Abstract
The repair of DNA damage in highly compact, transcriptionally silent heterochromatin requires that repair and chromatin packaging machineries be tightly coupled and regulated. KAP1 is a heterochromatin protein and co-repressor which binds to HP1 during gene silencing, but is also robustly phosphorylated by ATM at serine 824 in response to DNA damage. The interplay between HP1-KAP1 binding/ATM phosphorylation during DNA repair is not known. We show that HP1α and unmodified KAP1 are enriched in endogenous heterochromatic loci and at a silent transgene prior to damage. Following damage, γH2AX and pKAP1-s824 rapidly increase and persist at these loci. Cells which lack HP1 fail to form discreet pKAP1-s824 foci after damage but levels are higher and more persistent. KAP1 is phosphorylated at Serine 473 in response to DNA damage and its levels are also modulated by HP1. Unlike pKAP1-s824, pKAP1-s473 does not accumulate at damage foci but is diffusely localized in the nucleus. While HP1 association tempers KAP1 phosphorylation, this interaction also slows the resolution of γH2AX foci. Thus, HP1-dependent regulation of KAP1 influences DNA repair in heterochromatin.
Keywords: KAP1, HP1, Heterochromatin, ATM, Phosphorylation
Introduction
Cells must efficiently repair damaged DNA, which is packaged into chromatin. PI3-kinase family members, including Ataxia telangiectasia mutated (ATM) and DNA-dependent protein kinase (DNA-PK), become activated in response to DNA damage and phosphorylate the SQE motif found in many proteins including the core histones which package nucleosomes (1). For example, the ATM kinase phosphorylates histone H2AX at the site of damage, which acts as a scaffold for the assembly of the repair machinery. As most repair factors are highly mobile in the nucleus, this also promotes rapid recruitment and assembly at damage sites (2, 3).
An important requirement for efficient repair is that the regulatory machinery be able to gain access to damaged sites in a chromatin environment, including both nucleosomal as well as higher order chromatin architecture. Chromatin disassembly upon damage and chromatin reassembly after repair must be localized and efficient. Historically, chromatin can be broadly divided into heterochromatin, which is condensed and transcriptionally silent, and euchromatin, which has an open configuration and is associated with actively transcribing genes. Based on investigations of repair dynamics, it is thought that the timing and spatial regulation of the repair of these two types of chromatin are differentially regulated. Recent work by Noon et al. suggests that DNA repair, as measured by the number of H2AX Ser 139 phosphorylated (γH2AX) foci, is bimodal; the population of breaks in euchromatin is repaired within the first 2-3 hrs following irradiation, while heterochromatin is repaired later (4).
The H3-trimethyl K9 binding protein (HP1) and KRAB-associated protein 1 (KAP1) are two important components of heterochromatin (5, 6). KAP1 is best known as the obligate co-repressor for Krüppel-associated box zinc finger proteins (KRAB-ZFPs) (7). KRAB-ZFPs are sequence specific transcriptional repressors, which bind to DNA through their zinc fingers. KAP1 interacts with the KRAB domain and acts as a scaffold for the assembly of gene silencing factors including HP1, the H3-K9 methyltransferase Su(var)3-9 and ‘Enhancer of zeste’ domain protein 1 (SETDB1) and the chromatin remodeling factor Chromodomain-helicase-DNA-binding protein 3 (CHD3/Mi-2α), which promote transcriptional repression and chromatin condensation (8–11).
HP1 is abundant at sites of facultative and constitutive heterochromatin throughout the nucleus (12). There are three isoforms in mammals, HP1α, β, and γ. HP1α and HP1β predominantly localize to heterochromatin, while HP1γ has been observed in both euchromatin and heterochromatin (13, 14). HP1 exists as a homo-dimer and tetramer in vivo, and it is thought that HP1 multimerization contributes to chromatin condensation, which stabilizes constitutive heterochromatic structures including kinetochores, centromeres, and telomeres (15–17).
KAP1 interacts with HP1 through a well-defined peptide motif, PxVxL. Mutagenesis of this motif abolishes the ability of KAP1 to silence transcription (18). In undifferentiated cells, KAP1 is diffuse. Upon differentiation, KAP1 re-localizes to pericentromeric heterochromatic foci where it co-localizes with HP1. As the KAP1 HP1-binding mutant fails to co-localize with HP1 during differentiation, this supports the idea that its interaction with HP1 is required for its integration into the higher order architecture of heterochromatin (19, 20). Currently, it is not well understood how this interaction is dynamically regulated during the DNA damage response when the repair machinery must gain access to DNA assembled into heterochromatin.
In response to DNA damage, PI3-Kinase family members, like ATM, phosphorylate KAP1 at serine 824 and this modification co-localizes with γH2AX foci at damage sites (21–23). The hyperaccumulation of Mre11, Rad50 and Nbs1 (MRN) complex by Tumor suppressor p53-binding protein 1 (53BP1) helps retain activated ATM at sites of damage residing in heterochromatin, which require more time to repair. Without the recruitment of 53BP1, pKAP1 foci are not evident at these sites and DNA repair is less efficient suggesting that concentrated ATM activity, marked by KAP1 phosphorylation, facilitate DNA repair in damaged chromatin (4). Consistent with this, DNA breaks persist in the heterochromatin of cells lacking ATM. As this effect is abrogated by knocking down KAP1 or other components of the KAP1/KRAB-ZFP silencing complex, including HP1, phosphorylation of KAP1 at serine 824 may be critical for the repair of damaged chromatin (24). This idea is supported by the fact that reconstitution with a KAP1 mutant which mimics Ser 824 phosphorylation enhances DNA repair in heterochromatin while the null mutant S824A abrogates it (4, 24).
KAP1 is also targeted for phosphorylation at Serine 473 during the cell cycle. The highest quantities of Ser 473 phosphorylated KAP1 (pKAP1-s473) are evident in late G1, early S phase. Phosphorylation at this site is mediated by the Protein Kinase C delta pathway. The presence of this modification near the HP1 binding domain on KAP1 may affect its association with HP1 (25). It is not known whether Ser 473 phosphorylation is also dynamically regulated by the DNA damage response or whether this modification releases KAP1 from chromatin.
Here, we show that chromatin-bound KAP1 is phosphorylated in response to DNA damage. Like H2AX, phosphorylated KAP1 is not released from damaged chromatin but is organized in discreet foci surrounding DNA lesions residing in heterochromatin. We demonstrate that KAP1 is phosphorylated at Ser 473 in response to DNA damage with differing kinetics than Ser 824 and that HP1 regulates phosphorylation at both of these sites. While KAP1 phosphorylated at Ser 473 is always diffuse, HP1 interaction is critical for organizing pKAP1-s824 into DNA damage foci. These data suggest that the retention of phosphorylated KAP1 at DNA break sites in heterochromatin is not only dependent on the retention of activated ATM, but is also influenced by higher ordered chromatin organization.
Materials and Methods
Cell Lines
Early passage Mouse Embryonic Fibroblasts (MEFs) were transduced with a self-excisable, CRE-expressing lentiviral vector (CRE LV). PCR evaluation revealed that CRE-mediated recombination occurred with ≈90% efficiency to generate KAP1-/- MEFs. Lentiviruses expressing wild-type and Mut2-mutant KAP1 were used to reconstituted KAP1 knockout MEFs.
