Abstract
The SWI/SNF chromatin remodeling complex is a master regulator of developmental cell fate decisions, although the key target pathways are poorly characterized. Here, we interrogated the contribution of the SWI/SNF subunit and tumor suppressor SNF5 to the regulation of developmental pathways using conditional mouse and cell culture models. We find that loss of Snf5 phenocopies β-catenin hyperactivation and that SNF5 is essential for regulating Wnt/β-catenin pathway target expression. These data provide insight into chromatin-based mechanisms that underlie developmental regulation and elucidate the emerging theme that mutation of this tumor suppressor complex can activate developmental pathways by uncoupling them from upstream control.
INTRODUCTION
Dynamic modulation of chromatin structure plays an integral role in eukaryotic transcriptional regulation. SWI/SNF chromatin remodeling complexes utilize the energy of ATP hydrolysis to remodel chromatin and mobilize nucleosomes at target genes, where they can contribute to both gene activation and repression. Specific combinations of SWI/SNF subunits are essential for lineage specification and appropriate development of numerous tissues (1, 2). At least five subunits of SWI/SNF are specifically and recurrently mutated in a variety of cancers suggesting a broad role for the complex in tumor suppression (3). However, key target pathways and regulatory mechanisms that underlie the role of the SWI/SNF complex in development and tumor suppression are poorly understood.
SNF5 is a core subunit of SWI/SNF complexes that possesses potent tumor suppressor activity (4–6). Biallelic inactivation of SNF5 underlies the vast majority of malignant rhabdoid tumors (MRTs), highly aggressive pediatric cancers (7, 8). Inactivation of Snf5 in mice leads to rapid formation of cancers, demonstrating that SNF5 is a bona fide tumor suppressor gene (4, 9). Intriguingly, SNF5-deficient tumors are diploid and genomically stable, suggesting that oncogenesis may substantially be driven by epigenetically altered expression of target genes (10, 11).
SNF5 serves a critical role in lineage specification (9, 12–14), suggesting that its regulation of developmental pathways may contribute to its tumor suppressor activity. Interestingly, the SWI/SNF ATPase subunit BRG1 (SMARCA4) has been implicated in regulation of the Wnt pathway and can physically interact with its downstream effector, β-catenin (15–18). Here we investigated the mechanism by which SNF5 contributes to lineage specification and developmental patterning using a limb bud model and elucidate relevance to its role as a tumor suppressor.
RESULTS AND DISCUSSION
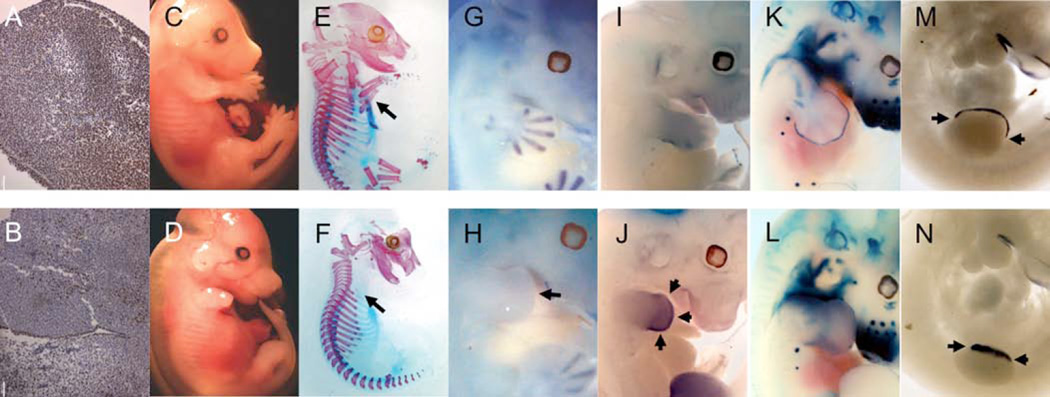
To evaluate the contribution of SNF5 to developmental regulation and patterning in vivo, we utilized Prx1-Cre to conditionally inactivate Snf5 in developing limb mesenchyme at the onset of limb morphogenesis (19). Immunohistochemistry at E11.5 revealed that SNF5 was ubiquitously expressed in wild-type limbs (Figure 1A) but was lost in the mesenchyme of Prx1-Cre; Snf5fl/fl (hereafter referred to as Snf5•/•) limb buds, with the residual positive cells likely representing surface epithelium and invading neural crest (Figure 1B). Since Prx1-Cre is expressed more uniformly in the forelimb than the hindlimb (19), we focused our subsequent analyses on forelimbs. Loss of SNF5 resulted in shortened, malformed limbs and agenesis of all limb bones from the scapula to the phalanges (Figure 1C–F). Sox9, a marker of the onset of cartilage development, from which bones of the limb ultimately form, was expressed correctly at E10.5, but was progressively lost over time in Snf5Δ/Δ limbs and was completely absent by E12.5 (Figure 1G, H).
