Abstract
Wolbachia pipientis are obligate intracellular bacteria commonly found in many arthropods. They can induce various reproductive alterations in hosts, including cytoplasmic incompatibility, male-killing, feminization, and parthenogenetic development, and can provide host protection against some viruses and other pathogens. Wolbachia differ from many other primary endosymbionts in arthropods because they undergo frequent horizontal transmission between hosts and are well known for an abundance of mobile elements and relatively high recombination rates. Here, we compare the genomes of two closely related Wolbachia (with 0.57% genome-wide synonymous divergence) that differ in their reproductive effects on hosts. wVitA induces a sperm–egg incompatibility (also known as cytoplasmic incompatibility) in the parasitoid insect Nasonia vitripennis, whereas wUni causes parthenogenetic development in a different parasitoid, Muscidifurax uniraptor. Although these bacteria are closely related, the genomic comparison reveals rampant rearrangements, protein truncations (particularly in proteins predicted to be secreted), and elevated substitution rates. These changes occur predominantly in the wUni lineage, and may be due in part to adaptations by wUni to a new host environment, or its phenotypic shift to parthenogenesis induction. However, we conclude that the approximately 8-fold elevated synonymous substitution rate in wUni is due to a either an elevated mutation rate or a greater number of generations per year in wUni, which occurs in semitropical host species. We identify a set of genes whose loss or pseudogenization in the wUni lineage implicates them in the phenotypic shift from cytoplasmic incompatibility to parthenogenesis induction. Finally, comparison of these closely related strains allows us to determine the fine-scale mutation patterns in Wolbachia. Although Wolbachia are AT rich, mutation probabilities estimated from 4-fold degenerate sites are not AT biased, and predict an equilibrium AT content much less biased than observed (57–50% AT predicted vs. 76% current content at degenerate sites genome wide). The contrast suggests selection for increased AT content within Wolbachia genomes.
Keywords: endosymbiont, reproductive manipulation, mutational bias
Introduction
Wolbachia pipientis is a ubiquitous alphaproteobacterial symbiont of arthropods and nematodes, distantly related to the Rickettsial pathogens Ehrlichia and Anaplasma (Werren et al. 2008). Within arthropods, these bacteria can induce a variety of reproductive alterations, including feminization of males, male-killing, sperm–egg incompatibility (known as cytoplasmic incompatibility or CI), and parthenogenetic development (Stouthamer et al. 1993; Werren et al. 2008). Other effects on arthropod hosts include protection against viruses (Hedges et al. 2008), suppression of sterile and lethal mutants in Drosophila (Starr and Cline 2002), supplementation of essential B-vitamins in bedbugs (Hosokawa et al. 2010), and ovarian development and reproduction in wasps (Dedeine et al. 2001). Wolbachia also infect nematodes, such as in Brugia, the causative agent of Elephantiasis, where the symbiont is required for embryogenesis and larval development (Casiraghi et al. 2002; Comandatore et al. 2013). Additionally, Wolbachia has received much recent attention due to its medical relevance (reviewed in [Hedges et al. 2008; LePage and Bordenstein 2013]). It is heavily studied as a potential drug target for filarial nematode infection (Taylor et al. 2000; Hoerauf et al. 2008) and is currently being tested as a means to reduce transmission of Dengue fever from mosquitoes to humans (Moreira et al. 2009; Turley et al. 2009).
Because Wolbachia is an obligate intracellular bacterium for which no genetic system exists, knowledge about its biology has stemmed from functional experiments in hosts or bacterial genome sequencing. The first complete Wolbachia genome was that of wMel, the parasite of Drosophila melanogaster and a CI-inducing strain (Wu et al. 2004). The genome sequence revealed that although Wolbachia are intracellular insect-associated bacteria, their genome architecture and evolution contrast sharply with other obligate symbionts such as Buchnera. Wolbachia genomes are littered with mobile elements (Wu et al. 2004; Kent and Bordenstein 2010), frequently subject to recombination (Jiggins et al. 2001; Baldo, Bordenstein, et al. 2006; Klasson et al. 2009), rearrangements, and gene losses and gains (Ishmael et al. 2009; Ellegaard, Klasson, Andersson, et al. 2013), and possess putative pathogenic determinants (such as a type IV secretion system) missing from many mutualistic endosymbionts (Masui et al. 2000; Rances et al. 2008). Wolbachia, like other host-switching obligate intracellular bacteria, differ from vertically transmitted obligate intracellular species in which reduced genomes, low recombination among strains, and reduced frequencies of mobile elements are the norm (Newton and Bordenstein 2011). Hypotheses as to why these bacteria do not follow these trends observed in many other symbionts include the fact that they show extensive horizontal movement between host species (Baldo, Hotopp, et al. 2006) and that different Wolbachia strains are sometimes found coinfecting the same host (Werren, Windsor, et al. 1995; Werren, Zhang, et al. 1995; Bordenstein and Wernegreen 2004; Kent, Salichos, et al. 2011), which can facilitate chromosomal recombination among strains (Werren and Bartos 2001; Baldo et al. 2005; Baldo, Bordenstein, et al. 2006) and rampant exchange of temperate bacteriophage WO (Masui et al. 2000; Bordenstein and Wernegreen 2004; Chafee et al. 2010; Kent, Salichos, et al. 2011).
Here, we present a comparative analysis of the evolution of two Wolbachia, wVitA and wUni, which have identical multilocus sequence type profiles (Baldo, Hotopp, et al. 2006) and relatively recent divergence (0.0057 pairwise synonymous difference). Importantly, these closely related Wolbachia infect different parasitoid wasps and cause very different reproductive phenotypes: wVitA infects Nasonia vitripennis where it induces CI, whereas wUni infects Muscidifurax uniraptor, where it causes parthenogenesis. We use the comparison between these closely related Wolbachia to address general questions about changes that occur during the initial stages of intracellular bacterial genome evolution, possible associations of genotypic changes with host and phenotypic shifts, and the mutational patterns in Wolbachia.
Materials and Methods
Wolbachia DNA Isolation
The same extraction and amplification procedures were used for both Wolbachia strains. The Nasonia strain IntG12.1 (Chafee et al 2011) was reared under standard laboratory conditions under low density with one female provided with two hosts for 48 h. Upon pupation, wasps were removed from hosts, placed in a clean tube, allowed to eclose, and aged for 1–3 days. They were then pooled and put on ice. Before transfer of wasps to 5.0 μm filter columns, each column was rinsed with 70% ethanol, immediately followed by a 2000 RPM spin for 2 min before transfer to a new tube. Wasps were then rinsed with 500 μl of sterile distilled water, followed by a 2000 RPM spin for 2 min. The insects are not homogenized prior to centrifugation. Rather, centrifugation releases hemolymph and cells from the intact insects. This method appears to provide cleaner Wolbachia preparations with lower amounts contamination from host or other bacterial DNA. Columns with wasps were then transferred to a new sterile tube and spun at 13,500 RPM for 20 min at 4°C. Columns were then removed and discarded. Supernatant was discarded and pellets resulting from the spin were suspended in sterile PBS. Resuspended pellets were then transferred to a new column filter (5.0 μm) and spun at 13,500 RPM for 30 s. The resulting pellet was resuspended in the PBS and transferred to a new column filter (5.0 μm) for a total passage through four columns total. For the final column, the spin was 5 min at 13,500 K to pellet the bacteria. The supernatant was removed and discarded. The pellet was frozen at −80 until DNA extraction. wUni was similarly harvested from Muscidifurax uniraptor.
DNA was extracted using the Qiagen DNA tissue extraction kit according to the manufacturers instructions for gram negative bacteria. Whole-genome amplification was performed using the Qiagen REPLI-g multiple displacement amplification kit as per the manufacturers instructions. Prior to sequencing, purity of Wolbachia preparation was evaluated in two ways. During bacterial purification, following passage through filter and resuspension, a 5 μl sample was collected, fixed on a lysine-coated microscope slide with 3.7% formaldehyde for 15 min, stained with DAPI, and examined with a Zeiss Axio-Imager Z1 Microscope. Each passage through the 5.0 μm filter reduced the numbers of nuclei present. Very few or no eukaryotic nuclei could be observed in the final bacterial sample. Purity was also examined by semiquantitative polymerase chain reaction (PCR) of DNA preparations. Following the DNA extraction, PCR was performed using Wolbachia 16S rRNA gene-specific primers (Werren et al 1995), nuclear gene wing-size 1 primers WS1-5 (Loehlin et al. 2010), and Nasonia mitochondrial COI (Hebert et al. 2003). Preparations were used only from samples that yielded no nuclear gene amplification in the undiluted Qiagen DNA and only faint or no COI amplification relative to the Wolbachia gene. Typically Wolbachia primers still provided a faint band at 1:10,000 dilution, whereas COI sometimes provided a faint band in undiluted and no band was visible at 1:10 dilution. Finally, general eubacterial 16S primers (Weisburg et al. 1991) were used to amplify DNA and then direct sequencing of the product revealed clean Wolbachia 16S sequences, indicating low contamination with other bacteria. The strong enrichment for Wolbachia DNA is supported by low levels of host and mitochondrial DNA recovered in high-throughput sequencing (only 8% of the reads mapping to either host nuclear or mitochondrial assemblies).
