Abstract
The KRAB-containing zinc finger (KRAB-ZF) proteins represent the largest family of transcription factors (TFs) in humans, yet for the great majority, their function and specific genomic target remain unknown. However, it has been shown that a large fraction of these genes arose from segmental duplications, and that they have expanded in gene and zinc finger number throughout vertebrate evolution. To determine whether this expansion is linked to selective pressures acting on different domains, we have manually curated all KRAB-ZF genes present in the human genome together with their orthologous genes in three closely related species and assessed the evolutionary forces acting at the sequence level as well as on their expression profiles. We provide evidence that KRAB-ZFs can be separated into two categories according to the polymorphism present in their DNA-contacting residues. Those carrying a nonsynonymous single nucleotide polymorphism (SNP) in their DNA-contacting amino acids exhibit significantly reduced expression in all tissues, have emerged in a recent lineage, and seem to be less strongly constrained evolutionarily than those without such a polymorphism. This work provides evidence for a link between age of the TF, as well as polymorphism in their DNA-contacting residues and expression levels—both of which may be jointly affected by selection.
Keywords: DNA-contacting residues, endogenous retroelements, KRAB-containing zinc finger genes, population genetics, regulatory evolution, transcription factors
Gene duplication can play a major role in species evolution: redundancy provides a medium for novelty while maintaining initial function. In the particular case of transcription factor (TF) genes, alterations in their expression profiles or binding properties can affect the expression of many target genes, often with a major functional impact. The KRAB-zinc finger (ZF) family of TFs, the largest family of TFs in the human genome, arose through tandem segmental duplications and contains arrays of C2H2 (also called Krüppel-type) ZFs combined with a KRAB (Krüppel-associated box) domain. Despite being so numerous, the function and specific genomic targets of the great majority of KRAB-containing ZF (KRAB-ZF) proteins remain unknown (Constantinou-Deltas et al. 1992; Huntley et al. 2006; Thomas and Emerson 2009).
KRAB-ZF regulatory specificity is determined by a ZF-DNA recognition code, implicating interaction between specific amino acids within the ZF motifs and nucleotides at the binding sites (Choo and Klug 1994; Kim and Berg 1996). The amino acids playing the most critical role in this DNA recognition are those at the -1, 2, 3, and 6 positions relative to the alpha-helical regions in each ZF domain (Pavletich and Pabo 1991; Elrod-Erickson et al. 1998). The strong conservation of some DNA-binding domains suggests that some genes have been stably integrated into essential regulatory relationships; however, in spite of this, little functional information from these genes is currently available (Liu et al. 2014).
In primates, KRAB-ZF genes duplicate at a higher rate than any other family. Paralogs diverge from the initial copy by a series of changes in the number and structure of ZF motifs, resulting in a dramatic diversity of binding specificities (Shannon et al. 2003; Hamilton et al. 2006). This DNA-binding diversity makes them ideal raw material for responding to newly emerging retrotransposons. Thomas and Schneider (2011) suggested that there is a continuous arms race between newly emerging retrotransposons and KRAB-ZFs acting as retrotransposon-specific repressors. Supporting this hypothesis, Jacobs et al. (2014) identified two KRAB-ZF genes involved in the repression of retrotransposons. They proposed a model where modifications to lineage-specific KRAB-ZFs result in repression of newly emerging families of retrotransposons, which in turn evolve to escape this repression. This evolutionary arms race may drive expansion and diversity of the KRAB-ZF genes and suggests a potential role for positive selection acting on affinity-modifying mutations in KRAB-ZFs. However, the extent to which positive selection has acted to shape this gene family is largely unknown.
One way to identify the relationships between sequence, function, and evolutionary process is to explore intraspecies (polymorphic) variation of functional elements—specifically, the relationship between observed polymorphism and measured function (Spivakov et al. 2012). Interestingly, Lockwood et al. (2014) assessed polymorphism in the ZF DNA-contacting amino acids and reported that the majority of missense SNPs in these DNA-contacting residues did not have any effect on fitness. This example suggests that relaxed selective constraint may potentially explain the diversity of binding amino acids of KRAB-ZFs.
The purpose of this study is to examine the underlying mechanisms behind the large expansion of the KRAB-ZF family in primates. By assessing the expression levels of KRAB-ZF genes in various tissues and taking into account polymorphism in the DNA-contacting amino acids, we link the sequence of the KRAB-ZFs with their underlying function. By manually curating all human KRAB-ZF genes and orthologous regions in three closely related species, and collecting polymorphism data from the 1000 Genomes Consortium, we were able to partition all human KRAB-ZF genes into two distinct categories according to the nature of SNPs occurring in the four DNA-contacting amino acids. Those two groups of genes differ significantly in their expression level for all tested tissues, the histone marks they bear in the gene body, and the time of emergence during primate evolution. This work thus represents a novel application of population genetic and transcriptomic data to an evolutionary study of a large family of TFs, resulting in insights that will allow future characterization of the regulatory role played by this family of genes.
Materials and Methods
Manual Curation of All Human KRAB-Containing ZF Genes
All human and mouse KRAB-ZF gene coordinates were obtained as described in Corsinotti et al. (2013). The resulting list was manually checked: from genes containing at least one ZF domain and one KRAB domain (based on PFAM annotation, http://pfam.xfam.org), the longest protein-coding transcript was selected (based on Ensembl release 71, http://www.ensembl.org), resulting in 346 human KRAB-ZF genes (Table S1). Genomic coordinates were downloaded from Ensembl for all genes as well as for all individual ZF and KRAB domains. The DNA sequences for the ZF domains were then translated into amino acid sequences using EMBOSS Transeq web-server (http://www.ebi.ac.uk/Tools/st/emboss_transeq/). As Ensembl annotation is automated, the start and end coordinates of the ZF domain may periodically be incorrect. We thus performed an extra check to ensure that the start and end of the well-characterized ZF domains correspond to the consensus sequence of a ZF (XX-C-XX-C-XXXXXXXXXXXX-H-XXX-H). If the protein sequence did not match the consensus sequence, we corrected the DNA coordinates in such a way that every ZF domain has the correct coordinate. Given that all further analyses depended on the accuracy of these datasets, annotation of the different domains was particularly rigorous.
In the ZF consensus sequence, positions -1, 2, 3, and 6 (marked in bold) are the putative DNA-binding amino acids and were therefore treated specially within the ZF domains.
We only kept complete (containing all 23 amino acids) and perfect (containing at least a C2 or H2 signature) ZFs. All degenerate and atypical ZF domains were removed for downstream analyses. In total, 733 KRAB and 3909 ZF domains were used.