Figure 1. Loss of Snf5 leads to skeletal agenesis, loss of chondrogenic markers, and upregulation of β-catenin targets in the developing limb.
The Prx1-Cre transgene was used to inactivate Snf5 in the developing limb. Immunohistochemistry for Snf5 from control (A) and Snf5•/• (B) limbs at E11.5. Gross morphology (C, D) and cartilage and skeletal preparations (E, F) of control and Snf5•/• embryos respectively. RNA in situ hybridization against Sox9 (G, H), Axin2 (I, J), and Fgf8 (M, N) in control and Snf5•/• embryos. Snf5fl/+; Prx1-Cre and Snf5fl/fl; Prx1-Cre were crossed to the Topgal reporter strain to monitor β-catenin activity in limbs during development (K, L).
β-catenin antagonizes Sox9 during chondrogenesis and, notably, the consequences of Snf5 loss phenocopied the skeletal agenesis caused by forced overexpression of β-catenin in the developing limb (20). We therefore investigated whether SNF5 loss affects expression of β-catenin/Tcf targets. Expression of Lmx1b, which is activated in part by mesenchymal β-catenin (21) and normally restricted to the dorsal mesenchyme, was expanded into the ventral mesenchyme in SNF5-deficient limbs (Supplemental Figure 1). Axin2, a direct transcriptional target of β-catenin and reported to be the most reliable indicator of β-catenin activity (22), was elevated in SNF5-deficient limbs compared to littermate controls (Figure 1I, J). We next crossed the Snf5fl/fl; Prx1-Cre mice to the Topgal reporter mice, in which TCF mediated repression of the β-galactosidase gene is relieved by binding of β-catenin (23). Unlike control limbs, where staining was restricted to the apical ectodermal ridge (AER) and developing cartilage, SNF5- deficient limbs instead exhibited diffuse staining throughout the limb field (Figure 1K, L). Overexpression of β-catenin in the limb bud mesenchyme leads to premature regression of the AER (21). In Snf5Δ/Δ limbs at E10.5, anterior/posterior expression of Fgf8, an AER marker was truncated and subsequently lost by E11.5 (Figure 1M,N), a pattern identical to that in limbs overexpressing β-catenin (21). Collectively, these results demonstrate that inactivation of Snf5 in developing limb mesenchyme leads to aberrant activation or the Wnt pathway and leads to phenotypic defects consistent with Wnt/β-catenin overexpression.
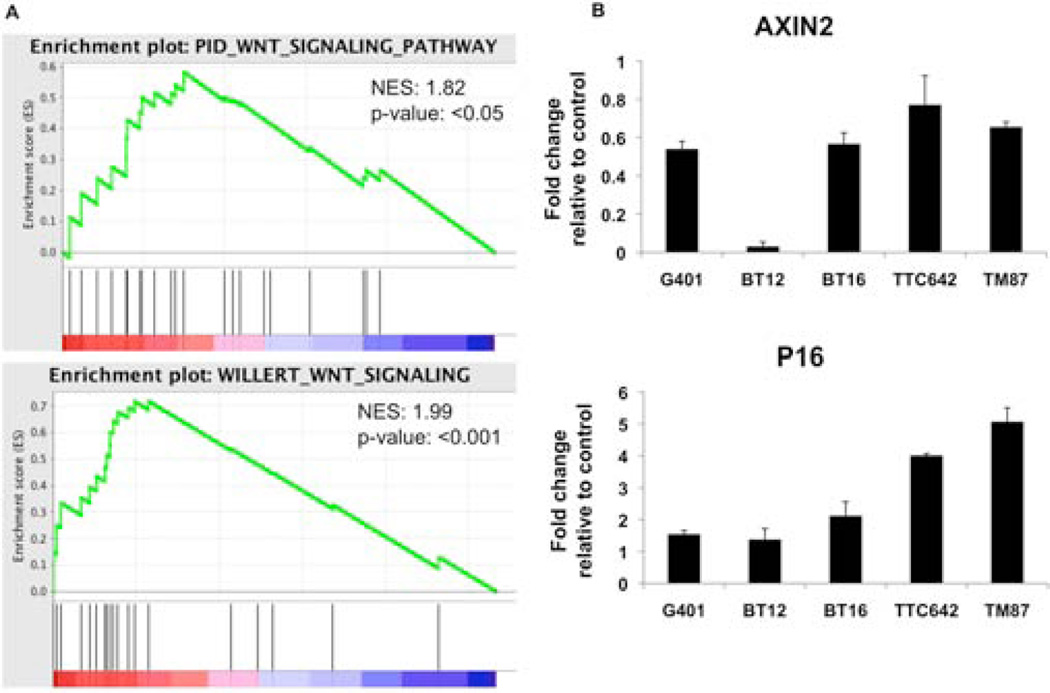