Short hairpins for HP1α, HP1β, HP1γ, and a scrambled sequence were expressed from a Doxycyline-regulatable TRIPZ Vector containing Puromycin resistance (1 ug/ml) and RFP. To obtain HP1 knockdown cells, H1299 cells were sequentially infected with the TRIPZ-HP1 targeting lentiviruses. To obtain the Vector control, H1299 cells were infected with a TRIPZ-scrambled sequence lentivirus. High Red fluorescent protein (RFP) expressing cells sorted by FACS were grown in doxycycline (1 ug/ml) for three days to diminish HP1 expression.
Green fluorescent protein (GFP) tagged short hairpins for KAP1 were expressed from a retroviral backbone and high GFP-expressing U2OS cells were isolated via FACS. A clone with the lowest expression of KAP1 was obtained by serial dilution. The GFP-tagged Vector lacking shRNA was used to create the U2OS-GFP cell line. KAP1 was restored through the stable expression of FLAG-tagged KAP1 maintained under Zeocin (200 ug/ml) selection (pcDNA3.1 vector).
Immunofluorescence
The method for immunostaining U2OS, MEFs, and H1299 cells is described in White et al 2006 (22). The following method was used for the KAP1, pKAP1, and for all HP1 antibody immunostaining of the NIH3T3 2/4 cells. For pre-extraction, cells grown on glass coverslips were washed once in PBS. Cells were then washed with CSK Buffer (10 mM PIPES pH 7.0, 100 mM NaCl, 300 mM Sucrose, 3 mM MgCl2, 1 mM PMSF, 1 μg/ml Leupeptin, 1 μg/ml Pepstatin, 1 μg/ml Aprotinin; Sigma Alderich) for 3 minutes. CSK Buffer containing 0.5% Triton X-100 was then added for 5 minutes. The cells were then washed with CSK Buffer for 3 minutes, followed by two washes in PBS. Pre-extracted cells were fixed in 4% formaldehyde in PBS for 10 minutes, and then blocked with Signal Enhancer solution (Invitrogen) as described.
For all U2OS, MEFs, and H1299 samples, coverslips were mounted in Fluoromount-G (Southern Biotech). For the NIH3T3 2/4 samples, coverslips were mounted in antifade fluorescence mounting medium (26).
Imaging
Images of the NIH3T3 2/4 cells were acquired using a Leica DMI 6000 B inverted automated microscope with HCX PL APO 100x/1.40-0.70 oil objective lens using a 457/488/514nm 30mW Argon-Ion laser for YFP, a 561nm/25mW diode laser for mCherry and a 442nm/70mW diode laser for CFP imaging. A Z-drive controlled by COMPIX SimplePCI software was used to collect image stacks using a Yokogawa CSU-10 real-time spinning disk confocal attachment with Nipkow and microlens disks. Image stacks were taken (0.4 μm increments) using Hamamatsu ORCA-AG camera (1×1 binning; 1344×1024 pixels). One image from the Z stack is represented in the figure. Image contrast adjustments were performed using COMPIX SimplePCI and Adobe Photoshop software.
Images of all other cell lines were acquired using a Leica DMIRBE TCS-Sp2 confocal laser scanning microscope with an inverted platform using a 100x objective lens with no additional zoom. A 488 Helium/Neon, a 568 Argon/Krypton, and a 633 Helium/Neon laser were used to obtain all confocal images.
Image analysis
Colocalization analyses were done using SimplePCI software. 2D intensity profiles across the transcription site were also obtained for some images.
The signal intensity of γH2AX immunofluorescence was analyzed using SimplePCI software. The nuclei of irradiated cells taken at 40x magnification were identified by imaging the Hoechst stain. A circle was manually drawn around each nucleus and the intensity of γH2AX signals were quantified from 50 cells for each timepoint using the “Hole Total Green” function (n=4). Similar methodology was utilized to quantify the γH2AX signal intensity of parental U2OS and reconstituted KAP1 knockdown cells. Here, parental U2OS cells were mixed 50:50 with the reconstituted cells which were GFP-tagged to distinguish them from the parental cells. Fifty parental and reconstituted KAP1 knockdown cells were counted with the parental cell acting as an internal control for γH2AX signal. The nuclear γH2AX signal was measured using the “Hole Total Red” function (n=3). All image settings and contrast adjustments were kept consistent within each timepoint.
Immunoprecipitation of Exogenously-expressed Proteins
Plasmids expressing MYC and FLAG-tagged genes were transfected into HEK293 cells using Lipofectamine 2000 (Invitrogen). N-ethylmaleimide(NEM)-based whole cell extracts were prepared as described in Ivanov et al (27). For immunoprecipitation, 250 μg of extracts were diluted 1/10 in NEM-Extraction Buffer and then incubated 3 hrs with FLAG Ab-conjugated beads (Sigma) at 4°C. Beads were washed three times in NEM-Extraction Buffer. Immunoprecipitated proteins were boiled and resolved on a precast gel (Invitrogen).
Following treatment with 100 nM NCS for one hour, proteins were isolated from reconstituted U2OS KAP1 knockdown cells using the NEM-Extraction method previously described. For immunoprecipitation, 500 μg of extract was immunoprecipitated as described in Ivanov et al (27).
Preparation and Western Blot Analysis of Protein Extracts
Cells were washed twice in PBS, then lysed in NP40-Extraction Buffer containing 50 mM Tris/Cl pH 7.6, 150 mM NaCl, 5 mM EDTA, 1% NP40, 2 mM DTT, 1x Phosphatase Inhibitor Cocktail (Sigma), and 1x Protease Inhibitor Cocktail (Roche). When indicated, nuclear extracts were prepared as described in Klenova et al (28) with 1% Phosphatase Inhibitor Cocktail 1 (Sigma) added to the lysis buffers.
The anti-KAP1 (C-terminal epitope, amino acids 618 - 835) and anti-pKAP1-Ser 824 Rabbit Polyclonal antibodies were generated by the Rauscher lab (22). The anti-pKAP1-Ser 473 Rabbit Polyclonal antibody was generated by the Lee lab (25). The anti-HP1α, HP1β, and HP1γ Rabbit Polyclonal antibodies were kind gifts from David C. Schultz. The anti-Actin Rabbit Polyclonal antibody (20–33, Sigma) and the Mouse Monoclonal antibodies anti-FLAG (F3165, Sigma), anti-γH2AX-Ser 139 (JBW301, Upstate), and anti-Myc (13-2500, Zymed Laboratories) were purchased.
Chromatin Fractionation
1x106 cells were plated onto 10 cm plates in duplicate and incubated overnight. Cells were scraped and washed twice with PBS. One replicate was lysed with 100 μl of SDS Loading Buffer and briefly sonicated. The other replicate was fractionated as described in Méndez et al (29). To extract KAP1 from the NP fraction in Figure 6C, Nuclear Extraction Buffer containing 500mM was substituted in the final extraction step (28).
