Abstract
Technology advances in genome-wide association studies (GWAS) has engendered optimism that we have entered a new age of precision medicine, in which the risk of breast cancer can be predicted on the basis of a person’s genetic variants. The goal of this study is to evaluate the discriminatory power of common genetic variants in breast cancer risk estimation. We conducted a retrospective case-control study drawing from an existing personalized medicine data repository. We collected variables that predict breast cancer risk: 153 high-frequency/low-penetrance genetic variants, reflecting the state-of-the-art GWAS on breast cancer, mammography descriptors and BI-RADS assessment categories in the Breast Imaging Reporting and Data System (BI-RADS) lexicon. We trained and tested naïve Bayes models by using these predictive variables. We generated ROC curves and used the area under the ROC curve (AUC) to quantify predictive performance. We found that genetic variants achieved comparable predictive performance to BI-RADS assessment categories in terms of AUC (0.650 vs. 0.659, p-value = 0.742), but significantly lower predictive performance than the combination of BI-RADS assessment categories and mammography descriptors (0.650 vs. 0.751, p-value < 0.001). A better understanding of relative predictive capability of genetic variants and mammography data may benefit clinicians and patients to make appropriate decisions about breast cancer screening, prevention, and treatment in the era of precision medicine.
Keywords: precision medicine, single-nucleotide polymorphisms, mammography, breast cancer
1. INTRODUCTION
Breast cancer is the most common non-skin malignancy affecting women. Stratification of women according to the risk of developing breast cancer could improve risk reduction and screening strategies by targeting those most likely to benefit. Technology advances in genome-wide association studies (GWAS) has engendered optimism that we have entered a new age of precision medicine, in which the risk of breast cancer can be predicted on the basis of a person’s genetic variants. However, early attempts to use a set of common genetic variants to predict breast cancer risk demonstrate only modest improvements over conventional demographic risk factors 1-3.
One of the most important questions of how much additional predictive power can be achieved by using more genetic variants remain uncertain. In our prior studies, we quantified predictive capability of 22 single-nucleotide polymorphisms (SNPs) in breast cancer risk estimation 4, 5. Recently, we consolidated a list of 77 SNPs and found that they demonstrated a significantly higher predictive performance than those 22 SNPs 6. With the rapid progress of genome-wide association studies, more and more new SNPs associated with breast cancer have been identified7, which has engendered the potential to improve predictive capability further by using a larger set of SNPs.
Theoretically the ability of SNPs to predict breast cancer risk has an upper bound 8, 9. However, practically the number of SNPs used to reach an upper bound of predictive capability is still unknown 10. Moreover, the implications of integrating SNPs into clinical practice along with other conventional diagnostic tests remain uncertain. In clinical practice, mammography is the most common breast cancer screening test, and the preeminent imaging modality supported by randomized trials demonstrating mortality reduction. A better understanding of relative predictive capability of SNPs in the context of mammography may help clinician and patients to make appropriate decisions of breast cancer screening, prevention, and treatment.
In this study, we assemble a list of breast cancer SNPs identified to date, which reflect the state-of-the-art breast cancer GWAS. We aim to evaluate the discriminatory power of genetic variants in personalized breast cancer diagnosis, using an existing personalized medicine data repository. We aim to reveal the relative predictive capability of genetic variants and mammography data in breast cancer risk estimation.
2. MATERIALS and METHODS
The Marshfield Clinic Institutional Review Board approved the use of Marshfield Clinic’s Personalized Medicine Research Project (PMRP) 11 cohort in the study.
