Summary
Damage repair is a fundamental requirement of all life as organisms find themselves in challenging and fluctuating environments. In particular, damage to the barrier between an organism and its environment (e.g., skin, plasma membrane, bacterial cell envelope) is frequent because these organs/organelles directly interact with the external world. Here we discuss the general strategies that bacteria use to cope with damage to their cell envelope and their repair limits. We then describe a novel damage-coping mechanism used by multicellular myxobacteria. We propose that cell-cell transfer of membrane material within a population serves as a wound-healing strategy and provide evidence for its utility. We suggest that – similar to how tissues in eukaryotes have evolved cooperative methods of damage repair – so too have some bacteria that live a multicellular lifestyle.
Keywords: damage dilution, damage repair, lipopolysaccharide, myxobacteria, outer membrane exchange, rejuvenation
Introduction
Multicellularity serves as the cornerstone for the specialization of cells and tissues that function synchronously for the benefit of the organism. Multicellularity also necessitates resource sharing between cells. Each cell contributes to the organism's success by the use of communication and cooperation networks with other cells. This includes a concerted response to cellular or tissue damage during which multiple cells, through signaling and response pathways, organize a wound repair program. Cooperation provides an advantage over independent cells responding to their own survival, as resources can be allocated from healthy to damaged tissue. Although the multicellular contribution to wound repair in higher eukaryotes is widely recognized [1,2], its contribution to damage repair in cooperative bacteria is largely unexplored.
Like all organisms, bacteria must maintain a highly controlled compartment that is separated from their environment and must sustain homeostasis in the face of external stresses. Successful organisms have evolved the ability to adapt to common stressors and to actively repair molecular- and macro-scale damage. Radiation, desiccation, osmolality, pH, temperature, metabolic errors, predators, reactive molecules and physical insults all threaten the integrity of the individual. Molecular and structural damage caused by such stressors must be either avoided or repaired to prevent fitness loss. For example, damage to DNA is an immediate threat to the fitness of the organism and is also heritable. For this reason, cells devote many resources to the repair of DNA, which has been well studied [3] and will not be discussed further here. However, damage can occur to every component of the cell, and maintaining health in response to this damage is an essential feature of the cell.
Although a number of self-repair mechanisms that address non-DNA damage have been described in bacteria, their ability to carry out repairs has limits. Moreover, it has been postulated that it would be impossible for a cell to address each type of damage with a dedicated repair system, because there are too many forms of damage [4]. As a result, damage slowly accumulates during an organism's lifetime. This notion helps to explain the observed phenomenon of cellular aging—the gradual loss of cell function and the inability to recover from damage inflicted by stress that ultimately leads to senescence or cell death. In recent years, aging has been described in single-celled organisms that reproduce asymmetrically or symmetrically by binary fission [5,6]. This finding implies that the known bacterial stress response systems that shield or repair cell damage are incomplete and thus damage accumulates over time. For single-celled organisms this necessitates other strategies such as damage segregation or shedding [7–9], although these strategies are not ideal (see below). Alternatively, multicellular bacteria can potentially share resources to aid damaged kin and to distribute a damage burden, making it plausible that multicellular repair mechanisms have evolved among certain bacteria. Until recently this possibility was unexplored.
We found that myxobacteria can use a cooperative strategy to cope with damage to their outer membrane (OM) [10]. Myxobacteria live in groups that move and function as a single unit with synchronized behaviors, including the formation of macroscopic multicellular fruiting body structures [11]. Because their main habitat is soil, they are exposed to many insults that can damage their cell envelope. We have reported the curious finding that myxobacteria fuse their OMs and exchange membrane proteins, lipids and lipopolysaccharide (LPS) [12]. Importantly, this fusion and exchange of contents can support the life of otherwise lethally damaged cells in a population [10]. Aside from the ability to exchange protein and LPS material to complement certain mutant phenotypes [13], we have also directly shown that fluorescently labeled lipids and proteins can be transferred between cells [14,15]. From these results we propose the following as a central thesis of this essay: OM exchange (OME) is used as a cooperative multicellular behavior to replenish cells with critical components and to dilute damaged factors among the population to allow cell damage to be buffered. In turn this might increase the efficiency of how damage components are repaired, recycled and/or removed. In other words, the nature of multicellularity provides a platform for ailing cells to be supplied with healthy components from other cells, and this facilitates repair. Indeed, direct cell-to-cell content sharing has been shown to play a role in eukaryotic stress and damage coping mechanisms [1,2]. By analogy, we propose that OME in myxobacteria acts as a multicellular wound-healing strategy by infusing healthy membrane and diluting damaged material at the wound site, and we discuss the implications in terms of multicellular bacterial repair.
Bacterial cell envelopes are susceptible to damage
Molecular damage can occur to all structures of a cell; however, in this essay we focus primarily on damage inflicted to the cell envelope of gram-negative bacteria. The cell envelope is a key organelle because it serves as a protective barrier to the cytoplasm, and its location puts the envelope in harm’s way as it is directly exposed to insults from the external world [16]. The envelope not only functions as a barrier but also senses and relays extracellular information to the cytoplasm, provides structural integrity to the cell and is the gateway for nutrient acquisition. The canonical envelope consists of the inner membrane, the cell wall and the OM [16]. The OM is an asymmetric bilayer made up of LPS in the outer leaflet and phospholipid in the inner leaflet. This membrane houses lipid-anchored lipoproteins and integral membrane proteins that typically assume a β-barrel fold. In addition, large protein complexes anchored in the cell envelope extend through the OM to function outside the cell (e.g. flagella, pili).
