Abstract
Mutations in spliceosomal genes are commonly found in patients with myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML)1–3. These mutations occur at highly recurrent amino acid residues and perturb normal splice site and exon recognition4–6. Spliceosomal mutations are always heterozygous and rarely co-occur with one another, suggesting that cells may only tolerate a partial deviation from normal splicing activity. To test this hypothesis, we engineered mice that express the SRSF2P95H mutation, which commonly occurs in MDS and AML, in an inducible hemizygous manner in hematopoietic cells. These mice developed lethal bone marrow failure, demonstrating that Srsf2-mutant cells depend on the wildtype Srsf2 allele for survival. In the context of leukemia, treatment with the spliceosome inhibitor E71077,8 resulted in significant reductions in leukemic burden specifically in isogenic mouse leukemias and patient-derived xenograft (PDX) AMLs carrying spliceosomal mutations. While in vivo E7107 exposure resulted in widespread intron retention and cassette exon skipping regardless of Srsf2 genotype, the magnitude of splicing inhibition following E7107 treatment was greater in Srsf2-mutant versus wildtype leukemias, consistent with its differential effect on survival in these two genotypes. Collectively, these data provide genetic and pharmacologic evidence that leukemias with spliceosomal mutations are preferentially susceptible to additional splicing perturbations in vivo compared with wildtype counterparts. Modulation of spliceosome function may provide a novel therapeutic avenue in genetically defined subsets of MDS and AML patients.
Mutations in the spliceosomal genes SRSF2, U2AF1 and SF3B1 are the most common class of mutations in patients with MDS1–3 and occur across the entire spectrum of myeloid malignancy, including 10–25% of patients with AML and a higher proportion of patients with AML transformed from an antecedent MDS9. Recent studies revealed that heterozygous mutations in SRSF25 as well as U2AF14 drive hematopoietic stem/progenitor cell (HSPC) expansion in mice in vivo and that these mutations alter mRNA recognition in a sequence-specific manner6. However, it is still unclear why spliceosomal gene mutations occur in an exclusively heterozygous state in myeloid malignancies, and why these mutations are mutually exclusive with one another. Moreover, given the frequency of these mutations and their early occurrence in myeloid malignancies10–12, strategies to therapeutically target spliceosome-mutant malignancies are urgently needed.
We first took a genetic approach to test the hypothesis that cells carrying spliceosomal mutations are sensitive to further perturbation of normal splicing. We engineered mice that express hemizygous Srsf2P95H mutant alleles in an inducible manner (upon polyI:C injection) in the hematopoietic system. These mice enabled study of the effects of wildtype Srsf2 deletion with concomitant activation of the Srsf2P95H allele. Mx1-Cre+Srsf2+/fl mice were crossed to Srsf2P95H/+ mice to generate Srsf2 wildtype (Mx1-Cre+Srsf2+/+), Srsf2 heterozygous knockout (Mx1-Cre+Srsf2+/fl), Srsf2 heterozygous P95H mutant (Mx1-Cre+Srsf2P95H/+), and Srsf2 hemizygous P95H mutant (Mx1-Cre+Srsf2P95H/fl) mice (Supplementary Fig. 1a). In non-competitive bone marrow (BM) transplantation assays, recipient mice reconstituted with hemizygous Mx1-Cre+Srsf2P95H/− BM mononuclear cells (MNCs) showed significantly shorter survival (p=0.004) and severe BM failure (Fig. 1a–c) due to loss of HSPCs (Supplementary Fig. 1b–f). Conversely, the loss of HSPCs and BM aplasia were not seen in Mx1-Cre+Srsf2P95H/+ or Mx1-Cre+Srsf2+/fl mice.
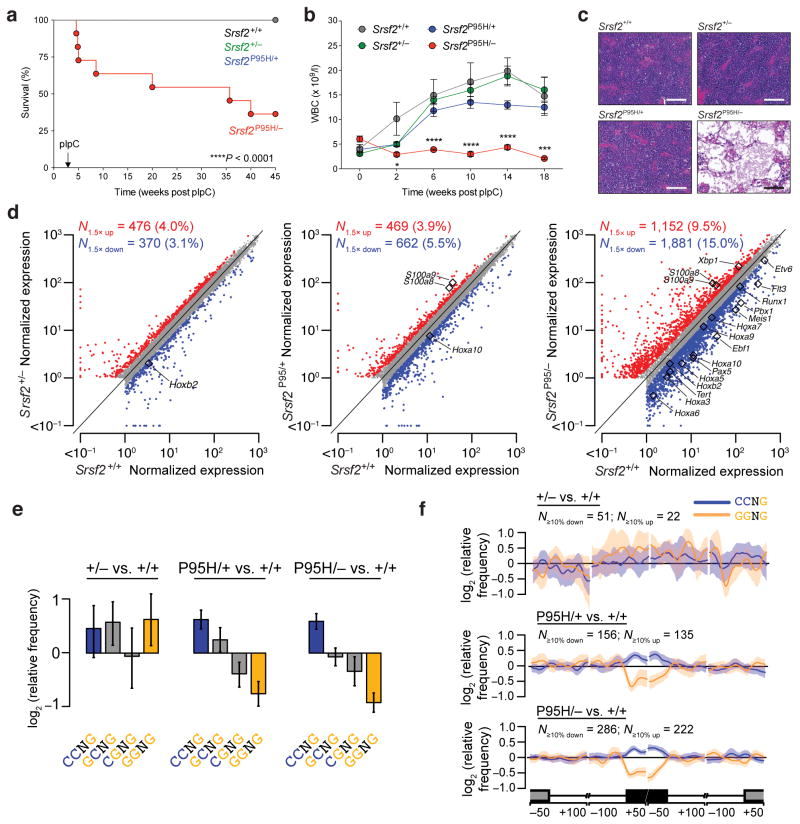
Figure 1. Spliceosome-mutant cells require the wildtype allele for survival.
(a) Kaplan-Meier survival curve of CD45.1 recipient mice transplanted with control (Mx1-Cre+Srsf2+/+), heterozygous knockout (Mx1-Cre+Srsf2+/−), heterozygous mutant (Mx1-Cre+Srsf2P95H/+), and hemizygous mutant (Mx1-Cre+Srsf2P95H/−) bone marrow (BM) cells. Administration of polyI:C (pIpC) was performed 4 weeks post-transplantation. Survival comparison by Mantel-Cox log-ranked test. (b) White blood cell (WBC) count of mice of each genotype over 18 weeks of noncompetitive transplantation (n = 10 mice per group for experiments in (a) and (b)). Error bars represent mean ± SD. ***P < 0.001; ****P < 0.0001. (c) H&E staining of BM of CD45.1 recipient mice 8 weeks post-transplantation (scale bars, 200 μm). (d) Scatter plots comparing normalized expression of individual genes in BM lineage-negative Sca1+ c-Kit+ (LSK) cells in heterozygous knockout (left), heterozygous mutant (middle), and hemizygous mutant (right) mice relative to wildtype control cells. Genes that are significantly dysregulated between comparisons (Bayes factor > 5; fold change > 1.5) are labeled in red (up-regulated) and blue (down-regulated), respectively. Differentially expressed genes of particular biological importance are highlighted in each plot. Units are transcripts per million. (e) Mean enrichment of all variants of the SSNG exonic splicing enhancer (ESE) motif in cassette exons that were differentially included versus excluded in LSK cells with Srsf2 heterozygous loss (+/−), heterozygous mutation (P95H/+), or hemizygous mutation (P95/−) relative to Srsf2 control (+/+). Error bars indicate 95% confidence intervals estimated by bootstrapping. (f) Spatial distribution of CCNG and GGNG motifs adjacent to the sets of differentially spliced cassette exons analyzed in (e). N≥10% down/up indicates the number of exons that exhibited decreases/increases in inclusion of absolute magnitude ≥10% in cells with the indicated genotypes relative to Srsf2 control (+/+) cells. Shading indicates 95% confidence intervals by bootstrapping. The schematic illustrates a portion of a metagene with the cassette exon (black box) separated from the upstream and downstream exons (grey boxes) by the beginning and end of the connecting introns. Horizontal axis, genomic coordinates defined with respect to the 5′ and 3′ splice sites where position 0 is the splice site itself. Vertical axis, relative frequency of the indicated motifs over genomic loci containing cassette exons included versus excluded in the indicated genotype comparisons (log scale).
