Abstract
Cytosine modification on DNA is variable among individuals, which could correlate with gene expression variation. The effect of cytosine modification on interindividual transcript isoform variation (TIV), however, remains unclear. In this study, we assessed the extent of cytosine modification-specific TIV in lymphoblastoid cell lines (LCLs) derived from unrelated individuals of European and African descent. Our study detected cytosine modification-specific TIVs for 17% of the analyzed genes at a 5% false discovery rate. Forty-five percent of the TIV-associated cytosine modifications correlated with the overall gene expression levels as well, with the corresponding CpG sites overrepresented in transcript initiation sites, transcription factor binding sites, and distinct histone modification peaks, suggesting that alternative isoform transcription underlies the TIVs. Our analysis also revealed 33% of the TIV-associated cytosine modifications that affected specific exons, with the corresponding CpG sites overrepresented in exon/intron junctions, splicing branching points, and transcript termination sites, implying that the TIVs are attributable to alternative splicing or transcription termination. Genetic and epigenetic regulation of TIV shared target preference but exerted independent effects on 61% of the common exon targets. Cytosine modification-specific TIVs detected from LCLs were differentially enriched in those detected from various tissues in The Cancer Genome Atlas, indicating their developmental dependency. Genes containing cytosine modification-specific TIVs were enriched in pathways of cancers and metabolic disorders. Our study demonstrated a prominent effect of cytosine modification variation on the transcript isoform spectrum over gross transcript abundance and revealed epigenetic contributions to diseases that were mediated through cytosine modification-specific TIV.
Keywords: DNA methylation, cytosine modification, alternative splicing, transcript isoform, lymphoblastoid cell line
CYTOSINE modification occurs at more than half of the CpG sites in the human genome (Ehrlich et al. 1982) and plays important roles in genome stability, cell lineage progression, and disease etiology (Feinberg and Tycko 2004). Recent genome-wide profiling in humans using gene-centered microarray platforms revealed abundant interindividual variations in cytosine modification (Heyn et al. 2013; Moen et al. 2013), a significant proportion of which were found to be associated with gene expression variation (Zhang et al. 2014). Although a causal relationship between cytosine modification and gene expression variation could not be readily established based on genomic correlation alone (Gutierrez-Arcelus et al. 2013), experimental studies of individual genes did demonstrate downstream effects of cytosine modification on gene transcription (Razin and Cedar 1991; Hmadcha et al. 1999; Rishi et al. 2010). Accumulating experimental evidence also indicated that cytosine modification-dependent gene regulation may execute on distinct phases of transcription and may coordinate with other epigenetic mechanisms (Yang et al. 2011; Rao et al. 2014).
Almost all multiexon human genes possess alternative transcript isoforms (Pan et al. 2008; Wang et al. 2008) that are under strong developmental regulation (Wang et al. 2008). Many of these transcript variants play distinct roles through biological regulation and functions (Muller et al. 2006; Pruunsild et al. 2007), abnormalities in which are frequently associated with diseases and cancers (Venables 2004; Tomasini et al. 2008; Dutertre et al. 2010). Alternative transcript isoforms can result from alternative transcription initiation, as well as alternative splicing and transcription termination. Splicing and transcription are intrinsically linked processes, which are demonstrated by the interactions of splicing machineries with RNA polymerase II (Pol II) (Beyer and Osheim 1988) and the discovery of splicing variants that depend on gene transcription (Cramer et al. 1997; Allemand et al. 2008; Sanchez et al. 2008). Recruitment of splicing factors to the Pol II complex can modulate splicing, whereas the kinetics of transcription elongation can affect the selection of competing splice sites (de la Mata et al. 2003; de la Mata and Kornblihtt 2006; Ip et al. 2011).
The relationship between cytosine modification and transcript isoform variation (TIV) is largely unknown. In this study we assessed cytosine modification-specific TIV in lymphoblastoid cell lines (LCLs) derived from two global populations. Our study demonstrated a prominent effect of cytosine modification on TIV, primarily through alternative isoform transcription and secondarily through alternative splicing. Our study uncovered the relative independence between genetic and epigenetic regulation on TIV and revealed the epigenetic contributions to disease etiology mediated through cytosine modification-specific TIV.
Materials and Methods
Cell lines, cytosine modification data processing, and validation
The raw cytosine modification data were downloaded from the NCBI Gene Expression Omnibus (GEO) (GEO accession no. GSE39672). Cell line preparation, DNA extraction, array hybridization, and related quality control procedures have been described in our previous publication (Moen et al. 2013). Briefly, genomic DNA samples for 60 unrelated Caucasian residents of Utah (CEU) (phase II) and 73 unrelated Yoruba people from Ibadan, Nigeria (YRI) (58 phase II plus 15 phase III samples) HapMap LCLs were purchased from Coriell Institute for Medical Research (Camden, NJ). The cell line identities were confirmed by genotyping 47 SNPs from the Sequenom iPLEX Sample ID Plus Panel in 24 randomly chosen LCLs maintained by the Pharmacogenetics of Anticancer Agents Research Group Cell Core at The University of Chicago.
Cytosine modification levels were then profiled with the Illumina HumanMethylation450 BeadChip (450K array) (Illumina, San Diego) (Moen et al. 2013). At least 150 ng DNA after bisulfite conversion was obtained for each sample, randomized by population identity, and run on the 450K array plates with the Illumina HiScan System. We excluded CpG probes ambiguously mapped to the human genome (Zhang et al. 2012) and CpG probes containing common single-nucleotide polymorphisms (SNPs) with minor allele frequency (MAF) >0.01 based on dbSNP (Sherry et al. 2001) v135. The final data set is composed of 283,540 autosomal CpGs with good hybridization quality (Moen et al. 2013). M values, defined as the log2 ratio of the intensities of modified probe vs. unmodified probe, were quantile normalized across 133 samples and adjusted for batch effect (Johnson et al. 2007). Cytosine modification levels at selected CpGs were validated by bisulfate sequencing (Moen et al. 2013).