2.1 Subjects
The subjects in this study were from the population-based PMRP cohort, details of which have been previously published 11. Western European women with an available plasma sample, a mammogram, and a breast biopsy within 12 months after the mammogram were included in the study. We decided to focus on high-frequency/low-penetrance SNPs that affect breast cancer risk as opposed to low frequency SNPs with high penetrance or intermediate penetrance. We excluded individuals who had a known high-penetrance genetic mutation. For this case/control study, Cases were defined as women having a confirmed diagnosis of breast cancer obtained from the institutional cancer registry. Controls were confirmed through the Marshfield Clinic electronic medical records as never having had a breast cancer diagnosis. Moreover, we selected a control whose age was within five years of the age of each case to make sure that case and control groups were similar in age distribution.
2.2 Genetic Variants
We consolidated a list of 153 common genetic variants which were identified by the recent large-scale GWAS studies or used to generate published predictive models (Table 1). The list included 77 SNPs used in our recent study to quantify predictive capability of genetic variants 6, in which 41 were identified by Collaborative Oncological Gene-environment Study (COGS) through a meta-analysis of 9 GWAS studies 12. The list also included some SNPs garnered from several other recent studies related to COGS 13-24. To the best of our knowledge, the list of 153 genetic variants provided the most comprehensive summary of SNPs identified in the major GWAS for breast cancer risk up to 2015.
Table 1.
The 153 SNPs identified to be associated to breast cancer.
| SNP | Chromosome | SNP | Chromosome | SNP | Chromosome | SNP | Chromosome |
|---|---|---|---|---|---|---|---|
| rs616488 | 1 | rs1017226 | 5 | rs10965163 | 9 | rs12422552 | 12 |
| rs11552449 | 1 | rs12655019 | 5 | rs865686 | 9 | rs6220 | 12 |
| rs11249433 | 1 | rs16886034 | 5 | rs1011970 | 9 | rs10771399 | 12 |
| rs2290854 | 1 | rs16886181 | 5 | rs7072776 | 10 | rs1292011 | 12 |
| rs4245739 | 1 | rs16886364 | 5 | rs7904519 | 10 | rs27633 | 12 |
| rs6678914 | 1 | rs16886397 | 5 | rs2981582 | 10 | rs17356907 | 12 |
| rs6682208 | 1 | rs16886448 | 5 | rs10995190 | 10 | rs11571833 | 13 |
| rs1550623 | 2 | rs2229882 | 5 | rs2380205 | 10 | rs2588809 | 14 |
| rs16857609 | 2 | rs2736108 | 5 | rs2981579 | 10 | rs941764 | 14 |
| rs2016394 | 2 | rs3822625 | 5 | rs704010 | 10 | rs999737 | 14 |
| rs4849887 | 2 | rs7726159 | 5 | rs11196174 | 10 | rs2236007 | 14 |
| rs1045485 | 2 | rs7726354 | 5 | rs1219648 | 10 | rs17817449 | 16 |
| rs13387042 | 2 | rs7716600 | 5 | rs16917302 | 10 | rs3803662 | 16 |
| rs17468277 | 2 | rs204247 | 6 | rs2420946 | 10 | rs12443621 | 16 |
| rs4666451 | 2 | rs2046210 | 6 | rs1243182 | 10 | rs8051542 | 16 |
| rs12710696 | 2 | rs2180341 | 6 | rs17221319 | 10 | rs4784227 | 16 |
| rs184577 | 2 | rs17530068 | 6 | rs17550038 | 10 | rs11075995 | 16 |
| rs1830298 | 2 | rs3757318 | 6 | rs2981575 | 10 | rs13329835 | 16 |
| rs2070959 | 2 | rs2253407 | 6 | rs2981578 | 10 | rs2075555 | 17 |
| rs36043647 | 2 | rs6569479 | 6 | rs45631563 | 10 | rs6504950 | 17 |
| rs4458204 | 2 | rs9348512 | 6 | rs11199914 | 10 | rs527616 | 18 |
| rs59278883 | 2 | rs9383938 | 6 | rs11814448 | 10 | rs1436904 | 18 |
| rs6759892 | 2 | rs9485372 | 6 | rs3903072 | 11 | rs4808801 | 19 |
| rs7558475 | 2 | rs12197388 | 6 | rs3817198 | 11 | rs8170 | 19 |
| rs12493607 | 3 | rs12662670 | 6 | rs2107425 | 11 | rs3745274 | 19 |