To determine the effect of hemizygous expression of mutant Srsf2 on the transcriptome, we performed RNA-seq on HSPCs (CD45.2+ lineage− Sca1+ c-Kit+ (LSK) cells) isolated two weeks after polyI:C injection from mice reconstituted with Mx1-Cre+Srsf2+/+, Mx1-Cre+Srsf2+/fl, Mx1-Cre+Srsf2P95H/+, or Mx1-Cre+Srsf2P95H/− BM cells. We observed >3,000 dysregulated genes in Srsf2P95H/− HSPCs relative to all other groups, including >1.5-fold repression of many genes involved in hematopoietic self-renewal, including Runx1, Erg, and the entire HoxA cluster (Fig. 1d and Supplementary Table 1). Gene Ontology (GO) analysis of the differentially expressed genes in HSPCs from all four groups revealed that pathways related to cell migration, chemotaxis, cytokine production and inflammatory responses were significantly overexpressed in Mx1-Cre+Srsf2P95H/− relative to Mx1-Cre+Srsf2P95H/+ HSPCs (Supplementary Fig. 1g). We next tested whether hemizygous expression of Srsf2P95H was associated with substantive alterations in splicing. We quantified differential splicing of ~44,000 annotated alternative splicing events and ~170,000 constitutive splice junctions using Bayesian statistical methods (the MISO algorithm13 and Wagenmakers’s framework14). We observed differential splicing of all classes of alternative splicing events, including competing 5′ and 3′ splice sites, cassette exons, and retained introns in Mx1-Cre+Srsf2P95H/− cells, as well as alternative splicing and intron retention affecting normally constitutively spliced junctions. Although most splicing events remained unchanged, Mx1-Cre+Srsf2P95H/− cells exhibited approximately two-fold more mis-splicing across all types of splicing events than did Mx1-Cre+Srsf2P95H/+ cells (Supplementary Tables 1–3).
We next tested whether increased mis-splicing in Mx1-Cre+Srsf2P95H/− cells was due to altered exon recognition. Heterozygous expression of SRSF2P95H/L/R mutations alters SRSF2 recognition of specific exonic splicing enhancer (ESE) motifs and drives recurrent mis-splicing of key hematopoietic regulators5,15. Quantification of the occurrence of ESE motifs within cassette exons that were differentially spliced in each genotype of LSK cells revealed a statistically significant preference for CCNG over GGNG ESE motifs in Mx1-Cre+Srsf2P95H/+ and Mx1-Cre+Srsf2P95H/− cells (Fig. 1e), consistent with previous reports. CCNG motifs were enriched and GGNG motifs were depleted within differentially spliced cassette exons and not within the flanking introns or exons (Fig. 1f).
To determine if the cell lethality seen with Srsf2P95H/− hemizygosity was also present in the setting of leukemogenesis, we transduced Srsf2P95H/fl (control) or Mx1-Cre+Srsf2P95H/fl (hemizygous) fetal liver cells with a retrovirus encoding the MLL-AF9 fusion oncogene (in an MSCV-IRES-GFP vector), and then transplanted them into lethally irradiated recipient mice (Supplementary Fig. 2a). While Srsf2P95H/fl control mice rapidly developed leukocytosis, anemia, thrombocytopenia and elevated levels of donor-derived GFP positive cells, these features were substantially diminished in the hemizygous Srsf2P95H/− background (Supplementary Fig. 2b–g). Consistent with this observation, all mice from the control group eventually developed leukemia, while only 5 out of 10 Srsf2P95H/− mice succumbed to disease (Supplementary Fig. 2h). Moreover, the leukemic cells in Srsf2P95H/− recipients all uniformly escaped polyI:C-mediated recombination of the wildtype Srsf2 allele compared to blood samples taken from the same animals in the pre-leukemic state (Supplementary Fig. 2i). Overall, these observations revealed that Srsf2P95H/+ cells specifically depend upon the presence of the wildtype Srsf2 allele for survival, even in the presence of a potent oncogene. These findings are consistent with the fact that mutations in genes encoding SRSF2 and other spliceosomal proteins are always heterozygous in MDS/AML patients and provide a potential explanation for the consistent heterozygous nature of spliceosomal gene mutations in cancer.
Having established that spliceosome-mutant cells depend on wildtype splicing function using mouse genetic models, we hypothesized that spliceosome-mutant hematopoietic cells might display an altered response to pharmacologic inhibition of pre-mRNA splicing relative to their wildtype counterparts. To test this, we treated recipient mice with the splicing inhibitor E71077,8. We first generated BM chimeras by transplantation of Srsf2+/+ and Srsf2P95H/+ BM MNCs into lethally irradiated recipient mice, followed by E7107 or vehicle treatment in vivo starting at 6 months post-transplant (a time point at which stable engraftment of long-term hematopoiesis is expected) (Supplementary Fig. 2j). After five daily treatments of vehicle or E7107, we purified HSPCs (CD45.2+ lineage− Sca1− c-Kit+ cells) by flow cytometry and analyzed splicing and gene expression by RNA-seq. Ordination analysis by multidimensional scaling based on global cassette exon inclusion and global gene expression revealed that all of the vehicle-treated samples clustered together irrespective of Srsf2 genotype, while the E7107-treated samples clustered based on Srsf2 genotype (Fig. 2a). These results indicated a differential gene expression and splicing response to E7107 in Srsf2P95H/+ hematopoietic cells compared to their Srsf2+/+ counterparts. We next examined whether this differential response to E7107 was due to the previously described altered ESE motif preference by mutant Srsf2. To do so, we performed a “two-factor” analysis across two experimental parameters (vehicle versus E7107, and Srsf2+/+ versus Srsf2P95H/+), in which we systematically tested for preferential recognition of exons with different variants of the core SSNG (S = C or G) motif between each comparison (Fig. 2b). This analysis revealed that exons that are differentially spliced between the vehicle-treated Srsf2+/+ and Srsf2P95H/+ samples showed the expected difference in ESE motif preference (Fig. 2b, left) as previously published5. In contrast, there was no motif enrichment associated with the exons that are affected by E7107 in either Srsf2+/+ or Srsf2P95H/+ cells (Fig. 2b, top and bottom). However, we did observe that preferential recognition of exons with CCNG versus GGNG motifs in Srsf2P95H/+ vs. Srsf2+/+ cells was weaker following E7107 treatment, and we identified a small subset of exons whose inclusion was affected in a genotype-dependent manner (Fig. 2b, right and Supplementary Table 3).
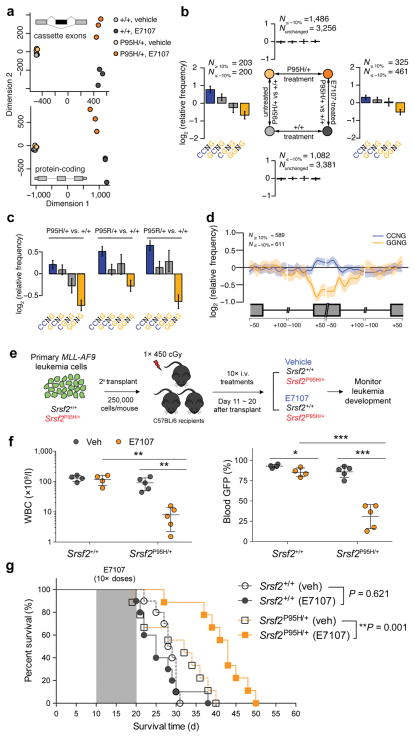
Figure 2. SRSF2-mutant myeloid leukemias are preferentially sensitive to pharmacologic modulation of splicing catalysis.