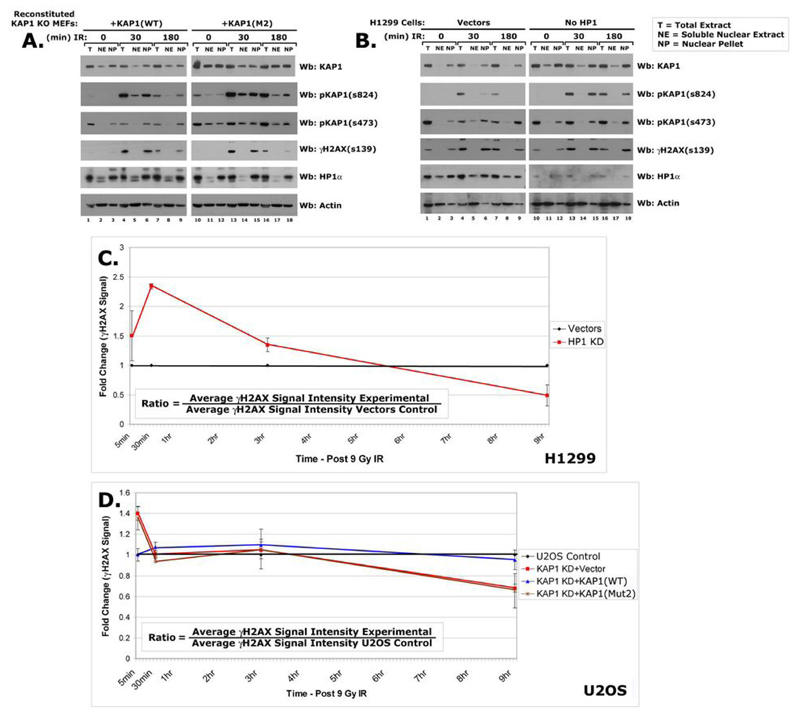
Figure 6.
Western blot of fractionated reconstituted KAP1 KO MEFs (A) HP1 knockdown cells (B) post-IR using KAP1, pKAP1-s473, pKAP1-s824, γH2AX-s139, HP1α and Actin abs. The γH2AX-s139 signal intensity was quantified at multiple timepoints post-IR (C-D). Treated cells were irradiated with 9 Gray IR. Fold change was determined (ratio in box) and plotted as line graphs.
Statistical Analyses
For comparative analyses between the Control and Experimental samples, Student’s unpaired two-tailed t tests were performed using on-line Graphpad software (http://www.graphpad.com/quickcalcs/ttest1.cfm?Format=SD).
Results
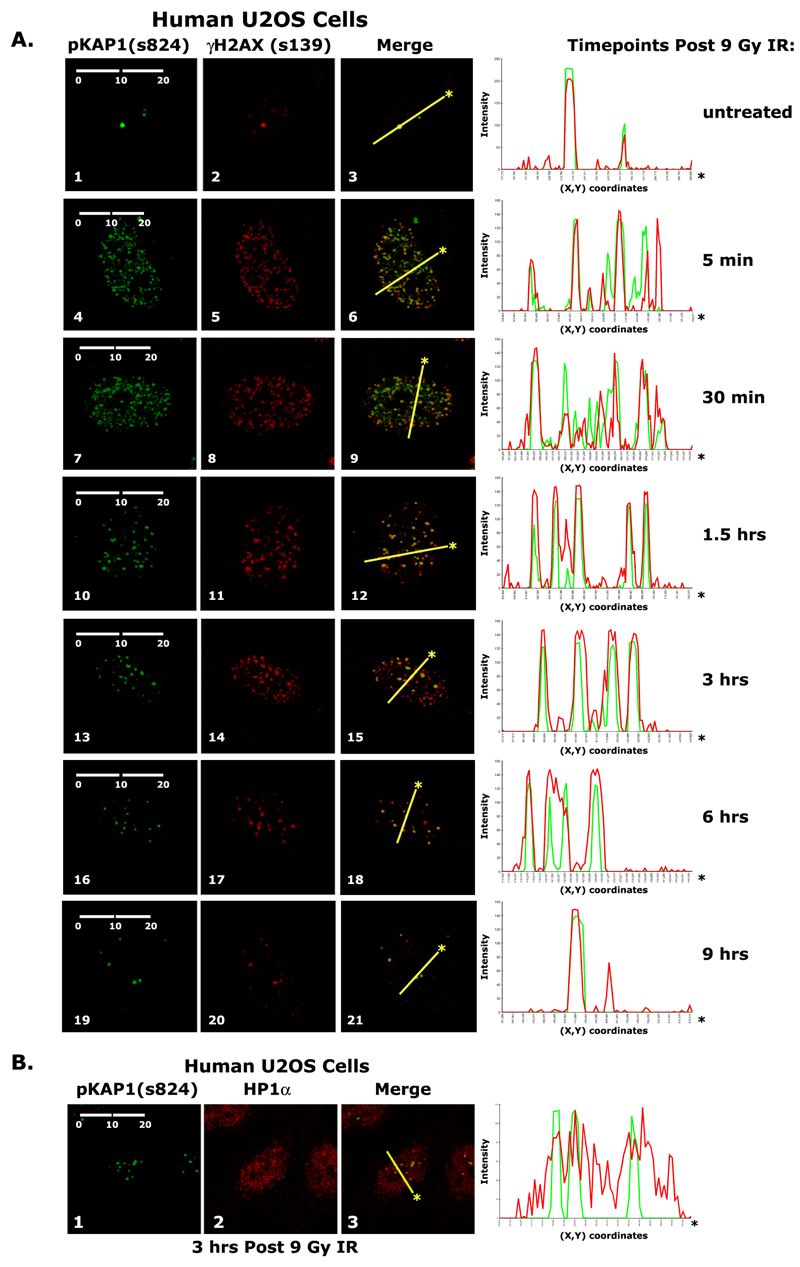
We first evaluated the localization pattern of KAP1 in human U2OS cells over the course of the cellular response to ionizing radiation (IR). At 5 minutes and 30 minutes post-irradiation, pKAP1-s824 and γH2AX appear to be in euchromatin as their foci are numerous, highly dispersed and only partially overlap with each other as determined by the line scanning diagrams of factor intensity levels (Fig. 1A: panels 6 & 9). The line in scanning diagram was arbitrarily drawn and represents a cross-section of the immunostained nucleus. In contrast, at 1.5 hour and later, most of the γH2AX and pKAP1-s824 foci completely co-localize (Fig. 1A: panels 12, 15, & 18). Nine hours post-IR, repair appears to be complete as only a few damage sites are seen, similar in number to those in undamaged proliferating cells (Fig. 1A: panel 3 & 21).
Figure 1.
A) Immunohistochemical detection of pKAP1-s824 and γH2AX-s139 in untreated and irradiated U2OS cells. B) Immunohistochemical detection of pKAP1-s824 and HP1α in irradiated U2OS cells. Treated cells were irradiated with 9 Gray IR. Line scans represent the colocalization of proteins within each image. Scale bars (μM, in white).
Since HP1α-rich heterochromatic regions do not have a distinct staining pattern in human cells (Fig. 1B), we examined γH2AX and pKAP1-s824 in mouse embryonic fibroblasts (MEFs) in which HP1α enrichments can be clearly seen at chromocenters (20). 6 hours after irradiation, γH2AX co-localizes in foci with both HP1α and pKAP1-s824 strongly suggesting that at the later time points of the DNA damage response γH2AX and pKAP1-s824 are enriched in heterochromatin (Fig. S1A and B;).