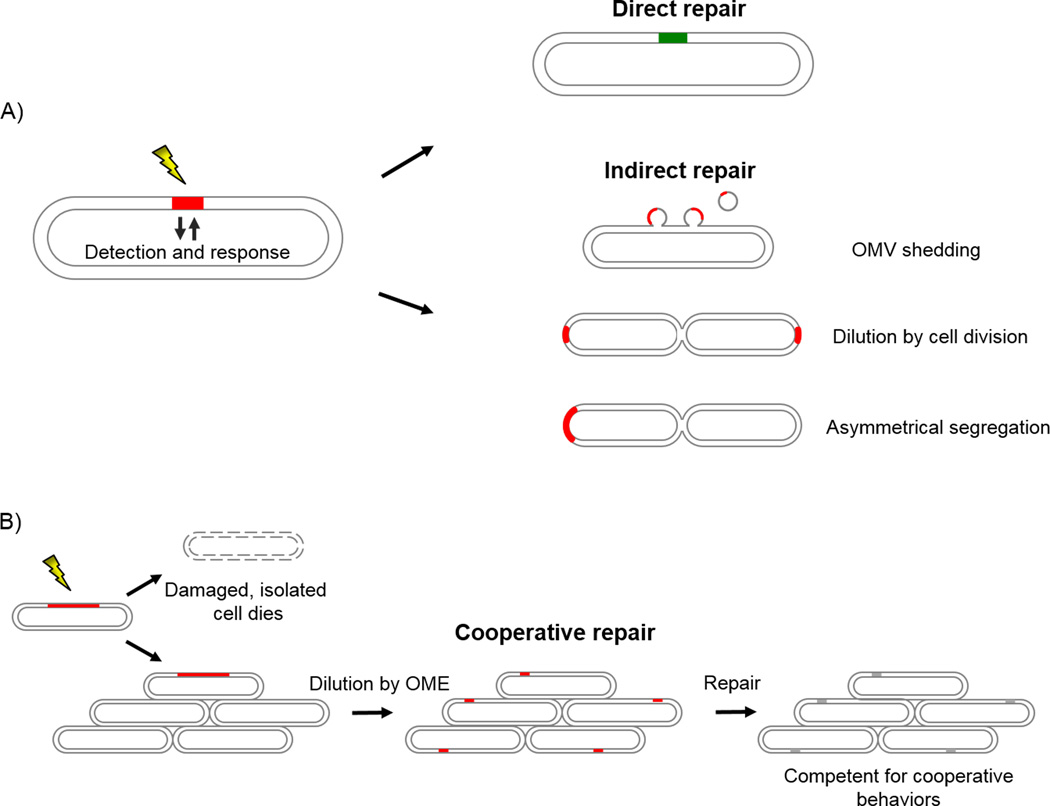
The components that make up the cell, which exist as complex assemblies of countless molecules, inevitably have imperfections and the cell envelope is no exception. Envelope damage, caused by environmental factors and internal errors in metabolism, can be manifest at a molecular or organizational (e.g. disruption of OM asymmetry) level. In particular, proteins are susceptible to damage by different means (reviewed in [17]). Physical stresses, such as heat, lead to misfolding and aggregation of envelope proteins. Oxidative reactions can result in protein and lipid oxidation [18,19]. Enzymatic, oxidative or hydrolytic reactions can damage the LPS sugars or acyl chains. Still other insults result in the release of OM proteins (OMPs) and LPS [20]. For instance, antibiotics or chelating agents can destabilize lateral LPS bridges, resulting in the release of proteins and LPS [21]. As a cell inevitably takes on and accumulates these and other forms of damage there are three possible outcomes: (i) cell senescence or death; (ii) cellular repair; or (iii) dilution, segregation or shedding of damage. Both restoration of function (direct repair) and loss of damage (indirect repair) are used by bacteria as single-cell recovery strategies (Fig. 1A).
Fig. 1.
Envelope repair strategies. A: A stress inflicts damage (red) to the cell envelope. The direct repair pathways sense this damage and respond by the induction of cellular machinery which results in repair (green). In indirect repair, damaged material is shed by OM vesicles (OMVs), diluted by cell division or asymmetrically segregated. B: Multicellular myxobacteria dilute cell envelope damage by OME. In some cases the dilution of damage may be sufficient in and of itself to alleviate the stress. In other cases, dilution makes damage more manageable for direct autonomous repair and/or indirect repair (grey; indicating alternative possibilities). An isolated damaged cell cannot participate in OME and dies.
Bacteria have evolved repair systems to directly address cell envelope damage
Perturbations to the cell envelope are sensed and responded to by signaling pathways in the individual cell. Of these pathways, the best studied are in the model organism Escherichia coli and related species. In E. coli, damage cues such as protein misfolding [22], abnormal LPS [23] and antimicrobial peptides [24] trigger responses including the Cpx [25], σE [26], Rcs [27] and Bae [28] regulons, which direct gene expression to enhance the repair programs [29]. These regulons include proteins that function as envelope chaperones or proteases or are involved in OMP and LPS transport, assembly and maintenance [29]. For instance, the periplasmic chaperone-protease DegP monitors the periplasm for unfolded proteins and can either refold or degrade them [30,31]. Oxidative damage in the cell envelope can be repaired by a number of reducing systems [32]. For example, the OM MsrA/B proteins of Neisseria provide protection against reactive oxygen species by reducing methionine sulfoxide residues in oxidatively damaged proteins [33]. Furthermore, proteases that reside in the OM, such as OmpT, can cleave foreign (antimicrobial) peptides that bind and inhibit LPS function [34]. Although these types of systems allow bacteria to respond and adapt to envelope stressors, they can nevertheless be overloaded, leading to permanent envelope damage and death. Indeed, cells that have lost their reproductive ability because of the accumulation of oxidative damage have upregulated stress responses, indicating that these mechanisms are not always sufficient for cell rejuvenation [35].
Certain stressors (temperature, antibiotics, EDTA) cause organizational damage to the OM such as the loss of protein and LPS molecules [21]. Damage to LPS or β-barrel proteins can cause phospholipids to mislocalize from the inner leaflet to the outer leaflet. The resulting OM loses its asymmetry and consequently reduces its permeability and protective properties [36]. In turn the cell responds to this damage. For instance, when phospholipids are mislocalized in the outer leaflet, some bacteria adapt by destroying them with PldA phospholipase [37,38]. In another example, the conserved OMP PagP of Salmonella can cleave stray outer leaflet phospholipids and in turn adds the resulting palmitoyl chain to lipid A of LPS, giving the molecule a hepta-acylated lipid anchor [39–41]. This palmitoylation also provides an adaptive response to enhance survival [42]. The Mla system provides yet another means for maintaining bilayer asymmetry, apparently by retrograde transport of excess phospholipids from the outer to the inner membrane [38]. These are examples of mechanisms that maintain an asymmetric and hence functional OM bilayer, but there are fundamental limits to their healing ability. Either the systems can become overwhelmed, or the cell lacks a repair pathway for the damage that has been acquired. In fact, cells often require more drastic and costly strategies—indirect repair mechanisms—that shed or segregate damaged material in the bilayer.
