Abstract
Both adaptive and acquired resistance significantly limits the efficacy of the epidermal growth factor receptor (EGFR) kinase inhibitors. However, the distinct or common mechanisms of adaptive and acquired resistance have not been fully characterized. Here, through systematic modeling of erlotinib resistance in lung cancer, we found that feedback reactivation of MAPK signaling following erlotinib treatment, which was dependent on the MET receptor, contributed to the adaptive resistance of EGFR inhibitors. Interestingly, acquired resistance to erlotinib was also associated with the MAPK pathway activation as a result of CRAF or NRAS amplification. Consequently, combined inhibition of EGFR and MAPK impeded the development of both adaptive and acquired resistance. These observations demonstrate that adaptive and acquired resistance to EGFR inhibitors can converge on the same pathway and credential cotargeting EGFR and MAPK as a promising therapeutic approach in EGFR mutant tumors.
Keywords: epidermal growth factor receptor, EGFR, MAPK
Introduction
Recent advances in molecular targeted therapies have changed the paradigm for the treatment of patients with non-small cell lung cancer (NSCLC) harboring somatic activating EGFR mutations 1-3. Several EGFR tyrosine kinase inhibitors (TKIs), such as erlotinib, gefitinib and afatinib, have shown remarkable clinical benefit and consequently been approved as the first-line therapy in advanced-stage EGFR mutant NSCLC 4-7. However, the development of drug resistance is inevitable and presents a great challenge to the durable success of TKIs treatment 8-10.
Over the last several years, extensive studies have elucidated a variety of molecular mechanisms that lead to acquired resistance to EGFR TKIs. For example, the emergence of a T790M gatekeeper mutation, occasionally accompanied by EGFR amplification, is detected in ~50% of EGFR mutant lung cancers with acquired resistance to gefitinib or erlotinib 11, 12. In other cases, “bypass track” signaling pathways, activated by amplification of related receptor tyrosine kinases or mutational activation of downstream kinases, may compensate the inhibitory effect of EGFR TKIs. These bypass tracks include amplification of MET or HER2, and mutation of BRAF or PIK3CA 13-16. Additionally, phenotypic changes to either small cell lung cancer or to NSCLC with evidence of epithelial-to-mesenchymal transformation (EMT) have been observed at the time of acquired resistance, although the biological underpinnings are by far elusive 14, 17. Despite these tremendous progressions, the mechanisms that contribute to resistance in the remaining 20% tumors are unknown 10, 14. Therefore, it remains important to study acquired resistance to EGFR TKIs for insights into additional resistance mechanisms and potential therapeutics.
Beyond the genetically defined and heritable acquired resistance, there is emerging evidence that adaptive resistance during initial therapy via feedback mechanisms results in tumor cell survival and residual disease, thus limiting EGFR inhibitor efficacy. We and others have reported that initial EGFR TKIs treatment could engage a Stat3 or NF-κB-mediated feedback loop as an adaptive event to promote NSCLC cell survival 18, 19. These feedback mechanisms enable a small population of oncogene-addicted cancer cells to survive the profound antagonistic effects of EGFR TKIs, and eventually develop acquired resistance 20, 21. The understanding of adaptive resistance could provide rationale for upfront polytherapies to eliminate residual tumor and achieve complete response.
Here, by systematically investigating the molecular basis of drug resistance in NSCLC cell line models, we aim to: 1) identify novel mechanisms of adaptive and acquired resistance to EGFR TKIs; 2) unveil distinct or common signaling pathways underlying adaptive and acquired resistance; and 3) nominate combination treatments to overcome resistance. We discovered that adaptive and acquired resistance to EGFR inhibitors converged on the activation of MAPK pathway, albeit through different mechanisms. Our findings suggest that concomitant EGFR and MAPK blockade is a promising strategy to enhance response magnitude and duration in EGFR mutant patients.