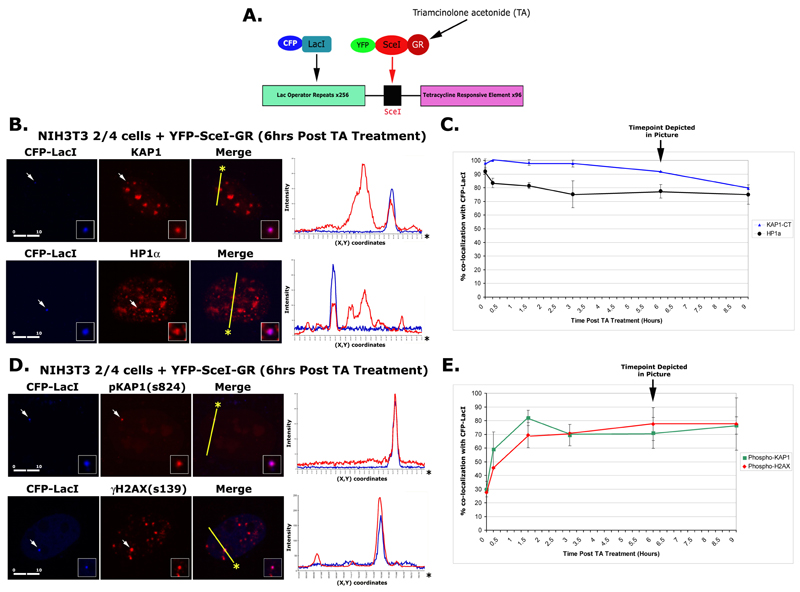
To define KAP1 and HP1 dynamics at a region of heterochromatin during DNA damage, we utilized a murine cell line, which contains a stably integrated multi-copy array of the L-SceI-T transgene integrated at a single locus (30) (Fig. 2A). The transgene contains a single SceI restriction site flanked by arrays of lac operator and Tetracycline Response Element repeats. The transgene array can be visualized by expressing CFP-lac repressor. A single double strand break site can be induced in the array by expressing a YFP-tagged SceI endonuclease-glucocorticoid receptor fusion protein (YFP-SceI-GR). Treatment of cells with triamcinolone acetonide (TA) triggers its translocation into the nucleus and cleavage of the SceI site. Since SceI sequences are not present in murine cells, DNA breaks only occur at the transgene array, which produces a localized site of DNA damage (30). Studies of this and other transgene arrays have shown that they are highly condensed and enriched in heterochromatic modifications and binding proteins, including histone H3 tri-meK9 and HP1, presumably due to mechanisms of repeat induced silencing (26). Thus, this SceI transgene array is an ideal model for understanding DNA repair dynamics in heterochromatin.
Figure 2.
A) Model depicting the L-ISceI-T transgene found in NIH3T3 2/4 cells. B) Immunofluorescent Confocal microscopy of NIH3T3 2/4 cells co-transfected with plasmids expressing CFP-LacI and YFP-SceI-GR and grown in the presence of TA using antibodies specific for KAP1 or HP1α (B) and pKAP1-s824 or γH2AX-s139 (D). Line scans represent the colocalization of proteins within each image. Scale bars (μM, in white). Line graphs depict the number of CFP-LacI foci that also exhibited positive KAP1 or HP1α (C) staining and pKAP1 or γH2AX staining (E) in transfected NIH3T3 2/4 cells grown in the presence or absence of TA.
Prior to damage, endogenous KAP1 and HP1α are enriched at the array. Both proteins are also enriched and co-localize at chromocenters which are easily identifiable in murine cells (Fig. 2B and S1C and D). Taken together, this indicates that the transgene array is a faithful model for endogenous heterochromatin and that KAP1 and HP1α are enriched at the array prior to the induction of damage.
To define the dynamics of KAP1 and HP1α in response to double strand breaks (DSBs) at the transgene array, we treated the cells with TA, which translocates the SceI-GR endonuclease into the nucleus so that it can target the SceI site. Introduction of a small number of highly localized DSBs is capable of activating the DNA damage response resulting in the phosphorylation of H2AX and KAP1 at this site (Fig. 2B). The number of cells with KAP1 and HP1α at the transgene array was counted to determine whether they were lost during the damage response. In the absence of damage, KAP1 and HP1α were detected at 98% and 91% of the arrays, respectively. After 9 hours, only ~80% contained KAP1 and HP1 (Fig. 2C). Thus, both were present at both early and late times in DNA repair response.
We next examined the kinetics of KAP1 and H2AX phosphorylation following SceI cleavage of the transgene. Phosphorylated KAP1 and H2AX were enriched on the array (Fig. 2D). In the absence of damage, both pKAP1-s824 and γH2AX were enriched at 28% of the arrays, which defines their background levels in this system. Upon addition of TA to the SceI-GR expressing cells, the number of loci enriched for pKAP1-s824 peaked by 1.5 hrs at ~82% and remained at ~70% for the rest of the time course. Phosphorylated H2AX levels also rapidly increased at the array after damage and persisted with kinetics similar to pKAP1-s824 (Fig. 2E). Taken together, these data indicate that KAP1 and H2AX are rapidly phosphorylated in response to the induction of SceI-mediated DSBs in heterochromatin. As total KAP1 is enriched at the site prior to damage, this also suggests that it is the heterochromatin-associated population, which is phosphorylated in response to damage.
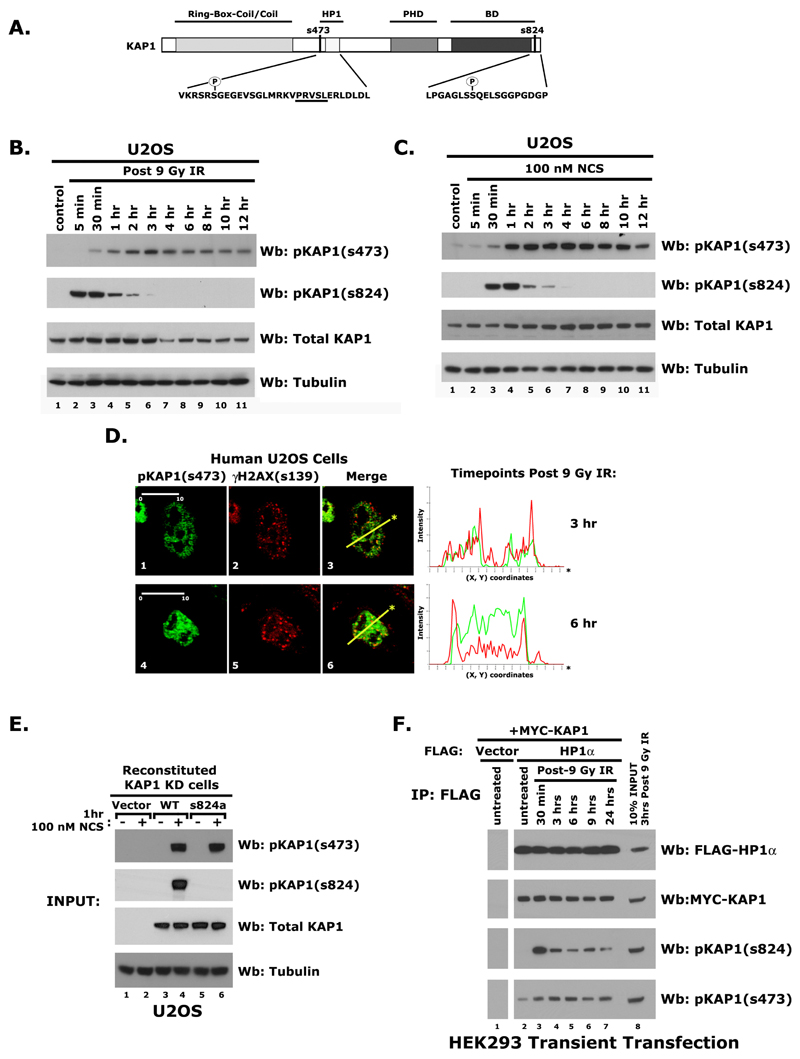
We next used a phosphomimetic mutant of KAP1, S473E, exhibited diminished HP1 association in vitro suggesting that the phosphorylation of KAP1 at Ser 473 may disrupt its interaction with HP1 (25). To determine whether this KAP1 modification plays a role in regulating HP1-KAP1 association in damaged chromatin, we examined Ser 473 phosphorylation in U2OS cells following irradiation or 100 nM Neocarzinostatin (NCS; a radiomimetic) treatment. High levels of pKAP1-s824 were prevalent within the first hour of exposure. In contrast, phosphorylation of KAP1 at Ser 473 peaked after 2 to 3 hrs of treatment with either IR or NCS (Fig. 3B-C) and was not ATM or DNA-PK dependent (Fig. S2A).
