopathology or not remains unclear 25-27, 29-31.
This study was aimed to identify the differences in gene expression between normal pulps and pulpitis samples and between mild and moderate to severe pulpitis pain, based on the diagnostic guidelines set by the AAE and on the patient-reported pain experience at the time of endodontic treatment, respectively. Gene Set Enrichment Analysis (GSEA) was employed for the analysis of the thousands of genes included in the microarray screening 32, 33. The Broad Institute defines GSEA as a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states.
Several applications are envisioned for the data that were gathered in this study. First, a biological basis at the molecular level can now be correlated not only with the current diagnostic testing procedures but also with the diagnosis of irreversible pulpitis itself. Molecular alterations precede histopathological changes and in the case of the dental pulp, there can be a wide array of histological presentations depending on the timing of pulp extirpation; thus, there are conflicting reports concerning the clinical presentation and actual histopathology of the pulp. Studies investigating inflammation at the molecular level may therefore provide a more accurate representation of the inflammatory changes in the dental pulp.
Next, the results may be used to cluster genes that are overexpressed or underexpressed in inflamed pulps and to use this information as biological basis for future studies on acute pain and on oral biology, especially those that focus on odontogenic pain and regeneration as these studies require a broad understanding of molecular changes that dictate clinical and biological outcomes. The difference in gene expression between normal and inflamed dental pulps (Figure 1) may have been the expected result considering that cytokines, chemokines and their receptors are known to be overexpressed in inflammatory conditions; however, this study is still an important addition to our current understanding of pulp biology as it provides a broad picture or a “snap-shot” of the molecular changes occurring in the dental pulp during the very moment that the patient is seeking endodontic treatment on the dental chair.
Another remarkable finding in the study is the overexpressed genes related to inflammation and immune response in pulps from patients who reported zero to mild pain (≤30mm on VAS) during their endodontic appointment compared to those who had moderate to severe pain (Figure 2). Moreover, looking at individual genes separately from the a priori gene sets in the GSEA software revealed that genes involved in key inflammatory response like IL8, TNFA, IL1A, ILIB and several immunoglobulins (i.e. IGHM, IGHV4-31, IGKV2D-40), among others (Table 1) are expressed at relatively lower levels in pulp samples from asymptomatic to mild pain patient compared to those with moderate to severe pain. IL8, TNFA and IL1B have been shown to be potent mediators of pain 34, 35.
IL8 has been recently associated with complex regional pain syndrome (CRPS) 36 and IL1B has been shown to contribute to the upregulation of nerve growth factor (NGF) during inflammation that consequently induces inflammatory hyperalgesia 37. Both IL8 and IL1B are expressed at relatively lower levelsin zero to mild pain pulpitis patients. Several genes that are upstream on the inflammatory cascade of both IL1B and IL8 (e.g. TLR2 and TLR44) are significantly increased in pulpitis samples (Table 1). Furthermore, most genes associated with structural formation and integrity (e.g. COL1-4 and MMPs) are expressed at relatively higher levels in pulpitis samples. In contrast however, several genes that are known to be pain mediators or associated with increased pain experience did not have significant discordance between mild and moderate to severe pain (Table 1). For example, Prostaglandin-Endoperoxide Synthase 2 or Cyclooxygenase-2 (PGS2 or COX2), an important inflammatory enzyme that has been shown to be significantly elevated in pulpitis tissues38 did not have expression difference in mild pain but is significantly elevated in pulpitis compared with normal controls (Table 1). IL6 showed the same results.