Bacteria discard damaged components during indirect repair
The mentioned direct repair mechanisms are involved in the degradation or restoration of defective material, but these repair strategies are not sufficient for all forms of damage. For example, these response pathways do not address the repair of mature OMP and LPS molecules. Whereas phospholipid turnover has been described in some detail [43], the question of how damaged, undesired or excessive integral OMP and LPS components are dealt with still remains. In E. coli it was recently shown that older OMPs are displaced to the cell poles, and thus the mother cell partitions old and new OMPs upon cell division [44,45]. This implies that, at least in E. coli, there is no mechanism to turn over OMPs. In addition, as LPS transport is apparently unidirectional [46], there is no known mechanism for recycling this component. This suggests that OMP and LPS genesis and insertion are in a sense irreversible and that the default repair mechanism for the OM involves shedding (OM vesicle or tube secretion [47,48]) and asymmetric distribution of damage to repository (aged) cells [7,9]. It has been shown in E. coli that the amount of vesicle formation correlates with the amount of protein accumulation in the cell envelope and increases in the absence of active stress response pathways. Furthermore, vesiculation enhances survival under stressful conditions and preferentially packages damaged proteins [47]. OMV production has been observed as a general stress response in a variety of bacteria, and has been shown to respond to misfolded proteins, accumulating peptidoglycan or LPS fragments and oxidative stress, all of which indicate cell damage or aging [49]. Aside from shedding, damaged molecules can be partitioned into one of two cell poles thereby creating a healthy daughter cell and a damage repository daughter cell after cell division. This was demonstrated by the observation of asymmetric division of protein aggregates [7], the reduced fitness of old-pole derived daughters [5,6], and that old OMPs migrate to only one pole [44]. These studies were mostly done in E. coli and more work is needed to determine if this is a general strategy to combat aging. We designate these healing strategies "indirect repair" to distinguish them from the restorative activity that takes place in direct repair (Fig. 1A). The existence of indirect repair strategies indicates that direct repair is not always adequate. During indirect repair, irreparable damage is intentionally discarded or segregated. In the case of segregation, the daughter cells that inherit the old pole accumulate damage following repeated cell divisions. Consequently these cells have decreased fitness and over time reach senescence [6,50].
All mechanisms of repair described so far have drawbacks if one considers a cell in a nutrient-limited habitat, as is typically found in nature [51]. Mechanisms that involve simple dilution or segregation of damage by cell division require growth to outpace damage accumulation (Fig. 1A). De novo synthesis of stress response pathways is similarly metabolically costly, and, although it may keep the cell alive for a period of time, it may hamper the ability of the cell to seek a more favorable habitat by exhausting local nutritional resources. Segregation of damage to a repository cell is particularly costly considering it demands the biosynthesis of a daughter cell. Similarly, OM shedding is sustainable only when the lost material is replaced with newly synthesized macromolecules.
Multicellular bacteria have the potential for unique solutions to these problems. For instance, damaged material that is shed could be re-metabolized by sibling cells akin to a bacterial autophagy-like process [52]. In addition, cannibalism or the sacrifice of some cells for the benefit of the population has been described [53,54]. If ‘old’ or senescent cells or vesicles were cannibalized, it would also provide a mechanism to remove damage and salvage community material. However, there are drawbacks to content recycling, considering competitors could also consume these external resources. To our knowledge, the recycling of released extracellular material specifically from damaged cells has not yet been demonstrated. Another multicellular damage coping mechanism could rely on direct content exchange between individuals to dilute a damage burden without the need for cell division or shedding. In direct content exchange a pool of healthy material would be available to be freely distributed to ailing cells to buffer and combat damage. This scenario works providing there is a mechanism to exchange content between cells and an incentive to redistribute resources.
Content exchange as a strategy for multicellular repair
Indeed, content exchange is observed as a strategy for eukaryotic damage repair both at the cell-to-cell and organelle-to-organelle levels. For example, transfer of mitochondria between cells is able to rescue respiration in recipient cells with defective mitochondria [55]. Transfer of organelles can occur through tunneling nanotubes (TNTs), which define a broad collection of cell-to-cell tube-like connections found in eukaryotes. TNTs formed between ischemically damaged cardiomyoblasts and mesenchymal stem cells facilitate the transfer of mitochondria to rescue injured cells [56]. Cytoplasmic and organelle transfer via cell-cell contacts is also involved in the restoration of cell function of renal tube cells [57]. Furthermore, selective transfer of lysosomes between endothelial progenitor cells and damaged endothelial cells via TNTs rescued lysosome recipients [58]. Recently it was demonstrated that stressed cells preferentially form specialized TNTs to facilitate rescue from apoptosis by mitochondrial transfer from healthy cells [59]. Aside from mitochondria and lysosomes, TNTs also transfer Golgi and endoplasmic reticulum (ER) [60]. In addition to organelles, TNTs are involved in the transfer of membrane proteins [61,62], endosomes [63] and RNA [64,65]. Stress appears to play a major role in inducing TNT formation between cells [66,67]. Though the in vivo contribution of TNTs to cell repair is not fully described, clearly cell-to-cell transfer of contents via TNTs can contribute to health and homeostasis in multicellular organisms through content exchange.
Like during OME, tunneling nanotubes form a continuous membrane between cells and can thus facilitate the transfer of lipids and lipid-anchored proteins [68]. Another process of plasma membrane exchange takes place during trogocytosis during which lymphocytes extract membrane patches and proteins from target cells. While trogocytosis certainly plays some role in immune modulation, it has been proposed that trogocytosis could have evolved from a primitive mechanism of specialized cells "feeding" off of other cells [69], or, put in other terms, kin cells donating lipid and protein components. Still other membrane-based content sharing platforms occur in plant cells in which a continuous ER transverses plasmodesmata [70] to share lipids and proteins via the ER membrane and lumen [71]. Intercellular protein transfer is not at all uncommon [72], but knowledge of its use as a platform for cell repair is limited.
Content sharing is used as a strategy to repair damage at the organelle level as well. For instance, metazoan cells repair lesions to their plasma membrane by the rapid fusion of cytoplasmic membrane-enclosed organelles to seal the hole at the site of damage, thereby sacrificing the organelle for the benefit of the cell as a whole [2,73]. In another example, ER-derived vesicles containing nuclear DNA-derived products are delivered to mitochondria to replenish mitochondrial membranes [74]. Also striking is the process of mitochondrial fusion and fission during which damaged material is diluted by content exchange [75,76]. Importantly, mitochondrial exchange dynamics allow the complementation of defective DNA, mRNA and proteins among individual organelles [77]. Disruption of mitochondria exchange and repair impedes homeostasis and leads to organelle dysfunction [78]. Mitochondria that display little motility and fusion such as those in cardiomyocyte cells can exchange contents via nanotubular extensions [79].
Maintaining health and homeostasis of the organism as a whole to preserve its fitness serves as the incentive for content exchange between individual cells and organelles in multicellular systems. The question then arises: can multicellular bacteria or bacteria that benefit from colonial growth use cell-to-cell content exchange to support the health of individual cells in their community? We hypothesize that this indeed can occur and that OME in myxobacteria is a content sharing platform that allows cell rescue and rejuvenation and helps maintain OM homeostasis in a population.
Myxobacteria use content exchange to repair their damaged kin through OME
Our appreciation for cooperation among bacteria has rapidly increased [80]. With cooperation comes the potential for multicellular responses to damage. Eukaryotes have evolved to include multicellular damage responses such as wound repair and immune responses [2,81]. In microbes, relatedness in a population of cells provides an evolutionary basis for assisting kin by sharing resources [82]. For instance, the collaboratively built biofilm matrix of a related microbial community provides protection from external stressors [83]; in essence sacrificing the health of some cells for the benefit of others and for the good of the community. Although there are many examples of cooperative behaviors among bacteria, multicellular damage repair mechanisms in bacteria have been explored little.