| rs6762644 | 3 | rs17529111 | 6 | rs614367 | 11 | rs2279343 | 19 |
| rs4973768 | 3 | rs9397435 | 6 | rs909116 | 11 | rs2363956 | 19 |
| rs6828523 | 4 | rs11242675 | 6 | rs12575120 | 11 | rs3760982 | 19 |
| rs9790517 | 4 | rs10235235 | 7 | rs494406 | 11 | rs2284378 | 20 |
| rs10472076 | 5 | rs720475 | 7 | rs537626 | 11 | rs13039229 | 20 |
| rs1353747 | 5 | rs2943559 | 8 | rs554219 | 11 | rs311499 | 20 |
| rs1432679 | 5 | rs6472903 | 8 | rs585568 | 11 | rs311498 | 20 |
| rs10941679 | 5 | rs9693444 | 8 | rs593679 | 11 | rs2823093 | 21 |
| rs889312 | 5 | rs13281615 | 8 | rs657686 | 11 | rs10483028 | 21 |
| rs30099 | 5 | rs1562430 | 8 | rs679162 | 11 | rs2242714 | 21 |
| rs981782 | 5 | rs4733664 | 8 | rs75915166 | 11 | rs6001930 | 22 |
| rs10069690 | 5 | rs799890 | 8 | rs78540526 | 11 | rs132390 | 22 |
| rs16886113 | 5 | rs11780156 | 8 | rs11820646 | 11 | ||
| rs4415084 | 5 | rs10759243 | 9 |
2.3 Mammography Features
The American College of Radiology developed the Breast Imaging Reporting and Data System (BI-RADS) lexicon 25 for mammography reporting. The BI-RADS lexicon consists of 49 descriptors 4, including the characteristics of masses and microcalcifications, special cases, associated findings, and breast composition. In this study, mammography data was recorded as free text reports in the electronic health record, from which we used a parser to extract these mammography features 26. After extraction, each mammography feature took the value “present” or “not present” except that the variable mass size was discretized into three values, “not present”, “small” and “large”, depending on whether there was a reported mass size and whether any dimension was larger than 30mm. In clinical practice, radiologists assign a BI-RADS assessment category to each mammogram, which indicates the radiologist’s assessment of the risk of breast cancer. In our study, the BI-RADS category prioritized values in the order of increasing probability of malignancy, 1, 2, 3, 0, 4a, 4, 4b, 4c and 5.
2.4 Study Design and Statistical Analysis
We built three breast cancer risk predictive models using Naïve Bayes implementation in WEKA 27.We developed a SNP153 model built on 153 SNPs. In this genetic model, we introduced one variable to represent the total count of risk alleles the person carries for those 153 SNPs in the DNA. This way of coding genetic variants was used in several models 3, 6, and is helpful to build risk models when each SNP only has a small contribution to the risk. To compare predictive power of SNPs with that of mammography, we developed a BI-RADS Category model (BCM) built on BI-RADS assessment categories only. We developed a BI-RADS Category and Descriptor model (BCDM) built on the combination of BI-RADS assessment categories and 49 mammography features. We generated receiver operator characteristic (ROC) curves using ROCKIT software 28, 29 based on the probabilities of malignancy predicted by each of the three models, and used the area under the curve (AUC) as a measure of performance. We compared predictive capability of the models using DeLong method 30, and evaluated the models using 10-fold cross-validation.
3. RESULTS
We identified 362 cases and 376 controls, details of which have been previously described 6. The age range for the subjects in this study was 29 to 90 years of age, with mean=62 and standard deviation=12.8. Among the cases, there were 358 Caucasians, three non-Caucasians and one case whose race information was unknown. Among the controls, there were 372 Caucasians and four non-Caucasians.
