Abstract
Introduction
Although the liver is less immunogenic than other solid organs, most liver transplant recipients receive lifelong immunosuppression. In both experimental models and clinical transplantation, total Lymphoid Irradiation (TLI) has been shown to induce allograft tolerance. Our goal was to identify the microRNAs (miRNAs) expressed in tolerant liver allograft recipients in an experimental model of TLI-induced tolerance.
Methods
To identify the miRNAs associated with TLI-induced tolerance we examined syngeneic recipients (Lewis→Lewis) and allogeneic recipients (DA→Lewis) of orthotropic liver transplants that received post-transplant TLI, allogeneic recipients that were not treated post-transplantation and experienced acute rejection, and native DA livers. QPCR miRNA array cards were used to profile liver grafts.
Results
We identified 12 miRNAs that were specifically and significantly increased during acute rejection. In early tolerance, 33 miRNAs were altered compared to syngeneic livers, with 80% of the miRNAs increased. In established tolerance 42 miRNAs were altered. In addition, miR-142-5p and miR-181a demonstrated increased expression in tolerant livers (both early and established tolerance) as compared to syngeneic livers. A principal component analysis of all miRNAs assayed, demonstrated a profile in established tolerance that was closely related to that seen in syngeneic livers.
Conclusions
The miRNA profile of established tolerant allografts is very similar to syngeneic grafts suggesting tolerance may be a return to an immunological state of quiescence.
Introduction
Orthotopic liver transplantation (OLT) remains the definitive treatment for many patients with end-stage liver disease. The liver is unique among transplanted organs in promoting a relatively tolerogenic microenvironment1,2. Although it is well established that liver allografts are less immunogenic that other solid organ allografts, life-long immunosuppression is still the standard course for recipients of liver allografts3. Numerous factors, including secretion of soluble MHC class I antigens, the unique cellular composition of the liver, and the circulation from the portal vein to the liver, have been proposed to explain the tolerance promoting capacity of the liver 4. In previous studies, we have demonstrated that induction of donor-specific tolerance can be achieved in a rodent model of OLT by post-transplant total lymphoid irradiation (TLI)5. Liver allograft tolerance was achieved, in the absence of donor cell infusion or anti-thymocyte reagents, through a mechanism involving apoptosis of graft-infiltrating T cells early after transplant and the subsequent accumulation of CD4+CD25+Foxp3+ T regulatory (Treg) in both the graft and periphery. It remains unclear precisely how Treg suppress the alloimmune response.
microRNAs (miRNAs; miR) are endogenously expressed non-coding RNAs that regulate a myriad of gene pathways 6-8. miRNAs bind to complementary target protein-encoding mRNAs in a RNA-dependent gene silencing process leading to translational repression or mRNA degradation. Many fundamental biological processes are regulated by miRNAs including cell differentiation, proliferation, maturation and cellular homeostasis; indeed, miRNAs regulate about 60% of the human genome 9,10. In an effort to further elucidate the immunoregulatory pathway that promotes and maintains liver allograft tolerance, we utilized our model of TLI-induced tolerance and quantitated intragraft miRNAs during early tolerance, established tolerance, graft rejection and compared the profile to TLI-treated synegenic and normal livers.
Materials and Methods
Study groups
Inbred male DA (RT1a) and Lewis (RT1l) rats weighing 220-299g, were purchased from Harlan (Indianapolis, IN). All animals were housed in accordance with institutional animal care policies and had access to water and standard laboratory chow ad libitum. To investigate the role of miRNA after transplantation, we transplanted DA livers into Lewis, a) with TLI protocol (tolerant, TOL), and b) without TLI treatment (acute rejection, AR). Lewis livers into Lewis recipients and DA livers into DA recipients (SYN) were similarly treated with TLI. The four experimental groups (n=3) are 1. Syngeneic (SYN), 2. Acute rejection (AR), 3. Early tolerance (TOL D7) and 4. Established tolerance (TOL D100) (Figure 1).
Figure 1. Study Design.
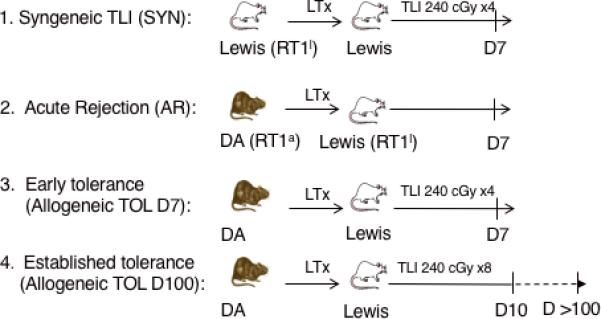
Groups (n=3) of syngeneic or allogeneic OLTs were left untreated (group 2) or treated with post-transplant TLI (groups 1,3,4). Allogeneic model: DA livers transplanted into Lewis recipients (groups 2-4). Syngeneic model: Lewis livers transplanted into Lewis recipients (group 1). TLI consisted of 240 cGy for 4 doses over 6 days (groups 1, 3,) or 8 doses over 10 days as (group 4) as described previously.
Donor and recipient surgeries were carried out aseptically, under anesthesia with isoflurane (Abbott Laboratories, North Chicago, IL). OLT was performed as we have previously published 5. Briefly, the liver was perfused with 15 mL of lactated Ringer's solution at 4°C through the catheter placed in the abdominal aorta, and the excised graft was stored in lactated Ringer's solution at 4°C. Cold ischemic time was around 60 min. The allograft was transplanted orthotopically. The anhepatic phase was ≤15 min. No immunosuppression was given to the recipient rats in this study. All animal experiments were approved by the Stanford Administrative Panel on Laboratory Animal Care.
Total Lymphoid Irradiation (TLI)
TLI was administered to liver graft recipients in eight treatments of 240 cGy each, over 10 days starting on day 1 post-transplant 5. For some experiments, rats were sacrificed on day seven and thus were administered only four doses of TLI over six days. TLI fields exposed the abdomen, peripheral lymph nodes, thymus, and spleen, but shielded the skull, liver graft, lungs, limbs, pelvis, and tail. Irradiation was administered using a Philips x-ray unit (200kV, 20mA; Philips Electronic Instruments, Inc., Rahway, NJ) at a rate of 60 cGy/minute with a 0.5 mm copper filter.