Myxobacteria are an ideal system to study multicellular behaviors in prokaryotes. They are typically found in soil communities that rely on communication networks to perform synchronized behaviors, analogous to how tissues function in eukaryotes. One such interaction platform is the constitutive exchange of large amounts of OM material between cells by OME [84]. In a cell contact–dependent manner, myxobacteria transiently fuse their OMs [10], allowing rapid exchange by lateral diffusion of lipids, proteins and LPS from partnering cells. As a consequence, they can phenotypically complement OMP and LPS mutants by efficiently transferring OM components from a wild-type cell to a mutant [10,85–87]. Two proteins, TraA and TraB, which are localized on the cell surface, are the only factors known to be necessary for OME [14,88]. These proteins are required in both contacting cells, and presumed homotypic interactions between TraA receptors help to ensure that transfer takes place between kin cells [89]. Although the biological utility of OME is not fully understood, we hypothesize that two consequences of OME are the cooperative repair of damage and the maintenance of OM homeostasis in a population.
LPS is the outermost structure of the cell and directly interacts with other cells and with the environment. As such, LPS functions in a number of behaviors involving cell-cell and cell-substrate interactions. In Myxococcus xanthus, truncated LPS molecules caused by mutations in the biosynthesis of distal LPS sugars impair both type IV pili–mediated (social) motility and adventurous motility [90,91]. The impairment might involve defective contacts at the interface of the LPS and the gliding surface. As motility is critical for the synchronized behaviors of myxobacteria, these mutants also have severe defects in development [10,90]. Like other OMP defects, mutants with truncated LPS can be complemented by OME from a wild-type cell, thereby restoring motility and development to the mutant (Fig. 2A) [10].
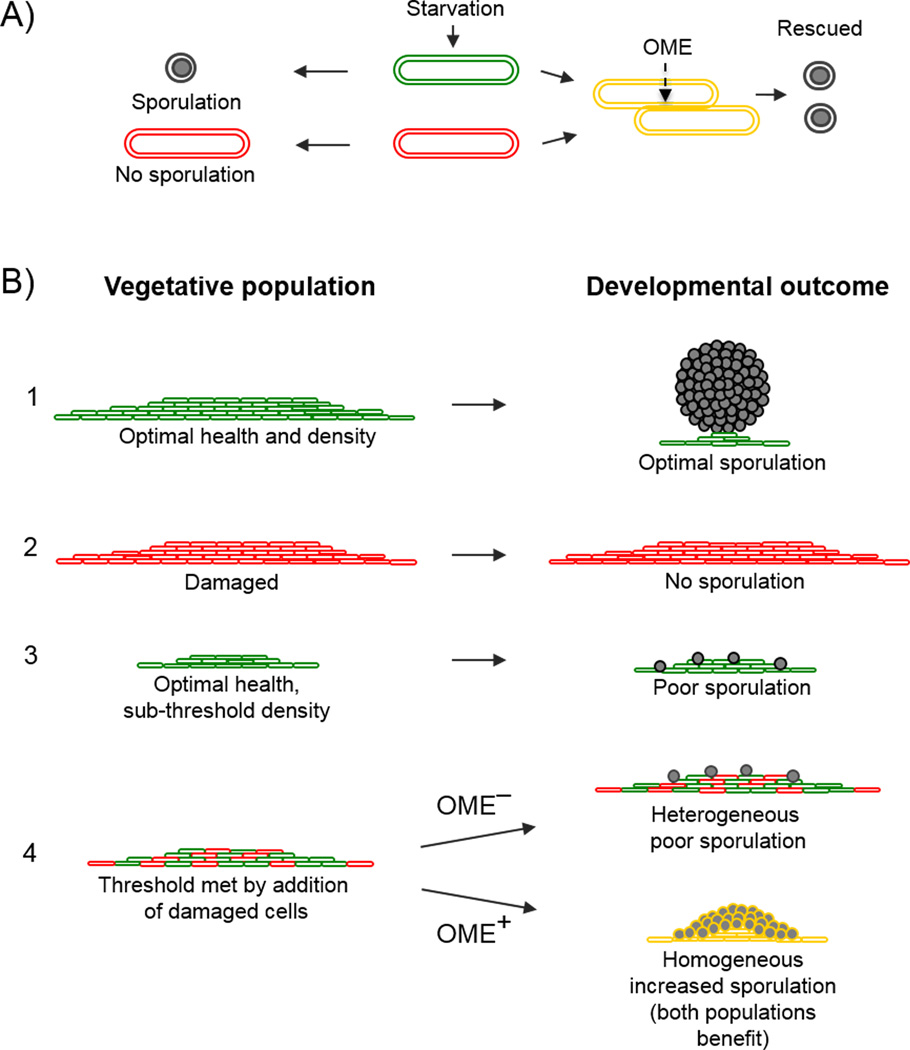
Fig. 2.
Multicellular damage repair by OME benefits the individual and the population. A: In response to starvation (center), a healthy cell (green) enters a developmental program and sporulates, whereas a cell with a damaged OM, e.g. truncated LPS (red), cannot (left). In contrast, when damage is exchanged, diluted and repaired by OME (yellow), the damaged cell is rescued (right). Note: development occurs in fruiting bodies that are not depicted. B: Schematic of vegetative myxobacteria populations. The corresponding developmental outcomes that depend on cell health and density are shown in rows one through three. Row four depicts how surpassing the density threshold by the addition of damaged cells leads to increased sporulation outcomes only when damaged cells can exchange membrane material and be repaired (OME+); otherwise sporulation does not improve (OME−).
To further investigate the ability of OME to repair damage, we created a genetic model of OM damage in M. xanthus. In gram-negative bacteria the acylated LPS component known as lipid A forms the essential hydrophobic module that anchors the distal polysaccharide moiety to the outer leaflet bilayer. To mimic OM damage, we constructed a strain that replaced the promoter of lpxC, a critical lipid A biosynthesis enzyme [92], with a heterologous inducible promoter. This strain grows like its parent in the presence of the inducer. However, in the absence of inducer, LpxC and hence lipid A are no longer produced, and LPS levels are reduced in the OM as the cells grow. Similar to what occurs when external stresses or metabolic errors cause a loss of OM function, the lpxC strain likely contains mislocalized phospholipids in the outer leaflet to compensate for the absence of LPS. This destroys the permeability barrier and the structural integrity that LPS provides and the cells lyse [10]. With this lpxC conditional strain, we then asked if OME with cells that make wild-type LPS could replenish the missing molecules and restore viability to the damaged cells. In the absence of inducer, the lpxC mutant lysed when placed in a one-to-one mixture with OME-deficient cells (traA mutant) with similar kinetics as an lpxC monoculture. However, in a one-to-one mixed culture with isogenic OME-competent cells, the lpxC strain survives and grows [10]. As the LpxC enzyme itself is not transferred, we interpret this result to mean that the rapid flux of OM material between mutant and healthy cells relieves the damage burden of the mutant cells by replenishing wild-type LPS, while distributing the damaged membrane among healthy cells (Fig. 1B). Because the membrane damage, presumably including mislocalized phospholipids, is distributed equally between strains, newly synthesized LPS from the wild-type strain in concert with the OM homeostasis machinery of both strains can then supply the population with a functional OM. In essence, we hypothesize that an indirect form of repair (damage dilution) combined with direct repair allows rejuvenation of damaged cells in a cooperative system. We propose that these findings can be extended to other forms of OM damage, as lipids, OMPs and LPS are all transferred efficiently by OME [10,14,84,86].