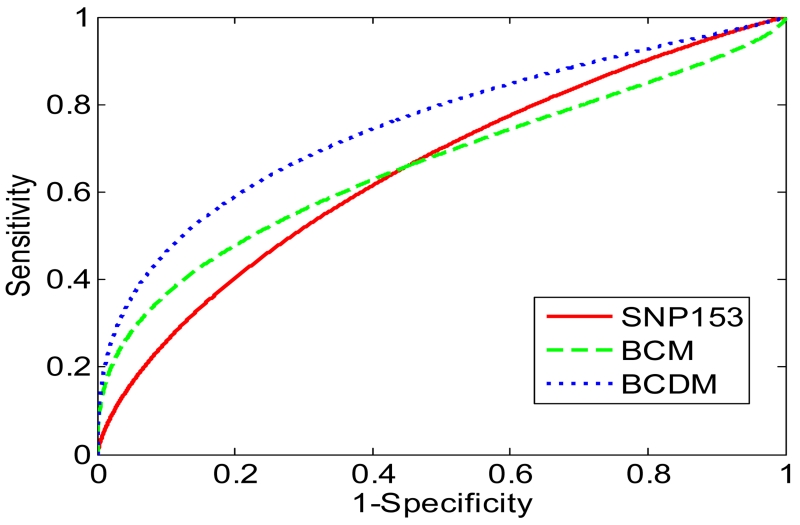
We observed that the SNP153 model can achieve comparable predictive performance to the BCM (Figure 1). The AUC of the SNP153 model was 0.650 and the AUC of the BCM was 0.659, with p-value = 0.742. We also observed that the SNP153 model demonstrated significantly lower predictive performance than the BCDM in terms of AUC (0.650 vs. 0.751, p-value < 0.001).
Figure 1.
ROC curves for different predictive models. Solid curve, the SNP153 model; dashed curve, the BI-RADS Category model (BCM); dotted curve, BI-RADS Category and Descriptor model (BCDM).
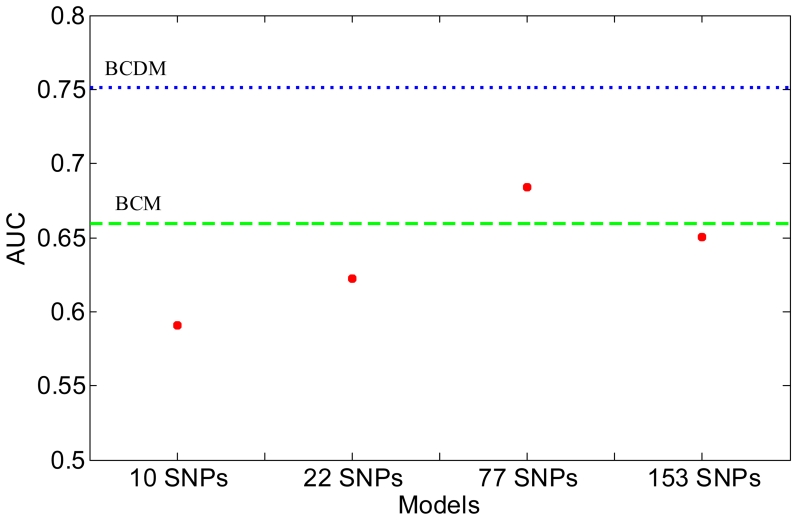
Our prior study quantified predictive performance of genetic variants 6. The AUCs for the models developed with 10, 22 and 77 SNPs were 0.591, 0.622 and 0.684, which indicated that the more associated SNPs the prediction model includes, the more discriminative the model becomes. The 10 SNPs identified at the early stage of GWAS show strong associations with breast cancer risk 3, 31, which have been validated by several large-scale GWAS 32, 33. The 22 SNPs reflects the breast cancer GWAS up to 2010 and the 77 SNPs demonstrate the progress of breast cancer GWAS up to 2013. In this study, we found that the AUC of the SNP153 model was 0.650, which was less than that of the model developed with 77 SNPs (Figure 2).