Sample preparation, global miRNA expression profiling and array data analysis
Total RNA was isolated from transplanted livers using the mirVana miRNA isolation kit (Applied Biosystems, Foster City, CA). Normal non-transplanted DA and Lewis livers were used as controls and calibrators for microarray data analysis. RNA samples from each group (n=3) of transplanted or normal animals were prepared using TaqMan microRNA reverse transcription kit and the corresponding Megaplex primer pools (Applied Biosystems) as previously described11. Pre-amplification of cDNA samples was performed with TaqMan PreAmp master mix and corresponding Megaplex PreAmp primers (Applied Biosystems). TaqMan array rodent microRNA A Card v2.0 (Applied Biosystems) were used. Complete profiles from three livers in each group were analyzed. Liver mononuclear cells were isolated as previously described12. Data was analyzed using SDS and RQ software (Applied Biosystems). Fold changes for each miRNA were normalized to the endogenous control RNU6. The expression fold changes were calculated using the comparative Ct method and compared to the relevant normal tissue. qPCR was used to determine the relative expression of specific miRNA as we have previously described6.
Statistics
The normalized miRNA Ct values were analyzed using an unpaired Student's T-test. Multiple hypothesis correction was performed using an FDR of 10%, to determine differentially altered miRNA. An adjusted p-value of 0.05 was considered significant for all statistical analyses. Deducer and its dependencies within the R statistical computing environment 13 and GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA) were used for statistical analysis and graphical representations of the data. Data are exhibited graphically as min to max with median, unless specified. Principal component analysis (PCA) was performed on the normalized miRNA expression data to assess the ability of selected differentially altered miRNA species to discriminate and separate the sample groups. PCA was performed using the relative fold-change values compared to RNU6 and with a DA native liver as control with the prcomp14-16 function and associated dependencies within the R v3.1.2 environment in R Studio v0.98.98713. PCA was performed on loge transformed data using a single value decomposition method of dimensionality reduction, that was zero centered and scaled. The first two or three principle components were plotted and visualized using devtools17 v1.6.1, ggplot218 v1.0.0, ggbiplot v0.55 and rgl19 v0.93.1098 and associated dependencies in the R environment.
Results
Specific miRNAs are Associated with Allograft Rejection
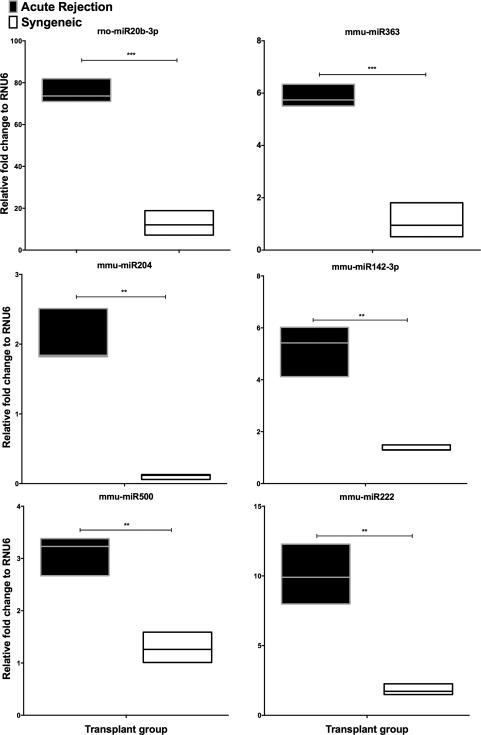
To establish the miRNA profile during liver allograft rejection, we isolated RNA from groups (n=3, each) of allogeneic (DA →Lewis) and syngeneic (SYN) (Lewis →Lewis) transplants on day seven post-transplant (D7) and normal DA livers. This time point was chosen since our previous studies clearly demonstrated that severe rejection was apparent in the allogeneic group12,20,21. Of the 226 rat mature miRNAs examined, 158 miRNAs with Ct <35 in all graft samples were included for analysis (Supplementary Table 1). Twenty-four miRNAs were significantly and specifically altered when comparing AR allografts vs. SYN livers using Student's T-test with an FDR adjusted p-value (Table 1). From these we identified twelve miRNAs that were increased specifically in AR compared to SYN. These significantly increased miRNA were rno-miR20-3p, mmu-miR363, mmu-miR204, mmu-miR142-3p, mmu-miR500, mmu-miR222, mmu-miR674, mmu-miR130b, rno-miR196c, mmu-miR17, mmu-miR7b and rno-miR17-3p (Table 1). Six significantly altered miRNA are shown in Figure 2. These data, directly comparing syngeneic and allogeneic livers on day 7 post-transplant demonstrate that several miRNAs are modulated by alloantigen and may contribute to rejection of the graft.
Table 1.