Implications of multicellular repair in myxobacteria
Damage repair by OME requires a sub-population of healthy cells in spatial proximity to damaged kin. Often biofilms exposed to external stress have an unequal distribution of damage, as cells at the periphery are more exposed to stressors than those in the center [93,94]. Similarly, myxobacteria form tightly aligned and aggregated populations, which may spatially establish a damage gradient that is greater toward the periphery of the population or microcolony. Since myxobacteria move by gliding motility [95,96], the population members are constantly repositioning themselves in the colony and hence exchanging OM material with different individuals. In addition, when microcolonies of myxobacteria encounter one another during migration [97], the populations may bear different damage loads as they are exposed to different environmental conditions along their migratory path. In these scenarios, OME offers a means to homogenize the differentially damaged populations. However, for this to occur there must be an incentive (positive selection) for the healthy cells to share material. In this regard, by directing repair function toward TraA-compatible cells (kin) [89], the fitness benefit of increasing the size and density of a functional population may exceed the fitness cost to healthy cells of acquiring damage [10,12]. In this scheme, damaged or aged cells acquire undamaged OM materials that are necessary for viability and to remain a productive member of their microbial community. In turn these cells remain competent for multicellular interactions, leading to improved outcomes for the whole population. For example, although OM damage impairs development (Fig. 2), we hypothesized that rejuvenated cells can participate effectively in development after OME. Indeed this idea was supported when a sub-threshold density of a healthy cell population, which cannot develop optimally, was mixed with a damaged population; the combined population effectively developed into spores, whereas a monoculture or OME-defective mixed population did not (Fig. 2B) [10]. Thus OME facilitated the formation of a developmentally competent population (size and density) by restoring function to damaged cells. This result explains how healthy cells can benefit by aiding ailing kin—ultimately the distribution of a damage load can increase the fitness of the entire population. Thus, like wound healing in multicellular eukaryotes, a part of the organism is healed to maintain the fitness of the whole organism, requiring resource sacrifice from kin cells. Although this sacrifice may establish a selective pressure to evolve cheating genotypes that do not participate in OME, the selective pressure to maintain traAB functionality, and hence receive aid and ‘private goods’ from kin cells, may be greater. In addition, there may be other benefits of traAB and OME that require myxobacteria to remain OME competent. For instance, OME may provide a competitive advantage since it has been demonstrated to be a platform to deliver toxins to competing myxobacteria [98]. Furthermore, TraA is an adhesin and may contribute to population viscosity, which limits migration away from the colony [99]. Therefore, the practical benefit to cheater genotypes may be outweighed by the selective pressure to maintain traAB.
An advantage of OME is that cells do not require de novo biosynthesis for damage dilution to take place; all that is needed is cell motility and the TraA/B proteins in partnering cells [14,84]. This makes the strategy of damage dilution by OME an efficient process in low-nutrient soil environments. In contrast, damage dilution by cell division or shedding mechanisms requires active metabolism and nutrients. We would like to add that although OME in myxobacteria does not directly exchange inner membrane or cytoplasm content, it is plausible that a secondary pathway(s) may facilitate the transport of specific cargo from the OM to proximal positions inside the cell.
Although from the literature it is unclear whether OMPs and LPS are ever turned over or recycled, in myxobacteria these components are distributed between cells [14,84]. In contrast, E. coli distributes old OMPs to the cell pole to be segregated upon cell division. As a result, cells have different aged poles, some of which are ‘old poles’, and with growth the population becomes heterogeneous [7]. Although there may be advantages and disadvantages to both strategies, the myxobacterial strategy supports a homogeneous population, which is likely important for their synchronous behaviors.
We have recently demonstrated that damaged or missing OMPs or LPS can be functionally restored to defective cells by OME from healthy donor cells. These findings are based on genetic approaches in which engineered mutants express defects in their OM. These genetic models serve as surrogates to mimic cellular damage inflicted by environmental insults. Future studies need to examine how OME can cope with specific damage to the OM imposed by physical or chemical insults. Such studies require an understanding of how particular insults damage the OM and how single-cell response pathways may lead to self-repair. Ideally these studies would ultimately determine whether OME simply buffers the population from damage or whether membrane sharing also augments autonomous cell repair pathways. It is also of interest to know whether (i) old and new OM components are transferred equally by OME, (ii) whether young cells can revitalize old cells, and (iii) under what conditions OME provides a fitness advantage to cells propagated in natural soil habitats.
Is multicellular repair unique to the myxobacteria?
Myxobacteria are remarkable in that they are at an extreme example of multicellularity in the prokaryotic kingdoms. However, as noted before, our appreciation for multicellular behaviors between otherwise unicellular organisms is expanding. With the development of more advanced microscopy techniques, research into new bacterial ultrastructures has emerged giving us fresh insight of bacterial cell-to-cell exchange platforms. Intriguingly, there is accumulating evidence that some bacterial groups respond to stress by triggering the exchange of cellular material [100,101]. As these other groups exchange cytoplasmic material, they may also engage in multicellular repair. Notably, cell-to-cell connections have been observed in pathogens such as Salmonella [102] and Francisella [103]. In fact, membrane fusion events similar to myxobacterial OME has been observed in Borellia, though content transfer was not tested [104]. Cell-to-cell transfer of electrons has also been documented [105], which sometimes occurs through membrane tube-like structures [106]. In addition, observation of large structures appearing to connect cells can be observed in natural environments with cryo-electron microscopy [107]. Other bacteria, such as actinomycetes, cyanobacteria and magnetotactic prokaryotes, exist as multicellular organisms, where cellular components may be exchanged among tightly associated cells. For example, in Anabaena filaments the cells are enclosed by a continuous OM and the cells exchange molecules [108,109]. Whether Anabaena or other multicellular bacteria engage in multicellular repair remains an open question.
Though bacterial cell-to-cell transfer of components has been noted as far back as the discovery of bacterial conjugation, research into the role of content exchange in damage repair is lacking. However, advances in our understanding of inter-microbial interactions may yet lead us to the discovery of analogous strategies in other bacteria. A better understanding of how bacterial content sharing contributes to buffering or repairing cellular damage will provide insight to community dynamics. These processes include infection, microbiome dynamics, microbial fermentation and bioremediation.