We next sought to determine whether the Wnt pathway was active in SNF5 deficient cancers. We performed gene expression analysis of SNF5-deficient primary human CNS rhabdoid tumors (RT) and compared their expression of known gene signatures of WNT activation to primary medulloblastomas and normal cerebellum using Gene Set Enrichment Analysis (GSEA) on a previously published data set (24)(Figure 2A and Supplemental Figure 3). This analysis identified that WNT targets are elevated in SNF5-deficient RTs compared to normal cerebellum. Further, this expression signature is similar to “Wnt type” medulloblastomas that contain activating mutations of the WNT pathway (Supplemental Figure 3).
Figure 2. The Wnt/β-catenin pathway is active in SNF5-deficient cancers.
Gene set enrichment analysis (GSEA) plot of genes upregulated in response to WNT activation (from (37) and (38), respectively) using expression data from MRT compared to primary medulloblastomas and normal cerebellum (A). Expression analysis of β-catenin/TCF target AXIN2 and known downstream effector of SNF5, P16, in G401, Bt12, BT16, TTC642, and TM87 MRT cell lines(B).
We next assessed whether upregulation of the β-catenin pathway was directly attributable to SNF5 loss. We re-expressed SNF5 in a panel of five SNF5-deficient RT cell lines by viral transduction. As a positive control for SNF5 function, we monitored the expression of P16INK4A, which has been shown to be upregulated when SNF5 is re-introduced into deficient cells (25). Expression of SNF5 resulted in downregulation of β-catenin target genes AXIN2, APC, βTRCP, LEF1, and HDAC4, while the control P16INK4A was upregulated (Figure 2B and Supplemental Figure 4).
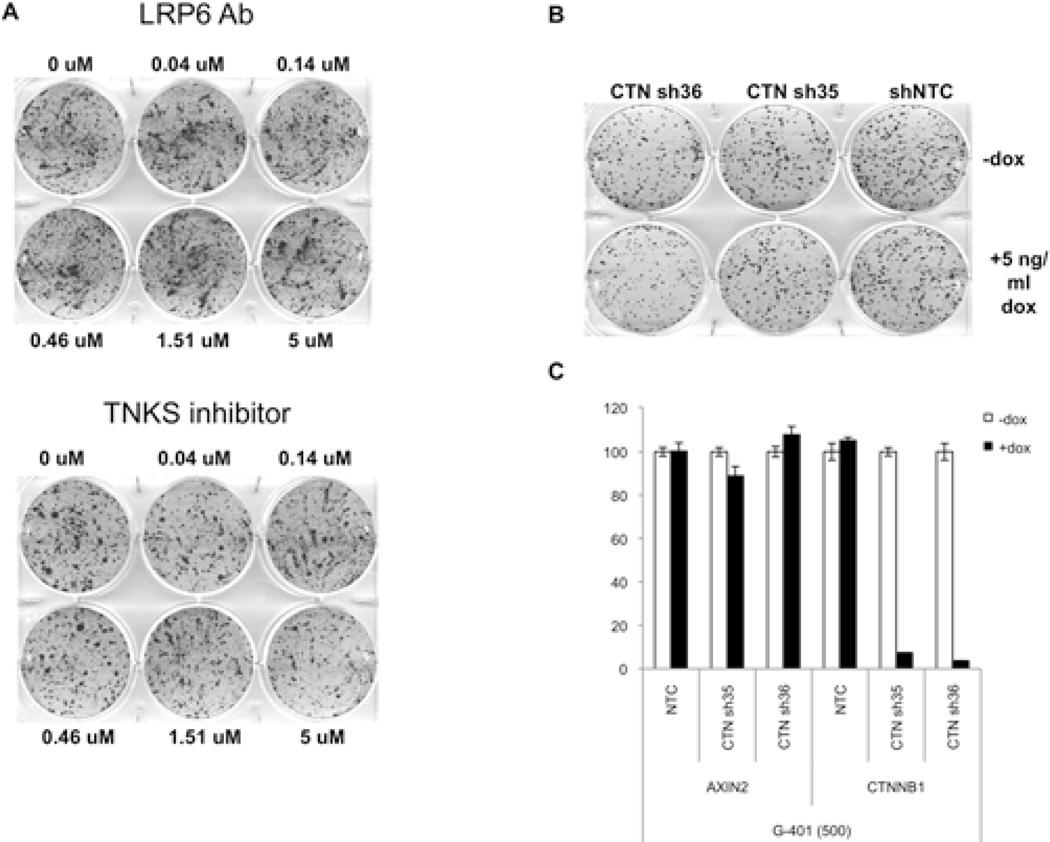
As a core member of the SWI/SNF complex, the direct contributions of SNF5 to pathway regulation putatively occur at the level of chromatin and transcription. To investigate the mechanism by which SNF5 loss activates the Wnt pathway, we first investigated whether inhibition of the upstream Wnt signaling would have a functional effect upon the growth of SNF5 deficient cancer cells. We therefore treated the G401 MRT cell line with three inhibitors previously shown to act directly upon the canonical Wnt pathway including 1) a Porcupine inhibitor to block the Wnt ligand processing and secretion; 2) an antibody against the LRP6 receptor to which the Wnt ligand binds; and 3) a Tankyrase inhibitor that