Polymorphism Data
Human SNP data were obtained from the 1000 Genomes Consortium phase 1, release version 3 (Consortium 2012). Variant Calling Format (.vcf) files aligned to the human reference genome (hg19) were downloaded for all KRAB-ZF genes with tabix-0.2.6. We included 1092 individuals from 14 populations. Only high-quality SNPs were kept and indels were removed, resulting in a total of 97,465 SNPs. Filtering was carried out using vcftools version 0.1.7 (Danecek et al. 2011) with the following parameters: minMQ = 10, minGQ = 40, minDP = 5, and minQ = 100. All variants marked as “SysErr” and “lowQual” were removed as well. The resulting SNPs were classified according to their correspondence in the KRAB domain, in the ZF domain, or as ZF-binding amino acids. Because of the repetitive nature of the ZF domains, it is feasible that the amount of polymorphism may have been over- or underestimated. To check for possible biases, we downloaded the mappability tracks available from the UCSC genome browser (hg19). Because the read lengths are a mixture of 36 to more than 100 base pairs (bp), we downloaded four tracks (of lengths 36, 50, 75, and 100 bp) according to their ability to uniquely align to different parts of the genome. In other words, each position in the genome has a mappability score (ranging from 0 to 1, 1 corresponding to a uniquely aligned read) that depends on the length of the short read (36 bp reads map less uniquely in the genome than 100 bp reads). We investigated whether there is a bias in read mapping and allele frequency. In Table S2, we calculated the Spearman correlation between the minor allele frequency (MAF) of the binding site SNPs and the mappability of the reads (for the four different read lengths used by the 1000 Genomes project for SNP calling). There is no significant correlation between the mappability score and the MAF (P > 0.15 in all cases). Furthermore, when comparing the mappability of synonymous versus nonsynonymous SNPs, there is no significant difference between them (Wilcoxon test, P-value = 0.7722).
Expression Data
RNA-Seq expression data for three species (humans, chimpanzees, and rhesus macaques) in six tissues (brain and cerebellum separately, heart, kidney, liver, and testis) were obtained from Brawand et al. (2011), in the form of FPKM values (processing steps described therein). Human embryonic stem cell RNA-Seq data were downloaded from the Gene Expression Omnibus with accession number GSE57989 and processed in a similar way.
Expression Breadth and Conservation
Expression conservation describes the degree of conservation of tissue-specific expression between two homologous genes, and was calculated between human–chimpanzee orthologous genes using the expression conservation index (ECI) according to Yang et al. (2005). More specifically, for a given gene, the ECI is equal to the number of tissues where the gene is expressed in both species (conserved expression) divided by the mean number of tissues with gene expression in humans and in chimpanzees. ECI values range from 0 and 1, where 1 corresponds to a gene with conserved expression in all tissues for the two species.
Expression breadth corresponds to the number of tissue types in which a given gene is expressed above some threshold value. We used a threshold of FPKM > 1 to define a gene as “expressed” in a given tissue.
Histone Data
We analyzed the H3K9me3 histone mark, which is marking an inactive chromatin state and therefore a repressed gene. Histone modification data, along with their input control for human adult kidney, liver, and heart tissues, were downloaded (in .wig format) from the Epigenomics Project (http://www.ncbi.nlm.nih.gov/epigenomics) with accessions codes: ESX000002152, ESX000002139, ESX000006561, ESX000006547, ESX000005777, ESX000005738. In order to extract only the significantly enriched regions for H3K9me3, only regions with a minimum twofold signal over the input control and an input signal greater than the cutoff were used (third quartile + 1.5 × IQR).
Orthologous Gene and Domain Annotation
The annotation of orthologous genes for humans, chimpanzees, and rhesus macaques was downloaded from the Ensembl Web Browser (http://www.ensembl.org). Only 1-to-1 orthologs were kept. Human–mouse orthologous genes were defined as described in Corsinotti et al. (2013).
All human ZF and KRAB domains were separately aligned to the chimpanzee (panTro4), rhesus macaque (rheMac2), and mouse (mm10) genomes using the blat software from the UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgBlat). From the resulting matches, only those belonging to orthologous genes were kept and in cases of multiple matches, manual inspection was used to confirm the correct corresponding ZF domain. Hence, only the best correspondences between the individual ZF and KRAB domains were used for the four species, providing exact 1-to-1 correspondence between all of the amino acids of the ZF domains (including the DNA-binding amino acids).
Tests for Selection
To evaluate the selection history of KRAB-ZF genes, we performed two types of analyses: McDonald—Kreitman (MK, 1991) tests and tests from the phylogenetic analysis by maximum likelihood (PAML) package (Yang 2007). We used all alignments of the ZF and KRAB domains for the orthologous genes of the four species, as described in the previous paragraph.
For the MK tests, synonymous and nonsynonymous divergence was calculated only for the fixed differences between two species (i.e., all human polymorphic positions as defined from the 1000 Genomes dataset were excluded). Statistical significance in each contingency table was determined using a chi-square test and a two-tailed Fisher’s exact test.
For the second analysis, the codeml package from the PAML suite (version 4.8, Yang 2007) was used to test different models (as described in Simkin et al., 2013). We used all KRAB-ZF genes having 1:1:1:1 orthologs in the four species: humans, chimpanzees, rhesus macaques, and mice (n = 52). Every ZF domain was used for the analysis by concatenating one after the other per gene (i.e., all ZF domains per gene were concatenated by excluding the linker residues existing between them). We evaluated several models: M0 (a site model with one omega for all branches) compared to the branch model (omega varying among lineages); site-model 7 (beta distribution with 0 < omega < 1) versus 8 (model M7 plus another site category assessing omega > 1), 8 versus 8a (an alternate null model for M8, with omega fixed at 1), and 1a (nearly neutral) versus 2a (positive selection). Sites evolving under positive selection were defined as having a posterior probability of >95% for omega being >1 using the Bayes empirical Bayes method. Lastly, we compared the branch-site neutral model versus the branch-site model (two or more omega values are accepted for the branches). The lineages are separated into two groups: one “background” lineage evolving neutrally or under negative selection and a “foreground” lineage that may contain some positively selected sites. In all cases, twice the difference of the two log-likelihood values (null vs. alternative model) has been compared to a chi-square distribution to assess significance.
The tree structure used for the analyses differed according to the tested model: for the M0, M1a, M2a, M7, M8, and M8a models, a rooted tree was utilized. For the branch model and branch-sites models, unrooted trees were used (three different trees according to the lineages tested: human specific, chimp specific, or human-chimp lineage specific).
GC Content
GC content data were downloaded from the UCSC genome table browser for the human genome assembly hg19 (http://genome.ucsc.edu/cgi-bin/hgTables?command=start).
Paralogs
Paralogs for the KRAB-ZF genes were obtained from the Ensembl website.