Conclusions and perspective
We propose that OME expands the paradigm for how cell damage repair occurs in bacteria by adding the dimension of cooperation. Many multicellular organisms have evolved the ability to coordinate cell behavior to combat damage [2], thus setting a precedent for multicellular repair in bacteria. As diverse forms of bacterial cooperative behaviors come to light, microbiologists should now consider how multicellular interactions are used to combat stress. This notion is especially relevant for organisms in which cell number and density correlate with fitness, because there is a selective advantage for individuals to assist their kin. In addition, understanding how groups of cells cope with stress is fundamental in elucidating bacterial survival strategies, including how they respond to antibiotics and the immune system of host organisms. We conclude that multicellular wound repair has evolved in prokaryotes, and this notion will hopefully stimulate and inform future studies on bacterial repair.
Acknowledgments
This work was supported by NIH grant GM101449 to D.W.
Abbreviations
- OM
outer membrane
- OME
outer membrane exchange
- LPS
lipopolysaccharide
- OMPs
outer membrane proteins
- OMVs
outer membrane vesicles
- TNTs
tunneling nanotubes
- ER
endoplasmic reticulum
References
- 1.Clark AG, Miller AL, Vaughan E, Yu HY, et al. Integration of single and multicellular wound responses. Curr Biol. 2009;19:1389–1395. doi: 10.1016/j.cub.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonnemann KJ, Bement WM. Wound repair: toward understanding and integration of single-cell and multicellular wound responses. Annu Rev Cell Dev Biol. 2011;27:237–263. doi: 10.1146/annurev-cellbio-092910-154251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 4.Gladyshev VN. On the cause of aging and control of lifespan: heterogeneity leads to inevitable damage accumulation, causing aging; control of damage composition and rate of accumulation define lifespan. Bioessays. 2012;34:925–929. doi: 10.1002/bies.201200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3:e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ackermann M, Stearns SC, Jenal U. Senescence in a bacterium with asymmetric division. Science. 2003;300:1920. doi: 10.1126/science.1083532. [DOI] [PubMed] [Google Scholar]
- 7.Lindner AB, Madden R, Demarez A, Stewart EJ, et al. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci U S A. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nystrom T. A bacterial kind of aging. PLoS Genet. 2007;3:e224. doi: 10.1371/journal.pgen.0030224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vassallo C, Pathak DT, Cao P, Zuckerman DM, et al. Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. Proc Natl Acad Sci U S A. 2015;112:E2939–E2946. doi: 10.1073/pnas.1503553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myxobacteria: Genomics, Cellular and Molecular Biology. Norfolk, UK: Caister Academic Press; 2014. [Google Scholar]
- 12.Cao P, Dey A, Vassallo CN, Wall D. How myxobacteria cooperate. J Mol Biol. 2015;427:3709–3721. doi: 10.1016/j.jmb.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathak DT, Wall D. Identification of the cglC, cglD, cglE and cglF genes and their role in cell contact-dependent gliding motility in Myxococcus xanthus. J Bacteriol. 2012;194:1940–1949. doi: 10.1128/JB.00055-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pathak DT, Wei X, Bucuvalas A, Haft DH, et al. Cell contact-dependent outer membrane exchange in myxobacteria: Genetic determinants and mechanism. PLoS Genet. 2012;8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei X, Pathak DT, Wall D. Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol. 2011;81:315–326. doi: 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- 16.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visick JE, Clarke S. Repair, refold, recycle: how bacteria can deal with spontaneous and environmental damage to proteins. Mol Microbiol. 1995;16:835–845. doi: 10.1111/j.1365-2958.1995.tb02311.x. [DOI] [PubMed] [Google Scholar]
- 18.Cabiscol E, Tamarit J, Ros J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol. 2000;3:3–8. [PubMed] [Google Scholar]
- 19.Tamarit J, Cabiscol E, Ros J. Identification of the major oxidatively damaged proteins in Escherichia coli cells exposed to oxidative stress. J Biol Chem. 1998;273:3027–3032. doi: 10.1074/jbc.273.5.3027. [DOI] [PubMed] [Google Scholar]
- 20.Hurst A, Nasim A. Repairable lesions in microorganisms: Academic Press. 1984 [Google Scholar]
- 21.Leive L. Release of lipopolysaccharide by EDTA treatment of E. coli. Biochem Biophys Res Commun. 1965;21:290–296. doi: 10.1016/0006-291x(65)90191-9. [DOI] [PubMed] [Google Scholar]
- 22.Raivio TL, Silhavy TJ. Periplasmic stress and ECF sigma factors. Annu Rev Microbiol. 2001;55:591–624. doi: 10.1146/annurev.micro.55.1.591. [DOI] [PubMed] [Google Scholar]
- 23.Lima S, Guo MS, Chaba R, Gross CA, et al. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science. 2013;340:837–841. doi: 10.1126/science.1235358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Detweiler CS, Monack DM, Brodsky IE, Mathew H, et al. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol Microbiol. 2003;48:385–400. doi: 10.1046/j.1365-2958.2003.03455.x. [DOI] [PubMed] [Google Scholar]
- 25.Vogt SL, Raivio TL. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett. 2012;326:2–11. doi: 10.1111/j.1574-6968.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- 26.Ades SE. Regulation by destruction: design of the sigmaE envelope stress response. Curr Opin Microbiol. 2008;11:535–540. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 28.Raffa RG, Raivio TL. A third envelope stress signal transduction pathway in Escherichia coli. Mol Microbiol. 2002;45:1599–1611. doi: 10.1046/j.1365-2958.2002.03112.x. [DOI] [PubMed] [Google Scholar]
- 29.Storz G, Hengge R. Bacterial stress responses: American Society for Microbiology Press. 2010 [Google Scholar]
- 30.Krojer T, Sawa J, Schafer E, Saibil HR, et al. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453:885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- 31.Merdanovic M, Clausen T, Kaiser M, Huber R, et al. Protein quality control in the bacterial periplasm. Annu Rev Microbiol. 2011;65:149–168. doi: 10.1146/annurev-micro-090110-102925. [DOI] [PubMed] [Google Scholar]
- 32.Arts IS, Gennaris A, Collet JF. Reducing systems protecting the bacterial cell envelope from oxidative damage. FEBS Lett. 2015;589:1559–1568. doi: 10.1016/j.febslet.2015.04.057. [DOI] [PubMed] [Google Scholar]