Gene expression data
The Affymetrix Human Exon Array 1.0ST was previously used to profile gene expression in the CEU and YRI samples (GEO accession no. GSE9703) (Zhang et al. 2009). Probe sequences were aligned to the human genome (GRCh37), allowing ≤1 mismatch. Probes with perfect, unique alignments were further filtered by excluding probes containing common SNPs (MAF ≥ 0.05) based on dbSNP (Sherry et al. 2001) v135. Flattened gene models were built based on Gencode release 19 (Harrow et al. 2012). Based on the gene models, probes mapped to intergenic regions, introns, and exon/exon and exon/intron junctions were removed. Probes interrogating multiple genes were also removed. Probe intensities were log2 transformed, background corrected (Zhang et al. 2008), and quantile normalized. Probe intensity was subtracted by the corresponding probe mean across samples. Gene-level and exon-level expression intensities were summarized as mean probe intensity within genes and within exons, respectively. The expression levels of several selected genes that correlated with cytosine modification levels have been validated in the LCLs, using quantitative RT-PCR (qRT-PCR) (Moen et al. 2013).
Detection of cytosine modification-specific TIV
To exclude genes not expressed in LCLs, genes for which >90% of the samples had absolute gene expression levels less than the 10% quantile of all gene expression levels were excluded from analysis. To exclude exons not expressed in LCLs, exons for which >95% of the samples had absolute exon expression levels less than the 20% quantile of all exon expression levels were excluded. The remaining exons interrogated by ≥2 probes, located within protein-coding genes with ≥2 exons, were analyzed. Within each gene, exon expression levels of exon for sample were modeled by a linear mixed-effects model:
| (1) |
Here y denotes length column vector of expression levels across I exons and J samples. denotes identity matrix for I exons. denotes length I column vector of exon main effects. denotes length N column vector of cytosine modification levels for J samples at a given CpG. denotes cytosine modification main effect. Z denotes identity matrix for J samples. γ denotes length J column vector of sample random effects. ε denotes length N vector of random errors. We tested exon-by-cytosine modification interaction effect, one at a time, by comparing the following model,
| (2) |
with model (1). Here denotes length N column vector of identity for the lth exon. denotes the interaction effect between the lth exon and cytosine modification at the CpG. P-values were obtained by a likelihood-ratio test with 1 d.f. Multiple comparisons were adjusted across all tests on exon-by-cytosine modification interaction, using the Benjamini and Yekutieli (BY) approach (Benjamini 2001). Exon-by-cytosine modification interactions with adjusted P < 0.05 were called significant. In the CEU samples, 218,463 unique exons within 15,933 genes were tested with 263,611 CpGs. In the YRI samples, 217,274 exons within 15,897 genes were tested with 263,126 CpGs.
Detection of cytosine modification-specific gene expression variation
Genes not expressed in LCLs were excluded as described above. Protein-coding genes interrogated by three or more probes were analyzed. Gene expression levels were linearly regressed on cytosine modification levels. Multiple comparisons were adjusted across all tests, using the BY approach (Benjamini 2001). CpG–gene associations with adjusted P < 0.05 were called significant. In the CEU samples, 17,232 unique genes were tested with 265,845 CpGs. In the YRI samples, 17,198 genes were tested with 265,393 CpGs.
Enrichment of the detected CpGs in proximal elements
Here we defined proximal regulatory regions based on all transcript isoforms of the protein-coding genes annotated in Gencode version 19, instead of the flattened gene models. For a given intron, a 5′-exon/intron junction region was defined as a 25-bp exon plus a 50-bp intron, a 3′-exon/intron junction region as a 50-bp intron plus a 25-bp exon, and a branching point region as a 101-bp region centered at the branching point (Mercer et al. 2015). In addition, a transcript initiation region was defined as a −50-bp to +50-bp region with a transcript initiation site at position +1, while a transcript termination region was defined as a −50-bp to +50-bp region with a transcript termination site at position −1. We then mapped to the defined proximal regions CpGs that were analyzed for cytosine modification-specific TIV and that were located <50 bp away from the genes. For each CpG we recorded whether it coincided in each of the proximal categories. Fold enrichment of TIV-associated cytosine modifications in a given proximal category was estimated as (#Tinside/#nonTinside)/(#Toutside/#nonTouside), where #T and #nonT represented the number of TIV-associated and non-TIV-associated cytosine modifications, respectively, with the subscripts denoting inside or outside of the category regions. Significance of enrichment was tested by Fisher’s exact test.
Enrichment of the detected CpGs in ENCODE peaks
We obtained uniformly processed narrow peaks for DNase hypersensitivity and transcription factor binding, both referred to as transcription factor binding peaks, and broad peaks for histone markers of cell line NA12878 from the Encyclopedia of DNA Elements ENCODE project (Encode Project Consortium 2012). Peaks for DNase hypersensitivity, each of the 90 canonical transcription factors, and histone modification markers were examined individually. We mapped all analyzed CpGs to positional bins including 5-kb bins along the upstream 100 kb from gene start sites, 10-percentile bins along gene regions, and 5-kb bins along the downstream 100 kb from gene end sites, using the flattened gene models. To estimate null distributions, we randomly sampled 10,000 times the same number of CpG–gene pairs as the true number of the CpG–gene pairs detected for cytosine modification-specific TIV, matching the positional bin distribution, and counted the number of CpGs colocalized with the peaks for the given marker at each bin. The true numbers of colocalization were then compared with the 95-percentile thresholds of the null distributions.
Detection of SNP-specific TIV
For the CEU and YRI samples, combined phases II and III HapMap genotypes were phased using SHAPIT2 (O’Connell et al. 2014) and imputed to the 1000 Genomes Project (1000 Genomes Project Consortium et al. 2012) phase 1 data, using IMPUTE2 (Howie et al. 2009). SNPs with imputation quality r2 > 0.8 and MAF > 0.1 within population were selected. Selection of exons for analysis is described above. The top 11 principal components estimated from the exon expression data were regressed out to adjust for hidden covariates. SNPs located <100 kb away from gene regions were associated with TIV in 58 CEU and in 59 YRI samples. Within each gene, exon-by-SNP interaction effect for the lth exon was tested using a linear regression model:
Here y denotes length column vector of expression levels of I exons in J samples. denotes identity matrix for I exons. denotes length I column vector of exon main effects. denotes length N column vector of alternative allele dosages at the SNP for J samples. denotes SNP main effect. denotes length N column vector of identity for the lth exon. denotes the interaction effect between the lth exon and the SNP genotype. Due to the large number of tests, the individual random effects included in the detection of cytosine modification-specific TIV were omitted here to speed up computation. In the CEU samples, 218,508 unique exons within 15,939 genes were tested with 3,396,831 SNPs. In the YRI samples, 217,633 exons within 15,912 genes were tested with 4,274,860 SNPs.