Pathogen–associated molecular pattern receptors, including TLRs, are key molecules in the response to pathological stimuli. Pulpitis is a disease that is mainly due to microbial insults. TLRs are expressed by both immune and non-immune cells in the pulp including neurons, fibroblasts, endothelial cells, epithelial cells and others. They recognize viral and microbial structures as well as self molecules (such as single stranded RNAs) that may accumulate in non-physiologic amounts or sites 39-43. TLR ligands stimulate the production of pro-pain molecules and, in this way, may contribute to the pathogenesis of pain 42-47. In this study, the expression of several TLRs (1,2,4,6, 8 and 9) were upregulated and TLR 3 was down regulated in inflamed pulps as compared to normal pulps. Our data also show that expression of TLR8 was higher in pulpits patients experiencing severe pain as compared to those who reported no pain or mild pain. This is of particular interest given the recent finding that murine TLR7 (which is phylogenetically and structurally related to human TLR8) interacts with TRPA1 and that specific miRNAs activate nociceptors via TLR7 and TRPA1 47.
Arriving at a reliable diagnosis that matches the histopathology of the dental pulp, especially during a pain episode is arguably the most challenging part of endodontics. The dental pulp is a dynamic tissue that is capable of responding to insults and of healing so that diagnosis may change over time 48. Furthermore, individual variations in immunologic and pain response further complicate not only the clinical but also the biological aspect of endodontic diagnosis. This may be the one of the main reasons for the observational differences among studies correlating clinical and histopathological diagnosis as pointed out in the earlier part of this paper. Understanding the molecular changes in pulpitis may help develop better diagnostic tools. For example, molecular markers collected from dentinal fluids may be used to determine the pulpal status. This would be a non-invasive and easy diagnostic tool.
The study of tissue molecular profiles to better understand and diagnose disease pathophysiology has been examined in another common oral disease- periodontitis. Using biopsies from patients diagnosed with either chronic periodontitis or aggressive periodontitis, Kebschull et al., explored whether molecular profiling can form the basis for a pathobiology based classification that correlates with the phenotypic features of the disease 49. They identified two de novo clusters with high similarity in transcriptional profiles. The clusters had distinct molecular signatures which did not align with the current classifications of periodontitis. However, they did translate into distinct phenotypic differences and can provide a basis for novel classification. This study along with current study supports the use of molecular profiling to classify disease.
While microarray analysis is a powerful tool to study the simultaneous expression of several genes it has some inherent limitations. These include technical factors such as limited dynamic range, signal saturations and cross-hybridization. It is important to also remember that the cellular profile of inflamed pulps differs from that of normal pulps. The residents cells in normal pulps include fibroblasts, odontoblasts and some immune cells such as macrophages, dendritic cells and mast cells. Inflamed pulps are characterized by an influx of immune cells and as such the results reported here may be in part, due to differences in the cellular makeup as well.
The present study shows that there are differences in gene expression between normal and inflamed pulps and; between zero to mild and moderate to severe pulpitis pain. Among the genes reported to have a strong association with pain and inflammation, IL8 showed the highest fold change (35.9, q-value = 0) in pulpitis samples and it is significantly under-expressed in mild pain compared with moderate to severe pain (0.14, q-value = 0). IL8 is increased in gingival crevicular fluid from patients with acute pulpitis and might be a useful measurement for staging patients with acute pulpitis 9. The same trend was also noted in IL1A, IL1B and IL10RA.
This study provides a molecular basis for the clinical diagnosis of pulpitis and for the differences or similarities between asymptomatic to mild pulpal pain vis-à-vis moderate to severe pain. With an enhanced understanding of pulpal inflammation, future studies on treatment and management of pulpitis and on pain associated with it can have a biological reference to bridge treatment strategies with pulpal biology.
Materials and Methods
The study was approved by the University of North Carolina Office of Human Ethics. Eligible patients seeking treatment at the School of Dentistry were recruited to participate in the study. Informed consent was obtained from each participant. Demographic details, history of odontogenic pain, vital signs (blood pressure, pulse rate), medical history (including full details of any medication) and patient's smoking and oral habits were recorded.