Figure 3.
A) Model depicting the Ser 473 and Ser 824 phosphorylation sites on KAP1. The PxVxL HP1 binding motif is underlined. Western blot of proteins isolated from U2OS cells at various timepoints following exposure to either 9 Gy IR (B) or 100 nM NCS (C) using antibodies specific for pKAP1-s473, pKAP1-s824, KAP1, and Tubulin. D) Immunohistochemical detection of pKAP1-s473 and γH2AX in U2OS cells following IR. Line scans represent the colocalization of proteins within each image. Scale bars (μM, in white). E) Reconstituted U2OS KAP1 Knockdown cells grown in the presence or absence of 100 nM NCS using antibodies specific for pKAP1-s473, pKAP1-s824, KAP1, and Tubulin: western blot. F) Immunoprecipitation of FLAG-HP1α co-expressed with a Myc-KAP1 in HEK293 cells. The levels of FLAG-HP1α, Myc-KAP1, pKAP1-s824, and pKAP1-s473 were examined via immunoblotting.
Since pKAP1-s824 accumulates specifically at sites of DNA damage, we next sought to determine whether pKAP1-s473 also co-localized with γH2AX in damage foci. While pKAP1-s473 was undetectable in untreated U2OS cells, it was detected after irradiation, but its pattern was diffuse (Fig. S2B and 3D). It was also not detected at the break site in NIH3T3 2/4 cells (Fig. S2C). These data suggest that although KAP1-s473 increases in response to DNA damage, only pKAP1-s824 is enriched at damage foci.
Since Ser 824 phosphorylation precedes Ser 473 phosphorylation in the damage response, we next examined whether pKAP1-s473 was dependent on Ser 824 phosphorylation. To address this question, we used a U2OS cell line stably expressing a KAP1 shRNA to deplete endogenous KAP1 protein (Fig. 3E and 4B). These cells were reconstituted with a FLAG-tagged wild type KAP1 or the KAP1-SS823/824AA mutant using shRNA-resistant cDNAs. Both serines were mutated to alanine in the KAP1-SS823/824AA construct to avoid the possibility that Ser 823 becomes phosphorylated when Ser 824 is mutated (Fig. 3E). As pKAP1-s473 was detected on immunoprecipitates of KAP1-SS823/824AA, this indicates that these events are not interdependent (Fig. 3F).
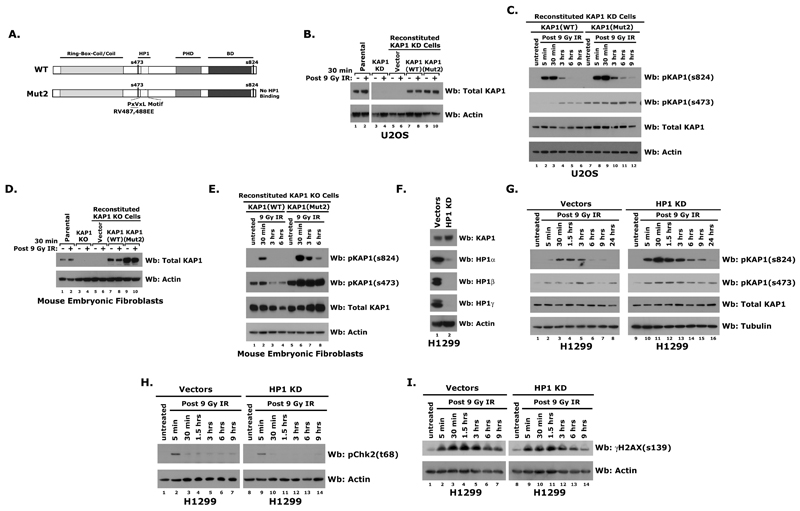
Figure 4.
A) Model depicting full length wild-type and Mut2 mutant KAP1. Western blots of proteins isolated from: (B-C) U2OS, U2OS KAP1 knockdown, and reconstituted knockdown cells post-9 Gy IR using pKAP1-s824, pKAP1-s473, KAP1, and Actin abs. (D-E) parental MEFs, KAP1 KO MEFs, and reconstituted KO MEFs post-IR using pKAP1-s824, pKAP1-s473, KAP1, and Actin antibodies. (F) Western blots of proteins isolated from H1299 Vector or HP1 knockdown cells using for KAP1, HP1α, HP1β, HP1γ, and Actin antibodies. Western blots of proteins isolated from H1299 Vector or HP1 knockdown cells post-9 Gy IR using antibodies specific for pKAP1-s824, pKAP1-s473, KAP1, and Tubulin (G), or pChk2-t68 and Actin (H), or γH2AX-s139 and Actin (I).
We directly tested the effects of Ser 473 phosphorylation on HP1 interaction in vitro. First, we generated biotinylated peptides encompassing the HP1 binding domain (HP1BD) of KAP1 with or without a phosphorylated Serine at position 473 (Fig. S2D). Then we tested whether the presence of a phosphoserine at 473 affected the ability of GST tagged HP1 isoforms to precipitate HP1BD peptides in vitro. Western blot analysis of the resulting GST-HP1 pull down assay revealed little difference in the ability of HP1 to associate with HP1BD peptides with or without the phosphorylated Ser 473 residue (Fig. S2E).
Next, we asked whether HP1 associates with pKAP1 in vivo. Similar to U2OS cells, HEK293 cells exhibit a temporal difference in Ser 473 and 824 phosphorylation (Fig. 3B and S2F). Following co-transfection and immunoprecipitation after damage, Western blot analyses revealed that HP1α, β, and γ robustly associate with KAP1 phosphorylated at Ser 473 and Ser 824 (Fig. 3F and SG-H). Taken together, these data suggest that the HP1-KAP1 association is not affected by phosphorylation at these sites.