Twenty-four differentially modulated miRNA's in acute rejection compared to syngeneic allografts. Data expressed as mean and standard error of the mean (SEM)
| miRNA | Acute rejection (mean±SEM) | Syngeneic (mean±SEM) | Adjusted P value | Direction in AR vs. SYN |
|---|---|---|---|---|
| rno-miR-20b-3p | 75.5 ± 3.3 | 12.7 ± 3.4 | 0.0002 | Up |
| mmu-miR-122 | 0.3 ± 0.0 | 0.8 ± 0.0 | 0.0002 | Down |
| mmu-miR-135b | 14.3 ± 3.9 | 100.5 ± 5.7 | 0.0002 | Down |
| mmu-miR-363 | 5.9 ± 0.2 | 1.1 ± 0.4 | 0.0005 | Up |
| mmu-let-7d | 0.7 ± 0.0 | 1.0 ± 0.0 | 0.0007 | Down |
| mmu-miR-204 | 2.1 ± 0.2 | 0.1 ± 0.0 | 0.0010 | Up |
| mmu-miR-194 | 0.2 ± 0.0 | 0.5 ± 0.0 | 0.0019 | Down |
| mmu-miR-142-3p | 5.2 ± 0.6 | 1.4 ± 0.1 | 0.0025 | Up |
| mmu-miR-500 | 3.1 ± 0.2 | 1.2 ± 0.2 | 0.0027 | Up |
| mmu-miR-222 | 10.1 ± 1.2 | 1.8 ± 0.2 | 0.0028 | Up |
| mmu-miR-200a | 0.6 ± 0.1 | 1.3 ± 0.1 | 0.0030 | Down |
| mmu-let-7a | 0.4 ± 0.0 | 0.8 ± 0.0 | 0.0034 | Down |
| mmu-miR-674 | 3.4 ± 0.2 | 1.7 ± 0.2 | 0.0041 | Up |
| mmu-miR-130b | 3.7 ± 0.3 | 1.8 ± 0.1 | 0.0048 | Up |
| mmu-miR-205 | 1.4 ± 0.4 | 4.0 ± 0.3 | 0.0049 | Down |
| mmu-let-7b | 0.4 ± 0.1 | 0.9 ± 0.0 | 0.0049 | Down |
| mmu-miR-183 | 0.5 ± 0.2 | 1.4 ± 0.1 | 0.0062 | Down |
| mmu-miR-375 | 0.2 ± 0.1 | 1.1 ± 0.2 | 0.0076 | Down |
| mmu-miR-192 | 0.2 ± 0.0 | 0.4 ± 0.0 | 0.0077 | Down |
| rno-miR-196c | 31.2 ± 5.8 | 2.3 ± 1.0 | 0.0078 | Up |
| mmu-miR-17 | 1.9 ± 0.2 | 1.0 ± 0.1 | 0.0090 | Up |
| mmu-miR-7b | 1.3 ± 0.0 | 0.9 ± 0.1 | 0.0109 | Up |
| mmu-let-7c | 0.5 ± 0.0 | 0.9 ± 0.1 | 0.0114 | Down |
| rno-miR-17-3p | 3.6 ± 0.4 | 1.5 ± 0.2 | 0.0131 | Up |
Figure 2. Intragraft miRNA are increased during AR.
Shown are the top six miRNA that were significantly and specifically differentially increased in AR as compared to SYN livers, ** P≤0.01, ***P≤0.001 via unpaired T-test, adjusted for FDR 10%.
Induction of Tolerance is Accompanied by an Increase in Specific miRNA
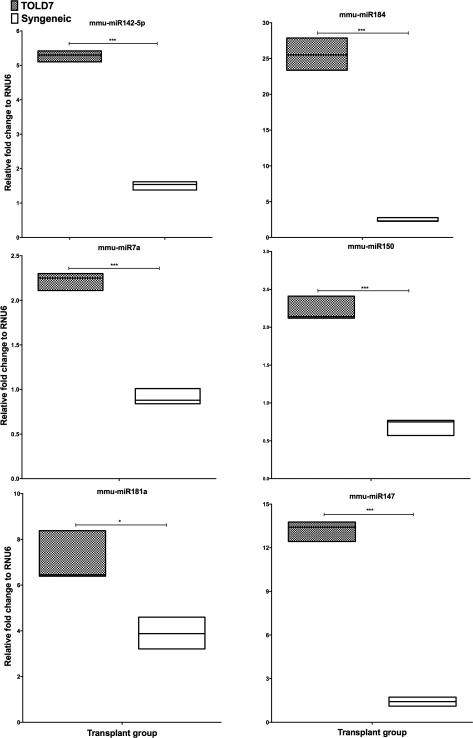
In previous studies we have demonstrated that robust donor-specific tolerance can be achieved in DA→Lewis liver allografts after TLI5. This tolerance was associated with an expansion of CD4+CD25+FoxP3+ Treg cells. To determine if specific miRNA were associated with this induction of tolerance we profiled miRNA from a group (n=3) of DA→Lewis transplants that were treated with TLI and compared them against SYN controls at D7 post-transplant. Of the 158 miRNA's investigated we observed 33 miRNAs that were differentially altered in TOL D7 compared to SYN allografts (Table 2). Of the 33 modulated miRNAs, over 87% (29 miRNA) of these were increased in TOL D7 allografts compared to SYN, with only four miRNA decreased in TOL D7. Six significantly altered miRNA are shown in Figure 3A. Three miRNA (mmu-miR203, mmu-miR9 and mmu-miR122) were significantly altered in TOL D7 compared to AR allografts. mmu-miR203 (P<0.0002 vs. AR) and mmu-miR9 (P<0.002 vs. AR) were significantly increased in TOL D7 compared to both AR and SYN allografts while mmu-miR122 was significantly increased when compared to SYN allografts and decreased compared to AR allografts (P<0.000004 vs. AR). As only minimal changes of mmu-miR122 were observed in lymphocytes isolated from AR and TOL livers (Supplementary Figure 1) this suggests that miR-122 is of hepatocyte origin in our model.
Table 2.
Thirty-three differentially modulated miRNA's in early tolerance, (TOL D7) as compared to syngeneic allografts. Data expressed as mean and standard error of the mean (SEM).