Patient selection and pulpal diagnosis
The inclusion criteria were adults (18 years or older) presenting for endodontic treatment with no evidence of periapical pathoses (i.e radiolucency, swelling, pressure sensitivity) and no previous pulp therapy (i.e., pulp capping, etc). Exclusion criteria were conditions requiring antibiotic prophylaxis or additional treatment procedures, debilitating disease, chronic pain, diabetes mellitus, hematological disorders or a history of taking centrally acting drugs (e.g. tricyclic antidepressants) known to interfere with the release of various pain mediators and/or modify pain experience within the previous 6 months or over-the-counter pain medicine within the last six hours, chronic use of medication known to affect the immune response and patients who were immunocompromised. Teeth with incompletely developed roots were also excluded.
Diagnosis of normal pulp (NP) and irreversible pulpitis (IP) was based on subjective and objective findings and was in accordance with the American Association of Endodontists (AAE) guidelines. In brief, NP is a clinical diagnostic category in which the pulp is symptom-free and normally responsive to diagnostic testing On the other hand, IP is clinical diagnosis indicating that the vital, inflamed pulp is incapable of healing. The symptoms include lingering and exacerbated pain in response to thermal stimuli, spontaneous pain, referred pain or no clinical symptoms but inflammation is evident after caries excavation or trauma. Pain history, cold test (Endo Ice® Refrigerant Spray, Coltene Inc, Cuyahoga Falls, OH) and electric pulp testing (Vitality Scanner 2006, Sybron Endodontics, Orange, CA) were used to determine pulpal diagnosis. Percussion, palpation, probing, mobility and radiographs were used to rule out periapical pathosis.
Pain levels were assessed using a visual analogue scale (VAS). If lingering pain is present or significantly evoked by a stimulus compared with a control tooth, the patient was asked to quantify the intensity of pain by placing a mark on a 10-cm VAS with anchors of “no pain” and “worst pain imaginable”. Using a modified version of the VAS recommended by Jensen et al 50, a VAS mark of ≤ 30mm was classified under mild, 31-74 mm under moderate and 75-100 under severe pain.
A total of 40 pulp samples from 28 patients with IP and from seven patients with NP were included in the study. Twelve of the IP samples presented with moderate to severe pain at the time of pulp extirpation. The total number of samples was based on a recently published paper by our group on the differences of micro-RNA expression between normal and inflamed pulps 51 and on a power analysis. With 20 samples in each group, we had 80% power to detect an effect size of 0.9 using a threshold of p<0.05 for significance. If we required a more stringent threshold of p<0.001, then we would have 80% power to detect an effect size of 1.4. Given that large effect sizes are common in microarray experiments 52, we had adequate power to detect the genes that exhibited the largest expression differences between the two groups. Moreover, we increased the power as much as needed by increasing the acceptable false discovery rate. Given that this initial analysis is intended to be exploratory, a higher false discovery rate is acceptable in this study. Thus, this study had adequate power to detect the genes with the largest group differences.
Sample collection
The inflamed pulps were extirpated during root canal treatment and the normal pulps were collected right after tooth extraction for orthodontic or restorative purposes or for third molar removal. Pulp collection methods were adapted from Awawdeh et al 53 with modifications. Briefly, local anesthesia (2% lidocaine with 1:100,000 epinephrine) was administered and an access cavity was prepared under rubber dam isolation. Pulp tissue was extirpated using a sterile barbed broach or Hedström-file, placed into a pre-weighed Eppendorf tube, immediately frozen in liquid nitrogen and stored at −70°C. The root canal treatment was then completed. For the controls and for some patients who chose to have extraction instead of root canal treatment, all extractions were performed under local anesthesia. Immediately after extraction, each tooth were split in a vice fitted with a cutting edge and the pulp tissue was removed using a barbed broach or Hedström-file. The pulp was then placed in an Eppendorf tube, immediately frozen in liquid nitrogen and stored at −70°C.