Results
EGFR TKIs trigger feedback activation of MAPK signaling in NSCLC cells
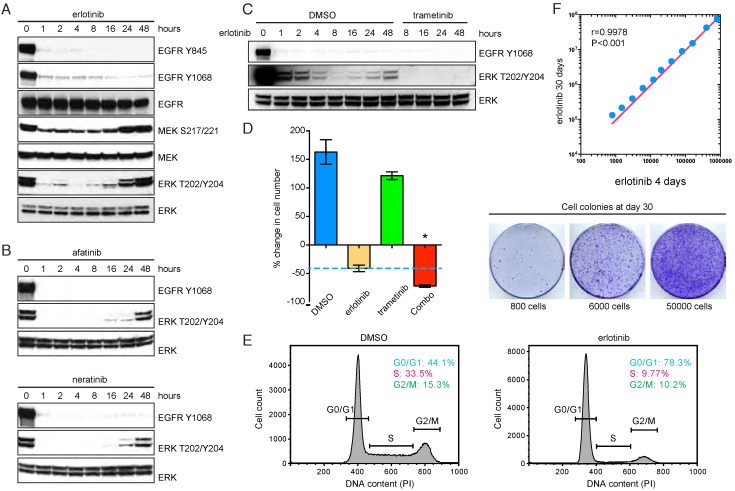
We used PC9, a human EGFR mutant NSCLC cell model bearing exon19 deletion (E746-A750del), to characterize adaptive resistance associated with EGFR TKIs. As expected, erlotinib treatment rapidly suppressed EGFR phosphorylation and downstream MAPK signaling, as indicated by decreased phospho-MEK and phospho-ERK (Figure 1A). However, prolonged erlotinib exposure was unable to produce sustained ERK inhibition, and there was a rebound in phospho-MEK and phospho-ERK after 24-48 hours (Figure 1A). The rebound phenomenon was also observed when PC9 cells were treated with afatinib or neratinib (Figure 1B), which are second-generation irreversible EGFR inhibitors 22, 23. These data imply that the adaptive reactivation of MAPK pathway may limit initial EGFR TKI response, reminiscent of recent findings using irreversible EGFR inhibitor WZ4002 24. Therefore, we evaluated pharmacologic inhibition of MAPK by using an approved MEK inhibitor trametinib (Mekinist®) in the context of EGFR TKI treatment. Concurrent administration of trametinib and erlotinib substantially attenuated the rebound in ERK phosphorylation (Figure 1C). As a result, the combination regimen significantly reduced the number of residual tumor cells compared to erlotinib treatment alone (Figure 1D). Similar data were obtained in two additional NSCLC cell lines harboring EGFR mutations, HCC827 (Supplementary Figure 1A) and HCC4006 (Supplementary Figure 1B). To formally investigate the pro-resistance role of residual cells surviving initial erlotinib inhibition, we cultured the cells at different concentrations in the presence of continuous erlotinib treatment. Although these cells were cell-cycle arrested upon erlotinib exposure (Figure 1E), we found that increased number of residual cells dramatically promoted the occurrence of cell colonies with acquired resistance to erlotinib after long-term treatment (Figure 1F). Therefore, the residual cells may provide a latent reservoir of cells for the emergence of drug-resistance mechanisms. These findings suggest that erlotinib-triggered adaptive MAPK reactivation may contribute to the incomplete response and ultimate relapse of EGFR mutant NSCLC.
Figure 1.
EGFR TKIs trigger feedback activation of MAPK signaling in NSCLC cells. A. Western blot analysis of EGFR, MEK and ERK phosphorylation in PC9 cells in the presence of erlotinib (1 μM). B. Western blot analysis of EGFR and ERK phosphorylation in PC9 cells treated with afatinib (1 μM) or neratinib (1 μM). C. Western blot analysis of EGFR and ERK phosphorylation in PC9 cells treated with erlotinib (1 μM) in combination with trametinib (0.5 μM). D. Cell viability assay of PC9 cells treated with erlotinib (1 μM), trametinb (0.5 μM) or combination (Combo). *P<0.05, ANOVA followed by Tukey's post-test. E. Cell cycle analysis of PC9 cells upon treatment with DMSO or erlotinib (1 μM). F. PC9 cells were treated with erlotinib for 4 days. The remaining cells were seeded at different concentrations and exposed to erlotinib for totally 30 days. Cell numbers were quantified at day 30.
MAPK feedback activation is dependent on the MET receptor
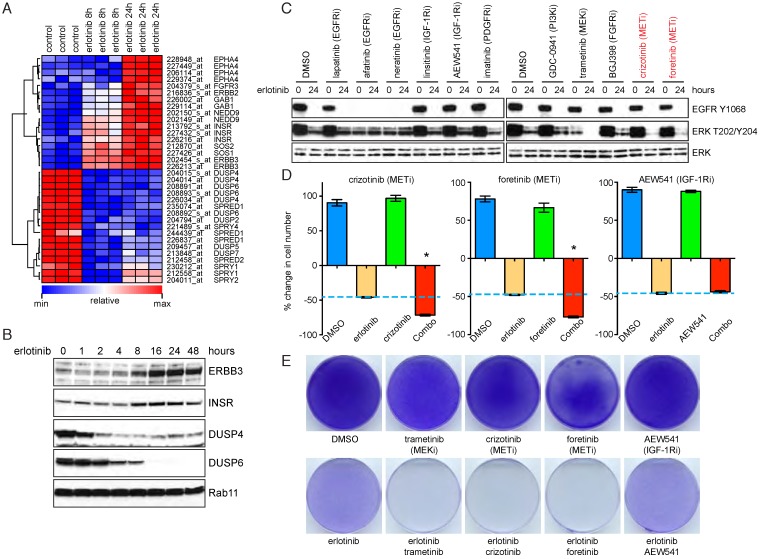
To address the molecular mechanism underlying the feedback activation of MAPK signaling elicited by EGFR inhibition, we perform kinetic microarray analyses on PC9 cells upon acute or prolonged erlotinib treatment. Among the genes differentially expressed in erlotinib-treated cells as compared with mock-treated cells, we found upregulation of various receptor tyrosine kinases (ERBB3, INSR, EPHA4 and FGFR3) and their adaptors (GAB1, NEDD9, SOS1 and SOS2). In contrast, genes involved in the negative regulation of receptor signaling and feedback inhibition of MAPK pathway (DUSPs, SPRYs and SPREDs) were significantly downregulated (Figure 2A). The transcriptional changes were further verified at the protein levels (Figure 2B). These data suggest that inhibition of EGFR by erlotinib may redirect downstream signals to depend on other receptor tyrosine kinases, which, in conjunction with decreased negative regulators including DUSPs and SPRYs, lead to feedback activation of MAPK.
Figure 2.