stabilizes the AXIN destruction complex that degrades β-catenin (26, 27). In order to capture effects upon either proliferation and colony forming ability of these cancer cells, we measured colony growth after treatment with each of the inhibitors. Notably, none of these Wnt pathway inhibitors impaired G401 colony formation (Figure 3A and data not shown). Further, depletion of β-catenin itself had no effect on colony formation or on expression of the β-catenin target AXIN2 (Figure 3B, C). These results indicated that aberrant activation of β-catenin target genes in SNF5-deficient cells occurs independently of canonical Wnt pathway activation.
Figure 3. Chemical inhibition of the Wnt pathway in SNF5-deficient MRTs.
Colony growth assay of MRT cells in the presence of the labeled Wnt pathway inhibitors (A). Colony growth assay of MRT cells expressing shRNAs against β-catenin (B). Knock-down of β-catenin by Doxycycline-inducible shRNA in MRT G401 cells (C).
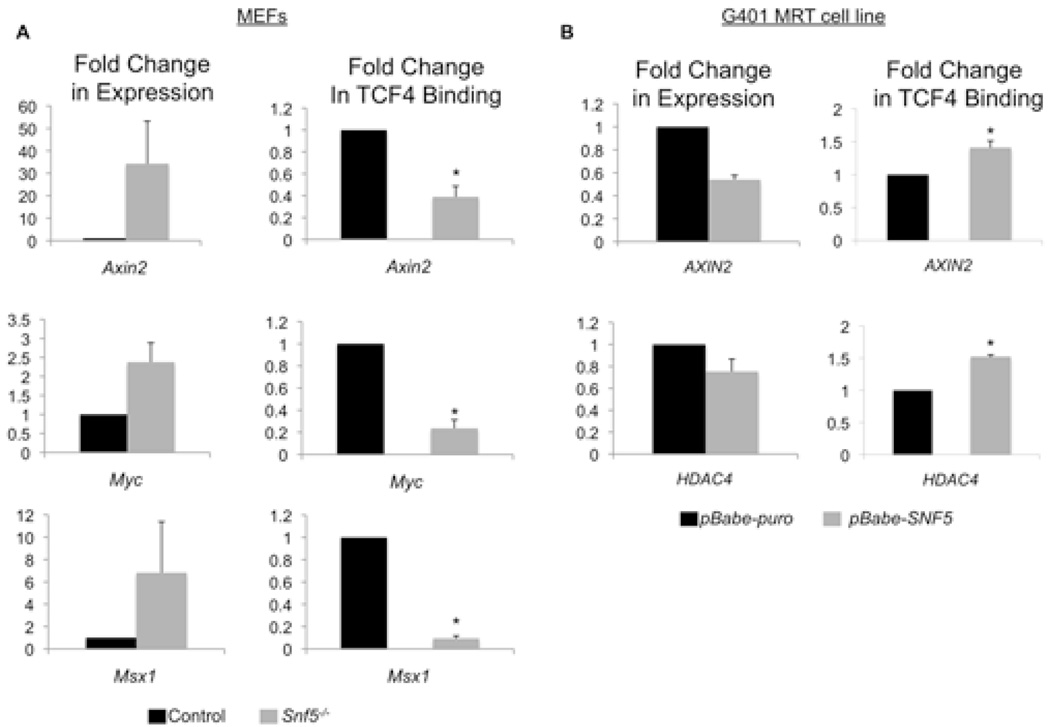
We therefore considered the possibility that β-catenin targets were being aberrantly de-repressed, rather than activated. In the absence of nuclear β-catenin the TCFs/LEF interact with co-repressor complexes to prevent permissive expression of β-catenin targets. We thus tested whether the co-factor TCFs are still capable of binding to their consensus sequence in the absence of SNF5. We inactivated Snf5 in Snf5fl/fl MEFs and performed chromatin immunoprecipitation (ChIP) analysis of TCF4 at well-characterized target promoters relevant in murine development (28, 29). Loss of SNF5 concomitantly decreased the ability of TCF4 to bind to the Axin2, Msx1, and Myc loci and increased gene expression of these targets, suggesting that SNF5 normally facilitates TCF4 binding to its target loci to induce repression (Figure 4A). As Wnt pathway targets are tissue and lineage-specific (with the exception of AXIN2 as a universal target) we examined whether target loci relevant in human cancer had altered TCF binding after re-introduction of SNF5 into G401 MRT cells by ChIP analysis (30). Introduction of SNF5 resulted in increased binding of TCF4 to targets assessed and was accompanied by a decrease in target gene expression (Figure 4B). To further interrogate TCF4 function in SNF5-deficient cells, we introduced a dominant negative form of TCF4 (TCF4ΔN) into the G401 MRT cell line. TCF4ΔN lacks the β-catenin interaction domain but retains the DNA binding domain and hence acts as a constitutive repressor. Expression of TCF4ΔN in G401 cells had no effect on the expression of TCF targets, further indicating that TCF4 cannot act as an efficient repressor in the absence of SNF5 (Supplemental Figure 2).