Sequencing and Assembly
The wVitA and wUni genomes were sequenced with a combination of 454 pyrosequencing, illumina sequencing, and targeted Sanger sequencing (to orient and link scaffolds). Libraries for 454 sequencing were generated by the Baylor College of Medicine Sequencing Center using standard Roche protocols whereas libraries for Illumina sequencing were generated at Tufts University Medical Center. We generated two 3 kb PE libraries for sequencing using the Roche 454 GS FLX Titanium platform. These libraries yielded a total of 38,372,568 bp (220 bp read length average) for wVitA and 238,654,439 bp (284 bp read length average) for wUni. In addition, we also generated a 500 bp PE library for sequencing on a single lane of the illumina Genome Analyzer II for wVitA (40 cycles, 1,282,539,680 total bp) and utilized the previously published and assembled contigs for wUni (867,873 bp) (Klasson, Westberg, et al. 2009). Short contigs from the illumina data were generated using the Velvet assembler; We used an iterative process where we ran velvet on all possible k-mers (between 11 and 21) and picked the k-mer length that yielded the assembly with the best N50 (v1.1.04; kmer = 15, using the closed wRi genome as a reference, generating a total of 194,160 contigs) and these short contigs were then merged with the 454 generated reads and used as input to Newbler (v2.5) for de novo assembly of the wVitA genome, excluding reads without proper mate pairs during the assembly process. In addition, we used PCR to orient the remaining scaffolds, linking 3 of the 11 scaffolds generated. In contrast, wUni was assembled using a combination of published contigs (accession: ACFP00000000) and 454 pyrosequencing data (both merged and used as input for the Newbler assembler, again only using proper mate pairs). The wUni genome proved more difficult to assemble than that of wVitA; the best assembly produced a larger number of resulting scaffolds for wUni than wVitA and in addition, this wUni assembly lacks a single ribosomal RNA gene (1) and a couple of transfer RNA genes (2). The presence of these catalytic RNA genes (all three rRNAs in addition to the 34 tRNAs identified in other Wolbachia strains) was confirmed through BLASTn against the wUni raw reads (BLAST+ v2.2.25) (supplementary text file, Supplementary Material online). We hypothesize that these reads did not assemble due to highly repetitive and complex flanking regions (as seen in other Wolbachia genome assemblies). Due to the difference in sequencing strategy between these two Wolbachia, we were careful to validate our observations of single nucleotide polymorphism (SNP) differences using coverage and targeted Sanger sequencing where applicable (supplementary fig. S2, Supplementary Material online), and as described below. We have validated the data in three ways: 1) We used PCR to orient scaffolds, 2) we used PCR (with targeted primer sets) and Sanger sequencing to confirm shortened predicted proteins, 3) we mapped the reads from wUni onto the wVitA assembly and vice versa, and 4) we mapped Illumina data from wVitA onto the 454 assembly generated for wVitA to confirm the alleles in that genome. In no case did the SNPs called by the Illumina data change the consensus sequence. All of our confirmations indicate that multiple displacement amplification bias has not significantly affected our assembly or analysis herein. Importantly, as we focus on SNP comparisons throughout our analysis, and confirm a subset through Sanger sequencing, assembly artifacts are not relevant to our conclusions.
Confirmation of Sequence Differences
To verify sequence differences between the genome assemblies of wVitA and wUni, we designed primers (supplementary table S2, Supplementary Material online) for a random sample of 11 different regions showing either SNPs (seven regions) or indels (four regions) between the bacteria, and amplified and sequenced the corresponding regions from both strains using Sanger chemistry. In each case, the predicted sequence difference was verified. In addition, we repeated the analysis using only 454 data for both genomes (wUni and wVitA) and again, the predicted differences (indels and SNPs) were confirmed. Therefore, the differences in sequence and patterns of divergence in sequence are not due to artifacts in assembly or sequencing chemistry. Confirmation of orientation for genomic scaffolds was performed for wVitA using PCR and custom primer sets.
Genome Analyses
Genomic scaffolds were submitted to the UMD IGS Annotation Engine (Galens et al. 2011) for structural and functional annotation. We verified the structural predictions through manual annotation—that is, observing the evidence for the genes, provided by UMD IGS, using the web interface Manatee. Resultant open reading frames (ORFs) were used as input for analyses of content and of molecular evolution. Comparative genomic analyses between the two Wolbachia genomes were performed using a variety of in house Perl scripts (generated by Newton and used for parsing flat file output), whole-genome alignment utilities (MUMmer v.3.22 using –filter and –fat flags [Kurtz et al. 2004]) and database searches (PFam, Genbank nr). Identification of candidate secreted substrates was performed using a combination of resulting searches between ORFs in the genomes and the PFam (version 26.0; hmmscan with defaults) and Genbank nr (searched mid April 2012 using TBLASTN 2.2.25+ and an e value threshold of 1E-3) databases. ORFs containing domains in PFam families with largely eukaryotic composition and/or regions of homology with eukaryotes (based on top BLAST hit outside of Wolbachia) and/or containing predicted secretion signals were included in the list of candidate secreted substrates. This type of bioinformatics approach has been used successfully in the past to identify proteins secreted by bacterial pathogens (de Felipe et al. 2005). Genomic alignments between wRi, wVitA, and wUni were generated by progressive Mauve (Darling et al. 2010) and output from this program used to calculate genomic rearrangements using two methods, one based on a Markov chain Monte Carlo algorithm (BADGER [Simon and Larget 2004]) and another based on the SPRING algorithm (Lin et al. 2006), which identifies the minimal number of rearrangement operations necessary to convert one genomic architecture to another (using locally collinear blocks as landmarks on the chromosomes). We used the predicted wVitA linear genome sequence, based on scaffold orientation generated by PCR, as the reference for comparison with wUni and wRi. Any rearrangements predicted at scaffold junctions were manually excluded.
Molecular Evolution
Evolutionary comparisons were made to wMel [NC_002978.6], wRi [NC_012416.1], and wHa [NC_021089.1], relatively closely related A group Wolbachia. Two other genome sequences reported to be Wolbachia (wSim and wAna) were not used in these comparative genomic analyses for the following reasons. First, these genomes were derived as a by-product of whole insect genome sequencing from their hosts (Salzberg et al. 2005). Second, the wSim genome was made from a composite of multiple infected Drosophila simulans hosts and therefore does not represent a single bacterial strain (Iturbe-Ormaetxe et al. 2005). Third, the wAna genome was also derived from a whole insect genome sequencing project (Salzberg et al. 2005), and has the added problem that a lateral insertion of the nearly complete Wolbachia genome is present on the fourth chromosome of the sequenced insect strain (Hotopp et al. 2007). Therefore, sequences cannot be reliably assigned to the bacterium. Furthermore, as the insect strain was cured of its Wolbachia prior to sequencing (Markow T, personal communication), the reported wAna assembly almost certainly does not represent Wolbachia, but rather the Wolbachia insertion in the nucleus of this insect host. For these reasons, neither wAna nor wSim assemblies are used here for the purposes of comparative bacterial genomics.
Orthologous gene sets between the type A Wolbachia were generated using reciprocal BLAST (wVitA: PRJDB1504, wUni: PRJDB1583, wMel: NC_002978, wRi: NC_012416). Two genes were considered orthologous between two genomes if they were reciprocal best hits. Additionally, to identify potential recombination between wVitA and wVitB, we performed a BLAST search (tBLASTn) between wVitA and the NCBI’s Genbank nt database (accessed Mach 2014) and did not find top BLAST hits to type B Wolbachia. Ancestral states were determined using outgroup genomes in the type A Wolbachia (wMel and wRi) for which orthologs could be identified and robust nucleotide alignments created. (nucleotide identity between the homologs was high). Comparisons between orthologs in the wVitA and wUni genomes were performed. Specifically, protein sequences for each ortholog were aligned using ClustalW and nucleotide sequences for each ortholog were aligned based on their amino acid translation (using in house scripts). The structural annotation for genes that appeared to have protein truncations (due to frameshifts) or indels in the comparison was manually verified. To identify positive selection on orthologous genes between wVitA and wUni as well as the frequency of nonsynonymous and synonymous changes, we utilized Phylogenetic Analysis by Maximum Likelihood (PAML v4.7) software suite using the Goldman and Yang (R1) substitution model (codeML) (Yang 2007). Site specific omega values were implemented using the beta distribution model (NSsites = 7) and equilibrium codon frequencies were estimated using average nucleotide frequencies at the three codon positions (CodonFreq = 2). We also iteratively ran the program (using NSsites 1, 2, 7, 8) to calculate maximum likelihood ratios for residues identified as under positive selection, but no such residues were identified. Codon usage statistics were generated using CodonW (Peden 2000). Statistical analyses of data sets (such as χ2) were performed using the SPSS software package. To identify significant differences in relation to enrichment of particular gene categories within different Wolbachia genomes, we identified the core genome as well as the accessory genome using reciprocal BLAST across the type A Wolbachia genomes included herein. This core genome is the gene set conserved across all type A Wolbachia whereas the accessory gene set includes those that are strain specific or found only in a subset of type A Wolbachia.
To predict the expected number of synonymous and nonsynonymous substitutions in the wVitA and wUni genomes, we generated a statistical probabilistic framework to account for the number and direction of changes from each nucleotide, compared with the predicted ancestral sequence based on the outgroups. We utilized mutational spectra based on third position, 4-fold degenerate sites (table 2) to generate a probability of direction of mutation for each SNP reflecting a change from the consensus ancestral sequence. For each codon, we then generated a nonsynonymous and a synonymous probability and determined the frequency of each codon within a set of orthologs used for SNP analysis (of 199 orthologous genes) as well as the entire genome.
Table 2.
Number of Single Substitutions at 4-Fold Degenerate Sites for the wVitA and wUni Wolbachia Relative to the Type A Wolbachia Outgroups (wRi and wMel)
| wUni | ||||
|---|---|---|---|---|
| A | C | G | T | |
| A to | 9,332 | 2 | 25 | 2 |
| C to | 3 | 3,056 | 0 | 4 |
| G to | 10 | 0 | 3,174 | 1 |
| T to | 1 | 28 | 1 | 10,198 |
| wVitA | ||||
| A | C | G | T | |
| A to | 9,353 | 0 | 8 | 0 |
| C to | 0 | 3,059 | 0 | 4 |
| G to | 2 | 0 | 3,183 | 0 |
| T to | 0 | 6 | 0 | 10,221 |
Note.—Direction of mutation was inferred based on allele frequency with the minor allele presumed to be the most recent mutation. Total numbers of each nucleotide at 4-fold degenerate sites was calculated and the number not exhibiting a mutation is shown in gray shading.
We calculated the predicted GC content at 4-fold degenerate sites at equilibrium, given the current mutational spectra in wUni and wVitA, in two different ways. First, we utilized the equation for GC4pred published by Hildebrand et al. (2010). Counts for the numbers of GC->AT and AT->GC mutations were based on comparisons between wVitA and wUni using wRi/wMel as outgroups, and 4-fold degenerate sites only (table 2). We also utilized a matrix-based calculation to predict GC content at equilibrium for 4-fold degenerate sites in these genomes. Essentially, we iteratively multiplied a vector (the current number of each nucleotide at 4-fold degenerate sites) by a matrix of probability and direction of change for each nucleotide (based on our observed frequencies of change, table 2). This process was repeated until equilibrium was reached (i.e., the number of predicted nucleotides no longer changed and the equilibrium was insensitive to starting frequencies).