MAPK feedback activation is dependent on the MET receptor. A. Heatmap of differentially expressed genes in DMSO versus erlotinib treated PC9 cells. B. Western blot analysis of ERBB3, INSR, DUSP4 and DUSP6 in erlotinib (1 μM) treated PC9 cells. C. Western blot analysis of EGFR and ERK activation in PC9 cells treated with erlotinib and inhibitors of EGFR, IGF-1R, PDGFR, FGFR or MET (0.5 μM). D. Cell viability assay of PC9 cells in the presence of erlotinib and inhibitors of MET or IGF-1R. Combo, erlotinib combined with IGF-1R or MET inhibitors (0.5 μM). *P<0.05, ANOVA followed by Tukey's post-test. E. PC9 cells were treated for 10 days as indicated and the cells were stained with crystal violet.
We sought to identify the receptors responsible for MAPK reactivation. To this end, PC9 cells were cotreated with erlotinib and inhibitors of EGFR, IGF-1R, PDGFR, FGFR or MET. We found that only MET inhibitors, in combination with erlotinib, completely suppressed phospho-ERK at 24 hour (Figure 2C). Consistently, the combination therapies of EGFR and MET inhibition, but not IGF-1R inhibition, significantly decreased the number of residual tumor cells (Figure 2D), and the effect was comparable to that of erlotinib and trametinib cotreatment (Figure 2E). Of note, MET inhibitor or IGF-1R inhibitor at the working concentration (500 nM) only exhibited modest inhibitory effects against PC9 cells as single agent (Supplementary Figure 2A). Based on these findings, we propose that MAPK feedback activation induced by EGFR inhibitors is dependent on the MET receptor.
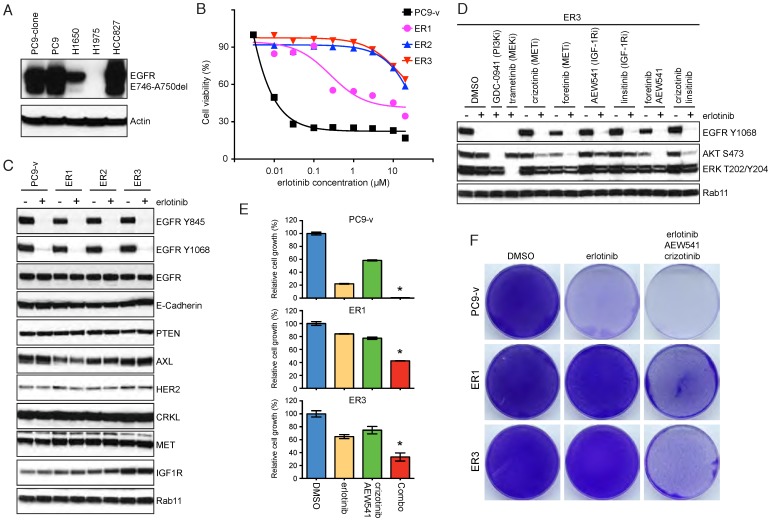
Acquired resistance to erlotinib treatment in a subclone of PC9 cells
PC9 cells that survived the initial erlotinib treatment may eventually develop acquired resistance. Previous studies revealed that PC9 cells usually exploited the T790M gatekeeper mutation to gain acquired resistance to EGFR TKIs 25, 26. To explore additional mechanisms of erlotinib resistance, we selected a subclone of PC9 cells, which expressed similar levels of mutant EGFR relative to parental cells (Figure 3A). By exposing the PC9 subclone to increasing concentrations of erlotinib for 5 months, we generated three independent resistant clones and termed them ER1-3 (Figure 3B). Both the PC9 subclone and erlotinib-resistant cells were submitted to short-tandem repeat analysis to verify the authenticity. Importantly, acquired resistance in ER1-3 could not be attributed to known mechanisms, such as amplification of CRKL, AXL and HER2, loss of PTEN or development of EMT 14, 15, 27, 28. EGFR phosphorylation was still efficiently inhibited by erlotinib, indicating that resistance was not due to T790M mutation (Figure 3C). Although we observed subtle increases in MET and IGF-1R expression, addition of MET and IGF-1R inhibitors failed to markedly reduce phospho-ERK in resistant cells (Figure 3D; Supplementary Figure 2B). Consequently, the combination therapy of EGFR, MET and IGF-1R inhibitors only resulted in partial growth inhibition of resistant cells (Figure 3E&F). We concluded that the resistant mechanisms for ER1-3 were yet to be identified.
Figure 3.
Acquired resistance to erlotinib treatment in a subclone of PC9 cells. A. Western blot assay of mutant EGFR (E746-A750del) in the PC9 subclone. B. Cell viability of PC9 subclone (PC9-v) and erlotinib-resistant cells (ER1, ER2 and ER3) treated with various concentrations of erlotinib. C. Western blot analysis of EGFR, E-Cadherin, PTEN, AXL, HER2, CRKL, MET and IGF-1R in erlotinib treated PC9-v, ER1, ER2 and ER3 cells. D. Effects of erlotinib and inhibitors of PI3K, MEK, MET or IGF-1R on phosphorylation of AKT and ERK in ER3 cells. E. Cell proliferation assay of PC9-v, ER1 and ER3 cells treated with erlotinib and inhibitors of MET and IGF-1R (0.5 μM). Combo, erlotinib combined with crizotinib and AEW-541. *P<0.05, ANOVA followed by Tukey's post-test. F. PC9-v, ER1 and ER3 cells were treated with DMSO, erlotinib, or erlotinib combined with crizotinib and AEW-541 for 10 days. The remaining cells were stained with crystal violet.