Overwhelming biochemical and immunocytological data indicate that KAP1 and HP1 exist as a complex, which plays a critical role in organizing higher order heterochromatin architecture and regulating gene silencing (11, 19, 20, 31). Therefore, we asked if the HP1-KAP1 interaction also regulates Ser 473 and 824 phosphorylation following DNA damage. To do this we used a KAP1 construct (KAP1-Mut2) with a mutation in the HP1-binding motif (RV487,488EE) which abolishes the HP1 interaction (Fig. 4A). KAP1-Mut2 does not co-localize with HP1 at pericentric heterochromatin and shows reduced transcriptional repressor activity (18). As expected, KAP1-Mut2 does not co-immunoprecipitate with HP1α in untreated or irradiated HEK293 cells (Fig. S3A).
To determine whether the HP1 interaction regulates KAP1 phosphorylation during damage, we stably depleted KAP1 in U2OS cells using short hairpins (shRNAs) and then reconstituted with shRNA-resistant wild-type KAP1 or the KAP1-Mut2 mutant (Fig. 4B). Wild-type KAP1 was efficiently phosphorylated in response to IR. Interestingly, KAP1-Mut2 was also rapidly phosphorylated and the levels of Ser 473 and Ser 824 were higher and persisted longer indicating that the interaction with the HP1s regulates it (Fig. 4C).
KAP1 was previously shown to be phosphorylated at serine 473 during the G1/S boundary (25). To determine whether the differences in pKAP1-s473 of KAP1-WT and Mut2 were due to alterations in the cell cycle, the cell cycle profiles of reconstituted U2OS KAP1 knockdown cells were examined using flow cytometry. As differences were not detected before or after irradiation (Fig. S3B), this suggests that the increase in KAP1-Mut2 phosphorylation was not due to cell cycle changes but to changes in the interaction with HP1.
To further explore this, we used mouse embryonic fibroblast (MEF) KAP1 knockout cells. The KAP1 knockout MEFs were infected with lentiviruses expressing KAP1-WT, KAP1-Mut2 and vector alone resulting in levels of KAP1 similar to endogenous expression (Fig. 4D). Similar to the U2OS cells, KAP1-Mut2 was more efficiently phosphorylated at both Ser 473 and Ser 824 in the KO MEFs (Fig. 4E). Together, these results suggest that the interaction of HP1 with KAP1 regulates its DNA damage-dependent phosphorylation. To further evaluate the role of HP1 in KAP1 Ser 473 and 824 phosphorylation, we used H1299 cells in which all three HP1 isoforms (α, β, γ) have been stably knocked down (Fig. 4F). In the HP1 knockdown H1299 cells, pKAP1-s 473 and s824 levels increase more rapidly and persist over the course of the damage response (Fig. 4G). Thus, the data from the three separate systems suggest that the HP1-KAP1 association regulates Serines 473 and 824 phosphorylation during the DNA damage response.
Like KAP1, H2AX and Chk2 are also phosphorylated by ATM in response to DNA damage (32, 33). Since HP1 affects KAP1 phosphorylation, we examined whether Chk2 and H2AX phosphorylation levels also increase in the absence of HP1. As Chk2 and H2AX phosphorylation were not enhanced in HP1 knockdown cells (Fig. 4H-I), this suggests that HP1 does not regulate the phosphorylation of all ATM substrates.
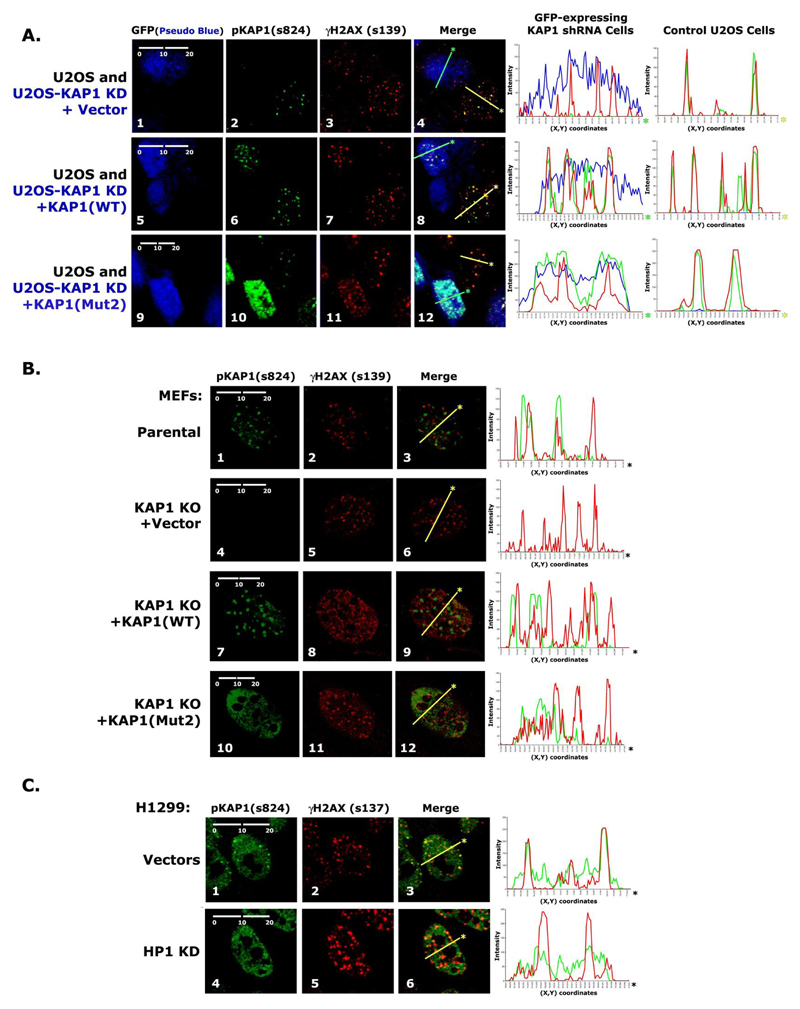
In a previous report as well as in this study, we showed that pKAP1-s824 co-localizes with γH2AX in discreet foci in response to DNA damage (22). Therefore, we next wanted to investigate the role of HP1 in regulating this organizational pattern. To do this, we used the U2OS KAP1 knockdown cells (Fig. 3E, and 4B-C) generated by stably expressing KAP1 shRNA as well as GFP. GFP-positive KAP1 knockdown cells were mixed in a 50:50 ratio with parental U2OS cells, which served as an internal control (Fig. 5A). In parental U2OS cells, pKAP1-s824 and γH2AX completely co-localized 3 hrs after IR-treatment. After KAP1 knockdown, γH2AX foci still form, but the pKAP1 signal is no longer detected (Fig. S3C and 5A: panel 4). In cells reconstituted with KAP1-WT, pKAP1-s824 was again seen in foci, which co-localized with γH2AX (Fig. 5A, panel 8). However, in KAP1-Mut2 reconstituted cells, pKAP1-s824 was both diffuse and more intense (Fig. 5A, panel 12). The increased signal intensity is consistent with the higher levels of Ser 824 phosphorylation detected in Mut2-reconstituted U2OS knockdown cells (Fig. 4C). The pKAP1-s824 pattern was also consistent with the MEF knockout cells reconstituted with KAP1-Mut2 (Fig. 5B, panel 12). These results indicate that the interaction of KAP1 with HP1 regulates the organization of pKAP1-s824 into discreet foci after DNA damage.