Results and Discussion
Comparisons with other A group Wolbachia indicate that wVitA and wUni are among the most closely related Wolbachia so far sequenced (wRi, wMel, wHa; supplementary table S1, Supplementary Material online). They share identical MLST sequences and have a synonymous divergence (SD) of 0.57% (supplementary table S1, Supplementary Material online). Other closely related Wolbachia strains for which genomes available are those of wRi-wMel (3.44% SD), and wMel-wHa (3.29% SD) and wRi-wHa (3.29% SD) (supplementary table S1, Supplementary Material online).
Estimating divergence time between the two Wolbachia strains requires some estimate of SD rate within these bacterial lineages. SD rate is more appropriate than total divergence rate because the former more closely approximates “neutral” rates of substitution, at least relative to rates that include nonsynonymous changes that are more subject to purifying selection. Raychoudhury et al. (2009) estimated molecular rates of evolution between two Wolbachia that codiverged with their Nasonia host species and calibrated these rates to two different estimates of divergence times between the hosts. They found 0.65–0.73% SD per million (106) years for Wolbachia (Raychoudhury et al. 2009), close to estimates for other endosymbiotic bacteria (Moran et al. 1993). Using this rate, the divergence time between wVitA and wUni is predicted to be 0.78–0.88 Myr (given our estimate of 0.57% SD). However, the mutation rates may differ between wVitA and wUni strains. Therefore, we calculated the divergence between each strain and the wRi outgroup to provide both a minimum and maximum estimate of divergence time, in case wUni shows an accelerated mutation rate. The divergence time estimate between wRi and wUni is 14–16 Myr (based on a dS of 0.1038 between wUni and wRi) whereas the divergence time between wRi and wVitA is 12–14 Myr (based on a dS of 0.0882 between wVitA and wRi). Because many more changes are predicted to have occurred on the wUni branch in the phylogeny, suggesting accelerated evolution on that branch, the wVitA estimate is likely the most parsimonious.
General Description of the wVitA and wUni Genomes
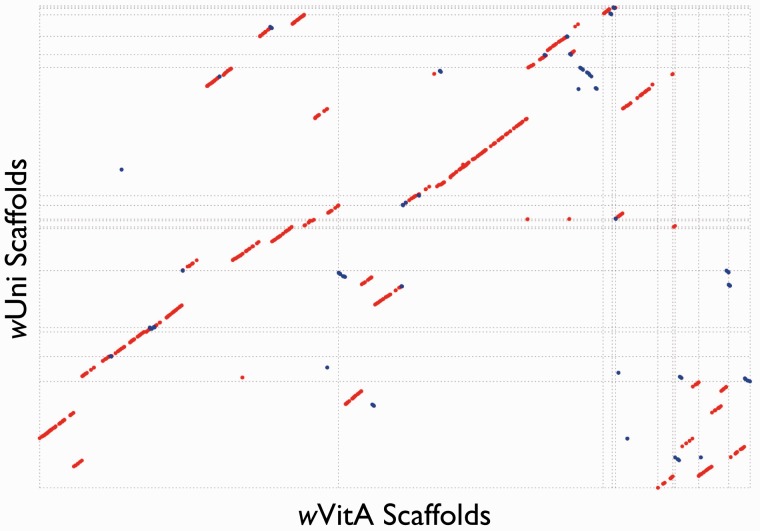
Predicted genome sizes for wVitA and wUni are similar to previously published Wolbachia genomes (wMel: 1.27 Mb, wRi: 1.45 Mb, wRec: 1.13 Mb; [Sun et al. 2001]). Using a combination of Illumina and 454 data for wVitA and published contigs plus 454 data for wUni, we were able to come close to the expected genome size for each (fig. 1). Sequence differences were verified by PCR and sequencing of 11 different regions, and therefore are unlikely to be due to artifacts of the assemblies (see Materials and Methods). However, differences in sequencing strategy as well as artifacts from the multiple displacement amplification used to generate the DNA sequenced may account for differences in N50 contigs and scaffolds sizes observed between wVitA and wUni. A larger portion of the wVitA genome was likely sequenced, as indicated by the larger number of rRNAs and tRNAs found in the wVitA assembly compared with wUni (fig. 1).
Fig. 1.—
Sequencing statistics for the two Wolbachia genomes presented here.
The GC content observed for these two genomes (32–35%) is well within the range expected for Wolbachia as is the number of ORFs (Wu et al. 2004; Klasson et al. 2008; Klasson, Westberg, et al. 2009). In total, 1,325 potential genes are predicted for the wVitA genome and 1,165 for wUni. Although the number of final scaffolds for each genome is relatively small (10 and 19 for wVitA and wUni, respectively), we were unable to completely close the genomes, likely due to repetitive elements, sequence polymorphisms, and recombination, as seen in other Wolbachia genome projects (Klasson et al. 2008).
Overall Patterns of Divergence between wVitA and wUni
By polarizing the changes in the branch leading to wVitA and wUni with the outgroups and predicting the ancestral sequences (table 1), we found highly elevated frequencies of rearrangements, gene truncations (particularly in genes predicted to be involved in host interaction), and both synonymous and nonsynonymous substitutions in wUni relative to wVitA. Details of these differences and possible explanations for the apparent accelerated rate of evolution in the wUni lineage are described in the sections below.
Table 1.
Numbers of Nonsynonymous and Nonsynonymous Substitutions and Rearrangements in wUni and wVitA Based on Comparisons to the Predicted Ancestral Sequence
| Non-Syn (NS) | Syn(S) | Rearrangements(R) | Duplication | Tri. | Quad. | |
|---|---|---|---|---|---|---|
| wVitA | 24 | 22 | 14 | 32 | 23 | 2 |
| wUni | 233 | 128 | 37 | 31 | 5 | 0 |
Note.—The numbers of NS and S and R are significantly higher in wUni, whereas proportions of NS to S are not significantly different and proportions of R relative to NS and S are significantly higher in wVitA (see text). Also, both wVitA and wUni genomes contain a large number of duplicated genes. Paralogous genes in each genome were identified by a BLASTp search against the resident genome. A large number of duplications, triplications, and quadruplications of genes were identified in this manner.
Extensive Rearrangements in Closely Related Wolbachia
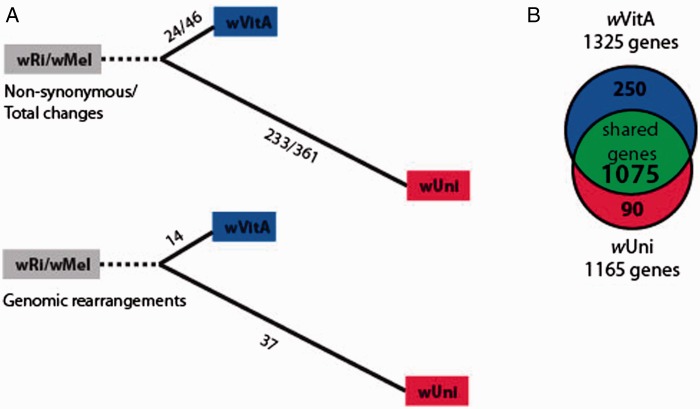
Wolbachia genomes are well known for undergoing numerous rearrangements (Salzberg et al. 2005; Klasson et al. 2008; Klasson, Kambris, et al. 2009; Klasson, Westberg, et al. 2009), which is in contrast to some Rickettsial relatives (McLeod et al. 2004) that maintain a high degree of genomic synteny. Importantly, all genomic assemblies are, to some extent, hypotheses, and artifacts introduced by the use of genome amplification strategies (such as MDA) before sequencing are known to bias assemblies (Ellegaard, Klasson, Naslund, et al. 2013). Although DNA from both species was subjected to MDA, comparisons of synteny presented below should be cautiously interpreted. To identify syntenic portions of genome scaffolds and possible gene inversions or rearrangements between wVitA and wUni, we used the program suite MUMmer. Scaffolds from the two genomes were used as input to Promer, and the genomic comparison was visualized using Mummerplot (fig. 2). When comparing wVitA and wUni, there is a significant amount of genome rearrangement despite that the two strains are closely related (fig. 2). Interestingly, although wVitA and wUni were identical in their MLST profiles (Baldo, Hotopp, et al. 2006), suggesting significant similarity in nucleotide identity between the two strains, to explain the difference in their genomic structure, between 51 and 233 rearrangements were estimated to have occurred in the lineages (Simon and Larget 2004; Lin et al. 2006; Darling et al. 2010). The estimate generated by Badger (51 rearrangements) does not account for gene gain and loss or reversals and is therefore a conservative, lower bound. The higher estimate of rearrangements predicted when comparing wVitA and wUni rivals that of single nucleotide polymorphisms in comparisons of orthologous genes for these same two genomes (270 SNPs between orthologous genes). Interestingly, these rearrangements are significantly enriched in the wUni lineage (χ2= 10.4, df =1, P = 0.0013), where 2.6-fold more rearrangements have occurred compared with the wVitA lineage relative to their most recent common ancestor (14 in wVitA vs. 37 in wUni; and 40 shared rearrangements relative to wRi). However, the relative proportion of rearrangements is also higher in wVitA when compared with either nonsynonymous (χ2 = 12.7, df = 1, P = 0.00036) changes or to synonymous (χ2 = 4.23, df = 1, P = 0.04) changes.
Fig. 2.—
Synteny analysis between the wVitA and wUni scaffolds reveals a large number of rearrangements and inversions. Syntenic portions of scaffolds are indicated by consecutive red dots whereas inversions are indicated in blue.
It is likely that the very large number of repetitive mobile elements found in Wolbachia genomes facilitate homologous recombination and rearrangements (Cordaux 2008; Cerveau et al. 2011; Leclercq et al. 2011). Like other sequenced A- and B-supergroup Wolbachia genomes, both wVitA (Kent, Funkhouser, et al. 2011; Kent, Salichos, et al. 2011) and wUni contain large prophage WO elements. In total, 151 genes are predicted to be mobile elements in wUni (13% of the genome, 68 transposons, 9 copies of the Wolbachia palindromic element, and 74 phage genes) whereas wVitA contains 137 mobile element genes (10% of the genome,18 transposons, 9 copies of the Wolbachia palindromic element, 110 phage genes, supplementary table S3, Supplementary Material online). These percentages of mobile elements are within the range for other type A Wolbachia (between 8.9% and 22% [Klasson, Westberg, et al. 2009]), although it is worth mentioning that this is likely an underestimate as repetitive regions and mobile elements are often difficult to assemble. Although wUni lacks numerous phage genes found in wVitA, the larger percentage of other mobile elements in this strain is due to an increase in the number of predicted transposons and includes nine Wolbachia palindromic elements and a diverse group of transposons from five different families including IS5, IS200, Tn5, IS116, and DDE proteins. This diversity of transposable elements has been observed before in Wolbachia genomes (Klasson et al. 2008), however the great difference in transposable element content in wUni compared with its close relative wVitA indicates a very recent expansion of these selfish genetic elements in the wUni genome, as these elements are not present in the same location in outgroup type A Wolbachia (wRi and wMel). In addition, a significant fraction of genes unique to wUni (5/14) and not found in other Wolbachia, are mobile elements.