| miRNA | Early tolerance (mean±SEM) | Syngeneic (mean±SEM) | Adjusted P value | Direction in TOL vs. SYN |
|---|---|---|---|---|
| mmu-miR-142-5p | 5.3 ± 0.1 | 1.5 ± 0.1 | 5.6 ×10−6 | Up |
| mmu-miR-184 | 25.6 ± 1.3 | 2.4 ± 0.2 | 6.0 ×10−5 | Up |
| mmu-miR-7a | 2.2 ± 0.1 | 0.9 ± 0.1 | 6.9 ×10−5 | Up |
| mmu-miR-150 | 2.2 ± 0.1 | 0.7 ± 0.1 | 1.7×10−4 | Up |
| mmu-miR-142-3p | 5.1 ± 0.3 | 1.4 ± 0.1 | 2.4 ×10−4 | Up |
| mmu-miR-147 | 13.2 ± 0.4 | 1.4 ± 0.3 | 2.4 ×10−4 | Up |
| mmu-miR-135b | 11.0 ± 5.5 | 100.5 ± 5.7 | 3.6 ×10−4 | Down |
| mmu-miR-342-3p | 6.8 ± 0.4 | 2.1 ± 0.2 | 3.7 ×10−4 | Up |
| mmu-miR-363 | 8.8 ± 0.6 | 1.1 ± 0.4 | 4.1 ×10−4 | Up |
| mmu-miR-128a | 2.6 ± 0.1 | 1.0 ± 0.2 | 0.0014 | Up |
| mmu-miR-7b | 1.8 ± 0.1 | 0.9 ± 0.1 | 0.0018 | Up |
| mmu-miR-204 | 2.0 ± 0.3 | 0.1 ± 0.0 | 0.0019 | Up |
| mmu-miR-16 | 2.6 ± 0.0 | 1.7 ± 0.1 | 0.0022 | Up |
| mmu-miR-183 | 0.8 ± 0.0 | 1.4 ± 0.1 | 0.0030 | Down |
| mmu-miR-222 | 4.6 ± 0.4 | 1.8 ± 0.2 | 0.0037 | Up |
| mmu-miR-674 | 3.2 ± 0.2 | 1.7 ± 0.2 | 0.0038 | Up |
| mmu-miR-203 | 1.1 ± 0.1 | 0.5 ± 0.1 | 0.0039 | Up |
| mmu-miR-9 | 10.6 ± 0.3 | 4.2 ± 1.0 | 0.0041 | Up |
| rno-miR-532-5p | 2.1 ± 0.1 | 1.3 ± 0.1 | 0.0042 | Up |
| mmu-miR-17 | 1.7 ± 0.1 | 1.0 ± 0.1 | 0.0043 | Up |
| mmu-miR-20a | 1.7 ± 0.0 | 1.1 ± 0.1 | 0.0050 | Up |
| rno-miR-20b-3p | 49.0 ± 5.6 | 12.7 ± 3.4 | 0.0052 | Up |
| mmu-miR-122 | 0.6 ± 0.0 | 0.8 ± 0.0 | 0.0053 | Down |
| mmu-miR-532-3p | 1.6 ± 0.1 | 1.0 ± 0.1 | 0.0068 | Up |
| mmu-miR-130b | 3.5 ± 0.3 | 1.8 ± 0.1 | 0.0083 | Up |
| mmu-miR-199a-3p | 1.7 ± 0.3 | 3.2 ± 0.1 | 0.0088 | Down |
| rno-miR-17-3p | 3.4 ± 0.3 | 1.5 ± 0.2 | 0.0097 | Up |
| mmu-miR-92a | 0.8 ± 0.8 | 0.6 ± 0.0 | 0.0100 | Up |
| mmu-miR-339-5p | 1.4 ± 0.1 | 0.9 ± 0.1 | 0.0107 | Up |
| mmu-miR-500 | 2.7 ± 2.7 | 1.3 ± 0.2 | 0.0119 | Up |
| mmu-miR-181a | 7.1 ± 0.7 | 3.9 ± 0.4 | 0.0144 | Up |
| mmu-miR-18a | 4.3 ± 0.2 | 2.1 ± 0.5 | 0.0177 | Up |
| mmu-miR-449a | 11.8 ± 1.5 | 6.0 ± 0.4 | 0.0182 | Up |
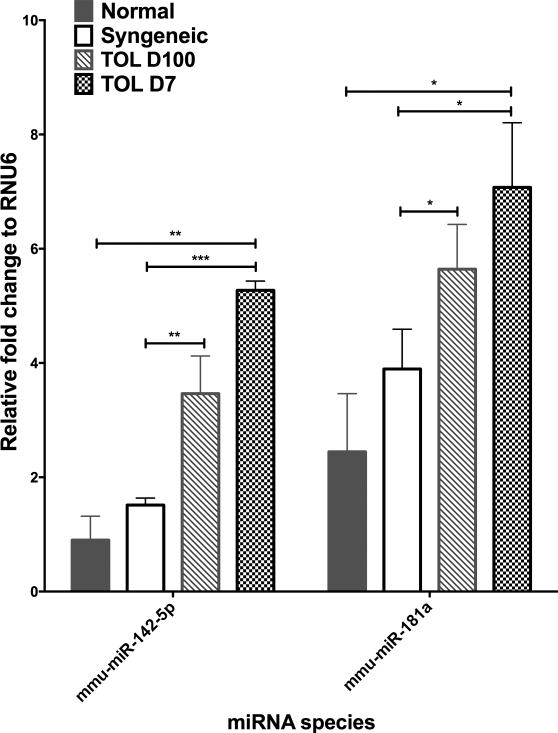
Figure 3. Intragraft miRNA are increased in tolerance.
(A) Shown are six miRNA that were significantly and specifically differentially increased in TOL D7 as compared to SYN, livers at D7 post-transplant. (B) Relative fold changes of mmu-miR142-5p, mmu-miR181 in the normal, SYN, TOL D100 and TOL D7 livers. *P≤0.05, ** P≤0.01, ***P≤0.001 via unpaired T-test, adjusted for FDR 10%.
In order to identify miRNA that are altered in established tolerance, we profiled miRNA from a group (n=3) of DA→Lewis transplants that were treated with TLI (TOL D100), and compared them at D100 post-transplant against healthy, nontransplanted DA liver controls. Of the 158 miRNAs investigated we observed 42 miRNAs that were differentially altered in TOL D100 compared to normal DA liver using a Student's T-test and FDR adjusted P≤0.05 (Table 3). Of the 42 differentially modulated miRNA RNA, we observed that 19 miRNAs were increased in established tolerance (TOL D100) as compared to nontransplanted DA livers (Table 3).
Table 3.
