Abstract
Pre-mRNA maturation frequently occurs at the same time and place as transcription by RNA polymerase II (pol II). The co-transcriptionality of mRNA processing has permitted the evolution of mechanisms that functionally couple transcription elongation with diverse events that occur on the nascent RNA. This review summarizes current understanding of the relationship between transcriptional elongation through a chromatin template and co-transcriptional splicing including alternative splicing decisions that affect the expression of most human genes.
Keywords: RNA polymerase II, CTD, transcription elongation, alternative splicing, kinetic coupling, histone modification, nucleosome occupancy
Graphical abstract
Introduction
An important shift occurred in our perception of eukaryotic transcription when it was realized that transcription is not a stand-alone process, but rather is functionally coupled to maturation of the RNA transcript. Thus the major mRNA processing steps of capping, splicing and cleavage/polyadenylation, as well as messenger ribonucleoprotein (mRNP) assembly, initiate co-transcriptionally rather than post-transcriptionally 1; 2; 3; 4. Co-transcriptionality permitted the co-evolution of transcription and processing factors with the result that in some cases they perform their functions in an interdependent or coupled fashion. Transcription and co-transcriptional RNA metabolism are integrated with one another by both spatial and kinetic coupling mechanisms. RNA polymerase II (pol II) is uniquely equipped with an essential appendage, the C-terminal heptad repeat domain (CTD) of the large subunit that is required for all three major mRNA processing reactions 5; 6; 7 and for recruitment of splicing factors to sites of transcription 8. The co-transcriptional nature of mRNA maturation means that the physiological substrate of the processing factors is not a full-length freely diffusible pre-mRNA, but a transcription elongation complex (TEC) with a growing RNA that is extruded at average rates of 0.5–4 kb/min on human genes9. A comprehensive understanding of mRNP maturation must therefore take into account the “co-transcriptionality” of this process. This perspective considers the cycle of transcription initiation, elongation and termination in the context of the processing of nascent transcripts. The “mRNA factory” is a useful model for how transcription and RNA processing occur at the same time and place within a dynamic macromolecular complex 6 that comprises both the RNA synthetic and processing machines (Fig. 1). In this review we focus on the elongation phase of the transcription cycle and its relation to splicing of the nascent transcript.
Figure 1.
The mRNA factory model. Coupling of transcription with pre-mRNA processing within a complex that contains both the synthetic and processing machines. Recruitment of RNA processing factors to the transcription elongation complex (TEC) occurs through dynamic interactions with the CTD “landing pad”. According to the CTD code hypothesis 30, interactions with the CTD are instructed by dynamic phosphorylation of the CTD heptad repeats including Ser2 and Ser5 residues (S2-P and S5-P, red and green dots) in ways that are synched with the transcription cycle. Capping enzyme and the cleavage/polyadenylation (poly(A)) complexes interact directly with S5-P and S2-P CTD isoforms that are enriched at 5’ and 3’ ends of genes respectively. Note that capping factors are detected also at 3’ ends and polyA factors at 5’ ends by ChIP. Whether splicing factors interact directly with the CTD has yet to be confirmed at the structural level. Whether different processing factors can simultaneously localize on the CTD is also not known but it is unlikely to be prohibited on steric grounds. The 7-methyl guanosine cap (7meG) is shown at the 5’ end of the nascent RNA (blue line).
Transcription elongation is far from a smooth ride for the RNA polymerase. Each journey made by a pol II TEC along a given gene follows a unique narrative punctuated by acceleration, deceleration, backtracking, pausing and release; and premature termination may sometimes end the journey before the 3’ end of the gene 9; 10; 11; 12. Each passage that pol II makes along a gene is influenced by numerous factors that govern elongation and ultimately determine how the nascent RNA grows. These effectors of elongation include sequence elements near the 3’ end of the RNA 13, nucleosomal barriers, and factors that bind and modify pol II such as positive transcription elongation factor PTEFb (Cdk9/CycT), negative elongation factors NELF, DRB (5,6-dichloro-1-β-D-ribofuranosyl-1H–benzimidazole) sensitivity inducing factor DSIF (Spt4/5), and TFIIS 12; 14. Average elongation rate is a function of the rates of catalysis of phosphodiester bonds and associated enzyme translocation as well as the duration of numerous pauses some of which can be several minutes long 15; 16; 17. Regulated polymerase pausing is used in prokaryotes to coordinate transcription with co-transcriptional translation 18; 19 and recent studies suggest that pausing may operate in eukaryotes to coordinate co-transcriptional pre-mRNA splicing 20; 21; 22; 23. The rate of nascent RNA growth can also affect the way it folds 24 and RNA structure is an important determinant of how the transcript will be processed by the splicing machinery.
Alternative splicing affects the expression of about 95% of human genes and abnormal proportions of alternatively spliced mRNA isoforms are hallmarks of the transcriptome in many diseases including cancer 25; 26,27. Most alternative splicing decisions are probably made co-transcriptionally 1; 2; 28,29 and how elongation rate affects co-transcriptional spliceosome assembly and function is an important challenge that promises to deliver a much deeper understanding of how normal and abnormal alternative splicing decisions are made.
Spatial coupling: recruitment of processing factors to the site of transcription
Coupling in space is achieved by recruitment of factors to the TEC often through interaction with the CTD “landing pad” (Fig. 1). The paradigm for recruitment coupling is that the instructions for processing factor association with the TEC are provided by dynamic phosphorylation of CTD heptad repeats (YS2PTS5PS) in a way that is synched with the transcription cycle 30; 31. Hence phosphorylation of the CTD repeats on Ser5 residues is essential for capping enzyme recruitment at 5’ ends of genes 32; 33,34 and phosphorylation on Ser2 facilitates recruitment of cleavage/polyadenylation factors at 3’ ends 35; 36. How the phospho-CTD code influences co-transcriptional splicing is less well understood, but recent work implicates CTD phosphorylation in recruitment of U2AF65 that binds polypyrimidine tracts and stabilizes U2snRNP binding 37; 38. Although pol II co-purifies with SR proteins and U1 snRNP 39; 40, and the CTD is required for the SR protein SRp20 to regulate alternative splicing 41, it is currently unclear whether recruitment of these splicing factors requires direct contacts with the polymerase. Expression of a pol II CTD mutant (S2A) that cannot be phosphorylated on Ser2 reduced co-transcriptional splicing in a promoter-specific way as well as U2AF65 and U2 snRNP recruitment 38. One should note however that mutation of one residue in the CTD heptad repeats could indirectly affect phosphorylation of other residues. Conversely, inhibition of splicing was reported to specifically reduce Ser2 CTD phosphorylation (Ser2-P) suggesting a role of splicing factors in maintenance of this modification 42. Consistent with this idea, the FUS protein, a regulator of alternative splicing 43 involved in the pathogenesis of ALS, binds the CTD and helps maintain Ser2-P 44. Furthermore, FUS acts as an adaptor for U1 snRNP binding to pol II 39. Whether FUS-mediated recruitment of U1 snRNP is affected by Ser2-P and how this recruitment might regulate alternative splicing remain to be resolved.