Figure 2.
Comparison of AUC for predictive models developed with different number of SNPs. Dashed line, the BI-RADS Category model (BCM); dotted line, BI-RADS Category and Descriptor model (BCDM).
4. DISCUSSION
This study demonstrates that the genetic variants can improve breast cancer risk prediction substantially but an upper bound of discriminatory power exists. We predict that some novel SNPs could be identified in the near future but their contribution to breast cancer risk estimation would likely be modest. In addition, we observe that genetic variants demonstrate significantly lower predictive performance than mammography features in terms of AUC for women undergoing breast biopsy.
For the first time, our study empirically demonstrates that prediction models developed with common genetic variants achieve a potential upper bound of predictive power from an existing personalized medicine data repository. The more associated SNPs the prediction model includes, the more discriminative the model becomes. The AUCs for the models developed with 10, 22 and 77 SNPs were 0.591, 0.622 and 0.684. However, the AUC of the SNP153 model was 0.650, which was less than that of the model developed with 77 SNPs. The upper bound of predictive performance for genetic variants can be achieved using SNPs selected from the set of 153 SNPs. Some prior studies recommended the number of SNPs for breast cancer risk prediction but those numbers are only illustrative 1, 34. Of note, we quantified predictive performance of a series of SNPs according to the progress of breast cancer GWAS; SNPs identified at the early stage of GWAS show strong associations with breast cancer risk while those discovered at the later stage were less likely to reach statistical significance 9. A possible line of future study is to seek the highest predictive performance of those 153 SNPs by using ranking algorithms such as mutual information analysis 35, 36, and constructing predictive models with ranked SNPs sequentially.
Genetic variants provide a lower predictive power than mammographic findings but they may still play an important role in risk stratification and breast cancer diagnosis. As one kind of so-called intermediate phenotypes, mammographic findings may both summarize breast cancer risk more powerfully and capture the interaction of genes and the environment 37, giving rise to sound performance in breast cancer diagnosis. Genetic variants could be used to augment diagnostic performance of mammography interpretation, as demonstrated in a series of prior studies 4-6. In summary, even though an upper bound of discriminatory power exists and a lower predictive power occurs for SNPs in breast cancer risk estimation, identification of common genetic variants may eventually allow improving breast cancer risk prediction and stratifying women according to their breast cancer risk.
There are several limitations to our study. The sample size is small compared with large-scale GWAS studies, due to the inherent difficulty of collecting a rich multi-modality dataset. Moreover, we do not explicitly model how individual SNPs function to alter breast cancer risk. Our current genetics model uses one feature to represent the total number of risk alleles for those 153 SNPs in the DNA, assuming that each individual SNP only confers a fairly mild relative risk and the genetic effect of the genetic variants is additive. Furthermore, we do not differentiate the different subtypes of breast cancers (for example, the estrogen-receptor status and progesterone-receptor status) in the current study. Breast cancer is a complex and heterogeneous disease with different subtypes, including two main subtypes of estrogen receptor (ER) negative tumors (basal-like and human epidermal growth factor receptor-2 positive/ER− subtype) and at least two types of ER positive tumors (luminal A and luminal B) 38. These molecular subtypes are important predictors of breast cancer mortality and have different genetic susceptibility. We plan to extend our study by quantifying predictive power of SNPs for different subsets of breast cancer. Finally, SNP associations may be specific to subsets of women with breast cancer, as defined by ethnicity 9. Our results cannot be generalized beyond western European populations.
5. CONCLUSION
We consolidate a list of the latest identified SNPs, which reflects the state of the art of breast cancer GWAS study and COGS analysis. For the first time, our study empirically demonstrates that prediction models developed with common genetic variants achieve a potential upper bound of predictive power from an existing personalized medicine data repository. Even though the upper bound exists for SNPs in breast cancer risk estimation, identification of common genetic variants may eventually allow understanding molecular mechanisms of breast cancer and stratifying women according to breast cancer risk, with a hope of improving breast cancer screening, prevention, and treatment strategies.