(a) Multidimensional scaling based on alternatively spliced cassette exons (top) and protein-coding genes (bottom) in BM lineage-negative Sca1− c-Kit+ (LK) cells from Mx1-Cre+Srsf2P95H/+ and Mx1-Cre+Srsf2+/+ mice treated with vehicle or E7107 (4 mg/kg) for 5 days (n = 3 mice per group). (b) Mean enrichment of all variants of the SSNG exonic splicing enhancer (ESE) motif in cassette exons that were differentially included versus excluded in a two-factor comparison across all 4 experimental groups. N≥10% and N≤ −10% indicate the numbers of exons that exhibited increases and decreases in inclusion, respectively, of absolute magnitude ≥10% for the illustrated comparisons. For each comparison, enrichment was computed by comparing the indicated sets of exons (e.g., N≥10% versus N≤−10% for the left- and right-hand comparisons). For sample comparisons where differentially spliced exons exhibiting increased inclusion were not present (top and bottom), cassette exons that exhibited differential splicing of absolute magnitude ≤10% were used as a background set to compute motif enrichment instead. The individual comparison used to generate each set of bar graphs is shown in the schema in the center of the figure. Error bars indicate 95% confidence intervals estimated by bootstrapping. (c) Mean enrichment of all variants of the SSNG ESE motif in individual SRSF2-mutant MLL-rearranged human AML samples relative to the median of 28 SRSF2-wildtype MLL-rearranged AML samples. Error bars indicate 95% confidence intervals estimated by bootstrapping. (d) Spatial distribution of CCNG and GGNG motifs along cassette exons included or excluded in the SRSF2P95H-mutant samples relative to the median wildtype control amongst MLL-rearranged AML samples. (e) Schematic of secondary transplantation experiments of mouse MLL-AF9 leukemias in Srsf2+/+ (Vav-Cre+Srsf2+/+) and Srsf2P95H/+ (Vav-Cre+Srsf2P95H/+) backgrounds to test the effects of E7107 in vivo. (f) WBC counts (left) and percentages of WBC cells expressing GFP (right) following 10 days of E7107 administration. (g) Kaplan-Meier survival curves of recipient mice treated with vehicle or E7107 (at 4 mg/kg) in Srsf2+/+ or Srsf2P95H/+ backgrounds. Shaded area represents period of vehicle or E7107 dosing. Error bars represent mean ± SD. *P < 0.05; **P < 0.005; ***P < 0.001.
Based on these observations, we hypothesized that spliceosome-mutant leukemias might display greater sensitivity to pharmacologic inhibition of splicing than wildtype leukemias. Recent work16 identified SRSF2 mutations in ~10% of adult MLL-rearranged AMLs, suggesting that MLL-rearranged leukemias constitute a relevant system to study SRSF2 mutations. By reanalyzing RNA-seq data16 from human subjects with MLL-rearranged AML, we observed global alterations in splicing and ESE motif preference in SRSF2-mutant MLL-rearranged AML transcriptomes that were similar to what we previously reported in SRSF2-mutant mouse MDS models and myeloid leukemia patients5 (Fig. 2c–d), suggesting that SRSF2 mutations alter exon recognition in MLL-rearranged AML as expected. We therefore created an isogenic mouse leukemia model by retroviral overexpression of the MLL-AF9 fusion oncogene in Vav-Cre+Srsf2+/+ or Vav-Cre+Srsf2P95H/+ BM cells followed by transplantation into lethally irradiated recipient mice (Supplementary Fig. 3a). Overexpression of MLL-AF9 in Vav-Cre+Srsf2+/+ or Vav-Cre+Srsf2P95H/+ BM cells resulted in fully penetrant AML with similar survival latencies and marked splenomegaly and hepatomegaly (Supplementary Fig. 3b–c). Although Srsf2-mutant leukemias exhibited altered gene expression related to processes such as cell migration and response to external stimuli relative to their wildtype counterparts (Supplemental Fig. 3d), immunophenotype or histological and cytological analyses of BM, spleen, and liver revealed no obvious differences between the two genotypes (Supplementary Fig. 3e–g). We next examined the effects of pharmacological spliceosomal inhibition in vivo. To accomplish this, equal numbers of primary MLL-AF9 leukemic cells from Srsf2+/+ or Srsf2P95H/+ mice were transplanted into secondary recipient mice to generate secondary leukemias, and secondary recipient mice were then treated with either E7107 or vehicle (Fig. 2e). Ten days of intravenous administration of E7107 at 4mg/kg/day resulted in decreased disease burden as assessed by peripheral blood leukocyte count and GFP percentage (Fig. 2f), histological analyses (Supplementary Fig. 3h) and survival benefit in Srsf2P95H/+ mice (p=0.001), whereas the same treatment regimen had no impact on overall survival of Srsf2+/+ mice (p=0.621) (Fig. 2g). E7107 also improved anemia and thrombocytopenia on both Srsf2+/+ and Srsf2P95H/+ backgrounds, with a slightly greater improvement in Srsf2P95H/+ mice (Supplementary Fig. 3i–j).
To determine the mechanistic origins of the Srsf2 mutant-selective effects of E7107, we analyzed transcriptional changes after five days of E7107 treatment in vivo (Fig. 3a). GFP+ Mac1+ cells were purified from the BM of recipient mice exactly three hours after the fifth dose of E7107 and were subjected to RNA-seq. E7107 exposure resulted in global splicing inhibition in both genotypes, typified by widespread intron retention and cassette exon skipping expected of inefficient splicing catalysis (Supplementary Fig. 4a and Fig. 3b–d, top and middle rows). While E7107-induced splicing dysregulation was highly variable across animals in both Srsf2-mutant and Srsf2-wildtype backgrounds, effects on global protein coding gene expression were highly distinct between genotypes (Fig. 3b–d, bottom panel, Supplementary Fig. 4b–c and Supplementary Table 1). Moreover, the magnitude of splicing inhibition following E7107 treatment was more severe in Srsf2P95H/+ versus Srsf2+/+ mice, consistent with its differential effect on survival in these two genotypes (Supplementary Fig. 4d). GO analysis revealed that differentially expressed genes in Srsf2P95H/+ relative to Srsf2+/+ leukemic cells after E7107 treatment were enriched in biological pathways related to cytokine and immune signaling and leukocyte activation and migration (Supplementary Fig. 4e).
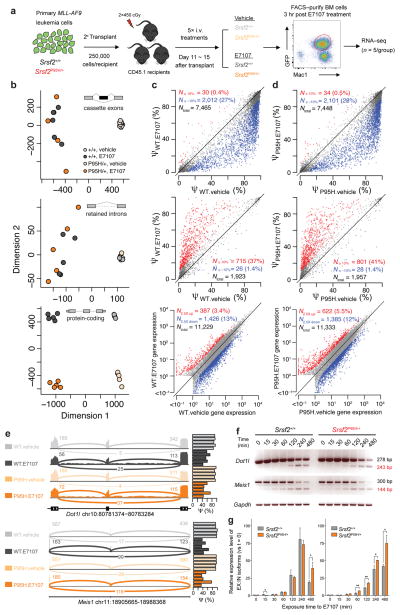
Figure 3. Splicing and gene expression changes in myeloid leukemias treated with E7107 in SRSF2-wildtype or mutant backgrounds.