RNA Extraction and Microarray analysis
Total RNA was isolated using a commercial kit (Qiagen RNEasy Kit, Valencia, CA) following the manufacturer's specifications. Quantity and purity of the total RNA was analyzed using Nano-Drop™ (Thermo Scientific, Wilmington, DE) and Bioanalyzer™ (Agilent Technologies, Sta. Clara, CA). Microarray experiments were conducted at the Microarray Core Facility of UNC-Chapel Hill Lineberger Cancer Center. The Nugen Ovation Pico WTA system V2 and the Nugen Encore Biotin module were used to prepare RNA for hybridization onto Affymetrix GeneChip Human Gene 2.0 arrays. Briefly, 25 ng of total RNA was converted into amplified SPIA cDNA using the Nugen Ovation Pico WTA system V2 and accompanying user guide. The amplified SPIA cDNA was purified using the Qiagen QIAquick PCR purification kit. 2.5 ug of SPIA cDNA was fragmented and labeled using the Nugen Encore Biotin module and accompanying user guide. The cocktail for hybridization onto an Affymetrix Human Gene 2.1 ST peg plate was prepared using the Nugen Ovation PicoSL WT System V2 user guide's cocktail assembly table. The hybridization cocktails were denatured and hybridized on the Human Gene 2.1 ST peg plate in the Affymetrix GeneTitan MC Instrument. Washing and scanning was also carried out on the Affymetrix GeneTitan MC Instrument. Individual sample RNAs were hybridized individually to microarrays. Basic data analysis of the arrays was carried out using the Affymetrix Expression Console software. Gene expression levels between the control and diseased pulps were compared. Changes in gene expressions were analyzed by Gene Set Enrichment analysis (GSEA) (Broad Institute, Cambridge, MA) and its leading edge analysis software that uses the permutation method.
The difference in gene expression levels between groups were determined by the Significance Analysis of Microarray (SAM) program using a false discovery rate (q-value) of 5%. Since this study analyzed over 53,600 genes, only those that were previously reported to play key roles in pain and inflammation will be discussed in this paper. In addition, only the key genes with a SAM fold change of over 1.5 (overexpressed) and under 0.5 (underexpressed) to signify a potentially relevant clinical effect are reported 36.
Acknowledgments
This study was financially supported by the American Association of Endodontists Foundation (AAEF) and T90DE021986.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- 1.Bender IB. Pulpal pain diagnosis--a review. J Endod. 2000;26(3):175–9. doi: 10.1097/00004770-200003000-00012. [DOI] [PubMed] [Google Scholar]
- 2.Hasselgren G, Calev D. Endodontics emergency treatment sound and simplified. N Y State Dent J. 1994;60(6):31–3. [PubMed] [Google Scholar]
- 3.Eriksen HM. Endodontology--epidemiologic considerations. Endod Dent Traumatol. 1991;7(5):189–95. doi: 10.1111/j.1600-9657.1991.tb00434.x. [DOI] [PubMed] [Google Scholar]
- 4.Allareddy V, Rampa S, Lee MK, Allareddy V, Nalliah RP. Hospital-based emergency department visits involving dental conditions: profile and predictors of poor outcomes and resource utilization. J Am Dent Assoc. 2014;145(4):331–7. doi: 10.14219/jada.2014.7. [DOI] [PubMed] [Google Scholar]
- 5.States TPCot. A Costly Dental Destination. Pew Children's Dental Campaign. 2012;(February) [Google Scholar]
- 6.Smith AJ. Pulpal responses to caries and dental repair. Caries Res. 2002;36(4):223–32. doi: 10.1159/000063930. [DOI] [PubMed] [Google Scholar]