Forty-two differentially modulated miRNA's in established tolerance (TOL as compared to normal livers. Data expressed as mean and standard error of the mean (SEM).
| miRNA | Established Tolerance (mean±SEM) | Normal liver (mean±SEM) | Adjusted P-value | Direction in TLID100 vs. NORM |
|---|---|---|---|---|
| mmu-miR-542-5p | 1.2 ± 0.0 | 0.1 ± 0.0 | 2.8×10−5 | Up |
| mmu-miR-872 | 1.1 ± 0.0 | 42.3 ± 3.1 | 1.8×10−4 | Down |
| mmu-miR-484 | 2.0 ± 0.2 | 0.1 ± 0.0 | 3.0×10−4 | Up |
| rno-miR-190b | 1.5 ± 0.2 | 4897.7 ± 493.0 | 5.8×10−4 | Down |
| mmu-miR-138 | 0.7 ± 0.0 | 1.6 ± 0.1 | 0.0012 | Down |
| rno-miR-466c | 2.9 ± 0.3 | 0.6 ± 0.2 | 0.0022 | Up |
| rno-miR-381 | 5.8 ± 0.7 | 1.2 ± 0.1 | 0.0029 | Up |
| mmu-miR-18a | 1.4 ± 0.2 | 0.2 ± 0.1 | 0.0030 | Up |
| mmu-miR-194 | 0.5 ± 0.1 | 1.5 ± 0.1 | 0.0034 | Down |
| mmu-miR-107 | 0.7 ± 0.3 | 15.4 ± 2.4 | 0.0036 | Down |
| mmu-miR-342-3p | 3.7 ± 0.6 | 0.3 ± 0.1 | 0.0041 | Up |
| mmu-miR-142-3p | 2.9 ± 0.2 | 1.2 ± 0.3 | 0.0047 | Up |
| mmu-miR-142-5p | 3.5 ± 0.4 | 0.9 ± 0.2 | 0.0057 | Up |
| mmu-miR-365 | 0.1 ± 0.0 | 1.4 ± 0.2 | 0.0057 | Down |
| mmu-miR-27b | 0.5 ± 0.0 | 1.2 ± 0.1 | 0.0063 | Down |
| mmu-miR-20a | 0.9 ± 0.1 | 1.7 ± 0.1 | 0.0071 | Down |
| mmu-miR-320 | 0.7 ± 0.1 | 1.8 ± 0.2 | 0.0076 | Down |
| mmu-miR-671-3p | 1.1 ± 0.0 | 1.4 ± 0.1 | 0.0080 | Down |
| mmu-miR-151-3p | 0.4 ± 0.1 | 3.0 ± 0.5 | 0.0092 | Down |
| mmu-miR-7a | 0.8 ± 0.1 | 0.2 ± 0.0 | 0.0101 | Up |
| mmu-miR-361 | 0.7 ± 0.1 | 1.2 ± 0.1 | 0.0110 | Down |
| mmu-miR-125a-5p | 5.1 ± 0.5 | 2.3 ± 0.4 | 0.0111 | Up |
| mmu-miR-672 | 0.4 ± 0.1 | 1.6 ± 0.2 | 0.0114 | Down |
| rno-miR-333 | 1.0 ± 0.3 | 4.7 ± 0.8 | 0.0115 | Down |
| mmu-miR-125a-3p | 5.6 ± 0.4 | 2.4 ± 0.6 | 0.0125 | Up |
| mmu-miR-181a | 5.6 ± 0.5 | 2.4 ± 0.6 | 0.0125 | Up |
| mmu-miR-30d | 1.2 ± 0.1 | 2.0 ± 0.2 | 0.0145 | Down |
| mmu-miR-96 | 1.3 ± 0.3 | 0.2 ± 0.1 | 0.0146 | Up |
| mmu-miR-31 | 0.7 ± 0.2 | 1.6 ± 0.1 | 0.0163 | Down |
| mmu-miR-503 | 2.7 ± 0.5 | 0.6 ± 0.1 | 0.0165 | Up |
| rno-miR-196c | 49.5 ± 10.6 | 8.0 ± 1.3 | 0.0178 | Up |
| mmu-miR-128a | 1.8 ± 0.3 | 0.7 ± 0.1 | 0.0182 | Up |
| mmu-miR-26a | 0.6 ± 0.0 | 2.1 ± 0.4 | 0.0184 | Down |
| mmu-miR-340-5p | 1.9 ± 0.1 | 2.6 ± 0.2 | 0.0187 | Down |
| mmu-let-7f | 1.0 ± 0.2 | 2.4 ± 0.3 | 0.0192 | Down |
| mmu-miR-322 | 1.5 ± 0.1 | 0.9 ± 0.1 | 0.0196 | Up |
| mmu-let-7b | 0.7 ± 0.1 | 1.4 ± 0.2 | 0.0209 | Down |
| mmu-miR-15b | 1.9 ± 0.2 | 1.0 ± 0.1 | 0.0214 | Up |
| mmu-miR-192 | 0.3 ± 0.0 | 1.3 ± 0.3 | 0.0238 | Down |
| mmu-miR-125b-5p | 0.7 ± 0.1 | 1.4 ± 0.2 | 0.0246 | Down |
| mmu-miR-99b | 4.2 ± 0.6 | 1.6 ± 0.3 | 0.0247 | Up |
To determine the miRNAs are associated with the induction and maintenance of tolerance we examined miRNA that were significantly differentially altered under three conditions, A) increased in established TOL (D100) compared to native livers, B) increased in early tolerance (D7) compared to syngeneic allografts, and C) increased in established TOL (D100) compared to syngeneic grafts. Two miRNA, mmu-miR142-5p and mmu-miR181a fit these criteria (Figure 3B). Expression of these miRNAs was increased in the tolerant group as compared to the syngeneic groups. Lymphocytes were isolated from one to three additional TOL graft recipients at 7, 35 and 100 days post-transplant and analyzed by q-PCR for mmu-miR142-5p and mmu-miR181a5,11. Both mmu-miR142-5p and mmu-miR181a were substantially elevated in the TOL lymphocytes as compared to lymphocytes isolated from normal liver suggesting that lymphocytes are the source of these miRNAs in our model (Supplementary Figure 1A). Further we show quite dramatically that mmu-miR142-5p and mmu-miR181a are significantly correlated with the numbers of infiltrating lymphocytes isolated from the liver grafts (Supplementary data 1B). Taken together, our results indicate that specific lymphocyte miRNAs can be identified that are associated with the establishment and maintenance of tolerance.