(a) Schematic of secondary transplantation experimentation with timed sacrifice following E7107 for RNA-seq analyses. Sub-lethally irradiated mice were transplanted with MLL-AF9;Srsf2+/+ or MLL-AF9;Srsf2P95H/+ primary leukemias followed by 5 days of E7107 (4 mg/kg/d) or vehicle treatment. GFP+ Mac1+ double-positive BM cells were then sorted 3 hours after the fifth treatment for RNA-seq (n = 5 mice per group). (b) Multidimensional scaling analysis of all 20 mice from (a) based on alternatively spliced cassette exons (top, N = 6,369), alternatively spliced retained introns (middle, N = 1,791), and expressed protein-coding genes (bottom, N = 9,339). Scatter plots of cassette exon splicing (top rows, isoform ratios represented as Percent Spliced In (PSI, Ψ) values), retained introns (middle rows, Ψ values), and gene expression (bottom rows, normalized expression values) from (c) MLL-AF9;Srsf2+/+ or (d) MLL-AF9;Srsf2P95H/+ mice treated with vehicle or E7107. Percentages indicate the fraction of differentially spliced cassette exons or retained introns (inclusion rates increased or decreased by absolute magnitude ≥ 10% with P < 0.01), or number of differentially expressed genes (fold change ≥ 2.5 with P < 0.01). Red and blue dots represent individual splicing events or coding genes that are promoted or repressed in E7107 versus vehicle-treated cells, respectively. (e) Sashimi plots28,29 across splice junctions surrounding differentially spliced cassette exons in Dot1l and Meis1, with reads summarized across five replicate mice. Barplots represent the percent spliced (Ψ) cassette exon inclusion ratios across all samples. (f) RT-PCR analysis of the effect of acute exposure to E7107 (10 nM) on splicing of Dot1l and Meis1 in MLL-AF9;Srsf2+/+ and MLL-AF9;Srsf2P95H/+ leukemia cells in vitro. (g) Quantitative RT-PCR (qRT-PCR) analysis quantifying the relative levels of exclusion (EX) and inclusion (IN) isoforms of Dot1l and Meis1 following acute exposure to E7107 (10 nM) in vitro. Error bars represent mean ± SD. *P < 0.05; **P < 0.01.
Interestingly, genes involved in maintaining the leukemogenic programs in MLL-rearranged leukemias, including Dot1l and Meis1, were among the most differentially spliced genes in Srsf2P95H/+ mice in response to E7107 (Fig. 3e and Supplementary Table 3). Given the known importance of DOT1L to MLL-mediated leukemogenesis17,18, we investigated the effects of E7107 on Dot1l splicing further. E7107 resulted in more pronounced exon skipping and intron retention within a region encoding the catalytic domain of Dot1l protein in MLL-AF9/Srsf2P95H/+ cells relative to their MLL-AF9/Srsf2+/+ counterparts, which was readily detectable by qRT-PCR during a time course of drug exposure (Fig. 3f–g). A similar increase in cassette exon skipping within Meis1 was also observed by qRT-PCR in MLL-AF9/Srsf2P95H/+ cells compared to MLL-AF9/Srsf2+/+ cells (Fig 3f–g). This increased mis-splicing in Dot1l and Meis1 in the Srsf2-mutant background post-E7107 exposure correlated with a mild decrease in Meis1 protein and Dot1l-mediated histone H3 lysine 79 di-methylation (H3K79me2), indicating a reduction in Dot1l catalytic activity (Supplementary Fig. 4f–g). Conversely, exogenous re-expression of DOT1L cDNA resulted in a mild but consistent restoration in cellular proliferation in MLL-AF9/Srsf2P95H/+ cells relative to MLL-AF9/Srsf2+/+ cells when exposed to E7107 (Supplementary Fig. 5a–b). In contrast, Meis1 overexpression was unable to rescue the inhibition of cell proliferation induced by E7107 (Supplementary Fig. 5c–d). Interestingly, stable introduction of a previously identified SF3B1 point mutation known to cause E7107 resistance (SF3B1R1074H)19 in MLL-AF9/Srsf2P95H/+ cells (Supplementary Fig. 5e–g) rendered these cells almost completely insensitive to E7107, with an IC50 nearly 300-fold greater than parental or SF3B1WT-overexpressing cells (Supplementary Fig. 5h). SF3B1R1074H-expressing cells were also impervious to E7107-mediated inhibition of splicing (Supplementary Fig. 5h–i). Collectively, our data suggest that E7107 induces phenotypic changes in MLL-AF9 leukemia through aberrant splicing of multiple downstream targets including Dot1l and Meis1, and provides genetic confirmation that E7107 affects cells through on-target inhibition of SF3B1.
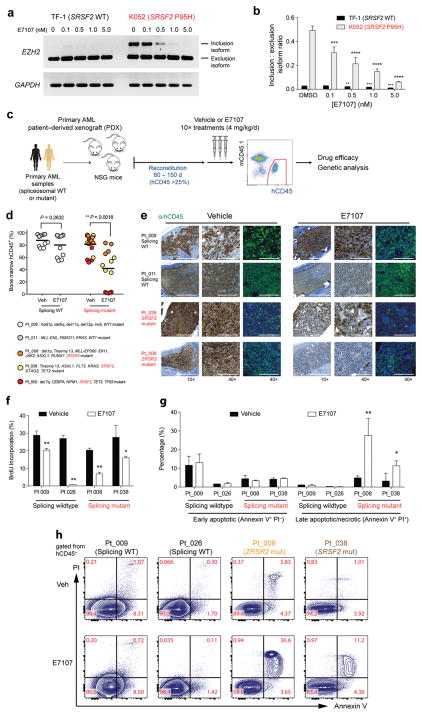
Given that the above data were generated in the specific context of MLL-rearranged mouse leukemias, we next sought to analyze the effect of splicing inhibition in the context of human AMLs with endogenous SRSF2 mutations co-occurring with a spectrum of different leukemia disease alleles. First, we tested the effect of E7107 on a previously described cassette exon inclusion event in EZH25 in SRSF2-wildtype (TF-1) and mutant (K052) human leukemia cell lines. qRT-PCR analyses revealed that E7107 inhibits SRSF2 mutant-specific EZH2 mis-splicing in a dose-dependent fashion (Fig. 4a–b). Next, to evaluate in vivo drug effects in primary human AMLs, we generated patient-derived xenografts (PDXs) from a cohort of primary AML patients (n=2 without a spliceosomal mutation, n=3 with a spliceosomal mutation; Supplementary Table 4). Primary leukemia cells from each individual patient were transplanted via tail-vein injection into 10 adult NOD-scid IL2rnull (NSG) mice. Mice were treated with E7107 (4mg/kg/day) or vehicle for 10 days once human CD45+ cells reached >25% in BM of recipient mice (median of 86 days post-transplantation; range from 62–148 days) (Fig. 4c). In each PDX model, targeted genomic analysis of purified human leukemic cells from the BM confirmed faithful engraftment of the major leukemic clones found in the primary patient samples (Supplementary Fig. 6a) and revealed that spliceosomal mutations were present in the major leukemic clones following E7107 treatment in vivo (Supplementary Fig. 6b). All spliceosome-mutant AML PDXs showed significant reductions in human leukemic burden in response to E7107, while the response in splicing wildtype AML cells was less robust (Fig. 4d–e). Further examination revealed that 2 of 3 spliceosome-mutant AMLs had a significant decrease in hCD45+ hCD34+ HSPC subsets. In contrast, spliceosome-wildtype AMLs showed less substantial reductions in leukemic cells as well as hCD45+ hCD34+ cell subsets (Figure 4d–e and Supplementary Fig. 6c–e). Although E7107 resulted in reduced cell proliferation 3 hours post-treatment in vivo regardless of spliceosome mutational status (Fig. 4f), the preferential sensitivity to E7107 in spliceosomal mutant AML was associated with substantially increased apoptosis only in spliceosome-mutant PDX samples (Fig. 4g–h and Supplementary Fig. 6f). These data establish a relationship between sensitivity to E7107 and spliceosome mutational status in primary AMLs.
Figure 4. Preferential sensitivity of primary human leukemias to pharmacologic modulation of splicing in vivo with E7107.