Gene Content Conservation and Change
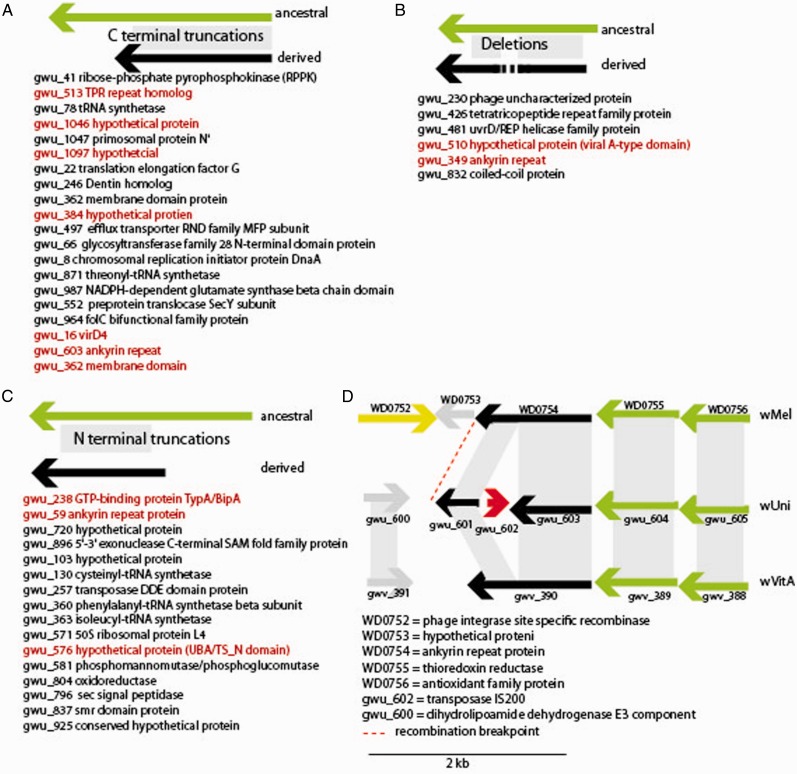
To explore changes in gene content between wVitA and wUni, homologous genes in wVitA, wUni, wRi, and wMel (identified using reciprocal BLAST) were aligned and compared. Both wVitA and wUni genomes contained comparable numbers of ORFs (wVitA = 1,325 and wUni = 1,165) and shared a substantial fraction of their genes (a total of 1,075, fig. 3B). A small fraction of these ORFs (N = 3) are unique to this pair of Wolbachia (based on BLASTn results). Moreover, wUni and wVitA harbor 90 and 250 unique genes, respectively, in comparison to each other (fig. 3B). Both genomes encode a large number of ankyrin repeat proteins (30 in wVitA and 18 in wUni) (supplementary table S4, Supplementary Material online). These ankyrin repeat proteins are homologous to proteins in other Wolbachia genomes although wVitA encodes three ankyrins, not found in wUni, with low, but closest similarity to B group Wolbachia genes (supplementary table S4, Supplementary Material online). Despite the fact that wVitA and wVitB coinfect N. vitripennis and have exchanged an entire genome of phage WO (Kent, Salichos, et al. 2011), we find no strong evidence for recombination between these A and B Wolbachia. Unique genes in the wUni genome are largely paralogs and duplications of genes with homologs in wVitA (table 1; supplementary table S3, Supplementary Material online). A total of 30 wUni genes, with orthologs in wVitA, have multiple paralogs in the wUni genome, many of which are transposon-related genes (13/30). Of note is an expansion in type IV secretion system components; there are a total of 4 virB4, 2 virB2, and 2 virB6 paralogs in wUni (supplementary table S3, Supplementary Material online).
Fig. 3.—
The wVitA and wUni genomes differ in regards to gene content and nucleotide divergence. (A) Most of the changes observed when comparing the two genomes have occurred on the branch leading to the wUni lineage, included rearrangements, and elevated nonsynonymous synonymous substitutions. (B) Both genomes share a substantial percentage (between 80% and 92%) of their gene content, although each harbors unique ORFs, when compared with the other.
A significant fraction (21/104 - χ2 = 23.43; df = 1; P < 0.0001) of genes unique to wVitA that are not found in wUni or in other type A Wolbachia are predicted to be involved in interactions with the host (see Materials and Methods). These wVitA-unique genes contain domains long hypothesized to be mediating the symbiosis (18 genes containing ankyrin repeats, supplementary table S5, Supplementary Material online) or domains not commonly found in bacterial genomes, but likely of eukaryotic or viral ancestry (ovarian tumor, PRANC; supplementary table S5, Supplementary Material online) (Wu et al. 2004; de Felipe et al. 2005; Siozios et al. 2013). Given the incomplete nature of the wUni genome it is possible that some of these genes involved in host interaction were missed in our sequencing. However, we have no reason to believe the sequencing process was somehow biased against the discovery of these specific ORFs. The data suggest that these genes were acquired by the common ancestor of wUni and wVitA and then selectively purged from the wUni genome, or were acquired in wVitA through horizontal gene transfer after the divergence with wUni.
Faster Evolution by ORF Truncations and Insertion/Deletions in wUni
In our comparison of wVitA and wUni orthologs, we noticed that 42 of the wUni ORFs appeared shortened or contained indels with respect to their counterparts in all other type A Wolbachia, including wVitA (fig. 4). In comparison, only three cases of wVitA homologs (gwv_46, gwv_937, gwv_909) show a similar truncation in ORF. A subset of these truncations were validated with PCR and sanger sequencing. Among the wUni shortened ORFs, there are six cases with small internal indels in the amino acid sequence in which they maintained the correct reading frame, either because multiple indel events occurred or an indel occurred in multiples of three nucleotides (fig. 4B). Of 42 shortened predicted proteins, 36 are due to changes in the predicted coding region resulting from a SNP or frameshift that generated a stop codon (in the case of C-terminal truncations) or altered the start of the ORF (in the case of N-terminal truncations; fig. 4A and C). This pool of shortened orthologs is not particularly divergent when comparing between these two related Wolbachia strains; they are between 95% and 100% identical based on nucleotide sequence. One might expect an accumulation of mutations downstream of introduced stop codons in wUni, but we similarly observe a range of percent identities in line with that for the actual ORF (100–95%), suggesting these orthologs were recently shortened, leaving insufficient time for the accumulation of additional substitutions.
Fig. 4.—
Schematic representation of gene evolution by truncations (A and C) and deletions (B) in the wUni genome with respect to the type A Wolbachia (wVitA, wMel, and wRi), some of which result from genomic rearrangements. Red text denotes those proteins predicted to be involved in host interaction based on secretion signal, a eukaryotic domain (such as an ankyrin repeat domain), and conservation of ancestral state with wHa, wRi, and wVitA. Only three wVitA genes (gwv_46, gwv_937, gwv_909) are predicted to have undergone similar truncations and deletions relative to the outgroup type A Wolbachia. (D) Illustrative example gene truncation due to transposon insertion within an ankyrin repeat protein. The wUni gene gwu_602 encodes an IS200 element that has inserted within an ankyrin repeat domain protein found intact in both the wVitA (gwv_390) and wMel genomes (WD0754). This segment of the genome is syntenic between these three type A Wolbachia strains.
This pool of shortened predicted proteins is enriched for proteins that are candidates for involvement in host interaction (fig. 4). We hypothesize that these proteins are likely involved in host interaction because they hold at least one of the following traits: they have a secretion signal (sec), harbor a eukaryotic domain (such as an ankyrin repeat domain), or are part of the type IV secretion machinery (vir proteins). Based on these criteria, a statistically significant enrichment of possible host-interaction proteins (16/42) was found in this pool of shortened predicted proteins in comparison to the core essential genes (χ2 = 31.65; df = 1; P < 0.0001). Out of the 42 shortened predicted wUni ORFs, 10 are disrupted by a mobile element (fig. 4), likely a consequence of the expansion of transposable elements observed in wUni. As has been observed for other Wolbachia genomes, ANK genes seem to evolve by insertions, duplications, and deletions (Klasson et al. 2008). These candidates (red text, fig. 4) are also found as full length homologs in the related Wolbachia strain wHa, suggesting their possible role in CI. We conclude that the wUni genome shows accumulation of truncations for ORFs involved in host interactions.
Faster Rate of Nucleotide and Protein Evolution in wUni
wUni also shows a faster rate of nucleotide substitution. To explore the nucleotide divergence between these two Wolbachia strains, we focused our analysis on the orthologous set of genes—the 1,075 shared ORFs. All ORFs were aligned based on their amino acid translations and the number of nucleotide substitutions, whether synonymous or nonsynonymous, was counted. A large fraction of this shared pool of genes did not show any sequence divergence between wVitA and wUni (N = 494).