Recruitment of processing factors to the site of transcription is not exclusively through the pol II CTD landing pad. The mediator complex that binds promoters and enhancers can influence alternative splicing through its Med23 subunit that contacts the splicing factors hnRNPL, SF3B and Eval1 45. This effect of mediator might in part account for the seminal observation that promoter sequences can determine how a transcript is spliced 46. In addition, as discussed below, a growing body of work has identified the chromatin template as a recruitment site for splicing factors.
Kinetic coupling: how elongation rate affects nascent pre-mRNA processing
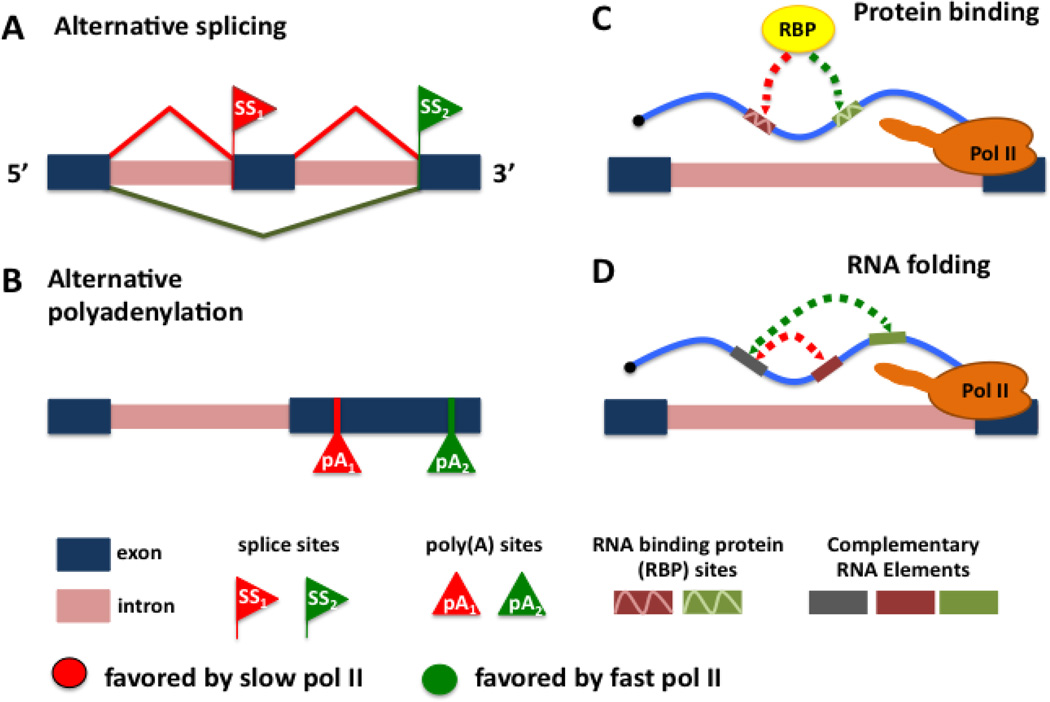
Kinetic coupling between transcription and co-transcriptional RNA transactions is less well understood than spatial coupling. The “window of opportunity” or “first come first served” model 47; 48; 49; 50 is a helpful way of thinking about this form of coupling. The idea is that when upstream and downstream events on the nascent transcript compete, then the upstream site will have a head start, and therefore a competitive advantage. That advantage is greater when elongation is slow, and smaller when elongation is fast. Potentially competing co-transcriptional events that could be modulated by elongation rate include alternative splice site and poly(A) site recognition, RNA binding site recognition by regulatory proteins, and base-pairing of competing sequences with a common complementary element as the transcript folds (Fig. 2).
Figure 2.
“Window of opportunity” model for kinetic coupling of nascent RNA metabolism with transcription elongation. When upstream and downstream events on the nascent transcript compete, then the upstream site will have a head start, and therefore a competitive advantage. Slow elongation lengthens the window of opportunity for upstream events to occur before they face competition from downstream sites. Competing upstream and downstream sites on the nascent RNA include: A. 3’ splice sites of alternative exons; B. polyadenylation sites; C. RNA binding protein (RBP) recognition sites; D. complementary RNA sequence elements that base pair as the RNA folds. Sites favored by slow and fast elongation are colored in red and green hues respectively.
Recent work has underscored the widespread importance of kinetic coupling in determining the outcome of co-transcriptional splicing reactions. The average pol II elongation rate in mammals is 0.5–4.0 kb/minute, however it varies extensively between genes and even between different regions within a gene 9; 11; 17; 51. The “window of opportunity” model predicts that elongation rate controls alternative splicing by modulating the competition between upstream and downstream 3’ splice sites (Fig. 2A) and it can explain why a slow mutant pol II enhanced inclusion of alternative exons in the fibronectin and NCAM genes 47; 52 and constitutive splicing in yeast 53. Changing the elongation rate can potentially alter the window of opportunity for binding of both positive and negative splicing factors, which accounts for why slow elongation can also favor exon skipping. For example slow elongation enhances CFTR exon 9 skipping by favoring association of the negative splicing factor, ETR-3 that competes with U2AF65 for binding to the polypyrimidine tract 54.
We used pol II rate mutants to investigate the impact of elongation rate on alternative splicing genome-wide in human cells. Both slow and fast transcription changed the alternative splicing of thousands of exons 55. Unexpectedly, while there were some cases where slow and fast elongation had opposite effects on splicing outcomes as predicted by the “window of opportunity” model, there were many more examples where slow and fast mutants both increased or both decreased inclusion of a particular alternative exon. This result suggests an optimal rate or “goldilocks” model in which most rate-sensitive splicing events require an elongation rate that is neither too fast nor too slow but “just right” to produce a proper balance of alternatively spliced RNA isoforms. It remains to be determined what differentiates rate-sensitive from rate-insensitive alternatively spliced exons but chromatin environment, RNA structure, or specific sequence motifs could be responsible. In future, it will be of interest to investigate whether elongation rate is regulated under physiological conditions to modulate alternative splicing programs. In addition, kinetic and spatial coupling of transcription with mRNA processing are likely to be interdependent because factors that control elongation rate could be differentially recruited to the TEC and conversely elongation rate could influence recruitment of processing factors.