Detection of expression QTL and modification QTL
Expression QTL (eQTL) were tested in 58 CEU and 59 YRI samples following previous publications (Zhang et al. 2014). Genes not expressed in LCLs were removed as described above. The top 11 principal components estimated from gene expression data were regressed out to adjust for hidden covariates. SNPs with imputation quality r2 > 0.8 and MAF > 0.1 within populations and located <100 kb away from gene regions were tested for association with gene expression levels, using linear modeling.
Modification QTL (mQTL) were tested in 60 CEU and 73 YRI samples following previous publications (Zhang et al. 2014). The top five and the top two principal components estimated from the cytosine modification data were regressed out to adjust for hidden covariates for the CEU and YRI samples, respectively. SNPs with imputation quality r2 > 0.8 and MAF > 0.1 within population and located <100 kb away from the CpGs were tested for association with cytosine modification levels, using linear modeling.
Enrichment in cancer tissues of genes containing cytosine modification-specific TIV detected in LCLs
The analyzed tissues were derived from seven cancer types from The Cancer Genome Atlas (TCGA), including blood (acute myeloid leukemia), brain (lower grade glioma), liver (liver hepatocellular carcinoma), lung (lung adenocarcinoma), kidney (kidney renal clear cell carcinoma), thyroid (thyroid carcinoma), and uterus (uterine corpus endometrial carcinoma). Exon junction counts were obtained from RNA-Seq version 2 level 3 data, and methylation M values were obtained from the human methylation 450K array level 2 data, at https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm. Exon junctions were mapped to transcript isoforms annotated by Gencode release 19. Exon junctions with ≥25% of the samples having zero count were removed from analysis. Exon junction counts were log2 transformed and quantile normalized. CpGs with ≥10% of the samples having a detection P-value ≥0.01 were removed from analysis. M values were quantile normalized. CpGs located <100 kb away from target gene regions were tested for exon junction-by-cytosine modification interaction effects, using a linear regression model:
Here y denotes length column vector of log2-transformed exon junction counts of I exon junctions in J samples. denotes identity matrix for I exon junctions. denotes length I column vector of exon junction main effects. denotes length N column vector of cytosine medication levels at a given CpG for J samples. denotes cytosine modification main effect. denotes length N column vector of identity for the lth exon junction. denotes the interaction effect between the lth exon junction and cytosine modification at the CpG. We omitted the random individual effects here to simplify the tests. Multiple comparisons were adjusted using the BY approach (Benjamini 2001). Exon junction-by-cytosine modification interactions with adjusted P < 0.05 were called significant.
Supplemental Material, Table S1 and Table S2 contain cytosine modification-specific TIVs detected in the CEU and YRI samples, respectively. Table S3 contains the enrichment of TIV-associated CpGs in proximal regulatory regions. Table S4 and Table S5 contain SNP-specific TIVs detected in the CEU and YRI samples, respectively. Table S6 contains the enrichment of cytosine modification-specific TIVs detected in LCLs of the YRI samples in those cytosine modification-specific TIVs detected in cancer tissues from TCGA. Table S7 contains Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched for cytosine modification-specific TIVs in the YRI samples. Cytosine modification data are available at GEO under accession no. GSE39672. Gene expression data are available at GEO under accession no. GSE9703.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Cytosine modification-specific TIV
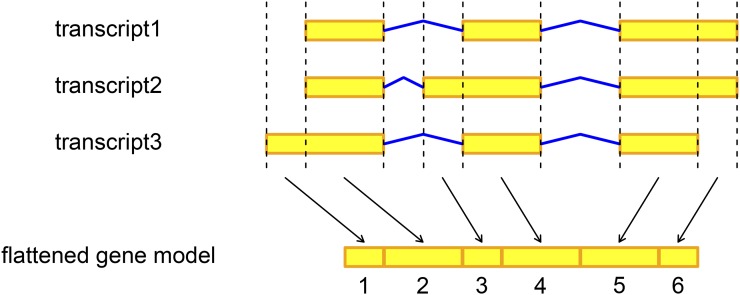
We used a flattening gene annotation approach, illustrated in Figure 1, that breaks down transcribed gene regions into a set of nonoverlapping units, referred to as exons hereafter (Zhang et al. 2008; Anders et al. 2012). We defined TIV as exon-level expression variation across individuals after accounting for gene-level expression variation. Because of the gene annotation approach, TIV detected here can potentially be attributed to a variety of molecular mechanisms, including alternative transcription, through different transcript initiation and termination sites, as well as alternative splicing in the post-transcription phase. To examine cytosine modification-specific TIV, we analyzed CpGs located <100 kb away from gene regions of 15,933 genes in 58 unrelated CEU samples. Within a gene, exon expression levels across individuals were modeled with exon main effects, cytosine modification main effect, and exon-by-cytosine modification interaction effect for the exon under consideration. The significance of cytosine modification-specific TIV, tested as exon-by-cytosine modification interaction effect, was assessed by a likelihood-ratio test comparing it with a reduced model without the interaction effect. Individual sample effects were treated as random effects to control for intrasample correlation across exons.
Figure 1.
Illustration of the flattened gene model for a gene with three transcript isoforms. The flattened gene model indexed six exons derived from the configuration of the three isoforms. Yellow block, exon; blue line, intron.