We next used outgroup taxa to determine whether individual substitutions occurred in the wUni or wVitA lineage. For genes with few substitutions (1–3), the allele at each position for the type A Wolbachia (wRi, wMel, wVitA, wUni) was used to identify the consensus ancestral state. For example, alleles present in wRi and wMel, are considered ancestral and changes in either wVitA or wUni, would be considered derived. Within this orthologous gene set, 604 substitutions (or 58% of the total number of substitutions) could be assigned an ancestral state. The overwhelming majority of changes (496 vs. 108) in the type A Wolbachia orthologs occurred in the branch leading to wUni. We then focused on the orthologs with one or two substitutions across the type A Wobachia, including only those orthologs for which we could create robust alignments with multiple outgroups of type A Wolbachia. The numbers of synonymous and nonsynonymous changes in the wVitA and wUni lineages are shown in figure 3A and Table 4. There are significantly more nonsynonymous than synonymous changes in wUni (nonsynonymous/Total 233/361 = 0.64, χ2 = 29.552; df = 1; P = 0.0001). Also, wUni shows a greater number of both substitution types relative to wVitA. There was significantly greater numbers in wUni of nonsynonymous (233/24 in wUni/wVitA or 9.6-fold higher, χ2 = 168.03; df = 1; P ∼ 0) and synonymous changes (128/22 or 5.8-fold higher, χ2 = 74.91; df = 1; P ∼ 0). However, the proportion of nonsynonymous substitutions was not significantly different between the two lineages (FET, P = 0.144, χ2 = 2.59; df = 1; P = 0.107). One explanation for the overall higher substitution rate in wUni could be smaller population sizes resulting in accelerated fixation of deleterious mutations (i.e., Muller’s Ratchet) in wUni. However, this should result in a higher proportion of nonsynonymous substitutions, because small population sizes disproportionately enhance fixation rates of mildly deleterious mutations while neutral mutations are relatively unaffected by population size (Ohta 1973).
Table 4.
Number of Synonymous and Nonsynonymous Changes for Orthologs in the Comparison between wVitA and wUni with the Ancestral Sequence with 1, 2, 3, or >3 SNPs
| 1 | 2 | 3 | >3 | |
|---|---|---|---|---|
| #S | 100 | 50 | 48 | 38 |
| #N | 155 (173) | 100 (102) | 90 | 97 |
Note.—Shown are orthologs for which we could produce robust alignments. For orthologs with 1 or 2 substitutions, we also show the expected number of nonsynonymous substitutions based on codon mutational probabilities (in parentheses).
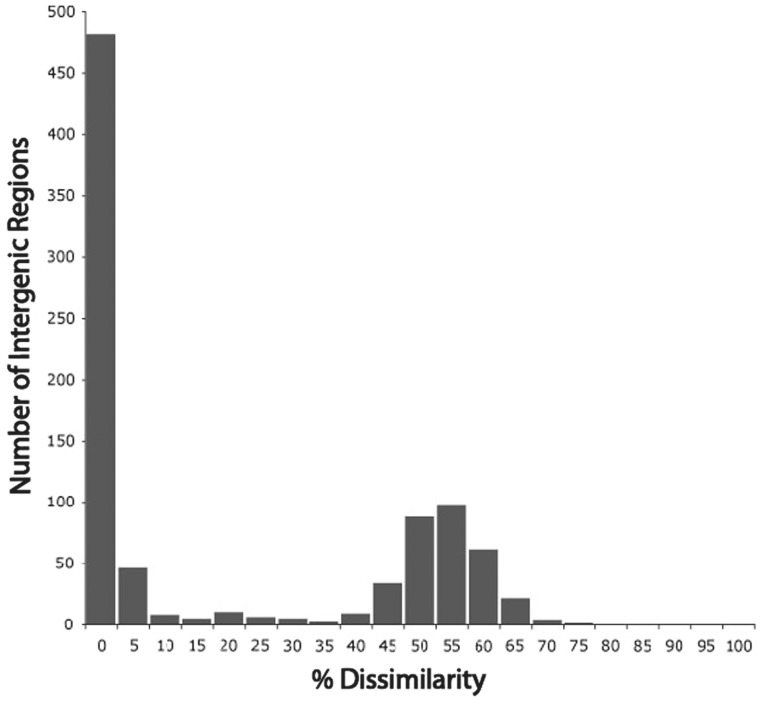
Because the ortholog set used in the above analysis is based on protein-coding genes, which may be subject to strong purifying selection, we also analyzed intergenic regions found to be homologous between wVitA and wUni. These regions were identified by a BLASTn search of the 50 bp upstream of orthologous genes in a reciprocal fashion. This region is likely to include promoters at the −10 and −35 positions upstream from transcriptional start as well as possible UP elements (a component of the promoter that enhances transcription) at −50 (Estrem et al. 1998). Although consensus binding sites for transcription machinery will likely be conserved, there are also a number of sites outside of these conserved sequence regions that can withstand substitutions and indels. Interestingly, about half (482/880) of the upstream untranslated regions compared between wVitA and wUni are identical (fig. 5). Also, the distribution of regions with regards to % identity is bimodal, with peaks at 100% and approximately 50% identity (fig. 5). To investigate whether or not these peaks reflect conservation between the two genomes with regards to synteny, we correlated these positions with rearrangements predicted via MUMmer and found that a small minority, only 71 out of 880, corresponds to rearrangement breakpoints. The intergenic regions that do occur within rearrangement breakpoints are, on average, 26% identical between wVitA and wUni. In contrast, the vast majority of intergenic regions (809/880) are syntenic in the comparison between these two strains (i.e., have the same flanking genes) and flank identical or nearly identical homologs in both genomes. Yet, many of these still show low levels (∼50%) of sequence identity, and therefore appear to be rapidly accumulating mutations.
Fig. 5.—
Comparison between wVitA and wUni in the upstream untranslated regions of orthologous genes (total of 880 compared). The 50 bp regions upstream of orthologous genes were compared using BLASTn and percent identity calculated. The vast majority of these sequences are syntenic in the two genomes (809/880) and therefore the level of divergence between the two strains represents the accumulation of nucleotide substitutions rather than intergenic recombination. Nevertheless, there is a striking bimodal distribution, with a second peak around 50% dissimilarity. This suggests that either some genes have very rapidly evolving 5′-intergenic regions, or that they were laterally transferred from other Wolbachia with divergent intergenic sequences. A gene ontology examination of those genes with divergent 5′-intergenic regions did not reveal significant enrichments in terms of function.
Mutation Spectra in wVitA and wUni
Given that wVitA and wUni have recently diverged and exhibit low substitution frequencies, they can be used to estimate the mutational spectra in Wolbachia. For this analysis, we examined orthologs with a single nucleotide substitution (singletons) at 4-fold degenerate sites. Given the low numbers of synonymous changes across the genome, synonymous differences between these closely related taxa are the best available proxy for mutational spectra. We used these SNPs to estimate the mutational patterns in these lineages and found a total of 97 orthologs with single substitutions difference between wVitA and wUni relative to the predicted ancestral sequences at 4-fold degenerate sites (table 2). We examined the allele frequencies at each position in the type A outgroup Wolbachia to infer which SNP was the new mutation.
As expected, there are a higher number of transitions versus transversions (more changes within than between purines and pyrimidines) in both genomes (table 2). Also, as observed earlier, there is a striking increase in substitutions in the wUni lineage relative to wVitA since their divergence. This result can be explained by either an increased mutation rate in the wUni lineage, by a shorter generation time, or by increased rates of fixation (either due to reduced purifying selection of deleterious mutations or to directional selection). However, note that these synonymous positions are not subject to strong purifying selection. Therefore, the pattern is not easily explained except by invoking an elevated mutation rate in wUni, or a greater number of generations per year in this bacterium, because it occurs in a semitropical host species.
Evolution of GC Content in Type a Wolbachia
In examining the SNP changes in these Wolbachia genomes, we noticed an enrichment in the number of changes towards GC relative to AT at 4-fold degenerate sites (table 2). For example, considering that we are more likely to observe AT->GC mutations (given the greater probability of an A or T at each nucleotide position), the counts of AT->GC single nucleotide substitutions exceed the expectation given an equal probability of mutation (for 77 4-fold degenerate SNPs in wUni, the expectation is ∼40 AT-> GC mutations but 56 are observed; for wVitA the expectation is ∼10 AT-> GC but 14 are observed).
Although we observe increased counts of AT->GC mutations in the wVitA and wUni genomes, an elevated frequency of AT to GC changes would imply evolution of a greater GC content in theses genomes, whereas these and other Wolbachia have AT rich genomes (Wolbachia genomes show a strong AT content bias [∼35% G + C] [Wu et al. 2004; Klasson et al. 2008; Klasson, Westberg, et al. 2009]). We therefore calculated the mutational probabilities, for each nucleotide, based on observed spectra at 4-fold degenerate sites (supplementary table S6, Supplementary Material online). Calculating the overall expected GC->AT and AT->GC probabilities based on 4-fold degenerate sites yields a GC ->AT of 0.00574 and 0.00193 and an AT->GC probability of 0.00572 and 0.00144 for wUni and wVitA, respectively. These conditional mutation rates are expected to produce equilibrium GC content much less biased than observed in 4-fold degenerate sites. To further confirm this finding using the per base mutational spectra (supplementary table S6, Supplementary Material online), we iterated the mutational spectra matrices for wUni and wVitA until an equilibrium GC content distribution was achieved. This method yielded predicted GC contents higher than the current content at 4-fold degenerate sites (44% GC predicted for wVitA and 49% GC for wUni, whereas the current GC content is 24%). Finally, based on the mutational spectra we observed here (table 2) and the observed GC content at 4-fold synonymous sites (GC4) we can estimate the predicted equilibrium G + C content assuming no selection (GCeq as in Hildebrand et al. [2010], table 3). The GCeq at each position indicates that if these genomes were allowed to reach equilibrium, their GC content at 4-fold synonymous sites would increase significantly (wUni: 50% GC; wVitA 43% GC) compared with the observed G + C content at 4-fold synonymous sites of 24% (table 3; two-tailed χ2 tests comparing observed versus expected GC content for wVitA χ2 = 14.729; df = 1; P < 0.0001; for wUni χ2 = 27.04; df = 1; P < 0.0001).
Table 3.
Table of Calculated Counts and Frequencies Used in the Generation of a GC4pred (Hildebrand et al. 2010)
| wUni | wVitA | |
|---|---|---|
| GC4 | 0.2418 | 0.2418 |
| V | 56 | 14 |
| U | 18 | 6 |
| GC4pred | 0.4981 | 0.4266 |
Note.—V, the number of AT->GC SNPs at 4-fold degenerate sites; U, the number of GC->AT SNPs at 4-fold degenerate sites; GC4, the GC content at 4-fold degenerate sites.
The finding that recent mutations in Wolbachia increase GC content is rather surprising; our results run counter to the assertion that AT mutation bias is universal in bacteria (Hershberg and Petrov 2010). In their meta-analysis of 149 bacterial species ranging in GC content from 20% to 70%, Hildebrand et al (2010) found evidence of at GC->AT mutational bias in nearly all taxa, and concluded that because all bacteria are not AT rich, there is selection for increased GC in most species. However, they did find an excess of AT->GC mutations in some taxa with AT rich genomes. They point out that interpretation of their data set could be compromised by the relatively large distances between some taxa, which violates the infinite sites assumption and can lead to multiple mutation events at individual sites. The data presented here support the finding of an increase in AT->GC mutations in the AT rich genome of Wolbachia, using recently diverged strains in which multiple substitutions at the same sites are unlikely.