Figure 4. SNF5 acts downstream in the Wnt pathway to regulate β-catenin/TCF targets.
Gene expession and chromatin immunoprecipitation of TCF4 and at the Axin2, Myc, and Msx1 loci in control (black bars) and Snf5-deleted (grey bars) MEFs (A). Gene expression and chromatin immunopcrecipitation of TCF4 at the AXIN2 and HDAC4 promoters in control (black bars) and SNF5-reintroduced (grey bars) G401 cells (B). Asterisk indicate P<0.05.
In this report, we demonstrate a novel mechanism of regulation of Wnt/β-catenin targets by the SWI/SNF chromatin remodeling subunit SNF5. Previous studies assessing the role of individual SWI/SNF subunits in Wnt signaling have yielded seemingly paradoxical results. In one study in Drosophila melanogaster, loss of the ortholog of mammalian BAF250, BRG1/BRM, or BAF155/170 proteins de-repressed Wnt/Wingless target genes without an increase in Armadillo, the fly β-catenin (31). However, in a different study that used the fly eye as a model system, it was found that Brm haploinsufficiency relieved the rough eye phenotype associated with overexpression of Armadillo (32). In T-cells, the role of Brg1 in the regulation of β-catenin/TCF targets has been similarly unclear (33). In endothelial cells Brg1 loss led to downregulation of the Wnt pathway through the transcriptional regulation of Wnt target genes and a subset Fzd receptors (15, 17). Collectively, these results begin to elucidate a model whereby the Swi/Snf complex can act on the same pathway in a tissue- and context-dependent manner to specifically regulate the WNT/β-catenin pathway by modulating both activation and repression. Disruption of this regulation via loss of the SNF5 tumor suppressor can then drive tumor formation. Given the recent finding that BRG1 (SMARCA4) is specifically mutated in human medulloblastomas, but only in the subgroup that also contain activating mutations in the WNT/β-catenin pathway (34), it is tempting to speculate that rather than preventing WNT signaling, mutation of this SWI/SNF subunit may cooperate to drive WNT signaling in these cancers..