- 33.Skaar EP, Tobiason DM, Quick J, Judd RC, et al. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc Natl Acad Sci U S A. 2002;99:10108–10113. doi: 10.1073/pnas.152334799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomassin JL, Brannon JR, Gibbs BF, Gruenheid S, et al. OmpT outer membrane proteases of enterohemorrhagic and enteropathogenic Escherichia coli contribute differently to the degradation of human LL-37. Infect Immun. 2012;80:483–492. doi: 10.1128/IAI.05674-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desnues B, Cuny C, Gregori G, Dukan S, et al. Differential oxidative damage and expression of stress defence regulons in culturable and non-culturable Escherichia coli cells. EMBO Rep. 2003;4:400–404. doi: 10.1038/sj.embor.embor799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikaido H. Restoring permeability barrier function to outer membrane. Chem Biol. 2005;12:507–509. doi: 10.1016/j.chembiol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Dekker N. Outer-membrane phospholipase A: known structure, unknown biological function. Mol Microbiol. 2000;35:711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 38.Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci U S A. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murata T, Tseng W, Guina T, Miller SI, et al. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bishop RE, Gibbons HS, Guina T, Trent MS, et al. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia W, El Zoeiby A, Petruzziello TN, Jayabalasingham B, et al. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem. 2004;279:44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- 42.Chalabaev S, Chauhan A, Novikov A, Iyer P, et al. Biofilms formed by gram-negative bacteria undergo increased lipid a palmitoylation, enhancing in vivo survival. MBio. 2014;5 doi: 10.1128/mBio.01116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 44.Rassam P, Copeland NA, Birkholz O, Toth C, et al. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature. 2015;523:333–336. doi: 10.1038/nature14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Misra R. Entry and exit of bacterial outer membrane proteins. Trends Microbiol. 2015;23:452–454. doi: 10.1016/j.tim.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 47.McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. 2007;63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei X, Vassallo CN, Pathak DT, Wall D. Myxobacteria produce outer membrane-enclosed tubes in unstructured environments. J Bacteriol. 2014;196:1807–1814. doi: 10.1128/JB.00850-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ericsson M, Hanstorp D, Hagberg P, Enger J, et al. Sorting out bacterial viability with optical tweezers. J Bacteriol. 2000;182:5551–5555. doi: 10.1128/jb.182.19.5551-5555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roszak DB, Colwell RR. Survival strategies of bacteria in the natural environment. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Starokadomskyy P, Dmytruk KV. A bird's-eye view of autophagy. Autophagy. 2013;9:1121–1126. doi: 10.4161/auto.24544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d'etre. Nature reviews Microbiology. 2007;5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- 54.Gonzalez-Pastor JE. Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol Rev. 2011;35:415–424. doi: 10.1111/j.1574-6976.2010.00253.x. [DOI] [PubMed] [Google Scholar]
- 55.Spees JL, Olson SD, Whitney MJ, Prockop DJ. Mitochondrial transfer between cells can rescue aerobic respiration. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1283–1288. doi: 10.1073/pnas.0510511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cselenyak A, Pankotai E, Horvath EM, Kiss L, et al. Mesenchymal stem cells rescue cardiomyoblasts from cell death in an in vitro ischemia model via direct cell-to-cell connections. BMC Cell Biol. 2010;11:29. doi: 10.1186/1471-2121-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plotnikov EY, Khryapenkova TG, Galkina SI, Sukhikh GT, et al. Cytoplasm and organelle transfer between mesenchymal multipotent stromal cells and renal tubular cells in co-culture. Experimental cell research. 2010;316:2447–2455. doi: 10.1016/j.yexcr.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 58.Yasuda K, Khandare A, Burianovskyy L, Maruyama S, et al. Tunneling nanotubes mediate rescue of prematurely senescent endothelial cells by endothelial progenitors: exchange of lysosomal pool. Aging (Albany NY) 2011;3:597–608. doi: 10.18632/aging.100341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang X, Gerdes HH. Transfer of mitochondria via tunneling nanotubes rescues apoptotic PC12 cells. Cell Death Differ. 2015;22:1181–1191. doi: 10.1038/cdd.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, Cui J, Sun X, Zhang Y. Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ. 2011;18:732–742. doi: 10.1038/cdd.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rainy N, Chetrit D, Rouger V, Vernitsky H, et al. H-Ras transfers from B to T cells via tunneling nanotubes. Cell Death Dis. 2013;4:e726. doi: 10.1038/cddis.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pasquier J, Galas L, Boulange-Lecomte C, Rioult D, et al. Different modalities of intercellular membrane exchanges mediate cell-to-cell p-glycoprotein transfers in MCF-7 breast cancer cells. J Biol Chem. 2012;287:7374–7387. doi: 10.1074/jbc.M111.312157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gurke S, Barroso JF, Hodneland E, Bukoreshtliev NV, et al. Tunneling nanotube (TNT)-like structures facilitate a constitutive, actomyosin-dependent exchange of endocytic organelles between normal rat kidney cells. Exp Cell Res. 2008;314:3669–3683. doi: 10.1016/j.yexcr.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 64.Thayanithy V, Dickson EL, Steer C, Subramanian S, et al. Tumor-stromal cross talk: direct cell-to-cell transfer of oncogenic microRNAs via tunneling nanotubes. Transl Res. 2014;164:359–365. doi: 10.1016/j.trsl.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J Immunol. 2013;191:6250–6260. doi: 10.4049/jimmunol.1301728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sisakhtnezhad S, Khosravi L. Emerging physiological and pathological implications of tunneling nanotubes formation between cells. Eur J Cell Biol. 2015;94:429–443. doi: 10.1016/j.ejcb.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 67.Marzo L, Gousset K, Zurzolo C. Multifaceted roles of tunneling nanotubes in intercellular communication. Front Physiol. 2012;3:72. doi: 10.3389/fphys.2012.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rustom A, Saffrich R, Markovic I, Walther P, et al. Nanotubular highways for intercellular organelle transport. Science. 2004;303:1007–1010. doi: 10.1126/science.1093133. [DOI] [PubMed] [Google Scholar]
- 69.Joly E, Hudrisier D. What is trogocytosis and what is its purpose? Nat Immunol. 2003;4:815. doi: 10.1038/ni0903-815. [DOI] [PubMed] [Google Scholar]
- 70.Grabski S, De Feijter AW, Schindler M. Endoplasmic Reticulum Forms a Dynamic Continuum for Lipid Diffusion between Contiguous Soybean Root Cells. Plant Cell. 1993;5:25–38. doi: 10.1105/tpc.5.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barton DA, Cole L, Collings DA, Liu DY, et al. Cell-to-cell transport via the lumen of the endoplasmic reticulum. Plant J. 2011;66:806–817. doi: 10.1111/j.1365-313X.2011.04545.x. [DOI] [PubMed] [Google Scholar]
- 72.Ahmed KA, Xiang J. Mechanisms of cellular communication through intercellular protein transfer. J Cell Mol Med. 2011;15:1458–1473. doi: 10.1111/j.1582-4934.2010.01008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Draeger A, Schoenauer R, Atanassoff AP, Wolfmeier H, et al. Dealing with damage: plasma membrane repair mechanisms. Biochimie. 2014;107(Pt A):66–72. doi: 10.1016/j.biochi.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 74.Amalia S, Bronislaw LS. Phosphatidylglycerol-containing ER-transport vesicles built and restore outer mitochondrial membrane and deliver nuclear DNA translation products to generate cardiolipin in the inner mitochondrial membrane. Advances in Biological Chemistry. 2012;2012 [Google Scholar]