It is possible that selection on codons based on tRNA abundances and gene expression levels could affect third position substitution probabilities ([Sharp and Li 1987a, 1987b] but see [Shah and Gilchrist 2010]). We are using the synonymous substitutions as a proxy for mutational spectra, but it is possible that selection has also influenced the spectra of substitutions observed, for example, through codon usage bias. tRNA abundances are not known in Wolbachia, and expression profiles for different genes are not well characterized. However, we can explore possible differences in translational selection in these genomes by analyzing codon usage. Selection for codon usage is inefficient in bacteria with unusually high GC or AT content (but see [Charles et al. 2006]). One test for translational selection in Wolbachia would be to identify codon usage patterns between the type A Wolbachia to determine if a difference in codon usage between wVitA and wUni could explain the data. We performed a factorial correspondence analysis of codon usage for four type A Wolbachia: wMel, wRi, wUni, and wVitA (supplementary fig. S1, Supplementary Material online). The analysis reveals that usage patterns within the type A Wolbachia are similar; neither of these intracellular symbiont genomes shows the typical pattern of nonrandom codon usage seen in some bacterial genomes (Gouy and Gautier 1982; Ikemura 1985; Shah and Gilchrist 2010; Karberg et al. 2011). In addition, no evidence of atypical codon usage was found between wUni and wVitA compared with the other type A Wolbachia genomes or each other.
An alternative explanation for the AT bias in Wolbachia genomes is that the energetics of DNA synthesis or the availability of nucleotides selects for G + C content genome-wide. It has been suggested that AT enrichment in bacterial endosymbionts may be due to the energetic costs of G + C nucleotides (or availability of A + T nucleotides in an intracellular environment) (Rocha and Danchin 2002). An argument for this is that intracellular bacteria are in a cellular environment relatively rich in ATP, and the availability of this nucleotide could affect the costs of DNA synthesis. However, this hypothesis is countered by the high G + C content of some intracellular symbionts, such as Hodgkinia and Tremblaya (Van Leuven and McCutcheon 2012). Interestingly, the high G + C content endosymbiont Hodgkinia exhibits a mutational bias toward AT (Van Leuven and McCutcheon 2012), as predicted by Hildebrand et al. (2010). Our study provides evidence that mutational bias alone cannot explain genome-wide A + T content in Wolbachia genomes, and that selection for greater A + T content (either on individual genes or genome wide) is a likely explanation.
Why Do wUni and wVitA Differ in Evolutionary Rates?
Since the divergence of these two bacteria, wUni shows an overall approximately 8-fold greater SNP substitution rate than does wVitA, as well as increased number of rearrangements, predicted protein truncations, insertion/deletions, and expansion of transposons (figs. 3 and 6, table 2; supplementary table S2, Supplementary Material online). Explanations for the differences fall into five broad categories (Hartl and Clark 2007): wUni has a significantly 1) higher mutation rate, 2) experiences a greater number of generations per year, 3) has a smaller effective population size leading to more rapid fixation of deleterious mutations, 4) experiences relaxed selection, or 5) is experiencing directional (adaptive) evolution. We cannot completely resolve these scenarios, but we briefly discuss each. In general, our data are most consistent with a faster generation time in wUni relative to wVitA because we can conservatively rule out the other four likely explanations (see below). Although an elevated mutation rate is possible, it would have to occur in single nucleotide substitutions, insertion/deletions, and rearrangements to explain the data. A shorter generation time can explain elevations in all these processes, assuming they are replication rather than time dependent. Furthermore, data suggest that wUni may be evolving by truncation and deletion of genes, possibly those no longer involved in the CI phenotype, whereas wVitA may be evolving adaptively by nonsynonymous substitutions.
Fig. 6.—
Model representation of evolution within the Wolbachia on short time scales. Gene loss, recombination and genomic rearrangements, evolution by point mutations as well as truncations and indels were evident.
(1) Higher mutation rate in wUni: The data are consistent with a higher wUni mutation rate, including the findings that 1) there are alterations in the DNA repair genes in wUni, 2) single nucleotide changes in wUni are elevated in both synonymous and nonsynonymous substitutions, and 3) the relative proportion of nonsynonymous substitution is not elevated relative to wVitA. In support of DNA repair pathway alterations causing mutation rate increases in wUni, the wUni genome has an approximately 4-fold greater synonymous substitution rate at third positions than wVitA. DNA repair processes play critical roles in determining the mutation rate of bacteria (Modrich 1987, 1991; Grilley et al. 1990) and more recent work has pointed to the importance of mismatch repair activity in generating mutations and defining mutational spectra (Acharya et al. 2003). In the absence of mismatch repair, mutations in Escherichia coli predominantly convert adenine and thymine to guanine and cytocine (AT -> GC) (Lee et al. 2012). Because Wolbachia genomes lack mutH, an endonuclease that nicks hemimethylated GATC sequences and maintains strand fidelity in mismatch repair (Horst et al. 1999), it is possible that new mutations are more likely to arise in the genome. Both wVitA and wUni contain most of the genetic elements required for mismatch repair (besides mutH), although wUni is missing two important DNA glycosylases: mutM (a glycosylase that removes 8-oxoG from 8-oxoG-C mispair) and mutY (that removes A from 8-oxoG-A or A-G mispairs), lesions known in other systems to increase mutation rates dramatically (Michaels et al. 1992; Horst et al. 1999; Debora et al. 2011). Besides the loss of these mismatch repair genes, our current data set reveals another major change in known DNA repair pathway genes in wUni compared with wVitA: RecR, a gene involved in recombinational repair is truncated in the wUni lineage. The difference in mutational spectra observed between these two bacteria may therefore be a result of differences in the existence of different DNA repair pathways. However, given that we see elevated rates of rearrangements and gene truncations, insertions, and deletions, then these mutation repair pathways would need to affect each of these processes.
(2) More generations in wUni: Many of the observed mutational patterns can be explained by a faster generation time in wUni, including elevated rates of SNPs and rearrangements if the rates are replication dependent. The insect host of wUni (Muscidifurax uniraptor) occurs in a subtropical region (Puerto Rico), whereas that of wVitA (N. vitripennis) is a largely temperate species. Although N. vitripennis occurs world-wide, this is likely due to dispersal by humans, as the Nasonia genus is holoarctic in distribution (Darling and Werren 1990). In temperate regions, N. vitripennis likely has only a few generations per year, and it has an overwinter diapause that can last approximately 8 months of the year, suggesting fewer generations per year for this species. However, N. vitripennis has a greater reproductive rate than M. uniraptor, which would likely require faster replication of Wolbachia within the ovaries. Therefore, the precise differences in average bacterial generation time of wVitA and wUni are unknown, but a possible explanation of our results.
(3) Smaller effective population size and Muller’s Ratchet in wUni: Previous work in insect endosymbionts suggested that Muller’s Ratchet in small effective populations is a driving force in fixing slightly deleterious mutations through genetic drift (Brynnel et al. 1998; Clark et al. 1999). In our data set, this hypothesis would predict that slightly deleterious mutations would be fixed as though nearly neutral, leading to an increase in the observed rate of evolution in wUni (Lynch 1996, 1997). Small population sizes would disproportionately enhance fixation rates of mildly deleterious mutations, leaving rates of fixation for neutral mutations relatively unaffected (Ohta 1973). However, in wUni we actually see a relatively lower proportion of nonsynonymous substitutions compared with wVitA, although the difference is not significant (FET P = 0.14). This observation is contrary to the expectations if the elevated rates were driven by Muller’s Ratchet. Therefore, these data are not consistent with the differences in substitution rates being due to smaller population size in wUni. Furthermore, with respect to population size, both Wolbachia are presumed to have undergone severe bottlenecks during transfer and establishment within their respective hosts. It is likely that the host N. vitripennis (worldwide distribution) has larger populations size than does M. uniraptor (so far found only in Puerto Rico), although it is unclear whether these differences are sufficient to drive the dramatic differences in genome evolution observed.
(4) Relaxed selection in wUni and insufficient time to purge deleterious mutations: The observed differences in substitution rate could be due to weaker purifying selection in wUni, or insufficient time for purging of deleterious mutations (Mira et al. 2001; Ogata et al. 2006). However, the observation that the proportion of synonymous substitutions in wUni is not significantly different from wVitA is not consistent with relaxed purifying selection (FET, P = 0.144, χ2 = 2.59; df = 1; P = 0.107). Furthermore, if relaxation of purifying selection is playing a role in the evolution of wUni, we expect an increase in dN/dS within genes or genome-wide. However, we were unable to detect any orthologs with elevated dN/dS ratios (that is dN/dS > 1) in the pairwise comparisons between wVitA and wUni.
Using the predicted mutation spectra, we find that the number of nonsynonymous mutations is significantly less than expected based on the mutational probabilities for each mutated codon relative to the ancestral (χ2 = 5.824, df = 1, P = 0.0158). That is, if we calculate the frequency with which we should observe a nonsynonymous or synonymous change for a given codon (using the mutational spectra based on 4-fold degenerate sites) we observe fewer nonsynonymous substitutions for genes with one SNP than expected. This result suggests that purifying selection is playing a role in the evolution of this gene set in both genomes.
However, it is possible that the shift in phenotype has resulted in relaxed purifying selection on the subset of genes involved in CI and related genes involved in host interactions. As a result, this set of genes would accumulate inactivating mutations “neutrally” by mutation and drift, or possibly by selection if their expression is deleterious in the new selective environment (see below). This process could also explain the observation of gene truncations that we have found as well as indels leading to frameshifts and stop codons. Overall, however, these results suggest that decreased purifying selection has not played a predominant role in the divergence of these Wolbachia.