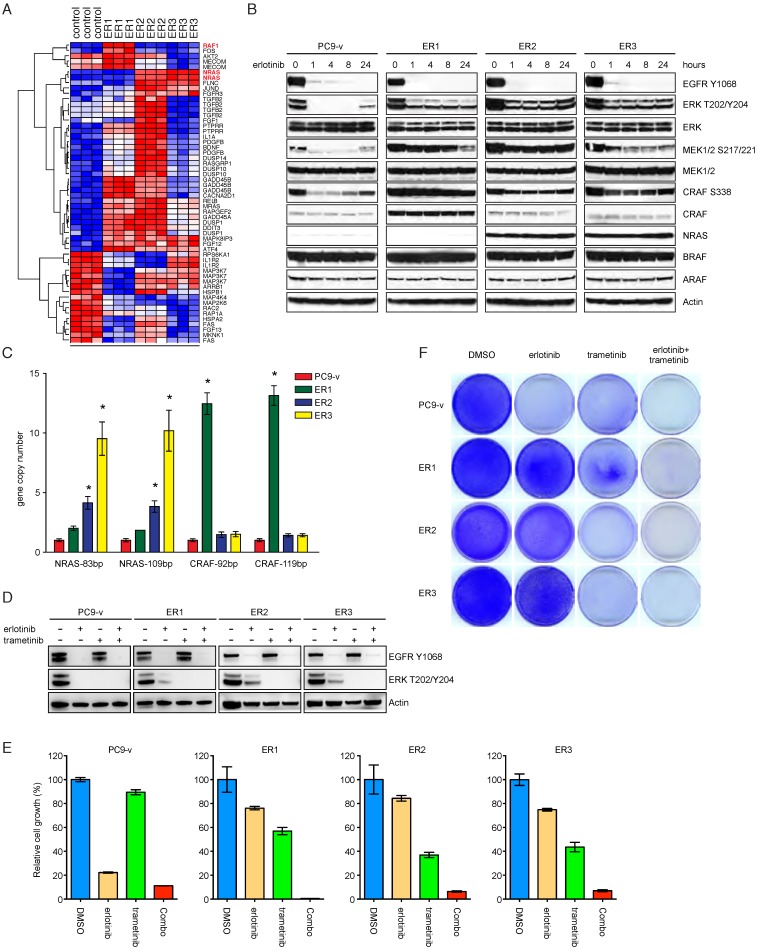
NRAS or CRAF amplification induces persistent activation of MAPK cascade in resistant cells
We next performed genome-wide mRNA profiling of PC9 ER1-3 and compared the gene expression with control PC9 subclone. Interestingly, many transcripts in the MAPK pathway were identified to be differentially expressed. Notably, RAF1 gene, which encodes CRAF, was significantly upregulated in ER1, while ER2 and ER3 exhibited appreciable increase of NRAS expression (Figure 4A). At the protein level, Western blot analysis validated remarkable overexpression of CRAF in ER1 and NRAS in ER2/ER3, respectively (Figure 4B). Consistent with these findings, phosphorylation of ERK, MEK and CRAF in PC9 ER1-3 cells was maintained in the presence of erlotinib. We did not observe obvious changes of ARAF and BRAF protein levels (Figure 4B). To determine whether there was a genomic basis for the increase in CRAF and NRAS, we quantified their copy numbers in the resistant cells and compared them to the parental PC9 subclone. Quantitative real-time PCR using two different primer sets indicated that CRAF was amplified in ER1 and NRAS was amplified in ER2 and ER3 (Figure 4C). To determine whether CRAF and NRAS amplification conferred acquired resistance to erlotinib, clustered regularly interspaced short palindromic repeats CRISPR-Cas9 system was employed to knock out CRAF or NRAS in resistant cell lines. We found that CRAF deletion in ER1 or NRAS deletion in ER3 resulted in impaired MAPK signaling and improved response to erlotinib treatment (Supplementary Figure 3). We next investigated whether inhibition of MAPK signaling would restore sensitivity to erlotinib in PC9 resistant cells. The MEK inhibitor trametinib completely inhibited phospho-ERK in all four cell lines (Figure 4D). When used in combination with erlotinib, trametinib significantly inhibited cell growth in ER1-3 to the similar level found in the PC9 subclone cell line (Figure 4E&F). Taken together, our results support that NRAS or CRAF amplification induces persistent activation of MAPK cascade in resistant cells and inhibition of MAPK signaling restores sensitivity to EGFR TKIs.
Figure 4.
NRAS or CRAF amplification induces persistent activation of MAPK cascade in resistant cells. A. Heatmap of differential gene expression in PC9-v versus resistant cells. B. Western blot analysis of the MAPK pathway components in PC9-v and resistant cells in the presence or absence of erlotinib. C. Copy number analysis of NRAS and CRAF in PC9-v and resistant cells. *P<0.05, ANOVA followed by Tukey's post-test. D. PC9-v, ER1, ER2 and ER3 cells were treated with DMSO, erlotinib (1 μM), trametinib (0.5 μM) or erlotinib combined with trametinib (Combo). Phospho-EGFR and phospho-ERK were analyzed by western blot. E. Cell viability assay of PC9-v, ER1, ER2 and ER3 cell treated with DMSO, erlotinib (1 μM), trametinib (0.5 μM) or erlotinib combined with trametinib (Combo). *P<0.05, ANOVA followed by Tukey's post-test. F. PC9-v, ER1, ER2 and ER3 cells were treated as indicated and stained with crystal violet.