ACKNOWLEDGEMENTS
The authors acknowledge the support of the Wisconsin Genomics Initiative, NCI grant R01CA127379-01 and its ARRA supplement 3R01CA127379-03S1, NIGMS grant R01GM097618-01, NLM grant R01LM011028-01, NIEHS grant 5R01ES017400-03, NIH grant 1U54AI117924-01, the UW Institute for Clinical and Translational Research (ICTR) and the UW Carbone Cancer Center.
REFERENCES
- [1].Gail MH. Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk. J Natl Cancer Inst. 2008;100(14):1037–41. doi: 10.1093/jnci/djn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gail MH. Value of adding single-nucleotide polymorphism genotypes to a breast cancer risk model. J Natl Cancer Inst. 2009;101(13):959–63. doi: 10.1093/jnci/djp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wacholder S, Hartge P, Prentice R, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 2010;362(11):986–93. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Liu J, Page D, Nassif H, et al. Genetic variants improve breast cancer risk prediction on mammograms; American Medical Informatics Association Symposium (AMIA); 2013; [PMC free article] [PubMed] [Google Scholar]
- [5].Wu Y, Liu J, Page D, et al. Comparing the value of mammographic features and genetic variants in breast cancer risk prediction; American Medical Informatics Association Symposium (AMIA); 2014; [PMC free article] [PubMed] [Google Scholar]
- [6].Liu J, Page D, Peissig P, et al. New genetic variants improve personalized breast cancer diagnosis; AMIA Summit on Translational Bioinformatics (AMIA-TBI); 2014; [PMC free article] [PubMed] [Google Scholar]
- [7].Bahcall OG. iCOGS collection provides a collaborative model. Nat Genet. 2013;45(4):343. doi: 10.1038/ng.2592. [DOI] [PubMed] [Google Scholar]
- [8].Couch FJ, Nathanson KL, Offit K. Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science. 2014;343(6178):1466. doi: 10.1126/science.1251827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Maxwell K, Nathanson K. Common breast cancer risk variants in the post-COGS era: a comprehensive review. Breast Cancer Res. 2013;15(6):212. doi: 10.1186/bcr3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Park J-H, Wacholder S, Gail MH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42(7):570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].McCarty CA, Wilke RA, Giampietro PF, et al. Marshfield Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Personalized Med. 2005;2(1):49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- [12].Michailidou K, Hall P, Gonzalez-Neira A, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–61. 361e1–2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ahsan H, Halpern J, Kibriya MG, et al. A genome-wide association study of early-onset breast cancer identifies PFKM as a novel breast cancer gene and supports a common genetic spectrum for breast cancer at any age. Cancer Epidemiol Biomarkers Prev. 2014;23(4):658–69. doi: 10.1158/1055-9965.EPI-13-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].French JD, Ghoussaini M, Edwards SL, et al. Functional variants at the 11q13 risk locus for breast cancer regulate cyclin D1 expression through long-range enhancers. Am J Hum Genet. 2013;92(4):489–503. doi: 10.1016/j.ajhg.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gaudet MM, Kuchenbaecker KB, Vijai J, et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9(3):e1003173. doi: 10.1371/journal.pgen.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Johnson N, Dudbridge F, Orr N, et al. Genetic variation at CYP3A is associated with age at menarche and breast cancer risk: a case-control study. Breast Cancer Res. 2014;16(3):R51. doi: 10.1186/bcr3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Justenhoven C, Obazee O, Winter S, et al. The UGT1A6_19_GG genotype is a breast cancer risk factor. Front Genet. 2013;4:104. doi: 10.3389/fgene.2013.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Justenhoven C, Pentimalli D, Rabstein S, et al. CYP2B6*6 is associated with increased breast cancer risk. Int J Cancer. 