(a) RT-PCR and (b) qRT-PCR analysis of the effect of E7107 exposure (6 hours) relative to DMSO treatment on expression of a cassette exon inclusion isoform (“poison” exon) of EZH2 in SRSF2 wildtype (TF-1) or mutant (K052) leukemia cell lines. (c) Schema of patient-derived xenograft (PDX) experiments using primary human acute myeloid leukemia (AML) samples wildtype or mutant for spliceosomal genes followed by engraftment into NOD-scid IL2Rnull (NSG) mice. (d) Percentage of BM human CD45 (hCD45) cells in NSG mice following 10 days of vehicle or E7107 (4 mg/kg/d) treatment based on spliceosome mutational status. Each point represents hCD45 values for one individual NSG mouse and each color represents PDX from a specific patient. Mutational data for each patient are listed below the graph. (e) Immunohistochemical and immunofluorescence analysis for hCD45 in BM sections of recipient mice as shown in (d) based on spliceosome mutational status. (f) Percentage of BM hCD45+ cells in S-phase based on in vivo BrdU-incorporation following 5 days of E7107 (4 mg/kg/d) or vehicle treatment. (g) Bar plots showing percentage of hCD45+ cells that are Annexin V+/PI−or Annexin V+/PI+. (h) Representative FACS plots of Annexin V/propidium iodide (PI) staining of hCD45 cells following 5 days of E1707 (4 mg/kg/d) or vehicle treatment in vivo. Error bars represent mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; **** P <0.0001.
While splicing is an essential process required for the normal function of all mammalian cells, here we provide both genetic and pharmacologic evidence that SRSF2-mutant leukemias are preferentially sensitive to splicing modulation in vivo relative to their wildtype counterparts. Genomic and biological analyses of cells expressing mutant SRSF2 in the absence of the wildtype protein indicated that the SRSF2 mutation, despite being selected for in human leukemia1,5, is unable to support gene expression and splicing patterns required for hematopoiesis on its own. The sensitivity of SRSF2-mutant leukemias to loss of the wildtype SRSF2 allele was mirrored by the exposure of leukemias bearing heterozygous SRSF2 mutations to E7107, an inhibitor of splicing8. As mutations affecting the spliceosomal proteins SF3B1 and U2AF1 are also found in an exclusively heterozygous context, it will be important to determine if the broad range of malignancies carrying diverse spliceosomal mutations may prove similarly sensitive to pharmacologic perturbation of normal splicing catalysis.
Given the high frequency of SRSF2 mutations across myeloid malignancies1,20,21, the adverse outcome associated with SRSF2 mutations20,22,23, and the need for novel therapeutic approaches for these disorders, the data provided in this study have important therapeutic implications for MDS and AML patients bearing genetic alterations in SRSF2. E7107 is one of a host of structurally distinct splicing inhibitors, which all hinder normal splicing in a similar manner by inhibiting SF3B1 function24,25. The only such compound that has been tested in humans to date is E7107, which has completed two phase I dose-escalation studies for patients with advanced solid tumors. In patients treated with E7107, inhibition of splicing was observed during dose escalation and the dose-limiting toxicity was primarily gastrointestinal-related; however, visual impairment was reported in 3/66 patients26,27. The data presented here demonstrate that SF3B1 inhibition has therapeutic potential for the treatment of malignancies with SRSF2 mutations; clinical studies with newly identified SF3B1 inhibitors will be essential to define the safety and therapeutic efficacy of this approach in patients. Moreover, ongoing efforts to understand how pharmacologic inhibitors of splicing alter the constellation of proteins that directly or indirectly bind to SF3B1, and how mutations affecting splicing factors might alter protein-protein interactions, will facilitate the future development of compounds with heightened specificity for spliceosome-mutant cells.
Online Methods
Animals
All animals were housed at Memorial Sloan Kettering Cancer Center (MSKCC). All animal procedures were completed in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees at MSKCC. The number of mice in each experiment was chosen to provide 90% statistical power with a 5% error level. Generation and genotyping of the Srsf2P95H/+ conditional knock-in mice as well as Srsf2 conditional knockout mice (both on C57BL/6 background) are as previously described5,30. For MLL-AF9 BM transplantation assays, Srsf2P95H/+ and littermate control mice were crossed to Vav-Cre transgenic mice31.
Peripheral blood analysis
Blood was collected by retro-orbital bleeding using heparinized microhematocrit capillary tubes (Thermo Fisher Scientific). Automated peripheral blood counts were obtained using an IDEXX ProCyte Dx Hematology Analyzer. Differential blood counts were scored on blood smears stained using Wright-Giemsa staining and visualized using an Axio Observer A1 microscope.
Histological analyses
Mice were sacrificed and autopsied, and dissected tissue samples were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Paraffin blocks were sectioned at 4μm and stained with hematoxylin and eosin (H&E). Images were acquired using an Axio Observer A1 microscope (Carl Zeiss) or scanned using a MIRAX Scanner (Zeiss).
Bone marrow (BM) transplantation assays
Freshly dissected femora and tibiae were isolated from Mx1-Cre+Srsf2+/+, Mx1-Cre+Srsf2+/fl, Mx1-Cre+Srsf2P95H/+ or Mx1-Cre+Srsf2P95H/fl CD45.2+ mice of both sexes. BM was flushed with a 3-cc insulin syringe into cold PBS (without Ca2+ and Mg2+) supplemented with 2% bovine serum albumin to generate single cell suspensions. BM cells were pelleted by centrifugation at 1,500 rpm for 5 min and red blood cells (RBCs) were lysed in ammonium chloride-potassium bicarbonate lysis (ACK) buffer for 5 min on ice. After centrifugation, cells were resuspended in PBS/2% BSA, passed through a 40μm cell strainer, and counted. For competitive transplantation experiments, 0.5 × 106 BM cells from Mx1-Cre+Srsf2+/+, Mx1-Cre+Srsf2+/f, Mx1-Cre+Srsf2P95H/+ or Mx1-Cre+Srsf2P95H/fl CD45.2+ mice were mixed with 0.5 × 106 wildtype CD45.1+ support BM and transplanted via tail vein injection into 6-week old lethally irradiated (900 cGy) female CD45.1+ recipient mice. To activate the conditional alleles, mice were treated with 3 doses of polyinosinic:polycytidylic acid (polyI:C; 12mg/kg/day; GE Healthcare) every second day via intra-peritoneal injection. Peripheral blood chimerism was assessed every 4 weeks by flow cytometry. For noncompetitive transplantation experiments, 1 × 106 total BM cells from Mx1-Cre+Srsf2+/+, Mx1-Cre+Srsf2+/f, Mx1-Cre+Srsf2P95H/+ or Mx1-Cre+Srsf2P95H/fl CD45.2+ mice were injected into lethally irradiated (900 cGy) CD45.1+ recipient mice. Peripheral blood chimerism was assessed as described in competitive transplantation experiments. Additionally, for each bleeding whole blood cell counts were measured on an automated blood analyzer. Animals that failed to engraft (<1% CD45.2 chimerism in peripheral blood) or were lost due to poly(I:C) toxicity were excluded from analysis.
Xenografts of primary human AML samples
Studies were approved by the Institutional Review Boards of Memorial Sloan Kettering Cancer Center and Fred Hutchinson Cancer Research Center and conducted in accordance to the Declaration of Helsinki protocol. Primary human AML samples derived from whole peripheral blood or BM MNCs were CD3-depleted, and transplanted via tail vein injection into 6-week old NOD-scid IL2rnull (NSG) mice (Jackson Laboratory) conditioned with 200 cGy of gamma-irradiation. Mice were bled monthly to assess the presence of human CD45+ cells in the blood. If hCD45+ was > 1%, BM aspiration was performed to assess BM hCD45 chimerism.
Retroviral transduction and transplantation of primary hematopoietic cells
Vav-Cre+Srsf2+/+ and Vav-Cre+Srsf2P95H/+ mice were treated with a single dose of 5-fluorouracil (150mg/kg) followed by BM harvest from the femora, tibiae and hipbones 6 days later. RBCs were removed by ACK lysis buffer, and nucleated BM cells were transduced with viral supernatants containing MSCV-MLL-AF9-IRES-GFP for 2 days in IMDM/15% FCS supplemented with mSCF (25ng/mL), mIL3 (10ng/mL) and mIL6 (10ng/mL), followed by injection of ~400,000 cells per recipient mouse via tail vein injection into lethally irradiated (900 cGy) CD45.1 mice. For secondary transplantation experiments, 6-week old, sub-lethally irradiated (450 cGy) C57/BL6 recipient mice were injected with 250,000 primary MLL-AF9 leukemic cells. For Srsf2 hemizygous rescue experiments, Srsf2P95H/fl and Mx1-Cre+Srsf2P95H/fl fetal liver cells (E12.5-14.5) were c-Kit-enriched using CD117 MicroBeads (MACS Miltenyl Biotec), and transduced with viral supernatants containing MSCV-MLL-AF9-IRES-GFP twice for 2 days in IMDM/15% FCS supplemented with mSCF (100 ng/mL), mTPO (50 ng/mL) mFLT3-L (5 ng/mL) and mIL6 (10 ng/mL), followed by tail vein injection of ~500,000 cells into lethally irradiated (900 cGy) CD45.1 recipient mice. All cytokines were purchased from R&D Systems.