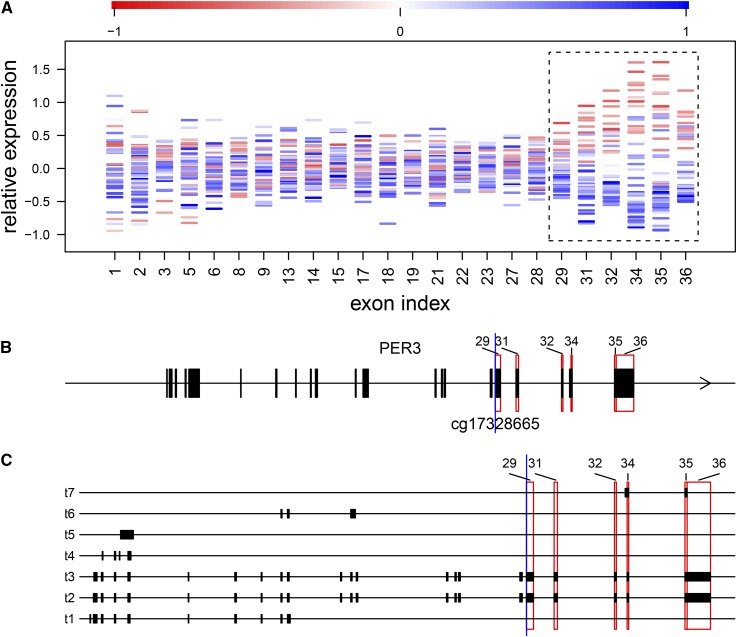
As listed in Table 1, at a 5% false discovery rate (FDR) we detected 9610 CpG–exon pairs for 2719 genes, which represented 17% of the analyzed genes. To eliminate redundancy due to correlation of cytosine modification levels among nearby CpGs, for each of the detected exons we pruned CpGs by Pearson’s r2 ≥ 0.1, resulting in 6039 CpG–exon pairs (Table S1). Figure 2 shows an example of cytosine modification-specific TIV for the PER3 gene that encodes period circadian clock 3, displaying exon expression levels and the associated cytosine modification levels across the CEU samples (Figure 2A), based on the flattened gene model (Figure 2B). Here elevated levels of cytosine modification at cg17328665 located at the 22nd base position of the exon indexed 29, associated with decreased expression levels of the exons indexed 29, 31, 32, and 34–36 that are specific to the 3′ ends of transcript 2 and transcript 3 of PER3 (Figure 2C).
Table 1. The associations detected at a 5% FDR for cytosine modification-specific TIV and SNP-specific TIV.
| Analysis | Population | Gene | Exon | CpG or SNP | CpG–exon or SNP–exon pair |
|---|---|---|---|---|---|
| CpG-specific TIV | CEU | 2,719 | 4,100 | 6,892 | 9,610 |
| YRI | 2,734 | 3,861 | 6,682 | 9,075 | |
| SNP-specific TIV | CEU | 1,153 | 1,343 | 45,907 | 57,747 |
| YRI | 1,865 | 2,208 | 55,955 | 63,632 |
The number of unique genes, exons, and CpGs or SNPs and the number of CpG–exon or SNP–exon pairs are shown.
Figure 2.
An example of cytosine modification-specific TIV, showing cytosine modification levels at cg17328665 associated with TIV of the PER3 gene. (A) For each of the analyzed exons indexed based on the flattened gene model (x-axis), the mean-subtracted expression levels are plotted for individual CEU samples (y-axis) and colored according to the relative cytosine modification levels at cg17328665. Highlighted are exons indexed 29, 31, 32, and 34–36 (blocked by the dashed line), whose expression levels increase with decreasing cytosine modification levels, suggesting that cytosine modification at cg17328665 suppresses the inclusion of these exons. Exons indexed 32 and 34–36 were called significant at the 5% FDR threshold. The discontinuity of exon indexes along the x-axis was due to exons interrogated by fewer than two probes and excluded from analysis. (B and C) The relative positions of cg17328665 (blue vertical line) and the highlighted exons (red rectangles) are marked in (B) the flattened gene model and (C) the transcripts annotated by Gencode release 19, shown as transcripts 1–7 (t1–t7) for simplicity.
We also examined cytosine modification-specific TIV in 57 unrelated YRI samples and detected 5724 CpG–exon pairs for 2734 genes after pruning correlated CpGs (Table 1 and Table S2). Across the CpG–exon pairs detected in the CEU and/or YRI samples, the exon-by-cytosine modification interaction effects showed moderate correlation between the two populations (Pearson’s r2 = 0.13, Figure S1), suggesting extensive population specificity of isoform regulation. For simplicity we describe only results of the pruned CpG–exon associations for the CEU samples, with results from the YRI samples provided in Table S3, Table S6, Table S7 and Figure S2, Figure S3, Figure S4. Our analysis was ensured by the observations that the exon expression levels and the overall gene expression levels showed different correlations with the cytosine modification levels across the detected CpG–exon pairs (Figure 3A and Figure S2A), while randomly sampled CpG–exon pairs in general showed similar cytosine modification correlations between the exons and the genes (Figure S3).
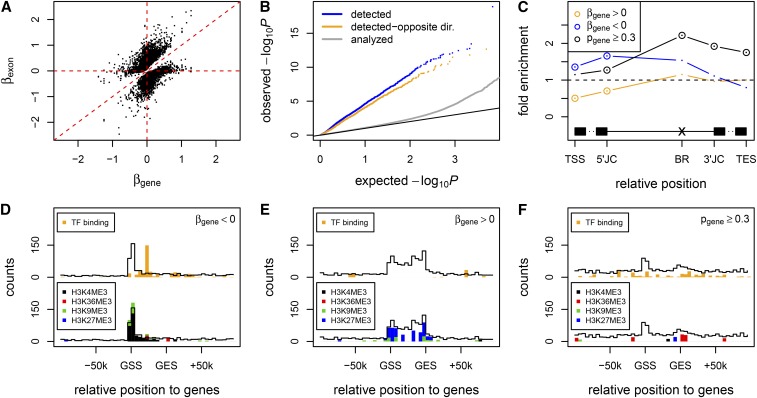
Figure 3.
Cytosine modification-specific TIVs attributable to alternative transcription and alternative splicing. (A) Across the detected CpG–exon pairs, the regression coefficients of exon expression levels ∼ cytosine modification levels (y-axis) are plotted against the regression coefficients of gene expression levels ∼ cytosine modification levels (x-axis). Here the gene expression level was estimated as the mean of exons excluding any detected exon. (B) The observed (y-axis) vs. expected (x-axis) –log10 P-values of cytosine modification–gene expression association, for all analyzed genes (gray), genes containing cytosine modification-specific TIV (blue), and those further varied to cytosine modification levels with opposite direction of the detected exons (orange). Black line denotes diagonal line. (C) Overrepresentation in proximal regulatory regions for TIV-associated CpGs whose cytosine modification levels show positive (orange), negative (blue), or no (black) correlation with gene expression variation. TSS, transcript start site; 5′JC, 5′ junction; BR, branching point (Mercer et al. 2015); 3′JC, 3′ junction; TES, transcript end site. Significant overrepresentation (P < 0.05) is marked by circles. (D–F) Overrepresentation in ENCODE transcription factor binding peaks (top) and histone modification peaks (bottom) for TIV-associated CpGs whose cytosine modification levels show negative (D), positive (E), or no (F) correlation with gene expression variation. The counts of overlapped CpGs in the positional bins are plotted as black outlines. The cumulative counts of CpGs overlapped with transcription factor binding peaks (orange) or with histone peaks (black, H3K4me3; red, H3K36me3; green, H3K9me3; blue, H3K27me3) are plotted if they exceeded the 95% quantile of the individual sampling distributions. GSS, gene start site; GES, gene end site.