- 7.Keller JF, Carrouel F, Staquet MJ, Kufer TA, Baudouin C, Msika P, et al. Expression of NOD2 is increased in inflamed human dental pulps and lipoteichoic acid-stimulated odontoblast-like cells. Innate Immun. 2011;17(1):29–34. doi: 10.1177/1753425909348527. [DOI] [PubMed] [Google Scholar]
- 8.Staquet MJ, Carrouel F, Keller JF, Baudouin C, Msika P, Bleicher F, et al. Pattern-recognition receptors in pulp defense. Adv Dent Res. 2011;23(3):296–301. doi: 10.1177/0022034511405390. [DOI] [PubMed] [Google Scholar]
- 9.Karapanou V, Kempuraj D, Theoharides TC. Interleukin-8 is increased in gingival crevicular fluid from patients with acute pulpitis. J Endod. 2008;34(2):148–51. doi: 10.1016/j.joen.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 10.Avellan NL, Sorsa T, Tervahartiala T, Forster C, Kemppainen P. Experimental tooth pain elevates substance P and matrix metalloproteinase-8 levels in human gingival crevice fluid. Acta Odontol Scand. 2008;66(1):18–22. doi: 10.1080/00016350701810658. [DOI] [PubMed] [Google Scholar]
- 11.Caviedes-Bucheli J, Arenas N, Guiza O, Moncada NA, Moreno GC, Diaz E, et al. Calcitonin gene-related peptide receptor expression in healthy and inflamed human pulp tissue. Int Endod J. 2005;38(10):712–7. doi: 10.1111/j.1365-2591.2005.01006.x. [DOI] [PubMed] [Google Scholar]
- 12.Caviedes-Bucheli J, Gutierrez-Guerra JE, Salazar F, Pichardo D, Moreno GC, Munoz HR. Substance P receptor expression in healthy and inflamed human pulp tissue. Int Endod J. 2007;40(2):106–11. doi: 10.1111/j.1365-2591.2006.01189.x. [DOI] [PubMed] [Google Scholar]
- 13.Caviedes-Bucheli J, Munoz HR, Azuero-Holguin MM, Ulate E. Neuropeptides in dental pulp: the silent protagonists. J Endod. 2008;34(7):773–88. doi: 10.1016/j.joen.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Esmaeili A, Akhavan A, Bouzari M, Mousavi SB, Torabinia N, Adibi S. Temporal expression pattern of sodium channel Nav 1. 8 messenger RNA in pulpitis Int Endod J. 2011;44(6):499–504. doi: 10.1111/j.1365-2591.2011.01853.x. [DOI] [PubMed] [Google Scholar]
- 15.Gong Q, Jiang H, Wei X, Ling J, Wang J. Expression of erythropoietin and erythropoietin receptor in human dental pulp. J Endod. 2010;36(12):1972–7. doi: 10.1016/j.joen.2010.08.041. [DOI] [PubMed] [Google Scholar]
- 16.Huang FM, Tsai CH, Yang SF, Chang YC. The upregulation of oncostatin M in inflamed human dental pulps. Int Endod J. 2009;42(7):627–31. doi: 10.1111/j.1365-2591.2009.01567.x. [DOI] [PubMed] [Google Scholar]
- 17.Jiang HW, Ling JQ, Gong QM. The expression of stromal cell-derived factor 1 (SDF-1) in inflamed human dental pulp. J Endod. 2008;34(11):1351–4. doi: 10.1016/j.joen.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 18.Korkmaz Y, Lang H, Beikler T, Cho B, Behrends S, Bloch W, et al. Irreversible inflammation is associated with decreased levels of the alpha1-, beta1-, and alpha2-subunits of sGC in human odontoblasts. J Dent Res. 2011;90(4):517–22. doi: 10.1177/0022034510390808. [DOI] [PubMed] [Google Scholar]
- 19.Lundy FT, About I, Curtis TM, McGahon MK, Linden GJ, Irwin CR, et al. PAR-2 regulates dental pulp inflammation associated with caries. J Dent Res. 2010;89(7):684–8. doi: 10.1177/0022034510365652. [DOI] [PubMed] [Google Scholar]
- 20.Mutoh N, Watabe H, Chieda K, Tani-Ishii N. Expression of Toll-like receptor 2 and 4 in inflamed pulp in severe combined immunodeficiency mice. J Endod. 2009;35(7):975–80. doi: 10.1016/j.joen.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Kokkas AB, Goulas A, Varsamidis K, Mirtsou V, Tziafas D. Irreversible but not reversible pulpitis is associated with up-regulation of tumour necrosis factor-alpha gene expression in human pulp. Int Endod J. 2007;40(3):198–203. doi: 10.1111/j.1365-2591.2007.01215.x. [DOI] [PubMed] [Google Scholar]