Results
Expression of Orthologous KRAB-ZF Genes is Species Specific
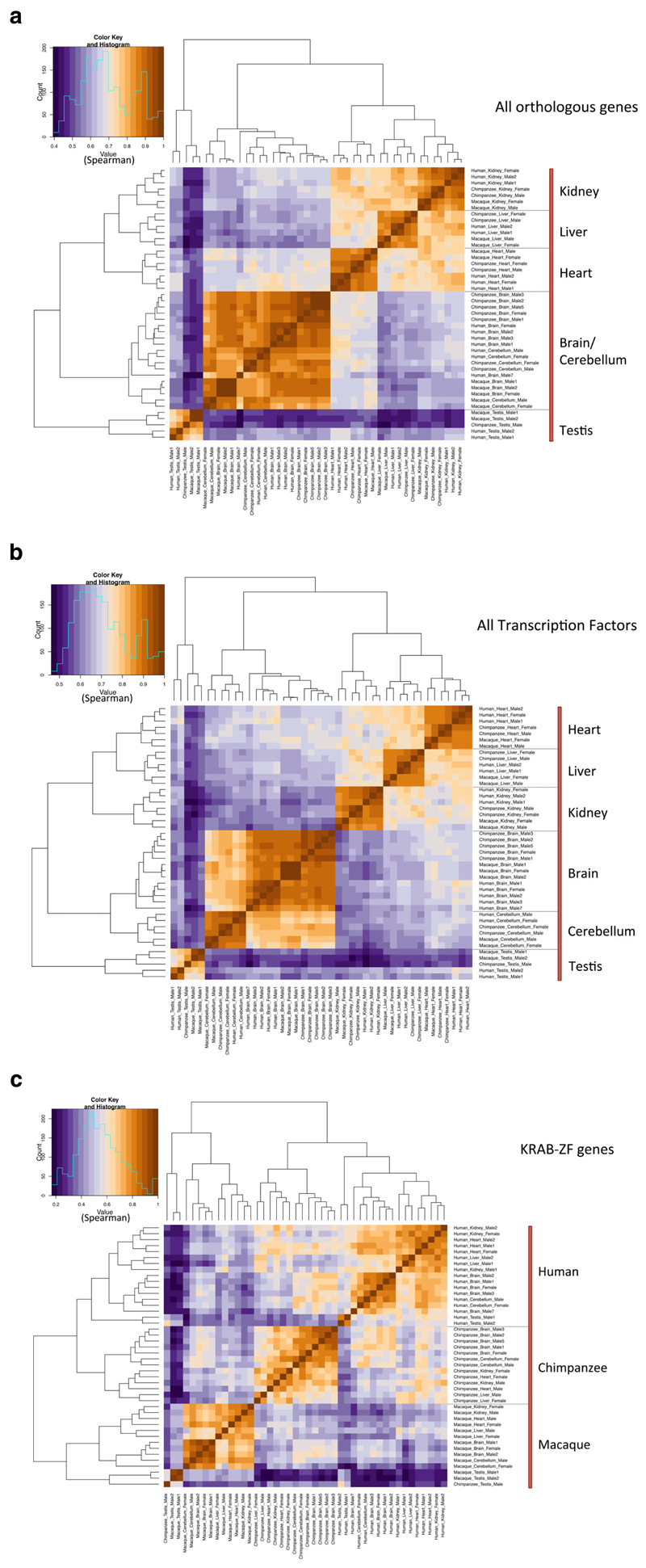
We investigated gene expression patterns for orthologous genes in six tissues (brain and cerebellum separately, heart, kidney, liver, and testis). Our analysis used RNA-Seq data from Brawand et al. (2011) and focused on three species (humans, chimpanzees, and rhesus macaques) for which we performed manual curation of all KRAB-ZF genes. Using hierarchical clustering (with Spearman correlation), we observe that expression levels of all orthologous genes from the whole transcriptome cluster in a tissue-specific manner (Fig. 1a). In other words, gene expression is conserved across the three species for a given tissue. This is fully in accordance with global patterns of gene expression among mammals demonstrated by Brawand et al. (2011), where data are arranged according to tissue. By contrast, when focusing only on KRAB-ZF gene orthologs (n = 238), the clustering becomes species specific (Fig.1c). The tissue-specific gene expression is lost, suggesting a rapid change in function for the KRAB-ZF family in primates. As a control, we did the same analysis using all TFs-orthologous genes for the three species (except ZFs, n = 726, downloaded from Animal Transcription Factor Database: http://www.bioguo.org/AnimalTFDB/index.php). Figure 1b reproduces results from Figure 1a: all orthologous genes, but KRAB-ZF, cluster in a tissue-specific manner, whereas KRAB-ZF gene expression clusters in a species-specific manner, indicating that this family of TFs has very different expression patterns than other TFs. Principal component analysis (Fig. S1) reached the same conclusions.
Figure 1. Correlations of mRNA levels for human, chimpanzee, and rhesus macaque orthologous genes.
Spearman correlation heatmaps and hierarchical clustering for (a) all orthologous genes, (b) all transcription factors orthologous genes (except ZFs), and (c) KRAB-ZF only. The highest Spearman correlation coefficients correspond to brown colors. (a) Expression of all orthologous genes and (b) expression of all human transcription factors cluster according to tissue, with a high Spearman correlation coefficient. (c) Expression of KRAB-ZF genes clusters according to species, with a high Spearman correlation coefficient.
Expression Breadth and Expression Conservation of KRAB-ZF Genes
Many studies highlight the importance of measuring the expression breadth and expression conservation across tissues and organisms when studying evolutionary rates (e.g., Yang et al. 2005; Park and Choi 2010). We calculated the number of genes expressed in all six tissues. Only 29% of KRAB-ZF genes were “expressed” in the six human tissues, whereas 47% of the totality of genes was expressed (with FPKM > 1) in all tissues. As an additional control, we used all human TFs (except the ZFs) to calculate how many are expressed in the six tissues (Table 1). There were significantly fewer KRAB-ZF genes with ubiquitous expression in all tested tissues when compared to either all TFs (χ2, P < 1.254 × 10−5) or all genes (χ2, P < 1.948 × 10−8), indicating a narrower pattern of expression for the KRAB-ZFs.
Table 1. Expression breadth of KRAB-ZFs, all TFs, and all genes for six human tissues.
| Broad expression (in all tissues, FPKM > 1) | Limited expression (in some tissues only) | Percentage (expressed/total) | |
|---|---|---|---|
| KRAB-ZFs | 68 | 170 | 29% |
| All TFs (except ZFs) | 578 | 736 | 44% |
| All genes | 7606 | 8548 | 47% |
The number of genes expressed in all tissues is reported.