Combination of erlotinib and trametinib leads to improved efficacy in vivo
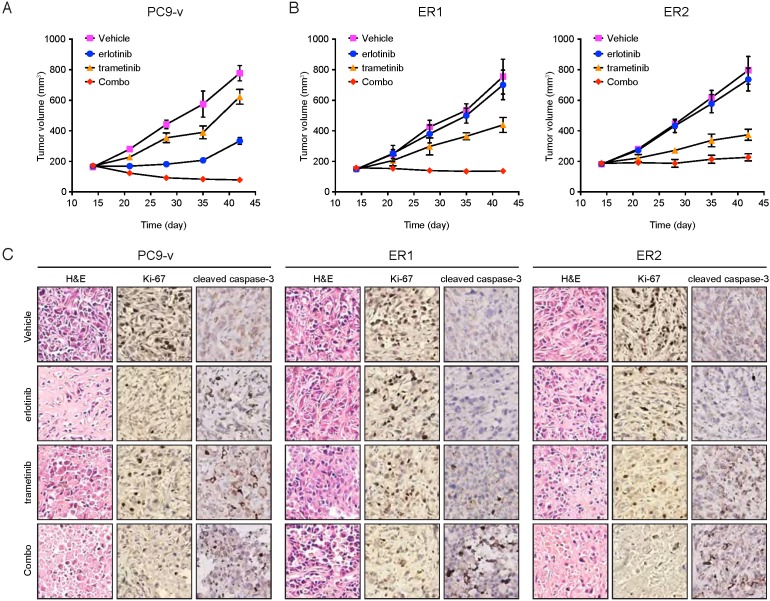
Our data implicated the MAPK pathway in mediating both adaptive and acquired resistance to EGFR inhibitors. To model adaptive and acquired resistance in vivo and evaluate the efficacy of targeting EGFR and MAPK simultaneously, we established xenografts of PC9 subclone and two of erlotinib-resistant cell lines. Because ER2 and ER3 displayed similar mechanism of acquired resistance, only ER2 was selected for in vivo studies. PC9 xenografts were treated with vehicle, erlotinib, and trametinib individually or in combination for four weeks. Although PC9 was sensitive to erlotinib in vivo, the erlotinib/trametinib polytherapy more effectively retarded tumor growth compared with single agent treated tumors. Notably, the combination therapy strongly induced tumor regression (Figure 5A), implying that MAPK inhibition of adaptive resistance could achieve a better efficacy for the management of EGFR mutant tumors. Likewise, in ER1 or ER2 cells that developed acquired resistance to erlotinib, erlotinib/trametinib cotreatment significantly suppressed tumor growth relative to single agent treatments (Figure 5B). Both cell proliferation and cell survival were synergistically inhibited by the combination therapy of erlotinib and trametinib, as assessed by Ki-67 and cleaved caspase-3 staining, respectively (Figure 5C). These findings confirm the in vitro observations and support the role of MAPK signaling pathway in promoting both adaptive and acquired resistance to EGFR TKIs.
Figure 5.
Combination of erlotinib and trametinib leads to improved efficacy in vivo. A. Mice bearing PC9-v tumors were treated with the indicated drugs for 4 weeks. Tumor growth was measured every week (ten mice per group). *P<0.05, ANOVA followed by Tukey's post-test. B. Mice bearing ER1 or ER2 tumors were treated with the indicated drugs for 4 weeks. Tumor growth was measured every week (ten mice per group). *P<0.05, ANOVA followed by Tukey's post-test. C. Immunohistochemical staining of Ki-67 and cleaved caspase-3 in tumor xenograft sections.
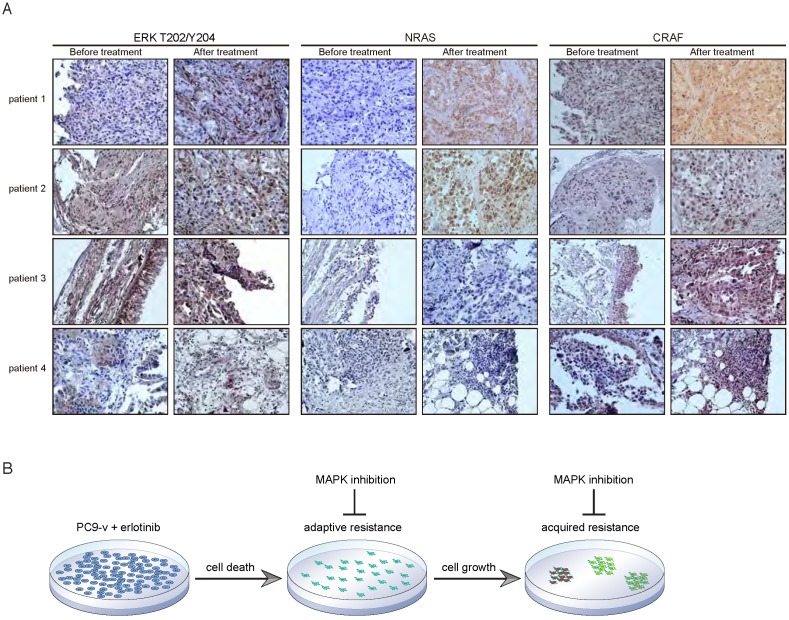
Post-erlotinib human lung tumor biopsies display increased CRAF and NRAS expression compared to pre-treatment specimens
Finally, we sought to determine the clinical relevance of our findings. We therefore evaluated the expression of phospho-ERK, CRAF and NRAS in primary human tumor specimens obtained from erlotinib-treated NSCLC patients that had developed drug resistance without the common resistant EGFR T790M mutations. We observed high levels of phospho-ERK in the majority of the tumor biopsies before and after erlotinib treatment, as measured by immunohistochemistry (Figure 6A), indicating that the MAPK pathway might be functionally important for these EGFR mutant tumors. Intriguingly, compared to erlotinib-naïve biopsies, NRAS expression was clearly upregulated in post-treatment tumor sections from patient 1 and patient 2, and patient 3 showed increased CRAF staining (Figure 6A). These findings suggest that increased CRAF or NRAS levels in these tumors might contribute to acquired resistance to EGFR inhibition.