Detailed understanding of kinetic coupling will require high-resolution analysis of elongation rate, pausing, and nascent RNA structure. Newly developed methods are helping to address this technical challenge (see 9 for a review). Elongation rate can be assayed in a genome-wide fashion by sequencing nascent pol II transcripts at time intervals after pol II is released from a block near the transcription start site induced by the drug, DRB 51; 56. In this way one can follow the progress of a wave of pol II as it progresses along genes. A low resolution way to detect pausing is by anti-pol II ChIP-seq that measures polymerase density within genes. A pause or slow down can be inferred from a local peak of pol II density, but there are other possible interpretations that are difficult to eliminate such as differential epitope availability (discussed in 57). Pausing is most directly measured by native elongating transcript sequencing (NET-seq), that precisely maps the 3’ ends of nascent transcripts enriched by high salt urea wash 21 58 or anti-pol II immunoprecipitation 59. There are potential caveats to NET-seq experiments however since precipitated pol II complexes may include RNAs that are not directly tethered to the template by pol II including excised introns 59. Furthermore anti-pol II immunoprecipitation could be biased by epitope availability. These reservations notwithstanding, an elevated frequency of NET-seq reads at a particular location strongly suggests that transcription has paused there. Recently long-range sequencing of nascent transcripts from their 3’ ends revealed that in yeasts, co-transcriptional splicing can be completed by the time pol II has extended only 25–30 bases beyond the 3’ splice site 58. This remarkable finding implies that splicing can be completed on the nascent RNA when only about 10 bases beyond the 3’ ss have emerged from the pol II RNA exit channel.
Relationship between pausing and splicing
Two important recent studies in human cells applied NET-seq to provide global maps of the 3’ ends of nascent transcripts at single nucleotide resolution 21; 22. Strikingly these studies revealed a high frequency of 3’ ends, very close to 3’ and 5’ splice sites that flank exons. Since splicing intermediates were computationally filtered from the NET-seq datasets, the results suggest that splice sites are potent pol II pausing signals. Pausing at 3’ splice sites at the beginning of exons is particularly remarkable because it occurs before exon definition 60 while the splice site is sequestered within the pol II ternary complex. These observations therefore suggest the surprising conclusion that the 3’ splice site is recognized by the transcription machinery as a pause site before it is recognized by the splicing machinery as a processing site. NET-seq employing immunoprecipitation with phospho-specific anti-CTD antibodies suggests that pol II paused at 3’ and 5’ splice sites is Ser5 hyperphosphorylated. In addition CTD Ser2-P appears to increase as pol II is released from the 3’ splice site and moves into the exon (Fig. 3). Release from splice site associated pauses might be stimulated by U2AF65 that associates with pol II elongation complexes and inhibits pausing in vitro 61. In summary, these NET-seq studies show that pausing of specific pol II phosphoisoforms is strongly correlated with splice sites although one potential caveat is that some antibodies against the phosphorylated CTD can cross react with abundant highly phosphorylated SR splicing factors 62. One attractive idea is that pausing at intron-exon boundaries or elsewhere in exons, provides a window of opportunity for some splicing steps to be completed co-transcriptionally, but whether the duration of transcriptional pauses is long enough to have a meaningful effect on co-transcriptional splicing has not been established 20; 23.
Figure 3.
Splicing-dependent pol II pausing. Commitment to splicing induces pol II pausing close to the 3’ splice site at the start of exons, which have higher nucleosome densities than introns. This pause is accompanied by CTD Ser5 phosphorylation. Pol II pauses with high CTD Ser5 phosphorylation (S2-P). Release from this pause is proposed to be contingent on a splicing-dependent checkpoint being satisfied (grey arrow, lower panel) that may be accompanied by increased CTD Ser2 phosphorylation (S2-P) reminiscent of the switch in CTD phosphorylation that occurs following release from the promoter-proximal pause at transcription start sites. This is a speculative model based on references 20–23, 63.
The remarkable coincidence of pause sites with splice sites begs the question of whether pausing at exon intron boundaries is a cause or consequence of splicing. Seminal work in budding yeast using anti-pol II ChIP-seq strongly suggests that at splicing-dependent transcription pause occurs at 3’ splice sites 20; 63 however these pause sites have yet to be confirmed by nascent RNA sequencing 58 Pausing near yeast 3’ splice sites was reduced by splice site mutations and restored by suppression of a branch point mutation with altered specificity U2 snRNA 20. Interestingly pol II paused at yeast 3’ splice sites has CTD Ser5-P 63 like the pause at mammalian 3’ splice sites 22. When splicing was blocked by prp5 or U2 snRNA mutations pausing of pol II in a high Ser5-P, low Ser2-P form occurred further upstream within introns suggesting that a splicing-dependent checkpoint must be satisfied before pol II is permitted to resume elongation 63. This model resembles the switch that occurs at 5’ ends of metazoan genes from paused Ser5-P pol II to actively elongating Ser2-P pol II 9; 10; a transition that may involve a capping-dependent checkpoint. The splicing-dependent pause model suggests some interesting questions for future studies: Does the relationship between splicing and pausing differ between organisms like yeast where splicing is specified by intron definition and humans where exon definition predominates? What happens if a splicing checkpoint is not satisfied? One possibility is that transcription could terminate prematurely aided by the Xrn2 5’-3’ exonuclease 64. How could a splicing-dependent checkpoint signal that is generated far from the pol II active site induce pausing? Intriguingly the yeast U2 snRNP-associated Cus2 protein is required for this pause 63 and its metazoan homologue, TAT-SF1, interacts with PTEFb that functions as a CTD Ser2-Ser5 kinase 65. Additional interactions between PTEFb and the SR protein, SC35 66, and the U5snRNP subunit, SKIP, 67 suggest further biochemical links between splicing and transcription elongation. In summary recent insights point to a remarkably precise connection between pol II pausing and recognition of exon-intron junctions and the underlying mechanism is a fascinating problem for future investigation.
Pol II elongation factors and co-transcriptional splicing
The kinetic coupling of co-transcriptional splicing suggests that factors which modulate pol II elongation could affect splicing decisions. As pol II moves through a gene, nucleosomes and other DNA binding proteins can cause it to arrest and backtrack, dislodging the 3’ end of the nascent transcript from the active site 68; 69; 70; 71. Backtracked pol II can be rescued by the transcription factor TFIIS, which inserts into a side channel in the enzyme and interacts with the active site to stimulate its intrinsic endonuclease activity 72; 73; 74. RNA cleavage realigns the 3’ end with the active site and allows transcription to continue. TFIIS reduces the duration of pol II pausing and accelerates elongation in vitro 75; 76 and in vivo 59. Remarkably, depletion of TFIIS in yeast 49 and expression of dominant negative TFIIS in Arabidopis 77 both enhanced exon inclusion. These results therefore suggest that pausing of backtracked pol II provides a window of opportunity to complete some splicing reactions co-transcriptionally. Further work will be required to determine whether TFIIS participates in normal control of alternative splicing. It will also be of interest to determine whether additional regulators of pol II pausing including TFIIF, G-Down1 78, Spt5 79, elongin 12, and components of the super elongator complex 14 affect co-transcriptional pre-mRNA splicing decisions.
Nucleosome occupancy and co-transcriptional splicing
A corollary of pervasive co-transcriptional splicing, is that this reaction occurs largely in a chromatin environment that can differ between different regions of a gene. The revelation that nucleosomes, and the histone modifications that mark them are unequally distributed between exons and introns 80; 81; 82 83 was an important conceptual breakthrough. Nucleosomes are more densely packed on exons than introns in many organisms because of their higher average G-C content 80; 83; 84 and nucleosome occupancy is higher on constitutive exons than alternative exons 80; 85. Furthermore exons flanked by weak splice sites have greater enrichment of nucleosomes than those with strong splice sites 83 suggesting a functional connection between chromatin structure and local splicing activity.