Figure 5.
A) Detection of pKAP1-s824 and γH2AX-s139 in U2OS cells mixed with reconstituted KAP1 knockdown cells 3 hrs post-9 Gy IR. B) Detection of pKAP1-s824 and γH2AX-s139 in parental MEFs, KAP1 KO MEFs, and reconstituted KAP1 KO MEFs 3 hrs post-9 Gy IR. C) Detection of pKAP1-s824 and γH2AX-s139 in H1299 Vectors and HP1 knockdown cells 3 hrs post-9 Gy IR.
To evaluate whether KAP1-Mut2 was able to accumulate at sites of damage, we knocked KAP1 down in NIH3T3 2/4 cells and reconstituted them with FLAG-tagged KAP1-WT or Mut2 (Fig. S3D). The percentage of NIH3T3 2/4 cells with pKAP1-s824 at the transgene was highest 1.5 hrs following SceI-cleavage (Fig. 2E). At this timepoint KAP1-WT was enriched at the cleaved transgene, while KAP1-Mut2 was not (Fig. S3E). These data suggest that DNA breaks residing in heterochromatin are not high affinity binding sites that enable KAP1 to accumulate in the absence of HP1 association.
To further confirm that KAP1 requires HP1 to form damage foci, we examined pKAP1-s824 foci formation in IR-treated H1299 HP1 knockdown cells (Fig. 5C). In parental cells, pKAP1-s824 and γH2AX co-localize in discreet foci (Fig. 5C, panel 3). In contrast, pKAP1 staining is diffuse in HP1 knockdown cells (Fig. 5C, panel 4). Taken together, these data support the hypothesis that HP1 regulates the nuclear organization of Ser 824 phosphorylation following DNA damage.
We next used cellular fractionation to determine the location of KAP1 before and after DNA damage. Slightly higher levels of KAP1-Mut2 were observed in the chromatin fraction (Nuclear Pellet, NP) of reconstituted MEF KAP1 knockout and U2OS KAP1 knockdown cells (Fig. 6A and data not shown). In addition, a slightly higher level of KAP1 was found in the chromatin fraction isolated from HP1 knockdown cells (Fig. 6B). These data revealed that HP1 is not required for the retention of KAP1 in detergent insoluble chromatin. In irradiated samples, DNA damage did not significantly affect the ability of KAP1 or HP1 to associate with chromatin (Fig. 6A-B). If HP1 association was required for KAP1 to be retained in chromatin following DNA damage, then an increase in soluble (Nuclear Extract, NE) pKAP1 would be evident. In all three cell systems used in this study, equivalent levels of phosphorylated KAP1 were observed in the NP fraction from irradiated cells (Fig. 6A-B and data not shown). These data suggest that HP1 association is not required for KAP1 to be anchored in chromatin in the presence or absence of 9 Gray IR.
We next sought to determine the effect of disrupting the HP1-KAP1 interaction on DNA repair. Formation of γH2AX foci is often used to identify sites of DNA damage (34) and the kinetics of their formation and resolution is a measure of DNA repair efficiency (24, 35). Therefore, we used quantitative image analysis to compare γH2AX staining in cells where the HP1-KAP1 interaction was disrupted by depletion of HP1, depletion of KAP1, or reconstitution with a HP1 binding mutant. At 30 min post-IR in H1299 cells without HP1, the γH2AX signal increased 2-fold over the vector control (p=>0.0001). At 9 hr post-IR the γH2AX signal intensity in HP1 knockdown cells was 50% less than in the vectors control (p=0.0079). These data suggest that DSBs in are repaired more rapidly when HP1 is not present (Fig. 6C). Similarly, U2OS KAP1 knockdown cells reconstituted with the vector (p=0.0005) or the KAP1-Mut2 mutant (p=0.0054) exhibited increased γH2AX staining at 5 minutes post-irradiation compared to the parental cells. These cells also displayed a 30% reduction in γH2AX staining 9 hours after damage (Vector p=0.0001; KAP1-Mut2 p=0.0226). Throughout the time course, the γH2AX signal intensity in the KAP1-WT reconstituted cells resembled the parental U2OS cells (Fig. 6D). Taken together, these data suggest that disruption of the HP1-KAP1 interaction promotes the rapid formation and resolution of γH2AX foci in response to DNA damage suggesting that repair is more efficient.
Discussion
The KAP1 co-repressor has turned out to be an interesting probe for defining mechanistic interfaces between gene silencing, heterochromatin and DNA repair. It was previously reported to be phosphorylated at Ser 824 by PIKK-family kinases, like ATM, in response to DNA damage suggesting that it is at the interface of these pathways (22, 23). In addition to the Ser 824 site, KAP1 is also phosphorylated at Ser 473 at the G1-S phase boundary, which may disrupt its ability to associate with HP1 (25). Here we report that KAP1 is phosphorylated at Ser 473 in response to DNA damage. While Ser 824 phosphorylation is rapidly induced within the first 30 minutes, phosphorylation of KAP1 at Ser 473 only becomes detectable ~1 hour following damage. In addition, Ser 824 phosphorylation is localized to sites of double stranded breaks, while pKAP1-s473 staining is diffuse.
Sustained phosphorylation of pKAP1-s824 at DNA breaks in heterochromatin is maintained by the 53BP1-dependent hyperaccumulation of MRN complex that facilitates the retention of activated ATM (4). In this context, KAP1 phosphorylation serves as a marker for sustained ATM activity at an unrepaired site of damage. Here we show that disruption of KAP1’s organization in chromatin by diminishing HP1 expression or mutating the HP1BD on KAP1 interferes with the ability of pKAP1-s824 to accumulate at sites of DNA damage. Confinement of this modification to DNA breaks seems to be unique to this site, since the pattern of damage-induced phosphorylation at Ser 473 is diffuse.
Initially, the diffuse staining pattern of pKAP1-s824 in both the KAP1-Mut2 reconstituted cells and the HP1 knockdown cells suggested that KAP1 was not chromatin bound in the absence of the HP1 interaction. Since fixation is required to look at the modification, this could not be tested using live cell techniques. Instead, we used cellular fractionation to evaluate its association with chromatin. Surprisingly, these results showed that even in the absence of HP1, KAP1 is chromatin bound. When the extraction was performed under high salt conditions (500 mM NaCl) KAP1, but not HP1 or H2AX, became extractable. No difference in its extractability was observed in the presence or absence of HP1 (Fig. S4). These data suggest that KAP1 is not an integral chromatin component like H2AX and HP1, but that it is associates with chromatin in an HP1-independent manner. Therefore, HP1 is not the only factor retaining it in chromatin. Consistent with this, KAP1 and pKAP1 can associate with the histone subunits H2B, H3.1, and H2AX in addition to chromatin components like 53BP1 and KRAB-ZFPs, like ZNF317 in vivo (Fig. S5A-E).