Established Tolerance is the Return to a Non-Activated State of miRNA Expression
In order to elucidate the overall pattern of miRNA expression in the different states of immune activation, we performed principal component analysis (PCA) and included every miRNA that had a full complement of expression values; thus 151 miRNAs were analyzed, as seven miRNA were removed for missing values for one or more samples in any of the study groups (Supplementary Figure 2). The study groups clearly separated with three principal components, describing 72% of the variance (Figure 4A).
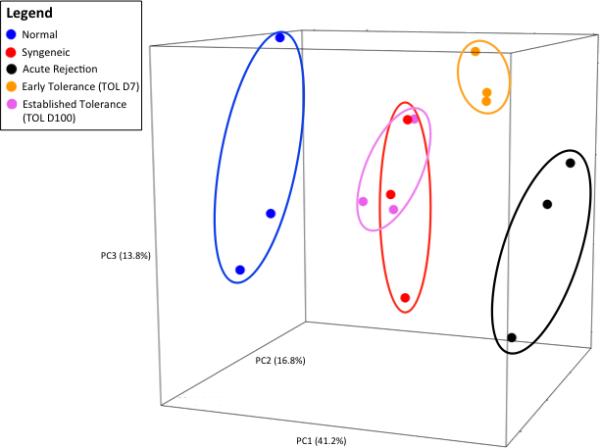
Figure 4. Established TOL livers group with SYN liver using miRNA expression in PCA.
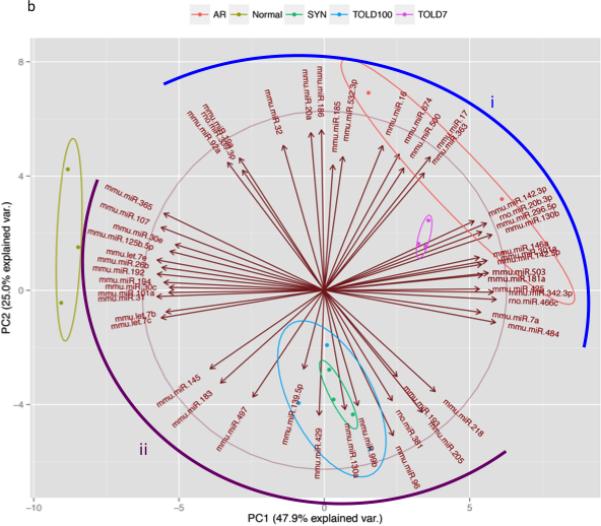
(A) PCA of the 151 miRNAs that had a full complement of values was able to discriminate the study groups from each other with the first 3 principal components (71.8% of variance). (B) PCA with the miRNA driving the first two principal components, the arrow heads demonstrate the positive or increased direction (1) miRNA influencing acute rejection and early immune activation, and (2) miRNA influencing an absence of immune activation or negative regulators of inflammation.
Using the first three principal components we observed that native livers were found in the left most portion of principal component one (PC1), in contrast to AR at the opposite right most portion of PC1 representing the extremes of the miRNA expression profiles. The TOL D7 group was positioned near the AR group, indicating an overall similar expression profile of miRNA. In contrast, TOL D100 and SYN groups were seen to occupy the same relative three-dimensional space, shifted towards the native DA liver groups, indicating extremely similar (almost identical) expression profiles of these two study groups. To more clearly identify the miRNAs driving the separation of our five experimental groups, a supervised PCA, using the 53 most informative miRNAs for PC1 and PC2 was performed (Figure 4B). Quite dramatically the miRNAs separated into two main groups, group i miRNAs that are expressed in hematopoietic cells and regulate immune responses were increased in early allografts (AR and early tolerance). Conversely, the group ii miRNAs are known tumor suppressor or negative regulators of inflammation.
Thus, our findings support a model whereby established, non-rejecting liver allografts express a profile resembling that of genetically matched, syngeneic, transplanted liver.
Discussion
We examined a well-characterized, high-responder model of OLT and demonstrate a distinct miRNA profile during graft rejection and upon the induction and maintenance of allograft tolerance. Of the miRNAs significantly increased in the tolerant groups, the miRNAs 142-5p and miR-181 are of particular interest since they were significantly increased in the TOL D7 group and continued to be significantly elevated (as compared to syngeneic grafts and native liver controls) in the TOL D100. miR-142-3p was similarly elevated in the TOL D7 and D100 groups however miR-142-3p was also significantly elevated during AR. The miR-142 gene family is of considerable interest as it has been reported to be associated with both rejection and tolerance after transplantation 22-26. Analysis of biopsy tissue and urine from renal allograft recipients with chronic allograft dysfunction demonstrated increased levels of miR-142-3p 26. Formalin fixed paraffin embedded sections from biopsies of intestinal allografts were examined and miR-142-3p was also found to be associated with acute cellular rejection 23. In contrast, analysis of PBMC from kidney graft recipients identified as drug-free, operationally tolerant identified miR-142-3p as being highly differentially expressed in OT patients as compared to immunosuppressed patients with stable graft function 24. B cells were the source of miR-142-3p. We demonstrate that another miR-142 family member, miR-142-5p correlated with both the induction and maintenance of tolerance to liver allografts. However, other studies indicate miR-142-5p as being diagnostic of AR of human renal allografts 22,25. miR-142 is one of the few miRNAs that are preferentially expressed in hematopoietic cells and its expression is abundant in immune cells. A recent study examining the biological function of miR-142 determined that this miRNA is a critical regulator of lymphopoiesis 27. Mice deficient in miR-142 had an enlarged splenic B-cell compartment and an abnormal expansion of marginal zone-like B cells. T cells and B1 B cells were decreased in the periphery of the miR-142−/− mice. Further, miR-142 is important in the generation of humoral and cellular immunity in mice. Given the important role of miR-142 in the immune response and its enhanced expression in immune cells, it is not surprising that increased expression is detected after transplantation. The association of miR-142 with both rejection and tolerance in many studies likely reflects the diverse role of B cells in regulation of the alloimmune response. Further, there are dozens of putative targets of miR-142 many that are involved in B cell function and other immune processes. Further studies are necessary to define, and biologically validate, the targets of miR-142 during rejection and the initiation and maintenance of tolerance.