- 22.Tete S, Mastrangelo F, Scioletti AP, Tranasi M, Raicu F, Paolantonio M, et al. Microarray expression profiling of human dental pulp from single subject. Clin Invest Med. 2008;31(2):E55–61. doi: 10.25011/cim.v31i2.3364. [DOI] [PubMed] [Google Scholar]
- 23.McLachlan JL, Smith AJ, Bujalska IJ, Cooper PR. Gene expression profiling of pulpal tissue reveals the molecular complexity of dental caries. Biochim Biophys Acta. 2005;1741(3):271–81. doi: 10.1016/j.bbadis.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Kaneko T, Okiji T, Kaneko R, Sunakawa M, Kaneko M, Suda H. Gene expression analysis of acutely traumatized pulps. J Endod. 2010;36(1):78–82. doi: 10.1016/j.joen.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Seltzer S, Bender IB, Ziontz M. The Dynamics of Pulp Inflammation: Correlations Between Diagnostic Data and Actual Histologic Findings in the Pulp. Oral Surg Oral Med Oral Pathol. 1963;16(7):846–71. doi: 10.1016/0030-4220(63)90323-2. [DOI] [PubMed] [Google Scholar]
- 26.Ricucci D, Loghin S, Siqueira JF., Jr Correlation between Clinical and Histologic Pulp Diagnoses. J Endod. 2014;40(12):1932–9. doi: 10.1016/j.joen.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Hasler JE, Mitchell DF. Painless pulpitis. J Am Dent Assoc. 1970;81(3):671–7. doi: 10.14219/jada.archive.1970.0293. [DOI] [PubMed] [Google Scholar]
- 28.Hirao K, Yumoto H, Takahashi K, Mukai K, Nakanishi T, Matsuo T. Roles of TLR2, TLR4, NOD2, and NOD1 in pulp fibroblasts. J Dent Res. 2009;88(8):762–7. doi: 10.1177/0022034509341779. [DOI] [PubMed] [Google Scholar]
- 29.Hyman JJ, Cohen ME. The predictive value of endodontic diagnostic tests. Oral Surg Oral Med Oral Pathol. 1984;58(3):343–6. doi: 10.1016/0030-4220(84)90065-3. [DOI] [PubMed] [Google Scholar]
- 30.Lundy T, Stanley HR. Correlation of pulpal histopathology and clinical symptoms in human teeth subjected to experimental irritation. Oral Surg Oral Med Oral Pathol. 1969;27(2):187–201. doi: 10.1016/0030-4220(69)90172-8. [DOI] [PubMed] [Google Scholar]
- 31.Johnson RH, Dachi SF, Haley JV. Pulpal hyperemia--a correlation of clinical and histologic data from 706 teeth. J Am Dent Assoc. 1970;81(1):108–17. doi: 10.14219/jada.archive.1970.0132. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 34.Cunha FQ, Lorenzetti BB, Poole S, Ferreira SH. Interleukin-8 as a mediator of sympathetic pain. Br J Pharmacol. 1991;104(3):765–7. doi: 10.1111/j.1476-5381.1991.tb12502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung L, Cahill CM. TNF-alpha and neuropathic pain--a review. J Neuroinflammation. 2010;7:27. doi: 10.1186/1742-2094-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin EH, Zhang E, Ko Y, Sim WS, Moon DE, Yoon KJ, et al. Genome-wide expression profiling of complex regional pain syndrome. PLoS One. 2013;8(11):e79435. doi: 10.1371/journal.pone.0079435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Safieh-Garabedian B, Poole S, Allchorne A, Winter J, Woolf CJ. Contribution of interleukin-1 beta to the inflammation-induced increase in nerve growth factor levels and inflammatory hyperalgesia. Br J Pharmacol. 1995;115(7):1265–75. doi: 10.1111/j.1476-5381.1995.tb15035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakanishi T, Shimizu H, Hosokawa Y, Matsuo T. An immunohistological study on cyclooxygenase-2 in human dental pulp. J Endod. 2001;27(6):385–8. doi: 10.1097/00004770-200106000-00003. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharyya S, Kelley K, Melichian DS, Tamaki Z, Fang F, Su Y, et al. Toll-like receptor 4 signaling augments transforming growth factor-beta responses: a novel mechanism for maintaining and amplifying fibrosis in scleroderma. Am J Pathol. 2013;182(1):192–205. doi: 10.1016/j.ajpath.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villalba M, Hott M, Martin C, Aguila B, Valdivia S, Quezada C, et al. Herpes simplex virus type 1 induces simultaneous activation of Toll-like receptors 2 and 4 and expression of the endogenous ligand serum amyloid A in astrocytes. Med Microbiol Immunol. 2012;201(3):371–9. doi: 10.1007/s00430-012-0247-0. [DOI] [PubMed] [Google Scholar]
- 41.Shi B, Huang Q, Tak PP, Vervoordeldonk MJ, Huang CC, Dorfleutner A, et al. SNAPIN: an endogenous Toll-like receptor ligand in rheumatoid arthritis. Ann Rheum Dis. 2012;71(8):1411–7. doi: 10.1136/annrheumdis-2011-200899. [DOI] [PubMed] [Google Scholar]
- 42.Goh FG, Piccinini AM, Krausgruber T, Udalova IA, Midwood KS. Transcriptional regulation of the endogenous danger signal tenascin-C: a novel autocrine loop in inflammation. J Immunol. 2010;184(5):2655–62. doi: 10.4049/jimmunol.0903359. [DOI] [PubMed] [Google Scholar]
- 43.Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhao JG, et al. Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and MicroRNA-21. Sci Signal. 2011;4(199):ra75. doi: 10.1126/scisignal.2001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109(31):E2110–6. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dasu MR, Devaraj S, Park S, Jialal I. Increased toll-like receptor (TLR) activation and TLR ligands in recently diagnosed type 2 diabetic subjects. Diabetes Care. 2010;33(4):861–8. doi: 10.2337/dc09-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jialal I, Kaur H. The Role of Toll-Like Receptors in Diabetes-Induced Inflammation: Implications for Vascular Complications. Curr Diab Rep. 2012 doi: 10.1007/s11892-012-0258-7. [DOI] [PubMed] [Google Scholar]
- 47.Devaraj S, Tobias P, Jialal I. Knockout of toll-like receptor-4 attenuates the pro-inflammatory state of diabetes. Cytokine. 2011;55(3):441–5. doi: 10.1016/j.cyto.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 48.Van Hassel HJ. Physiology of the human dental pulp. Oral Surg Oral Med Oral Pathol. 1971;32(1):126–34. doi: 10.1016/0030-4220(71)90258-1. [DOI] [PubMed] [Google Scholar]
- 49.Kebschull M, Demmer RT, Grun B, Guarnieri P, Pavlidis P, Papapanou PN. Gingival tissue transcriptomes identify distinct periodontitis phenotypes. J Dent Res. 2014;93(5):459–68. doi: 10.1177/0022034514527288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jensen MP, Chen C, Brugger AM. Interpretation of visual analog scale ratings and change scores: a reanalysis of two clinical trials of postoperative pain. J Pain. 2003;4(7):407–14. doi: 10.1016/s1526-5900(03)00716-8. [DOI] [PubMed] [Google Scholar]
- 51.Zhong S, Zhang S, Bair E, Nares S, Khan AA. Differential expression of microRNAs in normal and inflamed human pulps. J Endod. 2012;38(6):746–52. doi: 10.1016/j.joen.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 52.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Awawdeh L, Lundy FT, Shaw C, Lamey PJ, Linden GJ, Kennedy JG. Quantitative analysis of substance P, neurokinin A and calcitonin gene-related peptide in pulp tissue from painful and healthy human teeth. Int Endod J. 2002;35(1):30–6. doi: 10.1046/j.1365-2591.2002.00451.x. [DOI] [PubMed] [Google Scholar]