Mutations in genes encoding SWI/SNF subunits are increasingly being identified at high frequency in a wide variety of cancer types. However, the mechanisms underlying oncogenesis have been unclear. Via exome sequencing of 35 human SNF5-mutant primary rhabdoid tumors with matched normal controls, we recently showed that despite their highly aggressive nature, MRT contain a remarkably simple genome with loss of SNF5 being essentially the sole recurrent event (35). Indeed, in two of the cancers there were no other identified mutations at all. These results demonstrate that high mutation rates are dispensable for the genesis of cancers driven by SNF5 loss and suggest that the oncogenic drive provided by SNF5 loss is likely exerted at the epigenetic/chromatin level via perturbation of several pathways that ultimately cooperate to facilitate transformation. In addition to regulation of the WNT/β-catenin pathway, we have previously identified epigenetic antagonism between SWI/SNF and Polycomb complexes, and a role for SNF5 in regulation of the Hedgehog pathway. Collectively, this raises the possibility that the role of the complex in tumor suppression is derived from its contribution to the regulation of several caner related pathways. The mechanistic findings we report here also have substantial therapeutic implications. Our results reveal that mutation of a SWI/SNF subunit can uncouple pathway activation from dependence upon upstream molecules that otherwise modulate pathway activity. Consistent with these findings, we recently showed that activation of the Hedgehog pathway caused by SNF5 loss rendered a Smoothened antagonist targeting the upstream Hh pathway useless (14). Similarly, we have shown here that SNF5 loss uncouples Wnt pathway targets from canonical pathway control and renders existing inhibitors ineffective at blocking target gene expression. This likely occurs because mutation of SNF5 affects control of target genes directly at the level of chromatin, distal to the role of canonical pathway regulators. If the mechanism that drives tumor formation following mutation of other SWI/SNF subunits, including ARID1A, PBRM1, ARID2, BRD7 and BRG1 is similar, our work suggests that elevated target expression will be a poor predictor of response to targeted pathway inhibitors. Given that the sole detected genetic driver event in these cancers is the absence of the SNF5 tumor suppressor, development of effective targeted therapeutics will likely to require an understanding of the pathways activated by this loss and the mechanisms by which altered chromatin structure contributes to this activation.
MATERIALS AND METHODS
Snf5 Knockdown and Excision
Primary MEFs were harvested from E13.5 embryos. Cre was introduced into cells via retroviral infection with pBabe-puror-Cre retroviral supernatant two times at 4 h intervals. Cells were stably selected in medium containing puromycin (2.5 µg/ml) 48 h after infection.
Mouse Strains
Crosses were performed between strains carrying the floxed Snf5 allele (9), the Prx1-Cre transgene (19) and. the Topgal reporter (23). All mice were maintained on a mixed genetic background at the Harvard School of Public Health. All experiments were approved by the Institutional Animal Care and Use Committee (IACUC).
In Situ Hybridizations
Whole-mount in situ hybridizations were performed as described (36). Non-radioactive digoxygenin (DIG) labeled probes were generated according to the manufacturer (Roche). All probe templates used in this study were generously provided by Clifford Tabin (Harvard Medical School).
Cell lines
TTC642, and TM87 were the kind gift of Dr. Bernard Weissman, University of North Carolina. BT12 and BT16 were the kind gift of Dr. David James, University of San Francisco. G401 was obtained from ATCC.
X-gal Staining of Embryos
Dissected embryos were placed in 2% paraformaldehyde/Pipes buffer at 4°C for 30 minutes. Embryos were washed with cold PBS twice, then washed with concentrated rinse buffer (sodium phosphate, sodium deoxycholate, magnesium chloride and NP-40), Embryos were stained with X-gal (Roche) per manufacturers specifications.
RNA Extraction and Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using Trizol reagent (Invitrogen) according to the manufacturer’s instructions and reverse-transcribed by the Reverse Transcription System (Promega). Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad) and the iCycler thermocycler (Bio-Rad). Primer sequences are available upon request. Relative mRNA expression was calculated by the formula 2−(CT of sample − CT of β-actin), where CT (cycle count) is the threshold cycle value. The following primer pairs were used in the qPCR following chip: Mouse AXIN2 F 5’CTCGCATACCTCCCTTCC3’, R 5’TTCCAGCAGTCACTAGGC3’; Mouse MSX1 F 5’ GATCGGAGAATCCAAGTAGCTAC 3’, R 5’ GACAGTGGAGTTTGAGACCTACTC 3’; Mouse MYC F 5’ CAAGCTTTAATTAGCTTAACACA 3’, R 5’ GGAGCCTGCAG AGACCCTA 3’; Human AXIN2 F 5’TTTCCTCTCCTCCCAGTTG3’, F 5’AAGTTGA GCCTACAGTGATTAG3’; Human HDAC4 5’ TGAAAGCACCGCTCATTCTCTGTG 3’, R 5’ GCTGCCTTAAACTTGGCATCAAAGG 3’.