Flow cytometry analyses and antibodies
Surface marker staining of hematopoietic cells was performed by first lysing cells with ACK lysis buffer and washing cells with ice-cold PBS. Cells were stained with antibodies in PBS/2% BSA for 30 minutes on ice. For hematopoietic stem and progenitor staining, cells were stained with a lineage cocktail including B220 (RA3-6B2), CD3 (17A2), CD4 (RM4-5), Gr-1 (RB6-8C5), Mac1 (M1/70), NK1.1 (PK136) and Ter119, allowing for mature lineage exclusion from the analysis. Cells were also stained with monoclonal antibodies against c-Kit (2B8), Sca1 (D7), FcγRII/III (2.4G2), CD34 (RAM34), CD45.1 (A20), CD45.2 (104), CD48 (HM48-1) and CD150 (9D1). The composition of mature hematopoietic cell lineages in the BM, spleen and peripheral blood was assessed using a combination of Mac1, Gr-1, B220, CD19, CD4 and CD8. For analysis of human cell populations in mouse xenograft experiments, BM mononuclear cells and peripheral blood mononuclear cells were stained with a combination of antibodies against hCD3 (SK7), hCD19 (HIB19), hCD33 (WM-53), hCD34 (4H11), hCD38 (HIT2) and hCD45 (HI30). Antibodies against mouse CD45.1 (A20) and Ter119 were used to exclude host-derived cells. DAPI was used to exclude dead cells. The final staining volume was 100μl, and the final concentration for all antibodies used was 1:200, except for CD34 (1:50), c-Kit (1:100), CD150 (1:100). All flow cytometry antibodies were purchased from eBioscience and BioLegend. For in vivo apoptosis experiments, BM cells from were harvested from NSG mice 3 hours after E7107 treatment. Cells were stained with Annexin V-APC antibody in Annexin V binding buffer (BD Pharmingen) to detect cells undergoing apoptosis, according to manufacturer instructions. For assessment of the effect of E7107 on cell cycle status in vivo, BrdU (0.1mg/g) was administered via intra-peritoneally to NSG-S mice 24 hours prior to drug treatment. BM cells were harvested 3 hours after E7107 treatment, and the detection of BrdU incorporation was performed following manufacturer instructions (BD Pharmingen) Propidium iodide (PI) was used as counter stain in both Annexin V and BrdU experiments. All FACS sorting was performed on FACS Aria, and analysis was performed on an LSRII or LSR Fortessa (BD Biosciences). For western blotting, the following antibodies were used: H3K79me2 (Abcam; Ab-3594), Histone H3 (Cell Signaling Technologies; D1-H2), Meis1 (Abcam; Ab-124686), Sf3b1/Sap-155 (MBL; D221-3), Flag-M2 (Sigma-Aldrich; F-1084) and β-actin (Sigma-Aldrich; A-5441). All primary antibodies for western blotting were diluted to a final concentration of 1:1000, in either 5% BSA/TBS-T or 5% skim milk/TBS-T.
Administration of spliceosome modulator E7107 in vitro and in vivo
For all in vitro experiments E7107 was dissolved in DMSO. For drug sensitivity studies, cells were exposed to E7107 from a range of 10 μM to 0.05 nM. For in vivo administration, E7107 was dissolved in vehicle (10% ethanol and 4% Tween-80 in sterile PBS) and administered via IV injection at 4mg/kg/day. For drug efficacy studies, randomization was done by conducting CBC analysis prior to the start of drug administration and confirming that WBC count averages were equivalent in treatment and vehicle groups. All mice received 10 consecutive doses of E7107. No blinding was done in the in vivo drug studies or data analysis. For RNA-seq analysis in mouse MLL-AF9 leukemia model, 5 consecutive doses of E7107 were administered, and mice were sacrificed 3 hours after the last dose and BM Mac1+ GFP+ cells were purified by flow cytometry.
Cell culture
K052 leukemia cells (from JCRB Cell Bank; JCRB0123) were cultured in RPMI/10% FCS, and TF-1 (from DSMZ; ACC 344) cells were culture in RPMI/10% FCS plus hGM-CSF (R&D Systems; 5 ng/mL). Primary mouse MLL-AF9 leukemia cell lines were generated from BM cells of leukemia-bearing mice and maintained in IMDM/15% FCS supplemented with L-glutamine, mSCF (20 ng/mL), mIL3 (10 ng/mL) and mIL6 (10 ng/mL). All cell lines have been tested for mycoplasma contamination, and authenticated to confirm the status of SRSF2 mutation.
MSCV-Flag-Bio-DOT1L-Puro and MSCV-Meis1-IRES-Puro, and MSCV-IRES-Puro constructs were used for overexpression studies. Retroviral supernatants were produced by transfecting 293 GPII cells with cDNA constructs and the packaging plasmid VSV.G using XtremeGene9 (Roche), and were used to transduce MLL-AF9 leukemic cells in the presence of polybrene (5 μg/mL), followed by puromycin selection (1 μg/mL; Invitrogen) for successfully transduced cells. Cells were treated with E7107 (0.5 nM) or DMSO in 96-well plate format, and changed into fresh media containing E7107 or DMSO every two days. Relative cell growth was assessed on Day 6 after E7107 exposure using the LSR Canto.
Overexpression of SF3B1WT or SF3B1R1074H mutation was introduced into MLL-AF9/Srsf2P95H/+ leukemic cells using PiggyBac transposon system. 2 μg of PiggyBac Transposase construct (CMV-PB-Transposase-IRES-TK-HSV), and 6 μg of SF3B1WT (ITR-CAG-Flag-SF3B1WT-IRES-Puro-ITR) or SF3B1R1074H mutant (ITR-CAG-Flag-SF3B1R1074H-IRES-Puro-ITR) cDNA constructs were electroporated into 2 × 106 cells (in 200 μL volume) using the Amaxa Nucleofector Protocol (Program T-003) according to manufacturer instructions (Lonza). Puromycin selection (1 μg/mL) was initiated 4 days after electroporation to select for cells that successfully incorporated the constructs. Sanger sequencing was performed to confirm successful integration of the cDNA plasmid using the following primers – Fwd: TCCAATCAAAGATCTTCTTCCAA, Rev: GAGCAGGTTTCTGCAACGAT.
In vitro cell viability assays
Cells were seeded in white flat-well 96 well plates (Costar) at a density of 10,000 cells per well. ATP luminescence readings were taken 48 hours post E7107 treatment using Cell Titer Glo (Promega) according the manufacturer instructions.
Semi-quantitative and quantitative RT-PCR
Total RNA was isolated using RNeasy Mini or Micro Kit (Qiagen). For cDNA synthesis, total RNA was reverse transcribed with SuperScript VILO cDNA synthesis kit (Life Technologies). Primers used in the RT-PCR reactions were: Dot1l (Exon 11-13) – Fwd: ACTTGAGTGACATTGGCACCA, Rev: AGCACCAGAATCCGCGGGG; Meis1 (Exon 7-9) – Fwd: CGGCATCCACTCGTTCAG, Rev: TCACTTGAAGGATGGTAAGTCCT; Gapdh – Fwd: CCATGACAACTTTGGCATTG, Rev: CCTGCTTCACCACCTTCTTG; EZH2 (Exon 9-10) – Fwd: TTTCATGCAACACCCAACACT, Rev: CCCTGCTTCCCTATCACTGT. The PCR cycling conditions (33 cycles) chosen were as follows: (1) 45s at 95 °C (2) 45s at 52 °C (3) 60s at 72 °C with a subsequent 5 min extension at 72 °C. Reaction products were analyzed on 2 % agarose gels. The bands were visualized by ethidium bromide staining.