(5) Directional selection in wUni: In the transition to a new host and change in reproductive phenotype to parthenogenesis induction in wUni, there may have been selection on proteins involved in parthenogenesis induction and inactivation of genes that are no longer needed but cause deleterious effects in the new selective environment (e.g., by imposing fitness costs due to expression of genes unnecessary or detrimental to the bacterium in the new host context). Indeed, gene loss in other bacteria accompanies a large shift in environment and phenotype (Olson 1999; Moore et al. 2004; Ensminger et al. 2012). For example, adaptive gene loss in Yersinia pestis followed its transition to a flea host; the loss of rcsA—a negative regulator of biofilm formation—provides Y. pestis with the ability to form biofilms in fleas (Sun et al. 2008). Accompanying the loss of rcsA is the pseudogenization of 149 genes and the loss of 317 from Y pestis compared with its sister taxon and progenitor, Yersinia pseudotuberculosis (Chain et al. 2004; Sun et al. 2008; Zhang 2008). A similar story is found in the rapid evolution of lactic acid bacteria following adaptation to different environments such as meat, milk, and the GI tract (Makarova and Koonin 2007; Pfeiler and Klaenhammer 2007). As we have found in the wVitA–wUni comparison, truncations, rearrangements, duplications, IS element expansion, and pseudogenization of ORFs are observed in comparisons of the Lactobacilli. For example, the Lactobacillus species associated with yogurt have experienced substantial gene decay (between 12% and 19% of the ORFs are pseudogenes) (Callanan et al. 2008). Rapid evolution has also been observed within the major lineages of Lactobacillus casei specific to cheese, where a recently diverged sub cluster (∼50,000 years) has been found to be undergoing rapid, and significant gene loss (120 predicted coding regions) (Cai et al. 2007, 2009). In L. casei ATCC 334, inactivation or alteration of gene function by truncation (in the case of both lactocepin and aminopeptidase genes) was observed (Cai et al. 2009). In general, these results indicate that a large change in selective environment and resulting changes in fitness of the phenotypes will result in selection for “inactivation” and modification of many genes. Genes that are maladaptive in the new selective environment will accumulate inactivating mutations, through truncations, frameshifts, and stop codons. Consistent with this prediction, wRec in mushroom feeding Drosophila recens recently underwent genome reduction from its wMel ancestor in Drosophila melanogaster by a series of gene loss events and pseudogenezations (Metcalf et al. 2014). However, gene loss can also follow “relaxation” of selection and although we do not find evidence of relaxed purifying selection, it is hard to distinguish between these two possibilities.
Conclusions and Interpretation
Endosymbiont genomes have been proposed to experience a suite of genetic changes due to small effective population sizes, including increases in substitution rates (Wernegreen and Moran 1999), a mutational bias toward A + T content (Moran 1996), deletions leading to gene loss (Mira et al. 2001), IS element proliferation (early in an association with a host) (Moran and Plague 2004), and a general accumulation of deleterious mutations. Alternative explanations for at least some of these phenomena in wUni and wVitA include elevated mutation rates (due to lack of DNA repair mechanisms), directional evolution due to adaptation to new host environments and/or relaxed selection on genes no longer needed following host or phenotypic changes (Graur and Li 2000; Hartl and Clark 2007). To better understand the early stages of these processes, studies are needed of recently diverged bacteria that occur in different hosts and induce different phenotypic effects upon their hosts.
Here, we presented a genomic comparison of two recently diverged Wolbachia, which induce different reproductive effects on their respective hosts. One of these (wUni) has evolved parthenogenesis induction in the parasitoid host M. uniraptor, whereas the other infection retains the more common phenotype, CI. CI induction is likely the ancestral phenotype given that other closely related bacteria wRi and wMel are both CI inducers (Carrington et al. 2011; Penz et al. 2012). We observed an overall increase in evolution of wUni, including dramatically high rates of rearrangements, gene loss, increased gene truncations, insertions and deletions, and elevated substitutions, relative to wVitA. In addition, there is evidence of gene acquisition by wVitA relative to related outgroup wRi and wMel (apparently by lateral transfers from other bacteria) and IS element proliferation in wUni. This work also revealed an interesting mutational bias in the Wolbachia genomes toward a more balance AT content (57–50% AT) relative to that normally observed in AT rich Wolbachia genomes (∼76% at 4-fold degenerate sites). Therefore, there is presumably selection for genome-wide AT content that counteracts this mutational pressure.
It is tempting to suggest that as wUni entered its new host, the number and kind of proteins used for host interaction underwent a change in evolutionary pressure. A shift in the reproductive phenotype induced by the bacterium from CI (the common mode found in this bacterial clade) to parthenogenesis induction may have resulted in both relaxed selection on genes involved in CI induction, and possibly in positive selection for inactivation of these genes. Antagonistic coevolution between CI inducing Wolbachia and hosts is expected (Koehncke et al. 2009) and could involve maintenance of a suite of genes for modulating host responses to infection. In contrast, the wUni parthenogenesis inducing Wolbachia is fixed in its host (M. uniraptor), which therefore reproduces parthenogenetically. Therefore, the “genetic interests” of the bacterium and host are allied, and antagonistic coevolution is expected to be largely dissipated while mutualistic coevolution is enhanced. This, in turn, is expected to lead to rapid evolution of genes involved in host interaction, including inactivation of suites of genes involved in antagonistic coevolution. The reduction in likely secreted proteins and toxins may be indicative of a transition from a pathogenic to a more mutualistic association. Indeed, in the mutualistic nematode Wolbachia strains, a reduction was observed in ankyrin domain containing proteins, membrane associated proteins, and other genes predicted to be involved in pathogenesis (Foster et al. 2005). Because wUni has evolved from CI induction to parthenogenesis induction, the genes listed in figure 4, especially those highlighted in red, can be considered candidates for CI function and related host interactions. We surmise selection is more likely than degradation by Muller’s Ratchet to explain the changes observed once wUni moved into a drastically different selective environment. We conclude that the elevated mutation, deletion, and rearrangements observed in wUni are most likely due to a shorter generation time relative to wVitA, rather than due to an accelerated mutation rate, Muller’s Ratchet or relaxed selection.
Supplementary Material
Supplementary text, figures S1–S3, and tables S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank the IGS Annotation Engine staff, and specifically Michelle Giglio, for assistance with the structural functional annotation of the predicted ORFs. The authors also thank the Baylor College of Medicine, Human Genome Sequencing Center Production Team. I.L.G.N. was supported by generous startup funds from Indiana University. I.L.G.N. was also supported by NSF IOS 1456545. S.R.B. was supported by NSF DEB1046149 and IOS 1456778 and NIH R21 HD086833, S.R. and J.Q. were supported by NHGRI U54 HG003272 and J.H.W. was supported by NSF DEB0821936 and NSF DEB1257053.
Literature Cited
- Acharya S, Foster PL, Brooks P, Fishel R. 2003. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 12(1):233–246. [DOI] [PubMed] [Google Scholar]
- Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. 2006. Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 23(2):437–449. [DOI] [PubMed] [Google Scholar]
- Baldo L, Hotopp JCD, et al. 2006. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 72(11):7098–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Lo N, Werren JH. 2005. Mosaic nature of the Wolbachia surface protein. J Bacteriol. 187(15):5406–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR., Wernegreen JJ. 2004. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 21(10):1981–1991. [DOI] [PubMed] [Google Scholar]
- Brynnel EU, Kurland CG, Moran NA, Andersson SG. 1998. Evolutionary rates for tuf genes in endosymbionts of aphids. Mol Biol Evol. 15(5):574–582. [DOI] [PubMed] [Google Scholar]
- Cai H, Rodriguez BT,, Zhang W., Broadbent JR, Steele JL. 2007. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology 153:2655–2665. [DOI] [PubMed] [Google Scholar]
- Cai H, Thompson R,, Budinich MF,, Broadbent JR,, Steele JL. 2009. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol. 1:239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callanan M, et al. 2008. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J Bacteriol. 190(2):727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington LB, Lipkowitz JR, Hoffmann AA,, Turelli M. 2011. A Re-examination of Wolbachia-induced cytoplasmic incompatibility in California Drosophila simulans. PLoS One 6(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiraghi M, et al. 2002. Tetracycline treatment and sex-ratio distortion: a role for Wolbachia in the moulting of filarial nematodes? Int J Parasitol. 32(12):1457–1468. [DOI] [PubMed] [Google Scholar]
- Cerveau N, Leclercq S, Leroy E, Bouchon D, Cordaux R. 2011. Short- and long-term evolutionary dynamics of bacterial insertion sequences: insights from Wolbachia endosymbionts. Genome Biol Evol. 3:1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chafee ME, Funk DJ, Harrison RG, Bordenstein SR. 2010. Lateral phage transfer in obligate intracellular bacteria (Wolbachia): verification from natural populations. Mol Biol Evol. 27(3):501–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain PSG, et al. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 101(38):13826–13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles H, Calevro F, Vinuelas J, Fayard JM, Rahbe Y. 2006. Codon usage bias and tRNA over-expression in Buchnera aphidicola after aromatic amino acid nutritional stress on its host Acyrthosiphon pisum. Nucleic Acids Res. 34(16):4583–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MA, Moran NA, Baumann P. 1999. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol Biol Evol. 16(11):1586–1598. [DOI] [PubMed] [Google Scholar]
- Comandatore F, et al. 2013. Phylogenomics and analysis of shared genes suggest a single transition to mutualism in Wolbachia of nematodes. Genome Biol Evol. 5(9):1668–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R. 2008. ISWpi1 from Wolbachia pipientis defines a novel group of insertion sequences within the IS5 family. Gene 409(1-2):20–27. [DOI] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe KS, et al. 2005. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 187(22):7716–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debora BN, et al. 2011. Mismatch repair modulation of MutY activity drives Bacillus subtilis stationary-phase mutagenesis. J Bacteriol. 193(1):236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeine F, et al. 2001. Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc Natl Acad Sci U S A. 98(11):6247–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegaard KM, Klasson L, Andersson SG. 2013. Testing the reproducibility of multiple displacement amplification on genomes of clonal endosymbiont populations. PLoS One 8(11):e82319.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegaard KM, Klasson L, Naslund K, Bourtzis K, Andersson SG. 2013. Comparative genomics of Wolbachia and the bacterial species concept. PLoS Genet. 9(4):e1003381.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensminger AW, Yassin Y, Miron A, Isberg RR. 2012. Experimental evolution of Legionella pneumophila in mouse macrophages leads to strains with altered determinants of environmental survival. PLoS Pathog. 8(5):e1002731.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem ST, Gaal T, Ross W, Gourse RL. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci U S A. 95(17):9761–9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J, et al. 2005. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 3(4):e121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galens K, et al. 2011. The IGS standard operating procedure for automated prokaryotic annotation. Stand Genomic Sci. 4(2):244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Gautier C. 1982. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 10(22):7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Li WH. 2000. Fundamentals of molecular evolution. Sunderland, (MA: ): Sinauer Associates. [Google Scholar]