Similar to miR-142, miR-181 was elevated in tolerant liver allografts by D7 post-transplant and remained significantly elevated through D100. To our knowledge, our findings are the first report of a role for this miRNA in infiltrating lymphocytes after transplantation. The miR-181 family has been shown to regulate cellular growth, development, and activation including the immune response 28. Studies suggest that miR-181 acts independently in the T and B cell lineages, performing different functions and targeting different mRNA in T and B cells. In B cells, miR-181 acts as a positive regulator of development 29. miR-181a is important in T cell development and studies show that increasing miR-181a expression in CD4 T cells decreases TCR signaling threshold and increases antigen sensitivity 29,30. Further, miRNA expression profiling of subsets of self-antigen specific CD8 T cells demonstrated that miR-181a was highly expressed in tolerant CD8 T cells but was low in the memory subset 31. We recently demonstrated that miR-181a is increased in plasmacytoid dendritic cells (pDC) as compared to conventional DC (unpublished). Moreover, injection of miR-181a-expressing pDC to recipients of cardiac allografts prolonged graft survival whereas pDC from mice deficient in miR-181a did not. Taken together, these findings suggest miR-181a may be a novel key regulator of tolerance.
In our previous study that utilized this same OLT/TLI model, it was determined that the numbers of Tregs in in the allograft on D100 was similar to that seen in a naïve liver whereas the numbers of Treg detected in PBMC remained significantly increased 5. When Treg were prospectively analyzed in the peripheral blood and graft of pediatric liver transplant recipients it was determined that the mean level of circulating Tregs remained stable in patients in the absence of rejection or infection but decreased significantly during episodes of acute rejection 32. This decrease in peripheral Treg is accompanied by a concomitant increase of Treg levels in the allograft. It remains unclear as to whether Tregs alone are responsible for the alloimmune suppression in our model33. Treg cells have a diverse repertoire of mechanisms to inhibit and prevent immune cell activation including both cell-contact dependent and independent suppression 34. It is important to note that one critical miRNA, miR-155, that has been shown in several studies to be associated with Treg development was not included in the miRNA cards used in this study 33. A recent study has determined that Treg utilize miRNA-containing exosomes to suppress immune responses 35. Treg cells are thereby able to transfer miRNA to immune cells to suppress Th1 cell proliferation and cytokine secretion. Further studies will determine whether the miRNAs associated with tolerance in our study are associated with Treg cells or Treg cell-derived exosomes.
Our findings demonstrate, quite clearly, that that normal, nontransplanted livers are grouped distant from all the liver grafts when all miRNAs are surveyed by PCA. As would be expected, rejecting allografts (AR) cluster farthest from native livers further supporting that rejection does result in a distinct profile of miRNAs. The AR group and the early tolerance group differ only in that the D7 TOL group received the TLI-based tolerogenic regimen which we have previously shown results in diminished alloactivation hence D7 TOL shifts closer to the syngeneic grafts. There is however, a complete overlap between the syngeneic and the established tolerance, D100 TOL groups, suggesting established tolerance livers demonstrate minimal alteration of a normal miRNA profile consistent with little to no induction of alloactivation. Taken together our PCA of differentially expressed miRNAs suggest that by day 100 post-transplant, the liver graft may be in a quiescent state and that active regulation, by miRNAs associated with hematopoietic cells or regulation of the immune response, is no longer necessary. Indeed the microRNAs present in native nontransplanted liver, syngeneic liver grafts and liver allografts with established tolerance, are miRNA that have been identified as tumor suppressors in cancer studies, suggesting these are the miRNAs associated with homeostasis and abrogation of cellular proliferation. Studies that determine the in vivo targets and biological functions of the miRNAs identified in this study are necessary and warranted. Moreover, since only limited liver grafts were analyzed in this study, further experiments with additional numbers of samples, including grafts obtained at both earlier and later time points would be informative to establish if the stable allograft continues to shift towards the phenotype of a native liver. The liver is unique among transplanted organs in its well-established propensity towards a tolerogenic environment. The mechanism for this phenomenon has not been elucidated, although the unique composition of liver, the resident cells of the liver (NK and NKT subsets), immune exhaustion (through PD-1 interactions) and deletion or apoptosis of immune cells have all been proposed as plausible pathways leading to tolerance 36,37. Our findings suggest that the miRNAs present in liver allografts early after transplant are those associated with cells of the immune system and immune responses and that in the absence of immune events such as rejection and infection, these miRNAs decrease indicating the stable state post-transplantation.
Supplementary Material
Acknowledgments
MJV would like to sincerely thank Stephan Busque for his mentorship and manuscript advice.
Funding
This work was supported in part by the National Institutes of Health (AI044095 and AI084939) and by a generous gift from Robert and Patty Moore. LW and MF were supported by fellowships from the Transplant and Tissue Engineering Center of Excellence at Lucile Packard Children's Hospital. AHL was supported by an American Heart Association Early Career Award and a Pilot Early Career Award from Stanford Child Health Research Institute.
Abbreviations
- OLT
orthotropic liver transplant
- TLI
total lymphoid irradiation
- TOL
tolerance
- SYN
syngeneic
- AR
acute rejection
- PCA
principal component analysis
Footnotes
MJV contributed to study design, analyzed the data, data interpretation and manuscript preparation, LW designed the study and performed the experiments, MF performed the transplants, AHL and EL performed some experiments, COE and OMM were involved in project design, data interpretation, and manuscript preparation. SMK supervised the project, designed and interpreted all experiments, and wrote the manuscript.
Disclosures
The authors declare no conflicts of interest.