Figure 6.
Post-erlotinib human lung tumor biopsies display increased CRAF and NRAS expression compared to pre-treatment specimens. A. Immunohistochemical staining of phospho-ERK, NRAS and CRAF demonstrating increased CRAF and NRAS expression in post-erlotinib tumor sections compared to paired pre-treatment specimens. B. Schematic representation of targeting the MAPK pathway to overcome both adaptive resistance and acquired resistance to EGFR inhibitors.
Discussion
By comprehensively investigating the adaptive resistance and acquired resistance to EGFR TKIs (a schematic model was shown in Figure 6B), we have proposed that cotargeting EGFR and MAPK provides the opportunity for both prevention and treatment of drug resistance, and thus is a more effective combination strategy than just targeting EGFR alone. Our study illustrated the temporal dynamic evolution of EGFR mutant tumor cells in response to EGFR-targeted therapies and explored the mechanistic rationale for therapeutic intervention at different stages to maximize the efficacy of EGFR inhibitors. Novel resistant mechanisms have been identified including MET-dependent MAPK feedback activation and CRAF or NRAS amplification, both converging on the MAPK pathway. These findings can guide selection of appropriate therapy, such as MEK inhibitors, to combine with EGFR TKIs in order to overcome anticipated drug resistance, which warrants further clinical investigation in prospective studies.
It is a general theme that constitutively activated oncogenic drivers reprogram the signaling network and induce inhibition of certain pathways in tumors 29-35. Inhibition of these oncogenes in “addicted” cancer cells may relieve the feedbacks and lead to the activation of alternative pathways or reactivation of the same pathway. Molecular targeted therapies are often accompanied by this adaptive response of tumor cells, which hamper the initial effectiveness by sparing a subpopulation of surviving cells. This process may also facilitate the formation of tumor clones that harbor genetic alterations to confer acquired resistance. Therefore, understanding and inhibition of cell-protective feedback loops may not only improve the response rate but also prevent the emergence of drug resistance. Previous studies have identified Stat3 or NF-κB feedback activation in the context of EGFR TKIs 18, 19. Unfortunately, neither Stat3 nor NF-κB pathways are readily targetable in clinic. On the other hand, our discovery of MET-mediated MAPK reactivation has broader clinical applicability as both MET and MAPK pathway inhibitors are undergoing clinical development. Based on these findings, upfront combination therapies with EGFR inhibitors and MET or MAPK kinase inhibitors should be considered for NSCLC patients bearing EGFR mutations, given that the combined toxicity of these agents are manageable.
Acquired resistance has emerged as a major limitation of monotherapy with EGFR TKIs. Tremendous efforts have been made to unveil the molecular basis of acquired resistance. Along this line, many mechanisms of resistance to EGFR-targeted therapies have been proposed, including but not limited to T790M mutation, MET or HER2 amplification, and histological transformation. However, ~20% of acquired resistance cases lack favorable explanation 10, 14. We found CRAF or NRAS amplification and consequent MAPK hyperactivation as a new mechanism of acquired resistance. Accordingly, combination therapies with EGFR and MEK inhibitors remained effective in erlotinib-resistant tumors. Similarly, MAPK inhibition has been also reported to restore sensitivity to the irreversible EGFR kinase inhibitor WZ4002 36, 37. It is noteworthy that previously uncovered mechanisms such as MET or HER2 amplification may also rely on the MAPK pathway and could potentially be counteracted using the same approach. Importantly, we observed the upregulation of CRAF or NRAS in erlotinib-resistant patients, supporting the clinical relevance of our findings.
In summary, our findings have important implications for overcoming EGFR TKI resistance in NSCLC patients. Most previous studies aimed at understanding EGFR TKI resistance have focused on either adaptive resistance or acquired resistance and revealed a range of molecular mechanisms that require individually tailored therapy. Our data suggest that adaptive and acquired resistance to EGFR inhibitors may converge on a common pathway, and its functional delineation may offer unique therapeutic opportunities to combat drug resistance.
Materials and methods
Cell culture and reagents
Tumor cell lines were obtained from ATCC and were cultured in RPMI1640 (Invitrogen) supplemented with 10% fetal bovine serum (Millipore). Erlotinib, afatinib, neratinib, GDC-0941, trametinib, lapatinib, insitinib, AEW541, imatinib, BGJ398, crizotinib and foretinib were purchased from Selleck Chemicals. All inhibitors were reconstituted in DMSO (Sigma-Aldrich) at a stock concentration of 10 mM.