Nucleosomes are major barriers to transcription elongation in vitro, but in vivo they are efficiently evicted from transcribed chromatin. When pol II encounters a nucleosome, elongation pauses 59; 72; 86; 87; 88; 89; 90 preferentially at the entry point and 45 bases into the nucleosome at the contact point between DNA and the H3/H4 tetramer 68; 87. Nucleosome displacement from the template by pol II in vitro, is greater when elongation is faster suggesting a two-way relationship between elongation rate and the chromatin speedbumps91. Consistent with the speedbump effect of nucleosomes, ChIP-seq and NET-seq showed that pol II occupancy is higher 23; 82; 92; 93, and elongation rate is slower, in exons than introns 17; 94 with the possible exception of fission yeast 95. Kinetic coupling suggests that pausing at nucleosomes positioned near exons could tip the balance between competing splice sites thereby influencing alternative splicing decisions (Fig. 2). Two recent studies assessed the relationship between nucleosome occupancy and alternative splicing. One study found that near alternative exons that are preferentially included in progesterone treated breast cancer cells, the chromatin structure switched from poorly positioned to well-positioned nucleosomes and this rearrangement correlated with increased pol II occupancy consistent with pausing 96. A second study addressed whether changes in nucleosome occupancy can cause changes in splice site selection by restricting the histone supply in colon carcinoma cells through inhibition of histone mRNA 3’ end formation. In histone-depleted cells, chromatin accessibility and pol II elongation rate increased on several genes and many transcripts had elevated intron retention and altered alternative splicing 97. Together these studies suggest that positioned nucleosomes enhance splice site recognition and a likely mechanism is through kinetic coupling by nucleosome-induced pol II pausing.
Chromatin remodelers and co-transcriptional splicing
Histone chaperones and remodelers affect the movement of pol II by promoting the assembly or disassembly of nucleosomal roadblocks within transcribed genes. Kinetic coupling implies that histone dynamics could influence RNA processing decisions by affecting pol II pausing. The most important regulators of co-transcriptional histone dynamics within genes are the H2A/H2B chaperone FACT (facilitates chromatin transcription) and the H3 chaperone Spt6, that both increase the rate of pol II elongation through chromatin 98; 99; 100.
Chaperones may also influence pre-mRNA processing indirectly by controlling the deposition of histone variants that can affect the nucleosomal barrier to pol II elongation 101; 102. One such variant, H2A.Z, which is deposited by SWR-C, stimulates elongation rate in yeast 103 possibly due to enhanced nucleosome exchange and reduced pausing 68. Mammalian, H2A.Z may also influence splicing through recruitment of processing factors since it can can bind SF3B1, a component of the U2 snRNP 104.
The ATP-dependent chromatin remodelers SWI/SNF and CHD1 promote pol II progress through nucleosomes 105; 106. They also co-immunoprecipitate with splicing factors and their depletion changes alternative splicing patterns 105; 107; 108; 109; 110. The effects of the Brm SWI/SNF subunit on splicing correlate with altered pol II pausing, but unexpectedly they are independent of its ATPase activity 105; 108. It is therefore possible that Brm affects splicing in a chromatin-independent way through its incorporation into hnRNP particles on nascent transcripts 111. In summary histone chaperones and remodelers can exert influences over co-transcriptional splicing in at least two ways: 1) by modulating elongation through effects on nucleosome density and deposition of histone variants, and 2) by affecting recruitment of splicing factors to the chromatin template.
Covalent histone marks and splicing
Exons and introns differ in nucleosome occupancy and in the density covalent histone marks. After normalizing for total histone content, H3K27 me1, me2, and me3, H3K36me3, H3K79me1, H4K20me1, and H2BK5me1 are detectably enriched within exons 80; 81; 82; 92; 93; 112; 113 while introns are relatively enriched for H3K79me2, H2BK5me1, H2Bub, H3K4me1, H3K4me2, H3K9me1, H3K23ac, H3K79me1, H3K79me2, H3K79me3 and H4K20me1 93; 113. Which of these modifications actually affect splicing and by what mechanisms is an exciting area of investigation (reviewed in 114 115). Several histone marks associated with actively transcribed genes including H3K36me3, H2BK120 monoubiquitylation (H2Bub1) and H3, H4 N-terminal tail acetylation,are implicated in control of splicing.
The H3K36me3 mark is added co-transcriptionally by the SETD2 methyltransferase, that is recruited to the TEC, either directly by the Ser2-P CTD 116 or indirectly by Spt6 117. The importance of H3K36me3 for splicing is highlighted by the fact that SETD2 mutant kidney tumors have splicing defects in about 25% of expressed genes including extensive intron retention 118. The level of H3K36me3 within alternative exons correlates with their inclusion in spliced transcripts 81; 82 and inhibition of splicing re-positions this mark within genes suggesting that its deposition is sensitive to local splicing activity 119 probably through differential SETD2 recruitment 120. Not only is H3K36 methylation responsive to splicing, but it can also control splicing by recruiting polypyrimidine tract-binding protein (PTB) and Srsf1 to the chromatin template through contacts with the “chromatin readers” MRG15 and Psip1 121; 122. Other covalent histone marks may also recruit splicing factors including SR proteins and U2 snRNP to chromatin, from whence they are presumed to hop onto the nascent transcript 123; 124. An independent way that H3K36me3 could modulate co-transcriptional splicing is through kinetic coupling since this mark helps maintain nucleosome occupancy within genes 125; 126 and could thereby affect splicing by limiting elongation rate.
Another histone modification implicated in control of co-transcriptional splicing is H2Bub1, which is enriched on genes with fast elongation rates 127. H2Bub1 cooperates with FACT 128 and SWI/SNF 129 to facilitate elongation by mobilizing nucleosomes within genes. Depletion of the H2B deubiquitinase, USP49 caused extensive changes in alternative splicing marked by preferential skipping of exons with elevated H2Bub1 and reduced association of the U1 and U2 snRNPs with chromatin 130.
Histone H3, H4 hyperacetylation is associated with enhanced exon skipping and acceleration of transcription following depolarization of excitable cells 52; 131; 132. The proposed kinetic coupling mechanism is that nucleosomes with hyperacetylated N-terminal tails pose a lower barrier to transcription elongation, and indeed single molecule studies show slightly reduced pausing at a mock-acetylated nucleosome 89. Interestingly there may be a two-way interaction between the nascent transcript and histone acetylation since RNA-binding proteins, including splicing factors, that associate with the nascent transcript can recruit regulators of histone acetylation including a histone acetyl transferase 133, a deacetylases (HDAC) 134, and an HDAC inhibitor 132.