References
- 1.Lau AH, de Creus A, Lu L, Thomson AW. Liver tolerance mediated by antigen presenting cells: fact or fiction? Gut. 2003;52(8):1075–1078. doi: 10.1136/gut.52.8.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomson AW, Knolle PA. Antigen-presenting cell function in the tolerogenic liver environment. Nature reviews. Immunology. 2010;10(11):753–766. doi: 10.1038/nri2858. [DOI] [PubMed] [Google Scholar]
- 3.Feng S, Ekong UD, Lobritto SJ, et al. Complete immunosuppression withdrawal and subsequent allograft function among pediatric recipients of parental living donor liver transplants. Jama. 2012;307(3):283–293. doi: 10.1001/jama.2011.2014. [DOI] [PubMed] [Google Scholar]
- 4.Qian S, Demetris AJ, Murase N, Rao AS, Fung JJ, Starzl TE. Murine liver allograft transplantation: tolerance and donor cell chimerism. Hepatology. 1994;19(4):916–924. doi: 10.1002/hep.1840190418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujiki M, Esquivel CO, Martinez OM, Strober S, Uemoto S, Krams SM. Induced tolerance to rat liver allografts involves the apoptosis of intragraft T cells and the generation of CD4(+)CD25(+)FoxP3(+) T regulatory cells. Liver transplantation : official publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society. 2010;16(2):147–154. doi: 10.1002/lt.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris A, Krams SM, Martinez OM. MicroRNAs as immune regulators: implications for transplantation. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2010;10(4):713–719. doi: 10.1111/j.1600-6143.2010.03032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaul V, Krams S. Micro RNAs as master regulators of immune responses in transplant recipients. Current Opinion in Organ Transplantation. 2014 doi: 10.1097/MOT.0000000000000148. in press. [DOI] [PubMed] [Google Scholar]
- 8.Wei L, Gong X, Martinez OM, Krams SM. Differential expression and functions of microRNAs in liver transplantation and potential use as non-invasive biomarkers. Transplant immunology. 2013;29(1-4):123–129. doi: 10.1016/j.trim.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nature reviews. Immunology. 2008;8(2):120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 10.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nature reviews. Genetics. 2012;13(5):358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei L, Wang M, Qu X, et al. Differential expression of microRNAs during allograft rejection. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2012;12(5):1113–1123. doi: 10.1111/j.1600-6143.2011.03958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obara H, Nagasaki K, Hsieh CL, et al. IFN-gamma, produced by NK cells that infiltrate liver allografts early after transplantation, links the innate and adaptive immune responses. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2005;5(9):2094–2103. doi: 10.1111/j.1600-6143.2005.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.TeamR: A language and environment for statistical computing. [computer program] ComputingR Foundation for Statistical Computing R Foundation for Statistical Computing; 2007. [Google Scholar]
- 14.Becker RA, Chambers JM, Wilks AR. The New S Language:A Programming Environment for Data Analysis and Graphics. Wadsworth & Brooks/Cole; Pacific Grove, CA, USA: 1988. [Google Scholar]
- 15.Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. Academic Press; 1979. [Google Scholar]
- 16.Venables WN, Ripley BD. Modern Applied Statistics with S. Fourth edition Springer; 2002. [Google Scholar]
- 17.devtools: Tools to Make Developing R Packages Easier [computer program] 2015 [Google Scholar]
- 18.Wickham H. ggplot2Elegant Graphics for Data Analysis. 1 ed Springer-Verlag; New York: 2009. [Google Scholar]
- 19.Adler D, Murdoch D. 3D visualization device system. 2014 https://r-forge.r-project.org/projects/rgl/
- 20.Egawa H, Martinez OM, Quinn MB, et al. Acute liver allograft rejection in the rat. An analysis of the immune response. Transplantation. 1995;59(1):97–102. doi: 10.1097/00007890-199501150-00017. [DOI] [PubMed] [Google Scholar]
- 21.Krams SM, Egawa H, Quinn MB, Villanueva JC, Garcia-Kennedy R, Martinez OM. Apoptosis as a mechanism of cell death in liver allograft rejection. Transplantation. 1995;59(4):621–625. [PubMed] [Google Scholar]
- 22.Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A. 2009;106(13):5330–5335. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asaoka T, Sotolongo B, Island ER, et al. MicroRNA signature of intestinal acute cellular rejection in formalin-fixed paraffin-embedded mucosal biopsies. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2012;12(2):458–468. doi: 10.1111/j.1600-6143.2011.03807.x. [DOI] [PubMed] [Google Scholar]
- 24.Danger R, Pallier A, Giral M, et al. Upregulation of miR-142-3p in peripheral blood mononuclear cells of operationally tolerant patients with a renal transplant. Journal of the American Society of Nephrology : JASN. 2012;23(4):597–606. doi: 10.1681/ASN.2011060543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danger R, Paul C, Giral M, et al. Expression of miR-142-5p in peripheral blood mononuclear cells from renal transplant patients with chronic antibody-mediated rejection. PloS one. 2013;8(4):e60702. doi: 10.1371/journal.pone.0060702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scian MJ, Maluf DG, David KG, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2011;11(10):2110–2122. doi: 10.1111/j.1600-6143.2011.03666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer NJ, Wang WL, Reyes EY, et al. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood. 2015;125(24):3720–3730. doi: 10.1182/blood-2014-10-603951. [DOI] [PubMed] [Google Scholar]
- 28.Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nature reviews. Immunology. 2013;13(9):666–678. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 30.Ebert PJ, Jiang S, Xie J, Li QJ, Davis MM. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nature immunology. 2009;10(11):1162–1169. doi: 10.1038/ni.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schietinger A, Delrow JJ, Basom RS, Blattman JN, Greenberg PD. Rescued tolerant CD8 T cells are preprogrammed to reestablish the tolerant state. Science. 2012;335(6069):723–727. doi: 10.1126/science.1214277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stenard F, Nguyen C, Cox K, et al. Decreases in circulating CD4+CD25hiFOXP3+ cells and increases in intragraft FOXP3+ cells accompany allograft rejection in pediatric liver allograft recipients. Pediatric transplantation. 2009;13(1):70–80. doi: 10.1111/j.1399-3046.2008.00917.x. [DOI] [PubMed] [Google Scholar]
- 33.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. Journal of immunology. 2009;182(5):2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 34.Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30(5):636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Okoye IS, Coomes SM, Pelly VS, et al. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity. 2014;41(1):89–103. doi: 10.1016/j.immuni.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crispe IN. Immune tolerance in liver disease. Hepatology. 2014;60(6):2109–2117. doi: 10.1002/hep.27254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benseler V, Tay SS, Bowen DG, Bertolino P. Role of the hepatic parenchyma in liver transplant tolerance: a paradigm revisited. Digestive diseases. 2011;29(4):391–401. doi: 10.1159/000329802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.