Generation of resistant cells and CRAF/NRAS knockout cells
Erlotinib-resistant PC9 cells were established and maintained as described previously 25. In short, PC9-v cells were grown in culture medium containing escalating concentrations of erlotinib. After 5 months of passages, the remaining cells that could grow in the presence 5 μM erlotinib were considered as resistant cells. Three independent clones including ER1, ER2 and ER3 were obtained for the current study. CRISPR-Cas9 system was employed to knock out CRAF or NRAS in ER1 or ER3 cells, respectively 38. The sgRNA sequences were designed as following: CRAF-sgRNA1: 5'-CACCGGCTTGGAAGACGATCAGCAA-3' and 5'-AAACTTGCTGATCGTCTTCCAAGCC-3'; CRAF-sgRNA2: 5'-CACCGCAGCGCCGGGCATCAGATGA-3' and 5'-AAACTCATCTGATGCCCGGCGCTGC-3'; NRAS-sgRNA1: 5'-CACCGATTCATCTACAAAGTGGTTC-3' and 5'-AAACGAACCACTTTGTAGATGAATC-3'; NRAS-sgRNA2: 5'-CACCGCTTCGCCTGTCCTCATGTAT-3' and 5'-AAACATACATGAGGACAGGCGAAGC-3'.
Cell proliferation assays
Cells were seeded at 3,000 cells/well in growth media supplemented with 10% FBS and 2 mM L-glutamine, allowed to adhere overnight, and treated with a dilution series of erlotinib for 72 hours. Cell viability was determined by CellTiter Glo (Promega). Growth inhibition curves were plotted using GraphPad Prism software.
Cell cycle analysis
Cell cycle analysis was performed 96 hours after erlotinib treatment. Cells were fixed in cold ethanol and resuspended in Propidium Iodide (PI)/RNase Staining Solution (Cell Signaling Technology). After incubation for 15 minutes at room temperature in the dark, flow cytometric analysis was performed on a FACS AriaII cytometer (BD Biosciences). Flow cytometry data was analyzed by using FlowJo software and the cell cycle was plotted as histogram after excluding doublets.
Western blot
Cells were lysed in RIPA buffer (Tris pH 7.4 50 mM, NaCl 150 mM, NP-40 1%, SDS 0.1%, EDTA 2 μM) containing proteinase inhibitors (Roche) and phosphatase inhibitors (Roche). The cell lysates (20 μg protein) were subjected to SDS-PAGE and Western blot. Antibodies against the following proteins were used: phospho-EGFR (Y1068), phospho-EGFR (Y845), EGFR, phospho-MEK1/2 (S217/221), MEK1/2, phospho-ERK (T202/Y204), ERK, phospho-AKT (S473), AKT, ERBB3, INSR, DUSP4, DUSP6, Rab11, E-Cadherin, PTEN, AXL, HER2, CRKL, MET, IGF-1R, ARAF, BRAF, phospho-CRAF (S338), CRAF, NRAS, and Actin (Cell Signaling Technology).
Microarray analysis
RNA was prepared with RNeasy plus mini kit (Qiagen) according to the manufacturer's protocol. Total RNA was subjected to microarray analysis using Affymetrix human genome U133 Plus 2.0. Three biological replicates per treatment group were included for statistical analyses. Affymetrix microarray probe-level data were normalized by Robust Multi-array Average (RMA) procedure. Differential gene expression was analyzed with linear models for microarray data (Limma).
Copy number variation analysis
Quantitative PCR (qPCR) was performed to determine the status of NRAS and CRAF in DNA samples from resistant cells. NRAS, CRAF and MTHFR (endogenous control) levels were evaluated using the following primers: NRAS-83bp-sense: 5'-GGACATACTGGATACAGCTGGAC-3'; NRAS-83bp-anti-sense: 5'-CACAGAGGAAGCCTTCGCCTG-3'; NRAS-109bp-sense: 5'-CTCGGATGATGTACCTATGGTGC-3'; NRAS-109bp-anti-sense: 5'-GAATGGAATCCCGTAACTCTTGG-3'; CRAF-92bp-sense: 5'-GGACAACCTGGCAATTGTGACC-3'; CRAF-92bp-anti-sense: 5'-GCTGGAACATCTGAAACTTGGTC-3'; CRAF-119bp-sense: 5'-GGAGCACATACAGGGAGCTTGG-3'; CRAF-119bp-anti-sense: 5'-GGCGCTGATAGCCAAACTGCTG-3'; MTHFR-121bp-sense: 5'-CCATCTTCCTGCTGCTGTAACTG-3'; MTHFR-121bp-anti-sense: 5'-GCCTTCTCTGCCAACTGTCC-3'. For resistant cells (ER1, ER2 and ER3) and parental cell (PC9), 20 ng of genomic DNA was amplified for 40 cycles (15 sec 95°C, 60 sec 60°C) in an ABI 7500 Real-Time PCR system (ABI), using the SYBR Green I (Roche) and 400 nM primers.
Xenograft
Tumor cells (1×106) were mixed with Matrigel (BD Biosciences) and subcutaneously implanted in the dorsal flank of BALB/c Nude mice. When tumor sizes reached approximately 150 mm3, mice were randomized into 4 groups of 10 mice each. One group of mice was treated with vehicle control (0.5% methylcellulose and 0.2% Tween-80), and the other three groups were treated with erlotinib (50 mg/kg/day), trametinib (1 mg/kg/day) or erlotinib combined with trametinib, respectively. Tumor volumes (10 animals per group) were measured with digital caliper and calculated as length×width2×0.52. The animals were housed in a specific pathogen free (SPF) animal facility in accordance with the Guide for Care and Use of Laboratory Animals and the regulations of the Institutional Animal Care and Use Committee.