Immunoblots
The following antibodies were used: β-catenin (Sigma-Aldrich, C2206), SNF5 (Bethyl, A301-087A), β-Actin-HRP (Abcam, 20272-200), and HRP-conjugated secondary antibodies (Jackson Immunoresearch)
SNF5 Reintroduction-Expression Analysis
SNF5-deficient G401 cells were transduced using either pBabe-puror-FLAG-SNF5 (generously provided by Robert Kingston, Massachusetts General Hospital) or pBabe-puror-Empty two times at 4 h intervals followed by selection in puromycin (1µg/ml) for 48 h. Cells were then harvested 48 h later for RNA or protein analysis.
Chromatin immunoprecipitation
Chromatin immunoprecipitations (ChIP) was performed as described (13), using the anti-TCF4 antibody (sc-8631), Santa Cruz Biotechnology.
Colony Formation Assays
G401 cells were seeded in growth medium at 600 cells/well into 6-well plates. Sixteen hours after plating, compounds were added at the indicated concentrations. Medium was replenished every four days until colony formation was observed. Colonies were stained by a solution of 2 mg/ml crystal violet in buffered formalin and imaged using a HP Scanjet G4050 scanner.
Expression Analysis
Gene set enrichment analysis (GSEA) was performed as previously described (Subramanian and Tamayo et al PNAS 2005), using a previously published dataset of 11 ATRT samples, 194 primary medulloblastomas and 12 normal cerebellum samples(24).Three independent GSEA analyses were performed using the "CGP: chemical and genetic perturbations (3398 gene sets)", "CP: Canonical pathways (1452 gene sets)", and "C6: oncogenic signatures (189 gene sets)" (for more detailed description of gene sets, please refer to http://www.broad.mit.edu/gsea/msigdb). Medulloblastoma subgroups were identified as previously described (Cho et al. JCO 2011) and "WNT subgroup" medulloblastomas are highlighted in Supplemental Figure 3.
Supplementary Material
Acknowledgments
We thank E.S. McKenna for critical reading of the manuscript and A. Lassar for helpful discussions. The in situ probe constructs were generously provided by C. Tabin (Harvard Medical School).
Footnotes
AUTHOR CONTRIBUTIONS
E.L.M-B initiated the studies, conducted experiments, analyzed data and contributed to writing the manuscript. Y.M., E.J.T, Y-J.C. and C.S.T conducted experiments and analyzed data. S.L.P and W.S. supervised portions of the studies and assisted in the data analysis. C.W.M.R supervised the studies, assisted in the data analysis and contributed to the writing of the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet. 2006;7(6):461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 2.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463(7280):474–484. doi: 10.1038/nature08911. Epub 2010/01/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–492. doi: 10.1038/nrc3068. Epub 2011/06/10. [DOI] [PubMed] [Google Scholar]
- 4.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci USA. 2000;97(25):13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1(6):500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21(10):3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Versteege I, Sévenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394(6689):203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 8.Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59(1):74–79. Epub 1999/01/19. [PubMed] [Google Scholar]
- 9.Roberts CWM, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2(5):415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- 10.McKenna ES, Sansam CG, Cho Y-J, Greulich H, Evans JA, Thom CS, et al. Loss of the epigenetic tumor suppressor SNF5 leads to cancer without genomic instability. Mol Cell Biol. 2008;28(20):6223–6233. doi: 10.1128/MCB.00658-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna ES, Roberts CW. Epigenetics and cancer without genomic instability. Cell Cycle. 2009;8(1):23–26. doi: 10.4161/cc.8.1.7290. Epub 2008/12/23. [DOI] [PubMed] [Google Scholar]