Previous reports suggest that KAP1 is released from chromatin in response to DNA damage, which results in chromatin decondensation and increased repair factors accessibility (23). Here, we show that KAP1 is present at a region of heterochromatin prior to damage and that pKAP1-s824 foci form rapidly at the site upon the induction of DSBs. Additionally, our results indicate that KAP1 is not depleted from heterochromatin during the damage response suggesting that the population of phosphorylated KAP1 present at break sites is not recruited but present before damage. This is similar to H2AX, which is also an inherent component of chromatin. Damage induced phosphorylation of H2AX at Ser 139 by ATM spreads for kilo- to megabases on either side of DNA break sites (36, 37). Our data suggests that KAP1, like H2AX, is not recruited to sites of damage, but that resident proteins near a break are coordinately targeted for phosphorylation.
In this study, we also report a new role for HP1 in regulating the phosphorylation of KAP1 at Serines 473 and 824. While neither modification affects the ability of KAP1 to associate with HP1 or the chromatin infrastructure, interaction of KAP1 with HP1 either regulates its initial phosphorylation or its dephosphorylation.
Beyond increased phosphorylation, disruption of HP1-KAP1 association promoted a rapid formation and resolution of γH2AX foci (shown by increased γH2AX staining at early timepoints post-IR and by decreases in the level of γH2AX staining at the 9 hr timepoint). The temporal shifts in the level of γH2AX signal intensity occurred when HP1-KAP1 interaction was ablated in three different ways: stable knockdown of HP1, stable knockdown of KAP1, and the expression of the HP1-binding mutant of KAP1. The efficiency of γH2AX foci resolution in the absence of the KAP1-HP1 interaction is consistent with work by Goodarzi and colleagues (24). Here, our data indicate that HP1 restricts pKAP1-s824 to damage foci and suggest that higher levels of pKAP1-s824 and –s473 as well as the spread of pKAP1-s824 beyond γH2AX foci enhance the efficiency of γH2AX foci resolution in cells lacking the HP1-KAP1 interaction.
Signaling by damage response pathways to KAP1 Ser 824 seems to be one critical step for effective DNA repair in heterochromatin. Currently, KAP1 is thought to promote gene silencing in euchromatin and to maintain chromatin compaction in heterochromatin by condensing nucleosomes. The Ser 824 phosphorylation site is located in the extreme carboxyl terminus where KAP1 associates with SETDB1 and Mi2α/CHD3. Reconstitution experiments with a S824A mutant support the hypothesis that the decondensation of global chromatin in response to stress may result from Ser 824 phosphorylation which disrupts the ability of KAP1 to associate with this silencing machinery (4, 23, 24). However, the gene silencing activity of KAP1 does not diminish in irradiated cells (Fig. S5F). Moreover, pKAP1-Ser 473 and Ser 824 can still associate with SETDB1 in vivo following damage (Fig. S5G).
These studies begin to provide a portrait of the differences in the repair of DNA in euchromatin and heterochromatin. The bimodal repair kinetics of DSBs observed in euchromatin and heterochromatin make it likely that the outcome of ATM signaling on the regulation of these factors differs according to the transcriptional permissiveness of the chromatin in which they reside.
Supplementary Material
Acknowledgements
We thank Tom Misteli (NIH) for providing the NIH3T3 2/4 cells. We thank David C. Schultz (The Wistar Institute) and Louise Showe (The Wistar Institute) for comments and support during this study.
1. Financial Support:
Wistar Institute Cancer Center, NIH/NCI grants CA126283, CA129833, CA010815, and the Samuel Waxman Cancer Research Foundation.
Grant Support
This work was supported by, a pilot grant from the Wistar Institute Cancer Center, NIH/NCI grants CA126283, CA129833, CA010815, and the Samuel Waxman Cancer Research Foundation. We acknowledge the National Cancer Institute–supported Wistar Institute Cancer Center Shared Facilities: Genomics, Imaging, Flow Cytometry, Proteomics, and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
References
- 1.Yang J, Yu Y, Hamrick HE, Duerksen-Hughes PJ. ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinogenesis. 2003;24:1571–1580. doi: 10.1093/carcin/bgg137. [DOI] [PubMed] [Google Scholar]
- 2.Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. Embo J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 4.Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- 5.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 7.Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ. 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 8.Lechner MS, Begg GE, Speicher DW, Rauscher FJ. 3rd Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol Cell Biol. 2000;20:6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz DC, Friedman JR, Rauscher FJ. 3rd Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, Tanaka K, Torigoe K, Rauscher FJ. 3rd Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. Embo J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 14.Minc E, Courvalin JC, Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- 15.Kiyomitsu ET, Iwasaki O, Obuse C, Yanagida M. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J Cell Biol. 2010;188:791–807. doi: 10.1083/jcb.200908096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol Cell Biol. 2002;22:3204–3218. doi: 10.1128/MCB.22.9.3204-3218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cammas F, Herzog M, Lerouge T, Chambon P, Losson R. Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev. 2004;18:2147–2160. doi: 10.1101/gad.302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cammas F, Oulad-Abdelghani M, Vonesch JL, Huss-Garcia Y, Chambon P, Losson R. Cell differentiation induces TIF1beta association with centromeric heterochromatin via an HP1 interaction. J Cell Sci. 2002;115:3439–3448. doi: 10.1242/jcs.115.17.3439. [DOI] [PubMed] [Google Scholar]
- 21.Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, Nussenzweig M. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009;34:285–297. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White DE, Negorev D, Peng H, Ivanov AV, Maul GG, Rauscher FJ. 3rd KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 2006;66:11594–11599. doi: 10.1158/0008-5472.CAN-06-4138. [DOI] [PubMed] [Google Scholar]
- 23.Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 24.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Chang CW, Chou HY, Lin YS, Huang KH, Chang CJ, Hsu TC, Lee SC. Phosphorylation at Ser473 regulates heterochromatin protein 1 binding and corepressor function of TIF1beta/KAP1. BMC Mol Biol. 2008;9:61. doi: 10.1186/1471-2199-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, Sadofsky MJ, et al. 3rd PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klenova E, Chernukhin I, Inoue T, Shamsuddin S, Norton J. Immunoprecipitation techniques for the analysis of transcription factor complexes. Methods. 2002;26:254–259. doi: 10.1016/S1046-2023(02)00029-4. [DOI] [PubMed] [Google Scholar]
- 29.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ. 3rd KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 33.Zhou BB, Chaturvedi P, Spring K, Scott SP, Johanson RA, Mishra R, Mattern MR, Winkler JD, Khanna KK. Caffeine abolishes the mammalian G(2)/M DNA damage checkpoint by inhibiting ataxia-telangiectasia-mutated kinase activity. J Biol Chem. 2000;275:10342–10348. doi: 10.1074/jbc.275.14.10342. [DOI] [PubMed] [Google Scholar]
- 34.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahrhofer H, Burger S, Oppitz U, Flentje M, Djuzenova CS. Radiation induced DNA damage and damage repair in human tumor and fibroblast cell lines assessed by histone H2AX phosphorylation. Int J Radiat Oncol Biol Phys. 2006;64:573–580. doi: 10.1016/j.ijrobp.2005.09.037. [DOI] [PubMed] [Google Scholar]
- 36.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bewersdorf J, Bennett BT, Knight KL. H2AX chromatin structures and their response to DNA damage revealed by 4Pi microscopy. Proc Natl Acad Sci U S A. 2006;103:18137–18142. doi: 10.1073/pnas.0608709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.