Heterochromatin, Argonauts, and alternative splicing
Repressive histone modifications in heterochromatin can also influence specific alternative splicing decisions. Surprisingly Argonaut (AGO) proteins, best known as effectors of RNA-guided heterochromatin formation, can also function as regulators of alternative splicing (reviewed in 29; 114). This connection was discovered by synthetic siRNA targeting of AGO1 to an alternative exon in the fibronectin gene which caused an increase in its inclusion in spliced transcripts 135. Furthermore, knock down of AGOs changes alternative splicing of many transcripts in human and Drosophila cells 136; 137; 138. In some cases AGO-regulated alternative splicing of specific exons correlates with sites of H3K9 and H3K27 methylation 135 133. The latter heterochromatin mark is also implicated in control of FGFR2 alternative splicing by a long non-coding antisense RNA that directs polycomb-mediated H3K27 methylation and promotes a specific alternative splicing pathway for the FGFR2 mRNA 139. Heterochromatin marks could modulate splicing by kinetic coupling through a localized slow down in transcription elongation 135. Confirmation of this idea will require accurate determination of elongation rates at defined positions within genes, which remains an important technical challenge. Another potential mechanism of alternative splicing regulation by AGOs is suggested by the finding in C. elegans that an AGO complex appears to inhibit transcription elongation directly 140. Yet another regulatory mechanism is suggested by the observation that splicing factors can associate with AGO1/2 and the H3K9me binding protein HP1 136; 141; 142 143. While co-purification of RNA binding proteins with chromatin-associated factors should be interpreted with care because of the potential for indirect interaction through contaminating RNA, these studies suggest that splicing can be modulated by recruitment of splicing factors to the chromatin template through AGO and heterochromatin marks. How AGOs are targeted to alternatively spliced transcripts under normal conditions is poorly understood, but in Drosophila, at least, this mechanism appears to be independent of siRNAs 137.
In summary while much remains to be learned about the connections between co-transcriptional splicing and chromatin structure, a substantial body of evidence points to two mechanisms by which chromatin acts as an effector of splicing: 1) by inducing localized transcriptional pausing that could shift the balance between competing splicing reactions through kinetic coupling (Fig. 2) and 2) by serving as a scaffold for recruitment of splicing factors that might subsequently be handed off onto the nascent transcript.
DNA methylation and splicing
Another feature of the chromatin template that can exert a major influence on co-transcriptional splicing is DNA 5-methyl-CpG methylation (5meCpG), a modification that is depleted in alternative exons that are skipped relative to those that are included 144. Remarkably inclusion of about 20% of alternative exons is affected by DNA methylation based on experiments using methylation deficient ES cells 141. This mechanism of splicing control is mediated by methyl sensitive DNA binding proteins including MeCP2 144 and CTCF whose binding is inhibited by 5meCpG. CTCF binding to an oxidized hydroxymethyl C-modified sequence element in CD45 exon 5 induces pol II pausing and inclusion of this exon in the mRNA 145; 146. An alternative potential mechanism by which DNA methylation could affect co-transcriptional splicing is via interaction of RNA binding proteins with methyl sensitive DNA binding proteins. This idea is suggested by binding of the splicing regulator YB-1 to MeCP2, which might account for the splicing defects in Rett syndrome patients with MeCP2 mutations 147.
RNA structure and alternative splicing
A relatively little studied aspect of co-transcriptional pre-mRNA processing to date is the effect of nascent RNA folding. While nascent RNA is often depicted as a passive, linear structure it actually contains multiple layers of information in the form of secondary and tertiary structures 148. These structures can dictate important events in the life of an RNA molecule, including alternate splice site selection 149. For example, pre-mRNA structure could mask or unmask sites for RNA binding proteins (RBPs), sequester non-productive, cryptic binding sites and bring sequences such as 5’ and 3’ splice sites closer together to favor specific splicing events 150; 151; 152; 153 154. Folding can even impact RNA sequences by recruiting ADARs (Adenosine to Inosine Deaminases Acting on RNA) that catalyze conversion of adenosine to the non-canonical base inosine exclusively in double-stranded RNA. A–I RNA editing frequently affects the sequences of splice sites or splicing control elements 155. Further, inosine containing RNA is bound by Vigilin proteins 156 that are involved in heterochromatin formation 157, suggesting the possibility that RNA editing could influence transcription elongation rate by affecting chromatin structure.
Recent technological advances have provided the first glimpses into global in vivo mRNA structure by structure-seq a strategy that combines chemical probing of secondary structure with deep-sequencing 158; 159; 160. One observation made so far is that stable structures at 5’ splice sites in Arabidopsis correlate with unspliced introns 159. Future insights into the relationship between transcriptional elongation, RNA structure and co-transcriptional pre-mRNA processing should emerge from structure-seq studies of nascent RNA populations. Not only is pol II elongation rate likely to affect how nascent transcripts fold as previously observed in prokaryotes and in vitro 24; 161; 162; 163, but RNA structure may also feed back to control elongation rate. In vitro pol II synthesis of highly structured transcripts is faster than less structured transcripts, probably due to inhibition of backtracking by RNA hairpins 164. Future investigations of the relationship between pol II elongation and the structure of the nascent transcript seems likely to reveal novel mechanisms of co-transcriptional splicing regulation.
Conclusions and Future Directions
The functional relationship between transcriptional elongation and co-transcriptional RNA metabolism is a fascinating one that is beginning to yield up its secrets thanks to powerful new methods that permit genome-wide analysis of growing RNA ends at single nucleotide resolution. A major remaining challenge is to relate elongation at defined positions within genes to the splicing and folding of the nascent RNA. Important outstanding questions about how functional coupling of transcription and splicing works include:
Is the variation in pol II elongation rate between human genes a result of regulation, and what is its functional significance for mRNA processing?
What is the source of the signal(s) that pauses pol II at exon-intron boundaries and how does a splicing dependent checkpoint work? How does the CTD phosphorylation code influence co-transcriptional splicing?
How is nascent RNA structure affected by elongation rate and how important is RNA folding for co-transcriptional RNA processing decisions?
How does transcription elongation affect RNA binding protein association and spliceosome assembly on nascent transcripts?
What is the relative importance of chromatin-mediated regulation of alternative splicing by kinetic coupling versus splicing factor recruitment? Is there a mechanism that transfers splicing factors from chromatin to the nascent RNA?
Research Highlights.
Kinetic coupling of transcription elongation with pre-mRNA splicing is widespread.