We also calculated the ECI (cf. Methods) for orthologous genes between humans/chimpanzees, and tallied those with an ECI equal to one (i.e., conserved expression in all six tissues for humans and chimpanzees). Results are shown in Table 2. Roughly 16% of KRAB-ZF genes had a conserved expression (i.e., genes expressed in all six tissues in humans and in chimpanzees) whereas 39% of all orthologous genes were conserved (χ2, P < 8.22 × 10−13). Also, when compared with all TFs, the difference is also significant (χ2, P < 1.584 × 10−9) and is in accordance with previously reported conservation of tissue-specific gene expression for all orthologous genes (Ramsköld et al. 2009). However, we find that tissue-specific KRAB-ZF gene expression is not as well conserved between the two species. This result indicates that the KRAB-ZF gene family is more narrowly expressed than others and this pattern of expression is not conserved between two closely related species. This can be attributed to the fast evolving expression of KRAB-ZF genes.
Table 2. Expression conservation of KRAB-ZFs, all TFs, and all genes from human and chimpanzee tissues.
| Expressed in all tissues in humans and chimpanzees (ECI = 1) | Expression not conserved between humans and chimpanzees (ECI < 1) | Percentage (expressed/total) | |
|---|---|---|---|
| KRAB-ZFs | 38 | 200 | 16% |
| All TFs (except ZFs) | 476 | 838 | 36% |
| All genes | 6289 | 9865 | 39% |
The number of genes is reported.
Expression of KRAB-ZF Genes Correlates With Polymorphism in their ZF-Binding Amino Acids
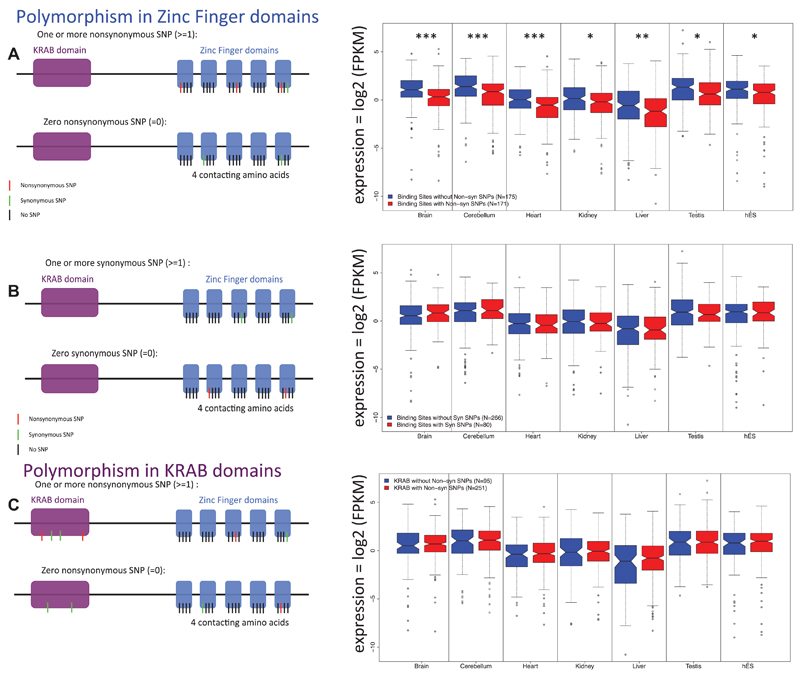
The ZF-contacting amino acids correspond to the three positions from the ZF domain contacting the primary strand of the DNA (positions -1, 3, and 6 of the alpha-helix) and one amino acid contacting the secondary strand of the DNA (position 2 of the alpha-helix; Elrod-Erickson et al. 1998). Those four amino acids are also called the ZF “fingerprint” (Liu et al. 2014). From the 1000 Genomes polymorphism data, we have extracted the SNPs occurring in those four amino acids, and separated the 346 human KRAB-ZF genes into two categories: KRAB-ZF genes with a nonsynonymous SNP in at least one of the four contacting amino acids, and KRAB-ZF genes without any nonsynonymous SNPs in any of the four contacting amino acids. Figure 2A shows the expression levels between these two categories of KRAB-ZF genes in the six adult tissues and in the human embryonic stem cells (hES).
Figure 2. Comparison of human mRNA levels for two categories of KRAB-ZF genes (with and without nonsynonymous SNP in their DNA-contacting residues).
Expression values of all KRAB-ZF genes with (red boxes) and without (blue boxes) nonsynonymous polymorphism(s) in at least one of the four binding amino acids (panel a). As a control, in panel b, expression values of all KRAB-ZF genes with (red boxes) and without (blue boxes) synonymous polymorphisms in at least one of the four binding amino acids are given. In panel c, expression values of all KRAB-ZF genes with (red boxes) and without (blue boxes) nonsynonymous polymorphism(s) in the KRAB domain. Accompanying cartoons illustrate examples of the corresponding two categories of KRAB-ZF genes compared. (A) Genes with nonsynonymous SNP(s) in their contacting residues are significantly less expressed in all tested tissues than genes without nonsynonymous SNP in their contacting residues. FDR: <0.05 (*), <0.01 (**), <0.001 (***). (B) There is no significant difference in expression level between genes with synonymous SNP(s) in their contacting residues when compared with genes without synonymous SNP(s) in their contacting residues. (C) There is no significant difference in expression level between genes with nonsynonymous SNP(s) in the KRAB domain when compared with genes without nonsynonymous SNP(s) in the KRAB domain.
Human KRAB-ZFs, having nonsynonymous polymorphism(s) located in their four binding amino acids, have significantly lower expression levels than those without such polymorphism (Wilcoxon’s rank sum test, Benjamini–Hochberg adjusted P-values < 0.05 for all comparisons, Fig. 2A). As a control, we also separated the 346 human KRAB-ZFs into two new categories: KRAB-ZF genes with a synonymous SNP in at least one of the four contacting amino acids and KRAB-ZF genes without any synonymous SNPs in any of the four contacting amino acids (this category contains both KRAB-ZF genes with nonsynonymous SNPs only and those without any SNPs). Figure 2B compares expression levels between these two categories of KRAB-ZF genes in the six adult tissues and the hES cells, observing no difference in expression levels (Wilcoxon’s rank sum test). As an additional control, KRAB-ZF genes were separated according to the presence or absence of nonsynonymous polymorphisms in their KRAB domains. Figure 2C illustrates that there is no significant difference in expression levels between the two categories. This re-enforces our conclusion that the presence of a nonsynonymous SNP in a binding site uniquely correlates with the reduced expression of the gene.
To test whether the observed difference in expression may be due to the number of nonsynonymous SNPs present in the genes, we separated the genes in two categories: only/mostly nonsynonymous SNPs and only/mostly synonymous SNPs. There is no significant difference between the two categories regarding their expression levels (Wilcoxon test P-value = 0.06), thus indicating that it is not the number of nonsynonymous SNPs per gene (i.e., nonsynonymous SNP density at the gene level) but the presence of a nonsynonymous SNP in the binding site only that correlates with the reduced expression.