Cytosine modification-specific TIVs attributable to alternative transcription
We found that a large number of TIV-associated cytosine modifications also correlated with the overall gene expression variations (Figure 3B and Figure S2B). To avoid confounding effects from the cytosine modification-specific exons, here overall gene expression levels were estimated by excluding these exons. Another fact worth consideration is that TIVs detected from microarray data could have resulted from probe effects (Zhang et al. 2008). For example, if cytosine modification increased the expression level of a gene, an exon interrogated by more sensitive probes may exhibit greater fold change compared to other exons, and vice versa. Probe effects, however, caused only the detection of cytosine modifications to which the levels of the exons and the genes varied in the same direction. When restricted to cytosine modifications to which the levels of the exons and the genes varied in opposite direction, therefore unlikely caused by probe effects, the enrichment in correlation with gene expression variations still remained (Figure 3B and Figure S2B).
Based on our flattening gene annotation approach, gene expression level was estimated as the mean expression level of exons that originated from multiple transcript isoforms. One explanation of the prominent coincidence of TIV and gene expression variation is that the associated cytosine modifications affected isoform transcription rather than specific exon splicing, thereby influencing the estimation of gene expression levels. To further investigate this, we analyzed CpGs located <100 kb away from gene regions of 17,232 genes, testing the correlation of cytosine modification levels with overall gene expression levels in the CEU samples. Using a similar approach to correct for multiple comparisons, 490 CpG–gene pairs for 237 genes were detected at a 5% FDR, representing 1.4% of the analyzed genes. Among these 237 genes, 175 (74%) contained TIV related to the corresponding CpGs. This suggests that the majority of the cytosine modification-specific gene expression variations detected by array were likely due to TIV through alternative isoform transcription.
To assess the underlying molecular mechanisms, we classified the TIV-associated cytosine modifications into two groups: 2176 CpG sites (45%) that correlated with the expression of the corresponding 1420 genes (P < 0.05) and 1615 CpG sites (33%) that showed no correlation with the expression of the corresponding 1043 genes (P ≥ 0.3). We examined the coincidence of the two groups of CpGs with cis elements potentially involved in transcript isoform configuration and with transcription factor binding peaks and histone modification peaks from the Encyclopedia of DNA Elements (ENCODE) (Encode Project Consortium 2012). CpGs whose modification levels negatively correlated with gene expression levels were significantly enriched in 5′ ends of transcripts (Figure 3C, Figure S2C, and Table S3), transcription factor binding peaks in gene regions, and H3K4me3 (trimethylation of lysine 4 in histone 3) peaks in the 5′ portion of genes (Figure 3D and Figure S2D), suggesting repressive effects of these cytosine modifications through interfering with transcription factor binding and transcription initiation. In contrast, CpGs whose modification levels positively correlated with gene expression levels were depleted in 5′ ends of transcripts but enriched in repressive H3K27me3 (trimethylation of lysine 27 in histone 3) peaks (Figure 3E and Figure S2E), implying their influences on local chromatin structure.
Cytosine modification-specific TIVs attributable to alternative splicing
As shown in Figure 3C, cytosine modifications that showed no correlation with gene expression variation (P ≥ 0.3) were significantly enriched in exon/intron junctions, intron branching points, and transcript termination sites (Table S3), implying their direct effects on splicing or transcription termination. Consistent with our hypothesis, here almost all of the CpGs causing alternative splicing (99%) associated with TIVs at a single exon in a given gene, whereas 21% of the CpGs causing alternative transcription associated with TIVs at more than one exon in a given gene. In contrast to the alternative transcription-associated CpGs that accumulated over gene regions (Figure 3, D and E, and Figure S2, D and E), the alternative splicing-associated CpGs spread along both genic and intergenic regions, where they showed certain enrichment in transcription factor binding peaks but no obvious enrichment pattern in histone modification peaks (Figure 3F and Figure S2F).
CCCTC-binding factor (CTCF) was shown to regulate splicing through promoting Pol II pausing (Shukla et al. 2011). We found that TIV-associated CpGs whose cytosine modification levels negatively correlated with gene expression variation significantly overlapped intragenic CTCF broad peaks (Figure S4), but not narrow peaks (data not shown), suggesting that these cytosine modifications affect TIV through modulating CTCF binding over a relatively broad intragenic region.
Genetic and epigenetic regulation of TIV
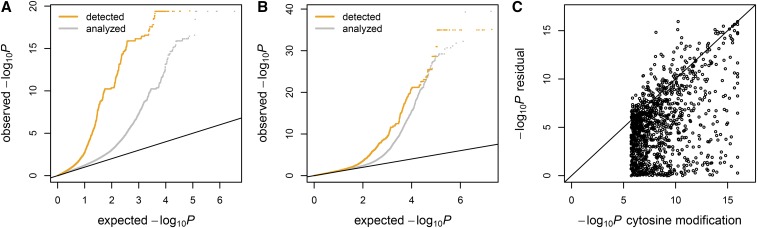
To assess genetic regulation of TIV, we analyzed TIV association for SNPs located <100 kb away from gene regions, using a similar approach to that for detecting cytosine modification-specific TIV, for 58 CEU samples. At a 5% FDR, we detected 57,747 SNP–exon pairs for 1153 genes in the CEU samples (Table 1), which represented 7.2% of the analyzed genes. Pruning for linkage disequilibrium (LD) at r2 ≥ 0.1 resulted in 1668 SNP–exon pairs (Table S4). We also detected SNP-specific TIVs for 12% of the analyzed genes in 59 YRI samples (Table 1 and Table S5). Overall, the extent of SNP-specific TIV was comparable to the extent of CpG-specific TIV (Figure S5). We observed an enrichment of the detected SNPs in association with overall gene expression variations (Figure 4A and Figure S6A), suggesting that alternative isoform transcription also underlies genetically regulated TIVs.
Figure 4.