- 75.Legros F, Lombes A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell. 2002;13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu X, Weaver D, Shirihai O, Hajnóczky G. Mitochondrial ‘kiss- and -run’: interplay between mitochondrial motility and fusion–fission dynamics. EMBO J. 2009;28:3074–3089. doi: 10.1038/emboj.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Labbe K, Murley A, Nunnari J. Determinants and functions of mitochondrial behavior. Annu Rev Cell Dev Biol. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- 78.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 79.Huang X, Sun L, Ji S, Zhao T, et al. Kissing and nanotunneling mediate intermitochondrial communication in the heart. Proc Natl Acad Sci U S A. 2013;110:2846–2851. doi: 10.1073/pnas.1300741110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Claessen D, Rozen DE, Kuipers OP, Sogaard-Andersen L, et al. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol. 2014;12:115–124. doi: 10.1038/nrmicro3178. [DOI] [PubMed] [Google Scholar]
- 81.Rechavi O, Goldstein I, Kloog Y. Intercellular exchange of proteins: the immune cell habit of sharing. FEBS Lett. 2009;583:1792–1799. doi: 10.1016/j.febslet.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 82.Bourke AFG. Principles of social evolution: Oxford University Press. 2011 [Google Scholar]
- 83.de la Fuente-Núñez C, Reffuveille F, Fernández L, Hancock RE. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol. 2013;16:580–589. doi: 10.1016/j.mib.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 84.Wei X, Pathak DT, Wall D. Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol. 2011;81:315–326. doi: 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- 85.Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nudleman E, Wall D, Kaiser D. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science. 2005;309:125–127. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 87.Wall D, Wu SS, Kaiser D. Contact Stimulation of Tgl and Type IV Pili in Myxococcus xanthus. J Bacteriol. 1998;180:759–761. doi: 10.1128/jb.180.3.759-761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dey A, Wall D. A genetic screen in Myxococcus xanthus identifies mutants that uncouple outer membrane exchange from a downstream cellular response. J Bacteriol. 2014;196:4324–4332. doi: 10.1128/JB.02217-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pathak DT, Wei X, Dey A, Wall D. Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLoS Genet. 2013;9:e1003891. doi: 10.1371/journal.pgen.1003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bowden MG, Kaplan HB. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol Microbiol. 1998;30:275–284. doi: 10.1046/j.1365-2958.1998.01060.x. [DOI] [PubMed] [Google Scholar]
- 91.Fink JM, Zissler JF. Defects in motility and development of Myxococcus xanthus lipopolysaccharide mutants. J Bacteriol. 1989;171:2042–2048. doi: 10.1128/jb.171.4.2042-2048.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Young K, Silver LL, Bramhill D, Cameron P, et al. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J Biol Chem. 1995;270:30384–30391. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 93.Bridier A, Briandet R, Thomas V, Dubois-Brissonnet F. Resistance of bacterial biofilms to disinfectants: a review. Biofouling. 2011;27:1017–1032. doi: 10.1080/08927014.2011.626899. [DOI] [PubMed] [Google Scholar]
- 94.Smukalla S, Caldara M, Pochet N, Beauvais A, et al. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 2008;135:726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nan B, Zusman DR. Uncovering the mystery of gliding motility in the myxobacteria. Annu Rev Genet. 2011;45:21–39. doi: 10.1146/annurev-genet-110410-132547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zusman DR, Scott AE, Yang Z, Kirby JR. Chemosensory pathways, motility and development in Myxococcus xanthus. Nature reviews Microbiology. 2007;5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 97.Vos M, Velicer GJ. Genetic population structure of the soil bacterium Myxococcus xanthus at the centimeter scale. Appl Environ Microbiol. 2006;72:3615–3625. doi: 10.1128/AEM.72.5.3615-3625.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dey A, Vassallo CN, Conklin AC, Pathak DT, et al. Sibling rivalry in Myxococcus xanthus is mediated by kin recognition and a polyploid prophage. J Bacteriol. 2016 doi: 10.1128/JB.00964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nature reviews Microbiology. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 100.Benomar S, Ranava D, Cardenas ML, Trably E, et al. Nutritional stress induces exchange of cell material and energetic coupling between bacterial species. Nature communications. 2015;6:6283. doi: 10.1038/ncomms7283. [DOI] [PubMed] [Google Scholar]
- 101.Pande S, Shitut S, Freund L, Westermann M, et al. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nature communications. 2015;6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 102.Galkina SI, Romanova JM, Bragina EE, Tiganova IG, et al. Membrane tubules attach Salmonella Typhimurium to eukaryotic cells and bacteria. FEMS Immunol Med Microbiol. 2011;61:114–124. doi: 10.1111/j.1574-695X.2010.00754.x. [DOI] [PubMed] [Google Scholar]
- 103.McCaig WD, Koller A, Thanassi DG. Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J Bacteriol. 2013;195:1120–1132. doi: 10.1128/JB.02007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kudryashev M, Cyrklaff M, Alex B, Lemgruber L, et al. Evidence of direct cell-cell fusion in Borrelia by cryogenic electron tomography. Cellular microbiology. 2011;13:731–741. doi: 10.1111/j.1462-5822.2011.01571.x. [DOI] [PubMed] [Google Scholar]
- 105.Summers ZM, Fogarty HE, Leang C, Franks AE, et al. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science. 2010;330:1413–1415. doi: 10.1126/science.1196526. [DOI] [PubMed] [Google Scholar]
- 106.Lovley DR, Malvankar NS. Seeing is believing: novel imaging techniques help clarify microbial nanowire structure and function. Environmental microbiology. 2015 doi: 10.1111/1462-2920.12708. [DOI] [PubMed] [Google Scholar]
- 107.Comolli LR, Banfield JF. Inter-species interconnections in acid mine drainage microbial communities. Front Microbiol. 2014;5:367. doi: 10.3389/fmicb.2014.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wilk L, Strauss M, Rudolf M, Nicolaisen K, et al. Outer membrane continuity and septosome formation between vegetative cells in the filaments of Anabaena sp. PCC 7120. Cellular microbiology. 2011;13:1744–1754. doi: 10.1111/j.1462-5822.2011.01655.x. [DOI] [PubMed] [Google Scholar]
- 109.Mullineaux CW, Mariscal V, Nenninger A, Khanum H, et al. Mechanism of intercellular molecular exchange in heterocyst-forming cyanobacteria. EMBO J. 2008;27:1299–1308. doi: 10.1038/emboj.2008.66. [DOI] [PMC free article] [PubMed] [Google Scholar]