Finally, we controlled for a possible relationship between the number of ZFs per gene and our observed expression differences. We did not found any significant correlation between the number of ZF domains per gene and their expression for the six tissues and human embryonic stem cells (hESC, Table 3).
Table 3. Spearman’s correlation coefficient (rho) and P-values between number of ZF per gene and gene expression for six tissues and hES cells.
| Tissue | Spearman’s rho | P-value |
|---|---|---|
| Brain | −0.0548 | 0.3093 |
| Cerebellum | −0.05 | 0.3538 |
| Heart | −0.0757 | 0.16 |
| Kidney | −0.0568 | 0.2923 |
| Liver | −0.082 | 0.1273 |
| Testis | −0.0876 | 0.1038 |
| hES | −0.038 | 0.4819 |
Histone Modification H3K9Me3 On ZF-Coding Exon Correlates With Polymorphism In Their ZF-Binding Amino Acids
Chromatin immunoprecipitation of histones followed by sequencing (ChIP-Seq) is used to identify chromatin states at very high resolution. The modification of histones changes the DNA compaction, resulting in differences in the accessibility of DNA fragments for TFs, and thus influences transcriptional regulation (Tollefsbol 2011). Using publicly available ChIP-Seq data, we analyzed one type of histone modification (H3K9me3, a marker of transcriptionally inactive chromatin) for presence or absence on the ZF-coding exon of all KRAB-ZF genes for the human kidney, liver, heart, and spleen. The 346 human KRAB-ZF genes were separated in the two categories described earlier (KRAB-ZF genes with/without a nonsynonymous SNP in at least one of the four contacting amino acids). Figure 3 compares the enrichment of H3K9me3 for the two groups of genes. Results indicate that KRAB-ZF genes bearing nonsynonymous SNP(s) in one of their four binding amino acids are significantly enriched for repressive histone marks (H3K9me3) than those without such polymorphism. Though this analysis is based on a different dataset (see Methods), it corresponds to the same three tissues used from the RNA-Seq expression results (Fig. 2).
Figure 3. H3K9me3 on ZF-coding exon.
Comparison of repressive (H3K9me3) histone mark for KRAB-ZF genes with (in red) and without (in green) nonsynonymous SNPs in their four DNA-contacting amino acids. There is a significant enrichment (Fisher’s exact test, two-tailed, P-values < 0.05) of H3K9me3 occupancy in the ZF-coding exon of KRAB-ZF genes carrying a nonsynonymous SNP in their contacting residues, indicating a repressed gene.
Expression Breadth and Expression Conservation of the Two Groups of KRAB-ZF Genes
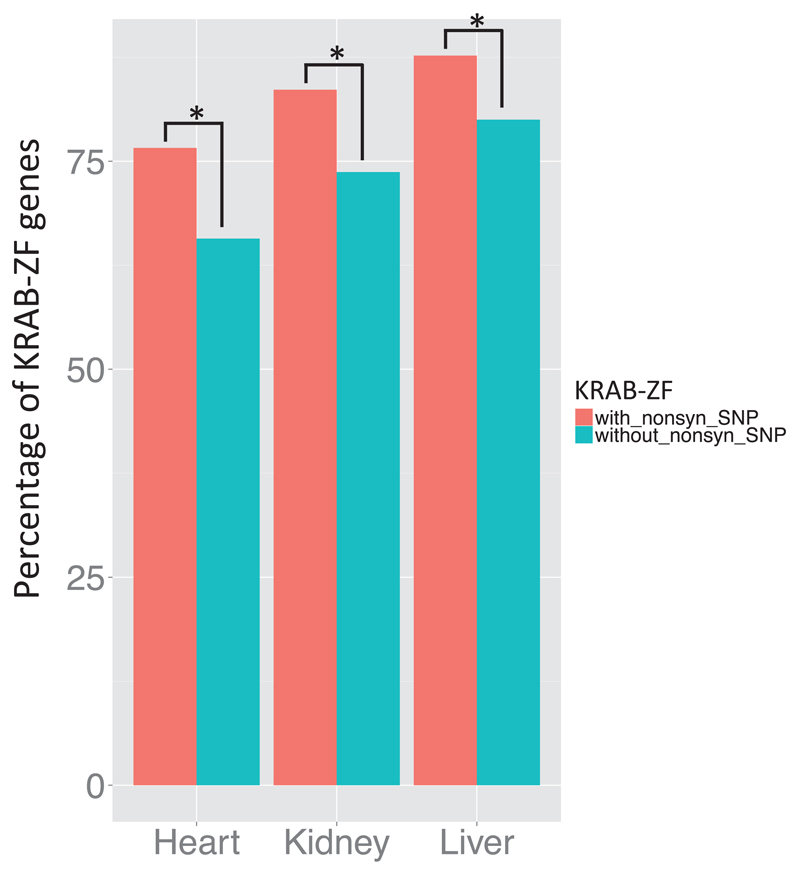
We investigated the expression breadth and conservation separately for the two groups of KRAB-ZF genes described above. Only 9/171 KRAB-ZF genes carrying a nonsynonymous SNP in their DNA-recognizing amino acids have conserved expression in all tissues for the two species (i.e., ECI = 1), whereas 28/175 genes without nonsynonymous SNPs meet this criterion (Fisher’s exact test, two-tailed, P-value = 0.0015). Similarly, there is a significant difference in the proportion of expression breadth between the two groups of KRAB-ZF genes, with those carrying nonsynonymous SNP(s) in their DNA-recognizing amino acids being less broadly expressed than the others (Fisher’s exact test, two-tailed, P-value = 0.00038).