Genetic and epigenetic regulation of TIV. (A) The observed (y-axis) vs. expected (x-axis) –log10 P-values of SNP genotype–gene expression association, for TIV-associated SNPs (orange) and all analyzed SNPs (gray). (B) The observed (y-axis) vs. expected (x-axis) –log10 P-values of SNP genotype–cytosine modification association, for TIV-associated SNPs (orange) and all analyzed SNPs (gray). (C) The extent of genetic regulation on TIV mediated through cytosine modification variation. For the 282 target exons detected in both SNP-specific TIV and CpG-specific TIV analyses, the –log10 P-values of exon-by-residual cytosine modification interaction (y-axis) are plotted against the –log10 P-values of exon-by-cytosine modification interaction (x-axis). Here the residual cytosine modification levels were cytosine modification levels regressed out of genetic variations at the corresponding exon-associated SNPs.
SNPs associated with TIV showed slight enrichment in cytosine mQTL (Figure 4B and Figure S6B), suggesting that genetic regulation on TIV may sometimes be mediated through cytosine modification variation. We found that the target exons of the genetically and epigenetically regulated TIVs were significantly overlapped: 282 of the analyzed exons were detected by both analyses, representing >11-fold enrichment (binomial test P < 2 × 10−16). To examine whether the SNP effects were indeed mediated through cytosine modification variations for these 282 exons, we regressed out the genetic variation at the SNPs for the corresponding CpGs and used the residual cytosine modification levels to test for TIV association at these exons. For a proportion of the CpGs, residual cytosine modification levels were no longer associated with the corresponding TIVs (Figure 4C and Figure S6C), indicating that for these TIVs, genetic regulation was mediated through cytosine modification variation. Nevertheless, residual cytosine modification-specific TIVs were significant at Bonferroni-corrected P < 0.05 for 744 of 1216 CpG–exon pairs (61%), corresponding to 172 of 282 exons (61%). This suggests that although genetic and epigenetic regulation of TIV share target preference, their effects are predominantly independent.
Developmental dependency of cytosine modification-specific TIV
To assess the developmental dependency of cytosine modification-specific TIV, we examined the relative enrichment of the genes detected from LCLs in those detected from seven types of cancer tissues in TCGA. For each cancer type, the counts of exon junctions derived from RNA sequencing were fitted with the cytosine modification levels profiled on an Illumina 450K array, to test for exon junction-by-cytosine modification interaction effect. Therefore, the analysis concerned cytosine modification-specific TIV across individuals in each type of cancer tissue, rather than cytosine modification-specific TIV associated with cancers. Genes detected from the seven types of cancer tissue all showed significant enrichment in genes detected from LCLs (Table 2 and Table S6), which may be viewed as a validation of the TIVs detected in the LCLs that were based on expression array measurements. Ranked by the fold enrichment were blood (3.0), lung (2.3), kidney (2.1), thyroid (2.0), brain (2.0), uterus (1.7), and liver (1.5) cancer tissues (Table 2 and Table S6). In LCLs genes containing cytosine modification-specific TIVs attributable to alternative splicing, compared to those attributable to alternative transcription, showed greater overlaps with cancer tissue data (Table 2 and Table S6), potentially because only the counts of exon junctions were analyzed for the cancer tissues.
Table 2. The enrichment of genes containing cytosine modification-specific TIVs detected in LCLs of the CEU samples (All) and those further attributable to alternative transcript or alternative splicing, in genes containing cytosine modification-specific TIVs detected in seven types of cancer tissues from TCGA.
| Type of gene | Brain (1533/8498)a | Liver (155/378) | Blood (317/907) | Thyroid (637/1650) | Lung (378/1192) | Kidney (695/1948) | Uterus (196/573) |
|---|---|---|---|---|---|---|---|
| All | 2.0 (3.8 × 10−26)b | 1.5 (0.062) | 3.0 (1.3 × 10−18) | 2.0 (7.8 × 10−14) | 2.3 (2.8 × 10−12) | 2.1 (6.3 × 10−17) | 1.7 (0.0015) |
| Alternative transcription | 1.8 (8.6 × 10−12) | 1.4 (0.19) | 2.0 (1.5 × 10−5) | 1.5 (0.0018) | 1.9 (5.2 × 10−5) | 1.8 (3.8 × 10−7) | 1.7 (0.016) |
| Alternative splicing | 2.3 (1.7 × 10−20) | 2.2 (0.0028) | 3.9 (1.6 × 10−17) | 2.7 (2.6 × 10−15) | 2.7 (1.2 × 10−9) | 2.7 (8.1 × 10−17) | 1.9 (0.0058) |
Cancer tissue type (no. unique genes/no. unique CpGs detected in the cancer tissue).
Fold enrichment (P-value of Fisher’s exact test).
Pathway enrichment of genes containing cytosine modification-specific TIVs
We further analyzed the 2719 genes containing cytosine modification-specific TIVs detected in LCLs for potential enrichment in biological pathways (Huang da et al. 2009). We identified 24 KEGG pathways significant at Benjamini-adjusted P < 0.05 (Table 3 and Table S7). Many cancer-related pathways were identified, including acute and chronic myeloid leukemia, small and nonsmall cell lung cancer, glioma, prostate cancer, pancreatic cancer, and endometrial cancer. Several pathways were related to metabolisms, for example type II diabetes mellitus and insulin signaling pathway.
Table 3. KEGG pathways enriched at adjusted P < 0.05 for genes containing cytosine modification-specific TIV detected in the CEU samples, ordered by fold enrichment.
| KEGG pathway | No. genesa | Fold enrichment | Nominal P |
|---|---|---|---|
| Type II diabetes mellitus | 19 | 2.5 | 2.2 × 10−4 |
| Phosphatidylinositol signaling system | 29 | 2.4 | 5.6 × 10−6 |
| Acute myeloid leukemia | 22 | 2.3 | 1.7 × 10−4 |
| Nonsmall cell lung cancer | 20 | 2.3 | 5.3 × 10−4 |
| Inositol phosphate metabolism | 19 | 2.2 | 1.5 × 10−3 |
| Glioma | 22 | 2.1 | 6.4 × 10−4 |
| mTOR signaling pathway | 18 | 2.1 | 2.6 × 10−3 |
| Endometrial cancer | 18 | 2.1 | 2.6 × 10−3 |
| ECM-receptor interaction | 29 | 2.1 | 8.4 × 10−5 |
| ErbB signaling pathway | 30 | 2.1 | 6.3 × 10−5 |
| Small cell lung cancer | 28 | 2.0 | 2.2 × 10−4 |
| B-cell receptor signaling pathway | 25 | 2.0 | 5.3 × 10−4 |
| VEGF signaling pathway | 25 | 2.0 | 5.3 × 10−4 |
| Chronic myeloid leukemia | 24 | 2.0 | 1.3 × 10−3 |
| T-cell receptor signaling pathway | 34 | 1.9 | 1.4 × 10−4 |
| Pancreatic cancer | 22 | 1.9 | 4.1 × 10−3 |
| Fc gamma R-mediated phagocytosis | 29 | 1.9 | 8.4 × 10−4 |
| Focal adhesion | 61 | 1.9 | 6.8 × 10−7 |
| Prostate cancer | 27 | 1.9 | 1.5 × 10−3 |
| Tight junction | 36 | 1.6 | 2.2 × 10−3 |
| Insulin signaling pathway | 35 | 1.6 | 4.8 × 10−3 |
| Pathways in cancer | 84 | 1.6 | 1.0 × 10−5 |
| Regulation of actin cytoskeleton | 54 | 1.5 | 8.3 × 10−4 |
| MAPK signaling pathway | 66 | 1.5 | 3.2 × 10−4 |
The number of genes containing cytosine modification-specific TIV that were in the pathway.