Transcriptional pausing and mRNA splicing appear to be functionally interdependent
Chromatin influences splicing through effects on pausing and splicing factor recruitment
Acknowledgments
Work in the authors’ laboratory is supported by NIH grants GM063873 and GM058613 to D.B. R.S. is supported by NIHT32-GM08730 and T.S. is supported by American Cancer Society fellowship PF-15-188-01 – RMC. We thank T. Blumenthal for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brugiolo M, Herzel L, Neugebauer KM. Counting on co-transcriptional splicing. F1000Prime Rep. 2013;5:9. doi: 10.12703/P5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 4.Daneholt B. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci U S A. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5'-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 7.Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 9.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell. 2013;50:212–222. doi: 10.1016/j.molcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, Nickels BE. Interactions between RNA polymerase and the "core recognition element" counteract pausing. Science. 2014;344:1285–1289. doi: 10.1126/science.1253458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13:543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- 15.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell. 2013;52:517–528. doi: 10.1016/j.molcel.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanofsky C. Transcription attenuation: once viewed as a novel regulatory strategy. J Bacteriol. 2000;182:1–8. doi: 10.1128/jb.182.1.1-8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, Block SM, Greenleaf WJ, Landick R, Weissman JS. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science. 2014;344:1042–1047. doi: 10.1126/science.1251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell. 2015;161:541–554. doi: 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nojima T, Gomes T, Grosso AR, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ. Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell. 2015;161:526–540. doi: 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Pan T, Sosnick T. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 25.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 27.Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol. 2016;212:13–27. doi: 10.1083/jcb.201510032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem. 2015;84:165–198. doi: 10.1146/annurev-biochem-060614-034242. [DOI] [PubMed] [Google Scholar]
- 30.Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 31.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell. 2011;43:311–318. doi: 10.1016/j.molcel.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3' end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 36.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3' end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 37.David CJ, Boyne AR, Millhouse SR, Manley JL. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011;25:972–983. doi: 10.1101/gad.2038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu B, Eick D, Bensaude O. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 2013;41:1591–1603. doi: 10.1093/nar/gks1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Y, Reed R. FUS functions in coupling transcription to splicing by mediating an interaction between RNAP II and U1 snRNP. Proc Natl Acad Sci U S A. 2015;112:8608–8613. doi: 10.1073/pnas.1506282112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26:867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 41.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 42.Koga M, Hayashi M, Kaida D. Splicing inhibition decreases phosphorylation level of Ser2 in Pol II CTD. Nucleic Acids Res. 2015;43:8258–8267. doi: 10.1093/nar/gkv740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogelj B, Easton LE, Bogu GK, Stanton LW, Rot G, Curk T, Zupan B, Sugimoto Y, Modic M, Haberman N, Tollervey J, Fujii R, Takumi T, Shaw CE, Ule J. Widespread binding of FUS along nascent RNA regulates alternative splicing in the brain. Sci Rep. 2012;2:603. doi: 10.1038/srep00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, Cech TR. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012;26:2690–2695. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y, Li W, Yao X, Lin QJ, Yin JW, Liang Y, Heiner M, Tian B, Hui J, Wang G. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell. 2012;45:459–469. doi: 10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci U S A. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Dujardin G, Lafaille C, Petrillo E, Buggiano V, Gomez Acuna LI, Fiszbein A, Godoy Herz MA, Nieto Moreno N, Munoz MJ, Allo M, Schor IE, Kornblihtt AR. Transcriptional elongation and alternative splicing. Biochim Biophys Acta. 2013;1829:134–140. doi: 10.1016/j.bbagrm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 49.Howe KJ, Kane CM, Ares M., Jr Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aebi M, Weissman C. Precision and orderliness in splicing. Trends in Genetics. 1987;3:102–107. [Google Scholar]
- 51.Fuchs G, Voichek Y, Benjamin S, Gilad S, Amit I, Oren M. 4sUDRB-seq: measuring genomewide transcriptional elongation rates and initiation frequencies within cells. Genome Biol. 2014;15:R69. doi: 10.1186/gb-2014-15-5-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci U S A. 2009;106:4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, Tang LK, Fraser JS, Holstege FC, Hieter P, Guthrie C, Kaplan CD, Krogan NJ. From Structure to Systems: High-Resolution, Quantitative Genetic Analysis of RNA Polymerase II. Cell. 2013;154:775–788. doi: 10.1016/j.cell.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dujardin G, Lafaille C, de la Mata M, Marasco LE, Munoz MJ, Le Jossic-Corcos C, Corcos L, Kornblihtt AR. How Slow RNA Polymerase II Elongation Favors Alternative Exon Skipping. Molecular Cell. 2014:1–8. doi: 10.1016/j.molcel.2014.03.044. [DOI] [PubMed] [Google Scholar]
- 55.Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu XD, Bentley DL. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014;28:2663–2676. doi: 10.1101/gad.252106.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev. 2013;113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 58.Oesterreich FC, Herzel L, Straube K, Hujer K, Howard J, Neugebauer KM. Splicing of Nascent RNA Coincides with Intron Exit from RNA Polymerase II. Cell. 2016;165 doi: 10.1016/j.cell.2016.02.045. http://dx.doi.org/10.1016/j.cell.2016.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schneider M, Will CL, Anokhina M, Tazi J, Urlaub H, Lührmann R. Exon Definition Complexes Contain the Tri-snRNP and Can Be Directly Converted into B-like Precatalytic Splicing Complexes. Molecular Cell. 2010;38:223–235. doi: 10.1016/j.molcel.2010.02.027. [DOI] [PubMed] [Google Scholar]
- 61.Ujvari A, Luse DS. Newly Initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J Biol Chem. 2004;279:49773–49779. doi: 10.1074/jbc.M409087200. [DOI] [PubMed] [Google Scholar]
- 62.Doyle O, Corden JL, Murphy C, Gall JG. The distribution of RNA polymerase II largest subunit (RPB1) in the Xenopus germinal vesicle. J Struct Biol. 2002;140:154–166. doi: 10.1016/s1047-8477(02)00547-6. [DOI] [PubMed] [Google Scholar]