Quantitative RT-PCR (qRT-PCR) analysis was performed on Applied Biosystems QuantStudio 6 Flex cycler using Power SYBR Green PCR Master Mix (ThermoFisher Scientific). The following primers were used:
EZH2 inclusion – Fwd: CAGCATTTGCCACTCCTACC, Rev: AGAGCAGCAGCAAACTCCTTT;
EZH2 exclusion – Fwd: CAGCATTTGGAGGGAGCA, Rev: GCTGGGCCTGCTACTGTTATT;
18s rRNA – Fwd: GTAACCCGTTGAACCCCATT, Rev: CCATCCAATCGGTAGTAGCG;
Dot1l exclusion – Fwd: GCAGGAACTTGAGTGCTTGAA, Rev: GGCAGCTGCTTTGCTCTC;
Dot1l inclusion – Fwd: CGGCAGAATCGTATCCTCAA, Rev: CAAGTATGGTGCGGTCAATG;
Meis1 exclusion – Fwd: TAACAGCAGTGAGCAAGCAC, Rev: AATAAACCAATTGTTCACTTGAAGG;
Meis1 inclusion – Fwd: AAGGTGATGGCTTGGACAAC, Rev: AGGGTGTGTTAGATGCTGGAA;
Gapdh – Fwd: TGGAGAAACCTGCCAAGTATG, Rev: GGAGACAACCTGGTCCTCAG.
All samples including the template controls were assayed in triplicate. The relative number of target transcripts was normalized to the housekeeping gene found in the same sample. The relative quantification of target gene expression was performed with the standard curve or comparative cycle threshold (CT) method.
Statistical analyses
Statistical significance was determined by Student’s t-test or analysis of variance (ANOVA) after testing for normal distribution. For Kaplan-Meier survival analysis, Mantel-Cox log-ranked test was used to determine statistical significance. Data were plotted as mean values with error bars representing standard deviation using GraphPad Prism 7 software.
Targeted genomic sequencing using MSKCC IMPACT
Genomic alterations from FACS-purified hCD45+ BM cells from NSG mice of PDX models were profiled using the MSKCC IMPACT assay as described previously32. Genomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen).
Replicates
RNA-Seq was conducted with 3–5 biological replicates from each group. Genetic phenotyping experiments were replicated three times independently. For in vivo experiments, the number of animals was chosen to ensure 90% power with 5% error based on observed standard deviation. Flow cytometric experiments were replicated independently two-three times. Pilot studies were conducted with drug studies and results were replicated in a larger study to achieve enough statistical power. In vitro experiments were replicated two-three times, with viability experiments being completed in triplicate.
mRNA isolation, sequencing, and analysis
RNA was extracted from sorted mouse cell populations using Qiagen RNeasy columns. Poly(A)-selected, unstranded Illumina libraries were prepared with a modified TruSeq protocol. 0.5X AMPure XP beads were added to the sample library to select for fragments <400 bp, followed by 1X beads to select for fragments >100 bp. These fragments were then amplified with PCR (15 cycles) and separated by gel electrophoresis (2% agarose). 300 bp DNA fragments were isolated and sequenced on the Illumina HiSeq 2000 (~100M 101 bp reads per sample).
Publicly available RNA-sequencing data
Unprocessed RNA-seq reads from 31 human AML patients with MLL rearrangements were downloaded from the NCBI Sequence Read Archive (SRA; accession numbers SRP028594, SRP033266, SRP048759 and SRP056295. The data consisted of paired-end 2×100 bp libraries, with an average read count of 102M per sample. The SRSF2 mutational status of the samples was obtained from Lavallée et al.16
Genome annotations
To create organism-specific gene and genome annotation files, information was combined across UCSC and Ensembl databases, using mouse assembly mm10 (NCBI GRCm38) and human assembly hg19 (NCBI GRCh37). To create splice junction annotation files, alternatively spliced cassette exons, competing 5′ or 3′ splice sites and retained introns were obtained from MISO v2.0.13 Constitutively spliced exons and introns were identified based on exon-exon junctions that were not alternatively spliced according to UCSC knownGene33 and the Ensembl 71 database34 using all possible combinations of splice sites, as described previously35.
RNA-seq read mapping
All human and mouse samples were processed using the same pipeline. Step 1: The reads were mapped to their respective genome assembly, using Bowtie v1.0.036 and RSEM v.1.2.4.37 The latter was internally modified to call Bowtie with -v 2, and was run on the gene annotation file with the parameters --bowtie-m 100 --bowtie-chunkmbs 500 --calc-ci --output-genome-bam. Step 2: BAM files from step 1 were filtered to remove reads where (i) the alignment mapq score was 0, and (ii) the splice junction overhang was less than 6 nucleotides. Step 3: All remaining unaligned reads were mapped to the splice junction annotation files using TopHat v2.0.8b38 called with the parameters --bowtie1 --read-mismatches 3 -- read-edit-dist 2 --no-mixed --no-discordant --min-anchor-length 6 --splice-mismatches 0 --min-intron-length 10 --max-intron-length 1000000 --min-isoform-fraction 0.0 --no-novel-juncs --no-novel-indels --raw-juncs. The --mate-inner-dist and --mate-std-dev arguments were calculated using the MISO exon_utils.py script, which maps reads to constitutively spliced exon junctions. Step 4: The reads aligned to splice junctions were filtered as in step 2. Step 5: All resulting BAM files were merged to create a combined file of all aligned RNA-seq reads.
Identification and quantification of differential splicing
Isoform ratios for all alternative splicing events were quantified using MISO v2.013. Constitutively spliced exons and introns were quantified using junction-spanning reads, as previously described35. The conditional knockin and knockout mice were compared in a pair-wise manner, and for each pair the analysis was restricted to splicing events with 20 or more reads supporting either or both isoforms, and where the event was alternatively spliced in the sample pair. From that subsets of events, those that fulfilled the following criteria were defined as differentially spliced: (i) they had at least 20 relevant reads in both samples, (ii) the change in absolute isoform ratio was ≥ 10%, and (iii) the statistical analysis of isoform ratios had a Bayes factor greater than or equal to 5, when calculated using Wagenmakers’s framework14. The human AML samples were analyzed by calculating the median isoform ratios across all 28 SRSF2 wildtype samples, and comparing that in a pair-wise manner to each SRSF2 mutant sample, using the same methodology as for the knockin and knockout mice. The Mx1-Cre+Srsf2+/+ or Mx1-Cre+Srsf2P95H/+ E7107-treated mice were compared in a pair-wise manner against the median isoform ratios of their vehicle-treated counterparts, using the same methodology. For the MLL-AF9 AML transformed mice there were sufficient replicates to do a group-based comparison within the Srsf2P95H and wildtype genotypes individually (N=5 for each genotype-treatment combination). E7107- and vehicle-treated mice were compared in a two-sided Wilcoxon rank-sum test, using the total number of isoform reads within each treatment group. Events were categorized as being differentially spliced if they fulfilled the following criteria: (i) they had at least 20 relevant reads in both samples, (ii) the change in median absolute isoform ratio was ≥ 10%, and (iii) they had a P-value < 0.01.
Gene expression analysis
All comparisons of gene levels were performed using RNA-seq read counts normalized with the trimmed mean of M values (TMM) method40. The scaling factors were calculated based on protein-coding genes only. For comparisons with fewer than five replicates per group, we used Wagenmakers’s Bayesian framework14 to compare samples in a pair-wise manner, as described above for the analysis of differential splicing. Differentially expressed genes had to have a Bayes factor greater than 100. For the MLL-AF9 leukemic mouse model, E7107- and vehicle-treated mice within the Srsf2+/+ or Srsf2P95H/+ genotype were analyzed using a two-sided Wilcoxon rank-sum test to compare the five replicates within each genotype-treatment group. Differentially expressed genes had to fulfill the following criteria: (i) a difference in abundance greater than a fold change of 2, and (ii) a P-value < 0.01.