Discussion
Our study of LCLs derived from European and African descents demonstrated a prominent effect of cytosine modification on interindividual TIV, primarily through alternative isoform transcription. More than 52% of the genes containing cytosine modification-specific TIV also showed evidence for cytosine modification-specific gene expression variation. On the other hand, 74% of the genes detected from genome-wide cytosine modification–gene expression correlations contained TIV associated with the corresponding CpGs. The linking of gene expression variation with alternative isoform transcription stemmed from our analysis approach that, instead of using a gene model composed of individual transcripts, uses a gene model of collapsed, transcribed units (Figure 1), by which varying levels of isoforms tend to affect the estimated level of overall gene expression.
The precise selection of alternative promoters allows transcript isoforms to be expressed in appropriate developmental contexts. In our study, suppressive cytosine modifications that frequently occur at the 5′ ends of transcripts were enriched with transcription factor binding peaks and H3K4me3 peaks that mark transcription initiation (Figure 3D), suggesting a widespread influence of epigenetically modified cis regulatory elements on the choice of alternative promoters (Archey et al. 1999; Ventura et al. 2002; Davuluri et al. 2008). Kinetics of transcription elongation may affect the isoform spectrum as well, through selecting competing splicing sites. We found significant coincidence of the detected cytosine modifications with H3K27me3 peaks (Figure 3E), which implies a role of these cytosine modifications in restricting repressive H3K27me3 deposition (Reddington et al. 2013) and favoring transcription elongation. A combinatorial role of H3K4me3 and H3K27me3 in regulating transcript isoform expression was previously indicated in a study of developing and adult cerebella (Pal et al. 2011).
Our study also revealed TIV-associated cytosine modifications that appeared to have no effect on transcription. These cytosine modifications affected a single exon within the genes and were significantly enriched in exon/intron junctions and splicing branching points. One explanation is that these cytosine modifications may cause localized fluctuation of the transcription elongation rate and affect only the proximal exon selection, as in the exclusion of exon 5 of CD45 during peripheral lymphocyte maturation (Shukla et al. 2011) and the skipping of exon 18 of NCAM1 during neuronal cell depolarization (Schor et al. 2009). Alternatively, these cytosine modifications may modulate the binding of splicing regulatory factors. For example, recruitment of CpG-binding proteins (Maunakea et al. 2013) and heterochromatin protein 1 (Yearim et al. 2015) at proximal regions may facilitate alternative exon recognizing, whereas long-range interaction of the basal transcriptional complex with the transcription factor bound at distal enhancers may recruit a splicing regulator that alters the specific exon inclusion level (Kornblihtt et al. 2013). The effect of cytosine modifications in altering positional distributions of transcription/splicing factors may in general influence TIV (Agirre et al. 2015).
The significant overlap of the target exons between the genetically and epigenetically regulated TIVs reflects an intrinsic difference in configuration variability among exons. Even for these common target exons the genetic and epigenetic regulation appeared to execute somewhat independent effects, which may imply different evolutionary constraints on genetic and epigenetic variations. The fact that >60% of the disease-associated genetic mutations cause abnormal transcript variants (Lopez-Bigas et al. 2005) raises the proposition that altered transcript variants due to epigenetic changes may also underlie many human diseases. Indeed, our study revealed significant enrichment of genes containing cytosine modification TIVs in disease-related pathways, suggesting the importance of nongenetic factors in disease etiology mediated through epigenetic variation.
TIV analysis using exon array data as in this study primarily tests the abundance of one exon relative to the other exons in the same gene, interpretable as exon skipping or retention events, depending on the context of transcript isoforms. RNA sequencing technology, with increasingly longer length of reads, will eventually allow direct comparison of individual transcript abundance (Trapnell et al. 2010; Li and Dewey 2011) and generate results more attractive for biological interpretation. Finally, alternative splicing is a complex process involved in both cis- and trans-acting factors. The local correlations observed between cytosine modification and TIV reflect only one aspect among various types of interindividual regulatory variations for TIV. Future molecular experiments are required to sufficiently establish causal relationships between cytosine modification and TIV beyond genomic associations.
Acknowledgments
The authors thank M. Eileen Dolan and Lucy A. Godley for helpful discussions during the preparation of this manuscript. This work was supported, in part, by grants from the National Institutes of Health: HG006367 (to W.Z.), CA187869 (to W.Z.), and The Robert H. Lurie Comprehensive Cancer Center–Developmental Funds P30CA060553 (to W.Z.).
Footnotes
Communicating editor: O. J. Rando
Supplemental material is available online at http://www.genetics.org/cgi/data/genetics.115.185504/DC1.