The Newest KRAB-ZF Genes are Enriched for Nonsynonymous SNPs in their Contacting Amino Acids Relative to Older KRAB-ZF Genes
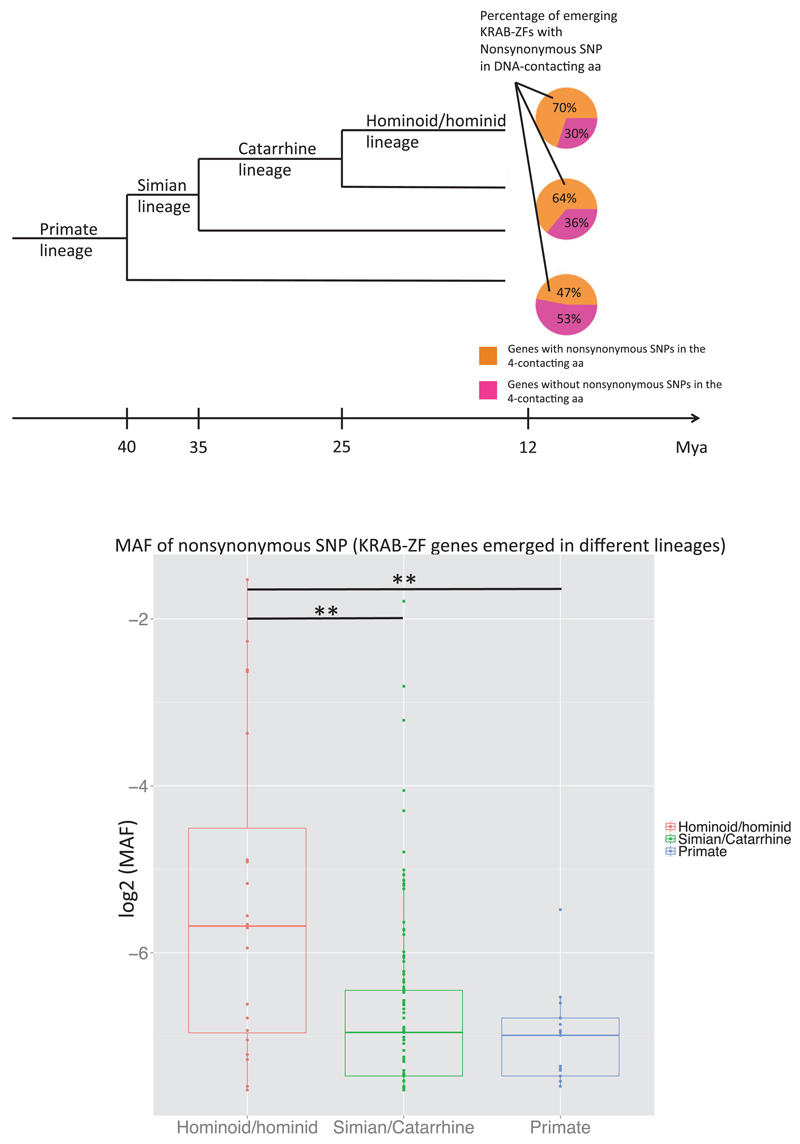
Jacobs et al. (2014) presented a phylogenetic tree with all KRAB-ZF genes and the lineages on which they emerge. We used these data to infer the number of genes emerging in the Primate, Simian/Catarrhine, and Hominoid/Hominid lineages having nonsynonymous polymorphism in their binding amino acids (Fig. 4a). Seventy percent of the total genes that emerged in the Hominoid/Hominid lineage have nonsynonymous SNPs in the binding amino acids, whereas genes that emerged during the primate lineage are more constrained (47% contain a nonsynonymous SNP). This indicates that older KRAB-ZF genes may be experiencing stronger purifying selection to maintain their four contacting amino acids. Another indicator of such constraint is their allele frequency; in Figure 4b, the MAFs of the nonsynonymous SNPs (only in the four contacting residues) for the three categories of KRAB-ZF genes are plotted according to the lineage on which they appear. Interestingly, nonsynonymous SNPs from KRAB-ZF genes emerging in the Hominoid/Hominid lineage have a significantly higher MAF than SNPs from genes emerging in older lineages (Wilcoxon Mann–Whitney P-values < 0.01). This result is consistent with stronger selective constraints acting on the oldest members of the KRAB-ZF family.
Figure 4. Minor allele frequency (MAF) and number of KRAB-ZF genes emerging in different lineages.
(a) Proportion of KRAB-ZF genes with/without a nonsynonymous SNP in their contacting residues emerging in recent lineages. In total, 70% of the genes emerging in the Hominoid/Hominid lineage carry a nonsynonymous SNP in their binding residues, 64% in the Simian/Catarrhine lineage, and only 47% in the primate lineage, indicating a potential relaxation of selective constraint for genes emerging in the most recent lineages. (b) MAFs of nonsynonymous SNPs (in the four contacting residues). SNPs from genes emerging in the Hominoid/Hominid lineage have a significantly higher MAF than SNPs from genes emerging in older lineages. Both (a) and (b) demonstrate the strong selective constraint acting on older genes to maintain their binding residues, thus indicating strong functional relevance. Conversely, contacting residues from younger genes seem to be under weaker purifying selection, potentially because of the lack of a specific target.
Evolutionary Analysis of Orthologous KRAB-ZF Genes
To investigate the selective pressures acting on the KRAB-ZF genes, we performed two different analyses. All amino acids present in the ZF domains were tested for positive selection using the codeml program implemented in the PAML suite. Three different approaches were implemented (see Materials and Methods).
First, we investigated the possibility that the ratio dN/dS (ratio of nonsynonymous changes to synonymous changes or omega) of a single branch was different from the rest of the phylogenetic tree (composed of four organisms: humans, chimpanzees, rhesus macaques, and mice). For this, we compared the null site model (one omega for all lineages) with the branch model (estimates of omega are produced for each lineage). No significant difference was found between the likelihood values of the two models; therefore, we assumed that the selective pressure for the ZF domains does not vary across the phylogeny.
Next, we used three different sites-model comparisons to estimate selective constraints on individual amino acids across the length of the ZFs. The comparison of model 7 versus model 8 identified only three genes rejecting neutrality in favor of positive selection (ZNF212, ZNF263, and ZNF473). ZNF212 had three individual amino acids with high probability of positive selection according to the Bayes empirical Bayes method, ZNF263 had four amino acids identified, and ZNF473 had no site localized. No sites from the four contacting amino acids were found to be experiencing positive selection. The other two site-model comparisons (M8 vs. M8a and M1a vs. M2a) did not identify specific sites undergoing positive selection.
Lastly, we tested the hypothesis that positively selected individual sites are present only in specific lineages. We used the comparison of the branch-site model against the branch-site neutral model. This test did not identify any positively selected site in any lineage.
To estimate levels of between-species divergence, we compared humans with closely related species (chimpanzees and rhesus macaques), as well as with mice. We separated the surveyed fragments into three categories that are likely to differ in the intensity and mode of selection acting on them, namely, the ZF domains, the KRAB domains, and the four DNA-contacting amino acids. The MK test is designed to distinguish neutrality in protein-coding genes from negative or positive selection by comparing levels of polymorphism within-species (humans) and divergence between species (human–chimpanzee, human–macaque, and human–mouse). If the sites evolve neutrally, the ratio of polymorphism to divergence for the nonsynonymous sites (dN/dS) should be similar to that for synonymous sites (pN/pS). Detailed results of each MK test are shown in Table S3. Using all genes pooled together for the ZF domains, there are fewer nonsynonymous substitutions between species than synonymous substitutions (dN/dS < pN/pS, χ2, P-value < 0.0001), indicating purifying selection, or the purging of deleterious mutations. However, as the ZF domain is highly conserved, comparing the average rate of synonymous and nonsynonymous substitutions for the whole ZF domain may mask specific positively selected sites. For this reason, we performed a separate MK test for the four DNA-contacting amino acids, pooling all genes together to gain statistical power. The results remain the same as for the ZF domain (dN/dS < pN/pS, Fisher’s exact test, two-tailed, P-value < 0.0001) for all three comparisons (human–chimpanzee, human–rhesus macaque, and human–mouse). For the KRAB domain, all MK tests indicate neutrality for the two comparisons (human/chimpanzee and human/rhesus macaque, Fisher’s exact test, two-tailed, P-value > 0.05, dN/dS ~ pN/pS). The pattern is different for the comparison with mice, where significant evidence of purifying selection is present (dN/dS < pN/pS, Fisher’s exact test, two-tailed, P-value < 0.05). Using only genes presenting a nonsynonymous SNP in the four contacting amino acids, the test is no longer significant, indicating that the KRAB domain is evolving neutrally for those genes. This result points toward weaker purifying selection acting on this group of genes.