- 12.Gresh L, Bourachot B, Reimann A, Guigas B, Fiette L, Garbay S, et al. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO J. 2005;24(18):3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson BG, Wang X, Shen X, Mckenna ES, Lemieux ME, Cho Y-J, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18(4):316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagani Z, Mora-Blanco EL, Sansam CG, McKenna ES, Wilson B, Chen D, et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat Med. 2010;16(12):1429–1433. doi: 10.1038/nm.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(6):2282–2287. doi: 10.1073/pnas.1013751108. Epub 2011/01/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J-I, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460(7251):66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis CD, Griffin CT. The chromatin-remodeling enzymes BRG1 and CHD4 antagonistically regulate vascular Wnt signaling. Mol Cell Biol. 2012;32(7):1312–1320. doi: 10.1128/MCB.06222-11. Epub 2012/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahmoudi T, Boj SF, Hatzis P, Li VS, Taouatas N, Vries RG, et al. The leukemia-associated Mllt10/Af10-Dot1l are Tcf4/beta-catenin coactivators essential for intestinal homeostasis. PLoS Biol. 2010;8(11):e1000539. doi: 10.1371/journal.pbio.1000539. Epub 2010/11/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33(2):77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 20.Hill TP, Später D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8(5):727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Hill TP, Taketo MM, Birchmeier W, Hartmann C. Multiple roles of mesenchymal beta-catenin during murine limb patterning. Development. 2006;133(7):1219–1229. doi: 10.1242/dev.02298. [DOI] [PubMed] [Google Scholar]
- 22.Chien AJ, Conrad WH, Moon RT. A Wnt survival guide: from flies to human disease. J Invest Dermatol. 2009;129(7):1614–1627. doi: 10.1038/jid.2008.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126(20):4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 24.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(11):1424–1430. doi: 10.1200/JCO.2010.28.5148. Epub 2010/11/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oruetxebarria I, Venturini F, Kekarainen T, Houweling A, Zuijderduijn LMP, Mohd-Sarip A, et al. P16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J Biol Chem. 2004;279(5):3807–3816. doi: 10.1074/jbc.M309333200. [DOI] [PubMed] [Google Scholar]
- 26.Ettenberg SA, Charlat O, Daley MP, Liu S, Vincent KJ, Stuart DD, et al. Inhibition of tumorigenesis driven by different Wnt proteins requires blockade of distinct ligand-binding regions by LRP6 antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(35):15473–15478. doi: 10.1073/pnas.1007428107. Epub 2010/08/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S-MA, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461(7264):614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 28.Miller KA, Barrow J, Collinson JM, Davidson S, Lear M, Hill RE, et al. A highly conserved Wnt-dependent TCF4 binding site within the proximal enhancer of the anti-myogenic Msx1 gene supports expression within Pax3-expressing limb bud muscle precursor cells. Dev Biol. 2007;311(2):665–678. doi: 10.1016/j.ydbio.2007.07.022. Epub 2007/08/31. [DOI] [PubMed] [Google Scholar]
- 29.Hu MC, Rosenblum ND. Smad1, beta-catenin and Tcf4 associate in a molecular complex with the Myc promoter in dysplastic renal tissue and cooperate to control Myc transcription. Development. 2005;132(1):215–225. doi: 10.1242/dev.01573. Epub 2004/12/04. [DOI] [PubMed] [Google Scholar]
- 30.Bottomly D, Kyler SL, McWeeney SK, Yochum GS. Identification of {beta}-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 2010;38(17):5735–5745. doi: 10.1093/nar/gkq363. Epub 2010/05/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins RT, Treisman JE. Osa-containing Brahma chromatin remodeling complexes are required for the repression of wingless target genes. Genes Dev. 2000;14(24):3140–3152. doi: 10.1101/gad.854300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 2001;20(17):4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, et al. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19(2):169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 34.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012 doi: 10.1038/nature11213. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez C, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. The Journal of clinical investigation. 2012;122(8):2983–2988. doi: 10.1172/JCI64400. Epub 2012/07/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ngo-Muller V, Muneoka K. Influence of FGF4 on digit morphogenesis during limb development in the mouse. Dev Biol. 2000;219(2):224–236. doi: 10.1006/dbio.2000.9612. [DOI] [PubMed] [Google Scholar]
- 37.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37(Database issue):D674–D679. doi: 10.1093/nar/gkn653. Epub 2008/10/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willert J, Epping M, Pollack JR, Brown PO, Nusse R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol. 2002;2:8. doi: 10.1186/1471-213x-2-8. Epub 2002/07/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.