- 63.Chathoth KT, Barrass JD, Webb S, Beggs JD. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol Cell. 2014;53:779–790. doi: 10.1016/j.molcel.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davidson L, Kerr A, West S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 2012;31:2566–2578. doi: 10.1038/emboj.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fong Y, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–932. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 66.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bres V, Gomes N, Pickle L, Jones KA. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 2005;19:1211–1226. doi: 10.1101/gad.1291705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 2014;53:819–830. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 69.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 70.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaykalova DA, Kulaeva OI, Volokh O, Shaytan AK, Hsieh FK, Kirpichnikov MP, Sokolova OS, Studitsky VM. Structural analysis of nucleosomal barrier to transcription. Proc Natl Acad Sci U S A. 2015;112:E5787–E5795. doi: 10.1073/pnas.1508371112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luse DS, Spangler LC, Ujvari A. Efficient and rapid nucleosome traversal by RNA polymerase II depends on a combination of transcript elongation factors. J Biol Chem. 2011;286:6040–6048. doi: 10.1074/jbc.M110.174722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3'-5' direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 74.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 75.Ishibashi T, Dangkulwanich M, Coello Y, Lionberger TA, Lubkowska L, Ponticelli AS, Kashlev M, Bustamante C. Transcription factors IIS and IIF enhance transcription efficiency by differentially modifying RNA polymerase pausing dynamics. Proc Natl Acad Sci U S A. 2014;111:3419–3424. doi: 10.1073/pnas.1401611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schweikhard V, Meng C, Murakami K, Kaplan CD, Kornberg RD, Block SM. Transcription factors TFIIF and TFIIS promote transcript elongation by RNA polymerase II by synergistic and independent mechanisms. Proc Natl Acad Sci U S A. 2014;111:6642–6647. doi: 10.1073/pnas.1405181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dolata J, Guo Y, Kolowerzo A, Smolinski D, Brzyzek G, Jarmolowski A, Swiezewski S. NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 2015;34:544–558. doi: 10.15252/embj.201489478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, Young RA, Price DH. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiao Y, Yang YH, Burckin TA, Shiue L, Hartzog GA, Segal MR. Analysis of a Splice Array Experiment Elucidates Roles of Chromatin Elongation Factor Spt4-5 in Splicing. PLoS Comput Biol. 2005;1:e39. doi: 10.1371/journal.pcbi.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 81.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 84.Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S, Lev-Maor G, Burstein D, Schwartz S, Postolsky B, Pupko T, Ast G. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep. 2012;1:543–556. doi: 10.1016/j.celrep.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 85.Huang H, Yu S, Liu H, Sun X. Nucleosome organization in sequences of alternative events in human genome. Biosystems. 2012;109:214–219. doi: 10.1016/j.biosystems.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 86.Teves SS, Weber CM, Henikoff S. Transcribing through the nucleosome. Trends Biochem Sci. 2014;39:577–586. doi: 10.1016/j.tibs.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 87.Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 88.Subtil-Rodriguez A, Reyes JC. BRG1 helps RNA polymerase II to overcome a nucleosomal barrier during elongation, in vivo. EMBO Rep. 2010;11:751–757. doi: 10.1038/embor.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bintu L, Ishibashi T, Dangkulwanich M, Wu YY, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal elements that control the topography of the barrier to transcription. Cell. 2012;151:738–749. doi: 10.1016/j.cell.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kulaeva OI, Hsieh FK, Chang HW, Luse DS, Studitsky VM. Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim Biophys Acta. 2013;1829:76–83. doi: 10.1016/j.bbagrm.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bintu L, Kopaczynska M, Hodges C, Lubkowska L, Kashlev M, Bustamante C. The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat Struct Mol Biol. 2011;18:1394–1399. doi: 10.1038/nsmb.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Kramer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dhami P, Saffrey P, Bruce AW, Dillon SC, Chiang K, Bonhoure N, Koch CM, Bye J, James K, Foad NS, Ellis P, Watkins NA, Ouwehand WH, Langford C, Andrews RM, Dunham I, Vetrie D. Complex exon-intron marking by histone modifications is not determined solely by nucleosome distribution. PLoS One. 2010;5:e12339. doi: 10.1371/journal.pone.0012339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 2014;24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilhelm BT, Marguerat S, Aligianni S, Codlin S, Watt S, Bahler J. Differential patterns of intronic and exonic DNA regions with respect to RNA polymerase II occupancy, nucleosome density and H3K36me3 marking in fission yeast. Genome Biol. 2011;12:R82. doi: 10.1186/gb-2011-12-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Iannone C, Pohl A, Papasaikas P, Soronellas D, Vicent GP, Beato M, ValcaRcel J. Relationship between nucleosome positioning and progesterone-induced alternative splicing in breast cancer cells. RNA. 2015;21:360–374. doi: 10.1261/rna.048843.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jimeno-Gonzalez S, Payan-Bravo L, Munoz-Cabello AM, Guijo M, Gutierrez G, Prado F, Reyes JC. Defective histone supply causes changes in RNA polymerase II elongation rate and cotranscriptional pre-mRNA splicing. Proc Natl Acad Sci U S A. 2015;112:14840–14845. doi: 10.1073/pnas.1506760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 99.Hsieh F-K, Kulaeva OI, Patel SS, Dyer PN, Luger K, Reinberg D, Studitsky VM. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc Natl Acad Sci U S A. 2013;110:7654–7659. doi: 10.1073/pnas.1222198110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009;28:1067–1077. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014;28:672–682. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 103.Santisteban MS, Hang M, Smith MM. Histone variant H2A.Z and RNA polymerase II transcription elongation. Mol Cell Biol. 2011;31:1848–1860. doi: 10.1128/MCB.01346-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fujimoto S, Seebart C, Guastafierro T, Prenni J, Caiafa P, Zlatanova J. Proteome analysis of protein partners to nucleosomes containing canonical H2A or the variant histones H2A.Z or H2A.X. Biol Chem. 2012;393:47–61. doi: 10.1515/BC-2011-216. [DOI] [PubMed] [Google Scholar]
- 105.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 106.Skene PJ, Hernandez AE, Groudine M, Henikoff S. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. Elife. 2014;3:e02042. doi: 10.7554/eLife.02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hnilicova J, Hozeifi S, Duskova E, Icha J, Tomankova T, Stanek D. Histone deacetylase activity modulates alternative splicing. PLoS One. 2011;6:e16727. doi: 10.1371/journal.pone.0016727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu S, Waldholm J, Bohm S, Visa N. Brahma regulates a specific trans-splicing event at the mod(mdg4) locus of Drosophila melanogaster. RNA Biol. 2014;11:134–145. doi: 10.4161/rna.27866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tai HH, Geisterfer M, Bell JC, Moniwa M, Davie JR, Boucher L, McBurney MW. CHD1 associates with NCoR and histone deacetylase as well as with RNA splicing proteins. Biochem Biophys Res Commun. 2003;308:170–176. doi: 10.1016/s0006-291x(03)01354-8. [DOI] [PubMed] [Google Scholar]
- 110.Patrick KL, Ryan CJ, Xu J, Lipp JJ, Nissen KE, Roguev A, Shales M, Krogan NJ, Guthrie C. Genetic interaction mapping reveals a role for the SWI/SNF nucleosome remodeler in spliceosome activation in fission yeast. PLoS Genet. 2015;11:e1005074. doi: 10.1371/journal.pgen.1005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tyagi A, Ryme J, Brodin D, Ostlund Farrants AK, Visa N. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet. 2009;5:e1000470. doi: 10.1371/journal.pgen.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Huff JT, Plocik AM, Guthrie C, Yamamoto KR. Reciprocal intronic and exonic histone modification regions in humans. Nat Struct Mol Biol. 2010;17:1495–1499. doi: 10.1038/nsmb.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou HL, Luo G, Wise JA, Lou H. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res. 2014;42:701–713. doi: 10.1093/nar/gkt875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iannone C, Valcarcel J. Chromatin's thread to alternative splicing regulation. Chromosoma. 2013;122:465–474. doi: 10.1007/s00412-013-0425-x. [DOI] [PubMed] [Google Scholar]