Discussion
The expression of many orthologous genes appears to be tissue specific. This has been previously demonstrated in a study of global patterns of gene expression differences among mammals (Brawand et al. 2011). From the same dataset, we focused on the cross-species, cross-tissue expression of KRAB-ZF genes. We found that the expression of orthologous KRAB-ZF genes follows a species-specific pattern rather than a tissue-specific pattern. This finding is in line with previous studies suggesting that KRAB-ZF genes have different tissue preferences in different species (Nowick et al. 2010) and supports the independent expansion and functional diversification of KRAB-ZFs in different vertebrate lineages (Liu et al. 2014). This loss of tissue-specific expression implies a rapid change in function for the KRAB-ZF family in primates, providing additional support for the hypothesis that this family of TFs plays a role in speciation by regulating evolutionarily divergent traits (see also Nowick et al., 2013).
Next, we analyzed the breadth and the conservation of expression for the KRAB-ZF genes. We confirmed that the KRAB-ZF genes do not have tissue-conserved expression among species, and are narrowly expressed in only a few tissues. Yang et al. (2005) and Park and Choi (2010) showed that gene expression evolves rapidly for genes expressed in only a limited number of tissues. They also demonstrated that, in many cases, tissue-specific gene expression may be transient and not evolutionarily stable. Our results support the hypothesis that the expression of KRAB-ZF genes is fast evolving in primates and this alteration in gene regulatory networks is playing a major role in primate evolution. New endogenous retroelements (EREs) are continuously emerging during evolution and their expression needs to be constrained in a tissue-specific manner. Thus, it is important for the organism to have a fast-evolving modular system capable of regulating retroelement expression at precise developmental stages and in a tissue-specific manner. Thus, KRAB-ZFs are good candidates to control aberrant expression of EREs.
Given that the expression of KRAB-ZF genes is rapidly evolving, we next evaluated models of selection at the nucleotide level. Both the MK test and PAML found that the KRAB and ZF domains are evolving under purifying selection. This conclusion aligns with previous results, which have demonstrated that orthologs of each KRAB-ZF are subject to negative constraint across the entire set of DNA-binding domains to retain its DNA-binding specificity (Thomas and Schneider 2011), with the nucleotide contacting residues being amongst the slowest evolving (Thomas and Schneider 2011). Also, there is evidence of selection against common SNPs at DNA-contacting amino acids given that substitutions in the DNA-contacting positions could alter the DNA-binding specificity of the KRAB-ZF protein and disrupt the TF function (Lockwood et al. 2014). However, studies on KRAB-ZF paralogous genes show evidence for a very short period of positive selection occurring just after duplication, followed by a long period of strong purifying selection (Thomas and Schneider 2011). Thus, signals of positive selection driving the acquisition of new DNA-binding specificities may be obscured by subsequent purifying selection to maintain those specificities (Emerson and Thomas 2009).
Since the expression divergence of KRAB-ZF genes seems to be an important parameter in their evolutionary process (Nowick et al. 2010), and because the drive for novelty in their function may be based on alterations of their DNA-contacting amino acids, we studied the expression of KRAB-ZF genes in the light of polymorphism in their four binding residues. We divided the 346 human KRAB-ZF genes into two categories: the ones bearing a nonsynonymous polymorphism in at least one of their DNA-contacting amino acids (171 genes in total) and the ones without nonsynonymous polymorphism(s) in any of their DNA-contacting amino acids (175 genes in total). We found that the average expression of the 171 genes having at least one nonsynonymous SNP was significantly lower. We extend this result using another dataset of histone ChIP-Seq that showed enrichment of repressive histone marks in the ZF region of the 171 KRAB-ZFs compared with genes without nonsynonymous SNPs. Comparison of global GC content also supports this result, where genes with lower expression have a smaller percentage of GCs. These findings shed light on the relationship between KRAB-ZF gene expression and the presence of polymorphisms in their ZF-binding amino acids.
By searching for more elements differentiating the two groups of KRAB-ZF genes (cf. Table 4), we discovered that the KRAB-ZFs with nonsynonymous SNP(s) in their binding site(s) have significantly fewer mouse orthologs than those without, which could be a consequence of their younger age. At the same time, they have more paralogs and ZF domains per gene on average, indicating formation by recent gene duplication (Emerson and Thomas 2009). Further investigation confirmed that KRAB-ZF genes emerging in the Simian, Catarrhine, and Hominoid/Hominid lineages were enriched for genes presenting a nonsynonymous SNP in their contacting residues (Fisher’s exact test, two-tailed, P-value = 6.4 × −5). Those SNPs have a significantly higher MAF, indicating a relaxation of strong purifying selection for the younger KRAB-ZF genes—as also observed by the nonsynonymous SNPs in their binding residues. In contrast, only 47% of genes emerging in the primate lineage bear a nonsynonymous SNP in their contacting amino acids and have a significantly lower MAF, strongly suggesting the action of purifying selection.
Table 4. Differences between the two groups of KRAB-ZF genes (with or without nonsynonymous SNP(s) in the four DNA-contacting amino acids).
| Comparison | KRAB-ZFs with nonsynonymous SNPs in their DNA-contacting amino acids | KRAB-ZFs without nonsynonymous SNPs in their DNA-contacting amino acids | P-value |
|---|---|---|---|
| Expression level (FPKM) | Less expressed | More expressed | <0.05 |
| H3K9me3 on the ZF-coding exon | More present | Less present | <0.05 |
| ECI and expression breadth | Narrowly expressed (i.e., tissue expression evolves rapidly) | Broadly expressed (i.e., tissue expression more conserved) | 0.0015 |
| GC content | Lower GC content (average = 42%, i.e., less expressed) | Higher GC content (average = 43%, i.e., more expressed) | 0.03 |
| Number of orthologous genes human/mouse | Fewer mouse orthologs (i.e., younger) | More mouse orthologs (i.e., older) | 0.00047 |
| Number of paralogs per gene | More paralogs (average = 25/gene) | Fewer paralogs (average = 21/gene) | 0.01 |
| Number of zinc-finger domains per gene | More ZF domains/gene (average = 12 ZFs/gene, i.e., more newly formed ZF domains) | Fewer ZF domains/gene (average = 10 ZFs/gene, i.e., older ZF domains) | 6.5 × 10−5 |
| Emergence in lineage | Simian, Catarrhine, or Hominoid/Hominid lineage | Primate lineage | 6.4 × 10−5 |
The group having nonsynonymous SNP(s) is globally less expressed, with repressive histone marks occupying their gene body, and less GC content. In addition, they appear to be younger, generally emerging in the Simian, Catarrhine, or Hominoid/Hominid lineage, thus having fewer mouse orthologs and more paralogs and zinc-finger domains per gene.