Gene Ontology (GO) enrichment analysis
For each genotype/treatment comparison, we identified genes that were differentially expressed in at least one of the replicates to test for enrichment of GO Biological Process terms using the R package goseq41. All protein-coding genes were included as the background gene set. The “Wallenius” method was used, and the resulting false discovery rates were corrected using the Benjamini-Hochberg approach. Only terms with at least two ancestors were tested, and terms with more than 500 genes associated with them were removed, to eliminate parent terms associated with generic biological processes.
Motif enrichment and distribution
The relative occurrences of sequence motifs in exons with increased inclusion versus exclusion rates in a given sample comparison were calculated. For the analysis of E7107 treatment in Srsf2+/+ or Srsf2P95H/+ mutant mice in the Mx1-Cre model, exons with decreased inclusion rates were compared to all exons with no changes in splicing, since the number of exons with increased inclusion rates was insufficient for statistical analysis. The 95% confidence interval for the enrichment ratios were computed based on bootstrapping, using 500 resampling steps for the enrichment ratios, or 100 steps for displaying the spatial distribution of motifs along a meta-exon.
Sample clustering
MLL-AF9 AML transformed mice were clustered using multidimensional scaling (also known as principal coordinates analysis) of distances calculated using the ‘canberra’ method, sum(|xi − yi|/|xi + yi|). For cassette exons and retained introns, only events that were alternatively spliced in the samples and had more than 20 reads in at least one sample were included. For protein-coding genes only genes with normalized expression >10 in at least one sample were included. Hierarchical clustering was performed on z-score standardized data with the ‘ward.D2’ method, using the most variable splicing events and genes across samples (standard deviation across the 20 mice > 0.25; splicing event or gene could be detected in at least 10 mice).
Mouse versus human splicing comparison
For the cumulative distribution function (CDF) comparison of cassette exon splicing and intron retention in MLL-AF9 myeloid leukemias following in vivo E7107 or vehicle treatment, only cassette exons and introns within mouse homologs of genes containing differentially spliced events in at least one of the human SRSF2 MLL-rearranged AML samples were included. Mouse homologs of the differentially spliced human genes were extracted from the Ensembl database version 8142 using BioMart.
Supplementary Material
Acknowledgments
This work was supported by the Leukemia and Lymphoma Society (S.C.-W.L., O.A.-W.), U.S. Department of Defense Breast Cancer Research Program grant W81XWH-14-1-0044 (H.D.), U.S. Dept. of Defense Bone Marrow Failure Research Program grants BM150092 (O.A.-W.) and W81XWH-12-1-0041 (R.K.B., O.A.-W.), the Worldwide Cancer Research Fund (E.K.), the Fondation de France (J.-B.M.), the American Society of Hematology (B.H.D., O.A.-W.), the Edward P. Evans Foundation (R.K.B., O.A.-W.), grant R01 HL128239 (R.K.B., O.A.-W.), grant 1K08CA160647-01 (O.A.-W.), the Ellison Medical Foundation grant AG-NS-1030-13 (R.K.B.), the Damon Runyon Foundation (R.K.B., O.A.-W.), grant R01 DK103854 (R.K.B.), the Starr Foundation grant I8-A8-075 (O.A.-W.), the Josie Robertson Investigator Program (O.A.-W.), the Mr. William H. Goodwin and Mrs. Alice Goodwin Commonwealth Foundation for Cancer Research (O.A.-W.), and The Experimental Therapeutics Center of MSKCC (O.A.-W.).
Footnotes
Accession codes
All newly generated RNA-seq data were deposited in the Gene Expression Omnibus database (GEO; accession number GSE74064).
Author Contributions
S.C.-W.L., H.D., E.K., R.K.B, and O.A.-W. designed the study. S.C.-W.L., E.K., H.C., Y.R.C., J.-B.M., B.D., A.Y., Y.J.K., C.-W.C., A.P., J.T., X.W., A.K., S.A.A, and O.A.-W. performed experiments. H.D. and R.K.B. performed RNA-seq analysis. M.T., J.P., S.B., and P.S. provided E7107 and advice with drug dosing experiments. S.C.-W.L., H.D., E.K., C.L., R.K.B, and O.A.-W. prepared the manuscript with help from all co-authors.
Competing financial interests
M.T., J.P., S.B., and P.G.S. are employees of H3 Biomedicine, Inc.
References
- 1.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2012;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shirai CL, et al. Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer cell. 2015;27:631–643. doi: 10.1016/j.ccell.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim E, et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilagan JO, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome research. 2014 doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Folco EG, Coil KE, Reed R. The anti-tumor drug E7107 reveals an essential role for SF3b in remodeling U2 snRNP to expose the branch point-binding region. Genes Dev. 2011;25:440–444. doi: 10.1101/gad.2009411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotake Y, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 9.Lindsley RC, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genovese G, Jaiswal S, Ebert BL, McCarroll SA. Clonal hematopoiesis and blood-cancer risk. N Engl J Med. 2015;372:1071–1072. doi: 10.1056/NEJMc1500684. [DOI] [PubMed] [Google Scholar]
- 11.Xie M, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature medicine. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nature methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagenmakers EJ, Lodewyckx T, Kuriyal H, Grasman R. Bayesian hypothesis testing for psychologists: a tutorial on the Savage-Dickey method. Cognitive psychology. 2010;60:158–189. doi: 10.1016/j.cogpsych.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci U S A. 2015;112:E4726–4734. doi: 10.1073/pnas.1514105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavallee VP, et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet. 2015;47:1030–1037. doi: 10.1038/ng.3371. [DOI] [PubMed] [Google Scholar]
- 17.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernt KM, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokoi A, et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J. 2011;278:4870–4880. doi: 10.1111/j.1742-4658.2011.08387.x. [DOI] [PubMed] [Google Scholar]
- 20.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bejar R, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang SJ, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood. 2012;119:4480–4485. doi: 10.1182/blood-2011-11-390252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vannucchi AM, et al. Mutations and prognosis in primary myelofibrosis. Leukemia. 2013;27:1861–1869. doi: 10.1038/leu.2013.119. [DOI] [PubMed] [Google Scholar]
- 24.Effenberger KA, Urabe VK, Prichard BE, Ghosh AK, Jurica MS. Interchangeable SF3B1 inhibitors interfere with pre-mRNA splicing at multiple stages. RNA. 2016;22:350–359. doi: 10.1261/rna.053108.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonnal S, Vigevani L, Valcárcel J. The spliceosome as a target of novel antitumour drugs. Nature reviews Drug discovery. 2012;11:847–859. doi: 10.1038/nrd3823. [DOI] [PubMed] [Google Scholar]
- 26.Eskens FA, et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:6296–6304. doi: 10.1158/1078-0432.CCR-13-0485. [DOI] [PubMed] [Google Scholar]
- 27.Hong DS, et al. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs. 2014;32:436–444. doi: 10.1007/s10637-013-0046-5. [DOI] [PubMed] [Google Scholar]
- 28.Katz Y, et al. Quantitative visualization of alternative exon expression from RNA-seq data. Bioinformatics. 2015;31:2400–2402. doi: 10.1093/bioinformatics/btv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang HY, Xu X, Ding JH, Bermingham JR, Jr, Fu XD. SC35 plays a role in T cell development and alternative splicing of CD45. Molecular cell. 2001;7:331–342. doi: 10.1016/s1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]
- 31.Georgiades P, et al. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis. 2002;34:251–256. doi: 10.1002/gene.10161. [DOI] [PubMed] [Google Scholar]
- 32.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic acids research. 2013;41:D64–69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flicek P, et al. Ensembl 2013. Nucleic acids research. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hubert CG, et al. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013;27:1032–1045. doi: 10.1101/gad.212548.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome biology. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guberman JM, et al. BioMart Central Portal: an open database network for the biological community. Database : the journal of biological databases and curation. 2011;2011:bar041. doi: 10.1093/database/bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.