2014;134(2):426–30. doi: 10.1002/ijc.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li J, Lindstrom LS, Foo JN, et al. 2q36.3 is associated with prognosis for oestrogen receptor-negative breast cancer patients treated with chemotherapy. Nat Commun. 2014;5:4051. doi: 10.1038/ncomms5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Meyer KB, O’Reilly M, Michailidou K, et al. Fine-scale mapping of the FGFR2 breast cancer risk locus: putative functional variants differentially bind FOXA1 and E2F1. Am J Hum Genet. 2013;93(6):1046–60. doi: 10.1016/j.ajhg.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Purrington KS, Slettedahl S, Bolla MK, et al. Genetic variation in mitotic regulatory pathway genes is associated with breast tumor grade. Hum Mol Genet. 2014;23(22):6034–46. doi: 10.1093/hmg/ddu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rudolph A, Milne RL, Truong T, et al. Investigation of gene-environment interactions between 47 newly identified breast cancer susceptibility loci and environmental risk factors. Int J Cancer. 2015;136(6):E685–95. doi: 10.1002/ijc.29188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sawyer E, Roylance R, Petridis C, et al. Genetic predisposition to in situ and invasive lobular carcinoma of the breast. PLoS Genet. 2014;10(4):e1004285. doi: 10.1371/journal.pgen.1004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schoeps A, Rudolph A, Seibold P, et al. Identification of new genetic susceptibility loci for breast cancer through consideration of gene-environment interactions. Genet Epidemiol. 2014;38(1):84–93. doi: 10.1002/gepi.21771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].American College of Radiology . Breast Imaging Reporting And Data System (BI-RADS®) Reston VA: 2003. [Google Scholar]
- [26].Nassif H, Woods R, Burnside E, et al. Information extraction for clinical data mining: a mammography case study; IEEE International Conference on Data Mining Workshops; 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hall M, Frank E, Holmes G, et al. The Weka data mining software: an update. SIGKDD Explorations. 2009;11(1) [Google Scholar]
- [28].Metz C. Basic principles of ROC analysis. Semin Nucl Med. 1978;8:283–298. doi: 10.1016/s0001-2998(78)80014-2. [DOI] [PubMed] [Google Scholar]
- [29].Metz C, Herman B, Shen J. Maximum likelihood estimation of receiver operating characteristic (ROC) curves from continuously-distributed data. Stat Med. 1998;17:1033–1053. doi: 10.1002/(sici)1097-0258(19980515)17:9<1033::aid-sim784>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- [30].DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–45. [PubMed] [Google Scholar]
- [31].Burnside ES, Liu J, Wu Y, et al. Comparing Mammography Abnormality Features to Genetic Variants in the Prediction of Breast Cancer in Women Recommended for Breast Biopsy. Acad Radiol. 2016;23(1):62–9. doi: 10.1016/j.acra.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39(7):870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kraft P, Hunter D. Genetic risk prediction--are we there yet? N Engl J Med. 2009;360(17):1701–3. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- [35].Wu Y, Alagoz O, Ayvaci MU, et al. A comprehensive methodology for determining the most informative mammographic features. J Digital Imaging. 2013;26(5):941–7. doi: 10.1007/s10278-013-9588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wu Y, Vanness DV, Burnside ES. Using multidimensional mutual information to prioritize mammographic features for breast cancer diagnosis; Ameican Medical Informatics Association Symposium (AMIA); 2013; [PMC free article] [PubMed] [Google Scholar]
- [37].Devilee P, Rookus MA. A tiny step closer to personalized risk prediction for breast cancer. N Engl J Med. 2010;362(11):1043–5. doi: 10.1056/NEJMe0912474. [DOI] [PubMed] [Google Scholar]
- [38].Perou C, Sørlie T, Eisen M, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]