In summary, through analyses combining transcriptomic data, histone-modification marks, and population genetics, we conclude that human KRAB-ZF genes can be separated into two categories according to the type of polymorphisms located within their four DNA-contacting residues. Genes without nonsynonymous polymorphism(s) seem to be the oldest members of this family and are significantly more expressed in humans, indicating that members of this subgroup are essential for the organism and therefore are highly conserved. The second category contains newer KRAB-ZFs, with significantly lower expression in all tested tissues and, in human populations, frequent polymorphisms present in their binding sites. Because EREs mutate to escape the KRAB-ZF control, slight changes in the four DNA-contacting residues provide the opportunity for the KRAB-ZF genes to re-create a new DNA-binding fingerprint able to control this newly generated binding site. Genetic diversity is generated very quickly from existing contacting residues, providing ground for fine-tuning of their DNA-binding specificity, without having a deleterious effect on the fitness of the organism. This reduced expression enables them to make slight modifications of their DNA-contacting residues and eventually establish high affinity between ZF residues and binding site. Since little is known about where these proteins bind, which ZFs they use or which genes they regulate, future results on their targets will reveal more about this family and its members’ putative function.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s website:
Table S1. Manually curated list of KRAB-ZF genes.
Table S2. Correlation coefficients between MAF and mappability.
Table S3. Details about all MK tests details and P-values.
Figure S1. Principal component analysis (PCA) on standardized expression values for (a) all orthologous genes and (b) KRAB-ZF only.
Acknowledgments
We are grateful to S. Laurent, K. Irwin, S. Quenneville, and A. Necsulea for discussions and manuscript comments. This study was supported by grants from the Swiss National Science Foundation and a European Research Council (ERC) Starting Grant to JDJ. AK, LM, AW, DT, and JDJ conceived and designed the experiments. AK and LM analyzed the data. AK and JDJ wrote the article.
Literature Cited
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Choo Y, Klug A. Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage. Proc Natl Acad Sci. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T. 1000 G. P. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou-Deltas CD, Gilbert J, Bartlett RJ, Herbstreith M, Roses AD, Lee JE. The identification and characterization of KRAB-domain-containing zinc finger proteins. Genomics. 1992;12:581–589. doi: 10.1016/0888-7543(92)90451-w. [DOI] [PubMed] [Google Scholar]
- Corsinotti A, Kapopoulou A, Gubelmann C, Imbeault M, Santoni de Sio FR, Rowe HM, Mouscaz Y, Deplancke B, Trono D. Global and stage specific patterns of Krüppel-associated-box zinc finger protein gene expression in murine early embryonic cells. PLoS ONE. 2013;8:e56721. doi: 10.1371/journal.pone.0056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod-Erickson M, Benson TE, Pabo CO. High-resolution structures of variant Zif268–DNA complexes: implications for understanding zinc finger-DNA recognition. Structure. 1998;6:451–464. doi: 10.1016/s0969-2126(98)00047-1. [DOI] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. Adaptive Evolution in Zinc Finger Transcription Factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AT, Huntley S, Tran-Gyamfi M, Baggott DM, Gordon L, Stubbs L. Evolutionary expansion and divergence in the ZNF91 subfamily of primate-specific zinc finger genes. Genome Res. 2006;16:584–594. doi: 10.1101/gr.4843906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–245. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Berg JM. A 2.2 A resolution crystal structure of a designed zinc finger protein bound to DNA. Nat Struct Biol. 1996;3:940–945. doi: 10.1038/nsb1196-940. [DOI] [PubMed] [Google Scholar]
- Liu H, Chang L-H, Sun Y, Lu X, Stubbs L. Deep vertebrate roots for mammalian zinc finger transcription factor subfamilies. Genome Biol Evol. 2014;6:510–525. doi: 10.1093/gbe/evu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood SH, Guan A, Yu AS, Zhang C, Zykovich A, Korf I, Rannala B, Segal DJ. The functional significance of common polymorphisms in zinc finger transcription factors. G3. 2014;4:1647–1655. doi: 10.1534/g3.114.012195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Nowick K, Carneiro M, Faria R. A prominent role of KRAB-ZNF transcription factors in mammalian speciation? Trends Genet. 2013;29:130–139. doi: 10.1016/j.tig.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Nowick K, Hamilton AT, Zhang H, Stubbs L. Rapid sequence and expression divergence suggest selection for novel function in primate-specific KRAB-ZNF genes. Mol Biol Evol. 2010;27:2606–2617. doi: 10.1093/molbev/msq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SG, Choi SS. Expression breadth and expression abundance behave differently in correlations with evolutionary rates. BMC Evol Biol. 2010;10:241. doi: 10.1186/1471-2148-10-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon M, Hamilton AT, Gordon L, Branscomb E, Stubbs L. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 2003;13:10971110. doi: 10.1101/gr.963903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A, Wong A, Poh Y-P, Theurkauf WE, Jensen JD. Recurrent and recent selective sweeps in the piRNA pathway. Evol Int J Org Evol. 2013;67:1081–1090. doi: 10.1111/evo.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivakov M, Akhtar J, Kheradpour P, Beal K, Girardot C, Koscielny G, Herrero J, Kellis M, Furlong EE, Birney E. Analysis of variation at transcription factor binding sites in Drosophila and humans. Genome Biol. 2012;13:R49. doi: 10.1186/gb-2012-13-9-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Emerson RO. Evolution of C2H2-zinc finger genes revisited. BMC Evol Biol. 2009;9:51. doi: 10.1186/1471-2148-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011;21:1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsbol TO, editor. Using ChIP-Seq technology to generate highresolution profiles of histone modifications. Springer; Humana Press; New York: 2011. [Google Scholar]
- Yang J, Su AI, Li W-H. Gene expression evolves faster in narrowly than in broadly expressed mammalian genes. Mol Biol Evol. 2005;22:2113–2118. doi: 10.1093/molbev/msi206. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.