- 116.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M, Ho TH, Kuan PF, Jonasch E, Furey TS, Prins JF, Lieb JD, Rathmell WK, Davis IJ. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 2014;24:241–250. doi: 10.1101/gr.158253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci U S A. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, Andrau JC, Ferrier P, Carmo-Fonseca M. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 121.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 126.Kraushaar DC, Jin W, Maunakea A, Abraham B, Ha M, Zhao K. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 2013;14:R121. doi: 10.1186/gb-2013-14-10-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fuchs G, Hollander D, Voichek Y, Ast G, Oren M. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res. 2014;24:1572–1583. doi: 10.1101/gr.176487.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B Monoubiquitination Functions Cooperatively with FACT to Regulate Elongation by RNA Polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 129.Shema-Yaacoby E, Nikolov M, Haj-Yahya M, Siman P, Allemand E, Yamaguchi Y, Muchardt C, Urlaub H, Brik A, Oren M, Fischle W. Systematic identification of proteins binding to chromatin-embedded ubiquitylated H2B reveals recruitment of SWI/SNF to regulate transcription. Cell Rep. 2013;4:601–608. doi: 10.1016/j.celrep.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 130.Zhang Z, Jones A, Joo HY, Zhou D, Cao Y, Chen S, Erdjument-Bromage H, Renfrow M, He H, Tempst P, Townes TM, Giles KE, Ma L, Wang H. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev. 2013;27:1581–1595. doi: 10.1101/gad.211037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sharma A, Nguyen H, Geng C, Hinman MN, Luo G, Lou H. Calcium-mediated histone modifications regulate alternative splicing in cardiomyocytes. Proc Natl Acad Sci U S A. 2014;111:E4920–E4928. doi: 10.1073/pnas.1408964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou HL, Hinman MN, Barron VA, Geng C, Zhou G, Luo G, Siegel RE, Lou H. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci U S A. 2011;108:E627–E635. doi: 10.1073/pnas.1103344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sjolinder M, Bjork P, Soderberg E, Sabri N, Farrants AK, Visa N. The growing pre-mRNA recruits actin and chromatin-modifying factors to transcriptionally active genes. Genes Dev. 2005;19:1871–1884. doi: 10.1101/gad.339405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Khan DH, Gonzalez C, Cooper C, Sun JM, Chen HY, Healy S, Xu W, Smith KT, Workman JL, Leygue E, Davie JR. RNA-dependent dynamic histone acetylation regulates MCL1 alternative splicing. Nucleic Acids Res. 2014;42:1656–1670. doi: 10.1093/nar/gkt1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, Klinck R, Chabot B, Kornblihtt AR. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 136.Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, Mathieu J, Hamiche A, Ait-Si-Ali S, Muchardt C, Batsche E, Harel-Bellan A. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 137.Taliaferro JM, Aspden JL, Bradley T, Marwha D, Blanchette M, Rio DC. Two new and distinct roles for Drosophila Argonaute-2 in the nucleus: alternative pre-mRNA splicing and transcriptional repression. Genes Dev. 2013;27:378–389. doi: 10.1101/gad.210708.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Allo M, Agirre E, Bessonov S, Bertucci P, Gomez Acuna L, Buggiano V, Bellora N, Singh B, Petrillo E, Blaustein M, Minana B, Dujardin G, Pozzi B, Pelisch F, Bechara E, Agafonov DE, Srebrow A, Luhrmann R, Valcarcel J, Eyras E, Kornblihtt AR. Argonaute-1 binds transcriptional enhancers and controls constitutive and alternative splicing in human cells. Proc Natl Acad Sci U S A. 2014;111:15622–15629. doi: 10.1073/pnas.1416858111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gonzalez I, Munita R, Agirre E, Dittmer TA, Gysling K, Misteli T, Luco RF. A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol. 2015;22:370–376. doi: 10.1038/nsmb.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yearim A, Gelfman S, Shayevitch R, Melcer S, Glaich O, Mallm JP, Nissim-Rafinia M, Cohen AH, Rippe K, Meshorer E, Ast G. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015;10:1122–1134. doi: 10.1016/j.celrep.2015.01.038. [DOI] [PubMed] [Google Scholar]
- 142.Smallwood A, Hon GC, Jin F, Henry RE, Espinosa JM, Ren B. CBX3 regulates efficient RNA processing genome-wide. Genome Res. 2012;22:1426–1436. doi: 10.1101/gr.124818.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Saint-Andre V, Batsche E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol. 2011;18:337–344. doi: 10.1038/nsmb.1995. [DOI] [PubMed] [Google Scholar]
- 144.Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Marina RJ, Sturgill D, Bailly MA, Thenoz M, Varma G, Prigge MF, Nanan KK, Shukla S, Haque N, Oberdoerffer S. TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing. EMBO J. 2015 doi: 10.15252/embj.201593235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Butcher SE, Pyle AM. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc Chem Res. 2011;44:1302–1311. doi: 10.1021/ar200098t. [DOI] [PubMed] [Google Scholar]
- 149.Wachter A. Gene regulation by structured mRNA elements. Trends Genet. 2014;30:172–181. doi: 10.1016/j.tig.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 150.Goguel V, Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993;72:893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- 151.Eperon LP, Graham IR, Griffiths AD, Eperon IC. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 152.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Meyer M, Plass M, Perez-Valle J, Eyras E, Vilardell J. Deciphering 3'ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol Cell. 2011;43:1033–1039. doi: 10.1016/j.molcel.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 154.Olson S, Blanchette M, Park J, Savva Y, Yeo GW, Yeakley JM, Rio DC, Graveley BR. A regulator of Dscam mutually exclusive splicing fidelity. Nat Struct Mol Biol. 2007;14:1134–1140. doi: 10.1038/nsmb1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Solomon O, Oren S, Safran M, Deshet-Unger N, Akiva P, Jacob-Hirsch J, Cesarkas K, Kabesa R, Amariglio N, Unger R, Rechavi G, Eyal E. Global regulation of alternative splicing by adenosine deaminase acting on RNA (ADAR) RNA. 2013;19:591–604. doi: 10.1261/rna.038042.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang Q, Zhang Z, Blackwell K, Carmichael GG. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr Biol. 2005;15:384–391. doi: 10.1016/j.cub.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 157.Zhou J, Wang Q, Chen LL, Carmichael GG. On the mechanism of induction of heterochromatin by the RNA-binding protein vigilin. RNA. 2008;14:1773–1781. doi: 10.1261/rna.1036308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Incarnato D, Neri F, Anselmi F, Oliviero S. Genome-wide profiling of mouse RNA secondary structures reveals key features of the mammalian transcriptome. Genome Biol. 2014;15:491. doi: 10.1186/s13059-014-0491-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 160.Wan Y, Qu K, Zhang QC, Flynn RA, Manor O, Ouyang Z, Zhang J, Spitale RC, Snyder MP, Segal E, Chang HY. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Heilman-Miller SL, Woodson SA. Effect of transcription on folding of the Tetrahymena ribozyme. RNA. 2003;9:722–733. doi: 10.1261/rna.5200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lewicki BT, Margus T, Remme J, Nierhaus KH. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J Mol Biol. 1993;231:581–593. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 163.Repsilber D, Wiese S, Rachen M, Schroder AW, Riesner D, Steger G. Formation of metastable RNA structures by sequential folding during transcription: time-resolved structural analysis of potato spindle tuber viroid (−)-stranded RNA by temperature-gradient gel electrophoresis. RNA. 1999;5:574–584. doi: 10.1017/s1355838299982018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zamft B, Bintu L, Ishibashi T, Bustamante C. Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc Natl Acad Sci U S A. 2012;109:8948–8953. doi: 10.1073/pnas.1205063109. [DOI] [PMC free article] [PubMed] [Google Scholar]