Immunohistochemistry
Immunohistochemistry was performed using 5 μm-thick, formalin-fixed, paraffin-embedded tissue sections. Slides were baked, deparaffinized in xylene, passed through graded alcohols, and antigen retrieved with 10 mM citrate buffer, pH 6.0 in a steam pressure cooker. Preprocessed tissues were treated with Peroxidase Block (Dako) to quench endogenous peroxidase activity, blocked using Protein Block (Dako), and subsequently incubated with Ki-67, cleaved caspase-3, NRAS or CRAF antibodies. Slides were then washed in 50 mM Tris-Cl, pH 7.4 and incubated with horseradish peroxidase-conjugated secondary antibody. Immunoperoxidase staining was developed using a 3,3'diaminobenzidine (DAB) chromogen. Slides were counterstained with hematoxylin, dehydrated in graded alcohol and xylene, and coverslipped using mounting solution.
Statistical analysis
Statistical analysis was performed with GraphPad Prism software. In all experiments, comparisons between two groups were based on two-sided Student's t-test and one-way analysis of variance (ANOVA) was used to test for differences among more groups. P-values of <0.05 were considered statistically significant.
Supplementary Material
Supplementary figures.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81472537 to G Zhuang and 81502597 to Y Jing), the Project Funded by China Postdoctoral Science Foundation (2015M580338 to P Ma), the Grants from the State Key Laboratory of Oncogenes and Related Genes (91-14-18 and 91-15-12 to G Zhuang), Collaborative Innovation Center for Translational Medicine at Shanghai Jiao Tong University School of Medicine, the Shanghai Institutions of Higher Learning (Eastern Scholar to G Zhuang), and Shanghai Rising-Star Program (16QA1403600 to G Zhuang).
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I. et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–11. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H. et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 5.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J. et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–8. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N. et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 7.Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y. et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:213–22. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 8.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–74. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19:1389–400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–81. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M. et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 12.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF. et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO. et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 14.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P. et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takezawa K, Pirazzoli V, Arcila ME, Nebhan CA, Song X, de Stanchina E. et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov. 2012;2:922–33. doi: 10.1158/2159-8290.CD-12-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL. et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–33. doi: 10.1073/pnas.1203530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N. et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–62. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 18.Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ, Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–21. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Blakely CM, Pazarentzos E, Olivas V, Asthana S, Yan JJ, Tan I. et al. NF-kappaB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell Rep. 2015;11:98–110. doi: 10.1016/j.celrep.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pazarentzos E, Bivona TG. Adaptive stress signaling in targeted cancer therapy resistance. Oncogene; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothenberg SM, Concannon K, Cullen S, Boulay G, Turke AB, Faber AC, Inhibition of mutant EGFR in lung cancer cells triggers SOX2-FOXO6-dependent survival pathways. Elife; 2015. p. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanderson K. Irreversible kinase inhibitors gain traction. Nat Rev Drug Discov. 2013;12:649–51. doi: 10.1038/nrd4103. [DOI] [PubMed] [Google Scholar]
- 23.Landi L, Cappuzzo F. Irreversible EGFR-TKIs: dreaming perfection. Transl Lung Cancer Res. 2013;2:40–9. doi: 10.3978/j.issn.2218-6751.2012.12.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tricker EM, Xu C, Uddin S, Capelletti M, Ercan D, Ogino A. et al. Combined EGFR/MEK Inhibition Prevents the Emergence of Resistance in EGFR-Mutant Lung Cancer. Cancer Discov. 2015;5:960–71. doi: 10.1158/2159-8290.CD-15-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogino A, Kitao H, Hirano S, Uchida A, Ishiai M, Kozuki T. et al. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non small cell lung cancer cell line. Cancer Res. 2007;67:7807–14. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- 26.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE. et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–9. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung HW, Du J, Boehm JS, He F, Weir BA, Wang X. et al. Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov. 2011;1:608–25. doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T. et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–60. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A. et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–74. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D. et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 31.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S. et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandarlapaty S, Sawai A, Scaltriti M, Rodrik-Outmezguine V, Grbovic-Huezo O, Serra V. et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E. et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov. 2011;1:248–59. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M. et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell. 2012;22:668–82. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartz S, Wongvipat J, Trigwell CB, Hancox U, Carver BS, Rodrik-Outmezguine V. et al. Feedback suppression of PI3Kalpha signaling in PTEN-mutated tumors is relieved by selective inhibition of PI3Kbeta. Cancer Cell. 2015;27:109–22. doi: 10.1016/j.ccell.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ercan D, Xu C, Yanagita M, Monast CS, Pratilas CA, Montero J. et al. Reactivation of ERK signaling causes resistance to EGFR kinase inhibitors. Cancer Discov. 2012;2:934–47. doi: 10.1158/2159-8290.CD-12-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortot AB, Repellin CE, Shimamura T, Capelletti M, Zejnullahu K, Ercan D. et al. Resistance to irreversible EGF receptor tyrosine kinase inhibitors through a multistep mechanism involving the IGF1R pathway. Cancer Res. 2013;73:834–43. doi: 10.1158/0008-5472.CAN-12-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.