Literature Cited
- Agirre E., Bellora N., Allo M., Pages A., Bertucci P., et al. , 2015. A chromatin code for alternative splicing involving a putative association between CTCF and HP1alpha proteins. BMC Biol. 13: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand E., Batsche E., Muchardt C., 2008. Splicing, transcription, and chromatin: a menage a trois. Curr. Opin. Genet. Dev. 18: 145–151. [DOI] [PubMed] [Google Scholar]
- Anders S., Reyes A., Huber W., 2012. Detecting differential usage of exons from RNA-seq data. Genome Res. 22: 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archey W. B., Sweet M. P., Alig G. C., Arrick B. A., 1999. Methylation of CpGs as a determinant of transcriptional activation at alternative promoters for transforming growth factor-beta3. Cancer Res. 59: 2292–2296. [PubMed] [Google Scholar]
- Benjamini Y. Y. D., 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29: 1165–1188. [Google Scholar]
- Beyer A. L., Osheim Y. N., 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2: 754–765. [DOI] [PubMed] [Google Scholar]
- Cramer P., Pesce C. G., Baralle F. E., Kornblihtt A. R., 1997. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. USA 94: 11456–11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri R. V., Suzuki Y., Sugano S., Plass C., Huang T. H., 2008. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 24: 167–177. [DOI] [PubMed] [Google Scholar]
- de la Mata M., Kornblihtt A. R., 2006. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat. Struct. Mol. Biol. 13: 973–980. [DOI] [PubMed] [Google Scholar]
- de la Mata M., Alonso C. R., Kadener S., Fededa J. P., Blaustein M., et al. , 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12: 525–532. [DOI] [PubMed] [Google Scholar]
- Dutertre M., Lacroix-Triki M., Driouch K., de la Grange P., Gratadou L., et al. , 2010. Exon-based clustering of murine breast tumor transcriptomes reveals alternative exons whose expression is associated with metastasis. Cancer Res. 70: 896–905. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Gama-Sosa M. A., Huang L. H., Midgett R. M., Kuo K. C., et al. , 1982. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 10: 2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium, 2012 An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed]
- Feinberg A. P., Tycko B., 2004. The history of cancer epigenetics. Nat. Rev. Cancer 4: 143–153. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M., Lappalainen T., Montgomery S. B., Buil A., Ongen H., et al. , 2013. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2: e00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J. M., Tapanari E., Diekhans M., et al. , 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H., Moran S., Hernando-Herraez I., Sayols S., Gomez A., et al. , 2013. DNA methylation contributes to natural human variation. Genome Res. 23: 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hmadcha A., Bedoya F. J., Sobrino F., Pintado E., 1999. Methylation-dependent gene silencing induced by interleukin 1beta via nitric oxide production. J. Exp. Med. 190: 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B. N., Donnelly P., Marchini J., 2009. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5: e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da, W., B. T. Sherman, and R. A. Lempicki, 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Ip J. Y., Schmidt D., Pan Q., Ramani A. K., Fraser A. G., et al. , 2011. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 21: 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. E., Li C., Rabinovic A., 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: 118–127. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A. R., Schor I. E., Allo M., Dujardin G., Petrillo E., et al. , 2013. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 14: 153–165. [DOI] [PubMed] [Google Scholar]
- Li B., Dewey C. N., 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N., Audit B., Ouzounis C., Parra G., Guigo R., 2005. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 579: 1900–1903. [DOI] [PubMed] [Google Scholar]
- Maunakea A. K., Chepelev I., Cui K., Zhao K., 2013. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 23: 1256–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Clark M. B., Andersen S. B., Brunck M. E., Haerty W., et al. , 2015. Genome-wide discovery of human splicing branchpoints. Genome Res. 25: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen E. L., Zhang X., Mu W., Delaney S. M., Wing C., et al. , 2013. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics 194: 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Schleithoff E. S., Stremmel W., Melino G., Krammer P. H., et al. , 2006. One, two, three–p53, p63, p73 and chemosensitivity. Drug Resist. Updat. 9: 288–306. [DOI] [PubMed] [Google Scholar]
- O’Connell J., Gurdasani D., Delaneau O., Pirastu N., Ulivi S., et al. , 2014. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 10: e1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium , G. R. Abecasis, A. Auton, L. D. Brooks, M. A. DePristo et al, 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Gupta R., Kim H., Wickramasinghe P., Baubet V., et al. , 2011. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 21: 1260–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J., 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40: 1413–1415. [DOI] [PubMed] [Google Scholar]
- Pruunsild P., Kazantseva A., Aid T., Palm K., Timmusk T., 2007. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics 90: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M. K., Matsumoto Y., Richardson M. E., Panneerdoss S., Bhardwaj A., et al. , 2014. Hormone-induced and DNA demethylation-induced relief of a tissue-specific and developmentally regulated block in transcriptional elongation. J. Biol. Chem. 289: 35087–35101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Cedar H., 1991. DNA methylation and gene expression. Microbiol. Rev. 55: 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddington J. P., Perricone S. M., Nestor C. E., Reichmann J., Youngson N. A., et al. , 2013. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 14: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishi V., Bhattacharya P., Chatterjee R., Rozenberg J., Zhao J., et al. , 2010. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc. Natl. Acad. Sci. USA 107: 20311–20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez G., Bittencourt D., Laud K., Barbier J., Delattre O., et al. , 2008. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc. Natl. Acad. Sci. USA 105: 6004–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor I. E., Rascovan N., Pelisch F., Allo M., Kornblihtt A. R., 2009. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc. Natl. Acad. Sci. USA 106: 4325–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., et al. , 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., et al. , 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R., Tsuchihara K., Wilhelm M., Fujitani M., Rufini A., et al. , 2008. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 22: 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables J. P., 2004. Aberrant and alternative splicing in cancer. Cancer Res. 64: 7647–7654. [DOI] [PubMed] [Google Scholar]
- Ventura A., Luzi L., Pacini S., Baldari C. T., Pelicci P. G., 2002. The p66Shc longevity gene is silenced through epigenetic modifications of an alternative promoter. J. Biol. Chem. 277: 22370–22376. [DOI] [PubMed] [Google Scholar]
- Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., et al. , 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B. T., Dayeh T. A., Kirkpatrick C. L., Taneera J., Kumar R., et al. , 2011. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia 54: 360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yearim A., Gelfman S., Shayevitch R., Melcer S., Glaich O., et al. , 2015. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 10: 1122–1134. [DOI] [PubMed] [Google Scholar]
- Zhang W., Duan S., Bleibel W. K., Wisel S. A., Huang R. S., et al. , 2009. Identification of common genetic variants that account for transcript isoform variation between human populations. Hum. Genet. 125: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Byrnes J. K., Gal T. S., Li W. H., Borevitz J. O., 2008. Whole genome transcriptome polymorphisms in Arabidopsis thaliana. Genome Biol. 9: R165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Mu W., Zhang W., 2012. On the analysis of the Illumina 450k array data: probes ambiguously mapped to the human genome. Front. Genet. 3: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Moen E. L., Liu C., Mu W., Gamazon E. R., et al. , 2014. Linking the genetic architecture of cytosine modifications with human complex traits. Hum. Mol. Genet. 23: 5893–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.