- Grilley M, Holmes J, Yashar B, Modrich P. 1990. Mechanisms of DNA-mismatch correction. Mutat Res. 236(2-3):253–267. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. 2007. Principles of population genetics. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Hebert PDN, Cywinska A,, Ball SL, DeWaard JR. 2003. Biological identifications through DNA barcodes. Proc R Soc B Biol Sci. 270(1512):313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. 2008. Wolbachia and virus protection in insects. Science 322(5902):702.. [DOI] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6(9):e1001115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F, Meyer A, Eyre-Walker A. 2010. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 6(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerauf A, et al. 2008. Wolbachia endobacteria depletion by doxycycline as antifilarial therapy has macrofilaricidal activity in onchocerciasis: a randomized placebo-controlled study. Med Microbiol Immunol. 197(3):295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst JP, Wu TH, Marinus MG. 1999. Escherichia coli mutator genes. Trends Microbiol. 7(1):29–36. [DOI] [PubMed] [Google Scholar]
- Hosokawa T, Koga R, Kikuchi Y, Meng XY, Fukatsu T. 2010. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci U S A. 107(2):769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotopp JCD, et al. 2007. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317(5845): 1753–1756. [DOI] [PubMed] [Google Scholar]
- Ikemura T. 1985. Codon usage and transfer-RNA content in unicellular and multicellular organisms. Mol Biol Evol. 2(1):13–34. [DOI] [PubMed] [Google Scholar]
- Ishmael N, et al. 2009. Extensive genomic diversity of closely related Wolbachia strains. Microbiology 155(Pt 7):2211–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, Riegler M, O'Neill SL. 2005. New names for old strains? Wolbachia wSim is actually wRi. Genome Biol. 6(7):401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, von Der Schulenburg JH, Hurst GD, Majerus ME. 2001. Recombination confounds interpretations of Wolbachia evolution. Proc R Soc B Biol Sci. 268(1474):1423–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karberg KA, Olsen GJ, Davis JJ. 2011. Similarity of genes horizontally acquired by Escherichia coli and Salmonella enterica is evidence of a supraspecies pangenome. Proc Natl Acad Sci U S A. 108(50): 20154–20159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent BN, Bordenstein SR. 2010. Phage WO of Wolbachia: lambda of the endosymbiont world. Trends Microbiol. 18(4):173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent BN, Funkhouser LJ,, Setia S,, Bordenstein SR. 2011. Evolutionary genomics of a temperate bacteriophage in an obligate intracellular bacteria (Wolbachia). PLoS One 6(9):e24984.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent BN, Salichos L, et al. 2011. Complete bacteriophage transfer in a bacterial endosymbiont (Wolbachia) determined by targeted genome capture. Genome Biol Evol. 3:209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, et al. 2008. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol. 25(9):1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, et al. 2009. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci U S A. 106(14):5725–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, Kambris Z, Cook PE, Walker T, Sinkins SP. 2009. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehncke A, Telschow A, Werren JH, Hammerstein P. 2009. Life and death of an influential passenger: Wolbachia and the evolution of CI-modifiers by their hosts. PLoS One 4(2). doi: 10.1371/journal.pone.0004425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, et al. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Giraud I, Cordaux R. 2011. Remarkable abundance and evolution of mobile group II introns in Wolbachia bacterial endosymbionts. Mol Biol Evol. 28(1):685–697. [DOI] [PubMed] [Google Scholar]
- Lee H, Popodi E, Tang HX, Foster PL. 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc Natl Acad Sci U S A. 109(41):E2774–E2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePage D, Bordenstein SR. 2013. Wolbachia: can we save lives with a great pandemic?. Trends Parasitol. 29(8):385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Lu CL, Liu YC, Tang CY. 2006. SPRING: a tool for the analysis of genome rearrangement using reversals and block-interchanges. Nucleic Acids Res. 34:W696–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehlin DW, et al. 2010. Non-coding changes cause sex-specific wing size differences between closely related species of Nasonia. PLoS Genet. 6(1). doi: 10.1371/journal.pgen.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 1996. Mutation accumulation in transfer RNAs: molecular evidence for Muller's Ratchet in mitochondrial genomes. Mol Biol Evol. 13(1):209–220. [DOI] [PubMed] [Google Scholar]
- Lynch M. 1997. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol Biol Evol. 14(9):914–925. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV. 2007. Evolutionary genomics of lactic acid bacteria. J Bacteriol. 189(4):1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Sasaki T, Ishikawa H. 2000. Genes for the type IV secretion system in an intracellular symbiont, Wolbachia, a causative agent of various sexual alterations in arthropods. J Bacteriol. 182(22):6529–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod MP, et al. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol. 186(17):5842–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf JA, Jo M, Bordenstein SR, Jaenike J, Bordenstein SR. 2014. Recent genome reduction of Wolbachia in Drosophila recens targets phage WO and narrows candidates for reproductive parasitism. Peer J. 2:e529.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels ML, Cruz C, Grollman AP, Miller JH. 1992. Evidence that Muty and Mutm combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci U S A. 89(15):7022–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17(10):589–596. [DOI] [PubMed] [Google Scholar]
- Modrich P. 1987. DNA mismatch correction. Annu Rev Biochem. 56:435–466. [DOI] [PubMed] [Google Scholar]
- Modrich P. 1991. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 25:229–253. [DOI] [PubMed] [Google Scholar]
- Moore RA, et al. 2004. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect Immun. 72(7):4172–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 93(7):2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc Lond B Biol Sci. 253:167–171. [Google Scholar]
- Moran NA, Plague GR. 2004. Genomic changes following host restriction in bacteria. Curr Opin Genet Dev. 14(6):627–633. [DOI] [PubMed] [Google Scholar]
- Moreira LA, et al. 2009. A Wolbachia symbiont in Aedes aegypti limits infection with Dengue, Chikungunya, and Plasmodium. Cell 139(7):1268–1278. [DOI] [PubMed] [Google Scholar]
- Newton IL, Bordenstein SR. 2011. Correlations between bacterial ecology and mobile DNA. Curr Microbiol. 62(1):198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, et al. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2(5):e76.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. 1973. Slightly deleterious mutant substitutions in evolution. Nature 246(5428):96–98. [DOI] [PubMed] [Google Scholar]
- Olson MV. 1999. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 64(1):18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden J. 2000. CodonW. [cited 2014 November]. Available from: http://codonw.sourceforge.net/.
- Penz T, et al. 2012. Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 8(10). doi: 10.1371/journal.pgen.1003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiler EA, Klaenhammer TR. 2007. The genomics of lactic acid bacteria. Trends Microbiol. 15(12):546–553. [DOI] [PubMed] [Google Scholar]
- Rances E, Voronin D, Tran-Van V, Mavingui P. 2008. Genetic and functional characterization of the type IV secretion system in Wolbachia. J Bacteriol. 190(14):5020–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychoudhury R, Baldo L, Oliveira DC, Werren JH. 2009. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution 63(1): 165–183. [DOI] [PubMed] [Google Scholar]
- Rocha EPC, Danchin A. 2002. Base composition bias might result from competition for metabolic resources. Trends Genet. 18(6): 291–294. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, et al. 2005. Serendipitous discovery of Wolbachia genomes in multiple Drosophila species (vol 6, art no R23, 2005). Genome Biol. 6(3):R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Gilchrist MA. 2010. Effect of correlated tRNA abundances on translation errors and evolution of codon usage bias. PLoS Genet. 6(9). doi: 10.1371/journal.pgen.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. 1987a. The codon Adaptation Index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15(3):1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li WH. 1987b. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 4(3):222–230. [DOI] [PubMed] [Google Scholar]
- Simon D, Larget B. 2004. Bayesian analysis to describe genomic evolution by rearrangement (BADGER), version 1.02 beta, Duquesne University. http://www.badger.duq.edu/.
- Siozios S, et al. 2013. The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS One 8(2):e55390.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DJ, Cline TW. 2002. A host parasite interaction rescues Drosophila oogenesis defects. Nature 418(6893):76–79. [DOI] [PubMed] [Google Scholar]
- Stouthamer R, Breeuwert JA, Luck RF, Werren JH. 1993. Molecular identification of microorganisms associated with parthenogenesis. Nature 361(6407):66–68. [DOI] [PubMed] [Google Scholar]
- Sun LV, et al. 2001. Determination of Wolbachia genome size by pulsed-field gel electrophoresis. J Bacteriol. 183(7):2219–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YC, Hinnebusch BJ, Darby C. 2008. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc Natl Acad Sci U S A. 105(23):8097–8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf AM, Lazdins J. 2000. Wolbachia bacteria of filarial nematodes: a target for control? Parasitol Today. 16(5): 179–180. [DOI] [PubMed] [Google Scholar]
- Turley AP, Moreira LA, O'Neill SL, McGraw EA. 2009. Wolbachia infection reduces blood-feeding success in the Dengue fever mosquito, Aedes aegypti. PLoS Negl Trop Dis. 3(9): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leuven JT, McCutcheon JP. 2012. An AT mutational bias in the tiny GC-rich endosymbiont genome of Hodgkinia. Genome Biol Evol. 4(1):24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16s Ribosomal DNA amplification for phylogenetic study. J Bacteriol. 173(2):697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ, Moran NA. 1999. Evidence for genetic drift in endosymbionts (Buchnera): analyses of protein-coding genes. Mol Biol Evol. 16(1):83–97. [DOI] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 6(10):741–751. [DOI] [PubMed] [Google Scholar]
- Werren JH, Bartos JD. 2001. Recombination in Wolbachia. Curr Biol. 11(6):431–435. [DOI] [PubMed] [Google Scholar]
- Werren JH, Windsor D, Guo LR. 1995. Distribution of Wolbachia among neotropical arthropods. Proc R Soc B Biol Sci. 262(1364): 197–204. [Google Scholar]
- Werren JH, Zhang W, Guo LR. 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc R Soc B Biol Sci. 261(1360):55–63. [DOI] [PubMed] [Google Scholar]
- Wu M, et al. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2(3):327–341. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J. 2008. Positive selection, not negative selection, in the pseudogenization of rcsA in Yersinia pestis. Proc Natl Acad Sci U S A. 105(42):E69; author reply E70.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.